Abstract
There is an urgent need to identify antivirals to curtail the COVID-19 pandemic. Herein, we report the sensitivity of SARS-CoV-2 to recombinant human interferons α and β (IFNα/β). Treatment with IFN-α or IFN-β at a concentration of 50 international units (IU) per milliliter reduces viral titers by 3.4 log or over 4 log, respectively, in Vero cells. The EC50 of IFN-α and IFN-β treatment is 1.35 IU/ml and 0.76 IU/ml, respectively, in Vero cells. These results suggest that SARS-CoV-2 is more sensitive than many other human pathogenic viruses, including SARS-CoV. Overall, our results demonstrate the potential efficacy of human Type I IFN in suppressing SARS-CoV-2 infection, a finding which could inform future treatment options for COVID-19.
Keywords: SARS-CoV-2, COVID-19, Interferon, Antiviral therapy, Innate immune
Highlights
-
•
Type I Interferons inhibit SARS-CoV-2, the causative agent for COVID-19, in cultured cells.
-
•
Treatment with IFN-α or IFN-β at 50 IU/ml reduces viral titers by 3.4-log or over 4-log, respectively, in Vero cells.
-
•
The EC50 of IFN-α and IFN-β treatment is 1.35 IU/ml and 0.76 IU/ml, respectively, in Vero cells.
-
•
Type I IFNs have been used in clinical therapies and thus could be repurposed in COVID-19 treatment.
1. Introduction
There is an urgent need to find treatments for COVID-19. Drugs already approved for the treatment of other diseases may offer the most expedient option for treating COVID-19, and several such drugs are already being tested in clinical trials.
Type I interferons (IFN-α/β) have broad spectrum antiviral activities against RNA viruses, which act by inducing an antiviral response across a wide range of cell types and mediating adaptive immune response. Humans produce 13 types of IFN-α and a singular IFN-β (Pestka et al., 2004). Type I IFNs ultimately induces a number of interferon-stimulated genes (ISGs) which encode for a variety of antiviral effectors (Schoggins et al., 2011). Notably, IFN-β production leads to a positive feedback loop that further stimulates the expression of many of the IFN-α genes (Honda et al., 2006). Clinically, Type I IFNs have already been approved for use in the treatment of certain cancers, autoimmune disorders, and viral infections (hepatitis B and hepatitis C). Importantly, type I IFNs are currently evaluated in a clinical trial to treat MERS-CoV and therefore have been proposed for the treatment of COVID-19 but without evidence from laboratory testing against SARS-CoV-2 (Sallard et al., 2020). We assessed the sensitivity of SARS-CoV-2 to both IFN-α and IFN-β in vitro. Herein, we report that type I IFNs exhibited potent anti-SARS-CoV-2 activities in cultured cells, demonstrating the therapeutic potency of type I IFNs for COVID-19.
2. Materials and methods
2.1. Virus and cells
The SARS-CoV-2 (USA-WA1/2020) was obtained from The World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), University of Texas Medical Branch, Galveston, TX. Stock virus was propagated by infecting Vero cells (ATCC CCL-81) at a low multiplicity of infection (MOI) of 0.0025. Three days after infection, supernatants were harvested and centrifuged at 2000 rpm for 5 min to remove cell debris. Stock virus was titrated with a 50% tissue culture infectious dose assay (TCID50) (Narayanan et al., 2008a). All experiments involving infectious virus were conducted at the University of Texas Medical Branch (Galveston, TX) in approved biosafety level 3 laboratories in accordance with institutional health and safety guidelines and federal regulations.
2.2. Virus growth curve
Vero cells were infected by SARS-CoV-2 at MOI 1 or 0.01 for 1 h. Then inoculum was removed, replaced with media (DMEM+5%FBS) and incubated at 37 °C and 5% CO2. At different time points after infection, supernatants were harvested. Virus titers were determined by a TCID50 assay on Vero cells.
2.3. Virus sensitivity to IFN treatment (infectious virus reduction assay)
Vero cells (2 × 104/well) were seeded into 48-well plates for 24 h and treated with human IFN-β1a (mammalian, cat# 11415-1, PBL) and IFN-α (Universal Type I alpha A/D (Bg III), PBL, cat# 11200–1) at different concentrations for 16 h. Cells were then infected with SARS-CoV-2 at an MOI of 0.01 TCID50/cell. IFNs were supplemented after virus infection. Supernatants were collected at 22 h post infection and assayed for virus titers.
