Abstract
Duplication plays an important role in creating drastic changes in genome evolution. In addition to well-known tandem duplication, duplication can occur such that a duplicated DNA fragment is inserted at another location in the genome. Here, we report several genomic regions in the human genome that could be best explained by two types of insertion-based duplication mechanisms, where a duplicated DNA fragment was modified structurally and then inserted into the genome. In one process, the DNA fragment is turned into an extrachromosomal circular DNA, cut somewhere in the circle, and reintegrated into another location in the genome. And in the other, the DNA fragment forms a “lariat structure” with a “knot”, the strand is swapped at the knot, and is then reintegrated into the genome. Our results suggest that insertion-based duplication may not be a simple process; it may involve a complicated procedures such as structural modification before reintegration. However, the molecular mechanism has yet to be fully understood.
Subject terms: Evolutionary genetics, Mimicry
Introduction
Various types of mutations can accumulate during genome evolution. Among these, duplication is thought to play an important role in providing drastic structural changes to the genome1–3. Segmental duplication accounts for up to 6% of the human genome4. Tandem duplication is the most well-known type of duplication and is generally initiated by non-allelic homologous recombination or by non-homologous end joining and replication-based mechanisms5,6. In addition, duplication can occur non-tandemly, that is, a duplicated region arises elsewhere in the genome. For this type of duplication, one may imagine that a duplicated fragment is inserted into an independent location some distance away from the corresponding segment, although the molecular mechanism not fully understood (see6–8).
Here, we report several interesting cases of insertion-based duplication in the human genome, where a duplicated DNA fragment is modified structurally, and then inserted into the genome. It has been reported that a DNA fragment can turn into extrachromosomal circular DNA (eccDNA) and can become reintegrated into the genome9–11. The existence of eccDNA in cancer cells (but not integrated in the nuclear genome) has been known for more than a half century12,13, but its presence in normal somatic cells has only been discovered recently14. It has also been reported that eccDNA is reintegrated back to the genome in cancer cells9, indicating that the reintegration of eccDNA may also occur in germ-line cells. Two clear demonstrations were reported in cattle10 and yeast11, and suggestive evidence was provided in human15. The findings in these cases provide fairly strong evidence for theories postulating the likelihood of eccDNA-mediated duplication in the human genome.
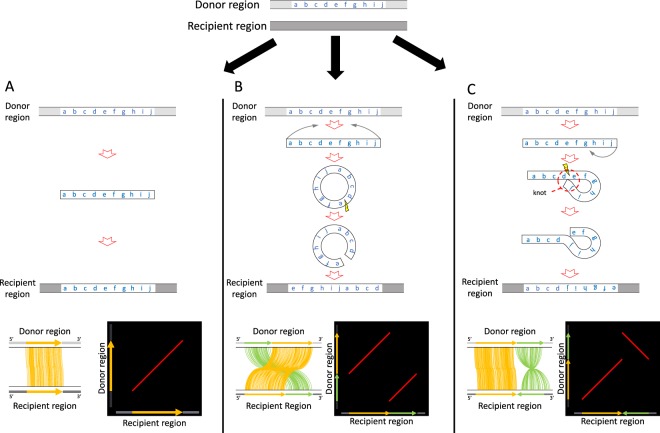
It is quite straightforward to detect eccDNA-mediated duplications in a genome, considering the process illustrated in Fig. 1B. Suppose that the region from markers a to j (donor region) is copied and turned into an eccDNA, cut somewhere in the circle (i.e., between markers d and e), and reintegrated into another independent location in the genome (recipient region). We could then observe a duplicated region in the syntenic order efghijabcd in the recipient region, which can be easily distinguished from the standard direct duplication that can be recognized as abcdefghij in the recipient region (Fig. 1A).
Figure 1.
Illustration of eccDNA- and eltDNA-mediated duplications, compared to normal duplication with no modification involved. (A) Normal non-tandem duplication through insertion. The region from makers a to j is copied and inserted back to the recipient region in the genome, so that the paralogous regions keep the synteny as shown in the alignment, and a straight line arises in the dot plot of identity. (B) eccDNA-mediated duplication. The region from markers a to j is copied, turned into an eccDNA, and inserted back to the recipient region in the genome. Two parallel lines arise in the dot plot. (C) eltDNA-mediated duplication. The region from markers a to j is copied, turned into an eltDNA, the strand is swapped at the knot, and inserted back to the recipient region in the genome. Two orthogonal lines arise in the dot plot.
