Coronavirus disease 2019 was first reported from Wuhan district of China and has spread to 210 countries across the world claiming over 97,602 human lives as on 10 April 2020 by 21:06 pm. Currently, there is no specific treatment for this virus and the treatment is mainly relied on controlling symptoms. Here we discuss our current understanding of SARS-CoV-2 with respect to its receptor recognition and how this knowledge could be useful in treatement using clinically known inhibitory drugs. We have also discussed the diagnosis, treatment and preventive measures that are currently being employed for controlling further spread of the virus.
Introduction
The ongoing pandemic caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) has been spreading rapidly across the world, infecting over 16 lakh people as on 10 April 2020. SARS-CoV-2 is less pathogenic in terms of mortality and severity than SARS-CoV-1, which had caused an epidemic in 2003 infecting about 8000 people (Yi et al. 2020). However, it is more transmissive and has spread at an alarming rate in the past few days. Middle East respiratory syndrome coronavirus (MERS-CoV) that emerged in 2012, has so far infected 2279 people claiming 806 lives (Mubarak et al. 2019). This century has already seen three coronavirus outbreaks in humans. All these viruses belong to the family Coronaviridae, which consists of two subfamilies: Coronavirinae and Torovirinae. Coronavirinae is further divided into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus, and SARS-CoV-2 belongs to Betacoronavirus. CoVs have very large, nonsegmented, positive-sense RNA genome, which is about 30 kb (Fehr and Perlman 2015). The nucleocapsid (N) is helical and covered by a spherical lipid bilayer envelope embedded with other structural proteins; spike-glycoprotein (S), membrane (M), and envelope (E) (figure 1) (De Wit et al. 2016).
Figure 1.
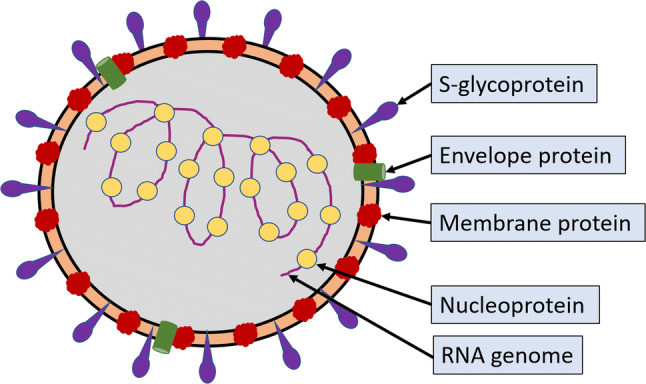
SARS-CoV-2 and related coronaviruses are enveloped positive sense RNA viruses. The structural proteins of the virus are: S-glycoprotein, envelope protein, membrane protein and nucleoprotein. The nucleocapsid is helical and is surrounded by a spherical envelope.
SARS-CoV-2 biology and possible evolution
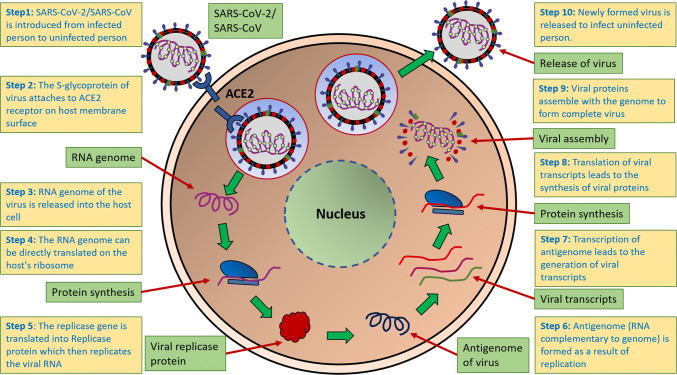
The CoV entry into its host is guided by the attachment of S-glycoprotein to a membrane protein that acts as a receptor on the host’s surface (figure 2). To date, several such receptors have been found for different CoVs-like angiotensin-converting enzyme 2 (ACE2) for SARS-CoV/SARS-CoV-2 and dipeptidyl peptidase4 (DPP4) for MERS-CoV (Raj et al. 2013). The receptor-binding domain (RBD) present in the S-glycoprotein takes part in the interaction with the host receptor for viral entry. A study has indicated that the mutations in the RBD have led to a higher binding affinity of SARS-CoV-2 to the human ACE2 receptor than SARS-CoV. Natural selection in humans or other organisms with human-like ACE2 has resulted in optimal binding of RBD which indicates that the virus has not been purposefully manipulated (Andersen et al. 2020). The same study also highlighted that a polybasic furin cleavage site in S-glycoprotein is present in SARS-CoV-2 and cleavage by host furin enzyme resulted in activation of spike protein (Andersen et al. 2020). The presence of furin in small intestine, liver and lungs partially explains the effective transmission rates of SARS-CoV-2 compared to other related viruses. ACE2 is