Cep57 has been characterized as a component of a pericentriolar complex containing Cep63 and Cep152. Interestingly, Cep63 and Cep152 self-assemble into a pericentriolar cylindrical architecture, and this event is critical for the orderly recruitment of Plk4, a key regulator of centriole duplication. However, the way in which Cep57 interacts with the Cep63-Cep152 complex and contributes to the structure and function of Cep63-Cep152 self-assembly remains unknown. We demonstrate that Cep57 interacts with Cep63 through N-terminal motifs and associates with Cep152 via Cep63.
KEYWORDS: Cep57, Cep63, Cep152, PCM, centriole biogenesis
ABSTRACT
Cep57 has been characterized as a component of a pericentriolar complex containing Cep63 and Cep152. Interestingly, Cep63 and Cep152 self-assemble into a pericentriolar cylindrical architecture, and this event is critical for the orderly recruitment of Plk4, a key regulator of centriole duplication. However, the way in which Cep57 interacts with the Cep63-Cep152 complex and contributes to the structure and function of Cep63-Cep152 self-assembly remains unknown. We demonstrate that Cep57 interacts with Cep63 through N-terminal motifs and associates with Cep152 via Cep63. Three-dimensional structured illumination microscopy (3D-SIM) analyses suggested that the Cep57-Cep63-Cep152 complex is concentrically arranged around a centriole in a Cep57-in and Cep152-out manner. Cep57 mutant cells defective in Cep63 binding exhibited improper Cep63 and Cep152 localization and impaired Sas6 recruitment for procentriole assembly, proving the significance of the Cep57-Cep63 interaction. Intriguingly, Cep63 fused to a microtubule (MT)-binding domain of Cep57 functioned in concert with Cep152 to assemble around stabilized MTs in vitro. Thus, Cep57 plays a key role in architecting the Cep63-Cep152 assembly around centriolar MTs and promoting centriole biogenesis. This study may offer a platform to investigate how the organization and function of the pericentriolar architecture are altered by disease-associated mutations found in the Cep57-Cep63-Cep152 complex.
INTRODUCTION
Mainly because of a featureless appearance by electron microscopy, pericentriolar material (PCM) has long been known as an amorphous mass of a proteinaceous matrix surrounding a pair of barrel-shaped centrioles (1–3). The way in which PCM proteins are organized around a centriole is beginning to emerge thanks largely to recent advances in imaging technologies. A growing body of evidence obtained from multiple organisms reveals that pericentriolar scaffold proteins are concentrically localized around a centriole in a highly organized manner (4–7), yet the molecular mechanisms underlying PCM organization remain elusive.
Other studies have shown that one of the pericentriolar scaffold proteins, Cep152, displays an open-ended cylinder-like localization pattern around a centriole (8). Moreover, it forms a heterotetrameric complex with another scaffold, Cep63, and self-assembles into a cylindrical architecture that appears to be critical for the proper recruitment of Sas6 (9), a major component of the centriolar cartwheel structure (10, 11). These findings suggest that the Cep63-Cep152 complex bears an intrinsic physicochemical property that enables it to form a higher-order structure without consuming any external energy such as ATP or GTP. Interestingly, the proper centrosomal localization of Cep63 and Cep152 requires an association with Cep57 (originally known as translokin) (12, 13). Since Cep152 is required for directly interacting with and properly positioning Plk4 (14), a key inducer of centriole biogenesis (15, 16), these observations suggest that Cep57 may function in concert with the Cep63-Cep152 complex to mediate Plk4-dependent downstream events critical for centriole duplication.
Although Cep57 is required for the proper pericentriolar localization of Cep63 and Cep152 (12, 17), the way in which Cep57 helps establish Cep63-Cep152 self-assembly remains elusive. Interestingly, a conserved N-terminal domain of Cep57 called PINC (present in the N terminus of Cep57) has been shown to interact with the conserved C-terminal pericentrin (PCNT)/AKAP9 centrosome-targeting (PACT) domain of PCNT (18), an integral component of PCM (19), suggesting that Cep57 may play a central role in organizing pericentriolar scaffold proteins. Not surprisingly, the loss of Cep57 function results in a reduction in the number of centrioles per centrosome (17, 20). Ironically, however, the loss of Cep57 also induces supernumerary centrosomes (17), presumably because Cep57 is required to prevent PCM fragmentation and premature centriole disengagement (18, 20) and to ensure proper cytokinesis (21). Other studies show that Cep57 interacts with kinetochore-localized Mis12 and a spindle assembly checkpoint component, Mad1, to ensure normal chromosome segregation (22). Just as Cep57 helps maintain spindle pole integrity and genomic stability, mutations in Cep57 are linked to the development of mosaic variegated aneuploidy syndrome (17, 23), an autosomal recessive disorder with a strong predisposition to cancer.
Here, we investigated whether and how Cep57 interacts with two other pericentriolar scaffold proteins, Cep63 and Cep152, and how it promotes Plk4-dependent centriole biogenesis. We found that the previously described PINC domain of Cep57 specifically interacts with a conserved N-terminal motif of Cep63 and that this interaction is critical for proper centriole duplication. Unlike the Cep57-Cep63 interaction, we did not find a stable interaction between Cep57 and the PCNT PACT domain. Intriguingly, ectopically expressed Cep57 exhibited microtubule (MT)-bundling activity, as described previously (24), and the coexpression of Cep57 C-terminal-domain (CTD)-fused Cep63 and Cep152 was sufficient to reconstitute a concentric cylinder-like Cep63-Cep152 arrangement around MT signals. Given that the C-terminal domain of Cep57 directly binds to MTs (24), we propose that Cep57 plays a pivotal role in linking the self-assembled Cep63-Cep152 architecture to centriolar MTs. Our data described here provide new insights into how the Cep57-Cep63-Cep152 complex is organized at the endogenous pericentriolar region and offer an underlying mechanism for the formation of a three-layered platform centrally required for PCM function.
RESULTS
Cep57 localized in the inner face of the Cep63 cylinder is critical for proper localization of Cep63 and induction of procentriole formation.
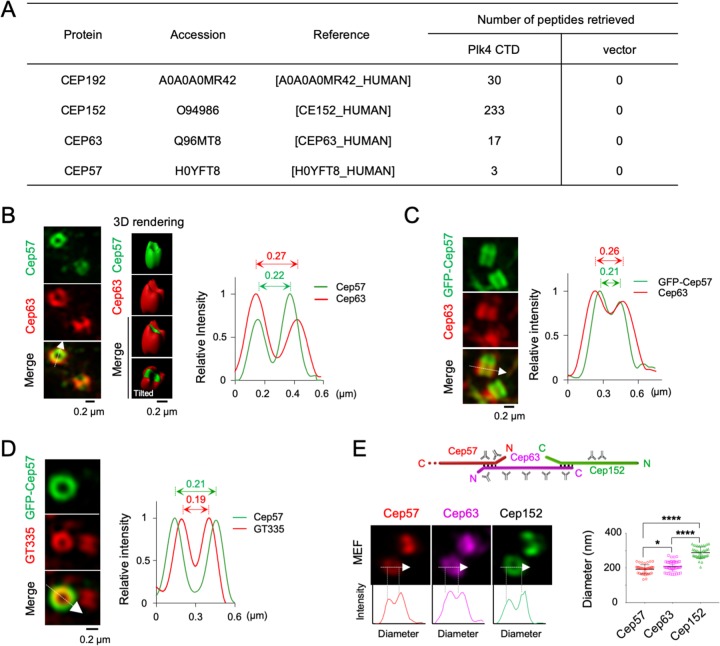
Although chemical cross-linking analysis suggested that Cep57 associates with Cep63 and Cep152 and localizes around the proximal end of centrioles (12), whether and how it contributes to the function of the Cep63-Cep152 complex in promoting centriole biogenesis remain unknown. In a mass spectrometry (MS) analysis carried out with coimmunoprecipitates of the Plk4 CTD (residues 581 to 970) as a ligand, we found that the Plk4 CTD precipitated not only Cep152 but also Cep63 and Cep57 from the lysates of asynchronously growing HEK293T cells (Fig. 1A). This finding supports the previous notion that Cep57, Cep63, and Cep152 form a centrosomal complex (12). The larger number of retrieved Cep152 peptides could be attributable to the fact that Cep152 directly interacts with the Plk4 CTD (14), while Cep63 associates with the latter via Cep152 (9). Cep192, another direct Plk4-binding target (14), was also effectively coprecipitated (Fig. 1A), demonstrating the validity of the coprecipitated sample.
FIG 1.
