Visual Abstract

Key Words: congenital heart disease, systems biology, translational genomics
Abbreviations and Acronyms: CHD, congenital heart disease; CORUM, Comprehensive Resource of Mammalian Protein Complexes; CRISPR, clustered regularly interspaced short palindromic repeats; CTD, conotruncal defect; GOBP, Gene Ontology biological processes; HHE, high heart expression; HLHS, hypoplastic left heart syndrome; HTX, heterotaxy; LVOTO, left ventricular outflow tract obstruction; MGI, Mouse Genome Informatics; PCGC, Pediatric Cardiac Genomics Consortium; PPI, protein-protein interaction
Highlights
-
•
Combining CHD phenotype–driven gene set enrichment and CRISPR knockdown screening in zebrafish is an effective approach to identifying novel CHD genes.
-
•
Mutations affecting genes coding for the WAVE2 protein complex and small GTPase-mediated signaling are associated with LVOTO lesions.
-
•
WAVE2 complex genes brk1, nckap1, and wasf2 and regulators of small GTPase signaling cul3a and racgap1 are critical to zebrafish heart development.
Summary
Genetic variants are the primary driver of congenital heart disease (CHD) pathogenesis. However, our ability to identify causative variants is limited. To identify causal CHD genes that are associated with specific molecular functions, the study used prior knowledge to filter de novo variants from 2,881 probands with sporadic severe CHD. This approach enabled the authors to identify an association between left ventricular outflow tract obstruction lesions and genes associated with the WAVE2 complex and regulation of small GTPase-mediated signal transduction. Using CRISPR zebrafish knockdowns, the study confirmed that WAVE2 complex proteins brk1, nckap1, and wasf2 and the regulators of small GTPase signaling cul3a and racgap1 are critical to cardiac development.
Congenital heart disease (CHD) is the most common clinically devastating birth defect (1). Among the multiple factors that drive CHD pathogenesis, genetic variants appear to be a primary driver (2,3). Despite greater understanding of the molecular mechanisms of heart development, our ability to definitively identify specific genetic causes for most patients has progressed more slowly.
A barrier to unraveling genotype-phenotype associations is the high degree of genetic heterogeneity, such that analysis of even a moderate-sized cohort results in a relatively low number of genes observed to be mutated recurrently (4,5). Hence, identifying novel CHD genes is well suited for network analysis, wherein discrete mutations and affected genes can be connected based on prior knowledge of gene-gene functional networks such as associations with developmental pathways and known protein-protein interactions. Together, these functional data can provide a platform to identify molecular pathways that link variants to prioritize identification and lead to the discovery of novel candidate causal CHD genes. Validation of selected genes in model systems will ultimately expand our ability to detect genetic etiologies for patients.
Initial whole exome sequencing from the Pediatric Cardiac Genomics Consortium (PCGC) identified a burden of de novo exonic mutations in genes with higher fetal heart expression (HHE) in sporadic, severe CHD accounting for 10% of cases (4). Gene Ontology enrichment analyses identified altered epigenetic regulators, particularly histone modifiers, as important for CHD pathogenesis. Subsequent PCGC whole exome sequencing studies have identified a significant overlap between HHE and high fetal brain expression for mutated genes identified in CHD probands—thus providing a potential mechanism for the frequent co-occurrence of extracardiac anomalies or neurodevelopmental delays (6). More recently, whole exome sequencing of this growing cohort of sporadic, severe CHD characterized the contribution of recessive variants in genes well established to cause CHD in human and mouse studies (7). To expand on these, we performed unbiased global analyses of phenotype-specific CHD-associated variants to prioritize candidate causal genes and identify pathways relevant for CHD pathogenesis.
Methods
Exome sequencing and variant filtering
Whole exome sequencing results from 2,881 sporadic, severe CHD trios enrolled in the PCGC or Pediatric Heart Network were compared with whole exome sequencing from 900 control trios in Simon’s Foundation Autism Research Initiative Simplex Collection (6). The research protocols were approved by the Institutional Review Boards at each participating center—Boston’s Children’s Hospital, Brigham and Women’s Hospital, Great Ormond Street Hospital, Children’s Hospital of Los Angeles, Children’s Hospital of Philadelphia, Columbia University Irving Medical Center, Icahn School of Medicine at Mount Sinai, Rochester School of Medicine and Dentistry, Steven and Alexandra Cohen Children’s Medical Center of New York, and Yale School of Medicine. Briefly, sequencing for case and control trios was performed at Yale Center for Genome Analysis using NimbleGen v2.0 exome capture reagent (Roche, Basel, Switzerland) and Illumina HiSeq 2000, 75-bp paired-end reads (Illumina, San Diego, California). Three independent analysis pipelines were used to process reads and were mapped to hg19 using Novoalign (Novocraft Technologies, Selangor, Malaysia) and Genome Analysis Toolkit 3.0 (Broad Institute, Cambridge, Massachusetts) best practices at Harvard Medical School and BWA-mem at Yale School of Medicine and Columbia University Medical Center (8,9), Variant calls were made using GATK HaplotypeCaller (8). De novo variants not meeting the following criteria after pooling from the 3 pipelines were filtered out: depth (minimum 10 reads total and 5 alternate allele reads), alternate allele balance (minimum 20% if alternate read depth ≥10 or minimum 28% if alternate read depth <10), and parental read characteristics (minimum depth of 10 reference reads and alternate allele balance <3.5%).
