Abstract
Phosphorylation of translational repressor eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) controls the initiation of cap-dependent translation, a type of protein synthesis that is frequently upregulated in human diseases such as cancer. Because of its critical cellular function, it is not surprising that multiple kinases can post-translationally modify 4E-BP1 to drive aberrant cap-dependent translation. We recently reported a site-selective chemoproteomic method for uncovering kinase-substrate interactions, and using this approach, we discovered the cyclin-dependent kinase CDK4 as a new 4E-BP1 kinase. Herein, we describe our extension of this work and reveal the role of CDK4 in modulating 4E-BP1 activity in the transition from mitosis to G1, thereby demonstrating a novel role for this kinase in cell cycle regulation.
Keywords: 4E-BP1, mitosis, CDK4, cap-dependent translation
INTRODUCTION
Cap-dependent translation is an important cellular process that controls the translation of select mRNAs typically encoding for growth factors and oncogenes.1–4 The initiation of cap-dependent mRNA translation is governed by the availability of eIF4E, the m7GpppX-cap-binding translation initiation factor.5,6 This protein is highly regulated, primarily through the work of the 4E-BPs, which sequester eIF4E from eIF4G and the eIF4F translation initiation complex.7–13 The activity of 4E-BP1 is in turn regulated by phosphorylation, where hypophosphorylated 4E-BP1 binds strongly to eIF4E to inhibit translation, while hyperphosphorylated 4E-BP1 releases eIF4E to initiate cap-dependent translation.13–16 For many years, the only validated kinase known to affect 4E-BP1 phosphorylation has been mechanistic target of rapamycin complex 1 (mTORC1), which was shown to hierarchically phosphorylate 4E-BP1 at T37 and T46 followed by T70 and S65.14,15,17,18 However, several findings have called into question the exclusivity of mTORC1 for each of these phosphorylation sites,19 namely reports demonstrating that other unknown kinases can also phosphorylate 4E-BP1 to stimulate cap-dependent translation,20–22 particularly in cases of mTOR inhibitor drug resistance.23–26
Recently, our laboratory has developed a chemoproteomic pipeline by which to identify site-specific kinase-substrate interactions, Phosphosite-Accurate kinase-substrate cross(X)linking Assay or PhAXA.27 Using this methodology, we discovered that cyclin-dependent kinase 4 (CDK4), which is primarily responsible for controlling the cell cycle checkpoint at the G1/S transition through phosphorylation of the retinoblastoma tumor suppressor protein (Rb),28 regulates cap-dependent translation via phosphorylation of 4E-BP1 at both canonical mTORC1 sites (T37, T46, T70) in addition to a non-canonical site (S101).27 Importantly, we found that CDK4 can promote rapamycin-resistant cap-dependent translation through this function, and inhibition of CDK4 using the clinically approved CDK4/6 inhibitor palbociclib led to a significant reduction in the expression of cap-dependent transcripts c-Myc and cyclins D2 and D3. 27 Moreover, we found that inhibition of both mTORC1 and CDK4 could cooperatively antagonize the initiation of cap-dependent translation. 27
While our report is the first to directly connect CDK4 to 4E-BP1 regulation, this is not the only cyclin-dependent kinase linked to phosphorylation of this translational repressor. CDK12, via phosphorylation of 4E-BP1 at S65 and T70, cooperates with mTORC1 to drive selective translation of proteins involved in maintenance of the mitotic genome.29 Additionally, CDK1, the master regulator of the G2/M transition, can substitute for mTORC1 to phosphorylate 4E-BP1 at the putative mTORC1 sites to activate cap-dependent translation during mitosis30,31 and meiosis.32 A mechanistic investigation of the mitotic phosphorylation of 4E-BP1 uncovered CDK1-mediated phosphorylation of the non-canonical site S83. This poorly understood phosphorylation site was demonstrated to function outside of the traditional context of eIF4E regulation and cap-dependent translation, and instead localizes 4E-BP1 to the mitotic spindle.33 Further evaluation of cell cycle-dependent cap-dependent translation has found that eIF4G interacts with eIF4E to similar degrees in interphase and mitotic cells.34 These findings are exciting, as it was previously thought that mitosis was associated with a decrease in cap-dependent translation,35,36 and together, hint at a larger role for phosphorylated 4E-BP1 and cap-dependent translation in regulation of the cell cycle.37 However, as the studies linking CDK1 and 4E-BP1 relied on the use of the CDK1 inhibitor RO-3308,38 which is typically used as a tool compound for inducing mitotic arrest at the precipice of prophase,39 they provide only a snapshot of the regulation of 4E-BP1 during a narrow window of mitosis. Thus, we became interested in using PhAXA to uncover potentially novel kinases that regulate 4E-BP1 hyperphosphorylation in later stages of mitosis once the anaphase-promoting complex mediates degradation of cyclin B1,rendering CDK1 inactive.40 Herein, we describe these efforts and reveal a novel function of CDK4 in driving mitotic cap-dependent translation and G2/M cell cycle progression.
