Abstract
Background
To investigate if supervised machine learning (ML) classifiers would be able to predict clinically significant cancer (sPC) from a set of quantitative image-features and to compare these results with established PI-RADS v2 assessment scores.
Methods
We retrospectively included 201, histopathologically-proven, peripheral zone (PZ) prostate cancer lesions. Gleason scores ≤3+3 were considered as clinically insignificant (inPC) and Gleason scores ≥3+4 as sPC and were encoded in a binary fashion, serving as ground-truth. MRI was performed at 3T with high spatiotemporal resolution DCE using Golden-angle RAdial SParse (GRASP) MRI. Perfusion maps (Ktrans, Kep, Ve), apparent diffusion coefficient (ADC), and absolute T2-signal intensities (SI) were determined in all lesions and served as input parameters for four supervised ML models: Gradient Boosting Machines (GBM), Neural Networks (NNet), Random Forest (RF) and Support Vector Machines (SVM). ML results and PI-RADS scores were compared with the ground-truth. Next ROC-curves and AUC values were calculated.
Results
All ML models outperformed PI-RADS v2 assessment scores in the prediction of sPC (RF, GBM, NNet and SVM vs. PI-RADS: AUC 0.899, 0.864, 0.884 and 0.874 vs. 0.595, all P<0.001).
Conclusions
Using quantitative imaging parameters as input, supervised ML models outperformed PI-RADS v2 assessment scores in the prediction of sPC. These results indicate that quantitative imagining parameters contain relevant information for the prediction of sPC from image features.
Keywords: Prostatic neoplasms, magnetic resonance imaging (MRI), supervised machine learning
Introduction
As a response to the growing importance of a noninvasive assessment of the prostate gland using magnetic resonance imaging and the need to distinguish between benign processes and prostate cancer based on image features, the Prostate Imaging – Reporting and Data System (PI-RADS) has been introduced in 2012 (1). This reporting system has been under constant evaluation and improvement since (2). The key goal of this reporting system, to improve “the detection of clinically significant caner” is yet unchanged.
In order to validate this scoring system, multiple attempts have been made in the past (3-6). The findings of these studies revealed one key limitations of the PI-RADS v2 assessment score, which is the false positive rate that lowers the cancer detection.
In summary, PI-RADS category 5 and 4 lesions are supposed to very likely and likely contain sPC while PI-RADS category 3 lesions are considered to equivocally contain sPC. Clinical trials like PRECISION (7) and MRI-FIRST (8) evaluated the performance of MRI targeted prostate biopsies and could demonstrate improved detection of clinically significant prostate cancer (sPC). Therefore, the European Association of Urology guidelines have adopted this approach and recommend combining targeted and systemic biopsy in case of positive mpMRI (PI RADS ≥3).
The PI-RADS score in theory equals a probability score for the detection of clinically significant cancer (sPC) based on the image findings. This turns out to be true for PI-RADS 5 lesions with detection rates of sPC in over 90% (9). For PI-RADS 4 lesions though, the detection rates of sPC after biopsy range between 22% (4) and 60% (7). For PI-RADS 3 lesions, sPC is found in 12% (4) of the cases or even not at all (3).
One possible explanation for this discrepancy between image-based likelihood of cancer and histopathologically-proven, sPC might be that the input features for the PI-RADS classification are solely qualitative, relative measures such as “moderately versus markedly hypointensity” of apparent diffusion coefficient (ADC) derived maps from diffusion-weighted imaging (DWI) or “heterogenous versus homogenous, moderate hypointensity” on T2-weighted imaging (10). Another potential limitation is the lack of implementation of perfusion information derived from acquisitions with high spatiotemporal resolution, e.g., golden-angle radial sparse MRI (GRASP). Several studies have shown an improved diagnostic performance in detecting primary cancer or local recurrence using such acquisition techniques (11,12).
In the past, multiple promising attempts have been made to use quantitative imaging parameters, including dynamic-contrast enhanced (DCE), with and without machine learning (ML) techniques, for the prediction of sPC (13-15). However, to the best of our knowledge, no study investigated the use of various ML techniques with high spatiotemporal perfusion data in a systematic manner, potentially addressing all the above-mentioned limitations.
The purpose of this study was therefore to investigate, whether or not supervised ML techniques, employing advanced feature analysis techniques and state-of-the art perfusion information, are able to predict clinically sPC from quantitative image-features and to compare these results with established PI-RADS v2 assessment scores.
Methods
Patients
This study was approved by the local ethics committees (ethics committee Northwest and Central Switzerland; EKNZ 2019-02364). We retrospectively analyzed patients meeting the following inclusion criteria: (I) clinically indicated 3T MRI of the prostate at our institution due to suspicious digital rectal examination and/or elevated prostate-specific antigen (PSA) level of ≥4 ng/mL or increased PSA velocity (>0.5 ng/mL/y), both biochemically determined within 30 days before the MRI examination, and (II) histopathologically-proven peripheral zone (PZ) prostate cancer with biopsies performed within 30 days after the imaging study. MRI examinations dates ranged from 04/2015 to 03/2019.
MRI examination
All examinations were performed on a single 3T MRI system (MAGNETOM Skyra, Siemens Healthcare GmbH, Erlangen, Germany) using a body phased-array coil with 60 channels. Relevant sequences for the study included: T2-weighted (T2w) fast spin-echo (FSE) acquisition in axial, coronal, and sagittal planes; fat-saturated T1-weighted (T1w) 3D GRE (“VIBE”) acquisition pre-contrast with flip angles of 2° and 5° for T1-map generation; DWI acquisition with b values of 0 and 800 mm2/s; and dynamic fat-saturated T1-weighted 3D GRE acquisition with radial stack-of-stars sampling (“GRASP”) after administration of 0.01 mmol/kg gadoterate meglumine (Dotarem, Guerbet, Villepint, France). Acquisition parameters for all sequences are summarized in Table 1.
Table 1. Multiparametric examination protocol.
| T2-weighted turbo-spin echo (TSE) (TR: 7.5 ms, TE: 101 ms, ST: 3 mm, acquisition matrix of 320×320, in-plane resolution 0.56×0.56 mm) |
| T1-mapping: Fat-saturated 3D GRE pre-contrast sequence, flip angles of 2° and 5° (TR: 4.13 ms, TE: 2.06 ms, ST: 3 mm, acquisition matrix of 288×202) |
| DWI with b values of 50 and 800 mm2/s (TR: 4.9 ms, TE: 59 ms, ST: 3.675 mm, acquisition matrix of 118×118, in-plane resolution 1.69×1.69 mm). ADC-Maps were generated |
| TR: 4 ms, TE: 2 ms, FA: 12º, ST: 2.5, acquisition matrix of 288×288, in-plane resolution 0.56×0.56 mm with temporal resolution of 2.5 s, based on 21 spokes/frame after administration of 0.01 mmol/kg gadoterate meglumine (Dotarem, Guerbet, Villepint, France) |
TR, repetition time; TE, echo time; ST, slice thickness; FA, flip angle; DWI, diffusion-weighted imaging; ADC, apparent diffusion coefficient; GRE, gradient-echo sequence.
