Abstract
Alzheimer’s disease and small vessel ischemic disease frequently co-exist in the aging brain. However, pathogenic links between these 2 disorders are yet to be identified. Therefore we used Taqman genotyping, exome and RNA sequencing to investigate Alzheimer’s disease known pathogenic variants and pathways: APOE ε4 allele, APP-Aβ metabolism and late-onset Alzheimer’s disease main genome-wide association loci (APOE, BIN1, CD33, MS4A6A, CD2AP, PICALM, CLU, CR1, EPHA1, ABCA7) in 96 early-onset small vessel ischemic disease Caucasian patients and 368 elderly neuropathologically proven controls (HEX database) and in a mouse model of cerebral hypoperfusion. Only a minority of patients (29%) carried APOE ε4 allele. We did not detect any pathogenic mutation in APP, PSEN1 and PSEN2 and report a burden of truncating mutations in APP-Aß degradation genes. The single-variant association test identified 3 common variants with a likely protective effect on small vessel ischemic disease (0.54>OR > 0.32, adj. p-value <0.05) (EPHA1 p.M900V and p.V160A and CD33 p.A14V). Moreover, 5/17 APP-Aß catabolism genes were significantly upregulated (LogFC > 1, adj. p-val<0.05) together with Apoe, Ms4a cluster and Cd33 during brain hypoperfusion and their overexpression correlated with the ischemic lesion size. Finally, the detection of Aβ oligomers in the hypoperfused hippocampus supported the link between brain ischemia and Alzheimer’s disease pathology.
Subject terms: Gene expression, Genetic association study, Genotype, Genetics, Neuroscience, Neurology
Introduction
Late-onset sporadic Alzheimer’s disease (LOAD) and small vessel ischemic disease (SVID) frequently influence each other and co-exist in the aging brain depicting a clinical, neuroradiological and neuropathological spectrum defined as ‘mixed dementia’. Although mixed dementia represents the second common form of dementia in the elderly, as over 45% of LOAD patients neuropathologically diagnosed displayed significant cerebrovascular pathology1, the nature and the pathogenic ground at the basis of AD-SVID interaction is poorly understood2. APOE ε4 allele is the strongest risk factor for sporadic LOAD3–5, however its role in SVID has not been extensively investigated. Common hallmark in small vessel disease is cerebral amyloid angiopathy (CAA), which is caused by excessive deposition of Aβ 40 and 42 on the walls of small vessels6,7, responsible both for its ischemic and hemorragic manifestations (SVID and intracerebral hemorrhage [ICH])8. Both rare familial and common sporadic small vessel disease cases pointed to the potential role of APP-Aß dysmetabolism as key pathogenic mechanism underlying CAA small vessel disease subtype. First, autosomal dominant fully penetrant mutations in the secretase domain of APP, APP duplication, CST3 and TTR rare mutations cause familial CAA9–11. Second, common variants in IDE and LRP1 have been associated with increased risk of diabetes type 2 and migraine, respectively, that frequently are co-morbidities in SVID patients12,13. Third, perivascular and parenchymal Aß deposits have been reported in genetically diagnosed CADASIL patients and vascular dementia cases14–17. Despite the growing body of evidence supporting an imbalance between Aß production and degradation, APP-Aß metabolism role in SVID remains unknown.
Finally, in the last 10 years 9 main LOAD genome-wide association study (GWAS) loci have been identified and replicated by at least 2 independent GWASs and present the strongest effect sizes after APOE (BIN1, CLU, CR1, PICALM, MS4A6A, ABCA7, EPHA1, CD33, CD2AP). They shed light on critical LOAD pathogenic pathways and these include: immune response (MSA4A cluster, CD33, CR1, EPHA1, CD2AP, ABCA7), Aβ40–42 clearance (PICALM, BIN1, CD33 and ABCA7), lipid metabolism (CLU, ABCA7) and vesicles trafficking (PICALM, BIN1) (http://www.alzgene.org/). Among the genetic mechanisms underlying an increased susceptibility for LOAD at these loci, coding variability is emerging as a critical factor18–21.
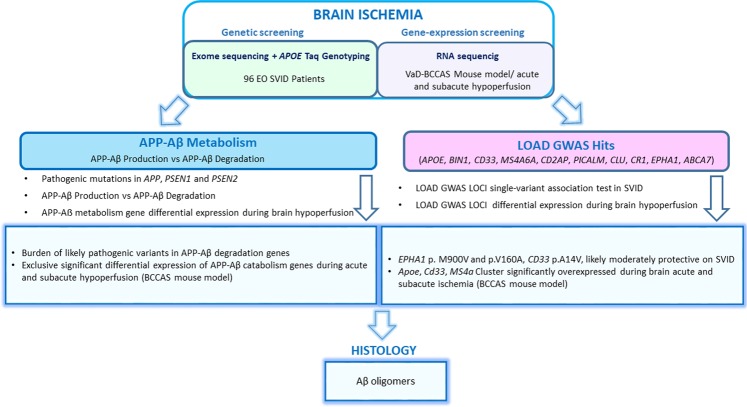
Therefore, in this study we investigated APOE ε2, ε3 and ε4 alleles, APP-Aβ metabolism genes and the most replicated AD GWAS loci through a genetic screening in 96 early-onset independent familial and apparently sporadic SVID Caucasian patients and 368 elderly neuropathological proven controls (HEX database) and through a differential gene expression study during acute and subacute brain ischemia in a mouse model of vascular dementia and subcortical ischemic stroke. Moreover, we analysed whether brain hypoperfusion may have contributed to the generation of AD neuropathological hallmarks (Fig. 1).
Figure 1.
Pipeline followed in the study. SVID, small vessel ischemic disease; VaD, vascular dementia; BCCAS, bilateral common carotid artery stenosis; LOAD, late-onset Alzheimer’s disease; GWAS, genome-wide association study.
We hypothesize that 1) coding variability together with significant differential gene expression in APP-Aß metabolism genes and LOAD GWAS loci may play a role in SVID and brain ischemia and 2) acute severe hypoperfusion-ischemia may prime APP misfolding, toxic soluble oligomers formation that may in the long term accumulate in the stable form of amyloid plaques, as described in elderly patients with vascular dementia22,23.
Materials and Methods
Gene selection
We studied APOE ε2, ε3 and ε4 genotype and 2 clusters of genes: 1) APP-Aβ metabolism genes: 31 genes involved in Aβ production (APP, PSEN1, PSEN2, ADAM9, ADAM10, ADAM17, BACE1, BACE2, NCSTN, PSENEN, APH1B, MEP1B, GPR3), APP stabilization (APLP1, APBA1), APP recycling (SORL1), Aβ deposition (TTR), intracellular degradation (ECE1, ECE2, IDE, CST3, CTSB, CTSD, LYZ, MME), extracellular degradation and clearance (ACE, MMP3, A2M, PLAT, KLK6, LRP1) and 2) LOAD GWAS mainly replicated loci: APOE, BIN1, CLU, CR1, PICALM, MS4A6A, ABCA7, EPHA1, CD33, CD2AP. Selection criteria for these genes has already been reported19,24. The pipeline followed in this study is described in Fig. 1.
Patient cohort
The cohort was composed of 96 independent familial and early-onset apparently sporadic SVID Caucasian non-Hispanic cases from the US, NINDS (National Institute of Neurological Disorders and Stroke), whose DNA was extracted and collected at the NINDS Repository.
All NINDS Repository samples were collected only after an IRB-approved, signed informed consent was secured by the submitter. All methods were carried out in accordance with relevant guidelines and regulations.
Inclusion criteria included small vessel ischemic disease diagnosis based on TOAST classification, early age at onset (<65 years [only 2 cases, whose age-at onset was 68 and 71 years old have been included in the study because familial]), absence of known pathogenic mutations in Mendelian small vessel disease genes (HTRA1, NOTCH3, ACTA2 and COL4A1) and no enrichment for vascular risk factors except for hypertension, which generally plays a critical role in elderly people12. The mean age at disease onset was 51.5 years (range 34–71 years). 82.3% of the cases were male and 44.8% of the cases were positive for a familial history of cerebrovascular disorders. Among the comorbidities and possible risk factors for SVID, hypertension was reported in 60.4% of the patients, diabetes type 2 in 30.2%, myocardial infarction in 7.3%. The majority of the patients (at least 88.54%) did not present atrial fibrillation (AF), which is among the most important risk factors for embolic small vessel occlusion25. In 4.1% and 7.3% of the patients the presence of AF was reported and unknown, respectively. Given the prevalent role of hypertension and type 2 diabetes in SVID in the elderly people12 and the young age at onset of the cohort, these patients were considered enriched for genetic risk factors (Table 1). Finally, 368 controls>70 years of age were selected from ‘HEALTHY EXOMES’, HEX, a publicly available database, which collects exome sequencing data from elderly neuropathologically proven controls (https://www.alzforum.org/exomes/hex)26.
