Abstract
The β-barrel assembly machinery (BAM) is responsible for the biogenesis of outer membrane proteins (OMPs) into the outer membranes of Gram-negative bacteria. These OMPs have a membrane-embedded domain consisting of a β-barrel fold which can vary from 8–36 β-strands, with each serving a diverse role in the cell such as nutrient uptake and virulence. BAM was first identified nearly two decades ago, but only recently has the molecular structure of the full complex been reported. Together with many years of functional characterization, we have a significantly clearer depiction of BAM’s structure, the intra-complex interactions, conformational changes that BAM may undergo during OMP biogenesis, and the role chaperones may play. But still, despite advances over the past two decades, the mechanism for BAM-mediated OMP biogenesis remains elusive. Over the years, several theories have been proposed that have varying degrees of support from the literature, but none has of yet been conclusive enough to be widely accepted as the sole mechanism. We will present a brief history of BAM, the recent work on the structures of BAM, and a critical analysis of the current theories for how it may function.
Keywords: BAM complex, membrane protein, conformational plasticity, crosslinking, outer membrane, protein folding, protein biogenesis, Gram-negative bacteria
Brief introduction to BAM
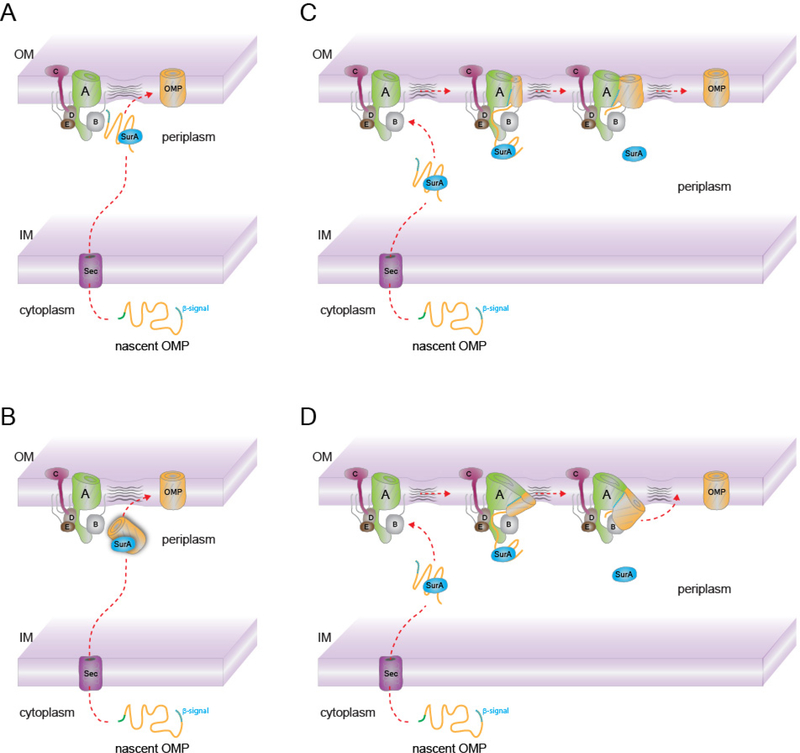
The outer membrane of a Gram-negative bacterium is a natural barrier that protects from the often-harsh extracellular milieu (1–3). It is composed of an asymmetric bilayer with phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet, and is host to a class of integral membrane proteins that are often referred as outer membrane proteins (OMPs) (4–6). These OMPs share a common membrane-embedded β-barrel domain which anchors them into the outer membrane (7, 8). Examples include OmpA, OmpF, OmpT, FepA, and PhoE which serve various roles for the cell including adhesion, signaling, and nutrient import (7–11). The biogenesis of these OMPs is mediated by a multi-component complex called the β-barrel assembly machinery (BAM) (4, 12, 13). As depicted in Figure 1, OMPs are first synthesized in the cytoplasm and then transported into the periplasm through the Sec translocation machinery. With the help of chaperones such as SurA, the OMPs are further escorted to the outer membrane where BAM mediates folding and insertion (4, 6, 14). For simplicity, for this review, we will focus on monomeric OMPs, where one polypeptide forms a single barrel domain. A recent review also discusses the biogenesis of OMPs with added focus on trimeric autotransporter adhesins (TAAs), where three polypeptides are required to form a signal barrel domain (15). BAM has been shown to play a critical role in folding TAAs, however, the exact mechanism here has also remained elusive (16–19).
Figure 1. Biogenesis of β-barrel outer membrane proteins.
β-barrel outer membrane proteins (OMPs) are synthesized in the cytoplasm and routed into the periplasm by the Sec pathway. Chaperones then further escort the OMPs to the β-barrel assembly machinery (BAM) for biogenesis into the outer membrane.
In E. coli, BAM consists of five components called BamA – E, yet the composition of BAM can vary from bacteria to bacteria (i.e. Neisseria lack a BamB ortholog) (20–27). BamA is an OMP itself while BamB-E are lipoproteins anchored into the periplasmic leaflet of the outer membrane. BamA (88 kDa) is a member of the Omp85 superfamily and plays a central role within BAM as both the scaffold which assembles the complex and as the workhorse which orchestrates OMP biogenesis (20, 28–31). BamA and BamD (26 kDa) are the core components of the complex that are essential for cell survival (20, 32). And while BamB (40 kDa), BamC (34 kDa), and BamE (11 kDa) are thought to play accessory roles in BAM, cells that lack these components are comprised and would not likely survive the harsh host conditions during an infection (33–36). Despite early reports that multiple BAMs may constitute the functional biological unit, it has been generally accepted that a single BAM is the active biological unit consisting of a 1:1:1:1:1 ratio of each component having a size of 200 kDa (12, 37–40). Recently though, it has been shown that multiple BAMs co-localize within the outer membrane in precincts which may be mediated by inter-complex BamB-BamB interactions, however, more work is needed to verify the role of BamB here (41).
Aside from OMP biogenesis, BAM also plays a role in the bacterial contact-dependent growth inhibition (CDI) system, a CdiA/CdiB family of two-partner secretion proteins, which suppresses the growth of neighboring target cells under nutrient-limited conditions (42, 43). Several studies have indicated that BamA is the receptor for CdiA while the accessory proteins BamB-E do not appear to be required. BAM’s role in OMP biogenesis does not appear to be necessary for its role in CDI, as the presence of only BamA seems sufficient and studies have localized the CdiAEC93 binding site to extracellular loops L6 and L7 within the β-barrel domain of BamA (43).
BAM as a promising therapeutic target
Multi-drug resistant (MDR) bacteria have quickly become a major concern in the United States and worldwide (44–46). According to a report by the CDC published in 2013, each year about 2 million people in the United States get an antibiotic-resistant infection with 23,000 eventually dying from the infection (44, 47). Antibiotic resistant bacteria are able to survive by undergoing genetic changes in which they become resistant to antibiotics used to treat them or they develop new mechanisms for evading the host’s immune defenses (44, 48, 49). Several studies have identified BAM and its components as promising targets for the development of new antibiotics and antimicrobial therapies against such threats (50–52). This is due to BAM’s essential role in the insertion of OMPs in the outer membrane and its localization at the cell surface. Essential cellular processes such as nutrient acquisition and host cell adhesion make OMPs critical for infection and pathogenesis, and targeting the function of BAM would down-regulate the production of these virulence factors, which would then limit a pathogen’s ability to combat the host’s immune responses.