MATERIALS AND METHODS
Reagents
Palbociclib isethionate (Selleckchem) was dissolved in water. Rapamycin (Alfa Aesar) and SP600125 (ApexBio) were dissolved in DMSO. Human recombinant insulin was purchased from Sigma. Nocodazole and 3XFLAG peptide were purchased from ApexBio. All reagents were used as received.
Cell Culture
HEK293T and HeLa cells were grown in DMEM (Corning) supplemented with 10% FBS, glutamine, penicillin, and streptomycin (Gibco). U2 OS cells were kindly provided by Dr. Beth Lawlor and cultured according to ATCC guidelines. MDA-MB-231 cells were a kind gift from Dr. Nouri Neamati and grown in RPMI-1640 media supplemented with 10% FBS and glutamine. MDA-MB-468 and MCF-7 cells were a kind gift from Dr. Max Wicha. MDA-MB-468 cells were grown in DMEM (Corning) supplemented with 10% FBS and glutamine. MCF-7 cells were cultured according to ATCC guidelines. Cells were grown at 37 °C with 5% CO2 in a humidified incubator, passaged at least twice before use for experiments and no more than 10 times before returning to low passage stocks. All cell lines were authenticated by STR profiling, and regularly tested for mycoplasma contamination.
Immunoblotting
Cells were lysed directly in-well using RIPA buffer (10 mM Tris-HCl, 150 mM NaCl, 1% Triton, 1% sodium deoxycholate, 0.1% SDS, pH 7.2) supplemented with 10 μg/mL aprotinin, 5 μg/mL leupeptin, 7 μg/mL pepstatin, 10 mM NaF, 2 mM sodium orthovanadate, 10 mM β-glycerophosphate, and 2 mM sodium pyrophosphate). Lysates were then sonicated thoroughly on ice. Protein concentrations were normalized by the BCA assay (Pierce), resolved on 4–20% Tris-glycine gels (Invitrogen), transferred to 0.45-μm PVDF (Thermo) using Towbin’s buffer (low amperage for ~4 h at 4 °C), blocked with 5% non-fat milk in TBST, then probed with primary antibodies overnight at 4 °C. Antibodies used in this study were the following: Actin-HRP (sc-47778) from Santa Cruz Biotechnology; CDK4 (12790), Cyclin D2, (3741), 4E-BP1 (9644), p4E-BP1 (T37/46) (2855), p4E-BP1 (S65/101) (9451), p4E-BP1 (T70) (9455), Rb (9313), pRb (S780) (3590), eIF4E (9742), eIF4G (2498), and pS6 (240/244) (2215) from Cell Signaling Technology.
Chemoproteomics
The Phosphosite-Accurate kinase-substrate cross(X)linking Assay (PhAXA) was carried out as previously described,27 with changes to the database search and relative protein quantification. Protein identification and quantification was performed using MaxQuant (version 1.6.7.0).41 MS/MS spectra were searched with Andromeda against the reference human database from Uniprot (02-02-2014 download) appended with common contaminants and the automatically generated reverse database for the decoy search, which was used to calculate the false-discovery rate (FDR). Carbamidomethylation of cysteine (57.021464 Da) was set as a fixed modification; acetylation of protein N-termini (42.010565 Da) and oxidation of methionine (15.994915 Da) were set as variable modifications. Other search parameters included fixed main-search MS1 error of 4.5ppm, with 0.5 Da mass deviation allowed for fragment ions; minimum peptide length of 7; two missed cleavages were allowed. Match between runs was enabled with a match time window of 0.5 min.
Data Analysis
Relative quantification of proteins was achieved with the MaxLFQ algorithm using default settings. Proteins identified with a FDR of <1% were further filtered by removal of known contaminants and decoy proteins, and those proteins identified by fewer than 3 PSMs in both samples, as well as those identified by a single peptide. This final list of proteins was loaded into Perseus (Version 1.6.5.0) and LFQ intensities were log2 transformed before imputing missing values column-wise, based on a normal distribution (downshift of 1.8 and a width of 0.3).
m7GDP cap affinity assay
The cap pull-down assay was carried out as previously described.42,43
NanoBiT assay
eIF4E was digested out of HaloTag-eIF4E using SgfI and PmeI, and then ligated into pFN33K (Promega) to obtain the LgBiT-eIF4E construct. 4E-BP1 was cloned into pFN35K (Promega) using the same method to obtain the SmBiT-4E-BP1 construct. HaloTag-4E-BP1 and -eIF4E have been described elsewhere. 27,44,45 pFN33K LgBiT-eIF4E and pFN35K SmBiT-4EBP1 (50 ng each) were reverse-transfected into MCF-7 and MDA-MB-468 cells in a 96-well white opaque plate using Lipfectamine LTX with PLUS reagent. After 16 h, cells were arrested with nocodazole (500 nM). 20 h later, cells were treated with rapamycin (100 nM) and/or palbociclib (5 μM). After 2 h incubation, Nano-Glo Live cell reagent (Promega #N2011; 25 μL) was added and total luminescence was read within 40 min on a BioTek Cytation 3 reader.