GRASP MRI
Dynamic imaging was done using the GRASP technique, which is based on continuous fat-saturated T1w 3D GRE acquisition with radial readout (16). This sequence samples k-space with a stack-of-stars scheme, in which radial “spokes” are stacked along the slice direction and rotated throughout the scan, resulting in a cylindrical spoke-wheel-like trajectory. The rotation angle is selected according to the Golden-Angle scheme, which rotates consecutive spokes by 111.25° and results in approximately uniform k-space coverage throughout the acquisition (17). The image reconstruction is done using an iterative compressed-sensing algorithm that exploits temporal correlations between successive time points to suppress undersampling artifacts (18), which allows obtaining images with higher temporal and spatial resolution compared to previous DCE-MRI techniques (in this study, a temporal resolution of 2.5 sec/frame at 0.56×0.56 mm spatial resolution was achieved by combining 21 k-space spokes into each frame). A detailed technical description of the GRASP technique and its imaging properties is provided in (19).
Transrectal prostate biopsy
Transrectal biopsies were performed by one of three board-certified urologists (C.W.). First, MRI transrectal US fusion-guided biopsies were performed after lesions suspicious for cancer (identified by two-board certified radiologists, with over 3 years subspecialty experience) were centrally marked on axial T2w MR images by using a crosshair fiducial marker; three cores per lesion were obtained. Afterwards, 12–18 conventional transrectal US–guided cluster biopsies were performed.
Perfusion processing
Image processing was performed by using a commercially available software application (Syngo.via VB30, MR Prostate and MR Tissue4D; Siemens Healthineers). First, the DCE-MRI datasets were corrected for residual motion by registering all volumes of the time series to a user-selected reference volume, which reduced data inconsistencies caused by patient and physiologic motion during the DCE-MRI acquisition. Next, registration of the morphologic images and T1-mapping series to the reference volume was performed. Last, a volume-of-interest (VOI) was defined encompassing the prostate and seminal vesicles. The VOI extended from the dorsal symphysis to the ventral rectal wall. Within this volume, perfusion maps were generated using a pixel-wise Tofts-modelling algorithm (20) and T1 fitting with restriction to pixel values above a noise level (fixed threshold value: >20 IU SD). For the Tofts modelling, a population-based arterial input function (AIF) was used. The following parameters from the Tofts model were used as perfusion maps: Ktrans, Kep, and Ve.
Histopathologic correlation, lesion annotation and automated measure extraction
The annotation process was as follows: in a first step, an anonymized, highly structured report as suggested by the PI-RADS guidelines was studied for each dataset by a radiology fellow with 2 years of subspecialty experience in prostate imaging and research (DJ Winkel). This report contained detailed information about the reported lesions per zone, especially mentioning series numbers on either T2w or ADC with accompanying image numbers. The reports have been created in a consensus read by two board-certified radiologists. Next, the histopathology results per patient was studied and all relevant metrics were extracted. In concordance with the histopathologically confirmed location, each lesion was carefully identified on the DWI series and corresponding ADC maps, using the T2-weighted images as morphological reference. Using this information, the lesions were annotated 3-dimensionally (3D) on the T2-weighted images—due to the higher spatial resolution—and labeled according to their PI-RADS scores using a proprietary software (Annotator Tool, V03_B41). From these annotations, we extracted the 3D volumes of the lesions and stored them separately as “masks”. In a next step, we registered the following sequences: T2w, ADC, Ktrans, Kep, and Ve and overlaid the beforehand extracted masks. Simple ITK toolbox (http://www.simpleitk.org) (21,22), a preprocessing script implemented in the programming languages Python (version 3.5; Python Software Foundation; https://www.python.org), was used to automatically load all the above mentioned sequences along with the lesion mask, to extract the mean ± standard deviation (SD) values from the lesion area for each sequence of all patients and to store the corresponding values in a comma-separated value (.csv) file for further analysis.
Input features
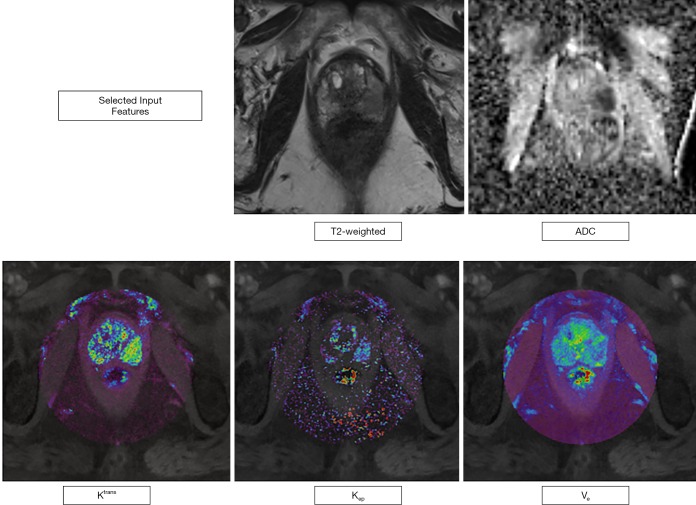
T2w images are mostly used to evaluate the prostate gland anatomy and to identify suspicious lesions with a concurrent morphological characterization. As the image information from T2w images cannot be considered as quantitative, we extracted signal intensities (SI) from the ROIs in order to obtain a semi-quantitative image parameter that could be compared within our dataset. Diffusion-weighted imaging (DWI) measures differences in the random motion of water molecules; ADC (in mm2/sec) maps can be computed from two b-Value images, often acquired between either 0–100 and 800–1,000 sec/mm2 (2) and represent a quantitative image parameter, that has been shown to inversely correlate with the histopathological grade of prostate tumors. Ktrans, Kep (both in min-1), and Ve (in %) maps are quantitative representations of the perfusion postprocessing steps outlined above as can be extracted using DCE MRI. Ktrans represents the contrast agent “wash-in”, Kep the “wash-out” component or rate constant (with the following formula and Ve the fractional volume of the extravascular, extracellular space (20,23). Figure 1 visually displays the input features in an exemplary case.
Figure 1.
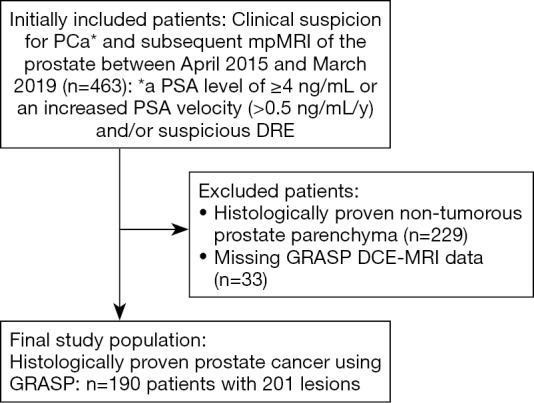
Flowchart outlining the selection of the final study population with utilized inclusion and exclusion criteria within the defined observation window. PSA, prostate-specific antigen; DRE, digital rectal examination; GRASP, golden-angle radial sparse MRI; DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging.
