Abstract
Flaviviruses encompass not only medically relevant arthropod-borne viruses (arboviruses) but also insect-specific flaviviruses (ISFs) that are presumably maintained primarily through vertical transmission in the insect host. Interestingly, ISFs are commonly found infecting important arbovirus vectors such as the mosquito Aedes aegypti. Cell-fusing agent virus (CFAV) was the first described ISF of mosquitoes more than four decades ago. Despite evidence for widespread CFAV infections in A.aegypti populations and for CFAV potential to interfere with arbovirus transmission, little is known about CFAV evolutionary history. Here, we generated six novel CFAV genome sequences by sequencing three new virus isolates and subjecting three mosquito samples to untargeted viral metagenomics. We used these new genome sequences together with published ones to perform a global phylogenetic analysis of CFAV genetic diversity. Although there was some degree of geographical clustering among CFAV sequences, there were also notable discrepancies between geography and phylogeny. In particular, CFAV sequences from Cambodia and Thailand diverged significantly, despite confirmation that A.aegypti populations from both locations are genetically close. The apparent phylogenetic discrepancy between CFAV and its A.aegypti host in Southeast Asia indicates that other factors than host population structure shape CFAV genetic diversity.
Keywords: insect-specific virus, Aedes aegypti, phylogenetic analysis
Introduction
Flaviviruses infect various vertebrate and invertebrate organisms (Gould and Solomon 2008; Blitvich and Firth 2015, 2017; Shi et al. 2018; Skoge et al. 2018; Parry and Asgari 2019). The most intensively studied members of the Flavivirus genus are medically relevant mosquito-borne flaviviruses (MBFs) and tick-borne flaviviruses (TBFs) that infect both hematophagous arthropods (mosquitoes and ticks, respectively) and vertebrate animals, including humans. For example, 390 million dengue virus infections are estimated to occur in the human population every year, of which 96 million results in clinically apparent symptoms such as fever, headache, joint pain, and rash (Bhatt et al. 2013). Such dual-host flaviviruses belong to arthropod-borne viruses (arboviruses) and their invertebrate host is generally referred to as a vector.
Interestingly, there also exist members of the Flavivirus genus that are considered to lack an invertebrate vector and are designated as ‘no known vector’ flaviviruses (Gould and Solomon 2008; Blitvich and Firth 2017; Shi et al. 2018). For example, Rio Bravo virus causes persistent but asymptomatic infections in bats and was also associated with symptomatic human infections acquired in the laboratory (Sulkin, Sims, and Allen 1966). Likewise, there are flaviviruses found in mosquitoes that are incapable of replicating in vertebrate cells (Blitvich and Firth 2015). Such insect-specific flaviviruses (ISFs) can be phylogenetically divided into two different groups: classical ISFs (cISFs), which form a divergent clade from vector-borne flaviviruses, and dual-host affiliated ISFs (dISFs), which are not monophyletic and are genetically close to MBFs (Guzman et al. 2018). ISFs have been shown to interact with arboviruses in vitro and in vivo (Goenaga et al. 2015; Hall-Mendelin et al. 2016; Baidaliuk et al. 2019) and can be ecologically associated with arboviruses in nature (Newman et al. 2011). ISFs are believed to be maintained primarily by vertical transmission from mother to offspring (Lutomiah et al. 2007; Bolling et al. 2012; Contreras-Gutierrez et al. 2017).
The genome of cISFs has a very similar structure to that of all flaviviruses. It consists of 5ʹ- and 3ʹ-untranslated regions (UTRs) and a single open reading frame (ORF), which encodes structural (C, prM, E) and nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) proteins. However, one remarkable difference with MBFs is the presence of an ORF called fifo, which results from a −1 ribosomal frameshift producing a protein of about 250 amino-acid residues in all cISFs (Firth et al. 2010). Interestingly, the fifo ORF is disrupted by premature stop codons in the cell-fusing agent virus (CFAV) strain that persistently infects the Aedes aegypti mosquito cell line Aag2 (Stollar and Thomas 1975; Cammisa-Parks et al. 1992; Firth et al. 2010; Maringer et al. 2017; Weger-Lucarelli et al. 2018).