In 2005, a study found that BamB plays a role in the invasive ability of E. coli strain LF82 (53). Mutants with the BamB gene deleted showed a 95% decrease in their invasive ability compared to the wild type strain. Additional studies performed in Klebsiella pneumoniae, Yersinia enterocolitica and Salmonella enterica revealed that deletion of the BamB gene resulted in changes in OMP levels in the outer membrane (34, 54), a decrease in virulence, and an increase in sensitivity to some antibiotics (34, 35, 54). Other studies focused on BamA found similar results. A study done with Acinetobacter baumannii found that mice immunized against BamA had a 60–80% survival rate when subsequently infected with a lethal dose of A. baumannii (55). A subsequent study found that the addition of MAB1, an antibody antagonistic to BamA, to a culture of E. coli disrupted outer membrane integrity, inhibited protein folding, and inhibited bacterial growth (52).
Overall, these studies point to BAM and its components as a promising target for the development of new antibiotics against pathogenic Gram-negative bacteria, particularly for those that have developed multi-drug resistance. BAM represents a novel target found on the surface of the bacteria, thereby circumventing the need to design compounds which must permeate the outer and inner membranes to reach their targets.
The BAM structures
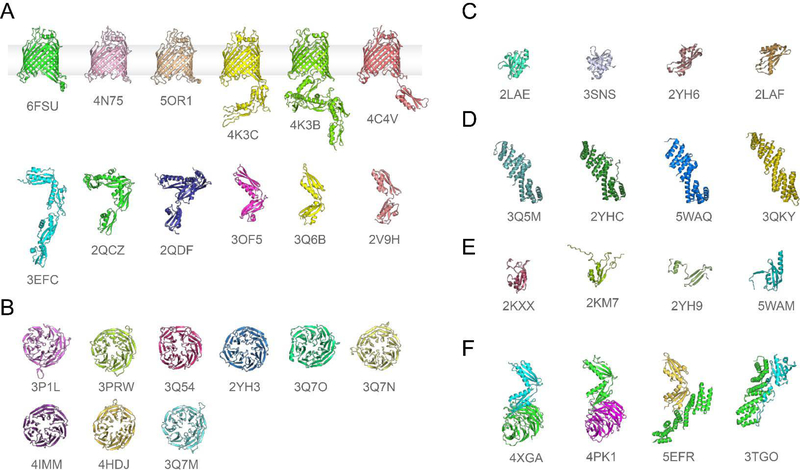
Over the past decade, all the structures of the individual domains of the Bam proteins have been reported from various bacteria (Figure 2), and more recently, structures of fully assembled BAM have been reported (56–77) (Tables 1 and 2). The first structures reported were of the polypeptide transport-associated (POTRA) domains of BamA, which revealed an elongated repeating structure which was thought to serve as the scaffold for the accessory proteins (Figure 2A) (60, 67, 75, 78). The POTRA domains have been observed in various conformations over the years and from SAXS and other experiments, it was concluded that the POTRAs are dynamic with varying regions of high/low flexibility; what role the flexibility of the POTRA domains may serve within BAM remains to be determined. The full length structure of BamA, along with several truncated versions, were later published revealing that BamA contained a membrane-embedded 16-stranded β-barrel domain (62, 79, 80). The barrel domain was found to have unique properties that were important for its role within BAM. The first was that the aromatic belt was thinned along the seam that closes the barrel domain, which was later shown to lead to a significant thinning of the local membrane accompanied by substantial destabilization of the lipid bilayer. And the second was that the seam between strands β1 and β16 undergoes a lateral separation that is required for BAM function (79, 81). Together, these observations offered clues that BamA was conformationally dynamic, particularly along the seam, and may be acting as a catalyst to destabilize the local membrane for OMP biogenesis, ideas later supported by other reports (82).
Figure 2. Structures of the Bam proteins.
A. BamA structures that include the β-barrel domain (top) and those that include just portions of the POTRA domains (bottom). B. All full length BamB structures. C. All BamC structures, which are fragments rather than the full structure. D. All full length BamD structures. E. All full length BamE structures; 2YH9 was found as a domain swap dimer, only one monomer is shown. F. Binary complexes of fragments of Bam proteins. See Table 1 for more information.
Table 1.
Summary of Bam protein structures.
| Protein name | Organism | Method | Resolution (Å) | PDB ID |
|---|---|---|---|---|
| Individual proteins and domains | ||||
| BamA (barrel + P5) | E. coli | X-ray | 3 | 4C4V |
| BamA (β-barrel) | E. coli | X-ray | 2.60 | 4N75 |
| BamA (β-barrel) | E. coli | X-ray | 2.6 | 6FSU |
| BamA (full length) | N. gonorrhoeae | X-ray | 3.2 | 4K3B |
| BamA (barrel + P45) | H. ducreyi | X-ray | 2.91 | 4K3C |
| BamA (β-barrel) | S. typhimurium | X-ray | 2.92 | 5OR1 |
| BamA (P1234) | E. coli | X-ray | 2.7 | 2QCZ |
| BamA (P1234) | E. coli | X-ray | 2.2 | 2QDF |
| BamA (P1234) | E. coli | X-ray | 3.3 | 3EFC |
| BamA (P45) | E. coli | X-ray | 2.69 | 3OG5 |
| BamA (P45) | E. coli | X-ray | 1.5 | 3Q6B |
| BamA (P12) | E. coli | NMR | 2V9H | |
| BamB | E. coli | X-ray | 1.65 | 3Q7M |
| BamB | E. coli | X-ray | 1.77 | 3Q7N |
| BamB | E. coli | X-ray | 2.09 | 3Q7O |
| BamB | E. coli | X-ray | 2.6 | 2YH3 |
| BamB | E. coli | X-ray | 2.00 | 3Q54 |
| BamB | E. coli | X-ray | 1.8 | 3PRW |
| BamB | E. coli | X-ray | 2.6 | 3P1L |
| BamB | P. aeruginosa | X-ray | 1.85 | 4HDJ |
| BamB | M. catarrhalis | X-ray | 2.33 | 4IMM |
| BamC N-terminal | E. coli | X-ray | 1.55 | 2YH6 |
| BamC C-terminal | E. coli | X-ray | 1.5 | 3SNS |
| BamC C-terminal | E. coli | NMR | 2LAE | |
| BamC N-terminal | E. coli | NMR | 2LAF | |
| BamD | E. coli | X-ray | 1.8 | 2YHC |
| BamD | E. coli | X-ray | 2.60 | 3Q5M |
| BamD | N. gonorrhoeae | X-ray | 2.50 | 5WAQ |
| BamD | R. marinus | X-ray | 2.15 | 3QKY |
| BamE | E. coli | X-ray | 1.8 | 2YH9 |
| BamE | E. coli | NMR | 2KM7 | |
| BamE | E. coli | NMR | 2KXX | |
| BamE | N. gonorrhoeae | X-ray | 2.45 | 5WAM |
| Binary complexes | ||||
| BamA(P3-5)B | E. coli | X-ray | 2.15 | 4XGA |
| BamA(POTRA3-5)B* | E. coli | X-ray | 3.1 | 4PK1 |
| BamA(POTRA45)D* | R. marinus | X-ray | 2 | 5EFR |
| BamCD | E. coli | X-ray | 2.9 | 3TGO |
fusion constructs
Table 2.