Reading process and data definition
All participants underwent a routine clinical reading process in an academic institution with two-board certified radiologists with at least 5 years of experience in prostate imaging reading; at our institution all women’s and men’s health imaging studies (mammographic imaging and prostate imaging) is read by 2 independent radiologists as consensus read. According to the PI-RADS guidelines (24), PI-RADS category 4 and 5 lesions were considered to be likely or highly likely linked with sPC, whereas PI-RADS category 3 lesions were considered equivocal with regards to the presence of sPC. According to the PI-RADS guidelines, PI-RADS category 3 lesions display a moderate hypointensity on T2w and are non-circumscribed or rounded. On ADC, they are focal mildly/moderately hypointense on ADC and isointense/mildly hyperintense on high b-value DWI.
Following these assumptions, we split the PI-RADS category lesions into two groups: low and high chance to predict sPC. We then proceeded with the histopathology results in a similar way by splitting the Gleason scores into two groups: Gleason scores 3+3 were assigned to be inPC, while all other Gleason patterns were considered as sPC. The Gleason score-based categories (3+3: negative; ≥3+4: positive) are used as the ground-truth labels for the following analysis and assessment.
Data preparation
In a first step, the rows of the study data, each corresponding to a feature set extracted from one mask, or region of interest (ROI), were randomly shuffled to avoid sample biases when splitting the study dataset into training and test data. Then, 80% of the rows were selected as training data and the residual 20% were used as test data. In the next step, the ground-truth column, corresponding to presence or absence of sPC, was isolated from the dataset and saved as separate labelling data, which has been utilized during the training and testing phase.
ML models
Four different ML techniques have been used for data analysis: Gradient Boosting Machines (GBM), Neural Networks (NNet), Random Forest (RF) and Support-Vector Machines (SVM) with a radial basis function (RBF) kernel. These ML algorithms are popular and well-established approaches for data classification and regression tasks (25) and have been implemented in various commercial data-analysis software products. SVMs are predictive models, where the main goal is to find a hyperplane in the N-dimensional space (given N-features) that is dividing two classes of data points with a maximum distance. In our approach, we used a RBF kernel due to the different scales of our input data and the demand for a highly flexible kernel. In contrast, other approaches such as RF, are based on a so-called “ensemble”. RF, for example, uses a multitude of deep, independent decision-trees with varying numbers of input variables for splitting at each node, leading to different predictions for each tree. The ensemble then averages the predictive value and a final, predicted outcome can be computed. The family of boosting methods, including GBMs, is based on a similar, but not identical approach. Here, new models are added to the ensemble sequentially. In this way, at each particular iteration, a new model is trained with respect to the error of the whole ensemble. The Neural Network model in our approach can be described as feedforward neural network with a single hidden layer and a variable number of nodes. The input from the selected features are first fed to this hidden layer. Depending on the set of weights, a mathematic calculation, the information from the input layer pass the hidden layer and are forwarded to the output layer. This last layer, representing a single node, outputs the classification result. Use cases for all of those approaches are both regression and classifications tasks. In this specific framework, we trained the four ML models to predict the presence or absence of sPC using the aforementioned input features.
Processing was performed using R [R Version 3.6.0, R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL, https://www.R-project.org/] using the “caret” package (short for: Classification And REgression Training, Version 6.0-84). The “caret” package, next to a multitude of alternative packages in R, allows to easily train a total of 238 ML models (see http://topepo.github.io/caret/available-models.html) with similar syntax. One of its strength relies in the streamlined process for creating predictive models, from data splitting and training to variable importance estimation (21). In this study, we used the GBM model to calculate the relative influence of each input parameter. For classification tasks as performed in the present study, ROC curve analysis was conducted on each predictor and the output represents each variable’s importance for the task. We used a 5-fold and 5-times repeated cross validation (CV) for all possible permutations of model-type and dataset (with and without perfusion parameters). K-fold, n-times repeated CV shuffles the training data prior to each repetition, resulting in a different split of the samples and, thus, lower bias. The code and package information are accessible at https://github.com/davidjeanwinkel/Quantitative.
Hyperparameter selection
A grid-search algorithm was used for finding the optimal hyperparameters for each ML model to achieve best possible performance (26). Using this approach, every possible permutation of hyperparameters is tested during training. Finally, the hyperparameter set with the best performance on the training data was selected for each ML model. Table 2 summarizes the mentioned hyperparameters and the parameters chosen for the grid search approach. A summary of all potential ML models available in the “caret” package in R, their type (classification, regression), the libraries they internally use and—most importantly—the parameters that can be possibly tuned can be found at https://topepo.github.io/caret/available-models.html.
Table 2. Hyperparameter selection.
| GBM | NNet | RF | SVM |
|---|---|---|---|
| Hyperparameter range per machine learning model during training | |||
| Interaction.depth =1,2,3,4,5 N.trees =1–350, by steps of 50 Shrinkage =0.1, 0.01 N.minobsinnode =1–10, by steps of 1 |
Size =1–10, by steps of 1 Decay =10−1−5 |
Mtry =1–5, by steps of 1 Splitrule = gini or splitrule Min.node.size =1–5, by steps of 1 |
Cost =0.1–1, by steps of 0.1 Sigma =0.1–1, by steps of 0.1 |
| Best hyperparameter selection for testing | |||
| Interaction.depth =5 N.trees =251 Shrinkage =0.1 N.minobsinnode =1 |
Size =10 Decay =10−1 |
Mtry =1 Splitrule = gini Min.node.size =2 |
Cost =0.9 Sigma =1 |
GBM, gradient boosting machines; NNet, neural network; RF, random forest; SVM, support vector machines.
Statistical analysis
Through comparison of the binary classifiers of the PI-RADS v2 assessment scores and the output of the four ML models—indicating the presence or absence of sPC—with the ground-truth—representing the histopathology results—AUC values of the corresponding receiver operating characteristics (ROC) curves were compared using the method of DeLong et al. (27) with 2,000 bootstrap iterations. ROC curves are a graphical illustration of the diagnostic ability of a binary classifier system as its discrimination threshold is varied. The AUC value is a measure of the accuracy of a diagnostic test. The AUC value represents the average true positive rate (i.e., sensitivity) across all possible false positive rates (i.e., the specificity). Therefore, the AUC values allow for a comparison of the diagnostic capabilities of two methods. Quantitative image parameters assessed. Confusion matrices were furthermore also calculated. All statistical analyses were performed using R.
Results
Patient results and clinical information
In the predefined observation window, 463 patients were referred to our institution for a prostate MRI examination due to a clinical suspicion for prostate cancer. After applying our inclusion criteria 190 patients with 201 lesions were included in the final analysis (see Figure 2). One hundred sixty-one lesions were randomly assigned to the training set (80%) and 40 lesions were randomly assigned to the training set (20%). Table 3 summarizes the demographic and clinical characteristics. The distribution of PI-RADS scores, Gleason scores and the presence or absence of sPC was similar between the two datasets, ensuring comparable results. Boxplots of all input features and their distribution with regards to the PI-RADS score and the Gleason pattern can be found in Figure 3.
Figure 2.
