The biological role of REEs has long been underestimated, and research has mainly focused on methanotrophic and methylotrophic bacteria. We have recently demonstrated that P. putida, a plant growth-promoting bacterium that thrives in the rhizosphere of various food crops, possesses a REE-dependent alcohol dehydrogenase (PedH), but knowledge about REE-specific effects on physiological traits in nonmethylotrophic bacteria is still scarce. This study demonstrates that the cellular response of P. putida to lanthanum (La3+) is mostly substrate specific and that La3+ availability highly affects the growth of cells on glycerol. Further, a novel route for glycerol metabolism is identified, which is initiated by PedE and/or PedH activity and provides a growth advantage to this biotechnologically relevant organism by allowing a faster onset of growth. Overall, these findings demonstrate that lanthanides can affect physiological traits in nonmethylotrophic bacteria and might influence their competitiveness in various environmental niches.
KEYWORDS: rare earth elements, proteomics, glycerol metabolism, Pseudomonas putida, PQQ, PedE, PedH, GarK, dehydrogenases, volatiles, lanthanides, alcohol dehydrogenases, carbon metabolism, metalloenzymes
ABSTRACT
Ever since the discovery of the first rare earth element (REE)-dependent enzyme, the physiological role of lanthanides has become an emerging field of research due to the environmental implications and biotechnological opportunities. In Pseudomonas putida KT2440, the two pyrroloquinoline quinone-dependent alcohol dehydrogenases (PQQ-ADHs) PedE and PedH are inversely regulated in response to REE availability. This transcriptional switch is orchestrated by a complex regulatory network that includes the PedR2/PedS2 two-component system and is important for efficient growth on several alcoholic volatiles. To study whether cellular responses beyond the REE switch exist, the differential proteomic responses that occur during growth on various model carbon sources were analyzed. Apart from the Ca2+-dependent enzyme PedE, the differential abundances of most identified proteins were conditional. During growth on glycerol—and concomitant with the proteomic changes—lanthanum (La3+) availability affected different growth parameters, including the onset of logarithmic growth and final optical densities. Studies with mutant strains revealed a novel metabolic route for glycerol utilization, initiated by PedE and/or PedH activity. Upon oxidation to glycerate via glyceraldehyde, phosphorylation by the glycerate kinase GarK most likely yields glycerate-2-phosphate, which is eventually channeled into the central metabolism of the cell. This new route functions in parallel with the main degradation pathway encoded by the glpFKRD operon and provides a growth advantage to the cells by allowing an earlier onset of growth with glycerol as the sole source of carbon and energy.
INTRODUCTION
The rhizosphere, defined as the narrow region of soil surrounding plant roots, is one of the most complex ecosystems on earth. It contains a multitude of organisms from different taxa, including fungi, oomycetes, nematodes, protozoa, algae, viruses, archaea, and arthropods as well as up to 108 soil-dwelling bacteria per gram of fresh root (1–3). Its diversity is predominantly shaped by root exudates, a complex mixture of organic compounds, including carbohydrates, amino acids, or carbon acids (4, 5) and plant-, fungus-, and bacterium-derived volatiles (VOCs) such as alkenes, alcohols, terpenes, or benzenoids (6, 7). As such, it is not surprising that the soil-dwelling organism Pseudomonas putida KT2440 is equipped with a broad diversity of metabolic pathways to maximize its cellular fitness in different environmental niches (8–10). For efficient growth on various alcoholic VOC substrates, P. putida uses a periplasmic oxidation system consisting of the pyrroloquinoline quinone-dependent alcohol dehydrogenases (PQQ-ADHs) PedE and PedH. These enzymes are inversely regulated on the transcriptional level depending on rare earth element (REE) availability and appear to be functionally redundant but differ in their metal cofactor dependencies (11–13). While PedE is a Ca2+-dependent enzyme, PedH requires REEs of the lanthanide series (Ln3+) for catalytic activity (12, 14).
Although they are among the most ubiquitous metals in the earth’s crust, REEs were long considered to be of no biological relevance due to their low solubility under environmental conditions (15). Indeed, the only known and characterized REE-dependent enzymes all belong to the family of PQQ-ADHs (12, 16–21). A conserved aspartic acid residue that is additionally present in the metal coordination sphere of these enzymes is a characteristic of the Ln3+-dependent activity and genes encoding such enzymes have been found in the genome of many bacteria from various origins (22–26). Very recently, a Ln3+-binding protein, called lanmodulin, was identified in Methylorubrum extorquens (formerly Methylobacterium extorquens) AM1 (27). This periplasmic protein, which shows structural similarities with the Ca2+-binding protein calmodulin, is able to bind up to four Ln3+ ions per protein with picomolar affinity and changes its conformation from a largely disordered to a compact ordered state upon REE binding. Although its exact cellular role is not clear, it most likely plays a role in Ln3+ uptake. Homologous genes exist in the genome of some other species of methylobacteria and bradyrhizobia.
In addition to their functional roles as metal cofactors, several recent studies investigated the effect of REEs on cellular physiology of different methano- and methylotrophic organisms (21, 28–33). Some of these studies found that Ln3+ availability can influence physiological traits, including changes in metabolite cross-feeding, growth rates, yields, or biofilm formation. In this context, it is further interesting that REEs have been used as microfertilizers, especially in China, for over 30 years, as Ln3+ supplementation can be associated with increased growth of different food crops, including rice, mung bean, maize, and coconut plants (34–38).
The aforementioned observations suggest that apart from the inverse transcriptional regulation of PQQ-ADHs, which has been described in detail for different organisms, including P. putida (12, 13, 20, 28, 39–43), additional cellular responses to REEs exist and could depend on the specific organism and/or environmental context. To investigate the existence of such conditional responses in the nonmethylotrophic organism P. putida KT2440, we used a differential proteomic approach during growth on various model carbon sources that reflect the metabolic diversity of the rhizosphere. From these experiments, we found that the vast majority of identified proteins are differentially abundant only under one specific set of growth conditions. Different physiological parameters, most importantly, the length of the lag phase (λ) of cultures, can be linked to the activity of PedE and PedH during growth on glycerol. On the basis of these results, we were able to identify and reconstruct a novel metabolic route for glycerol utilization, which is initiated by PedE and/or PedH activity. This route seems to operate in conjunction with the major degradation pathway initiated by the glycerol kinase GlpK and contributes to a reduced lag phase of P. putida cells growing on this polyol substrate.
RESULTS
To identify whether cellular responses of P. putida KT2440 to REEs beyond the regulation of the PQQ-ADHs exist, a comparative proteomic analysis was used during growth on four model carbon and energy sources, namely, 2-phenylethanol, glycerol, glucose, and citrate.
Evaluation of proteomics data.
Proteins were extracted from cells of P. putida KT2440*, representing the parental control for all mutant strains, by the use of SDS to enable extraction of cytoplasmic as well as transmembrane proteins followed by label-free (LF) nano-liquid chromatography–tandem mass spectrometry (nano-LC-MS/MS) quantification. In total, 2,771 proteins with at least two unique peptides and a false-discovery-rate (FDR) value of ≤1% were identified and quantified by our proteomics approach, corresponding to approximately 50% of the P. putida KT2440 proteome. Principal-component analysis (PCA) revealed high reproducibility for sample replicates and distinct patterns for the different carbon sources (see Fig. S1 in the supplemental material). The majority of proteins were increased or decreased in abundance in response to the different carbon sources. In contrast, La3+ availability during growth on the same carbon source caused only minor differences. Proteins that exhibited a 2-fold or higher change in abundance between different growth conditions and a P value of ≤0.01 were considered to represent differential abundance.
PCA comparing the four different carbon sources with and without La3+. Different carbon sources are indicated by squares (2-phenylethanol), circles (citrate), diamonds (glucose), and stars (glycerol). Samples with 10 μM La3+ and without La3+ in the medium are shown in blue and red, respectively. Biological replicates are indicated in the same color. Samples were separated according to the different carbon sources, while treatment with La3+ showed only minor effects. Download FIG S1, JPG file, 0.02 MB (20.1KB, jpg) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of lanthanum on protein abundance during growth on different substrates.