2.4. Virus sensitivity to IFN treatment (CPE inhibition assay)
Vero cells grown on 96-well plates (2 × 104/well) were treated with 2-fold serial diluted human IFN-β1a or IFN-α for 16 h (250 IU/ml to 0.49 IU/ml). Cells were then infected with SARS-CoV-2 at an MOI of 0.01 TCID50/cell or Vesicular stomatitis virus (VSV, Indiana strain) at MOI 0.1 PFU/cell for 1 h. The inoculums were removed and replaced with fresh media. As controls, cells were mock-infected, or infected without IFN treatment. All experiments were performed in quadruplicates. For VSV samples, the supernatants were aspirated at 12 hpi. The monolayers were washed with PBS for three times to remove dead cells, fixed with 10% formalin, and stained with crystal violet for cytopathic effect (CPE) observation. For SARS-CoV-2 samples, CPE was observed at 72 hpi.
3. Results
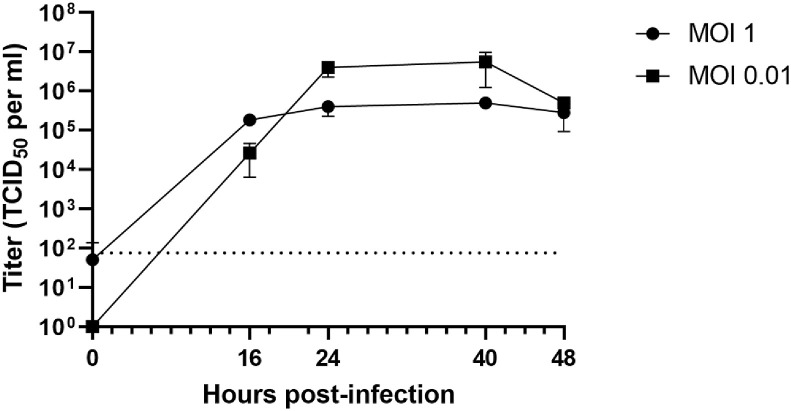
The growth kinetics of the newly identified SARS-CoV-2 in cultured cells had not been characterized. Thus, we first examined the growth kinetics of SARS-CoV-2 in Vero cells. Vero cells were infected at either a low MOI (MOI = 0.01) or high MOI (MOI = 1). Supernatant was collected every 8–16 h. At both conditions, viral titers peaked at approximately 24 h post-infection (hpi) and remained stable until 40 h post-infection before declining (Fig. 1 ). The peak virus titer was 5.5 × 106 TCID50/ml at MOI 0.01 and 3.75 × 105 TCID50/ml at MOI 1, indicating that viral replication was more efficient at a low MOI (MOI = 0.01) than a high MOI (MOI = 1). Additionally, virus infection caused strong cytopathic effect (CPE), which was evident at 48 hpi, much later than the peak of virus production (at 40 hpi).
Fig. 1.
Vero cells were infected by SARS-CoV-2 at MOI 1 or 0.01 for 1 h. At different time points after infection, virus titers were determined by a TCID50 assay on Vero cells. The average of triplicates and Standard deviation are shown. Dotted line indicates the detection limit.
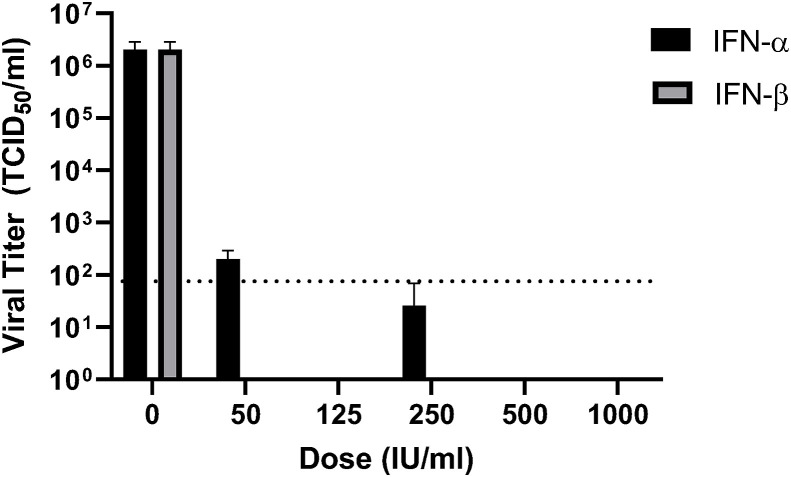
Next, we examined the effect of recombinant human IFN-α and IFN-β treatment on viral infection. Vero cells were pre-treated with different concentrations of IFN-α or IFN-β ranging from 50 to 1000 international units (IU) per milliliter for 16 h. After 1 h of infection with SARS-CoV-2 (MOI 0.01), media containing IFN was returned, and cells were incubated for a further 22 h. Supernatants were then collected, and viral titers were determined via TCID50 assay. The result indicated that IFN-α treatment potently inhibited SARS-CoV-2 infection. Virus titers were not detectable except at the lowest concentration tested (50 IU/ml), at which the viral titers were drastically reduced by 4 logs of magnitude (Fig. 2 ). For IFN-β, the virus titers were below the detection limit at all concentrations tested (50 u/ml-1000u/ml), indicating more potent anti-SARS-CoV-2 activity than IFN-α. Consistently, no CPE was observable under microscopic examination in all IFN-treated samples.
