Abstract
Human immunodeficiency virus type 1 (HIV-1) studies suggest that antibody-dependent cellular cytotoxicity (ADCC) influences both virus acquisition and subsequent disease outcome. Technical issues with currently available assays, however, have limited the ability to comprehensively assess the impact of ADCC on transmission and disease progression. Commonly used ADCC assays use a target cell line, CEM.NKr-CCR5-Luc, that often does not support replication of relevant HIV-1 variants. Thus, the extent of ADCC responses against a large panel of HIV-1 strains often cannot be assessed using the currently available methods. We developed two new reporter cell-lines (MT4-CCR5-Luc and PM1-CCR5-Luc) to overcome these issues. MT4-CCR5-Luc cells are resistant, whereas PM1-CCR5-Luc cells are susceptible, to killing by a natural killer cell line, CD16+KHYG-1, in the absence of antibody. Polyclonal HIVIG gave similar ADCC estimates against HIV-1 isolate, NL4–3, regardless of which of the three cell lines were used as the targets. In contrast to CEM.NKr-CCR5-Luc and PM1-CCR5-Luc, however, MT4-CCR5-Luc target cells produce significantly higher luciferase after exposure to various HIV-1 strains, including transmitted founder variants and viruses incorporating specific envelopes of interest. This higher luciferase expression does not yield spurious results because ADCC estimates are similar when killing is assessed by both reporter protein expression and flow cytometry. Furthermore, ADCC estimates derived from MT4-CCR5-Luc cells are not skewed by non-antibody contents present in human plasma. In aggregate, the MT4-CCR5-Luc cell line can be used to estimate monoclonal antibody or plasma-induced ADCC responses against a diverse range of HIV-1 envelopes relevant for transmission and disease progression studies.
Keywords: Human immunodeficiency virus type 1, antibody dependent cellular cytotoxicity, envelope
1. Introduction
The majority of human immunodeficiency virus type 1 (HIV-1) vaccine trials have failed to demonstrate any protection against viral acquisition. However, the RV144 trial showed modest efficacy, although some have argued that the statistically significant differences were potentially due to chance [1]. Secondary analyses implied that the observed protection correlated with antibody-dependent cellular cytotoxicity (ADCC) responses and not the presence of neutralizing antibodies (nAbs) [2]. Vaccine elicited antibodies may have induced killing of infected cells either present in the infectious source or at the initial site of invasion. In addition to this ability to prevent initial infection, ADCC function may also be important in eliminating infected cells that persist even after prolonged virus suppression. Thus, ADCC may be important for achieving viral remission in the absence of antiretroviral medications [3, 4]. There is a great interest in the field to thoroughly investigate ADCC capacity present in different plasma and various antibodies to fully understand its role in providing sterilizing protection and achieving viral remission.
While nAb responses have been thoroughly examined in various patient samples and among monoclonal antibodies (mAbs), ADCC assessments have been hindered due to assay limitations. Estimating nAb responses has been greatly facilitated by the development of the TZM-bl assay. In this assay, neutralization is estimated by the relative proportion of TZM-bl reporter cells that are infected in the presence as compared to the absence of either plasma or an antibody of interest [5]. Importantly, the TZM-bl assay is highly versatile because the target cell line is universally susceptible to diverse HIV variants. In addition, the level of infection can be easily monitored by measuring reporter gene expression, through either luciferase or beta-galactosidase readout. These reporters have large linear ranges, and this lends reliability to the TZM-bl assay. A similar method to measure ADCC would be of great utility for future studies to decipher the role of ADCC during initial infection and disease progression.
While nAb assays measure the ability of an antibody to prevent infection of a naïve susceptible cell, ADCC methods estimate killing of virus bearing cells. ADCC depends on both the antigen binding fragment (Fab) and the fragment crystallizable (Fc) portion of an antibody. The antibody Fab binds to an epitope present on infected cells, such as the HIV-1 envelope glycoprotein (Env), and the Fc interacts with the Fc receptor (FcR) on immune cells, such as monocytes, macrophages, neutrophils, dendritic cells, and natural killer (NK) cells [6]. NK cells have been found to be the predominant mediators of ADCC, and as a result, they are commonly used as effector cells in ADCC assays [7]. ADCC can be estimated by co-culturing HIV-1 infected primary cells with autologous immune cells in the presence as compared to the absence of antibody. This methodology, however, is labor intensive and highly susceptible to primary cell variability. Numerous techniques have been used to overcome some of the hurdles inherent to primary cell-based assays. For instance, the challenge of having Env present on the surface of target cells is often accomplished by pulsing cells with HIV-1 Env monomer, gp120 [8–10]. These non-infection-based assays are problematic for a number of reasons. First, accessible Env epitopes and Env density are likely different among productively infected cells as compared to gp120 pulsed target cells. Second, these assays are not amenable to high throughput testing against a large variety of HIV-1 isolates, such as the different transmitted-founder (T/F) strains, because it requires the generation of the gp120 protein. Finally, pulsing cells with gp120 often leads to killing of both cells with attached Env and bystander cells [11]. These potential shortcomings may account for observed differences in ADCC assessments from cell infection-based assays as compared to methods that use gp120 pulsed target cells [12].
Alpert et al. [13] developed an infection-based ADCC assay that uses CEM.NKr-CCR5-Luc as targets and the NK cell line CD16+KHYG-1 as effectors to overcome issues associated with both primary cell-based and gp120-using methods [14, 15]. The CEM.NKr-CCR5-Luc cells endogenously express the CD4 and CXCR4 receptors, and they have been engineered to have high levels of CCR5 receptors [16]. HIV requires a combination of these receptors to enter cells and initiate infection [17–19]. Furthermore, the CEM.NKr-CCR5-Luc cells express luciferase after HIV-1 infection, which aids in estimating infected cell levels [20]. The NK cell line KHYG-1 was also engineered to express FcγRIIIa (CD16), termed CD16+KHYG-1 [13, 21]. CD16 interaction with an antibody’s Fc portion is necessary for NK cell-mediated ADCC, and this cell line was shown to express CD16 at levels similar to what has been demonstrated on primary NKs [13, 22, 23]. In this ADCC assay, infected target cells are incubated with an antibody or plasma and then co-cultured with CD16+KHYG-1. ADCC activity is estimated as the decrease in luciferase expression in the presence as compared to the absence of antibody.
Although the Alpert et al. assay has become a standard in the field, the CEM.NKr-CCR5-Luc cells often do not support replication of diverse HIV-1 variants. Alpert et al. used spinoculation to “overcome the relative resistance of CEM.NKr-CCR5 cells to infection” [13]. A recent publication, and our studies, demonstrate relatively poor reporter gene expression in the CEM.NKr-CCR5-Luc cells after exposure to various HIV-1 variants [24]. This has limited the ability to assess ADCC responses against a broad range of primary strains, such as those isolated from acutely infected individuals (T/F) or the diverse variants present in chronically infected individuals.
We sought to overcome these issues with the CEM.NKr-CCR5-Luc cells by engineering novel lymphocyte cell lines that are both resistant to NK cell-mediated killing in the absence of antibody and more susceptible to a diverse range of HIV-1. The development of a new cell line to use as target cells in the Alpert et al. luciferase based ADCC assay will allow one to estimate ADCC breadth and potency in a high-throughput manner against diverse and clinically relevant variants, which will be useful for future prevention and virus remission efforts.
2. Materials and Methods
2.1. Cell Lines.
PM1 [25], 293T [26], and TZM-bl [27] cells were obtained from the NIH AIDS Reagent Program. CEM.NKr-CCR5-sLuc (subsequently referred to as CEM.NKr-CCR5-Luc) and the NK cell line, CD16+KHYG-1, were generously provided by Dr. David Evans [13]. MT4-CCR5-GFP cells were generously provided by Dr. Paul Bieniasz [28].
A Tat-inducible luciferase reporter gene was introduced into PM1 and MT4-CCR5-GFP cells using the following protocol. Briefly, 293T cells were co-transfected with: 1) 7.5μg of a plasmid LNSX-Luc-1 (generously provided by Dr. David Evans), which has a luciferase gene in 5’ position to a SIV long terminal repeat; 2) 6.75μg of plasmid pCL-10A1 (generously provided by Dr. Suryarahm Gummuluru), which expresses packaging proteins for murine leukemia virus (MuLV); and 3) 0.75μg of a vesicular stomatitis virus protein G (VSV-G) expression vector (obtained from NIH AIDS Reagent Program), after incubating for 30 minutes in the presence of 45μL of Fugene. The virus was collected 48 hours later. Approximately 1×106 PM1 and MT4-CCR5-GFP cells were exposed to concentrated viral supernatant in the presence of 20μg/mL diethylaminoethyl cellulose (DEAE) for 4 hours at 37°C. Cells were washed and plated in RPMI supplemented with 10% fetal bovine serum, 2mM L-glutamine, 100U/mL of penicillin, and 100 μg/mL of streptomycin (RMPI complete) for 72 hours. Infected cells were selected by incubating cells in complete RPMI supplemented with 0.8μg/mL G418 (Sigma). The resulting cells were named PM1-Luc and MT4-CCR5-Luc.
PM1-Luc cells were further transduced with a CCR5 expression plasmid. Briefly, 293T cells were co-transfected with: 1) 7.5μg of a CCR5 expression plasmid pCX4pur-synCCR5 (generously provided by Dr. Heinrich Gottlinger); 2) 6.75μg of pCL-10A1; and 3) 0.75μg of a VSV-G expression vector. Approximately 1×106 PM1-Luc cells were exposed to concentrated virus supernatant in the presence of 20 μg/mL diethylaminoethyl cellulose (DEAE) for 4h at 37°C. Cells were washed and plated in RPMI complete, and infected cells were selected by incubating cells in complete RPMI supplemented with 0.8μg/mL G418 and 1 μg/ml puromycin. The subsequent cell line was named PM1-CCR5-Luc.