CFAV was the first cISF discovered in A.aegypti mosquito cells in culture in 1975 and subsequently sequenced in 1992 (Stollar and Thomas 1975; Cammisa-Parks et al. 1992). Since its discovery, CFAV has been detected and/or isolated mainly from A.aegypti mosquitoes across a large number of locations around the world including Puerto Rico, Indonesia, Thailand, Mexico, Kenya, USA, Australia, Turkey, and Brazil (Cook 2006; Kihara et al. 2007; Hoshino et al. 2009; Espinoza-Gomez et al. 2011; Yamanaka et al. 2013; Bolling et al. 2015; Metsky et al. 2017; Ajamma et al. 2018; Fernandes et al. 2018; Iwashita et al. 2018; Zakrzewski et al. 2018; Akiner et al. 2019). Although CFAV was initially discovered more than four decades ago and is ubiquitous in mosquitoes worldwide, very little is known about its evolutionary history. Recently, we demonstrated that prior infection by CFAV reduces dengue virus and Zika virus dissemination in A.aegypti (Baidaliuk et al. 2019), supporting the idea that CFAV, and possibly other ISFs, may contribute to modulate arbovirus transmission in nature.
In this study, we generated six novel CFAV genome sequences and used them to perform a global phylogenetic analysis of CFAV genetic diversity. The six full or nearly full CFAV sequences were obtained from CFAV strains newly isolated from field-derived A.aegypti laboratory colonies (two from Cambodia and one from Uganda), and by untargeted viral metagenomics of a single colonized female from Cambodia and of two wild-caught A.aegypti specimens (one single female from Cambodia and one mosquito pool from Guadeloupe). Although we found some degree of geographical clustering among CFAV sequences, all four new CFAV genome sequences from Cambodia diverged significantly from published CFAV sequences from Thailand. Microsatellite genotyping of A.aegypti from both locations revealed a lack of phylogenetic congruence between CFAV and its A.aegypti host in Southeast Asia.
Materials and Methods
Ethics statement
Mosquito collections in Cambodia were approved by the Cambodian National Ethics Committee for Health Research (Protocol 063NECHR) and by the Institut Pasteur Ethics Board.
CFAV isolation
Three new CFAV strains were isolated from recently established colonies of A.aegypti from Cambodia and Uganda. The first CFAV strain, named CFAV-KC, was isolated from a mosquito colony initiated in Tratav village, Chi Ror 2 district, Tboung Khmum commune, Kampong Cham province, Cambodia in 2014. The second CFAV strain, named CFAV-PP, was isolated from a mosquito colony initiated in Tror Penang village, Kmouch district, Sen Sok commune, Phnom Penh city, Cambodia in 2015. The third CFAV strain, named CFAV-Zik, was isolated from a mosquito colony initiated in a village close to Zika forest, Wakiso district, Uganda in 2016. Prior to CFAV isolation, mosquito colonies were maintained under standard insectary conditions (27 °C, 70% relative humidity and 12:12 h light:dark cycle) for two and three generations for mosquitoes from Cambodia and Uganda, respectively. CFAV was isolated as previously described (Baidaliuk et al. 2019). In brief, adult mosquitoes were homogenized in pools of fifteen individuals (Uganda colony) or fifty individuals (Cambodia colonies) in 1 ml of Leibovitz’s L-15 medium (Gibco ThermoFisher Scientific). Homogenates were clarified by centrifugation (21,100 g, 4 °C, 5 min) and supernatants were filtered through 0.2-µm filters (Minisart, Merck). Sub confluent C6/36 (Aedes albopictus) cells in 25-cm2 flasks were inoculated with 500 µl of the filtered homogenate and incubated at 28 °C. After 1 h of incubation, 7 ml of Leibovitz’s L-15 medium complemented with 2 per cent fetal bovine serum (FBS, Gibco ThermoFisher Scientific), 2 per cent tryptose phosphate broth (TPB, Gibco ThermoFisher Scientific), 1× nonessential amino acids (NAA, Gibco ThermoFisher Scientific), 10 U/ml of penicillin (Gibco ThermoFisher Scientific), and 10 µg/ml of streptomycin (Gibco ThermoFisher Scientific) were added to the flask. After 7 days of virus amplification, the cell-culture supernatants were harvested and aliquoted with 10 per cent FBS. pH was adjusted with 0.075 per cent of sodium bicarbonate and the virus stocks were stored at −80 °C. The CFAV isolates were subsequently passaged once (CFAV-PP and CFAV-KC) or twice (CFAV-Zik) in C6/36 cells as described above to amplify virus stocks.