Cep57 associates with Plk4 immunoprecipitates, and the Cep57, Cep63, and Cep152 scaffolds display three concentric layers with distinct diameters. (A) Mass spectrometry analyses of anti-FLAG coimmunoprecipitates prepared from HEK293T cells stably expressing either FLAG3-Plk4 CTD or a control FLAG3 vector. Numbers of peptides retrieved from the analyses are shown. For Cep57, peptides LDLLEQEYNK (residues 189 to 198) and LIQESPTVELK (residues 384 to 394) (twice) were recovered. (B) 3D-SIM images of U2OS cells immunostained with anti-Cep57 and anti-Cep63 antibodies. M, mother centriole; D, daughter centriole. The intensity profile of fluorescent signals is shown by line scan plots generated in the arrowed direction. (C and D) 3D-SIM of immunostained U2OS cells transfected with GFP-Cep57. The intensity profile of fluorescent signals is shown by line scan plots generated in the arrowed direction. (E) 3D-SIM of primary mouse embryonic fibroblast (MEF) cells immunostained with the indicated antibodies. The schematic diagram shows the linearly arranged Cep57-Cep63-Cep152 complex (see Fig. 6G), with the positions of antibody epitopes marked. Line scan plots show the intensity profiles of fluorescent signals. The diameter was determined by measuring the longest peak-to-peak distance for ringlike signals. *, P < 0.05; ****, P < 0.0001 (unpaired two-tailed t test). Bars indicate means ± standard deviations (SD) (n = 40). Note that in part due to sample dehydration by MeOH fixation, the diameters of Cep63 and Cep152 cylinders are somewhat smaller than those obtained with paraformaldehyde fixation (9).
To closely investigate the interrelationship between Cep57 and the Cep63-Cep152 complex, we analyzed the pericentriolar localization patterns of Cep57 and Cep63 in U2OS cells. Three-dimensional structured illumination microscopy (3D-SIM) showed that both Cep57 and Cep63 were concentrically localized at the subcentrosomal region, exhibiting a hollow cylinder-like morphology around a centriole (Fig. 1B). Notably, the diameter of cylindrical Cep57 signals appeared to be reproducibly smaller than that of Cep63 signals (Fig. 1B, right). The diameter difference was also apparent when green fluorescent protein (GFP)-fused Cep57 was used for analysis (Fig. 1C), suggesting that these two proteins are discretely positioned in a Cep57-inside fashion. As expected, if Cep57 were a pericentriolar protein, it would be detected clearly outside centriolar glutamylated tubulins (Fig. 1D). Comparative analyses of the diameters of Cep57, Cep63, and Cep152 in primary mouse embryonic fibroblast (MEF) cells confirmed that these three proteins are arranged in three distinct layers, with Cep57 representing the layer with the smallest diameter and Cep152 constituting the outermost layer with the largest diameter (Fig. 1E).
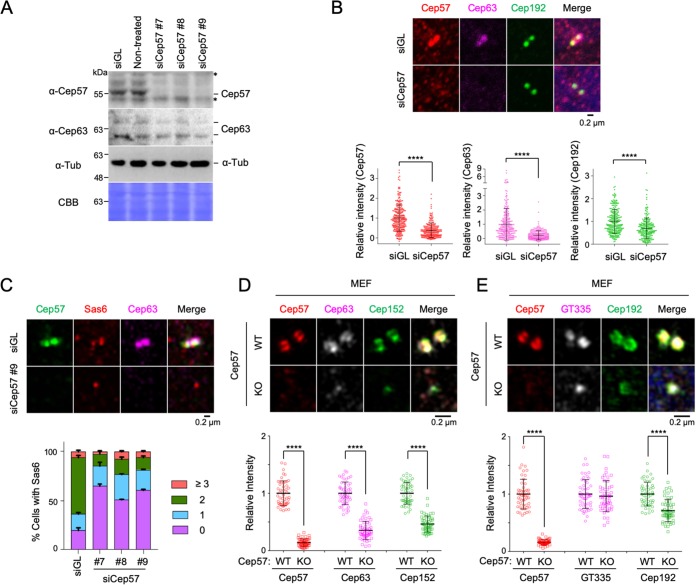
To investigate whether Cep57 plays a role in inducing centriole biogenesis, U2OS cells were silenced for endogenous Cep57 using small interfering RNAs (siRNAs) (siCep57#7 to siCep57#9) targeting the nonoverlapped regions of the 3′ untranslated region (UTR) of Cep57 (Fig. 2A). Immunoblot analysis showed that all three siRNAs effectively depleted Cep57. The depletion of Cep57 did not appear to alter the stability of Cep63 in all cases (Fig. 2A). Endogenous Cep63 was detected as a doublet. Therefore, we used Cep57#9 siRNA for further analyses unless specifically indicated otherwise. Subsequent immunostaining analyses showed that Cep57 depletion greatly diminished the level of Cep63 localized at centrosomes (Fig. 2B and C). In contrast, Cep192, one of the innermost pericentriolar scaffolds (4), exhibited a localization defect at a much-diminished level (Fig. 2B). Similar results were obtained from immunostainings with primary MEF cells obtained from either Cep57+/+ or Cep57–/– mice (17) (Fig. 2D and E). The signal intensities for centriolar glutamylated tubulins were unaltered by the Cep57–/– mutation (Fig. 2E). These findings suggest that Cep63 requires Cep57 to properly localize to centrosomes. In line with the critical role of Cep63 in recruiting Cep152 to pericentriolar regions and inducing Cep152-Plk4-dependent centriole biogenesis (9, 25), the depletion of Cep57 significantly increased the cell population without detectable Sas6, a major component of the procentriole’s cartwheel (10, 11). Consequently, the fraction of cells with properly duplicated Sas6 dot signals decreased (Fig. 2C).
FIG 2.
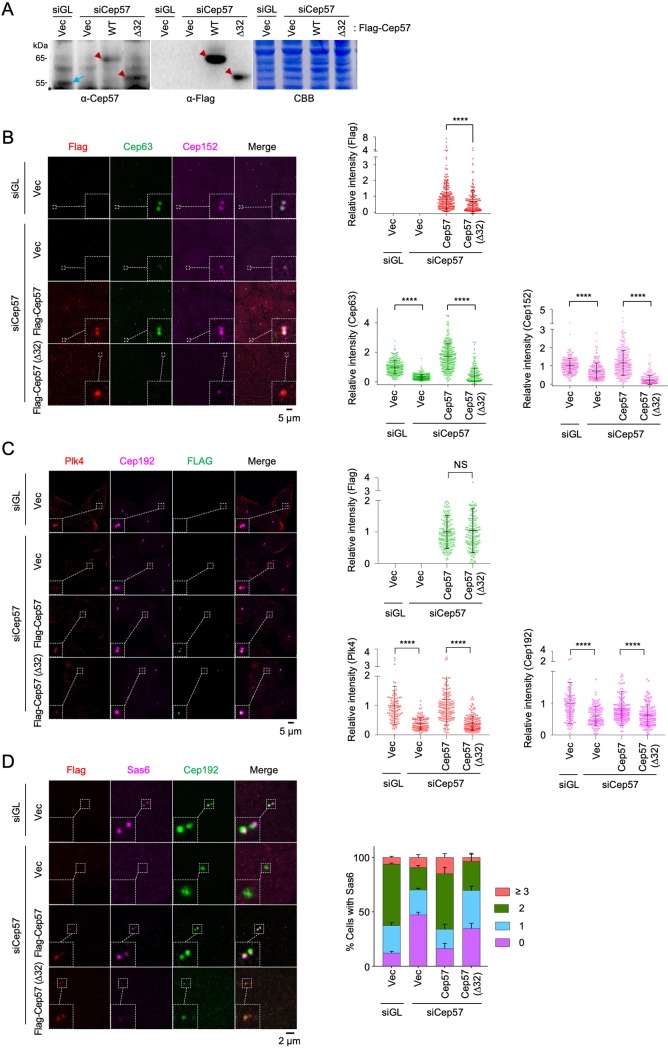
Cep57 is required for the centrosomal localization of Cep63 and Cep152 and for Sas6 recruitment to the procentriole assembly site. (A) Immunoblot analysis of U2OS cells silenced for either control luciferase (siGL) or Cep57 (siCep57#7 to siCep57#9). The same membrane was stained with Coomassie brilliant blue (CBB) for loading controls. Asterisks indicate cross-reacting proteins. Tub, tubulin. (B) Confocal images of immunostained U2OS cells after silencing with either control luciferase or Cep57 (siCep57#9) and quantification of signal intensities. ****, P < 0.0001 (unpaired two-tailed t test). Bars indicate means ± SD (n > 231 from three independent experiments). (C) Confocal images of immunostained U2OS cells analyzed similarly as described above for panel B. The number of centrosomal Sas6 signals was quantified. Bars indicate standard deviations (n > 110 from three independent experiments). (D and E) 3D-SIM images of immunostained primary MEF cells. Quantified relative fluorescence intensities are shown as means ± SD (n > 80 for each sample from three independent experiments). ****, P < 0.0001 (unpaired two-tailed t test). WT, wild type; KO, knockout.
The Cep57-Cep63 interaction occurs via the N-terminal regions.