The NK cell line, CD16+KHYG-1, was maintained at a density of 5 × 105 cells per mL in RPMI medium (Invitrogen) consisting of 10% fetal bovine serum (Invitrogen), 25 mM HEPES (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mg/ml Primocin (InvivoGen), 1 μg/ml cyclosporine (CsA)(Sigma), and 10U/mL interleukin-2 (IL-2)(NIH AIDS Reagent Program [29]).
2.2. Viruses.
Twelve global reference Envs were obtained from the NIH AIDS reagent program [30]. Replication competent viruses incorporating the envelopes were successfully generated using protocols detailed previously [31, 32]. Here we tested 6 of the viruses along with three T/F viruses. T/F IMCs were obtained from the NIH AIDS Reference Reagent program [33]. Furthermore, we tested two virus stocks that incorporate isolated Envs from strains circulating in infected mothers from a previously described mother-to-child-transmission cohort [34]. Viral stocks were generated by transfecting 293T cells. Virus supernatants were passaged on peripheral blood mononuclear cells (PBMC) or isolated CD4 T cells for 7 days. Viral supernatant was collected and stored at −80°C, prior to TZM-bl titration and subsequent use in assays.
2.3. Antibodies.
HIVIG was obtained from the NIH AIDS reagent program. HIVIG is prepared from pooled plasma of asymptomatic, HIV antibody positive donors with CD4+ counts above 400 cells/μL. Plasma from HIV-1 seronegative individuals was obtained from Corning (Cat# 35060Cl, Lot# 121). Plasma was heat inactivated at 56°C for one hour prior to use in the ADCC assays. Melon IgG columns (ThermoFisher Scientific 45206) were used to isolate IgG from HIV infected plasma samples per manufacturer’s protocol. Comparisons were among volumes of isolated IgG as that estimated to be present in different plasma dilutions.
2.4. ADCC assays.
Approximately 5 × 105 target cells were re-suspended in 500 μL RPMI complete in each well of a 24-well plate. Plated cells, exposed to the virus of interest, were centrifuged for 1.5 hours at 1200 × g at room temperature and then incubated for 30 minutes at 37°C. In some cases, target cells were exposed to virus in the presence of anions, 20 ug/ml DEAE or 30 ug/ml polybrene for 2 hours. Cells were washed twice with phosphate buffered saline (PBS) and cultured in RPMI complete at 37°C. Infections were monitored for luciferase expression over the next 7 days. Infected cells were used in the ADCC assay after relative light units (RLU) were approximately 10-fold higher compared to uninfected controls. Approximately 1 × 104 target cells were plated in 96-well plate (Corning #3610) and exposed to serially diluted or a fixed amount of antibody or plasma for 20 minutes at 37°C. Post incubation, the NK cell line CD16+KHYG-1, was added to each well at a target cell to effector cell ratio of 1:5. Co-cultures were incubated for 4 hours when using PM1-CCR5-Luc cells or 24 hours when using MT4-CCR5-Luc cells and then examined for luciferase expression using Bright Glo Luciferase Assay System (Promega E2620) per manufacturer’s instructions. The percent ADCC when using MT4-CCR5-Luc cells can be estimated anywhere between 8 and 24 hours, although the latter allows for a more convenient timeframe. When CEM.NKr-CCR5-Luc cells were used as targets, a 1:10 target cell to effector cell ratio was used and luciferase expression was examined after 4 hours of incubation. Uninfected cells in the presence of NK cells served as the negative background control, and RLU values from these wells were subtracted from each sample well. The positive control reference wells lacked antibodies but contained infected cells with NK effector cells. ADCC activity was estimated as the percent luciferase expression difference observed in the sample of interest as compared to that observed in positive control wells. To be confident with our results, we test the capacity of 500 μg/mL of HIVIG to induce ADCC against NL43-infected target cells for each assay. When CD16+KHYG-1 cells have sufficient killing performance, this control should yield high levels of ADCC. If CD16+KHYG-1 cells fail to demonstrate killing in this control we discard the data.
Flow cytometry was also used to analyze MT4-CCR5-Luc cells infected with NL4–3 virus. After 10-fold increase in RLU as compared to uninfected cells, target cells were stained with 1 μM eFlour670 (eBioscience Cat# 65-0840-85, Lot# E15450–106) diluted in OPTI-MEM (Gibco Cat# 31985–062) to distinguish target from effector cells in a co-culture. Approximately 5 × 104 MT4-CCR5-Luc cells were added to 96-well plate along with or without 500 μg/mL HIVIG. After 20 minutes, 2.5 × 105 NK cells were added to the well. After incubating at 37°C for 24 hours, RLU was assessed in 20 μL of the culture diluted to a total volume of 100 μL in RPMI complete. The remaining cells were stained using Zombie UV Live/Dead (BioLegend, Cat# 77474, Lot# B228494). Percentage of live-eFluor670+GFP+ lymphocytes were assessed using BD LSRII flow cytometer. ADCC was calculated as the decrease in percentage of live GFP-positive target cells in the presence as compared to absence of antibody. In each case, the number of live GFP+ target cells in the absence of virus exposure served as background, and this was subtracted from each well.
Non-specific CD16+KHYG-1 mediated cytotoxicity was estimated using two different methods. First, MT4-CCR5-Luc cells were infected with a single round pseudovirus, NL4–3-ΔEnv-Luc-vesicular stomatitis virus (VSV)-G, which does not express HIV Env upon infection. HIVIG induced cytotoxicity was assessed against these infected targets using CD16+KHYG-1 cells, and these estimates were deemed as being not specific for HIV-1 Env. Second, heat inactivated plasma from HIV-1 seronegative individuals (Corning Cat# 35060Cl, Lot# 121) was used to estimate cytotoxicity against NL4–3 infected MT4-CCR5-Luc. These ADCC estimates were deemed as mediated by antibodies not specific for HIV-1.
2.5. Flow Cytometry
Flow cytometry was performed using a BD FACSCanto II and analyzed with FlowJo software (Tree Star). CCR5 was probed with CCR5-PE Clone J418F1 (Coulter).
2.6. Statistical Analysis
Differences between cell lines were analyzed using the Wilcoxon matched-pair test. Spearman’s rank test was used to examine correlation between percent ADCC and percent neutralization. The area under the curve (AUC) was calculated by taking the average ADCC capacity within the range of dilutions tested [35].
3. Results
3.1. Generation of two new target cell lines for ADCC assessments.
We are specifically interested in examining ADCC responses against HIV-1 variants of clinical significance, such as T/F strains and the swarm encountered by infants that ingest breast milk from their infected mothers. We first proposed to employ the well-established Alpert et al. infection-based, luciferase ADCC assay that uses CEM.NKr-CCR5-Luc as targets [13]. We were unable to use this assay, as designed, because the infected target cells failed to generate significantly higher luciferase levels as compared to the uninfected target cells after exposure to a T/F or a swarm containing maternal envelopes (Fig. 1). A large luciferase increase was observed among cells exposed to a lab-adapted CXCR4-using strain, NL4–3, using various different protocols. Targets, exposed to T/F (REJO) or maternal envelope virus (1431), however, failed to generate significantly higher relative luminescence units as compared to uninfected cells regardless of the infection protocols, such as spinoculation and use of anions (Fig. 1A), the multiplicity of infection (MOI) (Fig. 1B), or the days post-infection (Fig. 1C). Others have also reported problems with reporter gene expression in the CEM.NKr-CCR5-Luc cell line [20, 24]. Thus, we needed to modify the existing infection-based ADCC assay to examine responses against strains of high interest.
Figure 1. CEM.NKr-CCR5-Luc fail to produce adequate luciferase levels after exposure to strains of interest.
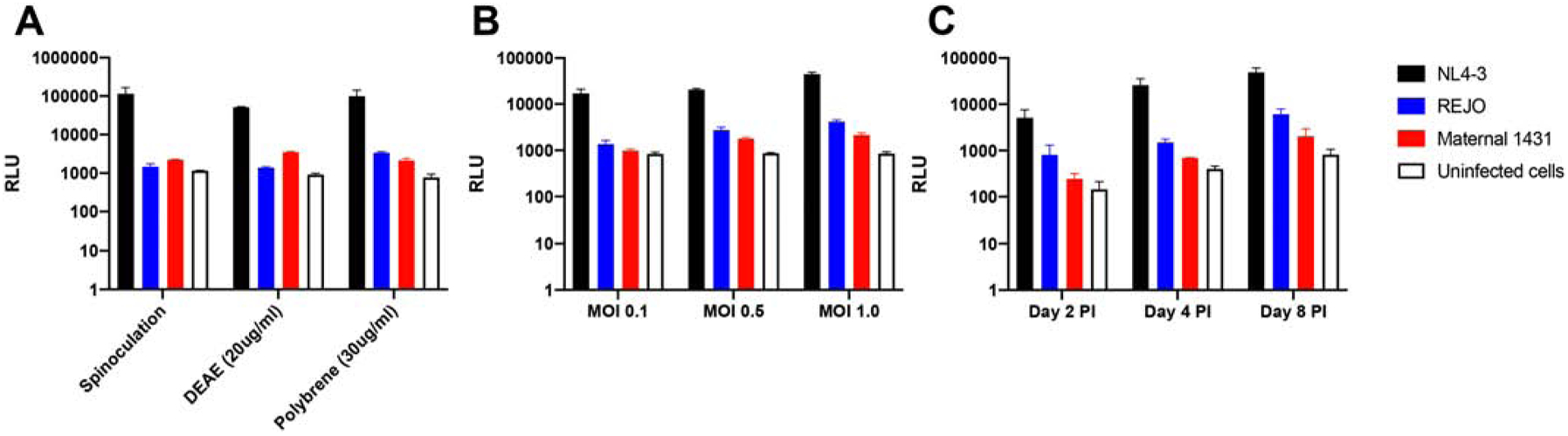
Representative examples of relative luminescence units (y-axis) after CEM.NKr-CCR5-Luc cells were exposed to virus using different protocols (A), varying multiplicity of infection (MOI) (B), and sampled at different days post infection (PI) (C). Each bar represents the mean and standard error mean from three replicates for NL4–3 (black bar), REJO (blue bar), swarm with maternal envelopes 1431 (red bar), and uninfected cells (white bar).