Table 1.
Discovery cohort.
| SVID | SEQUENCING STRATEGY | ORIGIN | AGE at onset (YRS) | MALE (%) | Familial(%) | APOE ε4* (%) | Hypertension | Diabetes | MI | AF +/-/NA | Hypercholesterolemia |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEAN ± SD(RANGE) | |||||||||||
| 96 | WES | Caucasian, non-Hispanic (US) | 51.5 (34–71) | 82.3 | 44.8 | 29 | 60.4 | 30.2 | 7.3 | 4 | 2 |
WES, whole exome sequencing; YRS, years; MI, myocardial infarction; AF, atrial fibrillation; *at least one APOE ε4 allele; NA, not available.
Exome sequencing in patients
We performed whole exome sequencing on a cohort of 96 independent familial and early-onset sporadic SVID cases. DNA was extracted from blood using standard protocols. Library preparation for next generation sequencing used 50 ng DNA. Exome libraries were prepared using Nextera® Rapid Capture Exome Kit (4 rxn × 12 plex, FC-140-1002). The DNA library was then hybridized to an exome capture library (Nextera, Illumina Inc.) and precipitated using streptavidin-coated magnetic beads (Nextera, Illumina). Exome-enriched libraries were PCR-amplified, and then DNA hybridized to paired-end flow cells using a cBot (Illumina, Inc.) cluster generation system. Samples were sequenced on the Illumina HiSeq™ 3000/4000 using 2×76 paired end reads cycles.We used exome sequencing data to identify common (minor allele frequency [MAF] > 3%), rare (MAF < 3%), and very rare (MAF < 1%) coding variants in 31 genes involved in APP-Aβ metabolism (A2M [NM_000014], ACE [NM_000789], ADAM9 [NM_003816], ADAM10 [NM_001110], ADAM17 [NM_003183], APBA1 [NM_001163], APH1B [NM_031301], APLP1 [NM_001024807], BACE1 [NM_012104], BACE2 [NM_012105], CST3 [NM_000099], CTSB [NM_001908], CTSD [NM_001909], ECE1 [NM_001397], ECE2 [NM_014693], GPR3 [NM_005281], IDE [NM_004969], LRP1 [NM_002332], KLK6 [NM_001012964], LYZ [NM_000239], MEP1B [NM_005925], MME [NM_000902], MMP3 [NM_002422], NCSTN [NM_015331], PLAT [NM_000930], PSENEN [NM_172341], SORL1 [NM_003105], TTR [NM_000371], APP [NM_000484.2], PSEN1 [NM_000021.2] and PSEN2 [NM_000447.1]) and 9 main LOAD candidate genes (ABCA7 [NM_019112]; CD2AP [NM_012120]; MS4A6A [NM_152851]; CR1 [NM_000573]; BIN1 [NM_139343]; PICALM [NM_001206946]; EPHA1 [NM_005232]; CLU [NM_001831]; CD33 [NM_001772]). The coding variants detected in these genes have been collected and analyzed. (Tables S1 and S2).
Bioinformatics, exome sequencing
The reads were aligned using BWA-MEM v0.7.1527 to the reference GRCh37 (hs37d5.fa), separate read groups were assigned for all reads from one lane, and duplicates were masked using Samblaster v0.1.2428. Standard QC was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). The variants were then called using GATK UnifiedGenotyper v3.729 and annotated using Jannovar v0.2430 using RefSeq v105 exons.
We used Gene Ontology (http://www.geneontology.org/) and the human lysosome gene database (http://lysosome.unipg.it/) to select lysosomal genes in our dataset.
APOE Genotyping
APOE genotypes comprising the APOE ɛ2, ɛ3 and ɛ4 alleles, were assayed using LightCycler 480 Instrument II (Roche). SNP-specific primers and probes were designed by Thermo Fisher (TaqMan genotyping assays). The polymorphisms distinguish the ɛ2 allele from the ɛ3 and ɛ4 alleles at amino acid position 158 (rs7412) and the ɛ4 allele from the ɛ2 and ɛ3 alleles at amino acid position 112 (rs429358).
Statistical analysis
Power calculation was performed for Fisher´s exact test based on allelic association. We had 80% power for the detection of common variants (minor allele frequency [MAF] > 3%) with strong effect (OR < 0.6 or>2), with a significance value of two-sided α = 0.05 (Fig. S1).
In the single-variant analysis, allele frequencies were calculated for each coding variant in cases and controls and Fisher’s exact test on allelic association was performed. Fisher’s exact test was also used for the statistical analysis of the truncating mutations in APP-Aß metabolism genes. Low frequency and rare variants were defined as having a 1%<MAF < 3% and MAF < 1%, respectively, either in cases or controls. MAF was based either on HEX database for elderly controls>70 years of age or EXac database version 0.3.1 database (http://exac.broadinstitute.org/). A p-value of 0.05 was set as a nominal significance threshold, after false discovery rate (FDR) correction. We report the complete list of coding variants detected in the APP-Aβ metabolism genes and LOAD GWAS loci in the supplementary tables (Table S1 and S2).
T-test performed was used to detect the statistical significance of the number of neurons and glial cells positive for Aß oligomers in hippocampus during acute (2d) and subacute (7d) hypoperfusion in BCCAS and naive mice.
R version 3.3.2 (2016-10-31) (https://www.r-project.org/) was used for computations and graphs, particularly statmod-package v1.4.32 (power calculation) and Ggplot2 (graphs).
BCCAS mouse model, experimental design and exclusion criteria
All experiments and experimental protocols were approved by the Landesamt für Gesundheit und Soziales and conducted according to the German Animal Welfare Act and institutional guidelines. 22 male C57BL/6 J mice, purchased at 8 weeks of age, Charles River, Germany, were housed in a temperature (22 ± 2 °C), humidity (55 ± 10%), and light (12/12-hour light/dark cycle) controlled environment. As previously described31, the animals underwent hypoperfusion between 9 and 13 weeks of age. Hypoperfusion was achieved by bilateral common carotid artery stenosis (BCCAS).
BCCAS mice were imaged before surgery, 24 hours and 1 week post-surgery. At 2 days and 7 days tissue was processed for immunohistochemistry and RNA sequencing.
The BCCAS surgery is further described in the supplementary.
RNA sequencing data: acute (2d) and subacute (7d) hypoperfusion in BCCAS mouse model
To study APP-Aβ metabolism and LOAD GWAS genes during brain acute and subacute hypoperfusion, we used a mouse model of vascular dementia, where brain hypoperfusion is achieved through the placement of microcoils around both common carotid arteries leading to a ≈ 70% stenosis (bilateral common carotid stenosis [BCCAS] mouse model)32. The main features of the model during severe acute and subacute hypoperfusion have been already described31.
In this study, 8 BCCAS mice, 8 sham and 4 naive mice were sacrificed with cervical dislocation 2 days and 7 days post coil insertion surgery, followed immediately by post-mortem dissection of the prefrontal cortex, striatum and hippocampus from one hemisphere. The other hemisphere was preserved for immunohistochemistry. The dissected tissues were immersed in RNA later and stored at −80 °C for later use for mRNA-Sequencing. Total RNA was extracted using miRNeasy Kit (Qiagen, Cat # 217004). Total RNA quality was assessed with the use of Bioanalyzer. Average RIN (RNA Integrity Number) of our samples was 9. Next Generation Sequencing mRNA libraries were prepared with Illumina TruSeq RNA Library Preparation Kit (Illumina, Cat # RS-122-2001).
Bioinformatics, RNA sequencing
Processing, quality assessment and analysis of RNAseq data was carried out using a custom pipeline. We aligned paired end reads with STAR33 against the GRCm38.p4 genome using gencode.vM12 annotation34 (http://www.gencodegenes.org/mouse_releases/12.html), excluding alternative scaffolds and patches. Gene counts were determined using HTSeq.35. Testing for differential gene expression and cerebral blood flow and gene-expression correlation was done using DESeq. 236. Genes were counted as differentially expressed where they had a moderated fold change of 2 or more, contrasting coil to shame samples and where their false discovery rate (FDR) adjusted p-value was below 0.05.