Fig. 2.
Vero cells were pretreated with human IFN-α or IFN-β (0, 50, 125, 250, 500, 1000 IU/ml) for 16 h, and then infected with SARS-CoV2 for 1 h at an MOI of 0.01. Viral inoculums were removed and replaced with fresh media containing listed concentrations of IFN-α or IFN-β. Media was collected at 22 hpi and titers were determined via TCID50 assay on Vero cells. The average of triplicates and Standard deviation are shown. Dotted line indicates the detection limit.
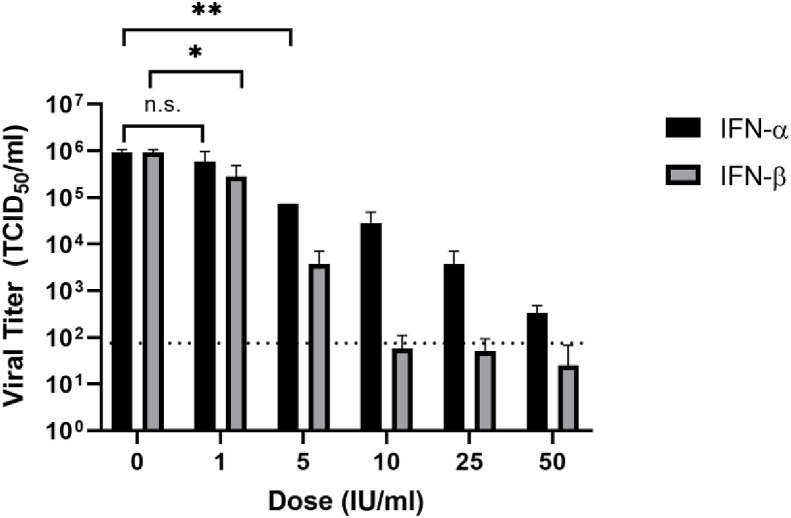
We next tested the antiviral efficacy of IFN-α and IFN-β at lower concentrations (1–50 IU/ml). Both IFN-α and IFN-β dose-dependently inhibited virus infection at these lower concentrations (Fig. 3 ). IFN-α exhibited anti-SARS-CoV-2 activity at a concentration as low as 5 IU/ml, resulting in a significant reduction of viral titer by over 1 log (P < 0.01). With increasing IFN-α concentrations, the virus titers steadily decreased. Treatment with IFN-α at 50 IU/ml drastically reduces viral titers by 3.4 log. Treatment with 1 IU/ml of IFN-β resulted in a moderate (approximately 70%) but significant decrease in virus titer (P < 0.05, Student t-test). Infectious virus was nearly undetectable upon treatment with 10, 25, and 50 IU/ml of IFN-β. The EC50 of IFN-α and IFN-β treatment is 1.35 IU/ml and 0.76 IU/ml, respectively. Taken together, these results indicate that treatment with low concentrations of both IFN-α and IFN-β significantly inhibited viral infection, with IFN-β being slightly more effective than IFN-α.
Fig. 3.