Variant pathogenicity was assessed using in silico prediction from PredictSNP2 (Loschmidt Laboratories, Brno, Czech Republic), which employs an ensemble approach by integrating data from multiple in silico tools, for all variants other than frameshift mutations (10). As PredictSNP2 does not score frameshift mutations, we used Combined Annotation Dependent Depletion v1.4 for these variants (11). Combined Annotation Dependent Depletion, 1 of the tools which contributes to PredictSNP2, yields PHRED-scaled C scores, such that a score of ≥30 correlates to the top 0.1% of all possible variants and in prior studies has been used to discriminate pathogenic from tolerated frameshift variants (11). Variants were also filtered for those affecting HHE genes (4). The HHE gene set was previously identified using RNA sequencing of isolated strain 129/SvEV mouse hearts including atria, ventricles, and all 4 valves at embryonic day 14.5 to dichotomize 16,676 genes with identified human-mouse orthologues with a minimum of 40 reads per million mapped reads into the top quartile of expression (4). The HHE gene list consists of 4,169 genes, and the low heart expression list consists of 12,507 genes.
We considered that the low heart expression genes may contain some genes critical to cardiogenesis, so in an orthogonal variant filtering, we excluded genes unlikely to have a role in cardiovascular development based on knockout mouse phenotypes using the Mouse Genome Informatics (MGI) knockout phenotype gene library (12). Specifically, a gene was excluded if its knockout had been phenotyped, and the knockout was not found to cause any cardiovascular related phenotype, embryogenesis phenotype, or embryonic or postnatal lethality.
Cardiac diagnoses were obtained from the PCGC Data Hub (13). Left ventricular outflow tract obstruction (LVOTO) (n = 802) included hypoplastic left heart syndrome, aortic coarctation, and aortic stenosis. Conotruncal defects (CTDs) (n = 1120) included D-transposition of the great arteries, tetralogy of Fallot, double outlet right ventricle, truncus arteriosus, ventricular septal defects, and abnormalities of the aortic arch patterning. Heterotaxy (HTX) (n = 274) included patients with left-right isomerism as the primary defect. The remaining patients not included in 1 of the 3 phenotype categories consisted of a heterogeneous group of defects including atrial septal defects and anomalous pulmonary venous connections, pulmonary valve lesions, atrioventricular canal defects, double inlet left ventricle, and tricuspid atresia.
Gene set enrichment analysis
We performed gene set enrichment analysis for Gene Ontology biological processes (GOBP) terms and Comprehensive Resource of Mammalian Protein Complexes (CORUM) using HHE in silico– and MGI library in silico–filtered variants and the hypergeometric test. Enrichment analyses were performed with Enrichr (Icahn School of Medicine at Mount Sinai, New York, New York), which, in addition to implementing the hypergeometric test, also employs a ranking method that combines the adjusted p value with a deviation from the expected rank based on enrichment analysis applied to random gene sets (z-score) (14). In addition, we repeated the enrichment analysis with the loading of 2 different background reference lists; 1 made of the 4,169 HHE genes and the other made of the entire human genome excluding the MGI library phenotype negative genes using WebGestalt (Vanderbilt University Medical Center, Nashville, Tennessee) with the default settings (15). The reference and background gene lists are available in supplemental information. A Benjamini-Hochberg–adjusted p < 0.05 using both tools, Enrichr and WebGestalt, was used for significance with significant terms ranked by Enrichr’s combined score. Gene Ontology enrichment analysis was performed for controls, all cases, and LVOTO, CTD, and HTX gene lists.
Given that among the CHD phenotype–specific groups, significant enrichment was only observed for the LVOTO group, we performed a protein-protein interaction (PPI) network analysis only for this group to prioritize candidate genes. We generated an LVOTO-specific PPI network, using GeNets (Broad Institute, Cambridge, Massachusetts), by inputting all LVOTO genes associated with all GOBP and CORUM terms identified as enriched including terms which were observed as enriched when using only Enrichr or WebGestalt enrichment analyses. Then we ranked genes associated with the consistently enriched terms by their number of connections identified in the LVOTO-specific PPI network.
A flow diagram of the variant filtering and enrichment analyses is provided in Supplemental Figure 1.
Modeling loss of candidate genes in zebrafish embryos
Clustered regularly interspaced short palindromic repeats (CRISPR)–mediated knockdown experiments were performed for candidate genes in zebrafish F0 embryos to assess morphological phenotypes, as described previously (16). Briefly, wild-type AB strain zebrafish embryos were injected at the 1- to 2-cell stage with zebrafish-optimized Cas9 protein and CRISPR RNAs targeting abi1, brk1, cyfip1 cul3a, nckap1, racgap1, or wasf2. CRISPR design and construction were performed by the University of Utah Mutation Generation and Detection Core using standard best practice procedures. Single guide RNAs were designed for high efficiency and low off-target effects, and concentrations for single guide RNA and cas9 protein were also titrated for optimal impact. The mutagenic efficiency of each CRISPR and validation of double-stand breaks was assessed using high-resolution melting analysis performed on genomic DNA from individual injected embryos (17). CRISPR target sequences and high-resolution melting analysis primer sequences are listed in Supplemental Table 1. Zebrafish embryos were phenotyped at 2 days past fertilization. To evaluate cardiac morphology and function, wild-type and mutant lines of zebrafish were evaluated on a cmlc2-GFP background and visualized with a fluorescent microscope.