involved in the cleavage of angiotensin II to angiotensin 1–7, leading to a lowering of blood pressure (Keidar et al. 2007). However, SARS-CoV-2 and related viruses have evolved to utilize such receptors for their own benefit by using them for initial attachment and entry. Several theories have been proposed for the putative origin of SARS-CoV-2. Phylogenetic analysis has revealed 89.1% similarity with a cluster of coronaviruses found in bats, suggesting that the virus might have naturally been selected in bats before zoonotic transfer to humans (Wu et al. 2020). Another study suggests Malayan pangolins as the origin, considering the high similarity in the major amino acid residues present in the RBD (Andersen et al. 2020). However, it is difficult to predict the exact origin of coronaviruses considering the inadequate sampling of bats, pangolins and other animals. The absence of polybasic furin cleavage site in the sampled coronaviruses also challenges the acceptability of the suggested theories. A recent phylogenetic analysis revealed that there are three major variants of SARS-CoV-2: A, B and C, among which A is the ancestral type; B is the most common type found in east Asia and the other two are found in America and Europe in significant proportion (Forster et al. 2020). This virus is quite different from other viruses that cause flu. Although flu is caused by RNA viruses, they belong to a different family called influenza viruses, which have a smaller genome compared to SARS-CoV-2. Some other coronaviruses are also reported to cause common cold with mild symptoms, but the similarity of SARS-CoV-2 with such viruses is only 50% at the nucleotide level. Nucleotide identity with SARS-CoV and MERS-CoV is 80% and 55%, respectively (Bar-on et al. 2020).
Figure 2.
SARS-CoV-2 and related viruses bind to receptors on the host membrane for entry. Viral RNA is released following entry, which is directly translated to replicase protein. This protein assists in viral replication. The antigenome is transcribed to produce viral transcripts which are translated into viral proteins. Viral assembly is followed by release of new virions to infect new cells.
Diagnosis, treatment and preventions
The most important step for devising treatment strategies for any disease is to diagnose it. SARS-CoV-2 nucleic acid isolated from the upper and lower respiratory tracts has been the main source of template for its detection by real-time (RT) PCR (Corman et al. 2020). There is strong positive correlation between leukocyte and lymphocyte count which can also be used as initial diagnosis (Jin et al. 2020). Symptoms associated with coronavirus disease 2019 (COVID-19) are quite diverse, ranging from fever, muscle aches, cough and shortness of breath. Sore throat and running nose, which is very prominent in case of human coronaviruses causing common cold, have been observed in less than 5% of patients. In severe cases, patients also suffer from pneumonia and SARS, leading to multiple-organ failure and death. Supplemented oxygen requirement has been seen in 75% of the cases (Poon and Peiris 2020). Chest x-rays have revealed interstitial changes and patchy shadows at multiple locations in the lungs, with maximum at the peripheral region (Chan et al. 2020; Huang et al. 2020). Apart from the respiratory fluids, viral RNA has also been detected in stool and blood; however, it is yet to be tested whether the virus can spread from these as well (Holshue et al. 2020).