Furthermore, we report another type of duplication that may also involve structural modification before reintegration. We hypothesize that a DNA fragment could form a “lariat structure” (referred to as an extrachromosomal lariat DNA, or eltDNA), in which one end of the fragment is attached to the middle of itself (the attached point is referred to as a knot). Then, the strands are swapped at the knot, and it is reintegrate into the genome (see Fig. 1C). Suppose a knot was formed between markers d and e; we could observe a duplicated region in the syntenic order abcdefghij. We found a number of regions with strong evidence for eltDNA-mediated duplications in the human genome.
It seems that none of these observed duplications created new duplicated copies of coding genes and therefore may not directly contribute to adaptive genome evolution. However, it would be important to recognize that these kinds of structural modification processes may be involved when a region is duplicated. Such a change could potentially provide a selective advantage or disadvantage, for example, through expressional and/or epigenetical changes.
Methods
We searched for eccDNA- and eltDNA-mediated duplications in the human genome. Segmental duplications (SDs) in the human genome were first comprehensively identified by Eichler and colleagues in GRCh3516. The authors defined SDs as duplicated regions with length >1 kb with nucleotide identity >90%.
Pu et al.15 updated the data of She et al.16 with the latest version, GRCh38, by developing the software SDquest. They identified 14,467 SDs (in total 198.3 Mb), which cover 95% (158.2 Mb) of the previously identified SDs by She et al.16 The sequence “homology” for most of them is >90%, but the data includes some SDs with 70–90% homology (see Table 1 in Pu et al.15), which seems lower compared to the results of She et al.16. The inconsistency could be due to the definition of homology; Pu et al.’s15 homology evaluates both nucleotide substitutions and indels. Throughout this article, we follow Pu et al.’s15 definition of homology, except when we construct NJ trees based on nucleotide substitutions.
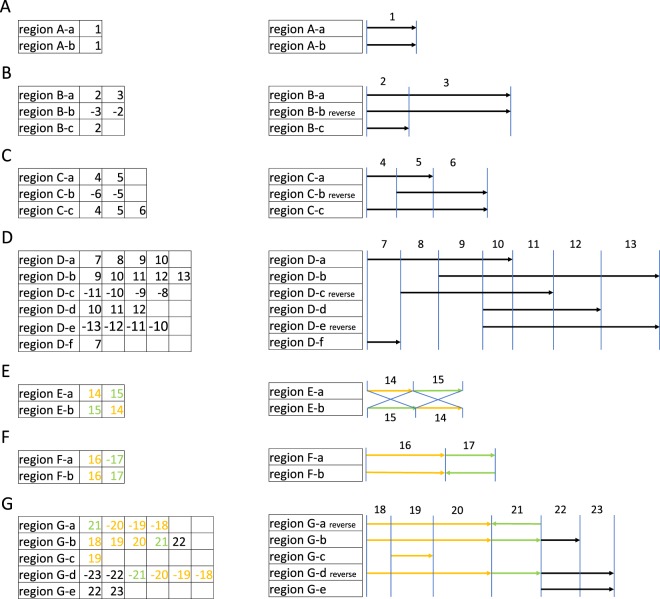
Figure 2 explains the structure of Pu et al.’s15 data in the MosaicSDs_Human_hg38.txt file, which can be downloaded at https://github.com/SDquest/SDquest. Figure 2A describes the simplest type of SD with a pair of paralogous regions. An index number is given to the entire duplicated region (1 in this hypothetical example). A slightly complicated case, as illustrated in Fig. 2B, involves three regions (labeled B-a, -b, -c), two of which (B-a and -b) have homology in an extended region. In such a case, the entire region would be divided into two subregions named elementary SDs (eSDs) with an index number given to each eSD (2 and 3 in Fig. 2B). The data can be tabulated as shown to the left of Fig. 2B. If homology is observed in the reverse strand, the index number is given as a negative value in the table (e.g., B-b in Fig. 2B, C-b in Fig. 2C). Figure 2D illustrates an example of complicated cases with many eSDs.
Figure 2.
Grouping of Pu et al.’s15 eSD data. Examples are shown with hypothetical eSDs #1 to #23. See text for details.