BCCAS mouse model, histology
The staining protocol for the mouse brain histological sections has been already described31. Briefly, for the 20 mice subjected to gene-expression study (8 BCCAS mice, 8 Sham mice and 4 naive mice) one hemisphere was used for RNA sequencing and the contralateral for histology. Fresh frozen hemispheres were cut into 20-µm-thick sections on a cryostat. Moreover, to histologically study both hemispheres, 2 mice were deeply anaesthetized with ketamine and xylazine and perfused through the heart with physiological saline followed by 4% paraformaldehyde, Alexa Fluor® 680 conjugate of WGA (Termofisher, W32465), 3% Gelatin (Sigma-Aldrich, G1890), 1% low melting agarose (Sigma Aldrich A4018) and 0.1% Evans Blue (Sigma Aldrich E2129). Subsequently, the brains were post-fixed for 24 hours in 4% PFA, and cryoprotected in 30% sucrose solution. PFA perfused brains were cut into 50-µm-thick on a cryostat. After washing with phosphate-buffered saline (PBS), free-floating sections were incubated with 10% normal goat serum (NGS, GeneTech, GTX27481) and 0.1% Triton-X-100 (Sigma-Aldrich, X100) in PBS for 1 h at room temperature to block unspecific binding. Primary and secondary antibodies were diluted in 1% NGS and 0.1% Triton-X-100 in PBS. Sections were incubated with rabbit anti-Aß oligomers primary antibody (abcam, Ab126892) and rat anti-GFAP primary antibody (Millipore, 345860) for astrocytes at 4 °C overnight. After thorough washing, sections were incubated at room temperature with AlexaFluor-594- conjugated goat anti-rat (Invitrogen, catalog #A11081) and AlexaFluor- 488-conjugated goat anti-rabbit (Invitrogen, catalog #A11034) secondary antibodies for 2 h at room temperature. Nuclei were counterstained with DAPI (Fluka, 32670). Sections were mounted with anti-fading mounting medium Shandon Immuno Mount (Thermo Scientific, 9990402) on Super Frost Plus glass slides (R.Langenbrinck, 03-0060). Microphotographs were taken with a confocal microscope (Leica TCS SPE; RRID: SciRes_000154).
Oligomers detection and counts
ImageJ version 1.52 A was used to count neuronal and glial cells positive for Aβ oligomers.
Methods to prevent bias, statistics
Mice were randomized to receive hypoperfusion. RNA library preparation and pooling were randomized and blinded, respectively. All methods were carried out in accordance with relevant guidelines and regulations.
Results
Genetic screening
APOE ε4 allele is not associated to increased risk for SVID
The majority of SVID cases were homozygous for ε3 allele (58.3%), around one third of the patients carried in heterozygosity the ε4 allele (27% and 1%, genotype frequencies for ε4/ε3 and ε4/ε2, respectively), whereas a minority of cases (1%) were homozygous either for APOE ε2 or ε4 allele (Table 2). Average age at onset for carriers was 51 years, which did not differ significantly compared to patients homozygous for ε3 allele (52 years). Finally, familial cases displayed a moderately higher APOE ε4 carrier frequency compared to sporadic ones (32.5% and 26.4%, respectively). APOE ε2 alelle was detected in 13/96 (13.5%) patients, 10/13 (77%) with a very young age at onset (≤55 years).
Table 2.
APOE genotype.
| COHORTS | N | ε4/ε4 | APOE GENOTYPE (%) | ||||
|---|---|---|---|---|---|---|---|
| ε4/ε3 | ε3/ε3 | ε3/ε2 | ε2/ ε4 | ε2/ ε2 | |||
| Caucasian SVID | 96 | 1 | 27 | 58.3 | 11.45 | 1 | 1 |
| Caucasian controls* | 6262 | 1.8 | 21.3 | 60.9 | 12.7 | 2.6 | 0.8 |
| Caucasian LOAD* | 5107 | 14.8 | 41.1 | 36.4 | 4.8 | 2.6 | 0.2 |
N, number; SVID, small vessel ischemic disease; LOAD, late-onset Alzheimer's disease. *Data for Caucasian controls and LOAD are taken from a previous publication (Farrer et al., 1997)
APP-Aβ metabolism genes
To study a possible role of APP-Aβ metabolism genes in SVID, we focused on 1) possible enrichment for pathogenic mutations in Mendelian genes (APP, PSEN1 and PSEN2), underpinning autosomal dominant AD (http://www.molgen.ua.ac.be/ADMutations/) and 2) burden of damaging mutations in APP-Aβ catabolism genes upon APP-Aβ production genes, underlying sporadic LOAD37.
We screened protein coding variability in 31 genes involved in APP-Aß metabolism and we identified 130 coding variants: 21 common and 88 rare. ADAM10 and PSENEN did not harbour any coding variant. Among the rare variants we report 21 novel variants, and 10 truncating mutations. The majority of the variants detected were singletons (86/130 [66.15%]). BACE1, BACE2, CST3, CTSB and CTSD did not harbour any rare damaging variant. The majority of patients, 75/96 (78.12%), carried at least one rare likely damaging variant and almost half of them, 43/96 (44.8%), harboured multiple likely pathogenic alleles in the studied genes.
ADMendelian genes:APP, PSEN1, PSEN2
We report a total of 5 rare coding variants in APP, PSEN1 and PSEN2. None of these are likely to be deleterious: APP p.V576I and p.T280del do not cluster in the conserved secretase domain; PSEN1 p.E318G and PSEN2 p.L2F and p.R62H map outside the alpha helix surface of the transmembrane domains (TMs), where all the pathogenic mutations have been reported (alpha-helix rule)38 (Table 3).
Table 3.
Coding variants detected in APP, PSEN1 and PSEN2 in the SVID cohort.
| Gene | Position | rsID | Ref/Alt | Genomic change | Aa change | PROVEAN | SIFT | Polyphen2 | Carrier freq (%) | MAF(%) | Carrier APOE ε4 | Hex70 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APP | 21: 27284236 | rs200769792 | C/T | c.1726G > A | p.V576I | Neutral | Tolerated | probably damaging | 1/96 (1) | 0.5 | e3/e2 | — |
| APP | 21: 27394181 | rs764406483 | TGTG/T | c.837_839del | p.T280del | NA | NA | NA | 2/96 (2) | 1 | e3/e3 e3/e3 | — |
| PSEN1 | 14: 73673178 | rs17125721 | A/G | c.953 A > G | p.E318G | Deleterious | Damaging | benign | 5/90 (5.5) | 2.7 |
e4/e3 e3/e3 e3/e3 e4/e3 e3/e3 |
2 |
| PSEN2 | 1: 227069612 | NOVEL | C/T | c.4 C > T | p.L2F | Neutral | Damaging | probably damaging | 1/93 (1) | 0.5 | e3/e3 | — |
| PSEN2 | 1: 227071449 | rs58973334 | G/A | c.185 G > A | p.R62H | Neutral | Tolerated | benign | 1/93 (1) | 0.5 | e3/e3 | — |
Aa, amino acid; freq, frequency, NA, not available.
Other genes playing a key role in APP-Aß metabolism. CST3 p.A25T in homozygosity and SORL1 variants clustering in VPS domain are known risk factors for LOAD and may influence SVID susceptibility
Among the variants detected in the other genes involved in APP-Aß metabolism, 3 missense mutations in genes playing a role in APP-Aß degradation were of particular interest: one polymorphism reported as pathogenic in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), CST3 p.A25T, and SORL1 p.E270K and p.A528T (Table 1).
Importantly, homozygosity for CST3 p.A25T has been significantly associated with AD39 and other neurodegenerative conditions such as macular degeneration40. In our cohort, we report 4/96 [4.16%] patients homozygous for the minor allele A, a carrier frequency which was 2.55 times higher when compared with HEX controls (6/368 [1.63%]). Notably, the homozygous carriers, 3 sporadic and 1 familial cases, displayed an average age at onset of 49.75 years (range: 39–60 y), 3/4 (75%) were homozygous for APOE ε3 allele and presented a MMSE score moderately lower (26 < MMSE < 28). By contrast, only one case carried in heterozygosity the APOE ε4 allele. Finally, among CST3 p.A25T homozygous carriers only 1/4 (25%) patients presented 3 moderate risk factors for SVID: hypertension, myocardial infarction and type 2 diabetes, 3/4 (75%) displayed hypertension and 1/4 (25%) did not present any risk factor for SVID (Table S3).
Interestingly, SORL1 p.E270K and p.A528T clustering in the vacuolar protein sorting (VPS10) domain (Aa 124–757) have been found in 2/96 SVID patients homozygous for APOE ε3 allele and have been reported pathogenic and to segregate within AD families41.
APP-Aβ genes, pooled variants. Analogously to LOAD, SVID patients are enriched for LoF mutations in genes involved in APP-Aß degradation rather than production
We then compared coding genetic variability between 14 genes mainly involved in APP-Aβ production (ADAM10, ADAM17, ADAM9, APBA1, APH1B, APLP1, APP, BACE1, BACE2, GPR3, NCSTN, PSEN1, PSEN2, PSENEN) and 17 genes taking part in APP-Aß degradation (A2M, ACE, CST3, CTSB, CTSD, ECE1, ECE2, IDE, KLK6, LRP1, LYZ, MEP1B, MME, MMP3, PLAT, SORL1, TTR).