Summary of BAM structures.
| PDB ID | Method; Resolution | Bam proteins | Detergent(s) | BamC region | BamA POTRA conformation | BamA β-barrel conformation |
|---|---|---|---|---|---|---|
| 5D0Q | X-ray; 3.5 Å | ACDE | C8E4/β-NG | N-term, HG1, HG2 | Closed | Outward-open |
| 5D0O* | X-ray; 2.9 Å | ABCDE | OG/LDAO | N-term | Open | Inward-open |
| 5AYW* | X-ray; 3.6 Å | ABCDE | C8E4/LDAO/OG | N-term | Open | Inward-open |
| 5EKQ | X-ray; 3.4 Å | ACDE | C8E4 | N-term, HG1 | Closed | Outward-open |
| 5LJO | cryo-EM; 4.9 Å | ABCDE | DDM | N-term, HG1† | Closed | Outward-open |
these structures are the same, having the same space group, cell parameters, conformations, and an overall RMSD of 0.46 Å.
observable only when density map is visualized at lower contour levels
BamB was determined to be have an eight-bladed β-propeller fold that shares structural homology with eukaryotic proteins containing WD40 repeat domains which often serve as scaffolds for larger protein complexes (Figure 2B) (63, 64). Therefore, BamB has been proposed to serve as a scaffold protein for optimally orienting the flexible periplasmic domain of BamA for interaction with other BAM components and/or substrates and for assisting in the handoff of substrates from chaperones to BAM (61, 63, 64, 83, 84). Structures of BamB bound to fragments of the POTRA domains of BamA show BamB interacts primarily along the hinge region between POTRA2 and POTRA3, which would indeed sterically restrict the conformations of the periplasmic domain of BamA (61, 77). Another scaffolding role for BamB was also recently reported for BAM (41, 85). Here, BamB was shown to be important for the formation of BamA-BamB inter-complex interactions, thereby leading to the formation of organized assembly precincts within the outer membrane. The molecular details of these interactions and why BAM may need to form these localized precincts remain unknown.
The structure of BamC has been solved in fragments with the N-terminal flexible domain only being observed while in complex with BamD, while the structures of the two C-terminal helix-grip domains have been determined individually (Figure 2C) (66, 68–70). While the N-terminal domain of BamC interacts directly with BamD, no stable interactions between BamC’s helix-grip domains with BamD (or any other Bam protein) have been reported. Therefore, the role of BamC alone or the role of BamC’s interaction with BamD remains unknown. Further, the topology of BamC remains a topic of controversy, as studies have shown portions of it to be surface exposed rather than being found solely within the periplasm; this will be addressed more later in this section as the full BAM structures are presented (41, 85, 86). Whether this observation is important for BamC’s role within BAM is not known.
BamD binds directly to POTRA5 of BamA and the lack of BamD or BamA is lethal (20, 68). The structure of BamD alone, which contains five tetratricopeptide repeat (TPR) domains, and structures in complex with POTRA5 have been reported (Figure 2D) (66, 71, 72, 87). Studies have reported that BamD recognizes unfolded OMP substrates specifically along a recognition sequence termed a ‘β-signal’, however, these studies have been focused on one or few substrates and will need to be studied more comprehensively to determine if such as a mechanism is true for all BAM substrates (88, 89). BamD has been suggested to activate BamA, yet exactly how remains unknown. In these studies, a suppressor mutant of BamD (R176L) was found that reversed the lethal phenotype of the E373K mutation in BamA. In this strain of E. coli, OMP biogenesis was fully restored and the phenotype was indistinguishable from wild type, however, there was no observable interaction between BamDR176L and BamAE373K; therefore, the nature of this interaction remains unknown (90). A similar finding was reported in studies of LptD/E biogenesis, but again, these studies were only suggestive and not conclusive of a BamA-activation mechanism by BamD (88).
BamE, which has an ααβββ fold, interacts directly with BamD and enhances the association of BamD with BamA, which is accomplished by bridging additional interactions as observed in the BAM structures discussed later (Figure 2E) (32, 56–58). Similar to BamC, though nonessential for viability, BamE may be required for certain substrates or other unidentified roles within BAM (91). For example, deletion of BamE abolishes the assembly of OMP/RcsF complexes, yet the role of BamE here remains unknown (92, 93).
The fully assembled structure of BAM has recently been reported by X-ray crystallography and cryo-EM (Figure 3) (56–59) (Table 2). BamB-E were all found interacting with BamA along the POTRA domains and base of the barrel domain, with a trimeric BamCDE complex interacting primarily with POTRA5 and portions of POTRA4, along with periplasmic turns of the barrel domain. BamB, however, was found interacting primarily with POTRA3 along the POTRA2/3 hinge. Two structures of BAM include BamB (PDB IDs 5D0O/5AYW and 5LJO) while two of the structures lack the BamB component (PDB IDs 5D0Q and 5EKQ) (12, 94). The conformational variations between the BAM structures is discussed in more detail in the following section.
Figure 3. Structures of assembled BAM.
A. The structures of BAM found in the outward-open state, including various different conformational of the POTRA domains and both in the presence and absence of BamB. B. The structures of BAM found in the inward-open state, in the presence of BamB. See Table 2 for more information.
Open and closed conformations of BAM
A comparison of the recently reported structures of BAM reveals a number of intriguing observations, which may offer clues to how BAM functions. These include (1) the conformation of the POTRA domains of BamA and (2) the conformation of the barrel domain of BamA. Initially, the X-ray crystal structures of BAM appeared to suggest that BamB may regulate the transition of these changes, however, the cryo-EM structure refuted this idea, leaving us the task of trying to decipher exactly how the observed conformational changes in BAM help drive OMP biogenesis (Figure 3). One caution here is that all structures of BAM to date have been reported in the presence of different detergents rather than in a lipid bilayer or in the presence of substrates (Table 2). Therefore, the field must be cognizant that one or more of the conformations may be artifacts; more work is needed to verify the true conformational states of BAM in a lipid bilayer and what role these conformational changes may play in the biogenesis of OMPs. To address this, NMR has recently been used to investigate BAM in proteolipsomes (95–100). Results from solid-state studies suggest that the barrel domain of BamA and POTRA5 movements are attributed to complex formation in membranes and that the lateral gate and POTRA5 are locally dynamic. While these results are consistent with other observations, more work is needed to decipher exactly how the movements and dynamics along the lateral gate are promoting OMP biogenesis.
In the BAM structures, the POTRA domain of BamA was found in two states which we will refer to as POTRACLOSED and POTRAOPEN (Figure 4 and Table 2). In the POTRACLOSED state, the POTRA domain is found along the base of the barrel domain fully occluding barrel access from the periplasm. In the POTRAOPEN state, the POTRA domain is found swung away from the barrel domain allowing full access to the lumen of the barrel from the periplasm. Crosslinking the POTRA domains in the closed state was shown to disrupt the function of BAM suggesting that cycling of the POTRA domains between open and closed states was necessary for function (57). However, the role of the conformational cycling of the POTRA domains has not been determined, although it has been suggested to possibly help drive OMP insertion into the outer membrane. Further, the spiral ring-shape of the POTRA domains in the BAM structures has been loosely suggested to somehow correlate with the fact that BAM’s substrates are barrel-shaped, however, this seems unlikely given that substrates range significantly in size and strand numbers and that there is no evidence to support such a mechanism (57). In addition, recent molecular dynamics (MD) simulations indicate that the POTRA domain of BamA interacts with the inner leaflet of the outer membrane, which may further restrict the conformational freedom required for POTRA cycling from open to closed and vice versa (101).
Figure 4. POTRA domain conformations in BamA observed within the BAM structures.
A. Top-down view of the POTRAOPEN state of BAM. BamA is shown in green, BamB in gray, BamC in cyan, BamD in magenta and BamE in blue. B. Top-down view of the POTRACLOSED state of BAM. Here, BamA is shown in dark gray. In panels A and B, the red dashed oval indicates the location and orientation of the barrel for reference. C. A top-down view of a structural comparison of the POTRA changes in BamA from the ‘open’ to ‘closed’ states. D. A side view depicting the motion and direction of the POTRA domains; only POTRA5 is shown for clarity. The red dashed arrows indicate the motion and direction of the conformational changes.