The data in MosaicSDs_Human_hg38.txt was grouped and tabulated, as illustrated in Fig. 2. By using the grouped eSD data, it was quite straightforward to search for eccDNA- and eltDNA-mediated duplications because they exhibit unique arrays of index numbers in the table. As shown in Fig. 2E, an eccDNA-mediated duplication should be detected as a pair of regions where the syntenic order of index numbers is different but the direction is the same (i.e., #14#15 vs. #15#14). An eltDNA-mediated duplication should be detected as a pair of regions with identical syntenic order of index numbers in the same direction in part, but in the opposite direction in the other (i.e., #16#17 vs. #16 #17) as show in the example of Fig. 2F. Figure 2G shows a complicated case that involves an eltDNA-mediated duplication (G-a) created from G-b. In addition, there are three regions (i.e., G-c, -d, and -e) exhibiting homology. In the following, when we find such a case, the full length regions of detected eltDNA- or eccDNA-mediated duplication will be shown (G-a, -b, and -d in the example of Fig. 2G, see below).
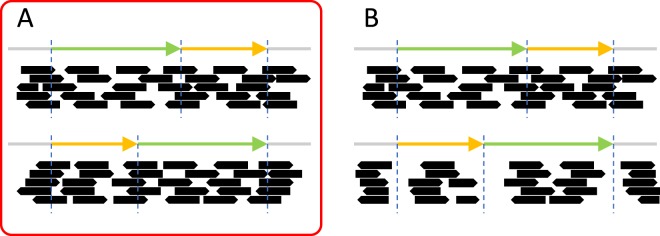
In order to confirm the presence of the detected eltDNA- or eccDNA-mediated duplications, we used NGS sequence data from the 1000 Genomes Project17. We arbitrarily chose 19 individuals representing the 19 “populations” defined in IGSR, The International Genome Sample Resource (see https://www.internationalgenome.org/faq/which-populations-are-part-your-study/). For these individual samples (summarized in Table S1), we downloaded high coverage whole genome sequence data from ftp://ftp.1000genomes.ebi.ac.uk, which were already mapped GRCh38 (hg38). For a structurally modified duplication, we predicted that both of the duplicated regions should be supported by the short-read data as illustrated in Fig. 3A (illustration for an eccDNA-mediated duplication), if they are really present in the genome. Alternatively, if it is an artifact, we should not be able to find reads supporting the breakpoints, as illustrated in Fig. 3B. As a result, we confirmed the presence of all detected eccDNA- and eltDNA-mediated duplications in all 19 individuals, indicating that they were not artifacts due to erroneous assembly of the reference genome.
Figure 3.
Illustration of predicted patterns of mapped NSG short-read data (A) when a detected eccDNA-mediated duplication is supported and (B) when not supported.
For all identified eccDNA- and eltDNA-mediated duplications in the human genome, we checked whether their orthologous regions are present in chimpanzee and macaque genomes (panTro3 and rheMac8). For a first choice of outgroup species, we chose chimpanzee as the closest relative to us, which has quite a reliable reference genome. We did not use gorilla because the evolutionary distance from human is similar that for chimpanzee and the quality is not as good. Macaque and Orangutan were among our candidates for a second outgroup species, and we chose macaque because the quality of the macaque reference genome is much better than the orangutan one. By using to the synteny information of flanking coding regions in VGNG (https://vertebrate.genenames.org/), we narrowed down the orthologous regions, which were subject to detailed sequence-based analysis with BLAST 2.9.018.
The codes used for our analyses are available at https://github.com/Kazuki526/circular_lariat.
Results
We searched for eccDNA- and eltDNA-mediated duplications using data from Pu et al.15, following the strategy illustrated in Fig. 2. The data consisted of 15,312 regions with 14,467 distinct eSDs. We first grouped them such that all regions in the same group shared at least one eSDs (see Fig. 2). We obtained 3,924 of such groups (mean 3.9, median 2, standard deviation (SD) 62.15) The majority of the groups (99.3%) had <20 regions, with maximum 89, if one obvious outlier (with 3890 regions) was excluded.
For detecting eccDNA- and eltDNA-mediated duplications, we found, respectively, 52 and 77 regions that were consistent with the patterns illustrated in Fig. 2E,F. However, we found that these regions included a number of false positives, which were easily excluded by looking at dot plots of surrounding regions.