Two new cell lines were generated to overcome this issue. CD4+ T cell lines, MT4-CCR5-GFP and PM1, were chosen because of documented ability to support replication of multiple R5 variants [24, 25, 28, 36]. We sought to develop an assay that could be used with viruses harboring diverse primary Envs and where infected cell killing could be estimated in an efficient manner. Thus, PM1 and MT4-CCR5-GFP were transduced with a plasmid harboring a SIV long terminal repeat (LTR) dependent firefly luciferase. Cell clones, subsequently named PM1-Luc and MT4-CCR5-Luc, were chosen that demonstrated low background and high luciferase expression after infection with a CCR5-tropic virus.
The PM1-Luc cells were further transduced with a CCR5 expression construct to enhance susceptibility to R5 strains, and then a single limiting dilution clone was selected to generate PM1-CCR5-Luc cells. CCR5 expression was assessed in all cell lines by flow cytometry after staining cells with an α-CCR5 antibody. Following transfection of the PM1 T-cell line with a CCR5 expression construct, the percentage of cells expressing CCR5 increased from 62% to 100%. Additionally, 93.3% of MT4-CCR5-Luc cells were found to express CCR5. Furthermore, PM1-CCR5-Luc, and more so MT4-CCR5-Luc, cells had higher CCR5 mean fluorescence intensity (MFI) compared to CEM.NKr-CCR5-Luc (Table 1). Higher expression of CCR5 should potentially enhance the susceptibility of these cell lines to HIV-1 strains that use the CCR5 receptor for cell entry.
Table 1.
Cell lines and CCR5 Expression.
| CCR5 Expression◊ | ||
|---|---|---|
| Cell Lines | Percentage* | MFI |
| CEM.NKr-CCR5-Luc | 59.1 | 72.5 |
| PM1 | 62.0 | 38.3 |
| PM1-CCR5-Luc | 100.0 | 3908.7 |
| MT4-CCR5-Luc | 93.3 | 1.49E4 |
Percentage of cells that are CCR5+
Mean fluorescent intensity (MFI) of CCR5 expression on individual cells
3.2. MT4-CCR5-Luc cells are resistant to CD16+KHYG-1 cell killing in the absence of antibodies.
Previous ADCC assays specifically used the CEM.NKr cells because they are resistant to NK cell mediated lysis in the absence of antibody [37]. PM1 and MT4 cells have not been selected for NK cell resistance, and thus, NK cell resistance was also assessed in the new PM1-CCR5-Luc and MT4-CCR5-Luc cell lines. Infected PM1-CCR5-Luc and MT4-CCR5-Luc were incubated with the NK cell line CD16+KHYG-1. CD16+KHYG-1 has been previously shown to induce CD16-dependent ADCC in the presence of appropriate antibodies [12, 13]. Percent ADCC was estimated by the decrease in luciferase levels in the presence relative to the absence of antibody. As opposed to the published resistance of CEM.NKr-CCR5-Luc to NK cells [13], the PM1-CCR5-Luc were more susceptible to background lysis in the absence of antibody (Fig. 2A). In contrast, MT4-CCR5-Luc were resistant to CD16+KHYG-1 cell killing in the absence of antibody (Fig. 2B). Importantly, all wells contained effector cells, thus any background lysis would be present in all wells. Percent ADCC can be estimated by both newly generated cell lines, although MT4-CCR5-Luc cells may be more ideal due to their natural resistance to the CD16+KHYG-1 cell line used in this assay.
Figure 2. MT4-CCR5-Luc, but not PM1-CCR5-Luc, cells are resistant to CD16+KHYG-1 cell killing in the absence of antibodies.
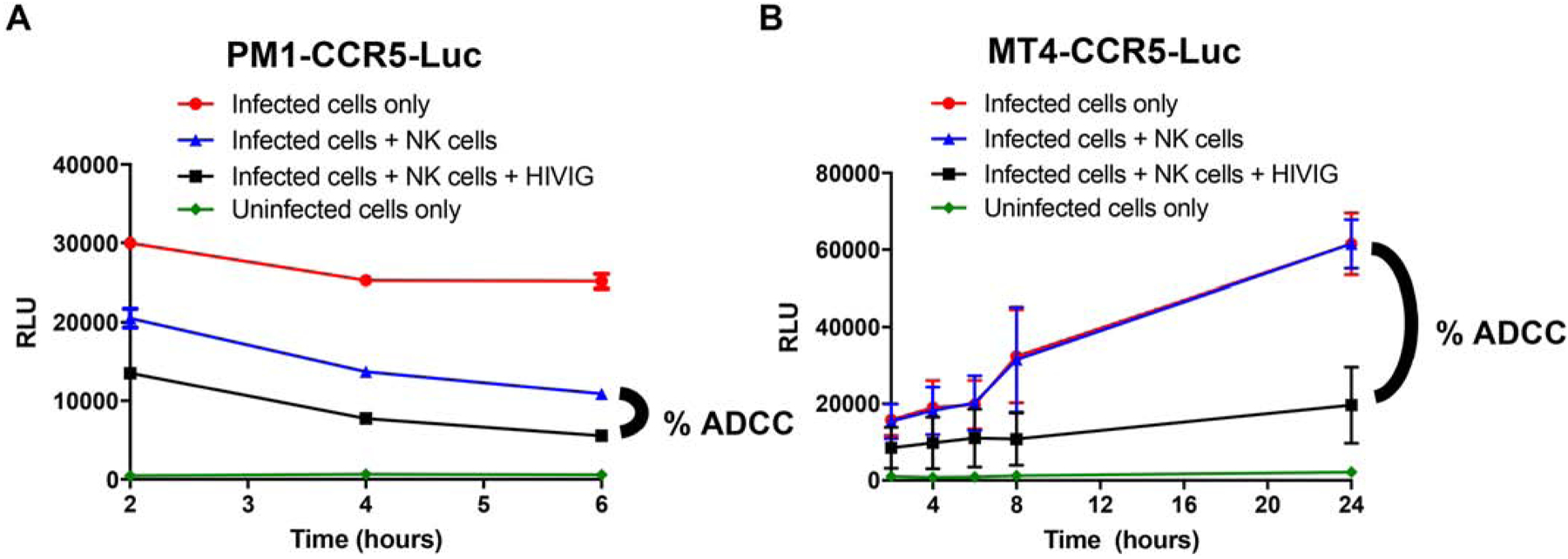
Relative light units (y-axis) over time (x-axis) in NL43-infected PM1-CCR5-Luc (A) and MT4-CCR5-Luc (B) cells without antibody or NK cells (red circle), with NK cells but no antibody (blue triangle), and with NK cells and 500 μg/mL of HIVIG (black square). ADCC is determined as the percent difference as graphed. Uninfected cells (green diamond) represents background RLU reading. Data from 2 independent experiments with 3 replicates each. Bars shows standard error mean (SEM).
3.3. PM1-CCR5-Luc, MT4-CCR5-Luc, and CEM.NKr-CCR5-Luc cells yield similar ADCC estimates.
All cell lines were infected with the same virus and incubated with the same antibody preparation to assess if the three cell lines yielded similar percent ADCC estimates under identical conditions. After infection with an exclusive CXCR4-using variant (termed X4), percent ADCC was similar for the known NK cell resistant cell line, CEM.NKr-CCR5-Luc, as well as PM1-CCR5-Luc and MT4-CCR5-Luc over a range of antibody concentrations (Fig. 3). The calculated AUC did not differ between CEM.NKr-CCR5-Luc (AUC = 0.579), MT4-CCR5-Luc cells (AUC = 0.614) and PM1-CCR5-Luc cells (AUC = 0.560) (p > 0.05 for all pairwise comparisons using Wilcoxon matched-pair test at each HIVIG dilution). The MT4-CCR5-Luc (r2 = 0.977), PM1-CCR5-Luc (r2 = 0.911), and CEM.NKr-CCR5-Luc (r2 = 0.903) also showed similar goodness of fit to a linear curve. The MT4-CCR5-Luc cell line, however, displayed a significantly lower coefficient of variation (%CV) across all six HIVIG concentrations as compared to the CEM.NKr-CCR5-Luc cell line (p = 0.031, Wilcoxon matched-pair test) but not PM1-CCR5-Luc (p = 0.156, Wilcoxon matched-pair test).
Figure 3. PM1-CCR5-Luc, MT4-CCR5-Luc, and CEM.NKr-CCR5-Luc cells yield similar ADCC estimates.