We report a significant enrichment for loss of function (LoF) mutations (stop gain/loss, inframe insertions/deletions, splice-site mutations) in genes regulating Aß degradation in SVID patients (9/96 [9.4%]), both when compared to APP-Aß production genes (1/96 [1%]) (Fisher p-value = 0.01837), and Aß degradation genes in the HEX cohort (6/368 [1.6%])(Fisher p-value= 3.496e-14) (Fig. 2, Table 4).
Figure 2.

Number of loss of function (LoF) mutations in APP-Aβ degradation and production genes detected in the HEX and SVID cohorts and number of individuals per each cohort. The SVID cohort presents a burden of truncating mutations, compared to the HEX cohort. SVID, small vessel ischemic disease.
Table 4.
Loss of function mutations detected in the SVID and HEX cohort.
| Cohort | Gene | Position | rsID | Ref/Alt | Genomic change | Aa change | Carrier freq (%) | MAF(%) | Hex70 (%) |
|---|---|---|---|---|---|---|---|---|---|
| SVID | MME | 3: 154834478 | rs749320057 | AC/A | c.467del | p.P156Lfs*14 | 1/96 (1) | 0.5 | 0 |
| SVID | PLAT | 8: 42033519 | rs1804182 | G/A | c.1681C > T | p.R561* | 1/96 (1) | 0.5 | — |
| SVID | IDE | 10: 94238404 | rs763710639 | G/GGT | c.1881_1882insAC | p.L628Tfs*5 | 1/96 (1) | 0.5 | — |
| SVID | IDE | 10: 94333763 | rs533083105 | AG/A | c.13del | p.L5fs | 1/96 (1) | 0.5 | 0.35 |
| SVID | MMP3 | 11: 102713159 | rs781898035 | TACC/T | c.499_499 + 2del | p.E167del | 1/96 (1) | 0.5 | — |
| SVID | LRP1 | 12: 57602503 | NOVEL | C/A | c.12048 C > A | p.Y4016* | 1/96 (1) | 0.5 | — |
| SVID | LRP1 | 12: 57603939 | rs757410385 | G/GC | c.12575dup | p.D4193Rfs*9 | 1/96 (1) | 0.5 | 0.14 |
| SVID | MEP1B | 18: 29784212 | rs200539508 | C/T | c.436 C > T | p.R146* | 1/96 (1) | 0.5 | 0 |
| SVID | APP | 21: 27394181 | rs764406483 | TGTG/T | c.837_839del | p.T280del | 2/96 (2) | 1 | — |
| SVID | LRP1 | 12:57605739 | rs759104743 | TTGC/T | c.13300_13302del | p.L4434del | 1/96 (1) | ||
| HEX | ADAM17 | 2:9630398 | GATC/G | p.Asp794del | 1/368 (0.3) | 0.0014 | 0.0014 | ||
| HEX | BACE1 | 11:117160453 | rs758335005 | G/T | p.Tyr445* | 1/324 (0.3) | 0.0015 | 0.0015 | |
| HEX | APP | 21:27394181 | rs768084853 | TGTGGTGGTG/TGTGGTG | p.Thr280del | 1/101 (1) | 0.005 | 0.005 | |
| HEX | APBA1 | 9:72131320 | rs768254638 | GTCC/G | p.Glu268del | 1/360 (0.2) | 0.0014 | 0.0014 | |
| HEX | ACE | 17:61562376 | C/T | p.Gln50* | 1/367 (0.2) | 0.0014 | 0.0014 | ||
| HEX | CST3 | 20:23615971 | C/A | p.Glu93* | 1/367 (0.2) | 0.0014 | 0.0014 | ||
| HEX | CST3 | 20:23615984 | rs760409425 | G/T | p.Tyr88* | 1/366 (0.2) | 0.0027 | 0.0027 | |
| HEX | LRP1 | 12:57605739 | rs759104743 | TTGC/T | p.Leu4432del | 1/368 (0.2) | 0.0014 | 0.0014 | |
| HEX | MEP1B | 18:29796983 | C/T | p.Gln597* | 1/366 (0.2) | 0.0014 | 0.0014 | ||
| HEX | ECE2 | 3:184002778 | rs769984677 | G/T | p.Glu463Ter | 1/368 (0.2) | 0.0014 | 0.0014 |
Aa, amino-acid change. Freq, frequencies; MAF, minor allele frequency; SVID, small vessel ischemic disease.
LOAD GWAS loci (BIN1, CD33, MS4A6A, PICALM, CLU, CR1, EPHA1, ABCA7)
Single coding variant association test. EPHA1 p.V160A, CD33 p.A14V, ABCA7 p.G1527A are LOAD GWAS hits or in LD with LOAD GWAS hits and may play a modest protective effect in SVID.
To investigate the role of the LOAD GWAS loci with the strongest effect sizes after APOE (BIN1, CD33, MS4A6A, PICALM, CLU, CR1, EPHA1, ABCA7), we focus on a possible significant association between coding variants detected in these loci and SVID.
We screened protein coding variability in 9 highly replicated LOAD GWAS loci in 96 SVID patients and we identified 69 coding variants: 26 common, 4 rare and 39 very rare. Among these, 24 were singletons, 6 were novel and 6 were truncating mutations. PICALM and CD2AP did harbour the lowest number of variants (0,0005 variants per Kb of coding sequence). By contrast, MS4A6A harboured the highest number of coding variants (7,43 variants per Kb of coding sequence). The majority of patients, 81/96 (84.3%), carried at least one rare likely damaging variant and almost half of them, 45/96 (46.8%), harboured multiple likely pathogenic alleles in the studied genes.
Among these variants, 36/69 (52.2%) have been also detected in the 368 controls>70 years of age in the HEX database and were selected for the single-variant-based analysis (Table 4).
The single variant association test identified 3 coding variants whose allelic frequency significantly differed between SVID cases and controls (EPHA1 p.M900V and p.V160A and CD33 p.A14V) (adj. p-value <0.05). Importantly, all of these are common variants, with a modest to no damaging effect (15.46 > CADD score> 5.284) with moderate to strong likely protective effect size (0.3 < OR < 0.6) and only a minority of carriers are at least heterozygous for the APOE ε4 or ε2 alleles (up to 28.2% and 21.2%, respectively) (Table 5).
Table 5.
Alzheimer’s disease GWAS hit single-variant association test in the SVID cohort.
| Gene | Position | rs ID | Alleles | cDNA change | Aa change | MAF HEX_70 | MAF SVD | CADD score | SVD_allele count/ allele number | HEX_allele count/ allele number | p-val | p-val adj | OR | CI | APOE ε4/ε2 (%) | Comment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EPHA1 | 7:143088867 | rs6967117 | T/C | c.2698 A > G | p.M900V | 0.919 | 0.786 | 15.46 | 151/192 | 658/716 | 1.414e-06 | 5.0e-05 | 0.325 | 0.205–0.517 | 27.5/15 | |
| CD33 | 19:51728477 | rs12459419 | C/T | c.41 C > T | p.A14V | 0.3251 | 0.208 | 13.71 | 40/192 | 238/732 | 1.4e-03 | 0.025 | 0.546 | 0.363–0.808 | 27.2/21.2 |
LD with LOAD GWAS hit rs3865444 (Raj et al., 2014, p. 33) Blood Protein Level GWAS hit (Suhre et al., 2017) Hematological trait GWAS hit (Astle et al., 2016) |
| EPHA1 | 7:143097100 | rs4725617 | A/G | c.479 T > C | p.V160A | 0.917 | 0.843 | 11.84 | 162/192 | 675/736 | 3.9e-03 | 0.04680 | 0.488 | 0.299–0.810 | 28.2/12.9 | Blood Protein Level GWAS hit (MacArthur et al., 2017) |
| ABCA7 | 19:1056492 | rs3752246 | G/C | c.4580 G > C | p.G1527A | 0.8388 | 0.755 | 5.284 | 145/192 | 609/726 | 1.06e-02 | 0.086 | 0.593 | 0.398–0.891 | 31.4/12.7 | LOAD GWAS hit (Cuyvers et al., 2015) |
| CR1 | 1:207726161 | rs200082366 | G/T | c.3066 G > T | p.Q1022H | 0.0936 | 0.036 | 15.51 | 7/192 | 38/406 | 0.012 | 0.086 | 0.366 | 0.135–0.852 | 16.6/50 | protecting against immunocomplex deposition (Birmingham et al., 2003) |
| CD33 | 19:51728641 | rs2455069 | A/G | c.205 A > G | p.R69G | 0.407 | 0.442 | 0.209 | 85/192 | 299/734 | 0.4108 | 1 | 1.155 | 0.826–1.611 | 32.3/15.38 |
Blood Protein Level GWAS hit (Suhre et al., 2017) Hematological trait GWAS hit (Astle et al., 2016) |
| CR1 | 1:207782916 | rs4844609 | A/T | c.4828 A > T | p.T1610S | 0.974 | 0.994 | 6.589 | 191/192 | 717/736 | 0.095 | 0.570 | 5.056 | 0.793–211.22 | 29.1/13.5 | |
| CR1 | 1:207795320 | rs2296160 | A/G | c.5905 A > G | p.T1969A | 0.834 | 0.880 | 0.001 | 169/192 | 599/718 | 0.145 | 0.745 | 1.459 | 0.893–2.468 | 29.78/13.8 | |
| CR1 | 1:207782931 | rs6691117 | A/G | c.4843 A > G | p.I1615V | 0.1943 | 0.239 | 8.406 | 46/192 | 143/736 | 0.190 | 0.820 | 1.306 | 0.873–1.931 | 25/10 | |
| ABCA7 | 19:1055191 | rs3745842 | G/A | c.4046 G > A | p.R1349Q | 0.354 | 0.357 | 0.367 | 68/192 | 217/536 | 0.228 | 0.820 | 0.806 | 0.562–1.149 | 31.5/15.78 |
ID, identification number; Aa, amino acid; MAF, minor allele frequency; SVID, small vessel ischemic disease; CADD, combined annotation dependent depletion; HEX, Healthy Exomes; p-val, p-value; adj, adjusted; OR, Odds Ratio; CI, Confidence Interval. LD, linkage disequilibrium, LOAD, late-onset alzheimer´s disease. GWAS, genome-wide association study.