Genome sequencing of CFAV isolates
The three CFAV strains isolated from A.aegypti laboratory colonies were sequenced as previously described (Baidaliuk et al. 2019). In brief, RNA was extracted from virus stock using QIAamp Viral RNA Mini Kit (QIAGEN) according to manufacturer’s instructions and treated with TURBO DNase (Ambion). cDNA was produced with random hexamer primers (Roche) using M-MLV reverse transcriptase (Invitrogen) following manufacturer’s instructions. It was incubated with Escherichia coli DNA ligase (New England BioLabs), E.coli DNA polymerase I (New England BioLabs), and E.coli RNase H (New England BioLabs) for second-strand synthesis with Second Strand Synthesis Buffer (New England BioLabs). dsDNA was purified with Agencourt AMPure XP beads (Beckman Coulter) and used for library preparation using Nextera XT DNA Kit (Illumina) according to the manufacturer’s instructions followed by a cDNA quality check with Bioanalyzer DNA 1000 kit (Agilent). The libraries were combined with other libraries from unrelated projects and sequenced on an Illumina NextSeq 500 instrument (150 bp cycles, paired ends). Raw sequencing datasets are available in the Sequence Read Archive (SRA) database under accession number PRJNA556544. The sequencing data were processed following a pipeline described elsewhere (Lequime et al. 2017). In brief, sequencing reads with a quality score less than thirty were trimmed using Trimmomatic v0.36 (Bolger, Lohse, and Usadel 2014). Reads were mapped to the host reference genome (GCA_001876365.2, VectorBase) using Bowtie2 v2.3.4.3, and remaining reads were subjected to de novo assembly with Ray v2.3.1-mpi followed by BLAST search and selection of viral contigs in Geneious v.10 (http://www.geneious.com/) (Boisvert, Laviolette, and Corbeil 2010; Langmead and Salzberg 2012). A seeded de novo assembly with IVA v1.0.3 was applied to verify previously obtained contigs (Hunt et al. 2015). Consensus CFAV genome sequences were constructed by scaffold mapping with Bowtie2 v2.3.4.3. The 3ʹ and 5ʹ ends of the consensus CFAV genome sequences from the isolated viruses were verified by rapid amplification of cDNA ends (RACE) using 5ʹ/3ʹ RACE kit, second Generation (Roche), following manufacturer’s instructions with prior Poly(A) addition to 3ʹ end of RNA using E.coli Poly(A) Polymerase (New England Biolabs). CFAV full-genome sequences are available in the European Nucleotide Archive (ENA) database (PRJEB33690: LR694076, LR694077, LR694078). For all viruses, single-nucleotide variant (SNV) frequencies and coverage were calculated using LoFreq v2.1.3.1 and bedtools v2.25.0, respectively (Quinlan and Hall 2010; Wilm et al. 2012). The coverage and SNV frequency results were visualized using ggplot2 v3.2.0 and cowplot v0.9.4 packages in R v3.5.2 (R Development Core Team 2013; Wickham 2016; Wilke 2019).