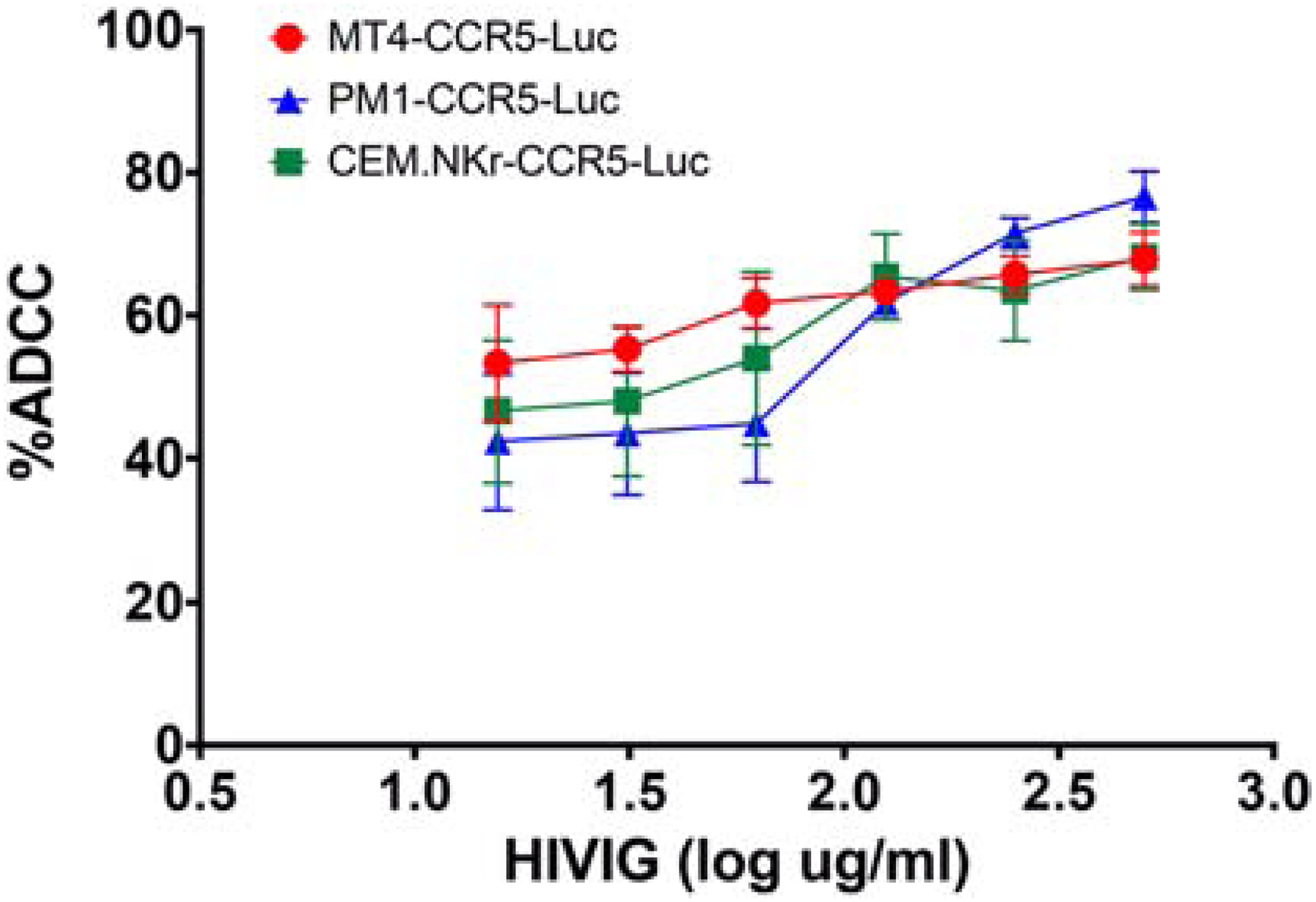
Percent ADCC (y-axis) observed against exclusively CXCR4-using virus NL4–3 infected MT4-CCR5-Luc (red), PM1-CCR5-Luc (blue), and CEM.NKr-CCR5-Luc (green) cells with serially diluted HIVIG (x-axis). Bars show SEM from 5 independent experiments with each condition tested in triplicate.
3.4. MT4-CCR5-Luc as compared to PM1-CCR5-Luc and CEM.NKr-CCR5-Luc demonstrates susceptibility to the widest range of HIV-1 variants.
The original description, a subsequent study, and our preliminary data showed that CEM.NKr-CCR5-Luc cells failed to yield high luciferase expression when exposed to a number of different exclusive CCR5-utilizing (termed R5) variants [20, 24] (Fig. 1). We compared the breadth of luciferase expression among CEM.NKr-CCR5-Luc, PM1-CCR5-Luc, and MT4-CCR5-Luc cells to establish if one cell line would be better for estimating ADCC against a large panel of viruses. Luciferase expression was examined after target cells were exposed to T/F strains and recombinant viruses containing Envs from a global reference panel or those isolated from chronically infected mothers that were part of a breastfeeding transmission study [30, 33, 34, 38]. Envs in this global reference panel are representative of a larger set of 219 variants. Neutralization against Envs in this global reference panel has been effectively used to estimate the neutralization spectrum of infected and vaccinated individuals. We have previously examined the capacity of infected mothers’ and exposed infants’ plasma to neutralize strains incorporating maternal Envs as a correlate of protection during mother-to-child-transmission. ADCC against these and other primary Envs can be examined in future studies if target cells yield acceptable reporter gene expression after virus exposure. Equal amounts of the same virus stock were used to infect each cell line by spinoculation at 1200 × g for 1.5 hours in the absence of any anions. Comparisons were made between cell lines using matched pairwise comparisons across all the different HIV-1 variants. Fold luciferase induction was significantly greater in the MT4-CCR5-Luc as compared to PM1-CCR5-Luc or CEM.NKr-CCR5-Luc cells (p < 0.001 for both comparisons, Wilcoxon matched-pair test). However, fold luciferase induction did not differ significantly between PM1-CCR5-Luc and CEM.NKr-CCR5-Luc cells (p = 0.850, Wilcoxon matched-pair test). MT4-CCR5-Luc cells were chosen as the ADCC target cell line for further validation because they may be amenable to estimating ADCC against a greater range of HIV-1 variants. The MT4-CCR5-Luc as compared to CEM.NKr-CCR5-Luc (p = 0.001) and PM1-CCR5-Luc cells (p = 0.007, Wilcoxon matched across each variant in both cases), however, did display significantly greater %CV in fold induction, suggesting that there is greater reporter gene expression variation in these cells after virus exposure. This is not necessarily surprising because CEM.NKr-CCR5-Luc and PM1-CCR5-Luc cells showed little to no fold induction with most variants.
3.5. HIV-1 Env and antibody ADCC specificity against infected MT4-CCR5-Luc cells.
We next determined the specificity of the observed ADCC activity in the MT4-CCR5-Luc cells using two independent methods. First, HIVIG mediated ADCC was assessed against MT4-CCR5-Luc cells infected with a VSV-G pseudotyped virus that did not express the HIV-1 Env after infection. Average ADCC of 10.990% (standard deviation (STD) 4.053, n = 3 independent experiments) was observed against these infected cells. Second, ADCC activity in HIV-1 seronegative plasma was assessed against NL4–3 infected MT4-CCR5-Luc cells. HIV-1 seronegative plasma showed a mean ADCC of 8.356 (STD 6.905, n = 6 independent experiments) against the NL4–3 infected MT4-CCR5-Luc cells. A combined average plus approximately three standard deviations of the ADCC observed with HIV-1 seronegative plasma and against infected cells that did not express HIV-1 Env (25%) was used as a threshold for ADCC specificity. Any ADCC estimate below 25% was deemed as HIV-1 antibody and HIV-1 Env independent cytotoxicity. This threshold was plotted on the subsequent ADCC graphs to clearly demonstrate positive ADCC activity.
3.6. ADCC estimates based on reduction in luciferase expression are similar to those obtained using flow cytometry.
The MT4-CCR5-Luc cells displayed exceptionally high luciferase after virus exposure as compared to the other two cell lines, and thus, flow cytometry was further used to validate the use of luciferase reduction in MT4-CCR5-Luc cells as an estimate for percent ADCC. ADCC was concurrently estimated by luciferase reduction and the loss of live HIV+ target cells after co-culture with CD16+KHYG-1 cells in the absence as compared to the presence of HIVIG. Zombie UV staining was used to distinguish live from dead target cells (Fig. 5A), and the tat-inducible GFP was used to differentiate infected from uninfected cells (Fig. 5A – 5C). At a single HIVIG dilution, similar ADCC estimates were obtained with the two methods against NL4–3 infected MT4-CCR5-Luc cells (p = 0.25, Wilcoxon matched-pair test) (Fig. 5D). As a further test, HIVIG induced ADCC was also concurrently estimated using both flow cytometry and luciferase decrease against HIV-VSV-G pseudovirus infected MT4-CCR5-Luc cells that did not express HIV-1 Env (Fig. 5E and 5F). Flow cytometry as compared to luciferase reduction yielded a not statistically significant lower ADCC estimate against cells that do not express HIV-1 Env (p = 0.25, Wilcoxon matched-pair test) (Fig. 5G). Importantly, these negative control estimates were below the pre-defined threshold (25%) for positive ADCC. This suggests that our reduction in reporter gene expression is a valid way to estimate percent ADCC in a high throughput manner.
Figure 5. Proposed ADCC assay has been validated via flow cytometry.