Interestingly, EPHA1 p.V160A, CD33 p.A14V and 2 missense mutations nominally associated with SVID (ABCA7 p.G1527A and CR1 p.Q1022H) have been already reported either as functional, as LOAD GWAS hit, in linkage disequlibrium (LD) with LOAD GWAS hits or significantly linked to different complex traits (blood protein levels and haematological traits)42,43, further supporting the critical effect of the amino acid substitution in these positions. EPHA1 p.V160A (rs4725617-G), was significantly associated with a moderate reduced risk for SVID (adj. p-value=0.046, OR = 0.488) and was located 12.04 kb proximal to rs11767557-T, significantly linked to LOAD and cognitive impairment44 and reported to be strongly associated to protein abundance levels42,43. In addition, CD33 p.A14V, associated to CD33 isoform lacking exon 2 and with a reduced inhibitory effect on microglia, has been reported to be in strong LD with the LOAD GWAS significant protective allele rs3865444-A45,46. CR1 p.Q1022H has been shown to lower CR1 expression and increase C4b binding, therefore protecting against immunocomplex deposition47. Moreover, ABCA7 p.1527 G (rs3752246-C) is likely to reduce SVID susceptibility and, analogously, represents a LOAD GWAS hit and a modest protective allele for LOAD48,49.
Gene expression screening during acute and subacute hypoperfusion in the BCCAS mouse model
APP-Aβmetabolism
We have used RNA sequencing data from prefrontal cortex, hippocampus and striatum of a mouse model of ischemia characterized by watershed and mainly subcortical infarcts (Fig. 3A–G), therefore a reliable model to study vascular dementia.
Figure 3.
A-G.Bilateral common carotid artery stenosis (BCCAS) mouse model, displaying monolateral right small subcortical lesions, mainly affecting striatum (A) and hippocampus (B) (blue arrows), during acute (2d) and subacute (7d) hypoperfusion. The most severly hypoperfused hemisphere, with ischemic lesions detected on T2 weighted MRI (right hemisphere) was used for RNA sequencing. The contralateral hypoperfused hemisphere, with no ischemic lesions detectable on T2-MRI (left hemisphere) was used for immunohistochemistry. C, hypoperfused brain stained with GFAP, presenting peri-infarct astrocytosis in hippocampus and striatum during subcute hypoperfusion (7d) (white arrows). The infarct area is delimited by white dashed lines. The left hemisphere is hypoperfused and although ischemic lesions in the left side are not detectable on T2-MRI, we report a significant gliosis (yellow arrow). D, hypoperfused brain stained with IBA1, presenting microglia infiltration of the infarct areas and to a lesser extent peri-infarct areas and in the most severe hypoperfused areas during subcute hypoperfusion (7d) (pink arrows). E. Hypoperfused hippocampus stained with GFAP, displaying astrocytosis. F. Hypoperfused hippocampus stained with IBA1, displaying gliosis. G. T2-MRI slides showing the extension of the ischemic lesion (red).
We detected a selective significant overexpression (up to 8-fold change and adj. p-value<0.05) of 5/17 genes (29.4%) involved in APP-Aß degradation (A2m, Plat, Ctsd, Ctsb and Klk6) and none of the genes controlling APP-Aß production during brain acute or subacute hypoperfusion (2d and 7d post-surgery, respectively). A2m was ubiquitously overexpressed both in prefrontal cortex, hippocampus and striatum, both 2 and 7 days post-surgery. By contrast, Plat, Ctsd and Klk6, displayed a specific time and region pattern of overexpression: Plat was overexpressed in prefrontal cortex and striatum 2 days post-ischemia and Ctsd and Klk6 in striatum and hippocampus 7 days post-surgery, both tissues characterized by the highest degree of tissue remodelling and overall differential expression (Table 6).
Table 6.
Differential gene expression during acute and subacute hypoperfusion in BCCAS mouse model.
| Gene | AD path | Region_Time | Log2FC | P-value FDR |
|---|---|---|---|---|
| A2m | Aß catabolism | Hippocampus_2d | 3.48 | 2.28E-14 |
| Prefrontal cortex_2d | 3.07 | 2.10E-11 | ||
| Striatum_2d | 2.57 | 3.56E-08 | ||
| Hippocampus_7d | 2.9 | 3.98E-11 | ||
| Prefrontal cortex_7d | 2.52 | 2.43E-07 | ||
| Striatum_7d | 2.94 | |||
| Plat | Aß catabolism | Prefrontal cortex_2d | 1.2 | 1.46E-09 |
| Striatum_2d | 1.07 | 8.95E-08 | ||
| Klk6 | Aß catabolism | Hippocampus_7d | 2.37 | 5.52E-08 |
| Striatum_7d | ||||
| Ctsb | Aß catabolism | Hippocampus_7d | 1.4 | 2.50E-10 |
| Striatum_7d | 1.3 | 8.37E-09 | ||
| Ctsd | Aß catabolism | Hippocampus_7d | 2.52 | 3.03E-14 |
| Striatum_7d | 2.16 | 1.69E-10 | ||
| Apoe | GWAS hit | Hippocampus_7d | 1.35 | 4.20E-10 |
| Ms4a4a | GWAS hit | Hippocampus_2d | 1.8 | 2.93E-04 |
| Hippocampus_7d | 1.85 | 6.08E-05 | ||
| Striatum_7d | 1.61 | 6.80E-04 | ||
| Ms4a4c | GWAS hit | Hippocampus_2d | 2.56 | 3.65E-07 |
| Hippocampus_7d | 2.08 | 1.48E-05 | ||
| Striatum_7d | 2.27 | 1.89E-06 | ||
| Ms4a6c | GWAS hit | Hippocampus_2d | 1.76 | 2.86E-04 |
| Hippocampus_7d | 2.8 | 4.64E-11 | ||
| Striatum_7d | 2.32 | 8.45E-08 | ||
| Ms4a6d | GWAS hit | Hippocampus_2d | 2.53 | 1.43E-09 |
| Hippocampus_7d | 2.33 | 3.98E-09 | ||
| Striatum_7d | 1.98 | 9.37E-07 | ||
| Ms4a14 | GWAS hit | Hippocampus_7d | 2.75 | 3.36E-09 |
| Striatum_7d | 1.66 | 8.88E-04 | ||
| Ms4a4b | GWAS hit | Hippocampus_7d | 2.11 | 8.51E-06 |
| Striatum_7d | 1.72 | 4.70E-04 | ||
| Ms4a6b | GWAS hit | Hippocampus_7d | 1.8 | 3.57E-07 |
| Striatum_7d | 1.48 | 4.41E-05 | ||
| Ms4a7 | GWAS hit | Hippocampus_7d | 2.65 | 1.50E-09 |
| Striatum_7d | 1.89 | 3.89E-05 | ||
| CD33 | GWAS hit | Hippocampus_7d | 2.18 | 1.52E-22 |
| Cd68 | Microglia marker | Hippocampus_2d | 1.46 | 6.68E-04 |
| Hippocampus_7d | 2.68 | 3.16E-13 | ||
| Striatum_7d | 2.33 | 5.05E-10 | ||
| Aif1 | Microglia marker | Hippocampus_7d | 2.25 | 5.64E-17 |
| Striatum_7d | 1.57 | 1.16E-08 |
AD, Alzheimer’s disease; FDR, false discovery rate; GWAS, genome-wide association study.