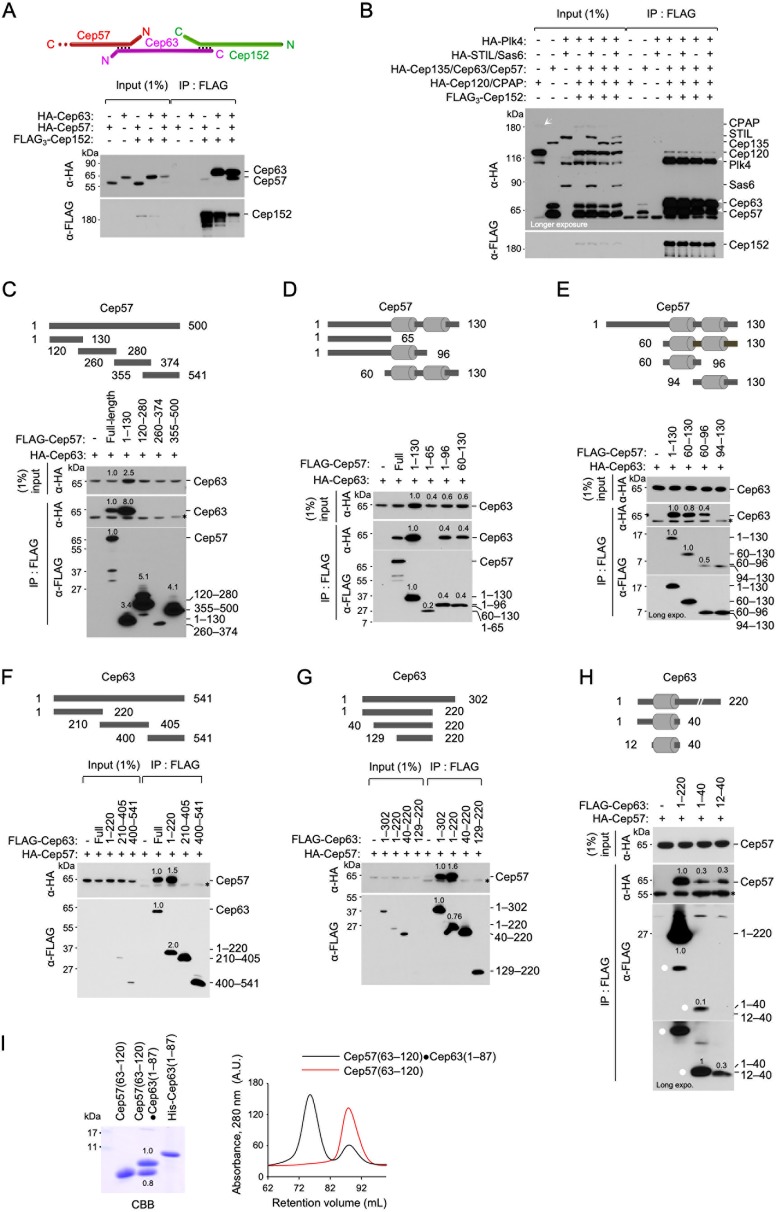
The three distinct layers of pericentriolar Cep57, Cep63, and Cep152 (Fig. 1E) suggest that the innermost Cep57 may directly interact with Cep63 but not the outermost Cep152. Consistent with this view, Cep152 efficiently coimmunoprecipitated Cep63 but not Cep57, and the coexpression of Cep63 allowed Cep152 to coprecipitate Cep57 (Fig. 3A). This finding and the data in Fig. 1E suggest that the Cep57-Cep63-Cep152 complex is orderly arranged around a centriole and that Cep63 bridges the interaction between Cep57 and Cep152. In a second experiment, we observed that immunoprecipitation (IP) of Cep152 efficiently coimmunoprecipitated not only its N-terminal and C-terminal binding targets (Plk4 [14] and Cep63 [9], respectively) but also Cep57 (Fig. 3B). Under these conditions, STIL, Sas6, CPAP, Cep135, and Cep120 were only weakly coimmunoprecipitated. Thus, Cep57, Cep63, and Cep152 form a stable complex that may effectively recruit Plk4.
FIG 3.
The Cep57 PINC motif directly interacts with the Cep63 CTB motif. (A and B) Immunoprecipitation (IP) and immunoblot analyses using HEK293T cells cotransfected with the indicated constructs. (A) Note that Cep152 coimmunoprecipitates Cep57 only in the presence of Cep63 (last lane). A schematic diagram depicts the trimeric Cep57-Cep63-Cep152 complex. (B) IP of Cep152 efficiently coprecipitated Cep57 and Cep63 along with its N-terminus-binding Plk4 (14). An arrow indicates weakly expressed CPAP. Three arrowheads (right) indicate Cep57, Cep63, and Plk4 efficiently coprecipitated with Cep152. (C to E) IP and immunoblot analyses using HEK293T cells cotransfected with full-length (FL) HA-Cep63 and various FLAG-Cep57 constructs. Gray bars in the schematic diagrams denote an α-helix predicted by the PSIPRED server. Numbers in blots are relative signal intensities. Asterisks indicate cross-reacting proteins. (F to H) IP and immunoblot analyses were carried out similarly as described above for panels C to E but with FL HA-Cep57 and various FLAG-Cep63 constructs. Gray bars indicate a predicted α-helix. Numbers in blots are relative signal intensities. Dots in panel H indicate matching signals with two different exposures. Note that Cep57(60–96) (i.e., the PINC motif) and Cep63(12–40) (i.e., the CTB motif) appear to possess the full capacity to interact with FL Cep63 and Cep57, respectively. Asterisks indicate a cross-reacting protein. (I) Size exclusion chromatography showing the Cep57(63–120)-Cep63(1–87) complex stably formed in E. coli. The sample was separated by 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (CBB). Overlaid chromatograms for the Cep57(63–120)-Cep63(1–87) complex and Cep57(63–120) are shown at the right. Longer forms were required to stably express Cep57 and Cep63 fragments. Purified His-tagged Cep63(1–87) was loaded for size comparison. A.U., arbitrary units.
Since the Cep63-Cep152 interaction has been investigated in detail (9), we then determined the regions critical for the Cep57-Cep63 interaction by carrying out coimmunoprecipitation (coIP) analyses with cotransfected HEK293 lysates. The results showed that the Cep57(1–130) fragment interacted with Cep63 at a level similar to that of full-length (FL) Cep57 (Fig. 3C), suggesting that the Cep57(1–130) region has the full capacity to interact with Cep63. Analysis of the primary sequence of Cep57(1–130) by using the PSIPRED server predicted the presence of two α-helical motifs within the fragment (Fig. 3D and E, rods in schematic diagrams). Subsequent IP analyses showed that the Cep57(60–96) fragment containing the first α-helix was important for a stable Cep57-Cep63 interaction (Fig. 3E).
In a converse experiment, we also found that the N terminus of Cep63(1–220) interacted with Cep57 as efficiently as FL Cep63 (Fig. 3F). Subsequent IP analyses showed that Cep63(1–40) containing a predicted α-helix appeared to be sufficient for Cep57 binding (Fig. 3G and H). These results suggest that the interaction between Cep63 and Cep57 is mediated mainly through a small α-helical fragment located at the respective N-terminal regions. This notion was confirmed by size exclusion chromatography, which showed that bacterially expressed Cep57(63–120) and Cep63(1–87) formed a stable complex with ∼1:1 stoichiometry under physiological buffer conditions (Fig. 3I).
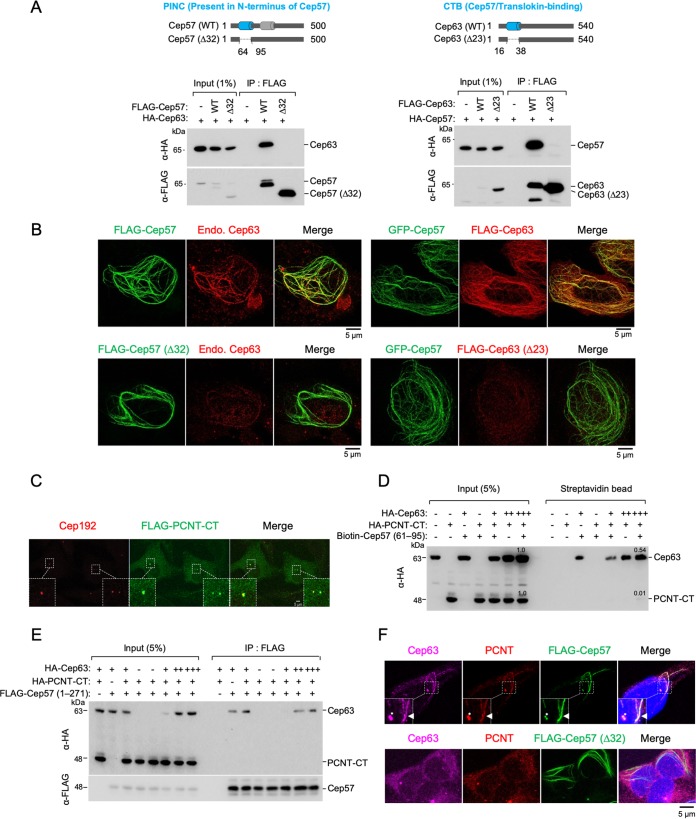
The Cep57 PINC and Cep63 CTB motifs are mutually required for the Cep57-Cep63 interaction and their colocalization.