Vero cells were pretreated with human IFN-α or IFN-β (0, 1, 5, 10, 25, 50 U/ml) for 16 h and then infected with SARS-CoV-2 at an MOI of 0.01. Viral inoculums were removed and replaced with fresh media containing listed concentrations of IFN-α or IFN-β. Virus titers at 22 hpi were determined via TCID50 assay. The average of triplicates and Standard deviation are shown. Dotted line indicates the detection limit. (*, P < 0.05; **, P < 0.01; n.s. not significant, one tail Student T test).
In addition, we compared the IFN sensitivity of SARS-CoV-2 with that of Vesicular stomatitis virus (VSV), an IFN-sensitive RNA virus. IFN-α or IFN-β were 2-fold serially diluted (250 IU/ml to 0.49 IU/ml) and added to Vero cells for 16 h. Then cells were infected by VSV (MOI 0.1) or SARS-CoV-2 (MOI 0.01). CPE were observed at 12 hpi for VSV and 72 hpi for SARS-CoV-2. In VSV-infected cells, IFN-α and IFN-β both inhibited CPE development at a concentration of 31.25 IU/ml, while at 15.6 IU/ml the CPE was not discernible from that of IFN-untreated samples. For SARS-CoV2, the lowest concentration that IFN-β or IFN-α inhibited CPE was 31.25 IU/ml and 62.5 IU/ml, respectively. The CPE inhibition data suggests that the IFN sensitivity of SARS-CoV-2 is comparable to that of VSV.
4. Discussion
Type I IFNs are currently evaluated in a clinical trial to treat MERS-CoV and have been proposed for the treatment of COVID-19 (Sallard et al., 2020). Our data clearly demonstrate that SARS-CoV-2 is highly sensitive to both IFN-α and IFN-β treatment in cultured cells, which is comparable to the IFN-sensitive VSV. Our discovery reveals a weakness of the new coronavirus, which may be informative to antiviral development. The experiment was performed in the IFN-α/β gene-defective Vero cells (Desmyter et al., 1968). It is plausible that in IFN-competent cells the efficacy of exogenous IFN-β treatment against SARS-CoV-2 infection is more potent, as IFN-β upregulates other subtypes of Type I IFN expression and augments the IFN-mediated antiviral response (Honda et al., 2006). It is also important to evaluate the antiviral effects of IFNs further in pathobiologically relevant human cell types. Our data may provide an explanation, at least in part, to the observation that approximately 80% of patients actually develop mild symptoms and recover (Wu and McGoogan, 2020). It is possible that many of them are able to mount IFN-α/β-mediated innate immune response upon SARS-CoV-2 infection, which helps to limit virus infection/dissemination at an early stage of disease. At a later stage, the adaptive immune response (antibody etc.) may eventually help patients recover from the COVID-19 disease.
Compared to SARS-CoV-2, it seems that SARS-CoV is relatively less sensitive to IFN treatment in vitro (Dahl et al., 2004; Moriguchi and Sato, 2003). One study reported that the EC50 of IFN-β for SARS-CoV is 95 or 105 IU/ml depending on virus strains (Cinatl et al., 2003). Many other highly pathogenic viruses are also resistant to exogenous IFN treatment. For Ebola virus, it has been reported that treatment with exogenous IFN-α does not affect viral replication and infectious virus production in cultured cells (Kash et al., 2006), probably as a result of antagonism of the IFN response by viral protein. Junín virus, an arenavirus that causes Argentine Hemorrhagic Fever, is likewise insensitive to IFN treatment: when treated with a high concentration of human IFN-α, β or γ (1000 U/ml), the titers of JUNV were reduced by less than 1-log in Vero cells (Huang et al., 2012). The antiviral function of type I IFNs are mediated by a spectrum of ISGs, including PKR, OASs, Mx proteins and RIG-I, which collectively reinforce virus detection and inhibition of viral replication, viral protein synthesis, and the assembly and release of progeny virus particles (Schoggins and Rice, 2011). The apparently higher sensitivity of SARS-CoV-2 to IFN pretreatment as compared to SARS-CoV suggests that the new coronavirus is more susceptible to ISG-mediated antiviral activities. Another possibility is that the new coronavirus may be less capable in suppressing IFN production and/or signaling than SARS-CoV. SARS-CoV encodes several IFN antagonists, including the structural protein NP and M protein (Kopecky-Bromberg et al., 2007; Lu et al., 2011; Siu et al., 2009), nonstructural protein nsp1 (Huang et al., 2011; Kamitani et al., 2006, 2009; Narayananj et al., 2008), nonstructural protein nsp3 (Devaraj et al., 2007; Frieman et al., 2009; Sun et al., 2012), and the accessory protein ORF3b, ORF6, ORF8a and 8 ab (Freundt et al., 2009; Frieman et al., 2007; Kopecky-Bromberg et al., 2007; Narayanan et al., 2008b; Wong et al., 2018). These SARS-CoV proteins are shown to suppress type I IFN production and the JAK/STAT IFN signaling pathway. The amino acid identity between SARS-CoV-2 and SARS-CoV counterparts are 91% (M), 94% (NP), 84% (nsp1), 76% (nsp3), 69% (ORF6) and 40% (ORF8) (Chan et al., 2020). For SARS-CoV-2, whether or not these putative IFN antagonists can interfere with IFN response, and to what extent if any, remains to be investigated. SARS-CoV-2 apparently does not encode ORF3b. Expression of SARS-CoV ORF3b has been shown to block IFN production and STAT1-mediated IFN signaling (Kopecky-Bromberg et al., 2007) and also induce AP-1 transcriptional activity (Varshney and Lal, 2011). Further work is warranted to investigate the IFN response during SARS-CoV-2 infection to better understand the underlying mechanism behind its IFN sensitivity.