Statistics
The burden of de novo mutations in controls was compared with that found in all CHD cases and then separately for the LVOTO, CTD, and HTX phenotype groups. A 2-tailed chi-square analysis was performed to generate the odds ratio with 95% confidence interval comparing the number of filter-selected variants (in silico, HHE in silico, or MGI library in silico) to filtered-out variants for each group compared with controls. As described previously, gene set enrichment was assessed using a hypergeometric test. All reported p values were adjusted using the Benjamini-Hochberg method. A p value <0.05 was considered statistically significant and R software version 3.5.2 (R Foundation for Statistical Computing, Vienna, Austria) was used for all statistical analyses.
Data availability
Whole exome sequencing data have been previously deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000571.v1.p1, phs000571.v2.p1, and phs000571.v3.p2.
Results
Exome sequencing and variant filtering
Consistent with the initial PCGC cohort and as published for this expanded PCGC cohort, the rate for all de novo variants in patients with CHD was not significantly different from healthy controls (1.04 vs. 1.05 variants/individual) (4,6,7). This was also true for the LVOTO (n = 802), CTD (n = 1120), and HTX (n = 274) groups (1.08, 1.03, and 0.90 de novo variants per individual, respectively). Variant pathogenicity was assessed using in silico prediction from PredictSNP2 and Combined Annotation Dependent Depletion (10,11). With the exception of HTX, which did not demonstrate significant burden with any combination of filters, all cases, LVOTOs, and CTDs exhibited increasing burden with each layer of filtering (Table 1). The HHE in silico filter demonstrated the greatest burden, with odds ratios of 1.7, 2.0, and 1.7 (p < 0.001, for all) for all cases, LVOTOs, and CTDs, respectively, with comparable burden identified using the MGI library in silico.
Table 1.
Mutation Burden by Variant Filter and Phenotype
| Mutation Type | Controls (n = 900) |
Cases (n = 2,881) |
LVOTO (n = 802) |
CTD (n = 1,120) |
HTX (n = 274) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Var | Var | OR (95% CI) | Adjusted p Value | Var | OR (95% CI) | Adjusted p Value | Var | OR (95% CI) | Adjusted p Value | Var | OR (95% CI) | Adjusted p Value | |
| All | 945 | 3010 | 863 | 1,150 | 247 | ||||||||
| In silico | 362 | 1,237 | 1.12 (0.96–1.30) | 0.145 | 375 | 1.24 (1.03–1.45) | 0.051 | 473 | 1.13 (0.94–1.34) | 0.273 | 89 | 0.91 (0.68–1.21) | 0.560 |
| HHE in silico | 116 | 583 | 1.73 (1.39–2.13) | <0.001 | 185 | 1.96 (1.51–2.51) | <0.001 | 223 | 1.73 (1.35–2.19) | <0.001 | 36 | 1.23 (0.81-1.83) | 0.469 |
| MGI library in silico | 178 | 819 | 1.61 (1.34–1.93) | <0.001 | 252 | 1.78 (1.43–2.21) | <0.001 | 304 | 1.55 (1.26–1.91) | <0.001 | 52 | 1.15 (0.81–1.63) | 0.531 |
Odds ratios (ORs) calculated comparing filter-selected with filtered-out variants between controls and cases or phenotype groups. All p values adjusted using Benjamini-Hochberg p < 0.05 for significance.
CI = confidence interval; CTD = conotruncal defect; HHE = high fetal heart expression; HTX = heterotaxy; MGI = Mouse Genome Informatics; Var = variants.
Gene set enrichment analyses
We performed GOBP and CORUM gene set enrichment analyses using the hypergeometric test with the MGI library in silico– and HHE in silico–filtered gene lists derived from all cases, CHD phenotypes, and control groups. Hypergeometric tests were performed using Enrichr and WebGestalt as described in the Methods (14,15). Using the MGI library in silico–filtered gene set from all cases, we identified 181 GOBP and 4 CORUM enriched terms. Similarly, using WebGestalt, we identified 140 GOBP and 3 CORUM enriched terms, of which 17 common GOBP and 2 common CORUM terms identified as significant with both tools (Supplemental Table 2). The relatively low overlap in enriched terms between Enrichr and WebGestalt is likely due to different methods to convert the Gene Ontology tree into a gene set library and different versions of the Gene Ontology tree. Notable enriched terms included the following developmental pathways and processes: heart morphogenesis, vasculogenesis, VEGF receptor signaling, beta-catenin-TCF complex, and regulation of actin filament–based processes. Using the same approach for the LVOTO phenotype genes, we identified enrichment for 23 GOBP and 3 CORUM enriched terms using Enrichr and 39 GOBP and 2 CORUM enriched terms using WebGestalt, with 2 from each library consistently enriched using both tools: regulation of actin filament-based processes, VEGF receptor signaling, WAVE2 complex, and ITGA6-ITGB4-Laminin 10/12 complex (Table 2).
Table 2.