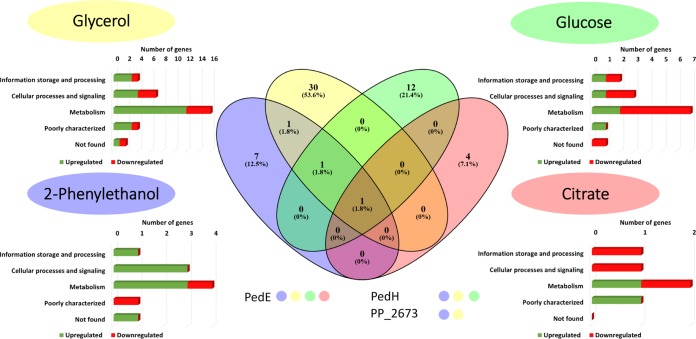
According to the aforementioned criteria, 56 proteins were differentially abundant in response to La3+ during growth on either of the different carbon sources (Fig. 1) (Table 1; see also Table S3 to S5 in the supplemental material). Among them, only the Ca2+-dependent PQQ-ADH PedE (PP_2674) showed decreased abundance in response to La3+ during growth on all tested carbon sources. The Ln3+-dependent PQQ-ADH PedH (PP_2679) showed increased abundance in response to La3+ during growth on glucose, glycerol, and 2-phenylethanol, whereas an uncharacterized pentapeptide repeat-containing protein (PP_2673) that is encoded by a gene directly upstream of pedE showed decreased abundance during growth on glycerol and 2-phenylethanol (Fig. 1). The remaining 53 proteins were identified under only one specific growth condition (Table 1; see also Table S3 to S5).
FIG 1.
Venn diagram (middle panel) of proteins in cells of KT2440* showing differential abundance in response to 10 μM La3+ during growth on glycerol, glucose, 2-phenylethanol, and citrate. Proteins that showed up under two or more sets of growth conditions are indicated below the diagram with color coding for classification as follows: yellow dot = glycerol; green dot = glucose; blue dot = 2-phenylethanol; red dot = citrate. Classifications of differentially abundant proteins according to the Cluster of Orthologous Groups database are depicted for each substrate.
TABLE 1.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ during growth on glycerol
| Locus tag |
Protein name |
Predicted protein functiona | Fold change (log2) | −log10 (P value) |
|---|---|---|---|---|
| PP_2426 | CalA | Coniferyl alcohol dehydrogenase | 6.28 | 4.03 |
| PP_2679 | PedH | Quinoprotein ethanol dehydrogenase | 4.75 | 3.80 |
| PP_3426 | MexF | Multidrug efflux RND transporter | 3.93 | 3.24 |
| PP_3425 | MexE | Efflux transporter RND family | 3.74 | 2.18 |
| PP_4921 | Transporter, NCS1 nucleoside transporter family | 3.65 | 2.46 | |
| PP_2440 | AhpF | Alkyl hydroperoxide reductase subunit F | 3.13 | 3.08 |
| PP_3745 | GlcD | Glycolate oxidase, putative FAD-linked subunit | 3.12 | 3.32 |
| PP_3747 | GlcF | Glycolate oxidase, iron-sulfur subunit | 2.67 | 3.50 |
| PP_3746 | GlcE | Glycolate oxidase, putative FAD-binding subunit | 2.64 | 2.98 |
| PP_4922 | ThiC | Phosphomethylpyrimidine synthase | 2.21 | 3.84 |
| PP_3748 | GlcG | Conserved hypothetical protein | 2.06 | 3.71 |
| PP_3622 | Isoquinoline 1-oxidoreductase, beta subunit | 1.97 | 2.82 | |
| PP_3178 | GarK | Glycerate kinase | 1.77 | 2.85 |
| PP_3621 | IorA-II | Isoquinoline 1-oxidoreductase subunit alpha (2Fe-2S clusters) | 1.60 | 2.34 |
| PP_0554 | AcoB | Acetoin:2,6-dichlorophenolindophenol oxidoreductase subunit beta | 1.54 | 2.27 |
| PP_3623 | AdhB | Alcohol dehydrogenase cytochrome c subunit | 1.54 | 3.67 |
| PP_2484 | Transcriptional regulator, ArsR family | 1.52 | 2.37 | |
| PP_0734 | HemK | Release factor-(glutamine-N5) methyltransferase | 1.51 | 2.67 |
| PP_2439 | AhpC | Peroxiredoxin, alkylhydroperoxide reductase (small subunit) | 1.39 | 2.02 |
| PP_0556 | Acetoin catabolism protein | 1.35 | 2.34 | |
| PP_1125 | Putative helicase | 1.30 | 3.01 | |
| PP_0555 | AcoA | Acetoin:2,6-dichlorophenolindophenol oxidoreductase subunit alpha | 1.20 | 2.52 |
| PP_1548 | Unknown function | 1.19 | 2.03 | |
| PP_1351 | PanE | Putative 2-dehydropantoate 2-reductase | −1.45 | 2.06 |
| PP_2258 | Sensory box protein | −1.84 | 2.40 | |
| PP_5658 | Unknown function | −1.99 | 2.93 | |
| PP_3557 | Methyl-accepting chemotaxis transducer | −2.41 | 2.33 | |
| PP_3603 | Transcriptional regulator, GntR family | −2.46 | 2.73 | |
| PP_4313 | Putative peptidylprolyl isomerase | −2.55 | 2.50 | |
| PP_0588 | Putative copper-binding chaperone | −2.75 | 2.62 | |
| PP_2674 | PedE | Quinoprotein ethanol dehydrogenase | −4.25 | 3.78 |
| PP_2673 | Pentapeptide repeat family protein | −5.37 | 3.71 | |
| PP_3732 | Enoyl-CoA hydratase/isomerase family protein | −5.78 | 3.07 |
CoA, coenzyme A; FAD, flavin adenine dinucleotide.
During growth on 2-phenylethanol and glycerol, the majority of the identified proteins increased in abundance (80% and 70%, respectively) in response to La3+ (Table 1; see also Table S3). In contrast, most of the proteins identified during growth on glucose and citrate were found to be less abundant in response to La3+ (36% and 40%, respectively) (Table S4 and Table S5). Notably, most identified proteins were related to metabolism, according to the cluster of orthologous protein groups (COG) database (44). To test whether the conditional proteomic responses correlate with PedE and/or PedH activity, the catalytic activities of the purified proteins with all four carbon sources were determined (Table 2). Apart from the already known substrate 2-phenylethanol, PedE and PedH also showed activity with glycerol, whereas no activity could be detected with citrate or glucose.
TABLE 2.
Specific enzyme activities of purified PedE and PedH with the four tested growth substrates at 10 mM measured with 2,6-dichlorophenolindophenol (DCPIP)-dependent colorimetric assay
| Substrate | Mean specific activity (U mg−1) ± SDa |
|
|---|---|---|
| PedE (1 mM Ca2+) | PedH (1 μM La3+) | |
| Citrate | n.d.b | n.d.b |
| Glucose | n.d.b | n.d.b |
| Glycerol | 0.3 ± 0.1 | 0.9 ± 0.1 |
| 2-Phenylethanol | 8.0 ± 0.4 | 6.3 ± 0.3 |
Data represent averages of results from biological triplicates with standard deviations (SD).
Activities below detection limit are indicated (n.d.).
Effect of lanthanum during growth on glycerol.