Watanabe et al. reported previously that the conserved N-terminal PINC motif of Cep57 (Fig. 4A) interacts with the C-terminal PACT domain of PCNT (i.e., PCNT-CT) (18). Notably, the Cep57 PINC motif is overlapped by the Cep57 region (residues 60 to 96), which appeared to be necessary and sufficient for binding to the Cep63 N-terminal domain (Fig. 3). Interestingly, like the Cep57 PINC motif, the Cep63 N-terminal domain also encodes an evolutionarily conserved motif predicted to form an α-helix. Therefore, because of its ability to bind to Cep57, the Cep63 N-terminal domain was named the CTB (Cep57/translokin-binding) motif (Fig. 4A).
FIG 4.
Mutual requirement of Cep57 PINC and Cep63 CTB for proper Cep57-Cep63 interaction and their colocalization. (A) Schematic diagram showing the Cep57(Δ32) and Cep63(Δ23) mutants lacking residues 64 to 95 (i.e., the PINC motif) of Cep57 and lacking residues 16 to 38 (i.e., the CTB motif) of Cep63, respectively. Gray bars indicate α-helices predicted by the PSIPRED server. (B) 3D-SIM images of U2OS cells transfected with the indicated constructs and immunostained with either anti-Cep63 antibody (left) or anti-FLAG antibody (right). Endo., endogenous. (C) Confocal analysis was performed with U2OS cells transfected with FLAG–PCNT-CT. The resulting cells were immunostained with anti-FLAG and anti-Cep192 antibodies. Boxes indicate areas of image enlargement. (D) Pulldown assays of biotinylated Cep57(61–95) with HEK293 lysates expressing HA-Cep63 and HA–PCNT-CT. Streptavidin-agarose-immobilized Cep57(61–95) and its coprecipitates were analyzed by anti-HA immunoblotting. Note that while Cep57(61–95) efficiently coprecipitates Cep63, it fails to significantly coprecipitate PCNT-CT containing the PACT domain, a suggested Cep57 PINC-binding region (18). (E) IP and immunoblot analyses using HEK293T cells cotransfected with the indicated constructs. Note that a Cep57 N-terminal construct containing the entire centrosomal localization domain (24) also preferentially binds to Cep63 but not PCNT-CT. (F) Confocal images of U2OS cells transfected with either FLAG-Cep57 WT or FLAG-Cep57(Δ32) and immunostained with either anti-PCNT, Alexa Fluor 647-conjugated anti-Cep63, or anti-FLAG antibodies. Nuclear DNA was labeled with DAPI (blue). Boxes indicate areas of enlargement. Asterisks indicate centrosomes. Arrowheads indicate endogenous Cep63 and PCNT recruited to a FLAG-Cep57 cable.
To investigate whether the interaction between Cep57 PINC and Cep63 CTB is mutually required for their interactions and colocalizations in vivo, we generated internal deletion constructs lacking their respective α-helices, Cep57(Δ64–95) and Cep63(Δ16–38) [referred to as Cep57(Δ32) and Cep63(Δ23) here]. In reciprocal immunoprecipitation analyses, Cep57(Δ32) and Cep63(Δ23) failed to interact with Cep63 and Cep57, respectively, which was expected if these fragments were essential for the interaction (Fig. 4A).
It has been shown that ectopically expressed full-length Cep57 or its MT-binding C-terminal domain induces MT bundles and generates long cable-like structures in the cytosol (24). Thus, we took advantage of the Cep57-induced cable-like structures as a platform to assess the functionality of Cep57. We observed that both Cep57 and Cep57(Δ32) efficiently induced the formation of a cable-like structure in the cytosol (Fig. 4B). However, like the coimmunoprecipitation data shown in Fig. 4A, while wild-type Cep57 (Cep57 WT) cables effectively recruited endogenous Cep63, the Cep57(Δ32) mutant cables lacking the PINC motif failed to recruit any detectable level of Cep63 (Fig. 4B, left). In a related experiment, GFP-Cep57-generated cables strongly recruited overexpressed Cep63 but not the Cep63(Δ23) mutant lacking the Cep57-interacting CTB motif (Fig. 4B, right). These observations suggest that the interactions between the Cep57 PINC and Cep63 CTB motifs are mutually required for Cep57 and Cep63 to interact with and colocalize at distinct subcellular sites.
Cep57 PINC efficiently interacts with Cep63 CTB but not PCNT-CT.
While our data described above (Fig. 3 and Fig. 4A and B) strongly suggest that Cep57 PINC efficiently interacts with Cep63 CTB, Watanabe et al. showed that it also interacts with PCNT-CT (18). If this were the case, then Cep63 and PCNT-CT may competitively bind to the Cep57 PINC region. Thus, we directly compared their efficiencies of binding to a biotinylated Cep57(61–95) polypeptide covering the core α-helix of the PINC motif using cell lysates expressing either Cep63, PCNT-CT, or both (full-length PCNT was not tested because of the difficulty in expressing the ∼350-kDa protein). As expected, PCNT-CT (residues 2953 to 3336) containing the entire centrosome-targeting PACT domain (26) efficiently localized to centrosomes (Fig. 4C), indicating that it is functional. The results showed that the bead-associated biotinylated Cep57(61–95) peptide efficiently precipitated Cep63 and that the levels of coprecipitated Cep63 were proportional to those of its expression in the lysates (Fig. 4D). In stark contrast, a marginal level of PCNT-CT was detectable only in the sample that had the highest level of Cep63 expression (Fig. 4D, last lane). This finding suggests that the PCNT-CT–Cep57 PINC interaction could be very weak and that PCNT-CT may not compete with Cep63 for Cep57 PINC binding.
Since the biotinylated Cep57(61–95) peptide may have an unanticipated structural deficiency, we then carried out a related experiment using a coexpressed N-terminal Cep57(1–271) fragment as the ligand. Under these conditions, we also observed that Cep63 (but not PCNT-CT) efficiently interacted with Cep57(1–271) (Fig. 4E). These data suggest that Cep63 is likely the main binding target of the Cep57 PINC motif in vivo and that PCNT could be binding to Cep57 at a very low level. Notably, however, Cep57 WT cables were able to recruit endogenous PCNT at a significant level, while the Cep57(Δ32) mutant cables were not (Fig. 4F). Since the in vitro Cep57–PCNT-CT interaction is inefficient (18), PCNT may require another component(s) or uncharacterized modifications to stably interact with Cep57.
The Cep57 PINC motif is required for proper recruitment of Cep63 and Cep152 and accumulation of Sas6 at the procentriole assembly site.
To examine the physiological function of the Cep57 PINC motif in centriole biogenesis, U2OS cells stably expressing either Cep57 WT or Cep57(Δ32) were generated using a lentiviral expression system and then silenced for endogenous Cep57 by RNA interference (RNAi). Immunoblot analysis showed that the level of exogenously expressed Cep57 or Cep57(Δ32) was similar to that of endogenous Cep57 (Fig. 5A). Under these conditions, we observed that Cep57(Δ32) was significantly localized to centrosomes, while its localization efficiency was clearly diminished compared to that of Cep57 WT (Fig. 5B). A diminished level of Cep57(Δ32) localization could be attributable to the loss of the PINC motif-dependent interaction with Cep63. Not surprisingly, the Cep63 and Cep152 intensities at the Cep57(Δ32)-associated centrosomes were greatly diminished (Fig. 5B). Consistent with these observations, Plk4 recruitment to centrosomes was drastically impaired, even when quantification was performed with the cell population among which the level of centrosome-localized Cep57(Δ32) was similar to that of Cep57 WT (Fig. 5C). Likewise, the Cep57(Δ32)-expressing cells failed to properly recruit Sas6, substantially decreasing the cell population with two distinct Sas6 signals compared with Cep57 WT-expressing cells (Fig. 5D).
FIG 5.