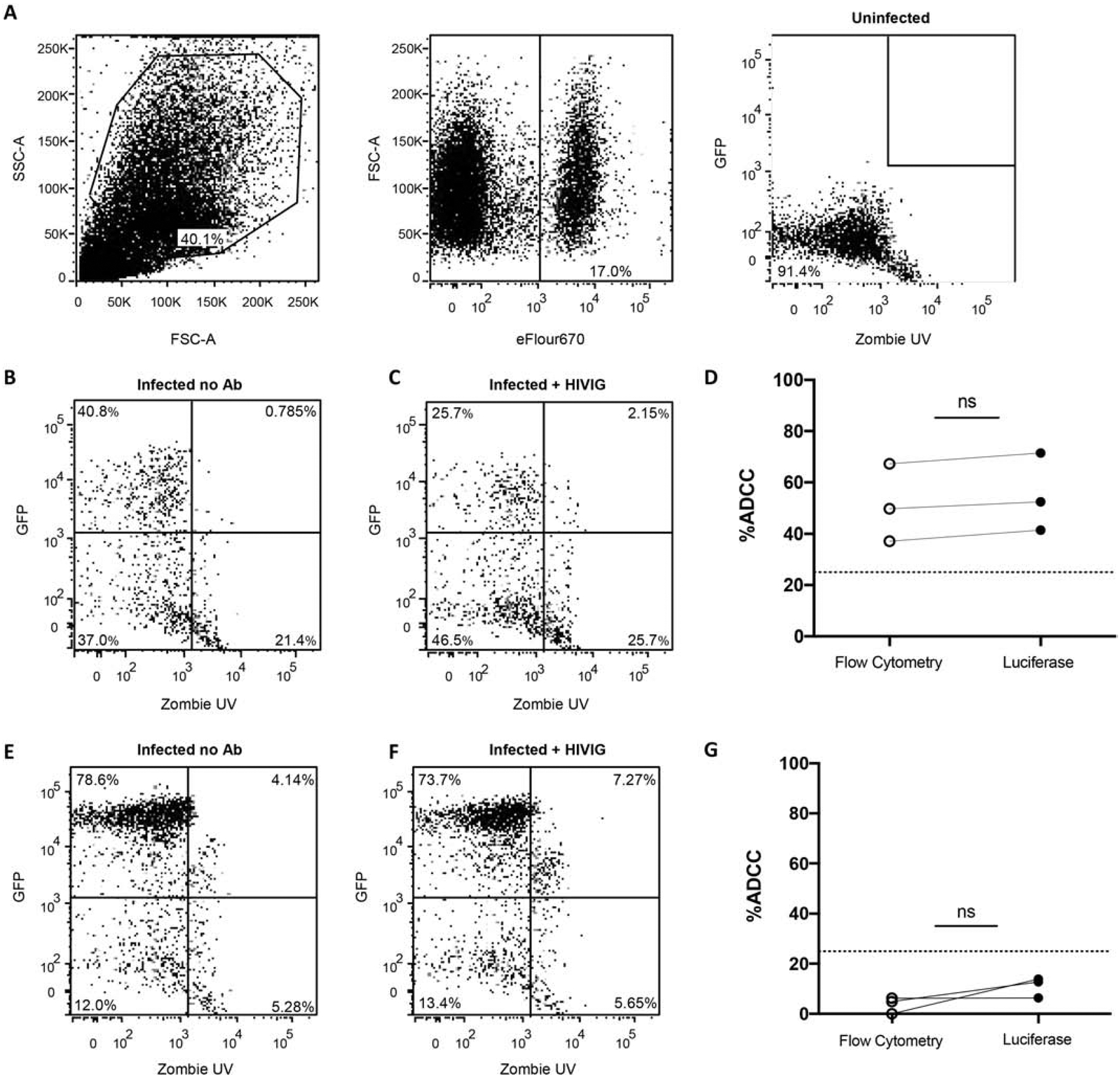
MT4-CCR5-Luc cells were stained with eFluor670 prior to use in assay. (A) Gating strategy is shown for the condition with uninfected MT4-CCR5-Luc cells and NK cells, which represents background signal. The final panel shows live GFP+ target cells (Q1). (B) NL43-infected MT4-CCR5-Luc cells and NK cells (no antibody added). (C) NL43-infected MT4-CCR5-Luc cells, NK cells, and HIVIG. (D) Percent ADCC calculated using luciferase readout and percent ADCC calculated as the change in percentage of live GFP+ target cells (Q1) in the flow cytometry analysis (p = 0.25, Wilcoxon matched-pair test). (E) NL43-ΔEnv-Luc-VSVG-infected MT4-CCR5-Luc cells and NK cells (no antibody added). (F) NL43-ΔEnv-Luc-VSVG-infected MT4-CCR5-Luc cells, NK cells, and HIV Ig. (G) Nonspecific cytotoxicity of NL43-ΔEnv-Luc-VSVG-infected MT4-CCR5-Luc cells observed using luciferase and flow cytometry readout.
3.7. ADCC estimates using MT4-CCR5-Luc cells are not biased from plasma contents.
A large number of individual’s plasma samples have been screened for neutralization breadth and potency using the TZM-bl assay, and bnAbs were engineered from the “elite neutralizers” cells [39, 40]. Similarly, future studies may screen individual’s plasma for ADCC breadth and potency to identify those that harbor “elite ADCC” activity, however, it is unclear as to whether plasma contents other than IgG skew ADCC estimates obtained from MT4-CCR5-Luc cells. IgG was isolated from HIV-1 positive plasma using Melon G columns, and ADCC estimates were compared among the isolated IgG and the original plasma sample. The amounts of isolated IgG used was estimated as the same as that present in a dilution of the original plasma sample. There was no significant difference between isolated IgG and whole plasma (p = 0.563, Wilcoxon matched-pair test) (Fig. 6A). Additionally, 500 μg/mL HIVIG and 500 μg/mL HIVIG spiked into HIV-1 negative heat inactivated plasma yielded similar ADCC activity against NL4–3 infected MT4-CCR5-Luc cells (mean 56.7% and 58.9%, respectively) (Fig. 6B). These observations suggest ADCC estimates from using MT4-CCR5-Luc cells as targets are not skewed by plasma contents aside from IgG, the variable of interest. Thus, future studies may be conducted with plasma as opposed to isolated IgG, which decreases both time and effort.
Figure 6. Plasma contents, besides IgG, do not impact ADCC estimates.
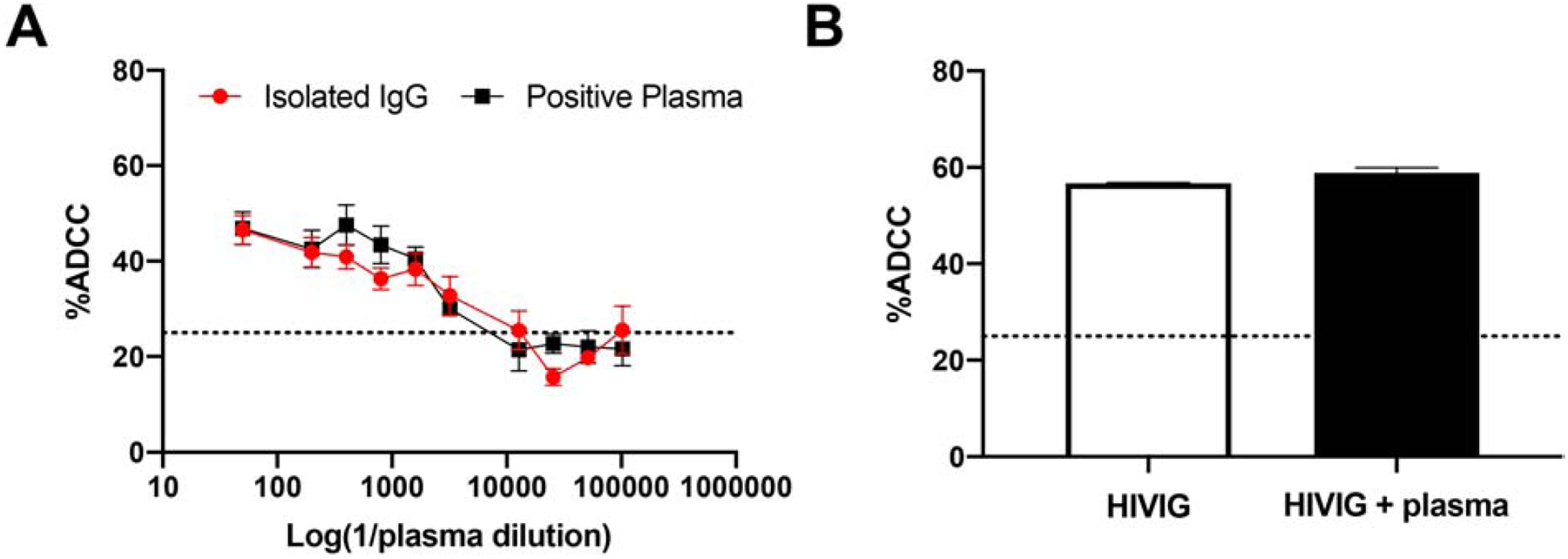
Percent ADCC (y-axis) observed against NL4–3 infected MT4-CCR5-Luc cells with plasma from an antiretroviral naïve individual versus IgG isolated from that plasma (top dilution 1:50) (A) and 500 μg/mL HIVIG versus 500 μg/mL HIVIG spiked into heat inactivated HIV-1 seronegative plasma (B). The bars show SEM and values are from 2–3 independent experiments with each condition done in triplicate. Dashed line at y = 25 denotes threshold for nonspecific cytotoxicity.