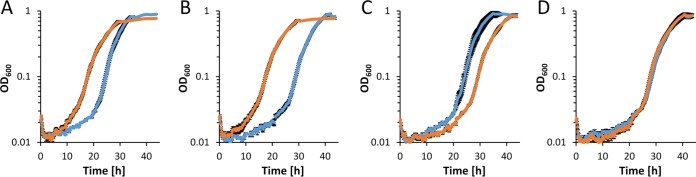
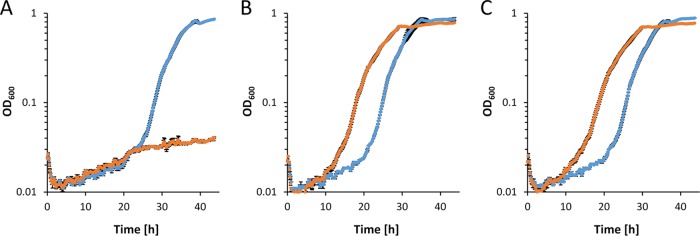
The proteomic and biochemical data indicate that the periplasmic oxidation system consisting of PedE and PedH could play a role in the metabolism of glycerol. As the degradation pathway and growth characteristics of P. putida have been recently characterized in great detail (45, 46), the effect of La3+ during growth on this specific substrate was examined. In experiments performed with the parental strain (Fig. 2A) (Table 3; see also Fig. S2), La3+ availability affected different physiological parameters. These parameters included the specific growth rates (maximum growth rate [μmax]; 0.277 ± 0.007 versus 0.351 ± 0.008 h−1), the maximal OD600 in stationary phase (OD600max; 0.771 ± 0.020 versus 0.894 ± 0.007), the CFU counts in stationary phase (1.1 ± 0.2 * 109 versus 1.6 ± 0.1 * 109 CFU/ml), and the lag phases (λ; 9.8 ± 0.2 versus 18.4 ± 0.3 h) of cultures. The observed decrease in the lag phase (λ) was of particular interest, as the purified PedH enzyme showed 3-fold higher specific activity than PedE for glycerol in vitro (0.9 ± 0.1 U mg−1 versus 0.3 ± 0.1 U mg−1; Table 2). Consequently, the differences in glycerol conversion rates of the two enzymes could be the underlying cause for the differences in the onset of logarithmic growth. To test this hypothesis, growth experiments with ΔpedE, ΔpedH, and ΔpedE ΔpedH strains were performed (Fig. 2B to D) (Table 3). In these experiments, the length of the lag phase correlated with the presence of functional PedE or PedH enzyme. Strains lacking the function of both enzymes (PedE and PedH), either due to gene deletions (strain ΔpedE ΔpedH) or due to combination of gene deletion and specific culture conditions (strain ΔpedE in the absence of La3+ and strain ΔpedH in the presence of La3+), exhibited similarly long lag phases of between 21.0 ± 0.2 and 21.8 ± 0.1 h. This was about twice as long as the lag phase observed for strains that can use the PedH enzyme (9.8 ± 0.2 and 10.5 ± 0.1 h for the parental strain and ΔpedE in the presence of La3+). Even though the difference was not as pronounced, it was also consistently longer than the lag phase observed for strains that could make use of the PedE enzyme (17.9 ± 0.3 to 18.4 ± 0.3 h for the ΔpedH strain and the parental strain in the absence of La3+).
FIG 2.
Growth of strains (A) KT2440*, (B) ΔpedE, (C) ΔpedH, and (D) ΔpedE ΔpedH on M9 minimal medium supplemented with 20 mM glycerol in the absence (blue dots) or presence (orange dots) of 10 μM La3+. Incubation was performed in 96-well microtiter plates in a microplate reader (Xenius; Safas, Monaco) at 30°C and 250 rpm. Data represent averages of results from biological triplicates with corresponding standard deviations.
TABLE 3.
Lag times (λ), maximal OD600 during stationary phase (OD600max), and maximal growth rates (μmax) of different P. putida strains during growth on M9 medium supplemented with 20 mM glycerol and 0 μM or 10 μM La3+ incubated in microtiter plates at 30°C and 250 rpm (see also Fig. 2 and Fig. 5)
| Strain | λ [h] ± SD |
OD600max ± SD |
μmax [h−1] ± SD |
|||
|---|---|---|---|---|---|---|
| 0 μM La3+ | 10 μM La3+ | 0 μM La3+ | 10 μM La3+ | 0 μM La3+ | 10 μM La3+ | |
| P. putida KT2440* | 18.4 ± 0.3 | 9.8 ± 0.2 | 0.894 ± 0.007 | 0.771 ± 0.020 | 0.351 ± 0.008 | 0.277 ± 0.007 |
| P. putida ΔpedE | 21.4 ± 0.4 | 10.5 ± 0.1 | 0.916 ± 0.007 | 0.774 ± 0.026 | 0.299 ± 0.008 | 0.289 ± 0.006 |
| P. putida ΔpedH | 17.9 ± 0.3 | 21.6 ± 0.5 | 0.849 ± 0.012 | 0.887 ± 0.001 | 0.352 ± 0.028 | 0.292 ± 0.006 |
| P. putida ΔpedE ΔpedH | 21.8 ± 0.1 | 21.0 ± 0.2 | 0.887 ± 0.001 | 0.880 ± 0.066 | 0.305 ± 0.006 | 0.315 ± 0.007 |
| P. putida ΔgarK | 21.4 ± 0.1 | n.d.a | 0.858 ± 0.005 | 0.042 ± 0.002 | 0.322 ± 0.004 | n.d.a |
| P. putida ΔcalA | 18.7 ± 0.5 | 10.6 ± 0.1 | 0.891 ± 0.001 | 0.783 ± 0.009 | 0.350 ± 0.013 | 0.307 ± 0.006 |
| P. putida ΔglcDEF | 19.6 ± 0.1 | 11.0 ± 0.1 | 0.885 ± 0.003 | 0.775 ± 0.006 | 0.335 ± 0.003 | 0.287 ± 0.001 |
No growth parameters were determined for cultures that did not reach stationary phase during incubation (n.d.).
(A) Growth of strain KT2440* in M9 medium supplemented with glycerol in the absence (blue points) or presence (red points) of 10 μM La3+. Cells were incubated in 96-well microtiter plates in a microplate reader (Xenius; Safas, Monaco) at 30°C and 250 rpm. (B) Upon 40 h of incubation (indicated with black arrow in panel A), OD600 (dark blue and dark orange bars) and CFU counts per milliliter (light blue and light orange bars) from incubations performed in the presence and absence of 10 μM La3+ were measured. Data represent averages of results from biological triplicates with corresponding standard deviations. Two asterisks (**; P < 0.01) and four asterisks (****; P < 0.0001) indicate statistically significant differences between conditions in the presence and absence of 10 μM La3+ in a two-tailed t test (n = 3). Download FIG S2, TIF file, 0.1 MB (138.2KB, tif) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
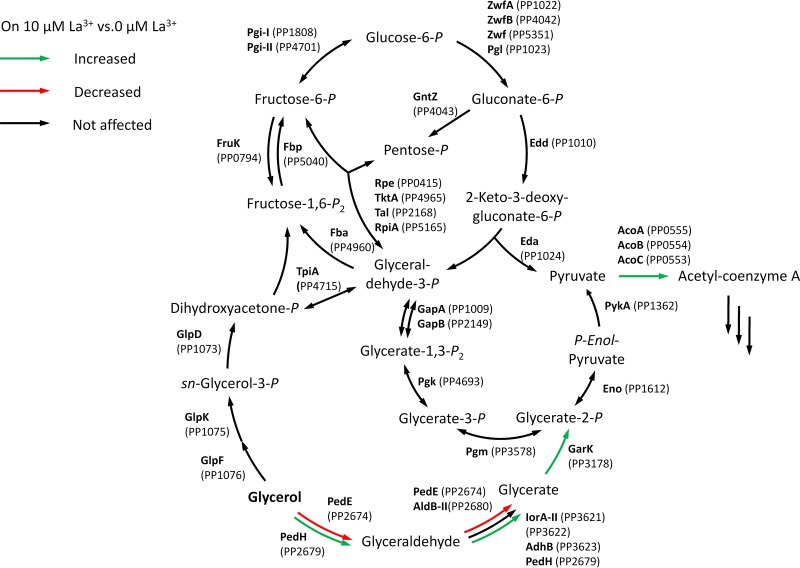
These results implied that the two PQQ-ADHs provide a benefit for growth on glycerol as the presence of functionally active PedH and/or PedE enzyme reduced the lag times of the cultures. Since PedE and PedH are not part of the described degradation pathway in P. putida KT2440 (45, 46), an additional metabolic route might exist (Fig. 3). On the basis of the proteomic data, PedE and/or PedH could initiate this route by oxidizing glycerol to glyceraldehyde. In the next steps, glyceraldehyde could be oxidized to glycerate by PedE and/or PedH, the aldehyde dehydrogenase AldB-II, or the aldehyde oxidase complex composed of proteins PP_3621 (IorA-II), PP_3622, and PP_3623 (AdhB). After phosphorylation by the glycerate kinase GarK, glycerate-2-phosphate could eventually enter the central metabolism.
FIG 3.
Metabolites and enzymes of the upstream central carbon metabolism of P. putida KT2440, including the proposed glycerol degradation pathway initiated by PQQ-ADH PedE or PedH or both. Enzymes involved in the specific metabolic steps that were differentially abundant in response to 10 μM La3+ during growth on glycerol are color coded (green = increased, red = decreased, black = not affected). The figure is inspired by a scheme originally published by Nikel et al. (46) and was redrawn to include the novel metabolic route(s) identified in this study.