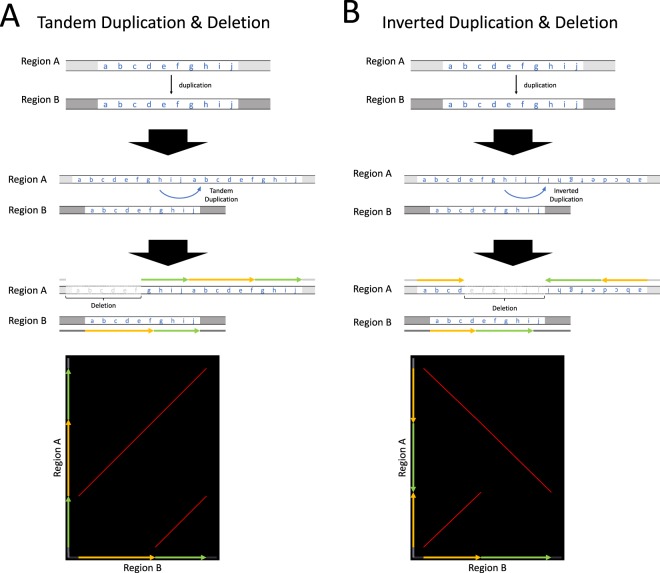
Typical examples of false positives are illustrated in Fig. 4. Figure 4A involves duplication events twice followed by a partial deletion. Region A (abcdefghij) is first duplicated into region B, and then region A undergoes a tandem duplication again, resulting in (abcdefghijabcdefghij). Next, a part of the tandemly duplicated region (abcdef) is deleted. Then, if we compare a part of region A (ghijabcdef) and region B (abcdefghij), the pattern is completely consistent with that expected by an eccDNA-mediated duplication (see Fig. 1B), that is, the order of the yellow and green regions (abcdef and ghij, respectively) is swapped. However, this pattern can be recognized as a false positive if we find that there are two green regions flanking the yellow region. Similarly, Fig. 4B shows a false positive pattern of an eltDNA-mediated duplication arising from head-to-head duplication, followed by duplication to an independent genomic location and a partial deletion. In all cases, if we look at a partial region, the pattern seems to be consistent with eccDNA- or eltDNA-mediated duplication, but not in the whole region. Although these patterns do not necessarily rule out the possibility of eccDNA- or eltDNA-mediated duplication, to be conservative, we excluded these cases. Then, we finally identified 3 and 20 fairly strong candidates for eccDNA- and eltDNA-mediated duplications (Table S2).
Figure 4.
Typical examples of false positive signatures for (A) eccDNA- and (B) eltDNA-mediated duplications. (A) A tandem duplication in the donor region, a duplication to the recipient region, followed by a partial deletion in the donor region, could create a similar pattern to that predicted by an eccDNA-mediated duplication in the dot plot. (B) A head-to-head duplication in the donor region, a duplication to the recipient region, followed by a partial deletion in the donor region, could create a similar pattern to that predicted by eltDNA-mediated duplication in the dot plot. See text for details.
We checked whether these detected regions were present in the whole genome sequence data in 19 human individuals in the 1000 Genomes Project (see Method). We confirmed that all of them were present, by mapping next-generation short-read data with special attention to the regions encompassing the breakpoints that specify the synteny of eSDs involved in eccDNA- and eltDNA-mediated duplications. It is indicated that all detected eccDNA- and eltDNA-mediated duplications are not due to erroneous assembly of the reference genome.
We also used the chimpanzee and macaque genomes to investigated whether their orthologous regions were present (see Method). For each region of the identified eccDNA- and eltDNA-mediated duplications, presence/absence was examined in its 5′ and 3′ regions and in the orthologous part in the chimpanzee and macaque genomes. The result is summarized in Table 1, where , ×, and Δ represent the presence, absence and partial presence of the region, respectively. For a pair of duplicated regions, the donor/recipient copies can be determined when we observe the pattern (, , )/(, ×, ), where the three symbols in each pair of parenthesis exhibit the presence/absence of the 5′ flanking, the focal duplicated region, and the 3′ flanking regions (see Table 1 for details).
Table 1.
Summary of coding overlap in the human genome, and presence/absence of the detected eccDNA- and eltDNA-mediated duplications in the chimpanzee and macaque genomes.