CFAV genomes obtained by untargeted viral metagenomics
Three novel nearly full CFAV genome sequences were obtained by untargeted viral metagenomics from A.aegypti specimens from Cambodia and Guadeloupe. The first sequence was obtained from a single wild-caught female in Kampong Cham, Cambodia in 2013. The second sequence was obtained from a single female of the third laboratory generation of an A.aegypti colony derived in 2013 from Kampong Cham, Cambodia. The third sequence was obtained from a pool of twenty-four wild-caught females in Les Abymes, Guadeloupe in 2016. For mosquito samples from Cambodia, carrier RNA and host ribosomal RNA were depleted from the RNA samples following a published protocol (Matranga et al. 2014) and RNA from selective depletion was used for cDNA synthesis. Libraries were prepared using the Nextera XT Library Preparation Kit (Illumina) and sequenced on an Illumina NextSeq 500 instrument (75 cycles, paired ends). Reads were assembled using SPAdes v3.7.0 and contigs were annotated using BLAST (Altschul 1997; Bankevich et al. 2012). The preassembled genome sequences were used in a second step to map the reads using CLC Genomics Assembly Cell v5.1 (https://www.qiagenbioinformatics.com). Read mapping files with CFAV reference sequences are available in the SRA database under accession number PRJNA556544. For the mosquito pool from Guadeloupe, the CFAV genome sequence was obtained using the NetoVIR protocol for viral metagenomics (Conceicação-Neto et al. 2015). In brief, whole mosquitoes were homogenized, centrifuged, and filtered to enrich for viral particles. The filtrate was treated with Benzonase (Novagen) and Micrococcal Nuclease (New England Biolabs) to digest free-floating nucleic acids. DNA and RNA were extracted (QIAGEN Viral RNA mini kit), reverse transcribed, and randomly amplified using a slightly modified Whole Transcriptome Amplification 2 (WTA2) Kit procedure (Sigma‐Aldrich). WTA2 products were purified and used to prepare libraries using the Nextera XT Library Preparation Kit (Illumina). Libraries were sequenced on an Illumina NextSeq 500 instrument (300 bp cycles, paired ends). The reads were cleaned with Trimmomatic (Bolger, Lohse, and Usadel 2014) and de novo assembled into contigs using metaSPAdes (Nurk et al. 2017), followed by annotation using DIAMMOND (Buchfink, Xie, and Huson 2015). Raw reads were remapped to the closest available CFAV genome sequence from Macapá, Brazil (CFAV-Macapa2 (Fernandes et al. 2018)) using BWA (Li and Durbin 2009). A 20-nucleotide gap and a few ambiguous nucleotides were resolved by PCR and Sanger sequencing. The primers and PCR-verified positions are provided in Supplementary material S1. The raw sequencing dataset of the mosquito pool from Guadeloupe is available in the SRA database under accession number SAMN10762280. The three consensus CFAV sequences obtained by untargeted viral metagenomics were ultimately verified by read mapping with Bowtie2 v2.3.4.3 (Langmead and Salzberg 2012). All three CFAV sequences from untargeted metagenomics of A.aegypti are available in the ENA under accession numbers (PRJEB33690: LR694079, LR694080, LR694081). SNV frequencies and coverage were calculated using LoFreq v2.1.3.1 and bedtools v2.25.0, respectively (Quinlan and Hall 2010; Wilm et al. 2012). The coverage and SNV frequency results were visualized using ggplot2 v3.2.0 and cowplot v0.9.4 packages in R v3.5.2 (R Development Core Team 2013; Wickham 2016; Wilke 2019).