Flow cytometry
Cells were harvested with trypsin, washed once with ice-cold 1× PBS containing 1% FBS, re-suspended in ice cold 1× PBS, and then 100% ice-cold ethanol was added dropwise to a final concentration of 70%. Cells were fixed at −20 °C for 4 h, and then stored at 4 °C for 18–96 h. Fixed cells were washed twice with 1× PBS containing 1% FBS, re-suspended in 1× PBS containing propidium iodide (50 μg/mL; Sigma) and RNase A (100 μg/mL, Fisher), and incubated at 37 °C for 30 min. Cells were then filtered and analyzed using a Cytoflex flow cytometer (Beckman). Cell cycle distributions were analyzed using FlowJo (v10).
Statistical Analysis
Two-sided t-tests were performed using Prism (v7); equal variance between samples being compared was established. Graphs show mean +/− S.E.M or +/− standard deviation as described in the figure legends.
RESULTS AND DISCUSSION
Given our limited knowledge regarding 4E-BP1 phosphorylation in mitosis, coupled with previous reports which demonstrated that this post-translational modification is mTOR independent,30,33 we used PhAXA-based chemoproteomic profiling 27 to identify candidate 4E-BP1 kinases. HEK293T cells were transiently transfected with wild-type (WT) or T46C mutant FLAG-4E-BP1 probes, arrested in prometaphase using nocodazole, and released into media containing insulin before harvesting and proceeding with PhAXA analysis. 27 Four protein kinases were identified: mTOR, CDK4, Erk2 and PRKDC, with mTOR and CDK4 showing the greatest level of enrichment, in line with our previous results using asynchronous cells (Figure 1A and 1B). 27 Pulldown of CDK4 from lysate was then confirmed via PhAXA and Western blot, 27 and enrichment was observed from samples expressing both T37C and T46C mutant probes (Figure 1C).
Figure 1. PhAXA identifies putative 4E-BP1 kinases in cells released from a nocodazole-arrest.
(A) Log2 fold change of LFQ ratios for the 3XFLAG-4E-BP1 (T46) probe relative to appropriate controls. ‘+’ refers to samples treated with crosslinker, while ‘-‘ refers to ATP only controls. Fold-changes were capped at 100 for this graph. Dotted lines represent 4-fold enrichment. (B) Table of all protein kinases identified by 3 or more peptide-spectrum-matches (PSMs) in at least one sample. (C) CDK4 is enriched from lysate generated from nocodazole arrested cells expressing 3XFLAG-4E-BP1 WT, T37C and T46C mutants. A representative input is included to show the mass shift of CDK4 upon crosslinking to 4E-BP1.
The discovery of CDK4 was unexpected given the relative lack of information linking CDK4 to regulation of mitosis and/or cytokinesis. Thus, we examined a panel of cell lines to determine the role of CDK4 in mediating mitotic phosphorylation of 4E-BP1. To probe this, prometaphase-arrested cells were released into media containing vehicle control, rapamycin, and/or palbociclib, and inhibition of 4E-BP1 phosphorylation was monitored by Western blot. As expected, rapamycin, the allosteric mTORC1 inhibitor, had no effect on nocodazole-induced mitotic phosphorylation of 4E-BP1 (Figure 2). However, cells released from nocodazole arrest in media containing the clinically approved CDK4/6 inhibitor palbociclib46,47 showed a marked decrease in phosphorylation, as demonstrated by the disappearance of the β and γ bands that indicate hyperphosphorylated 4E-BP1 (Figure 2). In MDA-MB-468 cells, 4E-BP1 phosphorylation was unaffected by CDK4 inhibition, similar to our previous findings in asynchronously proliferating cells. The lack of a response is likely due to the intrinsic resistance of this cell line to CDK4/6 inhibitors, resulting from an inability to assemble functional cyclin D-CDK4/6 complexes.48 Interestingly, in CDK4/6 inhibitor sensitive cells, palbociclib-induced downregulation of mitotic 4E-BP1 phosphorylation was markedly increased by combined treatment with rapamycin (Figure 2). These findings indicate that mTOR and CDK4 act in concert to regulate 4E-BP1 phosphorylation following release from prometaphase arrest.
Figure 2. Mitotic Phosphorylation of 4E-BP1 is Palbociclib-Sensitive.
Nocodazole-arrested cells (500 nM, 20 h) were treated with rapamycin (100 nM) and/or palbociclib (5 μM) in fresh media containing insulin (150 nM) for 2 h. Blots for pRb in HEK293T and MDA-MB-468 cells are not shown as they are Rb inactive and null, respectively.
To verify that palbociclib treatment was on-target, the phosphorylation state of 4E-BP1 was compared to that of Rb at S780, the canonical CDK4 substrate. A broad range of sensitivity to palbociclib was observed across the cell lines tested, ranging from <330 nM in U2OS cells to ~1.25 μM in HeLa cells (Figure 3A). In each case, however, the effective concentration of palbociclib required to inhibit both 4E-BP1 and Rb phosphorylation at S780 correlated with the relative expression of CDK4 in each cell line (Figure 3B). A similar effect was seen in HEK293T cells, which required a concentration of >1.25 μM to fully inhibit 4E-BP1 phosphorylation and, along with HeLa cells, have the highest relative expression of CDK4 at the protein level (Figure S1). While there are clearly other factors that affect CDK4 inhibitor sensitivity, including Rb status and D-cyclin expression levels among others,49 this correlation was noteworthy.