Figure illustrating the five different input features in an exemplary case of a 62-year-old men with a PSA level of 6.3 ng/mL. mpMRI showed a 16 mm lesion (PI-RADS category 5) in the peripheral zone, left midgland, posteromedial/posterolateral. Histopathology revealed a Gleason 4+3=7 pattern. The acquired values for this exemplary set of input features were: T2w: 138±10 SI, ADC: 617±126 mm2/s, Ktrans =0.98 min-1, Kep =0.78 min-1, Ve =36%. mpMRI, multiparametric magnetic resonance imaging; ADC, apparent diffusion coefficient; PI-RADS, Prostate Imaging Reporting and Data System; PSA, prostate-specific antigen.
Table 3. Demographic and clinical characteristics of 190 included men.
| Patient and clinical parameters | Values* |
|---|---|
| Age (in years, mean ± SD) | 69±8.7 |
| PSA (in ng/mL, mean ± SD) | 7±11.2 |
| Training dataset (n=161) | |
| PI-RADS category distribution | |
| PI-RADS 3 | 18 [11] |
| PI-RADS 4 | 76 [47] |
| PI-RADS 5 | 67 [42] |
| Gleason sum score distribution | |
| Gleason 3+3 | 47 [30] |
| Gleason 3+4 | 42 [26] |
| Gleason 4+3 | 36 [22] |
| Gleason 4+4 | 21 [13] |
| Gleason 4+5/5+4 | 15 [9] |
| Clinically significant cancer | |
| Yes | 114 [70] |
| No | 47 [30] |
| Test dataset (n=40) | |
| PI-RADS category distribution | |
| PI-RADS 3 | 4 [10] |
| PI-RADS 4 | 20 [50] |
| PI-RADS 5 | 16 [40] |
| Gleason sum score distribution | |
| Gleason 3+3 | 10 [25] |
| Gleason 3+4 | 11 [27] |
| Gleason 4+3 | 8 [20] |
| Gleason 4+4 | 8 [20] |
| Gleason 4+5/5+4 | 3 [8] |
| Clinically significant cancer | |
| Yes | 30 [75] |
| No | 10 [25] |
*, data in parentheses are percentages. PI-RADS, Prostate Imaging Reporting and Data System; PSA, prostate-specific antigen.
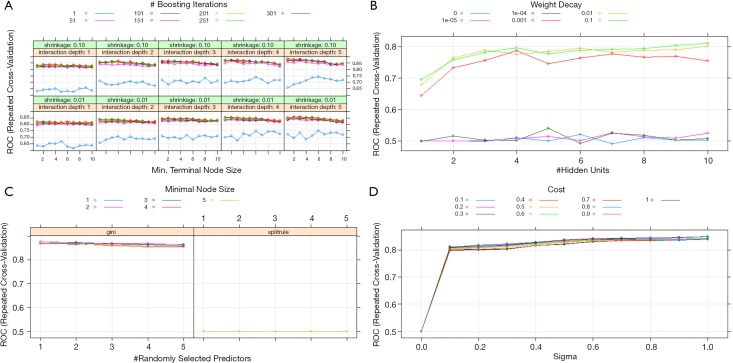
Figure 3.
Specific summary plots with ROC-AUC values given for all possible hyperparameter search permutations for (A) gradient boosting machines (B) neural networks, (C) random forest and (D) support vector machines models.
Hyperparameter selection and model output
The different values tested in our grid-search approach per model and the final selected parameters can be found in Table 2 and Figure 4. For the NNet model, a 5-10-1 (input layer—number of nodes in the hidden layer—output layer) architecture has been chosen with 71 weights.
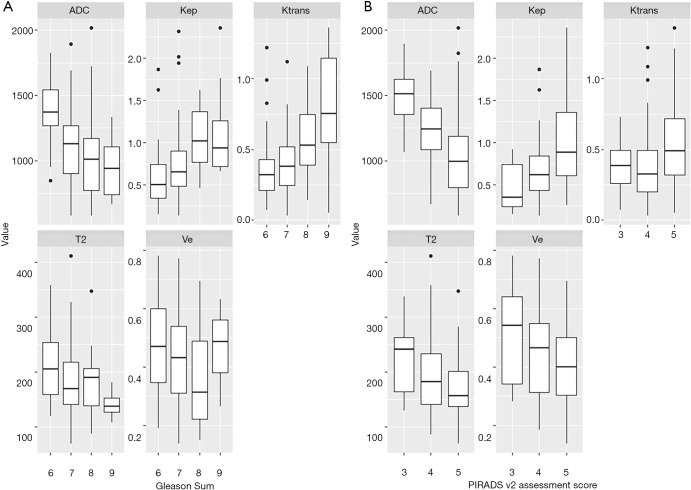
Figure 4.
Boxplots demonstrating the distribution of the quantitative imaging parameters (ADC, T2, Ktrans, Kep, Ve) depending on the (A) Gleason sum score and (B) on the PIRADS v2 assessment score. ADC, apparent diffusion coefficient.
For the GBM model the relative influence of each individual parameter for the final model predictions was automatically calculated, see Figure 5. The parameters with the highest influence was the mean ADC value with a relative influence of 32%, followed by Ktrans and Kep with 19% and 19% and Ve and the mean T2 SI with 17% and 13%, respectively.
Figure 5.
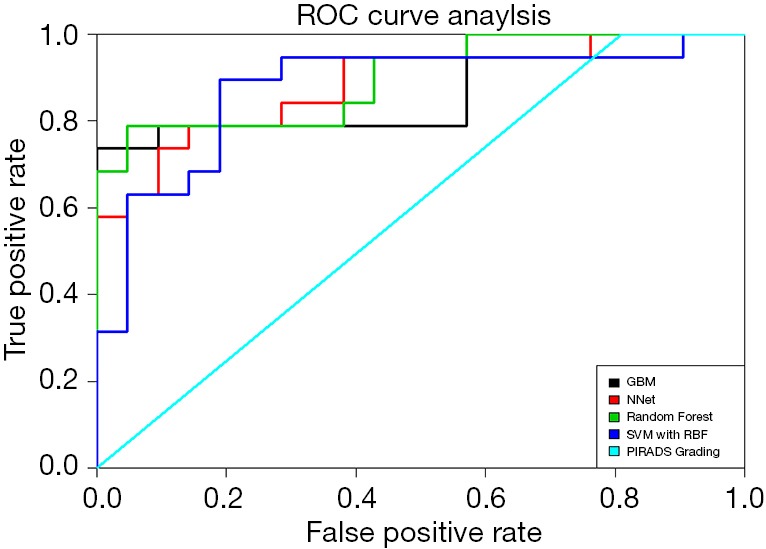
ROC Curve analyses of the different machine learning models and the PIRADS grading. GBM, gradient boosting machines; NNet, neural networks; SVM, support vector machines; RF, random forest.
Performance evaluation on training set
All relevant metrics of diagnostic accuracy and AUC values for the detection of clinically sPC within the training and test dataset can be found in Table 4. In the training dataset, the highest AUC value has been reached using the GBM and RF models with a value of 1 and a sensitivity and specificity of 100%. SVM models outperformed NNet models in terms of AUC values (0.969 vs. 0.863). Using the PI-RADS v2 assessment score and the discriminator between sPC and inPC showed an AUC value of 0.586 with a sensitivity of 100%, and a specificity of 39%. The P values for the ROC curve comparisons between the ML models and the PI-RADS v2 assessment score were all <0.001.