The other important conformational change observed was in the barrel domain of BamA, which was found in ‘inward-open’ and ‘outward-open’ states (Figure 5); this terminology is preferred over ‘lateral-open’ which can easily be confused with the unpaired C-terminal strand (unzipped or tucked) state. In the inward-open state, the base of the barrel domain is fully open to the periplasm, similar to what has been observed in nearly all other OMPs, except with a destabilized C-terminal strand which is found tucked inside the barrel domain (102, 103). In the outward-open state, the first half of the barrel domain undergoes a conformational twist where the periplasmic side constricts to close the base of the barrel by ~10 Å reducing the diameter of the barrel here by 25%, while the top of the barrel undergoes a significant opening by ~12 Å (Figures 5A and 5B). The outward-open conformation of the barrel domain has been observed in the presence and absence of BamB, countering the idea that this accessory protein is solely responsible for regulating the conformational state of the barrel domain of BamA (12). Therefore, it is still unclear what role the observed conformational states of BAM may serve in OMP biogenesis, or which are even important in vivo. While still preliminary, it seems clear that the conformation of the POTRA and barrel domains are coordinated such that when the POTRA domain is closed, the barrel is in the outward-open state, and when the POTRA domain is open, the barrel is in the inward-open state. Given the steric strain on the barrel caused from the closed state of the POTRA domain, it would be unfavorable for BAM to be found with a closed POTRA domain with the barrel in the inward-open state. If BAM has a preferred ‘active’ state or if cycling between the states truly is required remains to be determined, along with what role these conformational changes may be serving in OMP biogenesis.
Figure 5. Barrel domain conformations in BamA observed within the BAM structures.
A. Inward-open (gray) and outward-open (green) conformations were observed within the BAM structures, where the first eight strands of the barrel domain undergo a twisting conformation as depicted. The inset shows a zoomed view of the change, with the red arrow indicating the motion of the barrel from inward-open to outward-open. B. The barrel of BamA is shown looking up the periplasmic face. Here, the base of the barrel domain undergoes ~10 Å reduction in diameter to the outward-open state. C. A surface representation of the inward-open state in gray. D. A surface representation of the outward-open state in green.
With the structures of BAM available, the topology of BamC becomes even more confusing, as three of the structures showed at least the N-terminal domain and the first helix-grip domain interacting with BamD, with one of the structures (PDB ID 5D0Q) also including the second helix-grip domain (57). While seemingly contradictory to the studies showing surface-exposure (41, 85, 86), it is likely that the detergent-based solubilization and extraction method disrupts the native topology of BamC which cannot be reconstituted in detergents alone. Additional support for surface-exposure of BamC, and possibly other accessory proteins, was recently reported from work in Neisseria, where BamE was found to be surface-exposed, along with BamD when BamE was knocked out (87). It remains to be determined if these observations are important for the function of BAM.
Mechanisms for the function of BAM
Two major mechanisms or ‘theories’ for BAM-mediated OMP biogenesis have been proposed over the past two decades, each with varying degrees of literature support. We have reviewed these relatively recently and therefore will only briefly summarize here (12). In the first, which we refer to as the BamA-assisted mechanism, BAM serves as a trafficking complex to recruit chaperone-stabilized OMP substrates to the outer membrane where they undergo insertion, catalyzed in part by the locally destabilized membrane created by the barrel domain of BamA (Figure 6A). This mechanism is supported by a vast number of in vitro studies showing that some OMPs can be refolded efficiently into a lipid bilayer without the need of a separate insertase and by the fact that refolding efficiencies are increased when a bilayer is artificially thinned or perturbed (104–106). It has also been shown that BamA alone is able to catalyze OMP folding in an isolated system, yet exactly how this is facilitated wasn’t determined (82). The idea that the presence of a static BAM/BamA alone is necessary and sufficient for mediating insertion doesn’t align with studies definitively demonstrating that the barrel of BamA must undergo conformational changes for BAM to be active, particularly at the lateral seam. While recently reported in vitro experiments using a BamA lateral seam-crosslinked mutant still retains significant activity, this set of experiments did not report the efficiency of crosslink formation which could have been used to estimate residual activity from the non-crosslinked species (59). Even so, the activity of BAM increases significantly when the crosslinked species is chemically reduced, further indicating that a static BAM/BamA is likely insufficient to handle the task of folding cellular quantities of OMPs in vivo. Another study with tOmpA came to a similar conclusion indicating that conformational cycling of BamA may not be required for folding smaller OMPs (107). A variation of the BamA-assisted mechanism is that the substrate OMPs are partially or fully pre-formed within the periplasm, possibly mediated and stabilized by chaperones and/or other Bam proteins, prior to insertion into the membrane (Figure 2B) (6, 108–110).
Figure 6. Two leading theories for the role of BAM in the biogenesis of OMPs.
A. In the BamA-assisted model, BAM serves as a trafficking complex to localize OMPs at the outer membrane. The OMPs are then inserted directly into the membrane assisted by the local destabilization of the membrane by BamA. B. A variation of the BamA-assisted model is depicted where the nascent OMPs are partially or fully pre-folded prior to insertion by BAM. C. In the BamA-budding model, BAM serves to recruit the OMPs to the outer membrane and then the exposed edges of BamA’s barrel serve as a template for catalyzing the formation of a hybrid BamA:OMP barrel via β-augmentation. Each strand added nucleates formation of the next strand until strand exchange occurs within the new OMP, thereby completing the biogenesis process. To prevent the formation of a super-pore within the membrane, the new OMP is thought to ‘bud’ or bleb away from the BamA barrel. D. A recently proposed variation of the budding model, the ‘swing’ model proposes that the nascent OMPs are folded along the interface of the membrane and must undergo a BamA-mediated swinging motion to put them into the membrane.
In the second mechanism, which we refer to as the BamA-budding mechanism, chaperone-stabilized OMP substrates are delivered to BAM for systematic insertion, possibly initiated by the β-signal of the new OMP (Figure 6C) (12, 40, 111, 112). Rather than concerted insertion of the full barrel domain, the exposed edge of the first strand of the BamA barrel serves as a template for strand formation of the new OMP via strand templating, thereby forming a BamA:OMP hybrid barrel intermediate. Each added strand then nucleates formation of the next strand until folding of the new OMP is terminated when its β-signal undergoes dissociation from the first strand of the BamA barrel and binds with higher affinity and specificity to its own first strand during a process termed strand exchange. To prevent the formation of a super-pore in the outer membrane, the new OMP would undergo a ‘budding’ or blebbing away from the core barrel domain of BamA, such that the amphipathic strands of the new OMP form in the correct orientation with hydrophobic residues towards the outside of the barrel, mediating interactions with the membrane, and polar residues towards the inside. Support for this theory comes from several sources, including the observation that the barrel of BamA undergoes lateral opening and large conformational switches to outward-open and inward-open states. Preventing the lateral opening at the seam using crosslinking halts BAM’s function, suggesting that the role of BamA is more than just serving to locally destabilize the membrane (59, 81). Further, the β-barrel domain of all OMPs is organized as a linear arrangement of anti-parallel strands, which suggests a systematic folding process, rather than a stochastic one (6–8, 12). And recently, crosslinking studies with Sam50, a conserved ortholog of BamA in mitochondria, were consistent with the idea that a precursor protein enters the barrel domain of Sam50 and then gets inserted into the membrane through interaction with the lateral gate, in a systematic step-wise manner (113). Despite being proposed nearly a decade ago, even before the first structure of BamA was solved, this study represents the first direct observation in support of the budding mechanism. Recently, another study performed a similar study in bacteria demonstrated they could crosslink the barrel domain of EspP to the barrel of BamA, similar to what was shown for Sam50 (114). In these studies, a new ‘swing’ mechanism was proposed which aligns with the idea of barrel integration, however, differs in the proposed interactions with BamA and how the OMPs are inserted into the membrane; via a swinging motion mediated by BamA (Figure 6D). Still, more work is needed to verify if one or more of these mechanisms is truly used by bacteria, and to be conclusive, it should be demonstrated for a library of OMPs rather than for only one or a few to be accepted as a general mechanism for BAM-mediated OMP biogenesis.