There is no antiviral treatment or vaccine for SARS-CoV-2 or other coronaviruses, thus we need to rely on treatment of symptoms and intensive care unit support for seriously ill patients. Convalescent plasma (CP) from recovered patients has proved to be very promising to treat patients in serious conditions and improvement in symptoms has been seen within three days of transfusion (Duan et al. 2020). Chloroquine and hydroxychloroquine are in high demand as of now, as it is an effective drug for the treatment. However, the evidence for such an activity of the drug against the virus is limited and more studies should be undertaken before administering it to the patients as this may cause side effects in future (Gbinigie and Frie 2020). The mean incubation period of the virus is 5.2 days, and maximum infected individuals develop symptoms within 12.5 days (Li et al. 2020). This strongly supports the recommended idea of a 14-day quarantine period for medical observation. Social distancing, on the other hand, will help in limiting the spread of virus from carriers who are yet to develop symptoms but have already contracted the virus. N95 filtering facepiece respirators (N95 FFRs), which are being widely used as a precautionary measure, are efficient (99.8%) in filtering particulate matter of size up to 0.1 µm (Rengasamy et al. 2017). Since the size of SARS-CoV-2 is 0.1 µm in diameter, N95 FFRs can almost competently filter them out. The viability of the virus is quite variable with 3 h in aerosols and 72 h on plastic surfaces (van Doremalen et al. 2020). Thus, sterilizing surfaces and regularly washing hands with soap and water is recommended. Soap can disrupt the lipid bilayer envelope of the virus rendering it inviable. Several treatment methods have now been recommended in a series of research articles published recently. SARS-CoV-2 utilizes ACE2 and a serine protease TMPRSS2 for entry into its host. Blocking of TMPRSS2 activity by clinically proven inhibitors has the potential to prevent entry (Hoffmann et al. 2020). Another suggestion is the use of available angiotensin receptor 1 (ATR1) blockers considering the similarity between ACE2 and ATR1 structure (Gurwitz 2020).
In conclusion, our innate immunity is responsible for the adaptive immunity and antiviral defence. It is important that we further investigate the immune response of our body against SARS-CoV-2 so that effective therapeutics can be developed, and vaccine candidates can be mined. CP transfusion has proved to be effective, with no severe adverse effects and more studies must be conducted with a larger sample size to check its statistical significance (Duan et al. 2020). Development of vaccines might take some time, and presently, the best way to stay safe is to practice social distancing, keep surfaces disinfected, and reporting putative carriers and people with symptoms for medical help.
References
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat. Med. 2020;89:44–48. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-on YM, Flamholz A, Phillips R, Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9:e57309. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Morris DH, Holbrook MG, Gamble A, Williamson BN. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J. and Zhou M. 2020 Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA (10.1073/pnas.2004168117). [DOI] [PMC free article] [PubMed]
- Fehr AR, Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C. and Forster M. 2020 Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA (10.1073/pnas.2004999117). [DOI] [PMC free article] [PubMed]
- Gbinigie K. and Frie K. 2020 Should chloroquine and hydroxychloroquine be used to treat COVID-19? A rapid review. BJGP Open. (10.3399/bjgpopen20x101069). [DOI] [PMC free article] [PubMed]
- Gurwitz D. 2020 Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. (10.1002/ddr.21656). [DOI] [PMC free article] [PubMed]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP, et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keidar S, Kaplan M, Gamliel-Lazarovich A. ACE2 of the heart: from angiotensin I to angiotensin (1-7) Cardiovasc. Res. 2007;73:463–469. doi: 10.1016/j.cardiores.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission dynamics in Wuhan, China, of Novel Coronavirus-Infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubarak A, Alturaiki W, Hemida MG. Middle east respiratory syndrome coronavirus (mers-cov): Infection, immunological response, and vaccine development. J. Immunol. Res. 2019;2019:6491738. doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LLM, Peiris M. Emergence of a novel human coronavirus threatening human health. Nat. Med. 2020;26:317–319. doi: 10.1038/s41591-020-0796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengasamy S, Shaffer R, Williams B, Smit S. A comparison of facemask and respirator filtration test methods. J. Occup. Environ. Hyg. 2017;14:92–103. doi: 10.1080/15459624.2016.1225157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit E De, Van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Lagniton PNP, Ye S, Li E, Xu RH. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]