Figure 3. 4E-BP1 phosphorylation mirrors pRb in a dose-dependent manner.
(A) Nocodazole arrested cells were treated with rapamycin (100 nM) and/or palbociclib (5 μM – 313 nM) in fresh media containing insulin (150 nM) for 2 h. (B) Western blot of relative CDK4 expression in cell lines used in (A). Samples were run on the same gel and are from the same exposure, with unnecessary lanes removed for clarity.
While CDK4 inhibition clearly reduced mitotic phosphorylation of 4E-BP1, we were unsure of its impact on cap-dependent translation. Thus, we utilized the m7GDP cap-binding assay (Figure 4B) to measure assembly of the eIF4F translation initiation complex.50 Mitotic MCF-7 and HeLa cells treated with rapamycin or palbociclib alone showed a marginal increase in 4E-BP1 associated with cap-bound eIF4E; yet, the eIF4G-eIF4E interaction was only modestly affected by these conditions (Figure 4A). With the combination, however, inhibition of eIF4F complex formation was enhanced (Figure 4A). This benefit was observed only in cells released from nocodazole arrest, as short treatments with palbociclib had very little effect on eIF4F assembly in asynchronous cells (Figure 4A). As expected, MDA-MB-468 cells did not respond to CDK4 inhibition, but displayed a predictable response to rapamycin (Figure 4A).
Figure 4. mTOR and CDK4 Co-regulate Mitotic Cap-Dependent Translation.
(A) m7GDP cap affinity assay. Palbociclib and rapamycin cooperate to increase eIF4E-4E-BP1 association and decrease eIF4E-eIF4G association in palbociclib-sensitive cells. (B) Schematic for the cap-affinity assay. (C) Palbociclib results in the reduction of cap-dependent translation of cyclin D2 in nocodazole-arrested U2 OS cells. U2 OS cells were used for this experiment as they are one of the few cell lines in our panel to express satisfactory amounts of cyclin D2. All experiments were performed in biological triplicate; representative images are shown. Replicates of the data shown in (A) and (C) can be found in Figure S2.
As further confirmation of the impact of CDK4 on cap-dependent translation, we analyzed the expression of cyclin D2, an established cap-dependent transcript,51 in mitotic and asynchronous U2OS cells treated with palbociclib and/or rapamycin. In asynchronous cells, inhibition of CDK4 had no effect; whereas, rapamycin markedly reduced the levels of this protein (Figure 4C). Conversely, in cells recently released from a prometaphase block, rapamycin had negligible effect, while inhibition of CDK4 resulted in a reduction of cyclin D2 at the protein level (Figure 4C). Because cyclin D2 expression was found to be higher in nocodazole arrested cells than in asynchronous cells, this may allude to a requirement for CDK4/6 activity post-metaphase.
Although the m7GDP cap affinity assay is a robust way of analyzing the ratio of 4E-BP1- and eIF4G-bound eIF4E, it is performed in lysate, thus rendering any intricacies in sub-cellular localization of these proteins indeterminable. Therefore, a split-nanoluciferase-based assay was developed to quantify the eIF4E-4E-BP1 interaction in live cells (Figure 5A).52 Using this assay, we found that inhibition of CDK4 caused a large increase in this interaction in MCF-7 cells, an effect that was compounded by tandem inhibition of mTORC1 with rapamycin (Figure 5B). As expected, no change in the eIF4E-4E-BP1 interaction was observed in MDA-MB-468 cells upon CDK4/6 inhibition. Of note, attempts to develop a related assay for measuring the eIF4E−eIF4G interaction were unsuccessful, likely due to the large size of eIF4G (220 kDa) and the relative orientation of the N- and C-termini of the proteins. Together, these results demonstrate that CDK4-mediated phosphorylation of 4E-BP1 can promote mitotic cap-dependent translation in the absence of mTORC1 signaling.
Figure 5. mTOR and CDK4 Regulate eIF4E-4E-BP1 Association in Mitotic Cells.
A) Schematic for the nanoluciferase assay monitoring the eIF4E-4E-BP1 interaction in live cells. S=SmBit, LG=LargeBit. Data shown in panel B) Error is plotted as +/− SEM, n = 5.