3.8. MT4-CCR5-Luc cells can be used to estimate ADCC against viruses with primary Envs of interest
We have previously examined the plasma neutralization capacity of maternal infant dyads against HIV-1 variant’s circulating in the infected mother [34]. Plasma ADCC capacity against exposure strains has not been examined in these mother infant dyads primarily because previous assays used CEM.NKr-CCR5-Luc cells, and these cells failed to express adequate reporter levels after infection (Fig. 1 and 4). We assessed ADCC present in the maternal and infant samples against these primary exposure strains using our new cell line (Fig. 7A). All maternal and only one infant plasma dilution yielded ADCC estimates above the non-specific threshold implying that maternal but not the infant plasma harbored ADCC activity.
Figure 4. PM1-CCR5-Luc and MT4-CCR5-Luc as compared to CEM.NKr-CCR5-Luc support replication of diverse HIV-1 variants.
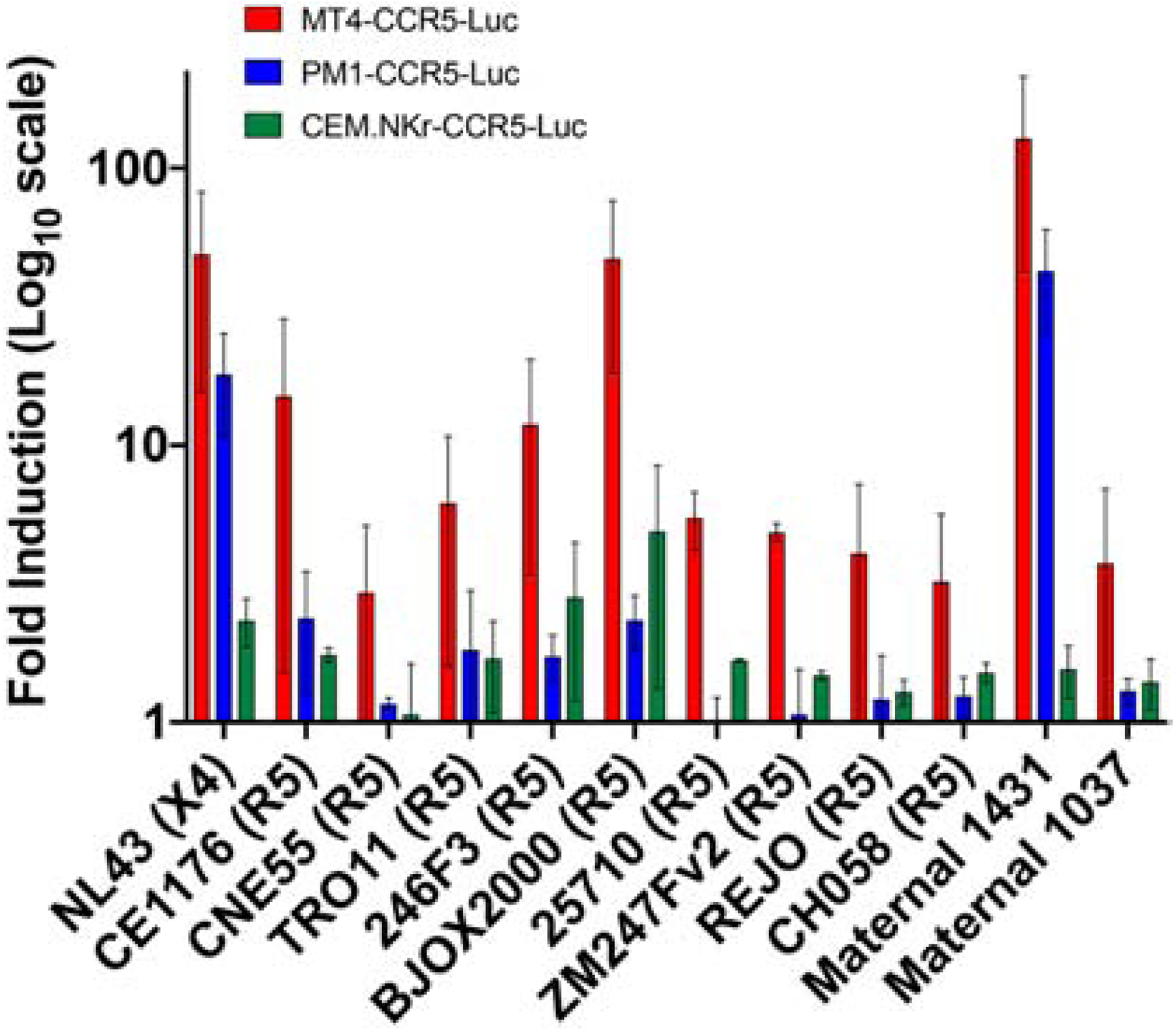
Relative light unit (RLU) fold induction 4 days post infection in MT4-CCR5-Luc (red), PM1-CCR5-Luc (blue), and CEM.NKr-CCR5-Luc cells (green) infected with various viruses compared to uninfected cells. Viruses and co-receptor usage are indicated at the bottom. Bars represent standard error of the mean (SEM) from minimum of 2 independent experiments.
Figure 7. MT4-CCR5-Luc cells can be used to estimate ADCC against viruses with primary Envs of interest.
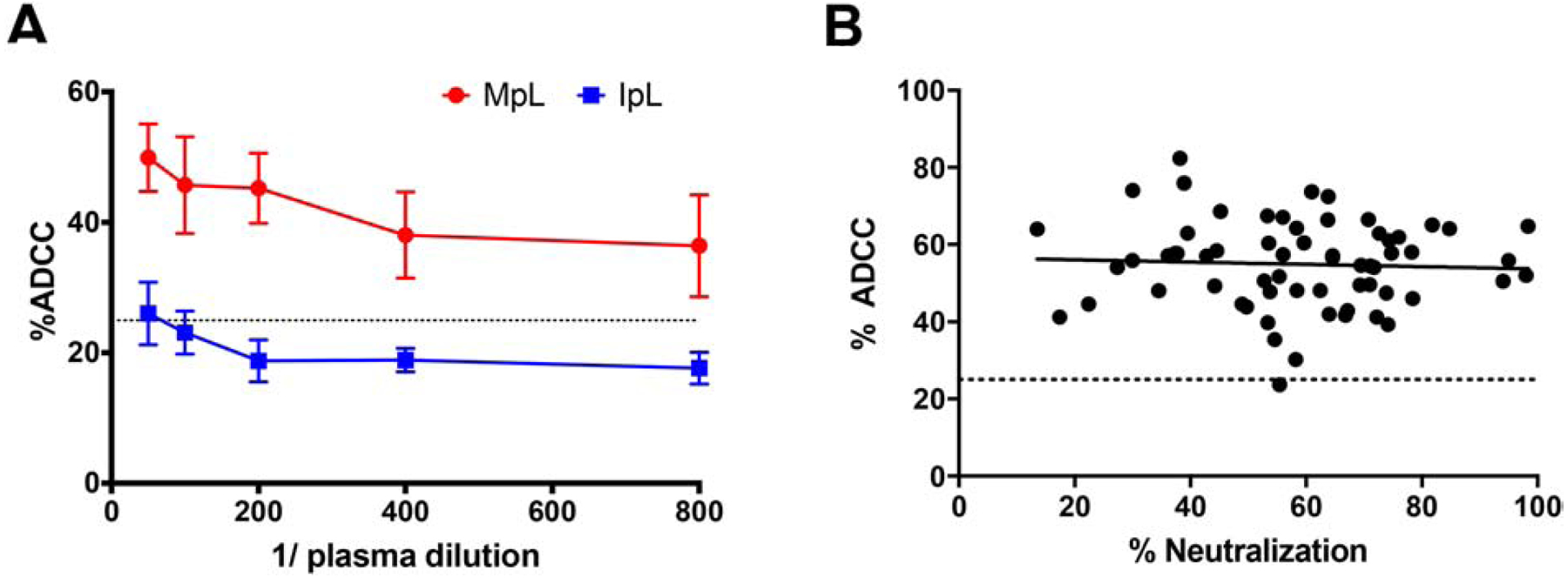
(A) MT4-CCR5-Luc cells were infected with viruses representing the circulating strains present in an infected mother. Percent ADCC (y-axis) was determined after incubating infected cells with NK cells and a serial dilution (x-axis) of either maternal plasma (MpL) or infant plasma (IpL). Top dilution was 1:50 of plasma. Bars show SEM from three independent experiments with each condition tested in triplicate. (B) Heterologous ADCC responses of maternal plasma against BJOX2000-infected cells (y-axis) were plotted against neutralization responses (x-axis) from the same samples against the same virus. Dashed line at y = 25 denotes threshold for nonspecific cytotoxicity.
We have also previously examined the ability of maternal and infant plasma to block viruses incorporating global reference Envs to estimate neutralization breadth and potency [34]. ADCC against viruses with global panel Envs has not been examined before because CEM.NKr-CCR5-Luc cells often fail to yield adequate luciferase levels after exposure to viruses incorporating global panel reference Envs (Fig. 4). We used our assay to examine ADCC responses from 63 maternal plasma samples against a virus with one of the reference global panel Env, BJOX2000 [30]. Interestingly, plasma ADCC activity did not correlate with neutralization capacity against BJOX2000 among these 63 maternal samples (p = 0.872, r = 0.021, Spearman’s rank test) (Fig. 7B), suggesting neutralization and ADCC are not always correlated, similar to what has been suggested in previous publications [12, 41, 42]. All but one maternal sample demonstrated ADCC activity against BJOX2000 above the non-specific threshold.