LOAD GWAS loci
We report a significant upregulation (up to 7-fold change, and adj. p-value<0.05) of Apoe, Cd33, Ms4a cluster (Ms4a4a, Ms4a4c, Ms4a6c, Ms4a6d, Ms4a14, Ms4a4b, Ms4a6b, Ms4a7), particularly in the most affected brain areas during subacute hypoperfusion (hippocampus and striatum, d7) (Fig. 3). Together with APP-Aβ degradation genes (A2m, Plat, Ctsd, Ctsb and Klk6), the overexpression of Apoe, Cd33, Ms4a likely relied on microglia infiltration in the infarct and peri-infarct area at day 7 as these genes shared the same expression pattern of other microglia markers such as Aif1 and Cd86 (up to 6-fold upregulation in hippocampus and striatum), was proportional to the hippocampal lesion volume detected at day 7 on T2-weighted MRI and caused by a severe drop of brain cerebral blood flow (CBF) (≈60–70% brain CBF reduction compared to naive mice) (Fig. 4). Thus strongly arguing for consequential rather than causal upregulation of these genes and a possible role in ischemic lesion resolution. This was further supported by the concomitant significant co-expression of 7 different matrix metallo proteases (Mmp2, Mmp8, Mmp11, Mmp12, Mmp13, Mmp19, Mmp25) and 51 lysosomal genes at day 7 (Tables S5 and S6, Fig. S2C,D).
Figure 4.
(A) Cerebral blood flow (CBF) reduction detected on MRI in hippocampus during acute (2d) and subacute (7d) hypoperfusion in BCCAS mice. The CBF drop at day 1 reaches 60–70% of the CBF values detected before the surgery (d0) or in naive mice and progressively recovers.CBF, cerebral blood flow; d, day; BCCAS, bilateral common carotid artery stenosis. (B) Differential gene expression, expressed in transcript per million (TPM) in APP-Aß metabolism genes and LOAD GWAS loci detected in BCCAS mice in hippocampus during subacute hypoperfusion (7d). C Percentage of ischemic lesion volume detected on T2-MRI in hippocampus of BCCAS mice during subacute hypoperfusion. H,hippocampus; L, left; R, right; d, day.
Aβ oligomers detection during brain acute-subacute hypoperfusion in BCCAS mice
We investigated the hypothesis that acute-subacute ischemia may have triggered the de novo misfolding of APP with a consequent generation of toxic Aβ oligomers. We indeed identified Aß oligomers mostly in CA1 region in the hippocampus of BCCAS mice 7 days post-surgery (Fig. 5A,B’). These were found both in pyramidal neurons (mainly axonal processes) (Fig. 5B-B’) and particularly in reactive astrocytes (Fig. 5C,D), analogously to those detected in APPPS1 mice at the age of 2 months (Fig. 3). These were present to a significant lower degree in the hippocampus of BCCAS mice 2 days post-surgery (t-test p-val=0.0321), characterized by a very moderate microglia/macrophage infiltration as suggested by Cd68 and Aif1 expression (Table 6), a modest overall tissue remodelling and gene differential expression (Fig S2) and almost absent in the hippocampus of naive mice (t-test p-val=0.0006) (Fig. 5E). Thus supporting the hypothesis that severe subacute brain hypoperfusion (60–70% CBF reduction), which however may not cause ischemic lesions detectable on T2-weighted MRI was necessary and sufficient to prime APP misfolding (Fig. 3A,B).
Figure 5.
Toxic Aβ oligomers are detected in hypoperfused hippocampus mostly in the CA1 region during subacute hypoperfusion (7d) (B,B’) and absent in naive hippocampus (A,A’). The Aβ oligomers co-localize with reactive astrocytes (C-D). The number of glial and neuronal cells positive for Aβ oligomers are significantly higher in hippocampus during subacute hypoperfusion (7d) compared to acute hypoperfusion (2d) and naive mice (E). BCCAS, bilateral common carotid artery stenosis; d, day; +, positive.
Discussion
In this study we aimed at investigating the role of AD known pathogenic alleles and pathways: APOE ε4 allele, APP-Aβ metabolism genes and LOAD most replicated GWAS hits both in terms of genetic variability in a cohort of 96 familial and early-onset SVID patients and differential gene expression during acute and subacute hypoperfusion in the BCCAS mouse model resembling vascular dementia (Fig. 1).
In our cohort, around one third of the patients (29%) carried APOE ε4 allele. E3/ε4 and ε4/ε4 genotype frequency (27% and 1%, respectively) approximated the one reported in Caucasian controls (21.3% and 1.8%, respectively) and was significantly lower compared to the frequency reported in Caucasian LOAD patients (41.1% and 14.8%, respectively) (p-value = 0.05192 and 0.0003155 for ε3/ε4 and ε4/ε4 genotypes, respectively) (Table 2)50. Therefore suggesting, in concert with previous studies, that APOE ε4 allele may not critically influence the susceptibility to SVID51,52.
Moreover we report the prevalent role of APP-Aβ degradation genes upon genes involved in APP-Aβ production as well as Mendelian genes causative for Alzheimer’s disease (APP, PSEN1 and PSEN2). We detected an enrichment for truncating mutations in genes playing a key role in APP-Aß catabolism, both when compared to genes controlling APP-Aß production in SVID patients or APP-Aß degradation genes in 368 neuropathologically confirmed elderly controls (9.4% and 1.6%, respectively) (Fisher p-value= 3.496e-14) (Fig. 2, Table 4). In addition, we report a common polymorphism in CST3 (p.A25T), whose homozygous carrier frequency was significantly higher compared to HEX controls (4.16% and 1.63%, respectively). Interestingly, this polymorphism has been already associated to macular degeneration and LOAD40. The presence of one or two minor alleles increases LOAD risk and lowered the age at onset, in a fashion described both independent and dependent from APOE ε4 allele39,53. Although the SVID patients carrying in homozygosity p.CST3 p.A25T displayed at the age at onset and diagnosis only a moderately lower MMSE score (MMSE 28 and 26), given the young age at onset (average 49.75years), we do not exclude that they may manifest a dementing phenotype later in life (average age at onset for the LOAD patients homozygous for CST3 p.A25T was> 75 years39,54) (Table S3). The polymorphism is supposed to influence CST3 intracellular processing with a reduced extracellular secretion55,56, leading to increased amyloid fibril formation and Aß deposition57. A similar effect to what has been described for the pathogenic mutation CST3 p.L68Q, causative for hereditary cerebral haemorrhage with amyloidosis, Icelandic type (HCHWA-I), resulting in increased intracellular localization of the mutant Cystatin C58.
Furthermore, we detected 2 rare coding variants (p.E270K and p.A528T) in the SORL1 VPS10 domain, reported to interact with Aβ and harbouring SORL1 pathogenic mutations59 (Table S1). The carrier frequency of SORL1 variants in the VPS10 domain detected in our SVID cohort was similar to the frequency detected in a Caucasian British and American LOAD cohort, where 8 variants in 323 patients have been identified (2% and 2.4%, carrier frequency in SVID and LOAD, respectively) whereas only 5 SORL1 variants in the VPS10 domain have been reported in 676 elderly controls in the same cohort (0.7%)24. This suggests that SORL1 mutations may influence the susceptibility also for SVID and may support the previously reported role of SORL1 in vascular dementia60.
By contrast, none of the 5 rare coding variants detected in APP, PSEN1 and PSEN2 are likely to be risk factors for SVID (Table 3). First, they have already been described as benign polymorphisms (PSEN1 [p.E318G] and PSEN2 [p.R62H]) (www.molgendatabase). Second, they all cluster outside the reported pathogenic domains (APP [p.V576I and p.T280del], PSEN1 [p.E318G] and PSEN2 [p.L2F and p.R62H]). This further shows that rare variants in APP, PSEN1 and PSEN2 are not common pathogenic factors in familial and early-onset apparently sporadic SVID cases. In line with this observation, 11/50 (22%) pathogenic mutations in APP have been reported as causative for AD and CAA, and only 2/50 (4%) lead exclusively to CAA. A smaller fraction of PSEN1 pathogenic mutations (4/219 [1.8%]) has been described as causative for both AD and CAA and none exclusively for CAA. On the other hand, PSEN2 harbours no causative mutation for CAA (www.molgendatabase).
The predominant role of APP-Aβ degradation genes was further confirmed by RNA sequencing data in a mouse model of mainly subcortical ischemia, mimicking small vessel ischemic disease, where only genes belonging to the APP-Aβ degradation path (A2m, Plat, Ctsd, Ctsb and Klk6) were significantly overexpressed in hippocampus and striatum during acute and subacute hypoperfusion (Table 6). Among these, KLK6 expression has been already reported restricted to endothelial cells and increased of approximately 2-fold in the frontal cortex of patients with vascular dementia61.
Importantly, we showed that genetic and gene expression variability in LOAD GWAS genes are also likely to influence the susceptibility to SVID and acute-subacute ischemia.