In vitro, we have demonstrated that SARS-CoV-2 replication is inhibited by IFN-α and IFN-β at concentrations that are clinically achievable in patients. Recombinant IFN-αs, Roferon-A and Intron-A, which have been approved for hepatitis B and C treatment, can reach concentrations of up to 330 IU/ml and 204 IU/ml, respectively, in serum (Strayer et al., 2014). Recombinant IFN-β drugs, Betaferon and Rebif, which have been approved for the treatment of multiple sclerosis, can reach concentrations of 40 IU/ml and 4.1 IU/ml, respectively, in serum (Strayer et al., 2014). Therefore, some of these drugs may have the potential to be repurposed for the treatment of COVID-19 either alone or in combination with other antiviral therapies.
Acknowledgments
We thank Drs. Kenneth Plante (The World Reference Center for Emerging Viruses and Arboviruses, UTMB) and Natalie Thornburg from the CDC for providing the SARS-CoV-2 stock virus. E.K.M was supported by NIH T32 training grant AI060549. Work in the Paessler laboratory was supported in parts by Public Health Service grants RO1AI093445 and RO1AI129198 and the John. S. Dunn Distinguished Chair in Biodefense endowment. C.H. was supported by UTMB Commitment Fund P84373 and Department of Pathology Pilot Grant and would like to acknowledge the Galveston National Laboratory (supported by the Public Health Service award 5UC7AI094660) for support of his research activity.
References
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl H., Linde A., Strannegard O. In vitro inhibition of SARS virus replication by human interferons. Scand. J. Infect. Dis. 2004;36:829–831. doi: 10.1080/00365540410021144. [DOI] [PubMed] [Google Scholar]
- Desmyter J., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J. Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Park E., Lenardo M.J., Xu X.N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Takaoka A., Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Huang Cheng. Junin virus infection Activates the type I interferon pathway in a RIG-I-dependent manner. PLoS Negl Trop Dis. 2012;6(5):e1659. doi: 10.1371/journal.pntd.0001659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16 doi: 10.1038/nsmb.1680. 1134-U1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J.C., Muhlberger E., Carter V., Grosch M., Perwitasari O., Proll S.C., Thomas M.J., Weber F., Klenk H.D., Katze M.G. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 2006;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Pan J.a., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Gene. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi H., Sato C. Treatment of SARS with human interferons. Lancet. 2003;362:1159. doi: 10.1016/S0140-6736(03)14484-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayananj K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.-T.K., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Sallard E., Lescure F.X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Kok K.H., Ng M.H., Poon V.K., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer D.R., Dickey R., Carter W.A. Sensitivity of SARS/MERS CoV to interferons and other drugs based on achievable serum concentrations in humans. Infect. Disord. - Drug Targets. 2014;14:37–43. doi: 10.2174/1871526514666140713152858. [DOI] [PubMed] [Google Scholar]
- Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C., Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PloS One. 2012;7 doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney B., Lal S.K. SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry. 2011;50:5419–5425. doi: 10.1021/bi200303r. [DOI] [PubMed] [Google Scholar]
- Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. J. Am. Med. Assoc. 2020;323(26):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]