The PINC domain of Cep57 is required for proper recruitment of Cep63 and Cep152 and normal centriole biogenesis. (A) Immunoblot analyses showing the levels of exogenous Cep57 and Cep57(Δ32) expression. Arrowheads indicate exogenous Cep57 and Cep57(Δ32), and the arrow shows endogenous Cep57. An asterisk indicates a cross-reacting protein. CBB, Coomassie brilliant blue-stained membrane; Vec, vector. (B) Confocal analysis of immunostained U2OS cells stably expressing the indicated constructs and silenced for control luciferase (siGL) or endogenous Cep57 (siCep57). Quantified relative fluorescence intensities for FLAG, Cep63, and Cep152 are shown as means ± SD (n > 170 for each time point from three independent experiments). ****, P < 0.0001 (unpaired two-tailed t test). (C) The PINC domain of Cep57 is required for the proper recruitment of Plk4 to centrosomes. Confocal analysis of immunostained U2OS cells stably expressing the indicated FLAG-fused constructs and silenced for control luciferase (siGL) or endogenous Cep57 (siCep57) was performed. Quantified relative fluorescence intensities for FLAG, Plk4, and Cep192 are shown as means ± SD (n > 150 for each sample from three independent experiments). NS, not significant (P > 0.05); ****, P < 0.0001 (unpaired two-tailed t test). Note that cells with similar levels of Cep57 or Cep57(Δ32) signals were chosen to comparatively analyze whether the Cep57(64–95) region is important for recruiting Plk4 and Cep192 to centrosomes. (D) Confocal analysis of immunostained U2OS cells prepared similarly as described above for panel B. The number of centrosomal Sas6 signals was quantified. Bars indicate SD (n > 118 from three independent experiments).
Cep57 overexpression induces tubular MT bundles capable of organizing Cep63 and Cep152 around its longitudinal axis.
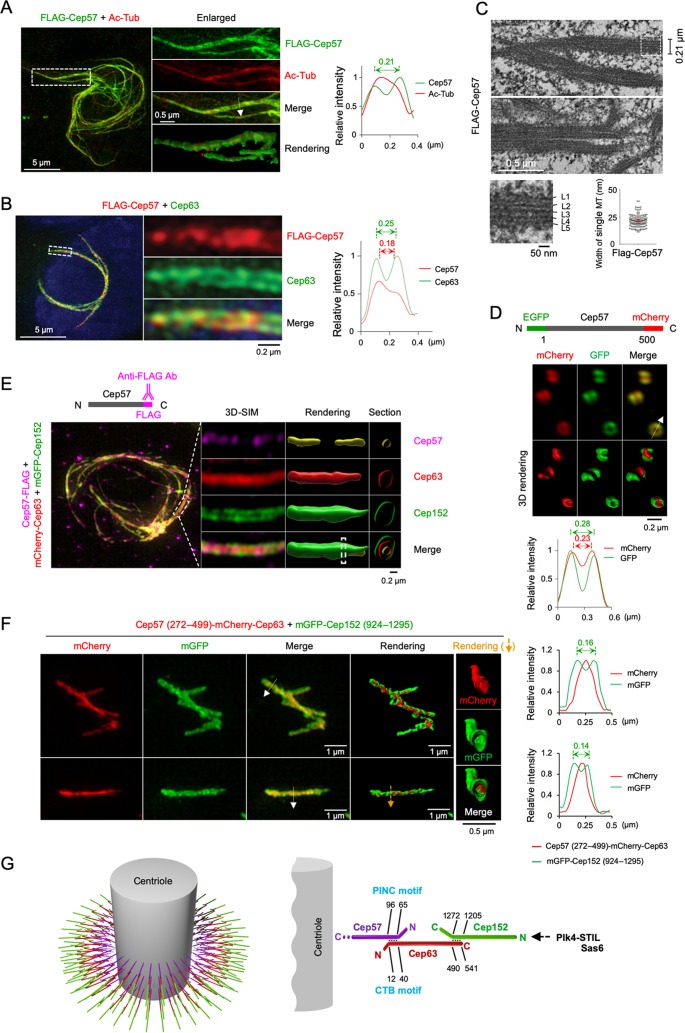
Similar to previous observations (24), the ectopic expression of Cep57 efficiently generated cable-like structures in the cytosol. Immunostaining analyses revealed that Cep57 localized at the outskirts of centrally located acetylated-tubulin signals, often exhibiting a tubular morphology (Fig. 6A; see also Movie S1 in the supplemental material). Measurement of the peak-to-peak distance revealed that some of the tubular Cep57 signals were as large as ∼200 nm in diameter. This is not surprising because Cep57 is capable of inducing MT bundles (24). In addition, Cep63 clearly localized at the periphery of Cep57 signals (Fig. 6B), which is reminiscent of the endogenous Cep63 arranged around Cep57 signals shown in Fig. 1B. Notably, thin-section transmission electron microscopy (TEM) revealed the generation of MT bundles made of tightly packed MT layers (Fig. 6C). This finding raises the possibility that Cep57 links the Cep63-Cep152 cylindrical structure (9) to the centriolar MTs.
FIG 6.
Cep57 may link the Cep63-Cep152 assemblies to centriolar MTs. (A and B) 3D-SIM analysis of U2OS cells transfected with FLAG-Cep57 and immunostained with anti-FLAG and either anti-acetylated tubulin (Ac-Tub) (A) or anti-Cep63 (B) antibodies. Nuclear DNA was labeled with DAPI (blue). indicate areas of enlargement. Double-headed arrows indicate the peak-to-peak distances for tubular Cep57 and Cep63 signals, and the arrow in the merged image represents the direction of line scan plots for the fluorescence intensity profiles. (C) Correlative light and electron microscopy (CLEM) analysis of U2OS cells expressing FLAG-Cep57. Thin-section transmission electron microscopy (TEM) images reveal the nature of Cep57-induced MT bundles. The widths of MT bundles and individual MTs are measured and provided. The dotted box indicates the area of enlargement for the bottom left image. (D) 3D-SIM imaging was performed using U2OS cells expressing a GFP-Cep57-mCherry construct. Note that the EGFP signal is positioned at the outskirts of the mCherry signal. (E) 3D-SIM analyses of U2OS cells transfected with Cep57-FLAG, infected with adenoviruses expressing mCherry-Cep63 and mGFP-Cep152, and immunostained with anti-FLAG antibody. Images were rendered, and cross sections at the indicated regions (dotted bars) are provided. Somewhat dotted Cep57 signals observed under both settings could be due to limited antibody accessibility in the presence of recruited Cep63 and Cep152. Cep152 exhibited the largest diameters, while Cep57 consistently displayed the smallest diameters (n > 30 cells). (F) 3D-SIM analysis showing in vitro-assembled mGFP-Cep152(924–1295) around the outskirts of Cep57(272–499)-fused mCherry-Cep63 that decorates a cell-free MT bundle induced by the MT-binding C-terminal domain (i.e., residues 272 to 499) of Cep57 (24). An MT was generated by the addition of 100 μM paclitaxel and then incubated with cell lysates expressing a Cep57(272–499)-mCherry-Cep63 fusion and mGFP-Cep152(924–1295). Two representative images from three independent experiments are shown. Arrows in the merged images indicate the direction of line scan plots for the fluorescence intensity profiles shown at the right. Rendered images at the right were generated from the cross section of the region marked by a yellow arrow. Note that Cep152 assembles around MT-binding Cep57(272–499)-mCherry-Cep63 signals. (G) Model illustrating how Cep57 links Cep63-Cep152 self-assembly to the centriolar MTs. The C-terminal domain of Cep57 has MT-binding activity (24), suggesting that it may bind to centriolar MTs directly. The N-terminal PINC motif (18) of Cep57 directly interacts with the N-terminal CTB motif of Cep63 to properly localize to the pericentriolar region. Cep63 has been shown to form a complex with Cep152 and generates a cylindrical architecture around a centriole with the Cep152 N terminus pointing outward (9). This organized assembly in turn recruits Plk4 through a Cep152 N-terminal motif (14), thus allowing Plk4-dependent STIL and Sas6 recruitment and procentriole assembly (15, 16, 35, 36).
Cep57’s ability to interact with centriolar MTs could be the basis of a Cep57-in and Cep63-out arrangement around a centriole (Fig. 1). This directional Cep57-Cep63 arrangement may function in concert with the Cep63-Cep152 complex’s ability to form Cep63-in and Cep152-out self-assembly, as reported previously (9). This view predicts that Cep57 is arranged around a centriole in a fashion with its C-terminal MT-binding domain pointing inward and its N-terminal Cep63-binding region pointing outward. Indeed, close-localization analysis with a Cep57 construct bearing an N-terminal enhanced GFP (EGFP) tag and a C-terminal mCherry tag demonstrated that Cep57’s N-terminal region is positioned at the outskirts of its C-terminal region (Fig. 6D). Furthermore, an additional analysis with cells expressing Cep57, Cep63, and Cep152 showed that Cep57 localizes at the innermost region of the cable-like structure, whereas Cep152 localizes at the outermost region of the structure (Fig. 6E and Movies S2 and S3).