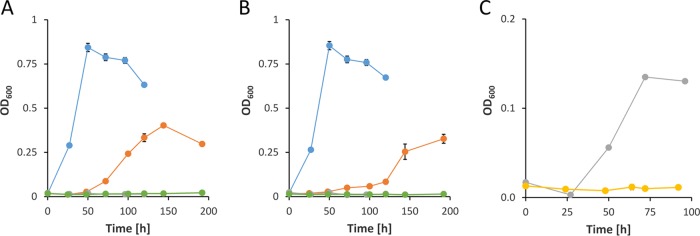
Given that such a metabolic route exists, a ΔglpFKRD deletion strain should still be able to grow on glycerol, whereas a ΔpedE ΔpedH ΔglpFKRD mutant and a ΔglpFKRD ΔgarK mutant should not (Fig. 3). Indeed, P. putida KT2440 grew on glycerol independently of the GlpFKRD pathway, although growth was dramatically impaired in comparison to that of the parental strain or strain ΔpedE ΔpedH (Fig. 2; see also Fig. 4A). Further, the ΔglpFKRD mutant strain also showed impaired growth in the presence of La3+ compared to conditions without La3+ (Fig. 4B). The additional deletion of pedE and pedH or garK in a ΔglpFKRD mutant led to no observable growth even after prolonged incubation times of ≥5 days. Ectopic complementation with either the pedE or the pedH gene from their native promoters successfully rescued growth of strain ΔpedE ΔpedH ΔglpFKRD on glycerol in the absence and presence of La3+ (Fig. 5). However, it has to be noted that growth of the pedE-complemented strain in the absence of La3+ appeared to be impaired in comparison to that of the ΔglpFKRD mutant (Fig. 4A and Fig. 5A). Due to the REE-dependent transcriptional regulation of PQQ-ADHs in P. putida KT2440 (12, 13), the pedE-complemented ΔpedE ΔpedH ΔglpFKRD strain did not grow in the presence of La3+ (Fig. 5B). In addition, pedH complementation did not restore growth in the absence of La3+ (Fig. 5A), presumably due to the metal cofactor dependency of the enzyme (12).
FIG 4.
(A and B) Growth of strains ΔpedE ΔpedH (blue dots), ΔglpFKRD (orange dots), ΔpedE ΔpedH ΔglpFKRD (gray dots), and ΔglpFKRD ΔgarK (green dots) in liquid M9 medium supplemented with 20 mM glycerol in the absence (A) or presence (B) of 10 μM La3+. (C) Growth of strains ΔpedE ΔpedH ΔglpFKRD (gray dots) and ΔgarK (yellow dots) in liquid M9 medium supplemented with 20 mM dl-glycerate in the absence of La3+. Incubation was performed in 96-well microtiter plates in a rotary shaker (Forma; Thermo Scientific) at 28°C and 220 rpm shaking. Data points represent averages of results from biological triplicates with the corresponding standard deviations.
FIG 5.
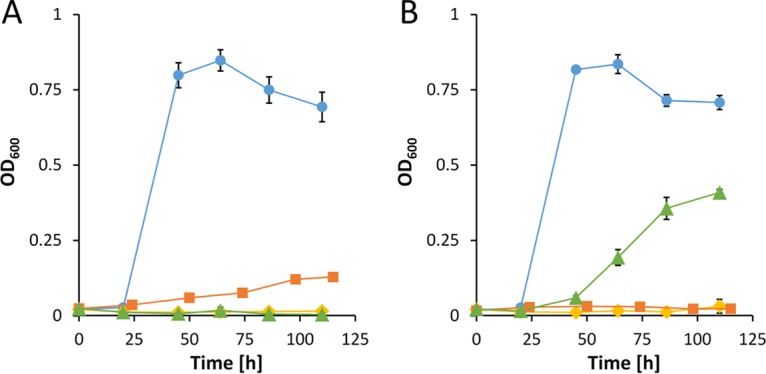
Growth of strains ΔpedE ΔpedH (blue circles), ΔpedE ΔpedH ΔglpFKRD (yellow diamonds), ΔpedE/H Δglp-Tn7M-pedE (orange squares), and ΔpedE/H Δglp-Tn7M-pedH (green triangles) in M9 minimal medium supplemented with 20 mM glycerol in the absence (A) or presence (B) of 10 μM La3+. Incubation was performed in 96-well microtiter plates in a rotary shaker (Forma; Thermo Scientific) at 28°C and 220 rpm. Data represent averages of results from biological triplicates with corresponding standard deviations.
These data are in support of the hypothesis that a metabolic route for glycerol exists, which is initiated by the PedE- and PedH-dependent oxidation of glycerol (Fig. 3). Given that the proposed route proceeds via glycerate and phosphorylation by GarK to glycerate-2-phosphate, a ΔgarK single-deletion strain should be incapable of channeling glycerate-2-phosphate into the central metabolism. Indeed, a ΔgarK mutant did not grow on glycerate even after incubation of up to 5 days, while strain ΔpedE ΔpedH ΔglpFKRD reached stationary phase within 72 h of incubation (Fig. 4C). In addition to the lack of growth on glycerate, growth of strain ΔgarK on glycerol in the presence of La3+ was also highly impaired with an OD600max of only 0.042 ± 0.002 after incubation for 44 h (Fig. 6A; see also Table 3). In the absence of La3+, the growth phenotype closely resembled that of the ΔpedE single mutant and the ΔpedE ΔpedH double mutant with λ = 21.4 ± 0.1 h, OD600max = 0.858 ± 0.005, and μmax = 0.322 ± 0.004 h−1.
FIG 6.
Growth of strains (A) ΔgarK, (B) ΔcalA, and (C) ΔglcDEF in M9 minimal medium supplemented with 20 mM glycerol in the absence (blue dots) or presence (orange dots) of 10 μM La3+. Incubation was performed in 96-well microtiter plates in a microplate reader (Xenius; Safas, Monaco) at 30°C and 250 rpm. Data represent averages of results from biological triplicates with corresponding standard deviations.
Notably, some of the proteins that were highly upregulated in response to La3+, namely, the multidrug efflux pump MexEF and the alkylhydroperoxide reductase subunits AhpC and AhpF, can be associated with stress responses. In addition, the predicted coniferyl-alcohol dehydrogenase CalA and the glycolate oxidase GlcDEF were also found to be upregulated in the presence of La3+. However, the corresponding ΔcalA and ΔglcDEF mutant strains showed no obvious differences in their growth patterns in the presence and the absence of La3+ compared to the parental strain (Fig. 6B and C; see also Table 3).
DISCUSSION
The present study investigated the cellular response of P. putida to La3+ availability during growth on several carbon sources. The only protein that showed a differential abundance independently of the substrate used for growth was the Ca2+-dependent PQQ-ADH PedE. This result is in line with data from our previous study (13), which demonstrated that the La3+-induced downregulation of pedE is dependent on the PedS2/PedR2 two-component system. It further indicates that this regulatory system is functional under all tested conditions. The only other two proteins (PedH and PP_2673) that showed differential abundance under more than one set of culture conditions are encoded by genes within the ped gene cluster, which encodes several crucial elements for regulation and function of PedE and PedH (13, 43, 47). Notably, the carbon sources provided under the conditions in which these proteins were identified either represent substrates of PedE and PedH (2-phenylethanol and glycerol) or can be converted by an enzyme depending on the same PQQ cofactor, namely, the glucose dehydrogenase Gcd (glucose). The remaining 53 proteins with differential abundance in the presence of La3+ were identified only during growth on one substrate, indicating a conditional response.
In P. putida, the known pathway of glycerol degradation starts with its uptake via GlpF, phosphorylation by GlpK, and subsequent GlpD-catalyzed oxidation of glycerol-3-phosphate to dihydroxyacetone-3-phosphate (46). In the next step, dihydroxyacetone-3-phosphate is converted to glyceraldehyde-3-phosphate and enters the central metabolism. Here, we identified a novel route for glycerol metabolism. This pathway is initiated by PedE and/or PedH, depending on La3+ availability (Fig. 3). Whether the two PQQ-ADHs oxidize glycerol only to glyceraldehyde or perform a second oxidation step to produce glycerate is currently unknown. However, it is worth noting that other enzymes can potentially also carry out the oxidation from glyceraldehyde to glycerate, such as the 3-hydroxybutyrate dehydrogenase (48), the aldehyde dehydrogenase AldB-II, or the aldehyde oxidase complex composed of proteins PP_3621 (IorA-II), PP_3622, and PP_3623 (AdhB). Interestingly, the abundance of the latter two enzymes was increased during growth in the presence of La3+.