Table 4. Diagnostic performance in training and test sets.
| Cohort and model | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | AUC | P value* |
|---|---|---|---|---|---|---|
| Training (n=161) | ||||||
| GBM | 100 | 100 | 100 | 100 | 1 | 0.001 |
| NNet | 87 | 64 | 82 | 72 | 0.863 | 0.001 |
| RF | 100 | 100 | 100 | 100 | 1 | 0.001 |
| SVM | 98 | 87 | 93 | 96 | 0.969 | 0.001 |
| PI-RADS v2 score | 100 | 39 | 17 | 100 | 0.586 | / |
| Testing (n=40) | ||||||
| GBM | 100 | 58 | 72 | 100 | 0.864 | 0.001 |
| NNet | 95 | 63 | 74 | 92 | 0.884 | 0.001 |
| RF | 100 | 52 | 70 | 100 | 0.899 | 0.001 |
| SVM | 95 | 47 | 66 | 90 | 0.874 | 0.001 |
| PI-RADS v2 score | 100 | 53 | 19 | 100 | 0.595 | / |
*, P values represent results from statistical comparison of ROC curves between the respective machine learning model predictions and the training and test set PI-RADS v2 labels using the method of DeLong with 2,000 bootstrap iterations. GBM, gradient boosting machines; NNet, neural network; RF, random forest; SVM, support vector machines.
Performance evaluation on test set
In the test dataset, a drop in the diagnostic accuracy of all ML models has been observed. The RF model reached the best performance with an AUC value of 0.899 and a sensitivity and specificity of 100% and 52%. This model is followed by the GBM, NNet and SVM models with AUC values of 0.864, 0.884 and 0.874, respectively. Again, the PI-RADS v2 assessment score showed the worst performance with an AUC value of 0.595 and a sensitivity and specificity of 100% and 53%. ROC curve comparisons showed a significant difference in the discriminatory power of GBM, NNet and RF models versus the PI-RADS v2 assessment score with P values <0.001. The ROC curves are displayed in Figure 6.
Figure 6.
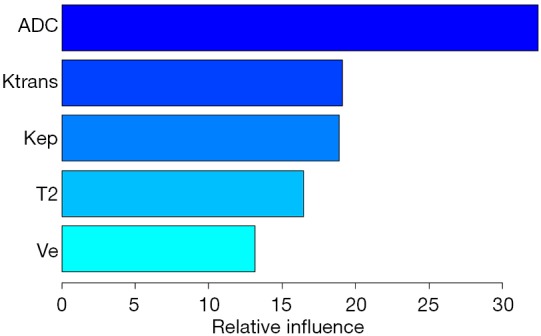
Relative influence of input features assessed with gradient boosting machine models.
Discussion
Supervised ML techniques with quantitative input features, including perfusion information from high-resolution DCE-MRI scans, improved the prediction accuracy of clinically sPC and outperformed established PI-RADS v2 assessment scores. These results indicate that both quantitative image data and perfusion information from high spatiotemporal DCE-MRI contain valuable information that go beyond qualitative image metrics. The latter finding may be explained with the technical advancement of DCE-MRI over the last years.
In the last decade, empowered by availability of healthcare data, increasing computational power and advancements in the field of computer science, medical image analysis has entered a new era: precision medicine. It has been recognized more and more that there is not one, e.g., “prostate cancer”, but rather multiple clusters of different tumor cells, so-called “habitats” within one conglomerate tumor, reflecting microstructure and tumor heterogeneity on a (epi) genetic level. It therefore seems perfectly understandable, that qualitative, ordinal scales of tumor prediction will not be able capture the whole picture. Based on this thoughts, multiple attempts have been made to quantitatively analyze prostate cancer-based image features. Langer et al. (28) were able to show that quantitatively acquired T2w and ADC image parameters correlated with the histopathology-based nuclear cell-density. Donati and Jung et al. (29,30) were able to show that quantitative image parameters correlate with the prostate cancer aggressiveness. Going even one step further, medical images can be analyzed using radiomics. Radiomics describe the conversion of images to higher-dimensional data, such as the extraction of shapes or the quantification of gray level dependencies in an image [so-called “gray level dependence matrix (GLDM)”]. Using these radiomics features, several groups were able to reveal their potential, e.g., for superior risk stratification of prostate cancer (31) or characterization of prostate cancer (32).
Interestingly, the input features (T2, ADC, Ktrans, Kep and Ve) in our data show similar trends in terms of correlation with the image-based PI-RADS v2 assessment score and the histopathology-based Gleason sum score; with higher PI-RADS score and higher Gleason sum score, T2, ADC and Ve values increasingly drop while Ktrans and Kep values increase.
Apparently though, these data trends do not influence the decision-making process with the PI-RADS v2 assessment score. One potential explanation is the well know overlap of image metrics, e.g., ADC, between different PI-RADS scores (33). In fact, supervised ML techniques have been shown to be able to utilize those subtle data differences and to find patterns, that might not seem obvious for the human reader. Bishop (34) formulated this as such: “The field of pattern recognition is concerned with the automatic discovery of regularities in data through the use of computer algorithms and with the use of these regularities to take actions such as classifying the data into different categories”.
In a recent work conducted by Antonelli et al. (13), the authors were able to show that ML classifiers can predict clinically significant tumor patterns in histopathology better than experienced radiologists. Dikaios et al. (35) demonstrated that a linear regression model had a similar performance as experienced radiologist in the classification of prostate cancer. Niaf et al. (36) successfully used a SVM model for the discrimination of benign and malignant lesions. Another important feature in our analysis was the use of perfusion data derived from high spatiotemporal DCE-MRI. In fact, the general usefulness of DCE-MRI is currently under investigation. While DCE-MRI has been an integral part in PI-RADS v1 and v2 (1,2), it has been declared not mandatory in PI-RADS v2.1 (10). One possible explanation for this decision is certainly the fact that DCE-MRI has shown less reproducible results as of yet (37-39). In fact, there is not “one” DCE-MRI workflow that is accepted in the scientific community. As a general rule, DCE-MRI workflows can be divided into three parts: (I) image acquisition and reconstruction, (II) extraction of tracer kinetics and (III) quantification of tracer kinetics. Concerning (I), DCE-MRI of the prostate has been performed with differing spatial and temporal resolutions, ranging from 15.8 down to 2 seconds and from 2.6–3.0×0.5×0.6 mm (40) to 4.0×2.8×2.8 mm (41). The specific k-space readout technique employed in this study, namely radial k-space sampling with a stack-of-stars scheme has been chosen because it has proven to be robust against motion artifacts. Furthermore, this technique allows a flexible reconstruction of DCE-MRI datasets with regards to temporal and spatial resolution. Compared to cartesian readout techniques, aliasing and ghosting artifacts associated with phase-offset errors can be robustly eliminated (42). In a recent study, it has been shown that GRASP DCE-MRI outperformed conventional DCE-MRI techniques (11), further highlighting the value of this technique. However, competing reconstruction schemes such as low-rank plus sparse matrix (43) or k-t FOCUSS (44) have equally proven their usefulness in the current literature. Next (II), the extraction of clinically useful metrics from these information ranges from qualitative to semi-quantitative or quantitative approaches. In our study, we opted for a quantitative approach with the extraction of tracer kinetics based on the Tofts model. A study conducted by Rosenkrantz et al. (45) has shown that the sensitivity for the detection of PZ prostate cancer was increased by the use of semiquantitative or quantitative metrics compared to a qualitative approach. Furthermore (III), the quantification of the tracer kinetics can be performed using different methodologies: one possible approach is numeric optimization, as performed in this study and in (11). Another approach is the use of Bayesian methods (46). The latter approach has proven to robustly quantify tracer kinetics when used for the detection of PZ prostate cancer. Furthermore, DCE-MRI metrics extracted using GRASP have shown to improve the diagnostic performance in detecting primary cancer or local recurrence (12,47). These findings are supported by our data in the sense that after ADC, the second and third most important contributor in the best-performing ML model, GBM, were Ktrans and Kep, followed by Ve and T2.