To directly investigate whether the MT-binding activity of Cep57 is critical for establishing a Cep63 arrangement that can recruit Cep152 at its periphery, we generated a Cep63 fusion construct that contains MT bundle-forming Cep57(272–499) (24) at its N terminus (to help avoid excessive aggregation of unevenly coexpressed Cep57, Cep63, and Cep152 proteins). The resulting construct [also known as Cep57(272–499)-mCherry-Cep63] was then coexpressed with the monomeric GFP (mGFP)-Cep152(924–1295) construct that was shown previously to effectively self-assemble with Cep63 (9). When the resulting lysates were incubated with paclitaxel-stabilized MTs in vitro, mGFP-Cep152 signals were reproducibly arranged at the outskirts of the backboned red fluorescent signals generated by the MT bundle-forming Cep57(272–499)-mCherry-Cep63 form (Fig. 6F and Movie S4). These results strongly suggest that the MT-binding capacity of the Cep57 C-terminal domain plays an important role in tethering Cep63 to the proximal region of bundled MTs (or centrioles) and that Cep63 and Cep152 have the capacity to assemble a Cep63-in and Cep152-out higher-order structure around Cep57-decorated centriolar MTs.
DISCUSSION
Requirement of the Cep57 PINC-Cep63 CTB interaction for proper centriole biogenesis.
It has been shown that two pericentriolar scaffold proteins, Cep63 and Cep152, generate a heterotetrameric complex that self-assembles into a higher-order cylindrical architecture (9). However, the mechanism that recruits Cep63 and Cep152 to the pericentriolar region remains unknown. Previous studies showed that Cep57 associated with Cep63 and Cep152 (12), although how it forms a functional complex with them has not been investigated. Here, we show that Cep57 localizes at the inner face of cylindrical Cep63 signals and is required for proper Cep63 recruitment to centrosomes (Fig. 1 and 2). Furthermore, Cep57 and Cep63 interacted with each other through their respective N-terminal PINC and CTB motifs (Fig. 3), and these motifs were required for their ability to colocalize at distinct subcellular structures in vivo (Fig. 4). In good agreement with these findings, cells expressing the Cep57(Δ32) mutant lacking its N-terminal PINC motif failed to properly promote Cep63 and Cep152 localization to centrosomes (Fig. 5B). Consequently, they were impaired in recruiting Plk4 and its downstream Sas6, a key component of cartwheel assembly (10, 11) (Fig. 5C and D). These observations suggest that Cep57 may play a pivotal role in the recruitment and organization of Cep63 and Cep152 around a centriole. Intriguingly, it has been demonstrated that the C-terminal domain of Cep57 exhibits MT-binding activity (24). Thus, we propose that Cep57 may serve as a structural link that tethers Cep63-Cep152 self-assembly to the outskirts of a centriole (Fig. 6G), presumably by directly binding to the centriolar MTs (see below). In the absence of the Cep57 tether, the recruitment and self-assembly of Cep63 and Cep152 as well as the stability of the Cep63-Cep152-generated architecture could be compromised, consequently failing to serve as a platform to promote Plk4-dependent centriole biogenesis. It should be noted that mutations in Cep57 have been implicated in mosaic variegated aneuploidy and Seckel syndrome (17, 23, 27, 28), and additional uncharacterized mutations have been identified (https://www.ncbi.nlm.nih.gov/snp/?term=cep57). Thus, understanding the structure and function of Cep57-mediated centriole biogenesis will likely be critical for not only deciphering the underlying mechanism of Plk4-dependent centriole biogenesis but also determining the etiology of these disorders.
The Cep57-Cep63 interaction versus the Cep57-PCNT interaction.
PCNT has been characterized as one of the pericentriolar scaffold proteins critical for PCM organization (29). A 3D-SIM-based positional mapping analysis showed that the C terminus of PCNT is anchored at or near the centriole, while its N terminus is extended toward the periphery (4). Interestingly, a recent study showed that the conserved C-terminal PACT domain of PCNT interacts with the Cep57 PINC motif (residues 68 to 103) and that this interaction is important for the proper localization of PCNT and organization of mitotic PCM (18). Since Cep63 also interacted with the same region in Cep57 (Fig. 3), it is possible that Cep63 and PCNT competitively bind to the Cep57 PINC motif. However, comparative coimmunoprecipitation and pulldown analyses revealed that, unlike the Cep57-Cep63 interaction, the interaction between Cep57 and PCNT was hardly detectable (Fig. 4D and E). Therefore, PCNT may not significantly interact with Cep57 to competitively alter the level of the Cep57-Cep63 interaction. However, Cep57 cable-like structures recruited both Cep63 and PCNT in a PINC motif-dependent manner (Fig. 4F). This finding hints that PCNT may either bind to the Cep57 PINC motif in a spatially regulated manner or require a posttranslational modification(s) or an unknown binding protein(s) to localize to Cep57-induced cables.
Structure and function of the Cep57-Cep63-Cep152 architecture around a centriole.
Analyses of pericentriolar localization patterns of Cep57, Cep63, and Cep152 (Fig. 1) (9, 30) suggest that these proteins constitute three distinct layers of a concentrically organized structure. Based on the Cep63-Cep152 cylindrical structure proposed previously (9), together with the data shown in this study, we envision that the trimeric Cep57-Cep63-Cep152 complex may assume a linearly aligned elongated structure that is then radially arranged from the axis of a centriole in a Cep57-in and Cep152-out manner (Fig. 6G). Based on the available 3D-SIM data (Fig. 1) (9, 14), the Cep57-Cep63-Cep152 complex could easily be stretched over the range of ∼150 nm in length so that the innermost Cep57 may not directly interact with the outermost Cep152 (Fig. 6G). Consistent with this view, Cep57 required Cep63 to associate with Cep152 (Fig. 3A). Notably, like Cep63 and Cep152, the primary sequence of Cep57 is composed of several coiled-coil domains that could adopt an elongated morphology.
Because of the high density of PCM, it is difficult to investigate how PCM is organized at the endogenous level. Therefore, a potential insight into the organization of its scaffold proteins could be gained from studies carried out under ectopically expressed conditions or in an in vitro cell-free system. Intriguingly, Cep57 appears to directly bind to MTs and can induce the formation of MT bundles as large as ∼200 nm in diameter (Fig. 6C), which is comparable to the diameter of centriolar MTs. Furthermore, in an in vitro assembly assay, MT bundles induced by the C-terminal MT-binding domain of Cep57 (24) were concentrically decorated with Cep57-fused Cep63 and Cep152 (Fig. 6F), reminiscent of the cylindrical organization of the Cep63-Cep152 complex around a centriole (9). These findings suggest that the inner-layered Cep57 may directly link Cep63-Cep152 self-assembly to the centriolar MTs via the Cep57 PINC-Cep63 CTB interaction and help organize the self-assembly around bundled MTs. Furthermore, since Cep63-Cep152 self-assemblies exhibit various cylindrical diameters in vitro (9), Cep57 may play a critical role in determining the exact dimension of the Cep63-Cep152 self-assembly around a centriole by binding to both centriolar MTs and Cep63.
The proper assembly and organization of pericentriolar scaffold proteins are likely critical for normally positioning client proteins and promoting their functions in a spatially regulated manner. In fact, aberrations in the functions of the Cep57, Cep63, and Cep152 scaffolds are closely associated with the development of many human diseases, including cancer, microcephaly, ciliopathy, and dwarfism (31, 32). What advantage, then, does a three-layered assembly offer for the structure and function of the pericentriolar architecture? One possibility is that a multilayered architecture confers structural flexibility with tunable functionality. It may also offer a platform whose structure can be highly dynamic yet resilient with regulatable refinement. In this regard, it will be interesting to investigate whether and how human disease-associated mutations found in the Cep57-Cep63-Cep152 complex (https://portal.gdc.cancer.gov/) alter the organization, dynamics, and function of the architecture generated by the complex.
MATERIALS AND METHODS
Plasmid construction.
To generate FLAG-tagged Cep57 constructs, a PmeI-NotI fragment containing FL Cep57 (pKM2797) from pCI-neo-HA-Cep57 (pKM1234) was cloned into the pCI-neo-FLAG3 vector (pKM2795) digested by the corresponding enzymes. For the construction of FLAG-tagged Cep57(1–130) (pKM5749), Cep57(120–280) (pKM5750), Cep57(260–374) (pKM5751), or Cep57(355–500) (pKM5752), a PCR-generated PmeI-XhoI fragment containing the respective Cep57 region was subcloned into the pCI-neo-FLAG3 vector digested by the corresponding enzymes. FLAG3-tagged Cep57(1–65) (pKM5782), Cep57(1–96) (pKM5783), Cep57(60–130) (pKM5784), Cep57(60–96) (pKM5815), or Cep57(94–130) (pKM5816) was created by inserting the corresponding PmeI-NotI fragment into the pCI-neo-FLAG3 vector digested by the same enzymes.