4. Discussion
We have developed a new cell line, MT4-CCR5-Luc to quantify HIV-1 ADCC using a high-throughput assay, originally developed by Alpert et al. [13]. There is great interest in examining antibody dependent Fc-mediated effector cell lysis because presence of ADCC antibodies have been deemed a correlate of protection in human vaccine studies [43–46]. Despite having a high-throughput assay available to detect ADCC activity [13], the field is still unable to evaluate a large number of samples against relevant virus strains, such as those circulating in the transmission source [5], due to issues with reporter gene expression in CEM.NKr-CCR5-Luc cells [20, 24]. We have engineered and validated the use of a new cell line in an ADCC protocol that overcomes this hurdle. Use of this new cell line in our assay will allow investigators to estimate ADCC breadth and potency among antibodies and plasma samples by examining activity against diverse Envs, such as those in the global reference panel [30]. Thus, this new method satisfies an area of need within the HIV antibody field.
The cornerstone of our assay is a target cell line capable of infection with a diverse range of CCR5-using viruses, which make up the majority of early stage infectious variants. The current standard target cell line, CEM.NKr-CCR5-Luc, did not yield adequate reporter expression over background for a large number of HIV-1 variants, limiting the ability to calculate ADCC capacity as a reduction in luciferase levels. Previous reports and communication with other investigators have reported similar difficulties, supporting the need to develop a new cell line for ADCC studies [20, 24]. To do this, we first chose to use human cells of lymphoid origin because these are most similar to primary CD4+ T-cells, which are responsible for >99% of HIV-1 replication in vivo [47]. Next, we chose and engineered cell lines that express CD4, CCR5, and CXCR4 receptors to permit replication of HIV-1 strains with diverse phenotypes. Finally, we introduced a tat-responsive luciferase reporter for rapid, convenient end-point assessment [16]. Similar considerations were made during the engineering of the CEM.NKr-CCR5-Luc cell line, yet in vitro analysis revealed sub-optimal CCR5 expression and low infection rates with a number of R5 strains [20] (Table 1, Figure 1 and 4). The new MT4-CCR5-Luc cell line showed significantly higher reporter gene expression after HIV-1 exposure as compared to CEM.NKr-CCR5-Luc and PM1-CCR5-Luc (Fig. 4). In addition, MT4-CCR5-Luc expressed a greater level of CCR5 receptors, suggesting that increased luciferase may be related to enhanced infection. CCR5 receptor levels, however, do not entirely explain the observed differences in reporter gene expression after virus exposure. For instance, MT4-CCR5-Luc showed significantly greater luciferase expression compared to PM1-CCR5-Luc even though percent of cells with CCR5 was not vastly different (Table 1). Furthermore, PM1-CCR5-Luc and CEM.NKr-CCR5-Luc did not have significant luciferase expression differences after virus exposure even though cell surface CCR5 percentage and mean fluorescence intensity were markedly different. Other factors, such as the density of the CD4 receptor and presence of attachment factors, may influence the observed differences. Our studies did not formally evaluate if CEM.NKr-CCR5-Luc cells fail to become infected after virus exposure, or if these cells induce less reporter gene expression after infection.
Natural killer (NK) cells expressing the low-affinity IgG receptor FcγRIIIa (CD16) are the primary effectors of ADCC in vivo [7]. We use cells derived from the immortalized cell line KHYG-1, as effector cells in this assay [21]. The KHYG-1 parent line was transduced with CD16, yielding CD16+KHYG-1 cells, and the new cell line was shown to express similar levels of CD16 as primary NK cells [13]. Furthermore, the use of cell lines for both targets and effectors reduces the time and resources needed for primary cell isolation and removes donor to donor variability.
CEM.NKr-CCR5 cells were engineered to be resistant to NK cells. NK cells can often kill T lymphocytes in the absence of antibody, which may skew ADCC estimates [37]. We found MT4-CCR5-Luc to be naturally resistant to the CD16+KHYG-1 NK cell line (Fig. 2B), whereas PM1-CCR5-Luc cells showed susceptibility to NK cell killing in the absence of antibodies (Fig. 2A). Although these new cell lines had differing NK susceptibility, ADCC estimates were similar, suggesting that ADCC estimates can still be assessed using targets lacking inherent NK-resistance (Fig. 3). ADCC estimations, however, will require relatively high reporter gene expression when using cells that are sensitive as opposed to resistant to NK cell killing in the absence of antibody. In our assay, we also estimated a threshold level that represents HIV-1 specific ADCC activity. Estimates below this threshold were deemed as non-specific cytotoxicity against HIV-1 infected cells that was not due to HIV-1 Env specific antibodies.
It is possible that excessively high reporter gene expression in the MT4-CCR5-Luc cells after virus exposure may yield skewed ADCC estimates that are based on decreases in luciferase. We confirmed that decreased luciferase reporter expression accurately represents cell killing using flow cytometry to estimate ADCC (Fig. 5). Our results indicate that ADCC calculated using our luciferase assay is analogous to calculating ADCC by the more tedious flow cytometric technique. To overcome the poor susceptibility of cell lines, ADCC assays will often pulse target cells with gp120. The Env monomer, however, can have both receptor dependent and non-specific binding, resulting in overestimation of ADCC responses due to bystander killing [15]. Congruity between the luciferase and flow cytometry-based results suggest that our cell-infection based assay is specifically measuring the elimination of infected virus producing cells.
Our primary motivation in developing a new ADCC assay was to assess whether ADCC is a correlate of protection in mother-to-child-transmission (MTCT) of HIV-1. We have previously shown that nAbs are not a correlate of protection in a cohort of 63 mother-infant pairs where transmission either did or did not occur during breastfeeding in the absence of antiretroviral therapy [34]. Although previous studies have implied that ADCC-inducing antibodies protect against HIV transmission [2, 9, 48], studies have not specifically examined ADCC activity against exposure strains, like those present in the infected mother. We first wanted to confirm that any ADCC activity we saw from plasma samples could be attributed to only the antibodies and not additional factors present in plasma. Our results suggest that only IgG is contributing to ADCC responses observed in plasma (Fig. 6), allowing us to confidently use plasma for high throughput analysis rather than having to first isolate IgG from subject samples.
We detected plasma-induced ADCC against viruses incorporating Envs isolated from a chronically infected mother (Fig. 7A). Importantly, uninfected infants are exposed to these Envs, and thus future studies using our assay will allow us to address if ADCC is a correlate of protection in MTCT of HIV-1. Furthermore, we have shown that ADCC does not always correlate with neutralization, against BJOX2000 (Fig. 7B), supporting the need to test ADCC activity in cohorts which may not have shown protective neutralizing responses [12, 41, 42].
Widespread assessment of ADCC activity in large cohorts against panels of HIV-1 variants has been limited due to the lack of a reliable, high throughput assay. Examining ADCC activity against a broad range of Envs may identify plasma samples that have “elite ADCC” capacity. Antibodies with the ability to mediate broad and potent cellular cytotoxicity may be isolated from these samples in the future. The use of our assay will facilitate these types of studies, which will aid vaccine development and antibody-based treatment efforts.
Highlights.
New cell line, MT4-CCR5-Luc, is resistant to natural killer cells in the absence of antibody.
MT4-CCR5-Luc yield much higher luciferase levels after exposure to viruses with a diverse range of envelopes as compared to other cell lines that could be used to estimate HIV antibody dependent cellular cytotoxicity (ADCC).
ADCC is similar when estimated from either luciferase reduction or decrease in live infected cells as assessed by flow cytometry.
This luciferase-based assay using this new cell line can be used to estimate ADCC against viruses with primary envelopes of interest in an efficient manner
ACKNOWLEGEMENTS
This work was supported by the National Institutes of Health (AI137119, AI122209). AST was supported by National Institutes of Health T32 Training Grant (5T32AI007309-28).