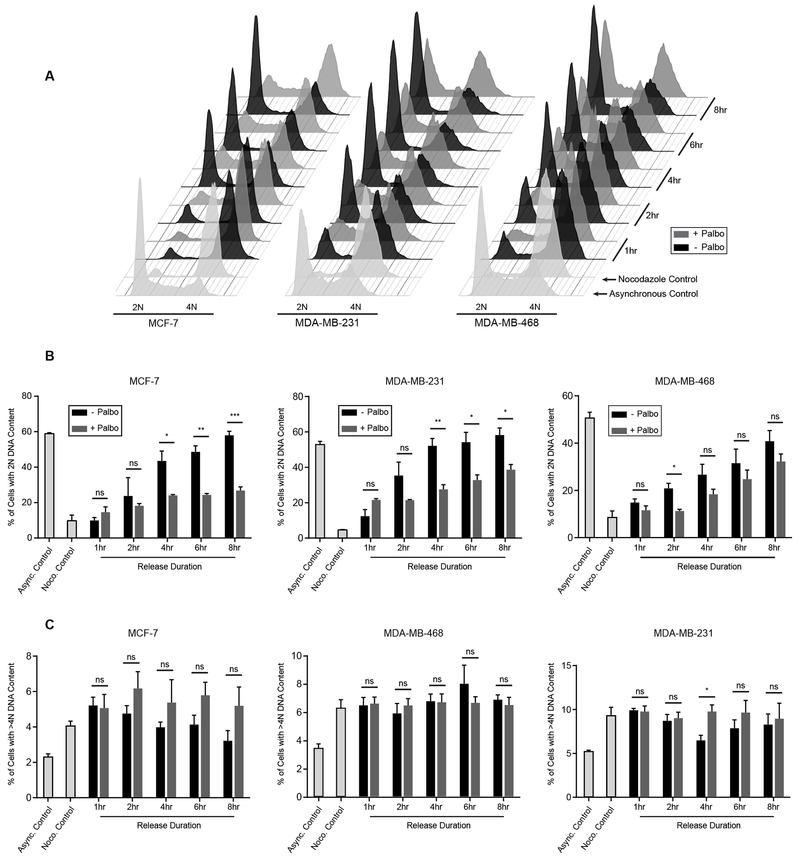
Finally, to characterize the effect of CDK4 inhibition on progression through mitosis into G1, the cell cycle distribution was analyzed at several time points following release from a prometaphase arrest. As cell lines, we chose MCF-7, MDA-MB-231 and MDA-MB-468 breast cancer cells that demonstrate high, medium or no sensitivity to palbociclib, respectively.43,51 MCF-7 cells treated with palbociclib showed a profound G2 block relative to control cells, as they were unable to fully transition into G1 (Figures 6A and 6B). In the CDK4 inhibitor-resistant MDA-MB-468 cell line, palbociclib had very little effect after an initial G2 delay. As expected, the MDA-MB-231 cell line showed an intermediate G2 block based on its intermediate potency for CDK4/6 inhibition.53 Of note, these results were not influenced by a change in polyploidy (Figure 6C).
Figure 6. CDK4 Inhibition Results in a G2 Block in Palbociclib-Sensitive cells.
(A) Full cell-cycle profiles of cells arrested with nocodazole, and then released into media with or without palbociclib (5 μM) for the indicated time points. (B) Quantification of the percentage of cells from (A) with 2N DNA content . (C) Quantification of the percentage of cells from (A) with >4N DNA content. Data are represented as the mean +/− standard error of three biological replicates.
In context of the canonical role of CDK4, our findings are quite interesting, as CDK4 is believed to be primarily responsible for controlling the checkpoint at the G1/S transition.28 However, several previous studies have demonstrated CDK4 activity outside of this phase of the cell cycle.54–57 First, cyclin D-CDK4 complexes have been observed to persist throughout the cell cycle although their function is entirely unknown.54,55 Moreover, it has previously been found that cyclin D3-CDK4 activity is essential for progression through G2,56,57 and that ionizing radiation-induced activation of p16 leads to a G2 phase delay via inhibition of CDK4.56 In fact, our own analysis of pRb phosphorylation at S780 showed an increase in CDK4 activity in the majority of cells released from a prometaphase arrest across a panel of cell lines (Figure S3). These data support an essential role of cyclin D-CDK4 in progression into G1 from G2/M, and provide evidence that pharmacological inhibition can result in a G2 block even in p16-null cancer cell lines, such as MCF-7.58 Although the exact mechanism of this G2 delay is not fully understood, it is possible that inhibition of Rb phosphorylation is the primary factor governing this phenotype. Alternatively, an inability to translate critical anabolic proteins due to inhibited cap-dependent translation may play a role. However, the fact that palbociclib treatment slows G1 entry in Rb-null MDA-MB-468 cells at early time points following release, suggests that another CDK4 activity may be important for regulation of this transition. This could be due to an altered transcriptional program induced by inhibition of phosphorylation of FOXM1, a master transcription factor that regulates expression of genes essential for mitosis.59 While it is unclear at what stage of the cell cycle these cells are being blocked, in later stages of mitosis or simply a failure to undergo cytokinesis, it is clear that a post-mitotic checkpoint is in part regulated by CDK4, and gaining a better understanding of the mechanism behind this warrants further consideration.