LVOTO Gene Set Enrichment
| Gene List | Library | Term | Combined∗ | Fold† | Adjusted p Value† | Genes |
|---|---|---|---|---|---|---|
| MGI library in silico | CORUM | Wave-2 complex | 1709.0 | 32.6 | 0.011 | ABI1;CYFIP1;NCKAP1 |
| MGI library in silico | CORUM | ITGA6-ITGB4-Laminin10/12 complex | 692.9 | 32.6 | 0.011 | LAMA5;LAMB1;LAMC1 |
| MGI library in silico | GOBP | Regulation of actin filament–based process | 77.1 | 3.5 | 0.003 | ABL2;DLC1;TENM1;ASAP3; SPTA1;SSH2;ANK2;TSC1;SMAD4; CYFIP1;NCKAP1;LRP1;RAC1; CDC42;RYR2;MTOR;ROCK1 |
| HHE in silico | GOBP | Regulation of small GTPase-mediated signal transduction | 73.6 | 3.8 | 0.002 | FOXM1;NF1;NOTCH2;NOTCH1; ARAP3;DLC1;CUL3;SIPA1L1; ITSN2;KALRN;TNFAIP1;RHOT2; RACGAP1;RAF1;RAC1;CDC42 |
| MGI library in silico | GOBP | Vascular endothelial growth factor receptor signaling pathway | 63.5 | 5.3 | 0.017 | MYOF;ABI1;CYFIP1;NCKAP1; RAC1;CDC42;ROCK1 |
Gene set enrichment was performed using HHE and MGI library in silico–filtered genes with the hypergeometric overrepresentation test implemented using Enrichr and WebGestalt. Terms were filtered for statistical significance if Benjamini-Hochberg–adjusted p < 0.05 using both tools.
CORUM = Comprehensive Resource of Mammalian Protein Complexes; GOBP = Gene Ontology biological processes; other abbreviations as in Table 1.
Combined score derived from Enrichr, which is a unique ranking system that combines the adjusted p value with a deviation from expected ranking for each term based on inputting random gene sets.
Fold enrichment and adjusted p values presented from WebGestalt using background gene list correction.
Similar themes emerged from enrichment results using HHE in silico–filtered gene sets for all cases and LVOTO groups but with overall narrower overlapping results. From all cases, 4 GOBP and 1 CORUM term were consistently identified as enriched: heart morphogenesis, ephrin receptor signaling, regulation of small GTPase-mediated signal transduction, actomyosin structure organization, and BAF complex (Supplemental Table 3). For LVOTO, the highest-ranked CORUM protein complex identified by Enrichr was WAVE2. One GOBP term was consistently enriched using both tools: regulation of small GTPase-mediated signal transduction (Table 2).
Much less enrichment was observed for the CTD, HTX, and control gene lists, with no terms reaching statistical significance for the control or the HTX MGI library in silico or HHE in silico gene lists using either tool. For CTDs, only 1 GOBP term—pericardium development—was consistently enriched using the MGI library in silico–filtered genes. No term was consistently enriched using HHE in silico–filtered CTD genes. Complete results of patient phenotypes and variants and complete enrichment analyses results are included in the Supplemental Appendix.
Across all enrichment analyses, LVOTO-driven WAVE2 complex enrichment demonstrated the highest Enrichr combined score and WebGestalt fold enrichment. Variants affecting 3 of the 5 genes encoding proteins within the WAVE2 complex and the direct regulator of WAVE2 (RAC1) were identified in LVOTO probands with none identified in other phenotypes: nonsense variants in ABI1 (p. R106X) and NCKAP1 (p. E1057X) and damaging missense variants in CYFIP1 (p. S35L) and RAC1 (p. R68C). PredictSNP2 scores ranged from 0.69 to 1.0 (maximum) and C scores were ≥32 (0.06% most damaging variants) for these 4 variants. The WAVE2 complex functions downstream of the small GTPase RAC1 to regulate branched actin synthesis and influence actin cytoskeleton organization in multiple cellular processes, including cell migration through formation of lamellipodia (18,19). In this context, it is notable that in addition to consistent enrichment observed for actin filament–based processes and regulation of small GTPase signaling, terms related to lamellipodium and actin cytoskeleton development were identified as enriched in at least 1 context.
We considered that visualizing protein network interactions between genes associated with the LVOTO enriched terms would allow us to better prioritize candidate genes for downstream validation. To that end, we used Metanetwork v1.0 (Broad Institute) predicted protein-protein interactions available from GeNets to interrogate for interactions between the 35 genes associated with the 5 consistently enriched GOBP and CORUM terms and all genes associated with all LVOTO enriched terms (Figure 1) (20). These 35 genes had a median of 3 connections in this LVOTO-specific protein network with CUL3, CDC42, CYFIP1, NCKAP1, NF1, NOTCH1, RAC1, RACGAP1, and RAF1 being in the top quartile with 7 to 14 connections each. Unsurprisingly, most of these genes are already strongly implicated in cardiac development and CHD (21, 22, 23, 24, 25). Notably, both Cul3–/– and Racgap1–/– result in embryonic lethality, suggesting a possible role in heart development, which is further supported by a more recent cardiomyocyte-specific Cul3–/– demonstrating a developmental cardiomyopathy and postnatal lethality (26, 27, 28).
Figure 1.
LVOTO Disease Gene Network
Bipartite graph generated using GeNets Metanetwork v1.0 protein-protein interactions connecting left ventricular outflow tract obstruction (LVOTO) genes associated with enriched terms using either high fetal heart expression (HHE) in silico or Mouse Genome Informatics (MGI) library in silico–filtered genes and either Enrichr or WebGestalt. Of the genes associated with consistently enriched terms and not previously implicated in structural heart defects, CUL3 and RACGAP1 are the most strongly connected.