Variations of these mechanisms have also been proposed. For example, studies have proposed that the N-terminus of nascent OMPs may interact with BAM immediately upon exiting the Sec translocon during translocation across the inner membrane, forming a super-complex spanning the periplasm (115). While attractive since it would be efficient and likely require a reduced role for chaperones, it opposes the concepts that the β-signal, which is typically at the C-terminus, of the new OMP initiates interaction with BAM (in both mechanisms) and that the β-signal may be the first strand to integrate into the barrel of BamA (in the BamA-budding mechanism). While still not well understood, studies suggest that the β-signals confer some species specificity and may directly modulate the conformational state of BamA itself (40, 105, 111, 112, 116–118). Therefore, this idea of a super-complex would also increase the risk of inserting an incompletely translated nascent OMP into the outer membrane, which could lead to compromised membrane integrity rendering the cell susceptible to environmental competition and host defenses. One advantage of having the β-signal initiate biogenesis is that it can also serve as a checkpoint to confirm that a fully translated nascent OMP will be inserted into the membrane, thereby ensuring membrane integrity is maintained.
Summary and Future Outlook
The simplest transmembrane domain of any membrane protein found in nature is a single α-helix which is composed entirely of hydrophobic residues. For decades, in vitro studies have been performed to show how efficiently a single transmembrane α-helix can be spontaneously inserted into an artificial lipid bilayer and how thinning the bilayer or perturbing it can further increase insertion efficiencies (119–121). Yet, in nature, rarely do spontaneous events occur in the cell, likely because these events are very challenging to regulate. And for something as critical as the biogenesis of a transmembrane protein, spontaneous insertion doesn’t occur in the cell, not even for a single transmembrane helix (121). N-terminal single transmembrane helix proteins are inserted during co-translation by complex machineries such as YidC and Sec, while C-terminal single transmembrane helix proteins (tail anchored) are inserted by a complex cascade of machineries via the GET pathway (122–125). Arguably more complex than a single transmembrane helix, β-barrel outer membrane proteins are composed of 8–36 amphipathic strands which must be inserted into the outer membrane in a systematic linear arrangement such that the hydrophobic residues are oriented towards the membrane (7, 8, 12). While the amphipathic nature of a nascent OMP may afford it a higher propensity to accommodate aqueous solution than a single transmembrane helix, there are added complexities including how to stabilize the large hydrophobic surface area prior to insertion and the amount of space and volume needed. This is complicated further when considering the biogenesis of trimeric autotransporter adhesins where three monomers make up the final barrel domain (15). Several theories for BAM-mediated OMP biogenesis are presented, however, we do not favor a spontaneous insertion mechanism for reasons noted previously, as evidence favors a more regulated and systematic mechanism. While it is possible that different OMPs may undergo different mechanisms during biogenesis, this isn’t observed for helical membrane proteins with varying numbers of transmembrane helices, although it is true that there are a host of accessory proteins which can assist Sec during biogenesis; a topic of debate and active research still within the helical membrane protein field. Admittedly, the idea that each strand of an OMP is individually inserted sounds messy and complex with many working parts that could potentially mess things up, especially in the absence of a known energy source. However, a similar process is observed for the biogenesis of α-helical membrane proteins with multiple transmembrane domains, which can range from a single helix to more than twenty. And yet, while also complex, the task is accomplished nearly flawlessly within the cell for nearly a third of the proteome.
Significant progress has been made over the past few years which has enabled a better understanding of the overall architecture of BAM, of the dynamics within BAM, and of how substrates may be recognized and inserted into the membrane by BAM. Still, the case of ‘BAM-mediated OMP biogenesis’ is not an open and closed case just yet, as there remain many aspects of OMP biogenesis that are yet to be fully deciphered. For example, the role chaperones such as SurA, Skp and FkpA play is still unclear; whether OMPs may be partially or mostly folded within the periplasm or not is yet to be conclusively determined; the process by which chaperones hand off OMPs to BAM remains mostly unexplored; the role of surface-exposed accessory proteins in OMP biogenesis remains a mystery; and what the conformational state of BAM is in a more native environment (i.e. nanodisc or lipid bilayer). To add clarity to these aspects of OMP biogenesis, future studies should aim (1) to decode the determinants for OMP substrate recognition by BAM and (2) to work out the mechanism for how BAM is able fold and insert new OMPs into the membrane and what role the observed conformational changes may serve. Since the structures of BAM have now been reported, we enter the era where the next breakthroughs in the field will require innovative experiments which aim to study the complex mechanism of BAM both at the biochemical level, but also at the near-atomic level using cryo-EM. And while it is easy to get lost in the science and wonderment of BAM and its complex role in OMP biogenesis, more studies targeting BAM for novel antibiotic discovery needs to be pursued, particularly against multi-drug resistant pathogens such as Neisseria, Pseudomonas, and Acinetobacter, where strains have been classified into the CDC’s highest threat levels.
Acknowledgements
We would like to acknowledge funding support from NIH grants 1R01GM127896 (NIGMS), 1R01GM127884 (NIGMS), and 1R01AI127793 (NIAID).