Upon phosphorylation by GarK, glycerate eventually enters the central metabolism, presumably as glycerate-2-phosphate. This conclusion is supported by the lack of growth of the ΔglpFKRD ΔgarK mutant and the results of a companion study (48), which used a comparative metabolome analysis of glycerol-growing cells of P. putida KT2440 and a ΔglpK deletion strain. The study found a dramatically increased glycerate concentration for the ΔglpK strain, whereas the concentrations of glyceraldehyde and glyceraldehyde-3-phosphate were in similar ranges for the two strains.
The PedE/PedH-dependent route is clearly not the main route for glycerol metabolism, as the growth on glycerol was more extensively affected by the deletion of the glpFKRD gene cluster than by the double deletion of pedE and pedH. However, the periplasmic oxidation system represents an important benefit for cells during growth on glycerol as it strongly reduces the lag phase, especially in the presence of La3+. Earlier studies showed that the long lag times observed during growth on glycerol are linked to the repression of the glpFKRD operon by the transcriptional regulator GlpR and argued that derepression of the glpFKRD operon depends on the intracellular concentration of glycerol-3-phosphate (45). As such, one could speculate that in addition to glycerol-3-phosphate, other phosphorylated glycerol derivatives, such as glycerate-2-phosphate derived from the PedE/PedH-GarK-dependent metabolic route, are also able to relieve the repression of glpFKRD by GlpR. An explanation for the La3+-dependent differences in lag times could hence be that these phosphorylated derivatives accumulate more rapidly in the presence of La3+. Whether this is the result of the higher specific activity of PedH than of PedE or is based on different expression levels of these and/or other enzymes involved in the formation of the phosphorylated derivatives is currently unknown.
Beside its positive effect on the lag times, growth in the presence of La3+ also correlates with reduced OD600max values for strains capable of utilizing PedH for glycerol oxidation. This observation might be a consequence of the higher specific activity or the higher protein production level of PedH compared to PedE, too. Cells using PedH for glycerol metabolism would produce more of the toxic intermediate glyceraldehyde leading to increased stress and as a consequence to lower optical densities and CFU counts (49, 50). This toxicity effect would further explain the elevated growth impairment of the ΔgarK mutant in the presence of La3+, as one can assume that even higher concentrations of glyceraldehyde accumulate in a mutant that is hampered in glycerate metabolism. Overall, the physiological changes in response to La3+ availability seem to represent a trade-off between an earlier onset of growth and lower absolute cell numbers (decreased OD600max and CFU counts). The notion that the RND-type transporter proteins MexEF, which are involved in efflux of various toxic compounds (51), and the alkylhydroperoxide reductase subunits AhpC and AhpF, which have been linked to ROS detoxification in P. putida (52), were also more abundant in the presence of La3+ during growth on glycerol is supportive of such a hypothesis.
The specific catalytic activities and/or production levels of PedH and PedE could also explain the proteomic differences observed during growth on other carbon sources that are known to be substrates for these enzymes, such as 2-phenylethanol. However, this reasoning fails to explain the differences during growth on glucose and citrate, as those two carbon sources do not represent substrates for both enzymes. Despite the fact that the underlying causes of the conditional proteomic changes seen under those conditions are still unknown, the results demonstrate that beside the interaction with PedH and PedS2, additional effects of REEs in P. putida cells do exist. Such effects could include the inhibition of protein functions by mismetallation (53, 54), changes in the physiology of the outer membrane (55), or the presence of thus far unknown REE-dependent enzymes and/or proteins with regulatory function. The latter explanation is of particular interest, as REE-specific import systems have recently been identified in different species, including P. putida KT2440 (41, 43, 56–58).
Altogether, the presented work demonstrates that during growth on glycerol, the activity of the periplasmic oxidation system leads to reduced lag times. This growth advantage is more pronounced in the presence of La3+ and might represent a competitive advantage in REE-rich environments such as the rhizosphere. Hence, the previously reported fertilizing effect of REEs on different food crops (34–38) could be a consequence of increased competitiveness of plant growth-promoting organisms such as P. putida during root colonization. This hypothesis is supported by a recent study which found that pseudomonads thrive predominantly on root exudates in vivo and are hence enriched in the rhizosphere of Arabidopsis thaliana (10). As such, it will be interesting to see what future research will add to the currently emerging theme of REEs being important micronutrients for methylotrophic and nonmethylotrophic organisms, in particular in regard to bacterium-plant interactions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Complete lists of all strains, plasmids, and primers used in this study can be found in Table S1 and Table S2 in the supplemental material. All Pseudomonas putida and Escherichia coli strains were stored at –80°C as glycerol stocks (30% glycerol [vol/vol]). For experiments, the bacterial stocks were used to inoculate LB-agar plates (59), which were stored at 4°C after an initial growth phase at 30°C overnight. Strains were maintained on LB-agar plates by regular transfer to new plates for up to 6 weeks. If necessary, 40 μg/ml kanamycin (Kan), 30 μg/ml gentamicin, or 20 μg/ml 5-fluorouracil (5-FU) was added for maintenance and/or selection. For growth, liquid LB medium or a modified M9 salt medium (12) supplemented with 5 mM 2-phenylethanol, 25 mM succinate, 10 mM glucose, 10 mM citrate, 20 mM dl-glycerate, or 20 mM glycerol as the sole source of carbon and energy was used. If not stated otherwise, precultures were grown overnight in test tubes with 3 ml M9 medium supplemented with succinate at 30°C and 180 rpm. The next day, cells were washed twice with M9 medium without supplemented C-source and were used to inoculate 200 μl of M9 medium supplemented with the desired C-source in a 96-well microtiter plate (Falcon, product no. 353047, or Sarstedt, product no. 83.3924) to an initial OD600 of 0.02. Cell cultures were incubated at 30°C and 250 rpm in a microplate reader (Xenius; Safas, Monaco) or, for long-term incubations (>4 days), at 28°C and 220 rpm in a rotary shaker (Forma; Thermo Scientific).
Strains and plasmids used in the study. Download Table S1, DOCX file, 0.03 MB (31.6KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in the study. Download Table S2, DOCX file, 0.02 MB (23.3KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ when grown on 2-phenylethanol. Download Table S3, DOCX file, 0.02 MB (23.8KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ when grown on glucose. Download Table S4, DOCX file, 0.02 MB (24.1KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ when grown on citrate. Download Table S5, DOCX file, 0.02 MB (23.5KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Calculation of growth parameters.
Maximum growth rates (μmax) and lag times (λ), defined as the intersection point between a horizontal line through the y-axis value at t = t0 and the tangent at the inclination point (the point of μmax), were derived by fitting the natural logarithm of the relative OD600 values [ln(N/N0), with N being the OD600 at time t] to the Richards growth model using the “grofit” package in R (60, 61). As the OD600 decreased directly upon beginning the experiment, t4 h was used as t0 and ln(N/Nt = 4 h) was used instead of ln(N/N0) for a better fit.
Measurement of CFU levels.
For CFU counts, cultures of KT2440* were grown in triplicate in a 96-well microplate in M9 medium with glycerol in the presence or absence of 10 μM La3+. Plates were incubated at 30°C and 250 rpm for 40 h. Samples were serially diluted in M9 medium with no added C-source, and 200-μl volumes of 10−6 dilutions were plated on LB agar plates, which were incubated at 30°C overnight. For statistical analysis, two-tailed t tests (n = 3, α = 0.05) were performed using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA).
Construction of plasmids.
Deletion plasmids pJOE-calA, pJOE-garK, pJOE-glp, and pMW08 were constructed as follows. The 650-bp to 1,000-bp regions upstream and downstream of the calA (PP_2426), garK (PP_3178), glpFKRD (PP_1076 to PP_1973), and glcDEF (PP_3745 to PP_3747) genes were amplified from genomic DNA of P. putida KT2440 using primer pairs PcalA1/2 and PcalA3/4, PgarK1/2 and PgarK3/4, Pglp1/2 and Pglp3/4, and MWH03/04 and MWH05/06, respectively (Table S2). The two upstream and downstream fragments and BamHI-digested pJOE6261.2 were then joined together using one-step isothermal assembly (62).
For complementation of the ΔpedE and ΔpedH mutations, DNA fragments comprising the complete pedE and pedH open reading frames and their respective promoters were amplified from KT2440 genomic DNA with primer pairs P2674-FSac/P2674-RHind and P2679-FSac/P2674-RHind, respectively, and cloned into pTn7-M as SacI-HindIII fragments to yield the complementation plasmids pTn7M-pedE and pTn7M-pedH.