Our results have clear implications for future clinical application, especially as current guidelines have adopted prostate MRI for primary prostate cancer diagnosis and the demand for MRI of the prostate is increasing (24), also due to prostate cancer screening programs. Given the steadily increasing associated health care costs, supervised ML techniques can be valuable tools to streamline the process of MRI diagnostics and to improve diagnostic accuracy. These improvements pave the way for precision diagnostics, in which supervised ML techniques can help to reduce the number sPCs missed and insignificant cancers detected. This may especially be the case for indeterminate lesions such as PI-RADS 3. Here, adding clinical information to the image-based features may prove beneficial (48).
Our study has limitations. First, only patients with lesions in the PZ were included, as DCE-MRI is typically assessed for PZ lesions and has shown low value for assessment of transition-zone lesions (49). Therefore, the results of our study are only applicable to PZ prostate cancers. Second, the analysis only investigated four different types of ML algorithms. However, all algorithms demonstrated the same tendency. Therefore, we considered this selection of algorithms as sufficient for verification of this hypothesis. Third, the locations of the ROIs, from which the feature values have been extracted, were selected on the T2w images. While the T2w input feature did not show much value for the subsequent ML-based differentiation between clinically significant and insignificant caner, the lesions were identified in the first place on the T2w sequence. Hence, the current study design does not allow answering the question if lesions could be detected (or segmented) based on only the input features without prior manual identification. Fourth, we did not include high b-value images (>1,400 sec/mm2) in our analysis. We believe that with regards to the evaluation of PZ prostate cancer lesions the ADC map may provide sufficient information on diffusion processes and that the added value of dedicated high b-value images is not relevant for the present study. A recent study has shown a very heterogenous adherence of the scientific community to the PI-RADS v2 guidelines, especially affecting DWI (50). In the light of these results, however, we cannot fully rule out that this has potentially influenced the performance of PI-RADS assessment scores. Fifth, the annotation process has been performed by one radiologist and no reproducibility measures of these annotations have been evaluated. However, utmost care has been applied to both match PI-RADS lesion and histopathologically-confirmed tumor locations and subsequent annotations, using all available sequences for orientation and confirmation. Furthermore, the annotations were performed in 3D. Because prostate cancer lesions are frequently heterogeneously vascularized, focused assessment on the most suspect lesion components will not necessarily reflect the biology of the whole tumor appropriately (51). Therefore, 3D segmentations seem advantageous for this purpose and by nature reduce variability compared to 2D approaches. Last, since this study retrospectively covered multiple years, this inherently causes a potential selection bias.
In conclusion, using quantitative imaging parameters, including perfusion maps from high spatiotemporal DCE-MRI, as input, supervised ML models outperformed PI-RADS v2 assessment scores in the prediction of sPC. These results indicate that quantitative imagining parameters contain useful information to predict sPC.
Acknowledgments
Funding: DJ Winkel receives research support from the Swiss Society of Radiology and the Research Fund Junior Researchers of the University Hospital Basel (grant number: 3MS1034).
Ethical Statement: This study was approved by the local ethics committees (ethics committee Northwest and Central Switzerland; EKNZ 2019-02364).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims.2020.03.08). The authors have no conflicts of interest to declare.
References
- 1.Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, Rouviere O, Logager V, Fütterer JJ. ESUR prostate MR guidelines 2012. Eur Radiol 2012;22:746-57. 10.1007/s00330-011-2377-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol 2016;69:16-40. 10.1016/j.eururo.2015.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mertan FV, Greer MD, Shih JH, George AK, Kongnyuy M, Muthigi A, Merino MJ, Wood BJ, Pinto PA, Choyke PL, Turkbey B. Prospective Evaluation of the Prostate Imaging Reporting and Data System Version 2 for Prostate Cancer Detection. J Urol 2016;196:690-6. 10.1016/j.juro.2016.04.057 [DOI] [PubMed] [Google Scholar]
- 4.Mehralivand S, Bednarova S, Shih JH, Mertan FV, Gaur S, Merino MJ, Wood BJ, Pinto PA, Choyke PL, Turkbey B. Prospective Evaluation of PI-RADSTM Version 2 Using the International Society of Urological Pathology Prostate Cancer Grade Group System. J Urol 2017;198:583-90. 10.1016/j.juro.2017.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofbauer SL, Maxeiner A, Kittner B, Heckmann R, Reimann M, Wiemer L, Asbach P, Haas M, Penzkofer T, Stephan C, Friedersdorff F, Fuller F, Miller K, Cash H. Validation of Prostate Imaging Reporting and Data System Version 2 for the Detection of Prostate Cancer. J Urol 2018;200:767-73. 10.1016/j.juro.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 6.Greer MD, Shih JH, Lay N, Barrett T, Kayat Bittencourt L, Borofsky S, Kabakus IM, Law YM, Marko J, Shebel H, Mertan FV, Merino MJ, Wood BJ, Pinto PA, Summers RM, Choyke PL, Turkbey B. Validation of the dominant sequence paradigm and role of dynamic contrast-enhanced imaging in Pi-RADS version 2. Radiology 2017;285:859-69. 10.1148/radiol.2017161316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, Briganti A, Budäus L, Hellawell G, Hindley RG, Roobol MJ, Eggener S, Ghei M, Villers A, Bladou F, Villeirs GM, Virdi J, Boxler S, Robert G, Singh PB, Venderink W, Hadaschik BA, Ruffion A, Hu JC, Margolis D, Crouzet S, Klotz L, Taneja SS, Pinto P, Gill I, Allen C, Giganti F, Freeman A, Morris S, Punwani S, Williams NR, Brew-Graves C, Deeks J, Takwoingi Y, Emberton M, Moore CM, PRECISION Study Group Collaborators MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med 2018;378:1767-77. 10.1056/NEJMoa1801993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouvière O, Puech P, Renard-Penna R, Claudon M, Roy C, Mège-Lechevallier F, Decaussin-Petrucci M, Dubreuil-Chambardel M, Magaud L, Remontet L, Ruffion A, Colombel M, Crouzet S, Schott AM, Lemaitre L, Rabilloud M, Grenier N; MRI-FIRST Investigators. Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsy-naive patients (MRI-FIRST): a prospective, multicentre, paired diagnostic study. Lancet Oncol 2019;20:100-9. 10.1016/S1470-2045(18)30569-2 [DOI] [PubMed] [Google Scholar]