To construct FLAG-tagged FL Cep63 (pKM2798), Cep63(1–220) (pKM5779), Cep63(210–405) (pKM5780), Cep63(400–541) (pKM5781), Cep63(1–302) (pKM5788), Cep63(40–220) (pKM5789), Cep63(129–220) (pKM5790), Cep63(1–40) (pKM5841), or Cep63(12–40) (pKM5842), a PCR-generated PmeI-NotI fragment containing the respective Cep63 region was subcloned into the pCI-neo-FLAG3 vector digested by the corresponding enzymes. The pCI-neo-HA-Cep63 (pKM1235) construct was generated by inserting an EcoRV-NotI fragment of the PCR product into the pCI-neo-HA vector (pKM1209) digested by SmaI and NotI. The internal deletion constructs Cep57(Δ32) [Cep57(Δ64–95)] (pKM5937) and Cep63(Δ23) [Cep63(Δ16–38)] (pKM5936) were subcloned by inserting a PmeI-AflIII fragment into the pCI-neo-FLAG3 vector digested by the same enzymes. pEGFP-C1-Cep57 (pKM7264) was created by inserting a BglII-SalI fragment of the Cep57 PCR product into the pEGFP-C1 vector digested by the corresponding enzymes.
Lentiviral FLAG-tagged Cep57 constructs (pKM7146 for the FL protein and pKM7147 for the Δ32 mutant) were generated by inserting an AscI-NotI fragment (end filled with Klenow fragment) containing FL Cep57 (from pKM2797) or Cep57(Δ32) (from pKM5937) into the pHRJ vector digested by AscI-PmeI.
To generate a pETDuet-1-based construct (pKM7290) expressing the Cep57(63–120) and Cep63(1–87) fragments in bacteria (Fig. 3I), a BamHI-NotI fragment containing Cep57(63–120) and an NdeI-XhoI fragment containing Cep63(1–87) were cloned into the corresponding restriction sites in pETDuet-1, in which a tobacco etch virus (TEV) protease cleavage site was introduced between the N-terminal His tag and the Cep57(63–120) fragment.
The FLAG3-Plk4 construct (pKM4591) was generated by inserting a BamHI-SalI fragment containing the Plk4 CTD (residues 581 to 970) into a pHR′.J-FLAG3 vector (pKM4067).
Cell culture transfection and lentivirus.
HEK293 and U2OS cells were cultured according to procedures recommended by the American Type Culture Collection (ATCC). For plasmids, U2OS cells were transfected by using Fugene HD (Promega), and HEK293 cells were transfected by using polyethylenimine (Polysciences). For siRNAs, cells were transfected with 20 nM the indicated siRNAs by using Lipofectamine RNAiMAX (Invitrogen) and then analyzed at least 48 h after transfection. To generate lentiviruses, HEK293 cells were cotransfected with pHR′-CMVΔR8.2Δvpr, pHR′-CMV-VSV-G, and a pHR′.J-CMV-SV-puro-based construct containing the gene of interest. The supernatant from each cotransfected culture was harvested 2 days after transfection. To generate a pool of stable cell lines, U2OS cells were infected with lentiviruses for 1 day and then selected with 2 μg/ml of puromycin for at least 1 day. For experiments in Fig. 5, puromycin-selected cells were then transfected with siCep57. The resulting cells were then immunostained 3 days after silencing endogenous Cep57. siRNAs used for silencing Cep57 expression are siCep57#7 (CACTTATGTTGGAATTAATTAAT), siCep57#8 (CTCAATGTTAAAGCATTTAAATG), and siCep57#9 (GTGTGATTTTTTAAAATAGTTCT).
Confocal and SIM imaging.
U2OS cells cultured on poly-l-lysine (Sigma-Aldrich)-coated no. 1.5 coverslips were fixed with cold methanol (MeOH) for 2 min, washed, and blocked with 3% bovine serum albumin (BSA) in a buffer containing 20 mM Tris-Cl, 150 mM NaCl, and 0.05% Tween 20 (TBST) for 1 h. Afterward, samples were incubated with a primary antibody diluted in 3% BSA in TBST overnight at 4°C, washed with TBST three times, and then incubated with a fluorescence-conjugated secondary antibody for 1 h at room temperature (RT). The resulting samples were washed with TBST three times, stained with Hoechst 33342 for 5 min, and then mounted in Prolong Gold antifade reagent (Thermo Fisher Scientific). Alexa Fluor-conjugated primary antibodies were used where indicated. Chromosomal DNA was decorated with 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired by using the Zeiss LSM880 Airyscan microscope with a 32-channel GaAsP Airyscan detector and a plan-apochromat 63× (numerical aperture [NA], 1.4) oil immersion lens objective or the Zeiss Elyra S1 3D-SIM system equipped with a 63× plan-apochromat (NA, 1.4) oil immersion lens objective, 405/488/561/641-nm solid-state lasers, and excitation and emission filter sets for imaging of x and y fluorescent labels. Acquired images were processed using the SIM processing algorithm in ZEN Black software. To quantify fluorescence signal intensities, images acquired under the same settings were analyzed after processing them to yield the maximum-intensity projection of z-stacks using ZEN Blue software (Carl Zeiss Microscopy, LLC). 3D surface renderings of 3D-SIM image stacks were generated using Imaris software version 8.4.1 (Bitplane).
Immunoprecipitation, biotin pulldown, and immunoblot analyses.
For immunoprecipitation, cells were lysed in a buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2.5 mM EDTA, 1% Triton X-100, 10% glycerol, and a protease inhibitor cocktail [Roche]) using a Precellys 24 homogenizer (Bertin Instruments). The resulting lysates were precleared by centrifugation at 16,000 × g for 20 min at 4°C before being subjected to immunoprecipitation using anti-FLAG antibody (Sigma), essentially as described previously (33).
To pull down proteins that bind to the Cep57 PINC motif, a chemically synthesized 35-mer biotinylated peptide (5′-YPESNSRAIFSALKNLQDKIRRLELERIQAEESVK-GGGSE-biotin [residues 61 to 95 of Cep57 are underlined]) was incubated with HEK293 lysates expressing hemagglutinin (HA)-Cep63 and HA–PACT-CT for 1 h. Biotinylated peptide-binding proteins were precipitated using streptavidin-agarose beads (Thermo Fisher Scientific).
Immunoblot analysis was performed according to standard procedures using an enhanced chemiluminescence detection system (Thermo Fisher Scientific), and immunoblotted signals were captured using a chemiluminescence imager (ChemiDoc imaging systems; Bio-Rad). Signal intensities were quantified using ImageJ (Fiji).
Mass spectrometry analysis for Plk4 CTD-binding proteins.
To identify cellular proteins binding to the Plk4 CTD (residues 581 to 970), HEK293T cells stably expressing FLAG3-Plk4 CTD or control FLAG3 alone were lysed and subjected to coimmunoprecipitation analyses. The samples were then processed essentially as described previously (34). To ensure the validity of the Cep57 peptides found in the CTD coprecipitates, the data were searched twice independently using different versions of Proteome Discoverer programs (Thermo Fisher Scientific) against the UniProt database.
Briefly, staining of gel bands was performed with in-gel tryptic digestion to extract the peptides. Next, each sample was loaded onto an Easy nLC II nanocapillary high-performance liquid chromatography (HPLC) system (Thermo Fisher Scientific) with a C18 Nano Trap column and a C18 Nano analytical column connected online with a Velos Pro dual-pressure linear ion trap mass spectrometer (Thermo Fisher Scientific) for MS analysis. Peptides were eluted using a linear gradient of 2% mobile phase B (acetonitrile with 0.1% formic acid) to 42% mobile phase B within 45 min at a constant flow rate of 200 nl/min. The 15 most intense molecular ions in the MS scan were sequentially selected for collision-induced dissociation (CID) using a normalized collision energy of 35%. The mass spectra were acquired in the mass range of m/z 350 to 2,000. The Nanospray Flex ion source (Thermo Fisher Scientific) capillary voltage and temperature were set at 1.7 kV and 300°C, respectively. The MS data were searched against the Homo sapiens protein database downloaded from the European Bioinformatics Institute website (http://www.ebi.ac.uk/integr8), utilizing SEQUEST HT (Thermo Fisher Scientific) interfaced with Proteome Discoverer version 1.4 (for Cep152, Cep63, and Cep57). The data for Cep57 were confirmed by another independent search with Proteome Discoverer version 2.3. Up to two missed tryptic cleavage sites were allowed, and oxidation (+15.9949 Da) of a methionyl residue was included as a dynamic modification.
Protein expression in E. coli and purification.
The pETDuet-1 construct expressing Cep57(63–120) and Cep63(1–87) (pKM7290) was transformed into Escherichia coli Rosetta(DE3) cells. After the bacterial culture was grown at 37°C to an optical density at 600 nm (OD600) of ∼0.8 to 1.0, cells were continuously cultured at 16°C for 20 h in the presence of 0.5 mM isopropyl-β-d-thiogalactoside. The resulting cells were lysed by sonication, and the protein was purified on Ni-nitrilotriacetic acid (NTA). The His tag was cleaved off by digestion overnight at 4°C with TEV protease. Uncut proteins and TEV protease were removed by a 2nd Ni-NTA step, and the passthrough was further purified in a Hiload 16/600 Superdex 200 column (GE Healthcare) equilibrated with a solution containing 20 mM HEPES (pH 7.5), 50 mM NaCl, 5% glycerol, and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP). Peak fractions containing the Cep57-Cep63 complex with a retention volume of 75 ml were pooled and analyzed by 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Correlative light and electron microscopy (CLEM).