Construction of strains.
Deletion mutant strains were constructed as previously described (63). Briefly, integration vector pJOE6261.2 harboring the upstream and downstream regions of the target gene(s) was transformed into P. putida KT2440 Δupp (KT2440*). Kanamycin (Kan)-resistant (Kanr) and 5-fluorouracil (5-FU)-sensitive (5-FUs) clones were selected, and one of these was incubated in LB medium at 30°C for 24 h. The cell suspension was then plated on M9 minimal agar plates containing 25 mM succinate and 20 μg ml−1 5-FU. Clones that carried the desired gene deletion were identified by colony PCR of the 5-FUr Kans clones using primer pair PcalA1/PcalA4, PgarK1/PgarK4, Pglp1/Pglp4, or MWH03/MWH06.
Delivery of the pTn7-M-based constructs into P. putida KT2440 was performed by tetraparental mating using PIR2/pTn7M-pedH or PIR2/pTn7M-pedE as the donor, E. coli CC118 λpir/pTNS1 and E. coli HB101/pRK600 as helper strains, and the appropriate KT2440 strain as the recipient (64). Briefly, cultures of the four strains grown under selective conditions were mixed, spotted on LB agar, and incubated overnight at 28°C. Transconjugants were selected on cetrimide agar (Sigma-Aldrich) containing gentamicin. Proper chromosomal insertion into the Tn7 att site was checked by colony PCR using Pput-glmSDN and PTn7R primers as described elsewhere (65).
Protein extraction for comparative proteome analysis.
For comparative proteome analysis, 50 ml M9 medium supplemented with citrate, glucose, glycerol, or 2-phenylethanol and 0 or 10 μM LaCl3 was inoculated with an OD600 of 0.05 from succinate precultures of strain P. putida KT2440* in 250-ml polycarbonate Erlenmeyer flasks and incubated at 30°C and 180 rpm. When cell cultures reached 0.4 < OD600 < 0.6, cells were harvested by centrifugation for 15 min at 6,000 × g and 4°C. Cell pellets were resuspended in 1 ml sample buffer (150 mM Tris-HCl [pH 6.8]; 2% SDS; 20 mM dithiothreitol [DTT]) and heated for 5 min at 95°C with gentle shaking. Subsequently, samples were centrifuged for 15 min at 21,000 × g and 4°C, and the supernatants were stored in new reaction tubes at –20°C. In the next step, proteins were precipitated using chloroform-methanol (66) and pellets were resuspended in Tris-buffered (50 mM, pH 8.5) urea (6 M). Protein concentrations were determined by the Bradford assay (67).
In-solution digestion of proteins and peptide purification with C18 stage tips.
To 25 μg protein in 60 μl Tris-buffered (50 mM, pH 8.5) urea (6 M), DTT was added to reach a final concentration of 10 mM to guarantee reduction of cysteines. Samples were incubated for 30 min at 56°C with shaking at 1,000 rpm. Alkylation of cysteines was performed by adding 30 mM iodoacetamide and incubating for 45 min at room temperature (RT) in the dark. Alkylation was stopped by adding 50 mM DTT, and samples were incubated for another 10 min at RT. A mixture of 500 ng LysC protease (Roche)–50 mM Tris buffer (pH 8.5) was added, and samples were digested overnight at 30°C. Next, the urea in the reaction mixture was diluted to 2 M by adding the appropriate amount of Tris buffer (50 mM, pH 8.5). A mixture of 1 μg trypsin (Roche)–Tris buffer (50 mM, pH 8.5) was added, and digestion was continued for 4 h at 37°C. The digestion was stopped by addition of 3 μl 10% (vol/vol) trifluoroacetic acid (TFA). Next, peptide mixtures were concentrated and desalted on C18 stage tips (68) and dried under vacuum. Samples were dissolved in 20 μl 0.1% (vol/vol) TFA. Aliquots of 1 μl were subjected to nano-LC-MS/MS analysis.
Mass spectrometry analysis.
Experiments employing nano-LC-ESI-MS/MS were performed on an EASY-nLC 1200 system (Thermo Fisher Scientific) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) using an EASY-Spray nanoelectrospray ion source (Thermo Fisher Scientific). Tryptic peptides were directly injected into an EASY-Spray analytical column (PepMapRSLC C18; Thermo Fisher Scientific) (2-μm pore size, 100 Å, 25 cm by 75 μm) operated at a constant temperature of 35°C. Peptides were separated at a flow rate of 250 nl/min using a 240-min gradient with the following profile: 2% to 10% solvent B for 100 min, 10% to 22% solvent B for 80 min, 22% to 45% solvent B for 55 min, 45% to 95% solvent B for 5 min, and isocratic conditions at 90% solvent B for 15 min. The solvents used were 0.5% acetic acid (solvent A) and 0.5% acetic acid–acetonitrile/H2O (80/20 [vol/vol]) (solvent B). The Q Exactive Plus mass spectrometer was operated under the control of XCalibur 3.0.63 software. MS spectra (m/z = 300 to 1,600) were detected in an Orbitrap analyzer at a resolution of 70,000 (m/z = 200) using a maximum injection time (MIT) of 100 ms and an automatic gain control (AGC) value of 1 × 106. Internal calibration of the Orbitrap analyzer was performed using lock-mass ions from ambient air as described elsewhere (69). Data-dependent MS/MS spectra were generated for the 10 most abundant peptide precursors in the Orbitrap analyzer using high-energy collision dissociation (HCD) fragmentation at a resolution of 17,500, a normalized collision energy value of 27, and an intensity threshold of 1.3 × 105. Only ions with charge states from +2 to +5 were selected for fragmentation using an isolation width of 1.6 Da. For each MS/MS scan, the AGC was set at 5 × 105 and the MIT was 100 ms. Fragmented precursor ions were dynamically excluded for 30 s within a 5-ppm mass window to avoid repeated fragmentation.
Protein quantification and data analysis.
Raw files were imported into MaxQuant (70) version 1.6.0.1 for protein identification and label-free quantification (LFQ) of proteins. Protein identification in MaxQuant was performed using the database search engine Andromeda (71). MS spectra and MS/MS spectra were searched against the P. putida KT2440 protein sequence database downloaded from UniProt (72). Reversed sequences as decoy database and common contaminant sequences were added automatically by MaxQuant. Mass tolerances of 4.5 ppm for MS spectra and 20 ppm for MS/MS spectra were used. Trypsin was specified as the enzyme, and two missed cleavages were allowed. Carbamidomethylation of cysteines was set as a fixed modification, and protein N-terminal acetylation and oxidation were allowed as variable modifications. The “match between runs” feature of MaxQuant was enabled with a match time window of 1 min and an alignment time window of 20 min. Peptide false-discovery-rate (FDR) and protein FDR thresholds were set to 0.01.
Statistical analyses, including t tests and principal-component analysis (PCA), were performed using Perseus software version 1.6.0.2 (73). Matches to contaminants (e.g., keratins, trypsin) and reverse databases identified by MaxQuant were excluded from further analysis. Proteins were considered for LFQ (label-free quantification) if they were identified by at least two peptides. First, normalized LFQ values from MaxQuant were log2 transformed. Missing values were replaced from normal distributions using a width value of 0.2 and a downshift value of 2.0. Statistically significant differences between two sample groups were determined using an unpaired t test, and a P value of <0.01 and a regulation factor of >2 (log2 fold change > 1) were considered representative of a significant change in protein abundance.
Purification and activity measurement of PQQ-ADHs PedE and PedH.
To measure the activity of the two PQQ-ADHs, PedE and PedH, the enzymes were expressed in E. coli BL21(DE3) cells using plasmids pMW09 and pMW10 and were purified by affinity chromatography as described elsewhere (12). The activities occurring with the four substrates 2-phenylethanol, citrate, glucose, and glycerol were determined at a concentration of 10 mM using a previously described colorimetric assay (12) with one minor modification. To represent the growth conditions, 1 μM La3+ instead of 1 μM Pr3+ was used as the metal cofactor for PedH.
Data availability.
The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the PRIDE (74) partner repository (identifier [ID]: PXD013011).
ACKNOWLEDGMENTS
The work of M.W. and J.K. was supported by an individual research grant from the Deutsche Forschungsgemeinschaft (DFG; KL 2340/2-1). The work of M.T. and P.B. was supported by Labex Ressources21 (ANR-10-LABX-21-01). M.W. and J.K. thank Bernhard Hauer for his continuous support.