The role of the WAVE2 complex, which consists of the 3 previously mentioned genes as well as BRICK1 and WASF2, in cardiogenesis is mostly unknown. Mouse knockout studies of Abi1, Brick1, and Cyfip1 resulted in embryonic lethality, with the only reported cardiac phenotype being hemorrhagic pericardial edema or discontinuous cardiac tissue layers from loss of Abi1 (29, 30, 31, 32). Additionally, loss of Wasf2 resulted in reversed cardiac looping in 1 of 2 mouse models and loss of Nckap1 resulted in cardia bifida in mice (33, 34, 35). Therefore, we selected the 5 WAVE2 complex genes, cul3a, and racgap1 for further validation in zebrafish embryos.
Modeling loss of candidate genes in zebrafish embryos
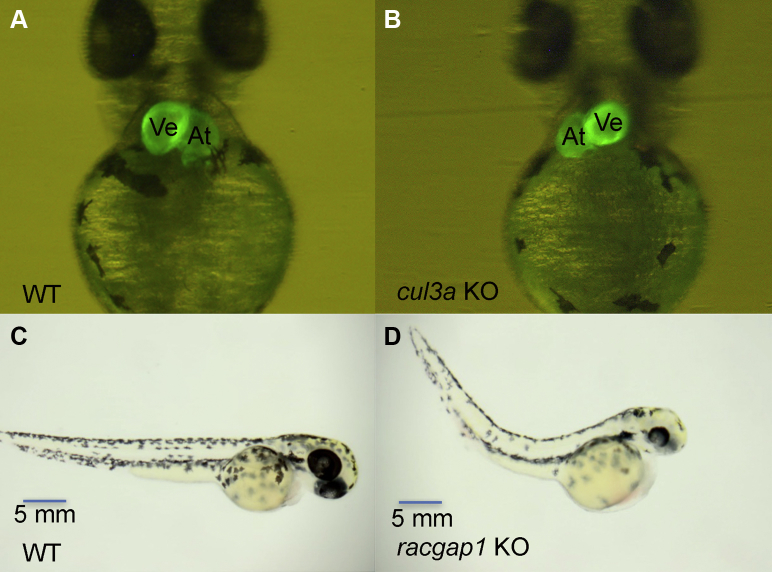
Zebrafish was selected for gene knockdown as its ability to survive via oxygen diffusion in the absence of a functional cardiovascular system permits the study of lethal cardiac defects later in development (36). Using CRISPR-guided knockdown, we observed cardiac phenotypes in F0 embryos with mosaic loss of brk1, cul3a, nckap1, racgap1, and wasf2 (Table 3). Reversed cardiac looping was observed in brk1, cul3a, nckap1, and wasf2 knockdown embryos, while racgap1 knockdown resulted in zebrafish embryos with a small, poorly contractile ventricle often with associated atrial dilation (Figure 2). Notably, all 7 of these genes are both highly expressed in the developing heart and brain of the mouse, but a brain phenotype was only observed in racgap1 knockdown embryos (6).
Table 3.
Phenotypes of CRISPR Gene Knockdown F0 Zebrafish
| Target Gene | Reversed Heart Looping | Atrial and Ventricular Size | Circulation | Head/Brain/Eye Structures |
|---|---|---|---|---|
| cul3a | 8/64 (13) | Normal | Normal | Normal |
| wasf2 | 19/102 (19) | Normal | Normal | Normal |
| racgap1 | 6/105 (6) | Dilated atria and small ventricle 42/105 (42) | Normal | Hypoplastic 99/105 (95) |
| brk1 | 23/122 (19) | Normal | Normal | Normal |
| nckap1 | 16/83 (19) | Normal | Normal | Normal |
| cyfpl1 | 1/75 (1.3) | Normal | Normal | Normal |
| abi1 | 1/72 (1.4) | Normal | Normal | Normal |
Values are n/N (%). Most (5 of 7) candidate genes demonstrate abnormal cardiac development in CRISPR-mediated knockdown, including reversed cardiac looping in 3 WAVE2 complex genes (brk1, nckap1, and wasf2) and cul3a. Loss of racgap1 also demonstrated extracardiac anomalies in addition to a small ventricle and atrial dilation.
CRISPR = clustered regularly interspaced short palindromic repeats.
Figure 2.
Crispr-Mediated Candidate Gene Knockdown in Zebrafish
(A, C) Wild-type (WT), (C)cul3a, and (D)racgap1 knockout (KO) zebrafish at 2 days past fertilization. Reversed cardiac looping illustrated in cul3a KO as compared with WT, both on a cmlc2-GFP background. The racgap1 KO zebrafish demonstrate atrial dilation and pericardial edema. At = atria; CRISPR = clustered regularly interspaced short palindromic repeats; Ve = ventricle.
Discussion
Human genetics
In this study, we used whole exome sequencing of 2,881 well-phenotyped, sporadic CHD trios and compared these with 900 control trios to identify de novo predicted damaging mutations using in silico, developmental heart expression, and previous implications in development or cardiac disease in knockout mouse phenotypes. As gene set enrichment analyses are sensitive to both the length of the genes list associated with each term and the background gene list, we performed enrichment analysis with different methods and considered not only statistical significance, but also consistency across tools and algorithms.