CONCLUSION
In conclusion, using a chemoproteomic approach, we have discovered that CDK4 phosphorylates 4E-BP1 during the M to G1 transition, thereby maintaining cap-dependent translation. These findings shed further light on the cell cycle dependent phosphorylation of 4E-BP1, the regulation of which was previously reported to be mediated by CDK1 and CDK12 in mitotic cells,29,60,61 and PLK1 and CDK1 in cells undergoing meiosis.22,32,62–64 Given this newly discovered role of CDK4, it is likely that inhibition of mitotic 4E-BP1 phosphorylation is a previously unknown function of CDK4/6 inhibitors such as palbociclib, and may explain the synergy between these drugs and mTOR inhibitors.65–71 It is important to try to reconcile these observations with those previously observed that report CDK1 as the principal kinase that phosphorylates 4E-BP1 during mitosis.30,33 It is possible that, given the differences in experimental design, CDK1 phosphorylates 4E-BP1 in prometaphase, but in later stages of mitosis and/or cytokinesis, CDK4 assumes that role. It is also possible that S101 phosphorylation, which we previously identified as strictly CDK4-dependent and important for the global phosphorylation of 4E-BP1, 27 renders CDK1 effectively null towards the reported CDK1 sites. Finally, although mTORC1 is inactive during mitosis,60,61,72,73 we have found that inhibition of both CDK4/6 and mTORC1 provides the most robust decrease of 4E-BP1 phosphorylation in prometaphase-released cells. To explain this cooperativity, we are investigating the possibility that CDK4 inhibitors induce mTORC1 reactivation in response to mitosis-associated dephosphorylation of 4E-BP1. Thus, our working hypothesis is that mTOR, CDK1 and CDK4 all act in concert to regulate cap-dependent translation during G2, mitosis and the transition into G1. Future efforts will be focused on the investigation of this interplay amongst kinases throughout the cell cycle.
Supplementary Material
Acknowledgements
This work was supported by the University of Michigan College of Pharmacy (A.L.G.), the University of Michigan Rogel Cancer Center John S. and Suzanne C. Munn Cancer Fund (A.L.G.), the University of Michigan Rackham Predoctoral Fellowship (D.C.M) and the NIH (R01 CA202018 to A.L.G. and T32 CA140044 to D.C.M.). We thank Dr. Venkatesha Basrur, Professor Alexey Nesvizhskii and the University of Michigan Proteomics Resource Facility for assistance with MS experiments.
REFERENCES
- 1.Willis AE Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell. Biol 31, 73–86, (1999). [DOI] [PubMed] [Google Scholar]
- 2.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW & Sonenberg N eIF4E-from translation to transformation. Oncogene 23, 3172–3179, (2004). [DOI] [PubMed] [Google Scholar]
- 3.Hsieh AC & Ruggero D Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin. Cancer Res 16, 4914–4920, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J, Graff J, Ruggero D & Sonenberg N Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 75, 250–263, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonenberg N, Morgan MA, Merrick WC & Shatkin AJ A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5’-terminal cap in mRNA. Proc. Natl. Acad. Sci., U. S. A 75, 4843–4847, (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von der Haar T, Gross JD, Wagner G & McCarthy JEG The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol 11, 503–511, (2004). [DOI] [PubMed] [Google Scholar]
- 7.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC Jr. & Sonenberg N Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature 371, 762–767, (1994). [DOI] [PubMed] [Google Scholar]
- 8.Lin T-A, Kong X, Haystead TA, Pause A, Belsham GJ, Sonenberg N & Lawrence JC Jr. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266, 653–656, (1994). [DOI] [PubMed] [Google Scholar]
- 9.Haghighat A, Mader S, Pause A & Sonenberg N Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14, 5701–5709, (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mader S, Lee H, Pause A & Sonenberg N The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol 15, 4990–4997, (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcotrigiano J, Gingras AC, Sonenberg N & Burley SK Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716, (1999). [DOI] [PubMed] [Google Scholar]
- 12.Ptushkina M, von der Haar T, Karim MM, Hughes JMX & McCarthy JEG Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 18, 4068–4075, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter JD & Sonenberg N Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480, (2005). [DOI] [PubMed] [Google Scholar]
- 14.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R & Sonenberg N Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13, 1422–1437, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R & Sonenberg N Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Sonenberg N, Gingras AC, Peterson M, Avdulov S, Polunovsky VA & Bitterman PB Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol. Cell. Biol 22, 2853–2861, (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras AC, Raught B & Sonenberg N Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15, 807–826, (2001). [DOI] [PubMed] [Google Scholar]
- 18.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC Jr. & Abraham RT Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277, 99–101, (1997). [DOI] [PubMed] [Google Scholar]
- 19.Batool A, Aashaq S & Andrabi KI Reappraisal to the study of 4E-BP1 as an mTOR substrate - a normative critique. Eur. J. Cell Biol 96, 325–336, (2017). [DOI] [PubMed] [Google Scholar]
- 20.Choo AY, Yoon SO, Kim SG, Roux PP & Blenis J Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci., U. S. A 105, 17414–17419, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choo AY & Blenis J Not all substrates are treated equally: implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle 8, 567–572, (2009). [DOI] [PubMed] [Google Scholar]