We further declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Citation Wehrmann M, Toussaint M, Pfannstiel J, Billard P, Klebensberger J. 2020. The cellular response to lanthanum is substrate specific and reveals a novel route for glycerol metabolism in Pseudomonas putida KT2440. mBio 11:e00516-20. https://doi.org/10.1128/mBio.00516-20.
REFERENCES
- 1.Mendes R, Garbeva P, Raaijmakers J. 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 2.Raaijmakers J, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y. 2009. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361. doi: 10.1007/s11104-008-9568-6. [DOI] [Google Scholar]
- 3.Berg G, Roskot N, Steidle A, Eberl L, Zock A, Smalla K. 2002. Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl Environ Microbiol 68:3328–3338. doi: 10.1128/AEM.68.7.3328-3338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalhais LC, Dennis PG, Fedoseyenko D, Hajirezaei MR, Borriss R, Von Wirén N. 2011. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. Z Pflanzenernähr Bodenk 174:3–11. doi: 10.1002/jpln.201000085. [DOI] [Google Scholar]
- 5.Dakora FD, Phillips DA. 2002. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245:35–47. doi: 10.1023/A:1020809400075. [DOI] [Google Scholar]
- 6.Schmidt R, Cordovez V, de Boer W, Raaijmakers J, Garbeva P. 2015. Volatile affairs in microbial interactions. ISME J 9:2329–2335. doi: 10.1038/ismej.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Insam H, Seewald M. 2010. Volatile organic compounds (VOCs) in soils. Biol Fertil Soils 46:199–213. doi: 10.1007/s00374-010-0442-3. [DOI] [Google Scholar]
- 8.Wackett LP. 2003. Pseudomonas putida—a versatile biocatalyst. Nat Biotechnol 21:136–138. doi: 10.1038/nbt0203-136. [DOI] [PubMed] [Google Scholar]
- 9.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP, Fouts DE, Gill R, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Lee PC, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen J, Timmis KN, Dusterhoft A, Tummler B, Fraser CM. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 10.Worsley SF, Macey MC, Newitt JT, Patrick E, Yu DW, Wilkinson B, Murrell C, Hutchings MI. 2019. Investigating the role of exudates in recruiting Streptomyces bacteria to the Arabidopsis thaliana root microbiome. bioRxiv doi: 10.1101/532309. [DOI] [PMC free article] [PubMed]
- 11.Mückschel B, Simon O, Klebensberger J, Graf N, Rosche B, Altenbuchner J, Pfannstiel J, Huber A, Hauer B. 2012. Ethylene glycol metabolism by Pseudomonas putida. Appl Environ Microbiol 78:8531–8539. doi: 10.1128/AEM.02062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. 2017. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio 8:e00570-17. doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wehrmann M, Berthelot C, Billard P, Klebensberger J. 2018. The PedS2/PedR2 two-component system is crucial for the rare earth element switch in Pseudomonas putida KT2440. mSphere 3:e00376-18. doi: 10.1128/mSphere.00376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, Matsumura H, Ishida T, Samejima M, Igarashi K, Nakamura N, Ohno H. 2013. The two-step electrochemical oxidation of alcohols using a novel recombinant PQQ alcohol dehydrogenase as a catalyst for a bioanode. Bioelectrochemistry 94:75–78. doi: 10.1016/j.bioelechem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Firsching FH, Brune SN. 1991. Solubility products of the trivalent rare-earth phosphates. J Chem Eng Data 36:93–95. doi: 10.1021/je00001a028. [DOI] [Google Scholar]
- 16.Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp H. 2014. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 17.Hibi Y, Asai K, Arafuka H, Hamajima M, Iwama T, Kawai K. 2011. Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J Biosci Bioeng 111:547–549. doi: 10.1016/j.jbiosc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Fitriyanto NA, Fushimi M, Matsunaga M, Pertiwiningrum A, Iwama T, Kawai K. 2011. Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111:613–617. doi: 10.1016/j.jbiosc.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Good NM, Vu HN, Suriano CJ, Subuyuj GA, Skovran E, Martinez-Gomez NC. 2016. Pyrroloquinoline quinone ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multicarbon substrates. J Bacteriol 198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez NC, Skovran E. 2016. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol 198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, Tani A. 2018. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere 3:e00462-17. doi: 10.1128/mSphere.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chistoserdova L. 2016. Lanthanides: new life metals? World J Microbiol Biotechnol 32:138. doi: 10.1007/s11274-016-2088-2. [DOI] [PubMed] [Google Scholar]
- 23.Keltjens JT, Pol A, Reimann J, Op den Camp H. 2014. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 24.Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF, Giovannoni SJ. 2011. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5:856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taubert M, Grob C, Howat AM, Burns OJ, Dixon JL, Chen Y, Murrell JC. 2015. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ Microbiol 17:3937–3948. doi: 10.1111/1462-2920.12896. [DOI] [PubMed] [Google Scholar]
- 26.Lv H, Sahin N, Tani A. 2018. Isolation and genomic characterization of Novimethylophilus kurashikiensis gen. nov. sp. nov., a new lanthanide-dependent methylotrophic species of Methylophilaceae. Environ Microbiol 20:1204–1223. doi: 10.1111/1462-2920.14062. [DOI] [PubMed] [Google Scholar]
- 27.Cotruvo JA, Featherston ER, Mattocks JA, Ho JV, Laremore TN. 2018. Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J Am Chem Soc 140:15056–15061. doi: 10.1021/jacs.8b09842. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Semrau JD. 2017. Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 101:8499–8516. doi: 10.1007/s00253-017-8572-2. [DOI] [PubMed] [Google Scholar]
- 29.Good NM, Walser ON, Moore RS, Suriano CJ, Huff AF, Martínez-Gómez NC. 2018. Investigation of lanthanide-dependent methylotrophy uncovers complementary roles for alcohol dehydrogenase enzymes. bioRxiv doi: 10.1101/329011. [DOI]
- 30.Fitriyanto NA, Nakamura M, Muto S, Kato K, Yabe T, Iwama T, Kawai K, Pertiwiningrum A. 2011. Ce3+-induced exopolysaccharide production by Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111:146–152. doi: 10.1016/j.jbiosc.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, Huang J, Zhao F, Chistoserdova L. 2018. Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy. mBio 9:e02430-17. doi: 10.1128/mBio.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause SMB, Johnson T, Samadhi Karunaratne Y, Fu Y, Beck DAC, Chistoserdova L, Lidstrom ME. 2017. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci U S A 114:358–363. doi: 10.1073/pnas.1619871114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akberdin IR, Collins DA, Hamilton R, Oshchepkov DY, Shukla AK, Nicora CD, Nakayasu ES, Adkins JN, Kalyuzhnaya MG. 27 November 2018, posting date Rare earth elements alter redox balance in Methylomicrobium alcaliphilum 20ZR. Front Microbiol doi: 10.3389/fmicb.2018.02735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diatloff E, Smith FW, Asher CJ. 1995. Rare earth elements and plant growth: III. Responses of corn and mungbean to low concentrations of cerium in dilute, continuously flowing nutrient solutions. J Plant Nutr 18:1991–2003. doi: 10.1080/01904169509365039. [DOI] [Google Scholar]
- 35.Diatloff E, Asher CJ, Smith FW. 1996. Rare earth elements and plant growth: I. Effects of lanthanum and cerium on root elongation of corn and mungbean. J Plant Nutr 18:1963–1976. doi: 10.1080/01904169509365037. [DOI] [Google Scholar]
- 36.Pang X, Li D, Peng A. 2002. Application of rare-earth elements in the agriculture of China and its environmental behavior in soil. Environ Sci Pollut Res 9:143–148. doi: 10.1007/BF02987462. [DOI] [PubMed] [Google Scholar]
- 37.Wahid PA, Valiathan MS, Kamalam NV, Eapen JT, Vijayalakshmi S, Prabhu RK, Mahalingam TR. 2000. Effect of rare earth elements on growth and nutrition of coconut palm and root competition for these elements between the palm and Calotropis gigantea. J Plant Nutr 23:329–338. doi: 10.1080/01904160009382019. [DOI] [Google Scholar]
- 38.Xie ZB, Zhu JG, Chu HY, Zhang YL, Zeng Q, Ma HL, Cao ZH. 2002. Effect of lanthanum on rice production, nutrient uptake, and distribution. J Plant Nutr 25:2315–2331. doi: 10.1081/PLN-120014078. [DOI] [Google Scholar]