- 9.Cash H, Maxeiner A, Stephan C, Fischer T, Durmus T, Holzmann J, Asbach P, Haas M, Hinz S, Neymeyer J, Miller K, Günzel K, Kempkensteffen C. The detection of significant prostate cancer is correlated with the Prostate Imaging Reporting and Data System (PI-RADS) in MRI/transrectal ultrasound fusion biopsy. World J Urol 2016;34:525-32. 10.1007/s00345-015-1671-8 [DOI] [PubMed] [Google Scholar]
- 10.Padhani AR, Weinreb J, Rosenkrantsz AB, Villeirs G, Turkbey B, Barentsz J, Prostate Imaging-Reporting and Data System Steering Committee : PI-RADS v2 Status Update and Future Directions. Eur Urol 2019;75:385-96. 10.1016/j.eururo.2018.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkel DJ, Heye TJ, Benz MR, Glessgen CG, Wetterauer C, Bubendorf L, Block TK, Boll DT. Compressed Sensing Radial Sampling MRI of Prostate Perfusion: Utility for Detection of Prostate Cancer. Radiology 2019;290:702-8. 10.1148/radiol.2018180556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenkrantz AB, Khasgiwala A, Doshi AM, Ream JM, Taneja SS, Lepor H. Detection of prostate cancer local recurrence following radical prostatectomy: assessment using a continuously acquired radial golden-angle compressed sensing acquisition. Abdom Radiol (NY) 2017;42:290-7. 10.1007/s00261-016-0881-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonelli M, Johnston EW, Dikaios N, Cheung KK, Sidhu HS, Appayya MB, Giganti F, Simmons LAM, Freeman A, Allen C, Ahmed HU, Atkinson D, Ourselin S, Punwani S. Machine learning classifiers can predict Gleason pattern 4 prostate cancer with greater accuracy than experienced radiologists. Eur Radiol 2019;29:4754-64. 10.1007/s00330-019-06244-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowak J, Malzahn U, Baur ADJ, Reichelt U, Franiel T, Hamm B, Durmus T. The value of ADC, T2 signal intensity, and a combination of both parameters to assess Gleason score and primary Gleason grades in patients with known prostate cancer. Acta Radiol 2016;57:107-14. 10.1177/0284185114561915 [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Wu CJ, Bao ML, Zhang J, Wang XN, Zhang YD. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur Radiol 2017;27:4082-90. 10.1007/s00330-017-4800-5 [DOI] [PubMed] [Google Scholar]
- 16.Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, Axel L, Sodickson DK, Otazo R. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med 2014;72:707-17. 10.1002/mrm.24980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the golden ratio for time-resolved MRI. IEEE Trans Med Imaging 2007;26:68-76. 10.1109/TMI.2006.885337 [DOI] [PubMed] [Google Scholar]
- 18.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58:1182-95. 10.1002/mrm.21391 [DOI] [PubMed] [Google Scholar]
- 19.Block KT, Chandarana H, Milla S, Bruno M, Mulholland T, Fatterpekar G, Hagiwara M, Grimm R, Geppert C, Kiefer B, Sodickson DK. Towards Routine Clinical Use of Radial Stack-of-Stars 3D Gradient-Echo Sequences for Reducing Motion Sensitivity. J Korean Soc Magn Reson Med 2014;18:87 10.13104/jksmrm.2014.18.2.87 [DOI] [Google Scholar]
- 20.Tofts PS, Brix G, Buckley DL, L Evelhoch J, Henderson E, Knopp M V, Larsson HBW, Lee T-Y, Mayr N a, Parker GJM, Port RE, Taylor J, Weisskoff RM. Estimating Kinetic Parameters From Dynamic Contrast-Enhanced T1-Weighted\tMRI of a Diffusable Tracer: Standardized Quantities and Symbols. J Magn Reson Imag 1999;10:223-32. [DOI] [PubMed] [Google Scholar]
- 21.Lowekamp BC, Chen DT, Ibáñez L, Blezek D. The Design of SimpleITK. Front Neuroinform. Frontiers Media 2013;7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaniv Z, Lowekamp BC, Johnson HJ, Beare R. SimpleITK Image-Analysis Notebooks: a Collaborative Environment for Education and Reproducible Research. J Digit Imaging 2018;31:290-303. 10.1007/s10278-017-0037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Cai Y, Moloney B, Chen Y, Huang W, Woods M, Coakley F V, Rooney WD, Garzotto MG, Springer CS., Jr Relative sensitivities of DCE-MRI pharmacokinetic parameters to arterial input function (AIF) scaling. J Magn Reson 2016;269:104-12. 10.1016/j.jmr.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany CM, Thoeny HC, Verma S. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol 2016;69:16-40. 10.1016/j.eururo.2015.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. RadioGraphics 2017;37:505-15. 10.1148/rg.2017160130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical Machine Learning Tools and Techniques. Available online: https://www.cs.waikato.ac.nz/ml/weka/book.html
- 27.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988;44:837-45. 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 28.Langer DL, van der Kwast TH, Evans AJ, Plotkin A, Trachtenberg J, Wilson BC, Haider MA. Prostate Tissue Composition and MR Measurements: Investigating the Relationships between ADC, T2, Ktrans, ve, and Corresponding Histologic Features. Radiology 2010;255:485-94. 10.1148/radiol.10091343 [DOI] [PubMed] [Google Scholar]
- 29.Donati OF, Mazaheri Y, Afaq A, Vargas HA, Zheng J, Moskowitz CS, Hricak H, Akin O. Prostate Cancer Aggressiveness: Assessment with Whole-Lesion Histogram Analysis of the Apparent Diffusion Coefficient. Radiology 2014;271:143-52. 10.1148/radiol.13130973 [DOI] [PubMed] [Google Scholar]
- 30.Jung SI, Donati OF, Vargas HA, Goldman D, Hricak H, Akin O. Transition zone prostate cancer: incremental value of diffusion-weighted endorectal MR imaging in tumor detection and assessment of aggressiveness. Radiology 2013;269:493. 10.1148/radiol.13130029 [DOI] [PubMed] [Google Scholar]