| Region | Overlap with coding genes in the human genome | Presence/absence in Chimpanzee | Presence/absence in Macaque | ||||||
|---|---|---|---|---|---|---|---|---|---|
| C1a | — | — | — | ||||||
| C1b | — | — | — | Δ | |||||
| C2a | — | TCAF1 | TCAF1 | ||||||
| C2b | — | TCAF2, TCAF2C | — | Δ | |||||
| C3a | — | — | — | ||||||
| C3b | — | — | — | ||||||
| C3c | — | — | — | ||||||
| C3d | — | — | — | ||||||
| C3e | — | — | — | ||||||
| C3f | — | — | — | ||||||
| C3g | — | — | — | ||||||
| C3h | — | — | — | ||||||
| C3i | — | — | — | ||||||
| C3j | — | — | — | ||||||
| C3k | — | — | — | ||||||
| C3l | — | — | — | ||||||
| C3m | — | — | — | ||||||
| C3n | — | — | — | ||||||
| C3o | PCMTD2 | — | — | Δ | |||||
| L1a | — | — | — | ||||||
| L1b | ATM | ATM | ATM | ||||||
| L2a | — | — | — | ||||||
| L2b | — | — | — | ||||||
| L2c | — | — | — | Δ | Δ | ||||
| L2d | — | — | — | ||||||
| L3a | — | — | — | ||||||
| L3b | — | — | — | ||||||
| L3c | — | — | — | ||||||
| L4a | — | — | — | Δ | |||||
| L4b | — | — | UGT2A1,UGT2A2 | ||||||
| L4c | ANKRD6, LYRM2 | ANKRD6, LYRM2 | ANKRD6, LYRM2, MDN1 | ||||||
| L5a | — | — | — | ||||||
| L5b | — | — | — | ||||||
| L6a | — | — | — | ||||||
| L6b | — | — | — | ||||||
| L6c | ARHGAP5 | ARHGAP5 | ARHGAP5 | ||||||
| L7a | — | — | — | ||||||
| L7b | — | — | — | Δ | |||||
| L7c | ERCC6 | ERCC6 | ERCC6 | ||||||
| L7d | — | — | — | Δ | |||||
| L8a | — | — | — | ||||||
| L8b | — | — | — | ||||||
| L8c | DNM1L | — | DNM1L, YARS2 | ||||||
| L8d | PHLDA1 | NAP1L1 | NAP1L1 | ||||||
| L9a | — | — | — | ||||||
| L9b | THSD1 | VPS36 | VPS36 | ||||||
| L10a | — | — | — | ||||||
| L10b | — | — | — | ||||||
| L11a | — | — | — | ||||||
| L11b | — | — | — | Δ | |||||
| L12a | — | — | — | ||||||
| L12b | HAUS2 | — | STARD9 | ||||||
| L13a | — | — | — | Δ | |||||
| L13b | ZDHHC20 | ZDHHC20 | ZDHHC20 | Δ | |||||
| L13c | — | — | — | ||||||
| L14a | — | — | — | ||||||
| L14b | MINDY2 | MINDY2 | MINDY2 | ||||||
| L15a | — | — | — | ||||||
| L15b | — | — | — | ||||||
| L15c | — | — | — | ||||||
| L16a | — | — | — | ||||||
| L16b | — | — | — | ||||||
| L17a | PRRC2C | PRRC2C | MYOCOS | Δ | |||||
| L17b | — | — | — | ||||||
| L17c | — | — | — | ||||||
| L18a | — | — | PDS5A | ||||||
| L18b | — | — | — | ||||||
| L18c | PABPC3 | PABPC3 | PABPC3 | ||||||
| L19a | STK31 | — | STK31 | ||||||
| L19b | PXDNL,PCMTD1 | PCMTD1 | — | ||||||
| L19c | — | — | — | ||||||
| L19d | — | — | — | ||||||
| L19e | — | — | — | ||||||
| L19f | — | — | — | % | |||||
| L19g | — | — | — | ||||||
| L19h | — | — | — | ||||||
| L20a | — | — | — | – | |||||
| L20a | — | — | — | ||||||
See text and Table S3 for details.
eccDNA-mediated duplication
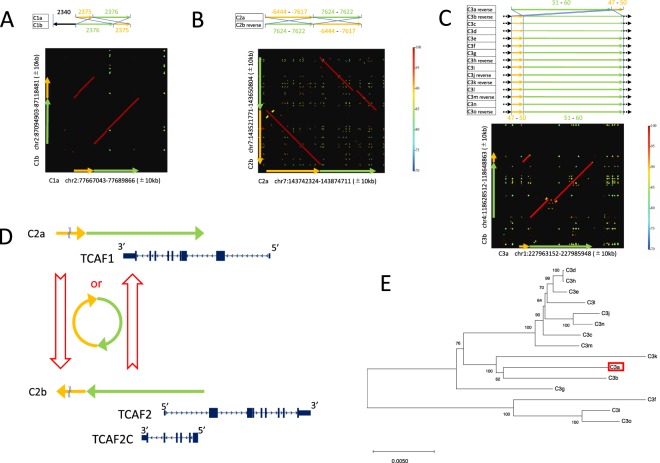
We found three cases with fairly strong signatures of eccDNA-mediated duplications. Figure 5A is a simple case (C1), where two distinct regions (C1a and C1b, both ~23 kb in length) on chromosome 2 were involved. The duplicated regions consist of two eSDs (2375 in yellow and #2376 in green in Fig. 5A), and the sequence homology between the two paralogous regions is 97.0%. We found orthologous regions of C1a and C1b as (, , ) and (, , ) in the chimpanzee genome, respectively, and (, , ) and (Δ, , ) in the macaque genome, respectively. However, from this observed pattern, it was difficult to determine their donor/recipient statuses. We found that these regions had no overlap with coding genes, which did not help to determine the donor/recipient statuses (see below for a successful case).