Through phenotype-driven elaborate gene set enrichment analyses, we identified a novel association between the LVOTO phenotype and genes associated with the WAVE2 complex, actin-filament based processes, and small GTPase signal transduction. The WAVE2 complex comprises 5 proteins (gene symbols in parentheses when different)—WAVE2 (WASF2), HSPC300 (BRK1), CYFIP1, NCKAP1, and ABI1—and is regulated by the small GTPase RAC1 to mediate branched actin synthesis, a key contributor to actin cytoskeleton organization, via Arp2/3-mediated actin polymerization (18). Enrichment analyses also identified CUL3 and RACGAP1 as mediators of small GTPase signaling. Placing the protein products of these genes within a PPI network we further highlight their strong connections within an LVOTO-specific network.
In contrast to LVOTO, the other phenotype groups demonstrated significantly less term enrichment. Although CTDs had similar variant burden to LVOTO, we hypothesize that genetic and phenotypic heterogeneity within this group may have limited our ability to identify consistent term enrichment with our approach. For HTX, we failed to identify a burden of mutations or identify any significant term enrichment using any combination of enrichment tools or gene filter. The smaller size of the HTX cohort compared with the other phenotypes may have limited our power for enrichment analysis, but limiting to de novo variants may also have missed significant genetic contributors. Recent results from the PCGC highlight that recessively inherited variants contribute substantially to pathogenesis of patients with CHD and laterality defects, suggesting that this model of inheritance needs to be incorporated into all future gene pathway enrichment studies for patients with HTX (7).
Candidate gene validation in zebrafish
Given consistency of LVOTO gene set enrichment, we selected candidate genes from this group for validation. We identified reversed cardiac looping with loss of brk1, cul3a, nckap1, and wasf2 and a small ventricle with atrial dilation and pericardial edema with loss of racgap1 in F0 zebrafish embryos. It is difficult to directly compare reversed cardiac looping in zebrafish with LVOTO defects in humans. Being comprised of only 2 chambers renders the developing zebrafish heart vulnerable to looping defects from mechanisms other than perturbed sidedness including altered myocardial cell polarity, cell number, or blood flow (37). Thus, while these results in zebrafish cannot definitively implicate these genes in LVOTO, we can conclude their critical nature to cardiac development and establish an association with mutations in these genes and LVOTO in humans.
Proposed molecular mechanisms
Recently, the Lo lab has developed a mouse forward genetics screen coupled with fetal imaging and whole exome sequencing of founder mice exhibiting a cardiovascular malformation to identify recessively inherited variants in novel CHD genes (38). This model has illustrated a role for complex genetic inheritance in multiple CHD phenotypes including the severest form of LVOTO, hypoplastic left heart syndrome, in the Ohia mouse line (Sap130m/m/Pcdha9m/m) (39). These genes were further validated in LVOTO pathogenesis after identifying mutations affecting SAP130 and a related gene, PCDHA13, in patients with hypoplastic left heart syndrome. Kyoto Encyclopedia of Genes and Genomes network analysis of differentially expressed genes in the Ohia right and left ventricular tissue by RNA sequencing and chromatin immunoprecipitation sequencing implicated multiple developmental pathways including Notch, Wnt, Tgfβ, and hedgehog signaling, as well as biological processes including extracellular matrix receptors, regulation of actin cytoskeleton, axon guidance, and metabolism. WAVE2 is a highly conserved regulator of actin cytoskeleton and cell morphology during development, a process that is critical to regulating cell polarity, cell migration, cytokinesis, and tissue architecture (35,40,41). Interestingly, the noncanonical Wnt planar cell polarity pathway, which regulates cell polarity during development via small RhoGTPase regulation including RAC1, was identified as enriched using Enrichr with both MGI library in silico– and HHE in silico–filtered LVOTO gene lists and could provide a mechanistic link between altered WAVE2 complex activity and CHD (42).
Knockout studies in mouse and zebrafish demonstrate that planar cell polarity genes influence heart development by regulating directional migration of progenitor cells, septation of the primitive heart tube, and patterning of cardiac structures (43). In loss of Wnt5a and mutants of Vangl2 and Dvl2 (which connect planar cell polarity to RhoGTPase signaling), abnormal outflow tract development, reduced cardiomyocyte polarity, and actin polymerization defects in cardiac progenitor cells are observed (44). The direct link between planar cell polarity and WAVE2 complex signaling is the RhoGTPase RAC1. Early lethality has prohibited studying global loss of Rac1, but second heart field-specific knockdown of Rac1 resulted in abnormal right ventricular cardiomyocyte polarity with inhibited second heart field progenitor cell migration and concomitant decreased expression of WAVE2 complex genes and Arp2/3 in embryonic right ventricular tissue (22). Taken together, these studies illustrate how altered signaling via planar cell polarity/Rac1/WAVE2 can disrupt normal mammalian heart development.
In addition to contributing to small GTPase-mediated signaling, loss of CUL3 may also contribute to CHD through its role in ubiquitination (45). In HEK293 cells, SAP130, a regulator of ubiquitination via cullin-RING ligases and 1 of 2 proteins implicated in the mouse Ohia line, binds CUL3 with higher affinity than other CUL proteins (46). Additionally, Lztr1 knockout in the mouse and loss of function LZTR1 mutations in humans have been implicated in Noonan syndrome through decreased CUL3-mediated RAS ubiquitination and ultimately increased RAS/MAPK signaling (47,48). Further, in the LVOTO-specific protein network generated in this study, the canonical ubiquitination regulator UBC demonstrated the highest number of connections including with CUL3. Thus, the mechanism of CUL3 loss leading to developmental cardiac defects might also be related to dysregulated ubiquitination.