- 31.Varghese B, Chen F, Hwang D, Palmer SL, De Castro Abreu AL, Ukimura O, Aron M, Aron M, Gill I, Duddalwar V, Pandey G. Objective risk stratification of prostate cancer using machine learning and radiomics applied to multiparametric magnetic resonance images. Sci Rep 2019;9:1570. 10.1038/s41598-018-38381-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonekamp D, Kohl S, Wiesenfarth M, Schelb P, Radtke JP, Götz M, Kickingereder P, Yaqubi K, Hitthaler B, Gählert N, Kuder TA, Deister F, Freitag M, Hohenfellner M, Hadaschik BA, Schlemmer HP, Maier-Hein KH. Radiomic machine learning for characterization of prostate lesions with MRI: Comparison to ADC values. Radiology 2018;289:128-37. 10.1148/radiol.2018173064 [DOI] [PubMed] [Google Scholar]
- 33.Bittencourt LK, Barentsz JO, de Miranda LCD, Gasparetto EL. Prostate MRI: diffusion-weighted imaging at 1.5T correlates better with prostatectomy Gleason grades than TRUS-guided biopsies in peripheral zone tumours. Eur Radiol 2012;22:468-75. 10.1007/s00330-011-2269-1 [DOI] [PubMed] [Google Scholar]
- 34.Bishop C. Pattern Recognition and Machine Learning. New York: Springer-Verlag, 2006. [Google Scholar]
- 35.Dikaios N, Giganti F, Sidhu HS, Johnston EW, Appayya MB, Simmons L, Freeman A, Ahmed HU, Atkinson D, Punwani S. Multi-parametric MRI zone-specific diagnostic model performance compared with experienced radiologists for detection of prostate cancer. Eur Radiol 2019;29:4150-9. 10.1007/s00330-018-5799-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niaf É, Flamary R, Rouvière O, Lartizien C, Canu S. Kernel-Based Learning From Both Qualitative and Quantitative Labels: Application to Prostate Cancer Diagnosis Based on Multiparametric MR Imaging. IEEE Trans Image Process 2014;23:979-91. 10.1109/TIP.2013.2295759 [DOI] [PubMed] [Google Scholar]
- 37.Lüdemann L, Prochnow D, Rohlfing T, Franiel T, Warmuth C, Taupitz M, Rehbein H, Beyersdorff D. Simultaneous Quantification of Perfusion and Permeability in the Prostate Using Dynamic Contrast-Enhanced Magnetic Resonance Imaging with an Inversion-Prepared Dual-Contrast Sequence. Ann Biomed Eng 2009;37:749-62. 10.1007/s10439-009-9645-x [DOI] [PubMed] [Google Scholar]
- 38.van Niekerk CG, van der Laak JAWM, Hambrock T, Huisman HJ, Witjes JA, Barentsz JO, de Kaa CAH. Correlation between dynamic contrast-enhanced MRI and quantitative histopathologic microvascular parameters in organ-confined prostate cancer. Eur Radiol 2014;24:2597-605. 10.1007/s00330-014-3301-z [DOI] [PubMed] [Google Scholar]
- 39.Langer DL, van der Kwast TH, Evans AJ, Trachtenberg J, Wilson BC, Haider MA. Prostate cancer detection with multi-parametric MRI: Logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J Magn Reson Imaging 2009;30:327-34. 10.1002/jmri.21824 [DOI] [PubMed] [Google Scholar]
- 40.Costa DN, Bloch BN, Yao DF, Sanda MG, Ngo L, Genega EM, Pedrosa I, DeWolf WC, Rofsky NM. Diagnosis of relevant prostate cancer using supplementary cores from magnetic resonance imaging-prompted areas following multiple failed biopsies. Magn Reson Imaging 2013;31:947-52. 10.1016/j.mri.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YJ, Chu WC, Pu YS, Chueh SC, Shun CT, Tseng WY. Washout gradient in dynamic contrast-enhanced MRI is associated with tumor aggressiveness of prostate cancer. J Magn Reson Imaging 2012;36:912-9. 10.1002/jmri.23723 [DOI] [PubMed] [Google Scholar]
- 42.Chandarana H, Feng L, Block TK, Rosenkrantz AB, Lim RP, Babb JS, Sodickson DK, Otazo R. Free-breathing contrast-enhanced multiphase MRI of the liver using a combination of compressed sensing, parallel imaging, and golden-angle radial sampling. Invest Radiol 2013;48:10-6. 10.1097/RLI.0b013e318271869c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trémoulhéac B, Dikaios N, Atkinson D, Arridge SR, Dynamic MR. Image Reconstruction-Separation From Undersampled (k,t)-space via Low-Rank Plus Sparse Prior. IEEE Trans Med Imaging 2014;33:1689-701. 10.1109/TMI.2014.2321190 [DOI] [PubMed] [Google Scholar]
- 44.Jung H, Sung K, Nayak KS, Kim EY, Ye JC. k-t FOCUSS: A general compressed sensing framework for high resolution dynamic MRI. Magn Reson Med 2009;61:103-16. 10.1002/mrm.21757 [DOI] [PubMed] [Google Scholar]
- 45.Rosenkrantz AB, Sabach A, Babb JS, Matza BW, Taneja SS, Deng FM. Prostate Cancer: Comparison of Dynamic Contrast-Enhanced MRI Techniques for Localization of Peripheral Zone Tumor. AJR Am J Roentgenol 2013;201:W471-8. 10.2214/AJR.12.9737 [DOI] [PubMed] [Google Scholar]
- 46.Dikaios N, Atkinson D, Tudisca C, Purpura P, Forster M, Ahmed H, Beale T, Emberton M, Punwani S. A comparison of Bayesian and non-linear regression methods for robust estimation of pharmacokinetics in DCE-MRI and how it affects cancer diagnosis. Comput Med Imaging Graph 2017;56:1-10. 10.1016/j.compmedimag.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 47.Rosenkrantz AB, Geppert C, Grimm R, Block TK, Glielmi C, Feng L, Otazo R, Ream JM, Romolo MM, Taneja SS, Sodickson DK, Chandarana H. Dynamic contrast-enhanced MRI of the prostate with high spatiotemporal resolution using compressed sensing, parallel imaging, and continuous golden-angle radial sampling: Preliminary experience. J Magn Reson Imaging 2015;41:1365-73. 10.1002/jmri.24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brizmohun Appayya M, Sidhu HS, Dikaios N, Johnston EW, Simmons LA, Freeman A, Kirkham AP, Ahmed HU, Punwani S. Characterizing indeterminate (Likert-score 3/5) peripheral zone prostate lesions with PSA density, PI-RADS scoring and qualitative descriptors on multiparametric MRI. Br J Radiol 2018;91:20170645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterjee A, Gallan AJ, He D, Fan X, Mustafi D, Yousuf A, Antic T, Karczmar GS, Oto A. Revisiting quantitative multi-parametric MRI of benign prostatic hyperplasia and its differentiation from transition zone cancer. Abdom Radiol (NY) 2019;44:2233-43. 10.1007/s00261-019-01936-1 [DOI] [PubMed] [Google Scholar]
- 50.Cuocolo R, Stanzione A, Ponsiglione A, Verde F, Ventimiglia A, Romeo V, Petretta M, Imbriaco M. Prostate MRI technical parameters standardization: A systematic review on adherence to PI-RADSv2 acquisition protocol. Eur J Radiol 2019;120:108662. 10.1016/j.ejrad.2019.108662 [DOI] [PubMed] [Google Scholar]
- 51.Mucci LA, Powolny A, Giovannucci E, Liao Z, Kenfield SA, Shen R, Stampfer MJ, Clinton SK. Prospective study of prostate tumor angiogenesis and cancer-specific mortality in the health professionals follow-up study. J Clin Oncol 2009;27:5627-33. 10.1200/JCO.2008.20.8876 [DOI] [PMC free article] [PubMed] [Google Scholar]