- 22.Severance AL & Latham KE PLK1 regulates spindle association of phosphorylated eukaryotic translation initiation factor 4E-binding protein and spindle function in mouse oocytes. Am J Physiol Cell Physiol 313, C501–C515, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y & Zheng XFS mTOR-independent 4E-BP1 phosphorylation is associated with cancer resistance to mTOR kinase inhibitors. Cell Cycle 11, 594–603, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ducker GS, Atreya CE, Simko JP, Hom YK, Matli MR, Benes CH, Hann B, Nakakura EK, Bergsland EK, Donner DB, Settleman J, Shokat KM & Warren RS Incomplete inhibition of phosphorylation of 4E-BP1 as a mechanism of primary resistance to ATP-competitive mTOR inhibitors. Oncogene 33, 1590–1600, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM & Gray NS An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem 284, 8023–8032, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D & Shokat KM Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PloS Biol. 7, e1000038, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell DC, Menon A & Garner AL Chemoproteomic Profiling Uncovers CDK4-Mediated Phosphorylation of the Translational Suppressor 4E-BP1. Cell Chemical Biology 26, 980–990.e988, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malumbres M & Barbacid M Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166, (2009). [DOI] [PubMed] [Google Scholar]
- 29.Choi SH, Martinez TF, Kim S, Donaldson C, Shokhirev MN, Saghatelian A & Jones KA CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes & development 33, 418–435, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuda M, Velasquez C, Cheng E, Cordek DG, Kwun HJ, Chang Y & Moore PS CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc. Natl. Acad. Sci., U. S. A 112, 5875–5882, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heesom KJ, Gampel A, Mellor H & Denton RM Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr. Biol 11, 1374–1379, (2001). [DOI] [PubMed] [Google Scholar]
- 32.Jansova D, Koncicka M, Tetkova A, Cerna R, Malik R, Del Llano E, Kubelka M & Susor A Regulation of 4E-BP1 activity in the mammalian oocyte. Cell Cycle 16, 927–939, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velasquez C, Cheng E, Shuda M, Lee-Oesterreich PJ, von Strandmann LP, Gritsenko MA, Jacobs JM, Moore PS & Chang Y Mitotic protein kinase CDK1 phosphorylation of mRNA translation regulator 4E-BP1 Ser83 may contribute to cell transformation. Proc. Natl. Acad. Sci., U. S. A 113, 8466–8471, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun R, Cheng E, Velasquez C, Chang Y & Moore PS Mitosis-related phosphorylation of the eukaryotic translation suppressor 4E-BP1 and its interaction with eukaryotic translation initiation factor 4E (eIF4E). The Journal of biological chemistry 294, 11840–11852, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyronnet S, Dostie J & Sonenberg N Suppression of cap-dependent translation in mitosis. Genes Dev. 15, 2083–2093, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonneau A-M & Sonenberg N Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem 262, 11134–11139, (1987). [PubMed] [Google Scholar]
- 37.Coldwell MJ, Cowan JL, Vlasak M, Mead A, Willett M, Perry LS & Morley SJ Phosphoyrlation of eIF4GII and 4E-BP1 in response to nocodazole treatment. Cell Cycle 12, 3615–3628, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorda R, Hendrychova D, Voller J, Reznickova E, Gucky T & Krystof V How selective are pharmacological inhibitors of cell-cycle-regulating cyclin-dependent kinases? J. Med. Chem 61, 9105–9120, (2018). [DOI] [PubMed] [Google Scholar]
- 39.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC & Chen L Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci., U. S. A 103, 10660–10665, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivakumar S & Gorbsky GJ Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol 16, 82–94, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367, (2008). [DOI] [PubMed] [Google Scholar]
- 42.Edery I, Altmann M & Sonenberg N High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene 74, 517–525, (1988). [DOI] [PubMed] [Google Scholar]
- 43.Kaur T, Menon A & Garner AL Synthesis of 7-benzylguanosine cap-analogue conjugates for eIF4E targeted degradation. Eur. J. Med. Chem 166, 339–350, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song JM, Menon A, Mitchell DC, Johnson OT & Garner AL High-throughput chemical probing of full-length protein-protein interactions. ACS Comb. Sci 19, 763–769, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell DC, Menon A & Garner AL Chemoproteomic profiling uncovers CDK4-mediated phosphorylation of the translational suppressor 4E-BP1. Cell Chem. Biol, accepted, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK & Toogood PL Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther 3, 1427–1437, (2004). [PubMed] [Google Scholar]
- 47.Toogood PL, Harvey PJ, Repine JT, Sheehan DJ, VanderWel SN, Zhou H, Keller PR, McNamara DJ, Sherry D, Zhu T, Brodfuehrer J, Choi C, Barvian MR & Fry DW Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J. Med. Chem 48, 2388–2406, (2005). [DOI] [PubMed] [Google Scholar]
- 48.Bates S, Parry D, Bonetta L, Vousden K, Dickson C & Peters G Absence of cyclin D/cdk complexes in cells lacking functional retinoblastoma protein. Oncogene 9, 1633–1640, (1994). [PubMed] [Google Scholar]
- 49.Knudsen ES & Witkiewicz AK The strange case of CDK4/6 inhibitors: mechanisms, resistance, and combination strategies. Trends Cancer 3, 39–55, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonenberg N, Rupprecht KM, Hecht SM & Shatkin AJ Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. Proc. Natl. Acad. Sci., U. S. A 76, 4345–4349, (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mainwaring LA & Kenney AM Divergent functions for eIF4E and S6 kinase by sonic hedgehog mitogenic signaling in the developing cerebellum. Oncogene 30, 1784–1797, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP & Wood KV NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol 11, 400–408, (2016). [DOI] [PubMed] [Google Scholar]