Figure 5.
(A–C) Three candidate regions for eccDNA-mediated duplications. For each, an alignment of eSD# and a dot plot are shown. Dot-plots were produced by using GenomeMatcher with its default setting19 (D) Overlapped coding genes in C2. (E) Unrooted NJ tree of the 15 regions involved in C3, which identified the donor and recipient copies (C3b and C3a, respectively). The copy created by eccDNA-mediated duplication is boxed in red. The tree was made using MEGA X20. The bootstrap support in percentage is presented for internal branches.
Figure 5B is another case (C2) with two distinct regions in chromosome 7 (C2a and C2b, both ~130 kb in length). The sequence homology between the two paralogous regions is 95.3%. We found that the orthologous regions of C2a were absent in the chimpanzee and macaque genomes, while the orthologous region of C2b was partially found in the chimpanzee genome, making it difficult to determine the donor/recipient status. The duplicates show an interesting pattern of coding gene overlap, as illustrated in Fig. 5D. C2a overlaps with a 3′ half of TCAF1, while C2b overlaps with 5′UTR of TCAF2. Furthermore, TCAF2C resides completely within C2b, which cannot be considered a partial duplicate of TCAF1 because they are on the different strands. See the DISCUSSION for this complex pattern.
The third case (C3) involves 15 regions with length about ~20 kb, where the synteny of C3-a is different from that of the others (Fig. 5C). An NJ tree of the 15 regions is shown in Fig. 5E, indicating that C3a was recently derived through an eccDNA-mediated duplication from C3b. The sequence homology C3a and the closet copy C3b is 96.7%.
eltDNA-mediated duplication
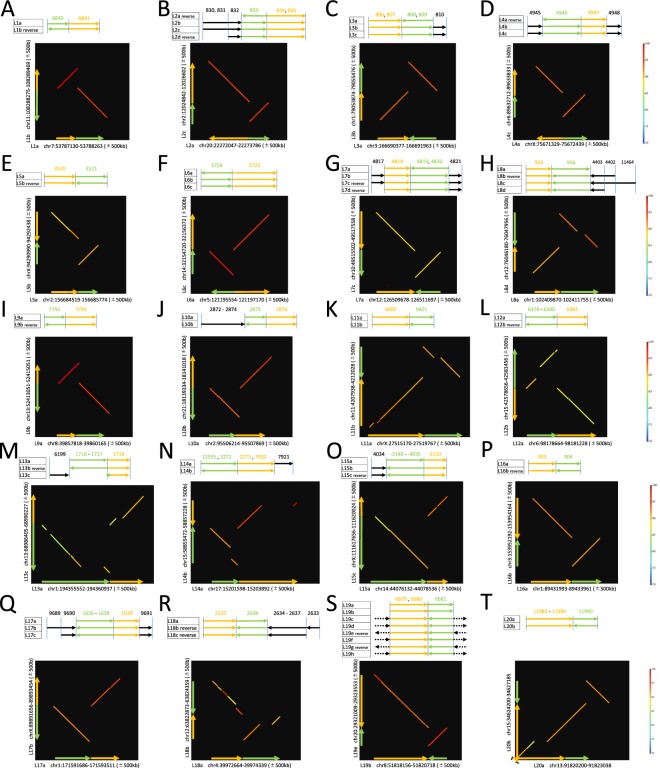
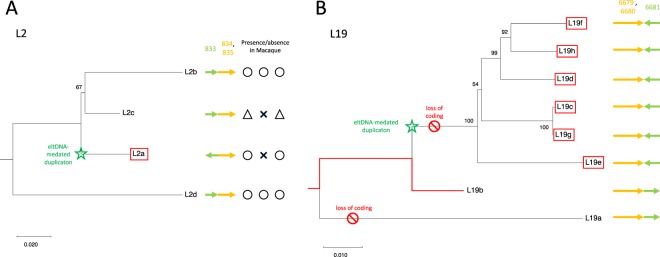
We found 20 cases with strong signatures of eltDNA-mediated duplication (Fig. 6). The lengths of duplicated regions distribute in a relatively narrow range of around 2–3 kb (mean 2,854 bp, median 1,934 bp, SD 4258), and if an exceptionally long one (L20, 25 kb) is excluded, and the average homology is 85.8% (SD 5.58%). Table 1 summarizes the overlap with coding genes and the presence/absence statuses in the chimpanzee and macaque genomes. In 11 cases (L1, L4, L6, L7, L8, L9, L13, L14, L17, L18, L19), at most, one region in each case exhibited an overlap with a coding gene. If we assume that duplication of a noncoding region is unlikely to create a novel coding gene, we may be able to assume that the copy with coding gene overlapped should be the donor. This prediction seems to be correct at least for L19 (see Fig. 7B for an NJ tree), where L19b overlaped with PCMTD1 and all others seemed to have lost the function after duplication. L19-a seems to be the recipient, which was inserted in an intronic region of STK31 (see Table 1).