We reported 3 common coding polymorphisms significantly associated to SVID and likely to play a mild protective role (adj. p-value <0.05 and 0.325<OR < 0.54): EPHA1 p.M900V and p.V160A, CD33 p.A14V (Table 5). Among these, CD33 rs12459419-T has been reported to be in high LD with LOAD GWAS hit rs3865444-A and was suggested to explain its effect, such as the alternative splicing of CD33 with increased production of isoforms lacking exon 2, which encodes the IgV domain that typically mediates binding of sialic acid in SIGLEC family members. This CD33 isoform counteracts the inhibitory effect of CD33 on TREM2 in microglia and would ultimately reduce amyloid deposition and thus exert a moderate protective effect on Alzheimer’s disease and likely SVID susceptibility46. In addition, the critical role of this polymorphism (CD33 rs12459419) was further reinforced by its significant association, together with EPHA1 p.V160A, with blood protein levels, and haematological traits in two different GWASs42,43.
Moreover, Apoe, Cd33 and the Ms4a cluster were significantly upregulated in hippocampus and striatum particularly during subacute hypoperfusion (up to 7-fold change at day 7)(Table 6). Together with APP-Aβ degradation genes, overexpression of Apoe, Cd33 and Ms4a cluster correlated with hippocampal lesion size at day 7 (Fig. 4B,C) and likely microglia infiltration of the infarct and peri-infarct areas (co-expression of microglia markers Cd68 and Aif1), suggesting that upregulation of these genes was tightly driven by and consequential to the severity of hypoperfusion, moreover, arguing for an active role of these genes in tissue remodelling and ischemic lesion resolution. This is further supported by the fact that significant overexpression of Cd33, Ms4a6d and Apoe (> 1.5 fold change and adj p-value <0.05) markedly correlated with AD pathology in 2 mouse models of Alzheimer’s disease characterized by severe Aβ plaques and tau tangle deposition (HOTASTPM and TAU mice, 18 months of age)62. Therefore implying that Cd33, Ms4a cluster and Apoe are not Aβ or tau specific.
Finally, the shared pathogenic pathway between LOAD-SVID-ischemic stroke was supported by histological findings of neurons and reactive astrocytes positive for Aβ oligomers in the main hypoperfused areas such as hippocampus at day 7 in the BCCAS mouse model (Fig. 5).
Previous studies in stroke experimental models (middle cerebral artery occlusion [MCAO]) reported Aβ in reactive astrocytes and neurons and Aβ plaque-like deposits in peri-infarct areas: particularly corpus callosum, CA1 hippocampal areas and mainly subcortical areas 7 days post-surgery63,64. The authors hypothesized that Aβ may have been the result of APP overexpression during ischemic stress65. Importantly, we show that acutely-subacutely hypoerfused brain areas and particularly reactive astrocytes and hippocampal neurons are positive for Aβ oligomers rather than β amyloid, that it is likely to represent a late and chronic event in the APP misfolding cascade. Moreover, we detected Aβ oligomers in the hypoperfused brain regions, displaying gliosis but not necessarily gray or white matter hyperintensities detectable on T2-weighted MRI (Fig. 5, Fig. 3A,B). Thus suggesting that a marked degree of hypoperfusion-ischemia that may remain however below the T2-weighted MRI detectability and does not lead to strokes, may trigger APP misfolding and may explain the link between brain microstructural changes detected on diffusion tensor imaging (DTI) and likely hypoxic-ischemic hyperintensities in white matter, detected decades before the onset of symptomps and autosomal dominant AD cases66,67 as well as common late-onset sporadic cases and elderly people68.
Therefore, our data may unveil at least some of the pathogenic mechanisms by which ischemic stroke may precipitate the progression of AD in experimental models and patients and why cerebrovascular accidents may accelerate AD onset particularly in asymptomatic elderly patients with AD pathology69. Indeed, the enrichment for genetic variability in APP-Aβ degradation genes has been reported playing a key role in sporadic late-onset AD whereas increases in Aβ production currently explain a minority of AD cases37,70.
In summary, we provide genetic, gene-expression and histological data supporting a shared pathogenic ground between LOAD and SVID-acute ischemia. Our genetic data in SVID patients, together with expression data in a vascular dementia mouse model show that 1) APOE transcriptional regulation but not ε4 allele may play a role in brain hypoperfusion and small vessel ischemic disease; 2) APP-Aβ degradation plays a prevalent role upon APP-Aβ production; 3) APP, PSEN1 and PSEN2 are not common pathogenic factors in SVID; 4) CD33, CR1, EPHA1 and the MS4A cluster may be involved in SVID and brain subacute hypoperfusion-ischemia and 4) acute and mainly subacute ischemia may trigger Aβ toxic oligomer formation. Thus suggesting that the vascular hypothesis71 and the amyloid cascade hypothesis72 in AD may complement each other, rather than being mutually exclusive. Our findings warrant further genetic screening in a larger cohort and functional studies.
Supplementary information
Acknowledgements
DNA samples from the NINDS Repository were used in this study, as well as clinical data. NeuroCure, Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Alexander von Humboldt Fellowship (to Celeste Sassi), German Research Foundation (DFG) and the Open Access Publication Funds of Charité – Universitätsmedizin Berlin.
Author contributions
S.B., M.F., C.S. and U.D. planned the experiments. S.B., M.F., C.S., D.B., S.M. performed the experiments. S.B., M.F., S.M., M.H., K.B., D.B., D.B., U.D. and C.S. analysed the data and drafted the manuscript.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sonja Blumenau and Marco Foddis.
Supplementary information
is available for this paper at 10.1038/s41598-020-63183-5.
References
- 1.Lim A, et al. Clinico-neuropathological correlation of Alzheimer’s disease in a community-based case series. J Am Geriatr Soc. 1999;47:564–569. doi: 10.1111/j.1532-5415.1999.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 2.Langa KM, Foster NL, Larson EB. Mixed dementia: emerging concepts and therapeutic implications. JAMA. 2004;292:2901–2908. doi: 10.1001/jama.292.23.2901. [DOI] [PubMed] [Google Scholar]
- 3.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 4.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulouis G, et al. Small vessel disease burden in cerebral amyloid angiopathy without symptomatic hemorrhage. Neurology. 2017;88:878–884. doi: 10.1212/WNL.0000000000003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol. 2010;9:689–701. doi: 10.1016/S1474-4422(10)70104-6. [DOI] [PubMed] [Google Scholar]
- 8.Reijmer YD, van Veluw SJ, Greenberg SM. Ischemic brain injury in cerebral amyloid angiopathy. J Cereb Blood Flow Metab. 2016;36:40–54. doi: 10.1038/jcbfm.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter S, Brayne C. Understanding the roles of mutations in the amyloid precursor protein in Alzheimer disease. Mol. Psychiatry. 2018;23:81–93. doi: 10.1038/mp.2017.218. [DOI] [PubMed] [Google Scholar]
- 10.Abrahamson M, Grubb A. Increased body temperature accelerates aggregation of the Leu-68–>Gln mutant cystatin C, the amyloid-forming protein in hereditary cystatin C amyloid angiopathy. Proc. Natl. Acad. Sci. USA. 1994;91:1416–1420. doi: 10.1073/pnas.91.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garzuly F, Vidal R, Wisniewski T, Brittig F, Budka H. Familial meningocerebrovascular amyloidosis, Hungarian type, with mutant transthyretin (TTR Asp18Gly) Neurology. 1996;47:1562–1567. doi: 10.1212/WNL.47.6.1562. [DOI] [PubMed] [Google Scholar]
- 12.Abraham HMA, et al. Cardiovascular risk factors and small vessel disease of the brain: Blood pressure, white matter lesions, and functional decline in older persons. J. Cereb. Blood Flow Metab. 2016;36:132–142. doi: 10.1038/jcbfm.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anttila V, et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat. Genet. 2013;45:912–917. doi: 10.1038/ng.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugiani O, et al. Hereditary cerebral hemorrhage with amyloidosis associated with the E693K mutation of APP. Arch. Neurol. 2010;67:987–995. doi: 10.1001/archneurol.2010.178. [DOI] [PubMed] [Google Scholar]
- 15.Levy E, et al. Mutation of the Alzheimer’s disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 16.Rossi G, et al. A family with Alzheimer disease and strokes associated with A713T mutation of the APP gene. Neurology. 2004;63:910–912. doi: 10.1212/01.WNL.0000137048.80666.86. [DOI] [PubMed] [Google Scholar]
- 17.Guerreiro RJ, et al. Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer’s disease. Neurobiol. Aging. 2012;33:1008.e17–23. doi: 10.1016/j.neurobiolaging.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinberg S, et al. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat. Genet. 2015;47:445–447. doi: 10.1038/ng.3246. [DOI] [PubMed] [Google Scholar]
- 19.Sassi, C. et al. ABCA7 p.G215S as potential protective factor for Alzheimer’s disease. Neurobiol. Aging, 10.1016/j.neurobiolaging.2016.04.004 (2016). [DOI] [PMC free article] [PubMed]
- 20.May P, et al. Rare ABCA7 variants in 2 German families with Alzheimer disease. Neurol Genet. 2018;4:e224. doi: 10.1212/NXG.0000000000000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keenan BT, et al. A coding variant in CR1 interacts with APOE-ε4 to influence cognitive decline. Hum. Mol. Genet. 2012;21:2377–2388. doi: 10.1093/hmg/dds054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501:45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mok VCT, et al. Early-onset and delayed-onset poststroke dementia - revisiting the mechanisms. Nat Rev Neurol. 2017;13:148–159. doi: 10.1038/nrneurol.2017.16. [DOI] [PubMed] [Google Scholar]
- 24.Sassi C, et al. Influence of Coding Variability in APP-Aβ Metabolism Genes in Sporadic Alzheimer’s Disease. PLoS ONE. 2016;11:e0150079. doi: 10.1371/journal.pone.0150079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leeuw FE, et al. Atrial fibrillation and the risk of cerebral white matter lesions. Neurology. 2000;54:1795–1801. doi: 10.1212/WNL.54.9.1795. [DOI] [PubMed] [Google Scholar]
- 26.Guerreiro, R. et al. A comprehensive assessment of benign genetic variability for neurodegenerative disorders. bioRxiv 270686, 10.1101/270686 (2018).