Finally, loss of cyfip1 and abi1a, unlike the other WAVE2 complex genes, was not found to impact zebrafish heart development. Zebrafish, unlike mouse or human, have 2 orthologs for ABI1 (abi1a and abi1b) with abi1b likely compensating to maintain normal cardiac development. Similarly, loss of cyfip1 may be compensated by irsp53, which in mice has been shown to also facilitate binding of activated Rac1 to WAVE2 (49).
Conclusions
Despite rigorous efforts to unravel the genetic mechanisms for severe forms of LVOTO pathogenesis, the etiology for most patients has largely remained elusive with only recent evidence confirming a role for SAP130 and PCDHA13 (39). Here, we exploited the strength of gene pathway enrichment analyses from whole exome sequencing results of sporadic, complex CHD trios to identify an association with LVOTO in humans and WAVE2 complex, actin-filament regulation, and small GTPase signaling genes. Furthermore, we confirmed a role for brk1, cul3a, nckap1, racgap1, and wasf2 in cardiac development using CRISPR mediated knockdown in zebrafish. Ultimately, we illustrate that combining phenotype-driven gene set enrichment analyses with validation in zebrafish is as an effective approach for identifying novel CHD genes. Given evidence linking planar cell polarity pathway to WAVE2 complex activity via small GTPase signaling, we propose this as a promising framework for future mechanistic investigation into LVOTO.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Genetic variants including single gene mutations or larger genomic changes, such as copy number variations or aneuploidy, contribute substantially to CHD pathogenesis. With the decreasing cost of next-generation sequencing, there is more widespread genetic testing, yet an unequivocal genetic cause is not identified in up to 80% of CHD patients. Closing this gap in our fund of knowledge has significant implications on genetic counseling for our patients and their families.
TRANSLATIONAL OUTLOOK: Predicted damaging mutations affecting genes in the WAVE2 complex and related genes are probably causative of CHD and, more specifically, LVOTO.
Footnotes
This work was supported by a grant from the National Center for Research Resources and the National Center for Advancing Translational Sciences (U01 HL098153), National Institutes of Health grants to the Pediatric Cardiac Genomics Consortium (U01-HL098188, U01-HL098147, U01-HL098153, U01-HL098163, U01-HL098123, U01-HL098162, and U01-HL098160), and the National Institutes of Health Centers for Mendelian Genomics (5U54HG006504). Dr. Edwards was supported by National Institutes of Health Grant No. 5T32HL007915. Drs. Lifton and Seidman were supported by the Howard Hughes Medical Institute. Dr. Chung was supported by the Simons Foundation. Dr. Srivastava is co-founder and has served on the scientific advisory board for Tenaya Therapeutics. Dr. Lifton is director of Roche; has served on the scientific advisory board for Regeneron; and has served as a consultant for Johnson and Johnson. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
Appendix
For supplemental databases, tables, and a figure, please see the online version of this paper.
Appendix
References
- 1.Razzaghi H., Oster M., Reefhuis J. Long-term outcomes in children with congenital heart disease: National Health Interview Survey. J Pediatr. 2015;166:119–124. doi: 10.1016/j.jpeds.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierpont M.E., Brueckner M., Chung W.K. Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association. Circulation. 2018;138:e653–e711. doi: 10.1161/CIR.0000000000000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oyen N., Poulsen G., Wohlfahrt J., Boyd H.A., Jensen P.K., Melbye M. Recurrence of discordant congenital heart defects in families. Circ Cardiovasc Genet. 2010;3:122–128. doi: 10.1161/CIRCGENETICS.109.890103. [DOI] [PubMed] [Google Scholar]
- 4.Zaidi S., Choi M., Wakimoto H. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson D.W., Sharkey A., Fatkin D. Reduced penetrance, variable expressivity, and genetic heterogeneity of familial atrial septal defects. Circulation. 1998;97:2043–2048. doi: 10.1161/01.cir.97.20.2043. [DOI] [PubMed] [Google Scholar]
- 6.Homsy J., Zaidi S., Shen Y. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin S.C., Homsy J., Zaidi S. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePristo M.A., Banks E., Poplin R. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bendl J., Musil M., Stourac J., Zendulka J., Damborsky J., Brezovsky J. PredictSNP2: a unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Mouse Genome Database G The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pediatric Cardiac Genomics C. Gelb B., Brueckner M. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ Res. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuleshov M.V., Jones M.R., Rouillard A.D. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankaran S.S., Dahlem T.J., Bisgrove B.W., Yost H.J., Tristani-Firouzi M. CRISPR/Cas9-Directed Gene Editing for the Generation of Loss-of-Function Mutants in High-Throughput Zebrafish F0 Screens. Curr Protoc Mol Biol. 2017;119:31.9.1–31.9.22. doi: 10.1002/cpmb.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parant J.M., George S.A., Pryor R., Wittwer C.T., Yost H.J. A rapid and efficient method of genotyping zebrafish mutants. Dev Dyn. 2009;238:3168–3174. doi: 10.1002/dvdy.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soderling S.H., Scott J.D. WAVE signalling: from biochemistry to biology. Biochem Soc Trans. 2006;34:73–76. doi: 10.1042/BST0340073. [DOI] [PubMed] [Google Scholar]