Survey of published CFAV genome sequences
The oldest wild-type CFAV full-genome sequence is available in GenBank under accession number GQ165810 and was obtained from a strain named Rio Piedras02 (Cook et al. 2009) initially detected by RT-PCR in mosquitoes from Puerto Rico in 2002 (Cook 2006). In 2011, CFAV was isolated from the first laboratory generation of A.aegypti mosquitoes from Axochiapan, Morelos, Mexico and the full polyprotein sequence is available in GenBank under accession number KJ476731. In 2012, a full-genome sequence was obtained from a CFAV isolate derived from an A.aegypti laboratory colony originating in Galveston, Texas, USA (Bolling et al. 2015) and is available in GenBank under accession number KJ741267. A full-genome sequence available in European Nucleotide Archive under accession number LR596014 was obtained from a CFAV strain recently isolated from an A.aegypti laboratory colony originating in Kamphaeng Phet, Thailand in 2013 (Baidaliuk et al. 2019). Two nearly full CFAV genome sequences derived from RNA-seq data of A.aegypti specimens caught in Cairns, Australia and Bangkok, Thailand in 2014 and 2015, respectively (Zakrzewski et al. 2018), were requested directly from the authors. CFAV genome sequences were remapped to obtain a consensus sequence and calculate SNV frequencies and coverage as described above. Two nearly full CFAV genomes were detected in wild-caught A.aegypti specimens from Miami, USA in 2016 (Metsky et al. 2017). Reads were de novo assembled to generate consensus genome sequences and calculate coverage and SNV frequencies as described above. The consensus sequences from both studies are now available in the ENA (PRJEB33801: LR694072, LR694073, LR694074, LR694075). Another set of nearly full-genome sequences of CFAV was detected in RNA-seq libraries of mosquitoes from Macapá, Amapá, Brazil, not only in A.aegypti but also in Culex spp. pools in 2017 (Fernandes et al. 2018) and were requested directly from the authors. In addition to mosquito-derived CFAV sequences, the CFAV strain persistently infecting the Aag2 cell line was sequenced on four separate occasions and the genome sequences are available in GenBank under accession numbers M91671, KU936054, MH237596, and MH310082 (Cammisa-Parks et al. 1992; Maringer et al. 2017; Di Giallonardo et al. 2018; Weger-Lucarelli et al. 2018). The main phylogenetic analyses relied on full or nearly full CFAV genomes, but additional analyses incorporated several partial CFAV genome sequences that are available from Github (https://github.com/artembaidaliuk/CFAV-cISF-phylogeny-2019.git).
CFAV phylogenetic analyses
All CFAV genome sequences used in this study and their genetic annotations according to Blitvich and Firth (2015) are available from Github (https://github.com/artembaidaliuk/CFAV-cISF-phylogeny-2019.git). Nucleotide alignments were prepared using local pairwise alignment (L-INS-i) in MAFFT v.7.407 (Katoh 2002) for the three subsets of sequences: (i) only sequences with full or nearly full ORF available, (ii) sequences that were used to test phylogenetic congruence in SE Asia, and (iii) full ORFs and partial sequences. Full-ORF alignment was subjected to recombination tests in RDP4 v.4.95 (Martin et al. 2015). Alignment positions with >20 per cent gaps, if any, were trimmed in the initial alignments using Goalign v.0.2.7 (https://github.com/evolbioinfo/goalign). Final single-gene alignments were concatenated into a single alignment. All single-gene alignments as well as a full-ORF alignment of the first subset of sequences were used for nucleotide substitution model search by ModelFinder in IQ-TREE v.1.6.3 and v.1.6.10 (Nguyen et al. 2015; Kalyaanamoorthy et al. 2017). The best model for each alignment was chosen based on the bayesian information criterion (BIC) score for the most comprehensive dataset (i) (Supplementary material 2). In data subsets (i) and (iii), an edge-unlinked partition model with a separate substitution model for each genome region was tested; however, none of the partition schemes resulted in a better model than a single model for the full ORF according to BIC scores. The test was performed using the −MF+MERGE option in IQ-TREE. The best substitution model for the ORF alignment of subset (i) and the concatenated alignment of subset (iii) was GTR+F+G4 and two almost equally best models for the ORF alignment of subset (ii) were GTR+F + I and GTR+F+G4, therefore, the latter was chosen for the following tree reconstruction of all three data subsets. Consensus phylogenetic trees were reconstructed in IQ-TREE from 