- 53.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G & Slamon DJ PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 11, R77, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsushime H, Quelle DE, Shurtleff SA, Shibuya M, Sherr CJ & Kato J-Y D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol 14, 2066–2076, (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Jenkins CW, Nichols MA & Xiong Y Cell cycle expression andn p53 regulation of the cyclin-dependent kinase inhibitor p21. Oncogene 9, 2261–2268, (1994). [PubMed] [Google Scholar]
- 56.Gabrielli BG, Sarcevic B, Sinnamon J, Walker G, Castellano M, Wang X-Q & Ellem KAO A cyclin D-Cdk4 activity required for G2 phase cell cycle progression is inhibited in ultraviolet radiation-induced G2 phase delay. J. Biol. Chem 274, 13961–13969, (1999). [DOI] [PubMed] [Google Scholar]
- 57.Burgess A, Wigan M, Giles N, DePinto W, Gillespie P, Stevens F & Gabrielli BG Inhibition of S/G2 phase CDK4 reduces mitotic fidelity. J. Biol. Chem 281, 9987–9995, (2006). [DOI] [PubMed] [Google Scholar]
- 58.Hui R, Macmillan D, Kenny FS, Musgrove EA, Blamey RW, Nicholson RI, Robertson JFR & Sutherland RL INK4a gene expression and methylation in primary breast cancer: overexpression of p16INK4a messenger RNA is a marker of poor prognosis. Clin. Cancer Res 6, 2777–2787, (2000). [PubMed] [Google Scholar]
- 59.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, Zhai H, Vidal M, Gygi SP, Braun P & Sicinski P A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 20, 620–634, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shuda M, Velasquez C, Cheng E, Cordek DG, Kwun HJ, Chang Y & Moore PS CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proceedings of the National Academy of Sciences of the United States of America 112, 5875–5882, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velasquez C, Cheng E, Shuda M, Lee-Oesterreich PJ, Pogge von Strandmann L, Gritsenko MA, Jacobs JM, Moore PS & Chang Y Mitotic protein kinase CDK1 phosphorylation of mRNA translation regulator 4E-BP1 Ser83 may contribute to cell transformation. Proceedings of the National Academy of Sciences of the United States of America 113, 8466–8471, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shang ZF, Yu L, Li B, Tu WZ, Wang Y, Liu XD, Guan H, Huang B, Rang WQ & Zhou PK 4E-BP1 participates in maintaining spindle integrity and genomic stability via interacting with PLK1. Cell Cycle 11, 3463–3471, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romasko EJ, Amarnath D, Midic U & Latham KE Association of maternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: localized translational control supporting female meiosis in mammals. Genetics 195, 349–358, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Susor A, Jansova D, Cerna R, Danylevska A, Anger M, Toralova T, Malik R, Supolikova J, Cook MS, Oh JS & Kubelka M Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat Commun 6, 6078, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, Lehar J, Wiesmann M, Wartmann M, Chen Y, Cao ZA, Pinzon-Ortiz M, Kim S, Schlegel R, Huang A & Engelman JA CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell 26, 136–149, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michaloglou C, Crafter C, Siersbaek R, Delpuech O, Curven J, Carnevalli L, Staniszweska A, Polanska U, Cheraghchi-Bashi A, Lawson M, Chernukhin I, McEwen R, Carroll J & Cosulich S Combined inhibition of mTOR and CDK4/6 is required for optimal blockade of E2F function and long term growth inhibition in estrogen receptor positive breast cancer. Cancer research 78, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olmez I, Brenneman B, Xiao A, Serbulea V, Benamar M, Zhang Y, Manigat L, Abbas T, Lee J, Nakano I, Godlewski J, Bronisz A, Abounader R, Leitinger N & Purow B Combined CDK4/6 and mTOR Inhibition Is Synergistic against Glioblastoma via Multiple Mechanisms. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 6958–6968, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pikman Y, Alexe G, Roti G, Conway AS, Furman A, Lee ES, Place AE, Kim S, Saran C, Modiste R, Weinstock DM, Harris M, Kung AL, Silverman LB & Stegmaier K Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-cell Acute Lymphoblastic Leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 1012–1024, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song X, Liu X, Wang H, Wang J, Qiao Y, Cigliano A, Utpatel K, Ribback S, Pilo MG, Serra M, Gordan JD, Che L, Zhang S, Cossu A, Porcu A, Pascale RM, Dombrowski F, Hu H, Calvisi DF, Evert M & Chen X Combined CDK4/6 and Pan-mTOR Inhibition Is Synergistic Against Intrahepatic Cholangiocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research 25, 403–413, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Divakar SK, Ramana Reddy MV, Cosenza SC, Baker SJ, Perumal D, Antonelli AC, Brody J, Akula B, Parekh S & Reddy EP Dual inhibition of CDK4/Rb and PI3K/AKT/mTOR pathways by ON123300 induces synthetic lethality in mantle cell lymphomas. Leukemia 30, 86–93, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franco J, Witkiewicz AK & Knudsen ES CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 5, 6512–6525, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanenbaum ME, Stern-Ginossar N, Weissman JS & Vale RD Regulation of mRNA translation during mitosis. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pyronnet S, Dostie J & Sonenberg N Suppression of cap-dependent translation in mitosis. Genes & development 15, 2083–2093, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.