Nonstandard Abbreviations
- BAM
β-barrel assembly machinery
- OMP
outer membrane protein
- CDI
contact-dependent inhibition
- LPS
lipopolysaccharide
- POTRA
polypeptide transport-associated
- TPR
tetratricopeptide repeat
- MDR
multi-drug resistance
- cryo-EM
cryo-electron microscopy
- RMSD
root-mean-square deviation
- MD
molecular dynamics
- GET
guided entry of tail-anchored proteins
References
- 1.Tokuda H (2009) Biogenesis of outer membranes in Gram-negative bacteria. Bioscience, biotechnology, and biochemistry 73, 465–473 [DOI] [PubMed] [Google Scholar]
- 2.Bos MP, Robert V, and Tommassen J (2007) Biogenesis of the gram-negative bacterial outer membrane. Annual review of microbiology 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 3.Delcour AH (2009) Outer membrane permeability and antibiotic resistance. Biochimica et biophysica acta 1794, 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knowles TJ, Scott-Tucker A, Overduin M, and Henderson IR (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nature reviews. Microbiology 7, 206–214 [DOI] [PubMed] [Google Scholar]
- 5.Schleiff E, and Soll J (2005) Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep 6, 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamm LK, Hong H, and Liang B (2004) Folding and assembly of beta-barrel membrane proteins. Biochimica et biophysica acta 1666, 250–263 [DOI] [PubMed] [Google Scholar]
- 7.Schulz GE (2000) beta-Barrel membrane proteins. Current opinion in structural biology 10, 443–447 [DOI] [PubMed] [Google Scholar]
- 8.Wimley WC (2003) The versatile β-barrel membrane protein. Current opinion in structural biology 13, 404–411 [DOI] [PubMed] [Google Scholar]
- 9.Kramer RA, Vandeputte-Rutten L, de Roon GJ, Gros P, Dekker N, and Egmond MR (2001) Identification of essential acidic residues of outer membrane protease OmpT supports a novel active site. FEBS Lett 505, 426–430 [DOI] [PubMed] [Google Scholar]
- 10.Noinaj N, Easley NC, Oke M, Mizuno N, Gumbart J, Boura E, Steere AN, Zak O, Aisen P, Tajkhorshid E, Evans RW, Gorringe AR, Mason AB, Steven AC, and Buchanan SK (2012) Structural basis for iron piracy by pathogenic Neisseria. Nature 483, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noinaj N, Guillier M, Barnard TJ, and Buchanan SK (2010) TonB-dependent transporters: regulation, structure, and function. Annual review of microbiology 64, 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noinaj N, Gumbart JC, and Buchanan SK (2017) The beta-barrel assembly machinery in motion. Nature reviews. Microbiology 15, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricci DP, and Silhavy TJ (2012) The Bam machine: A molecular cooper. Biochimica et biophysica acta 1818, 1067–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rollauer SE, Sooreshjani MA, Noinaj N, and Buchanan SK (2015) Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc Lond B Biol Sci 370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leo JC, and Linke D (2018) A unified model for BAM function that takes into account type Vc secretion and species differences in BAM composition. AIMS microbiology 4, 455–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain S, and Goldberg MB (2007) Requirement for YaeT in the outer membrane assembly of autotransporter proteins. Journal of bacteriology 189, 5393–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauri A, Soprova Z, Wickstrom D, de Gier JW, Van der Schors RC, Smit AB, Jong WS, and Luirink J (2009) The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155, 3982–3991 [DOI] [PubMed] [Google Scholar]
- 18.Lehr U, Schutz M, Oberhettinger P, Ruiz-Perez F, Donald JW, Palmer T, Linke D, Henderson IR, and Autenrieth IB (2010) C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol Microbiol 78, 932–946 [DOI] [PubMed] [Google Scholar]
- 19.Muhlenkamp M, Oberhettinger P, Leo JC, Linke D, and Schutz MS (2015) Yersinia adhesin A (YadA)--beauty & beast. International journal of medical microbiology : IJMM 305, 252–258 [DOI] [PubMed] [Google Scholar]
- 20.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, and Kahne D (2005) Identification of a Multicomponent Complex Required for Outer Membrane Biogenesis in Escherichia coli. Cell 121, 235–245 [DOI] [PubMed] [Google Scholar]
- 21.Volokhina EB, Beckers F, Tommassen J, and Bos MP (2009) The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. Journal of bacteriology 191, 7074–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neil PK, Rollauer SE, Noinaj N, and Buchanan SK (2015) Fitting the Pieces of the beta-Barrel Assembly Machinery Complex. Biochemistry 54, 6303–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb CT, Heinz E, and Lithgow T (2012) Evolution of the beta-barrel assembly machinery. Trends Microbiol 20, 612–620 [DOI] [PubMed] [Google Scholar]
- 24.Anwari K, Webb CT, Poggio S, Perry AJ, Belousoff M, Celik N, Ramm G, Lovering A, Sockett RE, Smit J, Jacobs-Wagner C, and Lithgow T (2012) The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol Microbiol 84, 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, and Lu L (2019) BamA is a pivotal protein in cell envelope synthesis and cell division in Deinococcus radiodurans. Biochimica et biophysica acta. Biomembranes 1861, 1365–1374 [DOI] [PubMed] [Google Scholar]
- 26.Iqbal H, Kenedy MR, Lybecker M, and Akins DR (2016) The TamB ortholog of Borrelia burgdorferi interacts with the beta-barrel assembly machine (BAM) complex protein BamA. Mol Microbiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenhart TR, Kenedy MR, Yang X, Pal U, and Akins DR (2012) BB0324 and BB0028 are constituents of the Borrelia burgdorferi beta-barrel assembly machine (BAM) complex. BMC microbiology 12, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentle IE, Burri L, and Lithgow T (2005) Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol 58, 1216–1225 [DOI] [PubMed] [Google Scholar]
- 29.Voulhoux R, and Tommassen J (2004) Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol 155, 129–135 [DOI] [PubMed] [Google Scholar]
- 30.Genevrois S, Steeghs L, Roholl P, Letesson JJ, and van der Ley P (2003) The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. The EMBO journal 22, 1780–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz E, and Lithgow T (2014) A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Frontiers in microbiology 5, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, and Silhavy TJ (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Molecular microbiology 61, 151–164 [DOI] [PubMed] [Google Scholar]
- 33.Lewis C, Skovierova H, Rowley G, Rezuchova B, Homerova D, Stevenson A, Sherry A, Kormanec J, and Roberts M (2008) Small outer-membrane lipoprotein, SmpA, is regulated by sigmaE and has a role in cell envelope integrity and virulence of Salmonella enterica serovar Typhimurium. Microbiology 154, 979–988 [DOI] [PubMed] [Google Scholar]
- 34.Hsieh PF, Hsu CR, Chen CT, Lin TL, and Wang JT (2016) The Klebsiella pneumoniae YfgL (BamB) lipoprotein contributes to outer membrane protein biogenesis, type-1 fimbriae expression, anti-phagocytosis, and in vivo virulence. Virulence 7, 587–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fardini Y, Chettab K, Grepinet O, Rochereau S, Trotereau J, Harvey P, Amy M, Bottreau E, Bumstead N, Barrow PA, and Virlogeux-Payant I (2007) The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica Serovar Enteritidis. Infection and immunity 75, 358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fardini Y, Trotereau J, Bottreau E, Souchard C, Velge P, and Virlogeux-Payant I (2009) Investigation of the role of the BAM complex and SurA chaperone in outer-membrane protein biogenesis and type III secretion system expression in Salmonella. Microbiology 155, 1613–1622 [DOI] [PubMed] [Google Scholar]
- 37.Eppens EF, Nouwen N, and Tommassen J (1997) Folding of a bacterial outer membrane protein during passage through the periplasm. The EMBO journal 16, 4295–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagan CL, Silhavy TJ, and Kahne D (2011) beta-Barrel membrane protein assembly by the Bam complex. Annual review of biochemistry 80, 189–210 [DOI] [PubMed] [Google Scholar]
- 39.Hagan CL, Kim S, and Kahne D (2010) Reconstitution of Outer Membrane Protein Assembly from Purified Components. Science 328, 890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robert V, Volokhina EB, Senf F, Bos MP, Gelder PV, and Tommassen J (2006) Assembly Factor Omp85 Recognizes Its Outer Membrane Protein Substrates by a Species-Specific C-Terminal Motif. PLoS Biol 4, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunasinghe SD, Shiota T, Stubenrauch CJ, Schulze KE, Webb CT, Fulcher AJ, Dunstan RA, Hay ID, Naderer T, Whelan DR, Bell TDM, Elgass KD, Strugnell RA, and Lithgow T (2018) The WD40 Protein BamB Mediates Coupling of BAM Complexes into Assembly Precincts in the Bacterial Outer Membrane. Cell reports 23, 2782–2794 [DOI] [PubMed] [Google Scholar]
- 42.Willett JL, Ruhe ZC, Goulding CW, Low DA, and Hayes CS (2015) Contact-Dependent Growth Inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins. J Mol Biol 427, 3754–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ, and Low DA (2014) Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harbor perspectives in medicine 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.CDC. (2013) Antibiotic Resistance Threats in the United States.