Glossary
- ADCC
Antibody-dependent cellular cytotoxicity
- Env
HIV-1 Envelope
- MTCT
Mother-to-child-transmission
- T/F
Transmitted founder strain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Desrosiers RC (2017) Protection against HIV Acquisition in the RV144 Trial. JVI 91:17–19. doi: 10.1128/JVI.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haynes BF, Gilbert PB, McElrath JM, et al. (2012) Immune-Correlates Analysis of an HIV-1 Vaccine Efficacy Trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruell H, Klein F (2018) Antibody ‑ mediated prevention and treatment of HIV ‑ 1 infection. Retrovirology 1–11. doi: 10.1186/s12977-018-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu C, Murakowski DK, Bournazos S, et al. (2016) Enhanced clearance of HIV-1-infected cells by anti-HIV-1 broadly neutralizing antibodies in vivo. Science (80- ) 352:1001–1004. doi: 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montefiori DC (2004) Evaluating Neutralizing Antibodies Against HIV, SIV, and SHIV in Luciferase Reporter Gene Assays. Curr Protoc Immunol 64:12.11.1–12.11.17. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 6.Boesch A, Brown E, Ackerman M (2015) The role of Fc receptors in HIV prevention and therapy. Immunol Rev 268:296–310. doi: 10.1111/imr.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scully E, Alter G (2016) NK Cells in HIV Disease. Curr HIV/AIDS Rep 13:85–94. doi: 10.1007/s11904-016-0310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlandi C, Flinko R, Lewis GK (2016) A new cell line for high throughput HIV-specific antibody-dependent cellular cytotoxicity (ADCC) and cell-to-cell virus transmission studies. J Immunol Methods 433:51–58. doi: 10.1016/j.jim.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mabuka J, Nduati R, Odem-Davis K, et al. (2012) HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog 8:. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milligan C, Richardson BA, John-Stewart G, et al. (2015) Passively Acquired Antibody-Dependent Cellular Cytotoxicity (ADCC) activity in HIV-infected infants is associated with reduced mortality. Cell Host Microbe 17:500–506. doi: 10.1016/j.chom.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard J, Prévost J, Baxter AE, et al. (2018) Uninfected Bystander Cells Impact the Measurement of HIV-Specific Antibody-Dependent Cellular Cytotoxicity Responses. MBio 9:e00358–18. doi: 10.1128/mBio.00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bredow B Von Arias JF, Heyer LN, et al. (2016) Comparison of Antibody-Dependent Cell-Mediated Cytotoxicity and Virus Neutralization by HIV-1 Env-Specific Monoclonal Antibodies. J Virol 90:6127–6139. doi: 10.1128/JVI.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alpert MD, Heyer LN, Williams DEJ, et al. (2012) A Novel Assay for Antibody-Dependent Cell-Mediated Cytotoxicity against HIV-1- or SIV-Infected Cells Reveals Incomplete Overlap with Antibodies Measured by Neutralization and Binding Assays. J Virol 86:12039–12052. doi: 10.1128/JVI.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruel T, Guivel-Benhassine F, Lorin V, et al. (2017) Lack of ADCC Breadth of Human Nonneutralizing Anti-HIV-1 Antibodies. J Virol 91:1–19. doi: 10.1128/JVI.02440-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard J, Prévost J, Baxter AE, von Bredow B, Ding S, Medjahed H, Delgado GG, Brassard N, Stürzel CM, Kirchhoff F, Hahn BH, Parsons MS, Kaufmann DE, Evans DT, Finzi A (2018) Uninfected Bystander Cells Impact the Measurement of HIV-Specific Antibody-Dependent Cellular Cytotoxicity Responses. MBio 9:e00358–18. doi: 10.1128/mBio.00358-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trkola A, Matthews J, Gordon C, et al. (1999) A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol 73:8966–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe H, Farzan M, Sun Y, et al. (1996) The NL-Chemokine Receptors CCR3 and CCR5 Facilitate Infection by Primary HIV-1 Isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 18.Deng H, Unutmaz D, Kewalramani VN, Littman DR (1997) Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296–300 [DOI] [PubMed] [Google Scholar]
- 19.Doranz BJ, Rucker J, Yi Y, et al. (1996) A Dual-Tropic Primary HIV-1 Isolate That Uses Fusin and the NL-Chemokine Receptors CKR-5, CKR-3, and CKR-2b as Fusion Cofactors. Cell 85:1149–1158 [DOI] [PubMed] [Google Scholar]
- 20.Spenlehauer C, Gordon CA, Trkola A, Moore JP (2001) A Luciferase-Reporter Gene-Expressing T-Cell Line Facilitates Neutralization and Drug-Sensitivity Assays That Use Either R5 or X4 Strains of Human Immunodeficiency Virus Type 1. Virology 280:292–300. doi: 10.1006/viro.2000.0780. [DOI] [PubMed] [Google Scholar]
- 21.Yagita M, Huang C, Umehara H, et al. (2000) A novel natural killer cell line (KHYG-1) from a patient with aggressive natural killer cell leukemia carrying a p53 point mutation. Leukemia 14:922–30 [DOI] [PubMed] [Google Scholar]
- 22.Bournazos S, Ravetch JV (2017) Anti- retroviral antibody FcγR- mediated effector functions. Immunol Rev 285–295. doi: 10.1111/imr.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimmerjahn F, Ravetch JV. (2008) Fcγ receptors as regulators of immune responses. Nat Rev Immunol 8:34–47. doi: 10.1038/nri2206 [DOI] [PubMed] [Google Scholar]
- 24.Li H, Chen BK (2019) Variable infectivity and conserved engagement in cell-to-cell viral transfer by HIV-1 Env from Clade B transmitted founder clones. Virology 526:189–202. doi: 10.1016/j.virol.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lusso P, Cocchi F, Balotta C, et al. (1995) Growth of Macrophage-Tropic and Primary Human Immunodeficiency Virus Type 1 (HIV-1) Isolates in a Unique CD4 ϩ T-Cell Clone (PM1): Failure To Downregulate CD4 and To Interfere with Cell-Line-Tropic HIV-1. J Virol 69:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham FL, Smiley J (1977) Characteristics of a Human Cell Line Transformed by D N A from Human Adenovirus Type 5. J gen Virol 36:59–74 [DOI] [PubMed] [Google Scholar]
- 27.Platt EJ, Bilska M, Kozak SL, et al. (2009) Evidence that Ecotropic Murine Leukemia Virus Contamination in TZM-bl Cells Does Not Affect the Outcome of Neutralizing Antibody Assays with Human Immunodeficiency Virus Type 1. JVI 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kane M, Zang TM, Rihn SJ, et al. (2016) Identification of Interferon-Stimulated Genes with Antiretroviral Activity Resource Identification of Interferon-Stimulated Genes with Antiretroviral Activity. Cell Host Microbe 20:392–405. doi: 10.1016/j.chom.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahm H-W, Stein S (1985) Characterization of recombinant human interleukin-2 with micromethods. J Chromatogr A 326:357–361. doi: 10.1016/S0021-9673(01)87461-6 [DOI] [PubMed] [Google Scholar]
- 30.deCamp A, Hraber P, Bailer RT, et al. (2014) Global Panel of HIV-1 Env Reference Strains for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J Virol 88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatziandreou N, Arauz AB, Freitas I, et al. (2012) Sensitivity Changes over the Course of Infection Increases the Likelihood of Resistance Against Fusion but Not CCR5 Receptor Blockers. AIDS Res Hum Retroviruses 28:1584–1593. doi: 10.1089/aid.2011.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etemad B, Fellows A, Kwambana B, et al. (2009) Human Immunodeficiency Virus Type 1 V1-to-V5 Envelope Variants from the Chronic Phase of Infection Use CCR5 and Fuse More Efficiently than Those from Early after Infection. J Virol 83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keele BF, Giorgi EE, Salazar-gonzalez JF, et al. (2008) Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. PNAS 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghulam-Smith M, Olson A, White LF, et al. (2017) Maternal but not infant anti-HIV-1 neutralizing antibody response associates with enhanced transmission and infant morbidity. MBio 8:1–19. doi: 10.1128/mBio.01373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, GIlbert PB, Hioe CE, et al. (2012) Statistical approaches to analyzing HIV-1 neutralizing antibody assay data. Stat Biopharm Res 4:1–13. doi: 10.1080/19466315.2011.633860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rihn SJ, Foster TL, Busnadiego I, et al. (2017) The Envelope Gene of Transmitted HIV-1 Resists a Late IFNγ-Induced Block. JVI. doi: 10.1128/JVI.02254-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howell DN, Andreotti PE, Dawson JR, Cresswell P (1985) Natural killing target antigens as inducers of interferon: studies with an immunoselected, natural killing-resistant human T lymphoblastoid cell line. J Immunol 134:971–976 [PubMed] [Google Scholar]
- 38.Salazar-Gonzalez JF, Salazar MG, Keele BF, et al. (2009) Genetic identity, biological phenotype, and evolutionary pathways of transmitted / founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simek MD, Rida W, Priddy FH, et al. (2009) Human Immunodeficiency Virus Type 1 Elite Neutralizers : Individuals with Broad and Potent Neutralizing Activity Identified by Using a High-Throughput Neutralization Assay together with an Analytical Selection Algorithm. JVI 83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landais E, Moore PL (2018) Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology 15:1–14. doi: 10.1186/s12977-018-0443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smalls-Mantey A, Doria-Rose N, Klein R, et al. (2012) Antibody-Dependent Cellular Cytotoxicity against Primary HIV-Infected CD4+ T Cells Is Directly Associated with the Magnitude of Surface IgG Binding. J Virol 86:8672–8680. doi: 10.1128/JVI.00287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Bredow B, Andrabi R, Grunst M, et al. (2019) Differences in the Binding Affinity of an HIV-1 V2 Apex- Specific Antibody for the SIV smm / mac Envelope Glycoprotein Uncouple Antibody-Dependent Cellular Cytotoxicity from Neutralization. MBio 10:1–12. doi: 10.1128/mBio.01255-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott-algara D, Truong LX, Versmisse P, et al. (2003) Cutting Edge: Increased NK Cell Activity in HIV-1-Exposed but Uninfected Vietnamese Intravascular Drug Users. J Immunol 5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 44.Lambotte O, Ferrari G, Moog C, et al. (2009) Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS 23:897–906. doi: 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wren LH, Chung AW, Isitman G, et al. (2013) Specific antibody-dependent cellular cytotoxicity responses associated with slow progression of HIV infection. Immunology 138:116–123. doi: 10.1111/imm.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madhavi V, Wines BD, Amin J, Emery S (2017) HIV-1 Env- and Vpu-Specific Antibody-Dependent Cellular Cytotoxicity Responses Associated with Elite Control of HIV. J Virol 91:e00700–17. doi: 10.1128/JVI.00700-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perelson AS, Neumann AU, Markowitz M, et al. (1996) HIV-1 Dynamics in Vivo : Virion Clearance Rate, Infected Cell Life-Span, and Viral Generation Time. Science (80- ) 271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 48.Permar SR, Fong Y, Vandergrift N, et al. (2015) Maternal HIV-1 envelope – specific antibody responses and reduced risk of perinatal transmission. J Clin Invest 125:0–4. doi: 10.1172/JCI81593. [DOI] [PMC free article] [PubMed] [Google Scholar]