- 19.Innocenti M., Zucconi A., Disanza A. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 20.Li T., Kim A., Rosenbluh J. GeNets: a unified web platform for network-based genomic analyses. Nat Methods. 2018;15:543–546. doi: 10.1038/s41592-018-0039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards A.A., Garg V. Genetics of congenital heart disease. Curr Cardiol Rev. 2010;6:91–97. doi: 10.2174/157340310791162703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung C., Lu X., Liu M., Feng Q. Rac1 signaling is critical to cardiomyocyte polarity and embryonic heart development. J Am Heart Assoc. 2014;3 doi: 10.1161/JAHA.114.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman J.M., Arbiser J., Epstein J.A. Cardiovascular disease in neurofibromatosis 1: report of the NF1 Cardiovascular Task Force. Genet Med. 2002;4:105–111. doi: 10.1097/00125817-200205000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Martinelli S., Krumbach O.H.F., Pantaleoni F. Functional dysregulation of CDC42 causes diverse developmental phenotypes. Am J Hum Genet. 2018;102:309–320. doi: 10.1016/j.ajhg.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Wang J., Li J. Deletion of Cdc42 in embryonic cardiomyocytes results in right ventricle hypoplasia. Clin Transl Med. 2017;6:40. doi: 10.1186/s40169-017-0171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warga R.M., Wicklund A., Webster S.E., Kane D.A. Progressive loss of RacGAP1/ogre activity has sequential effects on cytokinesis and zebrafish development. Dev Biol. 2016;418:307–322. doi: 10.1016/j.ydbio.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Singer J.D., Gurian-West M., Clurman B., Roberts J.M. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papizan J.B., Vidal A.H., Bezprozvannaya S., Bassel-Duby R., Olson E.N. Cullin-3-RING ubiquitin ligase activity is required for striated muscle function in mice. J Biol Chem. 2018;293:8802–8811. doi: 10.1074/jbc.RA118.002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubielecka P.M., Ladwein K.I., Xiong X. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc Natl Acad Sci U S A. 2011;108:7022–7027. doi: 10.1073/pnas.1016811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ring C., Ginsberg M.H., Haling J., Pendergast A.M. Abl-interactor-1 (Abi1) has a role in cardiovascular and placental development and is a binding partner of the alpha4 integrin. Proc Natl Acad Sci U S A. 2011;108:149–154. doi: 10.1073/pnas.1012316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escobar B., de Carcer G., Fernandez-Miranda G. Brick1 is an essential regulator of actin cytoskeleton required for embryonic development and cell transformation. Cancer Res. 2010;70:9349–9359. doi: 10.1158/0008-5472.CAN-09-4491. [DOI] [PubMed] [Google Scholar]
- 32.Pathania M., Davenport E.C., Muir J., Sheehan D.F., Lopez-Domenech G., Kittler J.T. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakeman A.S., Anderson K.V. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development. 2006;133:3075–3083. doi: 10.1242/dev.02473. [DOI] [PubMed] [Google Scholar]
- 34.Yan C., Martinez-Quiles N., Eden S. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamazaki D., Suetsugu S., Miki H. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- 36.Burggren W.W., Pinder A.W. Ontogeny of cardiovascular and respiratory physiology in lower vertebrates. Annu Rev Physiol. 1991;53:107–135. doi: 10.1146/annurev.ph.53.030191.000543. [DOI] [PubMed] [Google Scholar]
- 37.Miura G.I., Yelon D. A guide to analysis of cardiac phenotypes in the zebrafish embryo. Methods Cell Biol. 2011;101:161–180. doi: 10.1016/B978-0-12-387036-0.00007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Klena N.T., Gabriel G.C. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X., Yagi H., Saeed S. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49:1152–1159. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King J.S., Veltman D.M., Georgiou M., Baum B., Insall R.H. SCAR/WAVE is activated at mitosis and drives myosin-independent cytokinesis. J Cell Sci. 2010;123:2246–2255. doi: 10.1242/jcs.063735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel F.B., Bernadskaya Y.Y., Chen E. The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev Biol. 2008;324:297–309. doi: 10.1016/j.ydbio.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simons M., Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson D.J., Chaudhry B. Getting to the heart of planar cell polarity signaling. Birth Defects Res A Clin Mol Teratol. 2011;91:460–467. doi: 10.1002/bdra.20792. [DOI] [PubMed] [Google Scholar]
- 44.Sinha T., Wang B., Evans S., Wynshaw-Boris A., Wang J. Disheveled mediated planar cell polarity signaling is required in the second heart field lineage for outflow tract morphogenesis. Dev Biol. 2012;370:135–144. doi: 10.1016/j.ydbio.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wimuttisuk W., Singer J.D. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell. 2007;18:899–909. doi: 10.1091/mbc.E06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordero-Espinoza L., Hagen T. Regulation of Cullin-RING ubiquitin ligase 1 by Spliceosome-associated protein 130 (SAP130) Biol Open. 2013;2:838–844. doi: 10.1242/bio.20134374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motta M., Fidan M., Bellacchio E. Dominant Noonan syndrome-causing LZTR1 mutations specifically affect the Kelch domain substrate-recognition surface and enhance RAS-MAPK signaling. Hum Mol Genet. 2019;28:1007–1022. doi: 10.1093/hmg/ddy412. [DOI] [PubMed] [Google Scholar]
- 48.Steklov M., Pandolfi S., Baietti M.F. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science. 2018;362:1177–1182. doi: 10.1126/science.aap7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suetsugu S., Kurisu S., Oikawa T., Yamazaki D., Oda A., Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole exome sequencing data have been previously deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000571.v1.p1, phs000571.v2.p1, and phs000571.v3.p2.