Figure 6.
(A–T) 20 candidate regions of eltDNA-mediated duplications. For each, an alignment of eSD# and a dot plot are shown. For L20 (T), which is 26 kb long, only a part of the duplicated region is shown here. Dot-plots were produced by using GenomeMatcher with its default setting19.
Figure 7.
(A,B) NJ trees for L2 and L19. The trees were made by MEGA X20. The copy created by eltDNA-mediated duplication is boxed in red. The numbers on the tree are bootstrap values for internal lineages. The lineage is shown in red when the region overlaps with a coding gene.
Overall, the presence/absence information in the chimpanzee and macaque genomes was not very informative mainly because in many cases, large regions encompassing the focal duplicate regions are missing (i.e., (, , )). The only an exception was L2, where a convincing pattern (, , ) was observed for L2a in the macaque genome. It seems that there were two copies (L2b and L2d) in the genome of the ancestor of humans and macaques, and in the current macaque genome. After the split between humans and macaques, two duplications occurred in the lineage of humans to create L2a and L2b, one of which involved eltDNA-mediated modification (L2a). This is consistent with the NJ tree in Fig. 7A, which demonstrates L2a as a young copy, although the donor copy was not clearly determined due to a relatively low bootstrap value (66/100) for the internal branch.
Discussion
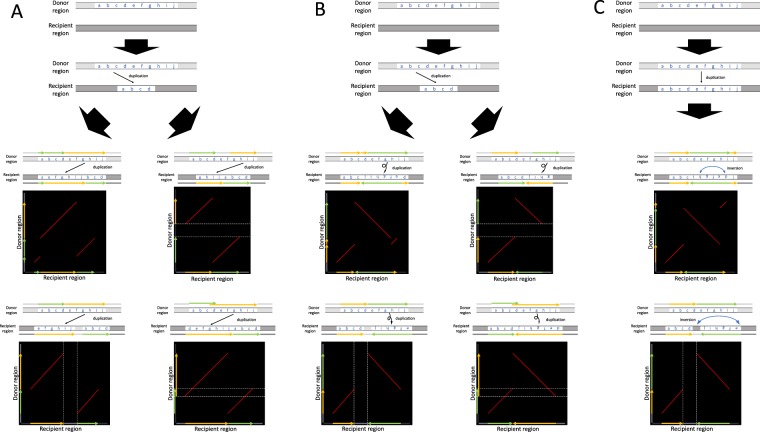
In this study, we found 3 and 20 cases with fairly strong evidence for eccDNA- and eltDNA-mediated duplications, respectively. We did not rule out other mutational scenarios that explain the detected regions. Indeed, one may think that the observed patterns may be explained by involving multiple duplication/deletion events. Nevertheless, we would emphasize that such a scenario would be quite unlikely, as illustrated in Fig. 8. Figure 8A examines whether two duplication events can explain an eccDNA-mediated duplication. Consider a donor region abcdefghij and suppose that a part of the region (abcd, presented by a yellow arrow) is first duplicated and inserted in the recipient region. If the remaining part (efghij, presented by a green arrow) is then duplicated and inserted exactly at the 5′ breakpoint of the former insertion, it could result in the pattern predicted by an eccDNA-mediated duplication (i.e., efghijabcd).
Figure 8.
Illustration of possible patterns involving multiple duplications and inversions. (A) Those showing similar patterns to after an eccDNA-mediated duplication. (B,C) Those showing similar patterns to after an eltDNA-mediated duplication. In (B), two duplication events are involved, while in (C) a duplication and an inversion event are considered.