U2OS cells infected with lentivirus expressing FLAG-Cep57 were grown on a 35-mm no. 1.5 glass-bottom gridded dish (MatTek Corporation) for 2 days, washed with phosphate-buffered saline (PBS), and fixed with cold methanol for 2 min. After immunostaining with anti-FLAG antibody and Alexa Fluor 594-conjugated secondary antibody, cells with Cep57 cables were located by 3D-SIM. After fixing the cells with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), the resulting samples were processed for thin-section transmission electron microscopy. The area containing the cells identified by light microscopy was sectioned, and the image was obtained using a Hitachi H-7650 microscope (Hitachi High-Tech). The distance between MT-like lines was measured with Image Pro Plus software (Media Cybernetics).
Cep57 C-supported Cep63-Cep152 assembly around an MT bundle in vitro.
Microtubules (Cytoskeleton, Inc.) were dissolved at 5 mg/ml in a buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5 mM TCEP) and incubated with 100 μM paclitaxel (Sigma-Aldrich) at RT for 30 min. The resulting sample was then further incubated with HEK293 lysates expressing a Cep57(272–499)-mCherry-Cep63 fusion and mGFP-Cep152(924–1295) to determine the ability of Cep63 and Cep152 to concentrically decorate the MT bundle induced by the C-terminal Cep57(272–499) domain.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Michael Kruhlak and Langston Lim for assistance with time-lapse microscopy, Kunio Nagashima for providing technical service with electron microscopy samples, Thorkell Andresson for carrying out the second phase of the mass spectrometry database search using Proteome Discoverer, Yanling Liu for generating 3D surface-rendered movies, and Kunsoo Rhee at Seoul National University, South Korea, for reagents.
This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health (K.S.L.); an NST grant (CAP-16-03-KRIBB) of South Korea (J.K.B.); a National Research Foundation grant (2019M3E5D6063955) funded by the Ministry of Science and ICT of South Korea (B.K.); and a Korean Biomedical Scientist fellowship (KGS1001911) from the KRIBB Research Initiative Program (J.I.A.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Woodruff JB, Wueseke O, Hyman AA. 2014. Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci 369:20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conduit PT, Wainman A, Raff JW. 2015. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol 16:611–624. doi: 10.1038/nrm4062. [DOI] [PubMed] [Google Scholar]
- 3.Vertii A, Hehnly H, Doxsey S. 2016. The centrosome, a multitalented renaissance organelle. Cold Spring Harb Perspect Biol 8:a025049. doi: 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawo S, Hasegan M, Gupta GD, Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 5.Mennella VK, Eszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu J, Glover DM. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol 2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivier N, Keller D, Gonczy P, Manley S. 2013. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One 8:e69004. doi: 10.1371/journal.pone.0069004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TS, Zhang L, Il Ahn J, Meng L, Chen Y, Lee E, Bang JK, Lim JM, Ghirlando R, Fan L, Wang YX, Kim BY, Park JE, Lee KS. 2019. Molecular architecture of a cylindrical self-assembly at human centrosomes. Nat Commun 10:1151. doi: 10.1038/s41467-019-08838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, Johnson CM, Veprintsev D, Zuber B. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, Steinmetz MO. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukinavicius G, Lavogina D, Orpinell M, Umezawa K, Reymond L, Garin N, Gonczy P, Johnsson K. 2013. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr Biol 23:265–270. doi: 10.1016/j.cub.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Bossard C, Laurell H, Van den Berghe L, Meunier S, Zanibellato C, Prats H. 2003. Translokin is an intracellular mediator of FGF-2 trafficking. Nat Cell Biol 5:433–439. doi: 10.1038/ncb979. [DOI] [PubMed] [Google Scholar]
- 14.Park SY, Park JE, Kim TS, Kim JH, Kwak MJ, Ku B, Tian L, Murugan RN, Ahn M, Komiya S, Hojo H, Kim NH, Kim BY, Bang JK, Erikson RL, Lee KW, Kim SJ, Oh BH, Yang W, Lee KS. 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat Struct Mol Biol 21:696–703. doi: 10.1038/nsmb.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banterle N, Gonczy P. 2017. Centriole biogenesis: from identifying the characters to understanding the plot. Annu Rev Cell Dev Biol 33:23–49. doi: 10.1146/annurev-cellbio-100616-060454. [DOI] [PubMed] [Google Scholar]
- 16.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. 2014. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol 15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 17.Aziz K, Sieben CJ, Jeganathan KB, Hamada M, Davies BA, Velasco ROF, Rahman N, Katzmann DJ, van Deursen JM. 2018. Mosaic-variegated aneuploidy syndrome mutation or haploinsufficiency in Cep57 impairs tumor suppression. J Clin Invest 128:3517–3534. doi: 10.1172/JCI120316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe K, Takao D, Ito KK, Takahashi M, Kitagawa D. 2019. The Cep57-pericentrin module organizes PCM expansion and centriole engagement. Nat Commun 10:931. doi: 10.1038/s41467-019-08862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaval B, Doxsey SJ. 2010. Pericentrin in cellular function and disease. J Cell Biol 188:181–190. doi: 10.1083/jcb.200908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, He R, Zhou H, Yu AC, Zhang B, Teng J, Chen J. 2012. Cep57, a NEDD1-binding pericentriolar material component, is essential for spindle pole integrity. Cell Res 22:1390–1401. doi: 10.1038/cr.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He R, Wu Q, Zhou H, Huang N, Chen J, Teng J. 2013. Cep57 protein is required for cytokinesis by facilitating central spindle microtubule organization. J Biol Chem 288:14384–14390. doi: 10.1074/jbc.M112.441501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, Wang T, Zheng T, Teng J, Chen J. 2016. Cep57 is a Mis12-interacting kinetochore protein involved in kinetochore targeting of Mad1-Mad2. Nat Commun 7:10151. doi: 10.1038/ncomms10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snape K, Hanks S, Ruark E, Barros-Nunez P, Elliott A, Murray A, Lane AH, Shannon N, Callier P, Chitayat D, Clayton-Smith J, Fitzpatrick DR, Gisselsson D, Jacquemont S, Asakura-Hay K, Micale MA, Tolmie J, Turnpenny PD, Wright M, Douglas J, Rahman N. 2011. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet 43:527–529. doi: 10.1038/ng.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Momotani K, Khromov AS, Miyake T, Stukenberg PT, Somlyo AV. 2008. Cep57, a multidomain protein with unique microtubule and centrosomal localization domains. Biochem J 412:265–273. doi: 10.1042/BJ20071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown NJ, Marjanovic M, Luders J, Stracker TH, Costanzo V. 2013. Cep63 and Cep152 cooperate to ensure centriole duplication. PLoS One 8:e69986. doi: 10.1371/journal.pone.0069986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillingham AK, Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep 1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinson L, Mannini L, Willems M, Cucco F, Sirvent N, Frebourg T, Quarantotti V, Collet C, Schneider A, Sarda P, Genevieve D, Puechberty J, Lefort G, Musio A. 2014. CEP57 mutation in a girl with mosaic variegated aneuploidy syndrome. Am J Med Genet A 164A:177–181. doi: 10.1002/ajmg.a.36166. [DOI] [PubMed] [Google Scholar]
- 28.Brightman DS, Ejaz S, Dauber A. 2018. Mosaic variegated aneuploidy syndrome caused by a CEP57 mutation diagnosed by whole exome sequencing. Clin Case Rep 6:1531–1534. doi: 10.1002/ccr3.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Kim J, Rhee K. 2019. PCNT is critical for the association and conversion of centrioles to centrosomes during mitosis. J Cell Sci 132:jcs225789. doi: 10.1242/jcs.225789. [DOI] [PubMed] [Google Scholar]
- 30.Sieben C, Banterle N, Douglass KM, Gonczy P, Manley S. 2018. Multicolor single-particle reconstruction of protein complexes. Nat Methods 15:777–780. doi: 10.1038/s41592-018-0140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonczy P. 2015. Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer 15:639–652. doi: 10.1038/nrc3995. [DOI] [PubMed] [Google Scholar]
- 32.Nigg EA, Holland AJ. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19:297–312. doi: 10.1038/nrm.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KS, Yuan Y-L, Kuriyama R, Erikson RL. 1995. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol 15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JE, Zhang L, Bang JK, Andresson T, DiMaio F, Lee KS. 2019. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat Commun 10:4959. doi: 10.1038/s41467-019-12619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arquint C, Nigg EA. 2016. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem Soc Trans 44:1253–1263. doi: 10.1042/BST20160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu J, Hagan IM, Glover DM. 2015. The centrosome and its duplication cycle. Cold Spring Harb Perspect Biol 7:a015800. doi: 10.1101/cshperspect.a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.