- 45.Nikaido H (2009) Multidrug resistance in bacteria. Annual review of biochemistry 78, 119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Duin D, and Paterson DL (2016) Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infectious disease clinics of North America 30, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon SL, and Oliver KB (2014) Antibiotic resistance threats in the United States: stepping back from the brink. American family physician 89, 938–941 [PubMed] [Google Scholar]
- 48.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, and Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nature reviews. Microbiology 8, 251–259 [DOI] [PubMed] [Google Scholar]
- 49.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, and Bartlett J (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48, 1–12 [DOI] [PubMed] [Google Scholar]
- 50.Choi U, and Lee CR (2019) Antimicrobial Agents That Inhibit the Outer Membrane Assembly Machines of Gram-Negative Bacteria. Journal of microbiology and biotechnology 29, 1–10 [DOI] [PubMed] [Google Scholar]
- 51.Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, Ahrens CH, Pessi G, Eberl L, and Robinson JA (2016) A Peptidomimetic Antibiotic Targets Outer Membrane Proteins and Disrupts Selectively the Outer Membrane in Escherichia coli. The Journal of biological chemistry 291, 1921–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Storek KM, Auerbach MR, Shi H, Garcia NK, Sun D, Nickerson NN, Vij R, Lin Z, Chiang N, Schneider K, Wecksler AT, Skippington E, Nakamura G, Seshasayee D, Koerber JT, Payandeh J, Smith PA, and Rutherford ST (2018) Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal. Proceedings of the National Academy of Sciences of the United States of America 115, 3692–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rolhion N, Barnich N, Claret L, and Darfeuille-Michaud A (2005) Strong decrease in invasive ability and outer membrane vesicle release in Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. Journal of bacteriology 187, 2286–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weirich J, Brautigam C, Muhlenkamp M, Franz-Wachtel M, Macek B, Meuskens I, Skurnik M, Leskinen K, Bohn E, Autenrieth I, and Schutz M (2017) Identifying components required for OMP biogenesis as novel targets for antiinfective drugs. Virulence 8, 1170–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh R, Capalash N, and Sharma P (2017) Immunoprotective potential of BamA, the outer membrane protein assembly factor, against MDR Acinetobacter baumannii. Scientific reports 7, 12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakelar J, Buchanan SK, and Noinaj N (2016) The structure of the beta-barrel assembly machinery complex. Science 351, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, and Dong C (2016) Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 [DOI] [PubMed] [Google Scholar]
- 58.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, and Huang Y (2016) Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23, 192–196 [DOI] [PubMed] [Google Scholar]
- 59.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, and Ranson NA (2016) Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nature communications 7, 12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gatzeva-Topalova PZ, Walton TA, and Sousa MC (2008) Crystal Structure of YaeT: Conformational Flexibility and Substrate Recognition. Structure 16, 1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jansen KB, Baker SL, and Sousa MC (2015) Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the beta-barrel assembly machine. The Journal of biological chemistry 290, 2126–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albrecht R, Schutz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, and Zeth K (2014) Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallographica Section D 70, 1779–1789 [DOI] [PubMed] [Google Scholar]
- 63.Jansen KB, Baker SL, and Sousa MC (2012) Crystal structure of BamB from Pseudomonas aeruginosa and functional evaluation of its conserved structural features. PloS one 7, e49749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noinaj N, Fairman JW, and Buchanan SK (2011) The Crystal Structure of BamB Suggests Interactions with BamA and Its Role within the BAM Complex. J Mol Biol 407, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heuck A, Schleiffer A, and Clausen T (2011) Augmenting beta-Augmentation: Structural Basis of How BamB Binds BamA and May Support Folding of Outer Membrane Proteins. J Mol Biol 406, 659–666 [DOI] [PubMed] [Google Scholar]
- 66.Albrecht R, and Zeth K (2011) Structural basis of outer membrane protein biogenesis in bacteria. The Journal of biological chemistry 286, 27792–27803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gatzeva-Topalova PZ, Warner LR, Pardi A, and Sousa MC (2010) Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim KH, Aulakh S, and Paetzel M (2011) Crystal structure of beta-barrel assembly machinery BamCD protein complex. The Journal of biological chemistry 286, 39116–39121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim KH, Aulakh S, Tan W, and Paetzel M (2011) Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC. Acta Crystallogr Sect F Struct Biol Cryst Commun 67, 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warner LR, Varga K, Lange OF, Baker SL, Baker D, Sousa MC, and Pardi A (2011) Structure of the BamC two-domain protein obtained by Rosetta with a limited NMR data set. J Mol Biol 411, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong C, Hou HF, Yang X, Shen YQ, and Dong YH (2012) Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta crystallographica. Section D, Biological crystallography 68, 95–101 [DOI] [PubMed] [Google Scholar]
- 72.Sandoval CM, Baker SL, Jansen K, Metzner SI, and Sousa MC (2011) Crystal structure of BamD: an essential component of the beta-Barrel assembly machinery of gram-negative bacteria. J Mol Biol 409, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim KH, Kang HS, Okon M, Escobar-Cabrera E, McIntosh LP, and Paetzel M (2011) Structural characterization of Escherichia coli BamE, a lipoprotein component of the beta-barrel assembly machinery complex. Biochemistry 50, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 74.Knowles TJ, Browning DF, Jeeves M, Maderbocus R, Rajesh S, Sridhar P, Manoli E, Emery D, Sommer U, Spencer A, Leyton DL, Squire D, Chaudhuri RR, Viant MR, Cunningham AF, Henderson IR, and Overduin M (2011) Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep 12, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, and Kahne D (2007) Structure and Function of an Essential Component of the Outer Membrane Protein Assembly Machine. Science 317, 961–964 [DOI] [PubMed] [Google Scholar]
- 76.Bergal HT, Hopkins AH, Metzner SI, and Sousa MC (2016) The Structure of a BamA-BamD Fusion Illuminates the Architecture of the beta-Barrel Assembly Machine Core. Structure 24, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Z, Zhan LH, Hou HF, Gao ZQ, Xu JH, Dong C, and Dong YH (2016) Structural basis for the interaction of BamB with the POTRA3–4 domains of BamA. Acta Crystallogr D Struct Biol 72, 236–244 [DOI] [PubMed] [Google Scholar]
- 78.Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, and Henderson IR (2008) Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol 68, 1216–1227 [DOI] [PubMed] [Google Scholar]
- 79.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, and Buchanan SK (2013) Structural insight into the biogenesis of beta-barrel membrane proteins. Nature 501, 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, and Huang Y (2014) Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. The FASEB Journal 28, 2677–2685 [DOI] [PubMed] [Google Scholar]