- 39.Farhan Ul Haque M, Kalidass B, Bandow N, Turpin EA, DiSpirito AA, Semrau JD. 2015. Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl Environ Microbiol 81:7546–7552. doi: 10.1128/AEM.02542-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu F, Beck DAC, Lidstrom ME. 2016. MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense. PeerJ 4:e2435. doi: 10.7717/peerj.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochsner AM, Hemmerle L, Vonderach T, Nüssli R, Bortfeld-Miller M, Hattendorf B, Vorholt JA. 2019. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol 111:1152–1166. doi: 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu F, Lidstrom ME. 2016. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol 198:1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wehrmann M, Berthelot C, Billard P, Klebensberger J. 2019. Rare earth element (REE)-dependent growth of Pseudomonas putida KT2440 relies on the ABC-transporter PedA1A2BC and is influenced by iron availability. Front Microbiol 10:670216. doi: 10.3389/fmicb.2019.02494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikel PI, Romero-Campero FJ, Zeidman JA, Goñi-Moreno Á, de Lorenzo V. 2015. The glycerol-dependent metabolic persistence of Pseudomonas putida KT2440 reflects the regulatory logic of the GlpR repressor. mBio 6:e00340-15. doi: 10.1128/mBio.00340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikel PI, Kim J, de Lorenzo V. 2014. Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ Microbiol 16:239–254. doi: 10.1111/1462-2920.12224. [DOI] [PubMed] [Google Scholar]
- 47.Arias S, Olivera ER, Arcos M, Naharro G, Luengo JM. 2008. Genetic analyses and molecular characterization of the pathways involved in the conversion of 2-phenylethylamine and 2-phenylethanol into phenylacetic acid in Pseudomonas putida U. Environ Microbiol 10:413–432. doi: 10.1111/j.1462-2920.2007.01464.x. [DOI] [PubMed] [Google Scholar]
- 48.Walsh M, Casey W, Kenny ST, Narancic T, Blank LM, Wierckx N, Ballerstedt H, Connor K. 2019. Insights into an alternative pathway for glycerol metabolism in a glycerol kinase deficient Pseudomonas putida KT2440. bioRxiv doi: 10.1101/567230. [DOI]
- 49.Tessier FJ, Monnier VM, Sayre LM, Kornfield JA. 2003. Triosidines: novel Maillard reaction products and cross-links from the reaction of triose sugars with lysine and arginine residues. Biochem J 369:705–719. doi: 10.1042/bj20020668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benov L, Beema AF. 2003. Superoxide-dependence of the short chain sugars-induced mutagenesis. Free Radic Biol Med 34:429–433. doi: 10.1016/S0891-5849(02)01331-X. [DOI] [PubMed] [Google Scholar]
- 51.Li XZ, Zhang L, Poole K. 1998. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol 180:2987–2991. doi: 10.1128/JB.180.11.2987-2991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hishinuma S, Yuki M, Fujimura M, Fukumori F. 2006. OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ Microbiol 8:2115–2124. doi: 10.1111/j.1462-2920.2006.01088.x. [DOI] [PubMed] [Google Scholar]
- 53.Agarwal N, Kalra VK. 1983. Interaction of lanthanide cations and uranyl ion with the calcium/proton antiport system in Mycobacterium phlei. Biochim Biophys Acta 727:285–292. doi: 10.1016/0005-2736(83)90414-5. [DOI] [PubMed] [Google Scholar]
- 54.Brown PH, Rathjen AH, Graham RD, Tribe DE. 1990. Chapter 92 rare earth elements in biological systems, p 423–452. In Handbook on the physics and chemistry of rare earths, vol 11 https://www.sciencedirect.com/science/article/pii/S0168127305801357. [Google Scholar]
- 55.Peng L, Yi L, Zhexue L, Juncheng Z, Jiaxin D, Daiwen P, Ping S, Songsheng Q. 2004. Study on biological effect of La3+ on Escherichia coli by atomic force microscopy. J Inorg Biochem 98:68–72. doi: 10.1016/j.jinorgbio.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Mattocks JA, Ho JV, Cotruvo JA. 2019. A selective, protein-based fluorescent sensor with picomolar affinity for rare earth elements. J Am Chem Soc 141:2857–2861. doi: 10.1021/jacs.8b12155. [DOI] [PubMed] [Google Scholar]
- 57.Roszczenko-Jasińska P, Vu HN, Subuyuj GA, Crisostomo RV, Ayala EM, Cai J, Lien NF, Clippard EJ, Ngo RT, Yarza F, Wingett JP, Raghuraman C, Hoeber CA, Martinez-Gomez NC, Skovran E. 2020. Lanthanide transport, storage, and beyond: gene products and processes contributing to lanthanide and methanol metabolism in Methylorubrum extorquens AM1. bioRxiv doi: 10.1101/647677. [DOI] [PMC free article] [PubMed]
- 58.Groom J, Ford SM, Pesesky MW, Lidstrom ME. 2019. A mutagenic screen identifies a TonB-dependent receptor required for the lanthanide metal switch in the type I methanotroph “Methylotuvimicrobium buryatense” 5GB1C. J Bacteriol 201:e00120-19. doi: 10.1128/JB.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maniatis T, Sambrook J, Fritsch E. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 60.Kahm M, Kschischo M, Ludwig J, Lichtenberg-Fraté H, Hasenbrink G. 2015. grofit: fitting biological growth curves with R. J Stat Softw 33(7):1–21. 10.18637/jss.v033.i07. [DOI] [Google Scholar]
- 61.Richards FJ. 1959. A flexible growth function for empirical use. J Exp Bot 10:290–301. doi: 10.1093/jxb/10.2.290. [DOI] [Google Scholar]
- 62.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graf N, Altenbuchner J. 2011. Development of a method for markerless gene deletion in Pseudomonas putida. Appl Environ Microbiol 77:5549–5552. doi: 10.1128/AEM.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zobel S, Benedetti I, Eisenbach L, de Lorenzo V, Wierckx N, Blank LM. 2015. Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth Biol 4:1341–1351. doi: 10.1021/acssynbio.5b00058. [DOI] [PubMed] [Google Scholar]
- 65.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 66.Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 67.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 68.Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 69.de Godoy LMF, Mortensen P, Makarov A, Li G, Macek B, Horning S, Pesch R, Mann M, Olsen JV, Lange O. 2005. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 71.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 72.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 74.Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCA comparing the four different carbon sources with and without La3+. Different carbon sources are indicated by squares (2-phenylethanol), circles (citrate), diamonds (glucose), and stars (glycerol). Samples with 10 μM La3+ and without La3+ in the medium are shown in blue and red, respectively. Biological replicates are indicated in the same color. Samples were separated according to the different carbon sources, while treatment with La3+ showed only minor effects. Download FIG S1, JPG file, 0.02 MB (20.1KB, jpg) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Growth of strain KT2440* in M9 medium supplemented with glycerol in the absence (blue points) or presence (red points) of 10 μM La3+. Cells were incubated in 96-well microtiter plates in a microplate reader (Xenius; Safas, Monaco) at 30°C and 250 rpm. (B) Upon 40 h of incubation (indicated with black arrow in panel A), OD600 (dark blue and dark orange bars) and CFU counts per milliliter (light blue and light orange bars) from incubations performed in the presence and absence of 10 μM La3+ were measured. Data represent averages of results from biological triplicates with corresponding standard deviations. Two asterisks (**; P < 0.01) and four asterisks (****; P < 0.0001) indicate statistically significant differences between conditions in the presence and absence of 10 μM La3+ in a two-tailed t test (n = 3). Download FIG S2, TIF file, 0.1 MB (138.2KB, tif) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in the study. Download Table S1, DOCX file, 0.03 MB (31.6KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in the study. Download Table S2, DOCX file, 0.02 MB (23.3KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ when grown on 2-phenylethanol. Download Table S3, DOCX file, 0.02 MB (23.8KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ when grown on glucose. Download Table S4, DOCX file, 0.02 MB (24.1KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially abundant proteins in cells of KT2440* in the presence of 10 μM La3+ compared to the absence of La3+ when grown on citrate. Download Table S5, DOCX file, 0.02 MB (23.5KB, docx) .
Copyright © 2020 Wehrmann et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the PRIDE (74) partner repository (identifier [ID]: PXD013011).