- 27.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio] (2013).
- 28.Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäger M, et al. Jannovar: a java library for exome annotation. Hum. Mutat. 2014;35:548–555. doi: 10.1002/humu.22531. [DOI] [PubMed] [Google Scholar]
- 31.Foddis, M. et al. An exploratory investigation of brain collateral circulation plasticity after cerebral ischemia in two experimental C57BL/6 mouse models. J Cereb Blood Flow Metab 0271678X19827251, 10.1177/0271678X19827251 (2019). [DOI] [PMC free article] [PubMed]
- 32.Nishio K, et al. A mouse model characterizing features of vascular dementia with hippocampal atrophy. Stroke. 2010;41:1278–1284. doi: 10.1161/STROKEAHA.110.581686. [DOI] [PubMed] [Google Scholar]
- 33.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selkoe DJ. Clearing the brain’s amyloid cobwebs. Neuron. 2001;32:177–180. doi: 10.1016/S0896-6273(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 38.Hardy J, Crook R. Presenilin mutations line up along transmembrane alpha-helices. Neurosci. Lett. 2001;306:203–205. doi: 10.1016/S0304-3940(01)01910-3. [DOI] [PubMed] [Google Scholar]
- 39.Finckh U, et al. Genetic association of a cystatin C gene polymorphism with late-onset alzheimer disease. Arch. Neurol. 2000;57:1579–1583. doi: 10.1001/archneur.57.11.1579. [DOI] [PubMed] [Google Scholar]
- 40.Butler JM, et al. A missense variant in CST3 exerts a recessive effect on susceptibility to age-related macular degeneration resembling its association with Alzheimer’s disease. Hum. Genet. 2015;134:705–715. doi: 10.1007/s00439-015-1552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vardarajan BN, et al. Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 2015;77:215–227. doi: 10.1002/ana.24305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrasquillo MM, et al. Late-onset alzheimer’s risk variants in memory decline, incident mild cognitive impairment, and Alzheimer’s disease. Neurobiol. Aging. 2015;36:60–67. doi: 10.1016/j.neurobiolaging.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik M, et al. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raj T, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum. Mol. Genet. 2014;23:2729–2736. doi: 10.1093/hmg/ddt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birmingham DJ, et al. A CR1 polymorphism associated with constitutive erythrocyte CR1 levels affects binding to C4b but not C3b. Immunology. 2003;108:531–538. doi: 10.1046/j.1365-2567.2003.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naj AC, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset alzheimer’s disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farrer LA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 51.Groot C, et al. Clinical phenotype, atrophy, and small vessel disease in APOEε2 carriers with Alzheimer disease. Neurology. 2018;91:e1851–e1859. doi: 10.1212/WNL.0000000000006503. [DOI] [PubMed] [Google Scholar]
- 52.Gesierich B, et al. APOE ɛ2 is associated with white matter hyperintensity volume in CADASIL. J. Cereb. Blood Flow Metab. 2016;36:199–203. doi: 10.1038/jcbfm.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cathcart HM, Huang R, Lanham IS, Corder EH, Poduslo SE. Cystatin C as a risk factor for Alzheimer disease. Neurology. 2005;64:755–757. doi: 10.1212/01.WNL.0000151980.42337.44. [DOI] [PubMed] [Google Scholar]
- 54.Lin C, Wang ST, Wu CW, Chuo LJ, Kuo YM. The association of a cystatin C gene polymorphism with late-onset alzheimer’s disease and vascular dementia. Chin J Physiol. 2003;46:111–115. [PubMed] [Google Scholar]
- 55.Benussi L, et al. Alzheimer disease-associated cystatin C variant undergoes impaired secretion. Neurobiol. Dis. 2003;13:15–21. doi: 10.1016/S0969-9961(03)00012-3. [DOI] [PubMed] [Google Scholar]
- 56.Paraoan L, et al. Unexpected intracellular localization of the AMD-associated cystatin C variant. Traffic. 2004;5:884–895. doi: 10.1111/j.1600-0854.2004.00230.x. [DOI] [PubMed] [Google Scholar]
- 57.Kaeser SA, et al. Cystatin C modulates cerebral beta-amyloidosis. Nat. Genet. 2007;39:1437–1439. doi: 10.1038/ng.2007.23. [DOI] [PubMed] [Google Scholar]
- 58.Bjarnadottir M, et al. Intracellular accumulation of the amyloidogenic L68Q variant of human cystatin C in NIH/3T3 cells. MP. Mol. Pathol. 1998;51:317–326. doi: 10.1136/mp.51.6.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pottier C, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset alzheimer disease. Mol. Psychiatry. 2012;17:875–879. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- 60.Cuenco T. K. et al. Association of distinct variants in SORL1 with cerebrovascular and neurodegenerative changes related to Alzheimer disease. Arch. Neurol. 2008;65:1640–1648. doi: 10.1001/archneur.65.12.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashby EL, Kehoe PG, Love S. Kallikrein-related peptidase 6 in Alzheimer’s disease and vascular dementia. Brain Res. 2010;1363:1–10. doi: 10.1016/j.brainres.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 62.Matarin M, et al. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell Rep. 2015;10:633–644. doi: 10.1016/j.celrep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 63.Nihashi T, et al. Expression and distribution of beta amyloid precursor protein and beta amyloid peptide in reactive astrocytes after transient middle cerebral artery occlusion. Acta Neurochir (Wien) 2001;143:287–295. doi: 10.1007/s007010170109. [DOI] [PubMed] [Google Scholar]
- 64.Yokota M, Saido TC, Tani E, Yamaura I, Minami N. Cytotoxic fragment of amyloid precursor protein accumulates in hippocampus after global forebrain ischemia. J. Cereb. Blood Flow Metab. 1996;16:1219–1223. doi: 10.1097/00004647-199611000-00016. [DOI] [PubMed] [Google Scholar]
- 65.Banati RB, Gehrmann J, Wiessner C, Hossmann KA, Kreutzberg GW. Glial expression of the beta-amyloid precursor protein (APP) in global ischemia. J. Cereb. Blood Flow Metab. 1995;15:647–654. doi: 10.1038/jcbfm.1995.80. [DOI] [PubMed] [Google Scholar]
- 66.Lee S, et al. White matter hyperintensities are a core feature of Alzheimer’s disease: Evidence from the dominantly inherited Alzheimer network. Ann. Neurol. 2016;79:929–939. doi: 10.1002/ana.24647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Araque Caballero MÁ, et al. White matter diffusion alterations precede symptom onset in autosomal dominant Alzheimer’s disease. Brain. 2018;141:3065–3080. doi: 10.1093/brain/awy229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rabin JS, et al. Interactive Associations of Vascular Risk and β-Amyloid Burden With Cognitive Decline in Clinically Normal Elderly Individuals: Findings From the Harvard Aging Brain Study. JAMA Neurol. 2018;75:1124–1131. doi: 10.1001/jamaneurol.2018.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Snowdon DA, et al. Brain infarction and the clinical expression of Alzheimer disease. The Nun Study. JAMA. 1997;277:813–817. doi: 10.1001/jama.1997.03540340047031. [DOI] [PubMed] [Google Scholar]
- 70.Miners JS, et al. Abeta-degrading enzymes in Alzheimer’s disease. Brain Pathol. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat. Rev. Neurosci. 2011;12:723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).