- 81.Noinaj N, Kuszak Adam J., Balusek C, Gumbart James C., and Buchanan Susan K. (2014) Lateral Opening and Exit Pore Formation Are Required for BamA Function. Structure 22, 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Plummer AM, and Fleming KG (2015) BamA Alone Accelerates Outer Membrane Protein Folding In Vitro through a Catalytic Mechanism. Biochemistry 54, 6009–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vuong P, Bennion D, Mantei J, Frost D, and Misra R (2008) Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. Journal of bacteriology 190, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ureta AR, Endres RG, Wingreen NS, and Silhavy TJ (2007) Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. Journal of bacteriology 189, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rassam P, Copeland NA, Birkholz O, Toth C, Chavent M, Duncan AL, Cross SJ, Housden NG, Kaminska R, Seger U, Quinn DM, Garrod TJ, Sansom MS, Piehler J, Baumann CG, and Kleanthous C (2015) Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature 523, 333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Webb CT, Selkrig J, Perry AJ, Noinaj N, Buchanan SK, and Lithgow T (2012) Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J Mol Biol 422, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sikora AE, Wierzbicki IH, Zielke RA, Ryner RF, Korotkov KV, Buchanan SK, and Noinaj N (2018) Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae. The Journal of biological chemistry 293, 1106–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee J, Sutterlin HA, Wzorek JS, Mandler MD, Hagan CL, Grabowicz M, Tomasek D, May MD, Hart EM, Silhavy TJ, and Kahne D (2018) Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proceedings of the National Academy of Sciences of the United States of America 115, 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, and Kahne DE (2016) Characterization of a stalled complex on the beta-barrel assembly machine. Proceedings of the National Academy of Sciences of the United States of America 113, 8717–8722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ricci DP, Hagan CL, Kahne D, and Silhavy TJ (2012) Activation of the Escherichia coli beta-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proceedings of the National Academy of Sciences of the United States of America 109, 3487–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, and Silhavy TJ (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proceedings of the National Academy of Sciences 104, 6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hart EM, Gupta M, Wuhr M, and Silhavy TJ (2019) The Synthetic Phenotype of DeltabamB DeltabamE Double Mutants Results from a Lethal Jamming of the Bam Complex by the Lipoprotein RcsF. mBio 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tata M, and Konovalova A (2019) Improper Coordination of BamA and BamD Results in Bam Complex Jamming by a Lipoprotein Substrate. mBio 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bakelar J, Buchanan SK, and Noinaj N (2017) Structural snapshots of the beta-barrel assembly machinery. FEBS J 284, 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pinto C, Mance D, Sinnige T, Daniels M, Weingarth M, and Baldus M (2018) Formation of the beta-barrel assembly machinery complex in lipid bilayers as seen by solid-state NMR. Nature communications 9, 4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pinto C, Mance D, Julien M, Daniels M, Weingarth M, and Baldus M (2019) Studying assembly of the BAM complex in native membranes by cellular solid-state NMR spectroscopy. Journal of structural biology 206, 1–11 [DOI] [PubMed] [Google Scholar]
- 97.Hartmann JB, Zahn M, Burmann IM, Bibow S, and Hiller S (2018) Sequence-Specific Solution NMR Assignments of the beta-Barrel Insertase BamA to Monitor Its Conformational Ensemble at the Atomic Level. Journal of the American Chemical Society 140, 11252–11260 [DOI] [PubMed] [Google Scholar]
- 98.Sinnige T, Weingarth M, Renault M, Baker L, Tommassen J, and Baldus M (2014) Solid-State NMR Studies of Full-Length BamA in Lipid Bilayers Suggest Limited Overall POTRA Mobility. J Mol Biol [DOI] [PubMed] [Google Scholar]
- 99.Sinnige T, Houben K, Pritisanac I, Renault M, Boelens R, and Baldus M (2015) Insight into the conformational stability of membrane-embedded BamA using a combined solution and solid-state NMR approach. Journal of biomolecular NMR 61, 321–332 [DOI] [PubMed] [Google Scholar]
- 100.Sinnige T, Weingarth M, Daniels M, Boelens R, Bonvin AM, Houben K, and Baldus M (2015) Conformational Plasticity of the POTRA 5 Domain in the Outer Membrane Protein Assembly Factor BamA. Structure 23, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 101.Fleming PJ, Patel DS, Wu EL, Qi Y, Yeom MS, Sousa MC, Fleming KG, and Im W (2016) BamA POTRA Domain Interacts with a Native Lipid Membrane Surface. Biophys J 110, 2698–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Noinaj N, Rollauer SE, and Buchanan SK (2015) The beta-barrel membrane protein insertase machinery from Gram-negative bacteria. Current opinion in structural biology 31, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lundquist K, Bakelar J, Noinaj N, and Gumbart JC (2018) C-terminal kink formation is required for lateral gating in BamA. Proceedings of the National Academy of Sciences of the United States of America 115, E7942–E7949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burgess NK, Dao TP, Stanley AM, and Fleming KG (2008) Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. The Journal of biological chemistry 283, 26748–26758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, and Fleming KG (2014) Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proceedings of the National Academy of Sciences of the United States of America 111, 5878–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Danoff EJ, and Fleming KG (2017) Novel Kinetic Intermediates Populated along the Folding Pathway of the Transmembrane beta-Barrel OmpA. Biochemistry 56, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schiffrin B, Calabrese AN, Higgins AJ, Humes JR, Ashcroft AE, Kalli AC, Brockwell DJ, and Radford SE (2017) Effects of Periplasmic Chaperones and Membrane Thickness on BamA-Catalyzed Outer-Membrane Protein Folding. J Mol Biol 429, 3776–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tamm LK, Arora A, and Kleinschmidt JH (2001) Structure and assembly of beta-barrel membrane proteins. The Journal of biological chemistry 276, 32399–32402 [DOI] [PubMed] [Google Scholar]
- 109.Gatsos X, Perry AJ, Anwari K, Dolezal P, Wolynec PP, Likic VA, Purcell AW, Buchanan SK, and Lithgow T (2008) Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS microbiology reviews 32, 995–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Plummer AM, and Fleming KG (2016) From Chaperones to the Membrane with a BAM! Trends in biochemical sciences 41, 872–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paramasivam N, Habeck M, and Linke D (2012) Is the C-terminal insertional signal in Gram-negative bacterial outer membrane proteins species-specific or not? BMC Genomics 13, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Volokhina EB, Grijpstra J, Beckers F, Lindh E, Robert V, Tommassen J, and Bos MP (2013) Species-specificity of the BamA component of the bacterial outer membrane protein-assembly machinery. PloS one 8, e85799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hohr AIC, Lindau C, Wirth C, Qiu J, Stroud DA, Kutik S, Guiard B, Hunte C, Becker T, Pfanner N, and Wiedemann N (2018) Membrane protein insertion through a mitochondrial beta-barrel gate. Science 359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Doyle MT, and Bernstein HD (2019) Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA beta-barrel. Nature communications 10, 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sara Alvira DWW, Lucy Troman, James Lorriman, Bertram Daum, Vicki A.M. Gold, Ian Collinson. (2019) Trans-membrane association of the Sec and BAM complexes for bacterial outer-membrane biogenesis. BioRxiv [Google Scholar]
- 116.Struyvé M, Moons M, and Tommassen J (1991) Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. Journal of Molecular Biology 218, 141–148 [DOI] [PubMed] [Google Scholar]
- 117.Walther DM, Rapaport D, and Tommassen J (2009) Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cellular and molecular life sciences : CMLS 66, 2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tommassen J (2010) Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156, 2587–2596 [DOI] [PubMed] [Google Scholar]
- 119.Ulmschneider MB, Ulmschneider JP, Schiller N, Wallace BA, von Heijne G, and White SH (2014) Spontaneous transmembrane helix insertion thermodynamically mimics translocon-guided insertion. Nature communications 5, 4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Engelman DM, and Steitz TA (1981) The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell 23, 411–422 [DOI] [PubMed] [Google Scholar]
- 121.Renthal R (2010) Helix insertion into bilayers and the evolution of membrane proteins. Cellular and molecular life sciences : CMLS 67, 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mateja A, and Keenan RJ (2018) A structural perspective on tail-anchored protein biogenesis by the GET pathway. Current opinion in structural biology 51, 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Denic V (2012) A portrait of the GET pathway as a surprisingly complicated young man. Trends in biochemical sciences 37, 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tsukazaki T (2019) Structural Basis of the Sec Translocon and YidC Revealed Through X-ray Crystallography. The protein journal [DOI] [PubMed] [Google Scholar]
- 125.Kuhn A, Koch HG, and Dalbey RE (2017) Targeting and Insertion of Membrane Proteins. EcoSal Plus 7 [DOI] [PMC free article] [PubMed] [Google Scholar]