However, the odds that the secondary insertion comes at this exact breakpoint should be extremely low. If an insertion is randomly placed in the genome, the odds would be assuming the genome size to be . Although we did not find any evidence that insertion occurred with local homology, if an insertion requires a 6 bp match, the odds would be (assuming equal frequencies of four nucleotides). It should be noted that, if the secondary insertion comes slightly upstream or downstream, the outcome will be different from that of an eccDNA-mediated duplication, so that we can distinguish them. Figure 8A clearly demonstrates that such cases would be distinguished in dot plots, where there is a gap or an overlap between the yellow and green arrows. The same logic also applies to an eltDNA-mediated duplication. A duplication (abcd, yellow arrow) followed by an inverted duplication at the 3′ breakpoint of the former duplication could exhibit the same pattern as an eltDNA-mediated duplication, but we emphasize that this is only true when the secondary duplication is inserted at the 3′ breakpoint of the primary duplication (Fig. 8B). Similarly, it is difficult to explain by a duplication followed by an inversion as shown in Fig. 8C. Thus, the likelihood that multiple duplication/deletion/inversion events explain the 23 identified duplicated regions should be low.
A caveat applies to the two cases, C1 and C2, where the duplicates are located on the same chromosome with 10 Mb and 200 kb intervals, respectively. For these cases, complex nested duplications and structural changes may have created the observed patterns. Indeed, there are many copies of the same eSDs in the surrounding region of C2. There also are multiple copies of the TCAF gene family, which could explain the unique observation that both donor and recipient regions overlap with coding genes. Nevertheless, we demonstrate that duplication involving structural modification should play role in the human genome evolution.
It is found that all identified cases have flipped regions with length more than 500 bp, which is identical to the minimum length of eSDs15. It is indicated that there may be a number of undetected eccDNA- and eltDNA-mediated duplications in the human genome, most likely smaller than 500 bp long.
The presence of eccDNAs that are not integrated in the host chromosome is well-known, particularly in cancer cells, and there is some evidence that eccDNA can be reintegrated into the genome in cattle10 and yeast11. We have demonstrated that reintegration of eccDNA could occur in the human genome, indicating that duplication may occasionally involve structural modifications before reintegration, rather than simply inserting a copied region. An eltDNA could be another type of such structural modification. Our results suggest that insertion-based duplication may not be a simple process; it may involve a complicated process such as structural modification before reintegration, although the molecular mechanism is not yet fully understood. Such modifications may potentially contribute to adaptive genome evolution, although we did not find any clear evidence for this in our data.
Supplementary information
Author contributions
K.K.T. and H.I. designed the research, K.K.T. performed the research, and K.K.T. and H.I. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-63665-6.
References
- 1.Samonte RV, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- 2.Bailey JA, Eichler EE. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 3.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 4.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 5.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ottaviani D, LeCain M, Sheer D. The role of microhomology in genomic structural variation. Trends Genet. 2014;30:85–94. doi: 10.1016/j.tig.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Reams AB, Roth JR. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect Biol. 2015;7:a016592. doi: 10.1101/cshperspect.a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt N, et al. Amplicon rearrangements during the extrachromosomal and intrachromosomal amplification process in a glioma. Nucleic Acids Res. 2014;42:13194–13205. doi: 10.1093/nar/gku1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durkin K, et al. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature. 2012;482:81–84. doi: 10.1038/nature10757. [DOI] [PubMed] [Google Scholar]
- 11.Galeote V, et al. Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. Plos One. 2011;6:1–10. doi: 10.1371/journal.pone.0017872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox D, Yuncken C, Spriggs AI. Minute Chromatin Bodies in Malignant Tumours of Childhood. Lancet. 1965;286:55–58. doi: 10.1016/S0140-6736(65)90131-5. [DOI] [PubMed] [Google Scholar]
- 13.Radloff R, Bauer W, Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex dna: the closed circular dna in hela cells. Proc. Natl. Acad. Sci. 1967;57:1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Møller HD, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-03369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu L, Lin Y, Pevzner PA. Detection and analysis of ancient segmental duplications in mammalian genomes. Genome Res. 2018;28:901–909. doi: 10.1101/gr.228718.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.She, X. et al. Shotgun sequence assembly and recent segmental duplications within the human genome (2004). [DOI] [PubMed]
- 17.Fairley S, Lowy-Gallego E, Perry E, Flicek P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2019;48:D941–D947. doi: 10.1093/nar/gkz836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho C, et al. Blast+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. Genomematcher: a graphical user interface for dna sequence comparison. BMC bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.