1,000 ultrafast bootstrap maximum likelihood (ML) tree replicates. The trees were built as unrooted and visualized with midpoint rooting because including the closest possible outgroup sequences did not allow reliable rooting. In data subset (ii), constrained trees were built following different clustering scenarios and tested for the differences in topology with an unconstrained tree using approximately unbiased (AU) test implemented in IQ-TREE (Shimodaira 2002). In addition, alignments for data subsets (i) and (ii) were used for reconstruction of Bayesian phylogenetic trees with BEAST v1.10.4 (Suchard et al. 2018). A Markov chain Monte Carlo of 108 steps was run in three independent replicates for each data subset. A GTR+G4 model with uncorrelated relaxed lognormal clock was used. The population size was considered constant due to sparse geographical and temporal sampling of the virus. Effective sample sizes were estimated and confirmed to exceed 200 for all three replicates for both data subsets using Tracer v1.7.1 (Rambaut et al. 2018). The data from the three replicates were merged after removing 10 per cent of burn-in states. Maximum clade credibility (MCC) consensus trees were built with median node heights from the total of 2,700 trees for each data subset. Since the tree topologies of MCC and consensus ML trees matched, we provided posterior probability values as node support mapped onto the consensus ML trees together with the ultrafast bootstrap proportion values. The consensus tree topology of data subset (ii), the tree topology of A.aegypti, and the alternative tree topologies of posterior Bayesian trees of data subset (ii) were compared using the ape package in R v.3.6.1 (Paradis, Claude, and Strimmer 2004; R Development Core Team 2013).
cISF phylogenetic analyses
All full or nearly full-ORF cISF sequences available in GenBank (March 2019) and the novel CFAV sequences were used for recombination and phylogenetic analyses. The full data subset of cISF sequences, their annotations based on available sources, and metadata are available from Github (https://github.com/artembaidaliuk/CFAV-cISF-phylogeny-2019.git). Amino-acid alignments of each protein were obtained by pairwise alignment (L-INS-I, BLOSUM62) in MAFFT v.7.407. After trimming positions containing >20 per cent of gaps with Goalign v.0.2.7, both the amino-acid and back-translated nucleotide alignments were kept for further analyses. Amino-acid guided nucleotide alignments of all genes were concatenated into a full-ORF alignment and subjected to recombination tests in RDP4 v4.95. The protein alignments containing three outgroup sequences (dengue virus 1, tick-borne encephalitis virus, and Rio Bravo virus) are available from Github (https://github.com/artembaidaliuk/CFAV-cISF-phylogeny-2019.git). They were concatenated into structural (C-prM-E) and nonstructural (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) protein alignments as well as a full polyprotein alignment. The best substitution model according to BIC scores was selected by ModelFinder in IQ-TREE v.1.6.12. For all three alignments, LG+F+R5 was the best model, and it was used to reconstruct consensus ML trees from 1,000 ultrafast bootstrap replicates in IQ-TREE based on structural and nonstructural fractions of the polyprotein. In all phylogenetic analyses, Biopython was used for data manipulation and visualization (Cock et al. 2009) in Python 3.6.0 (https://www.python.org), BMGE v1.12 (Criscuolo and Gribaldo 2010), FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree), and Geneious v.10.2.3 (https://www.geneious.com).
Aedes aegypti population genetics
To assess the genetic homogeneity of A.aegypti populations in Southeast Asia, twelve male and thirty female A.aegypti adults trapped in various locations in Kampong Cham and Phnom Penh provinces of Cambodia were individually used for A.aegypti genotyping assays. Initial species identification was done by visual inspection. Total DNA from individual mosquitoes was extracted using the DNeasy Blood and Tissue kit (QIAGEN) according to the manufacturer’s instructions. Species identification was verified by Sanger sequencing of the cytochrome c oxidase subunit I gene. Newly genotyped specimens from Cambodia were analyzed together with previously published data for A.aegypti populations from Lunyo, Uganda (n = 42); Bangkok, Thailand (n = 42); Amacuzac, Morelos, Mexico (n = 42); Houston, Texas, USA (n = 19); Patillas, Puerto Rico (n = 42); Hanoi, Vietnam (n = 54); Ho Chi Minh City, Vietnam (n = 54); Attock, Pakistan (n = 49); Riyadh, Saudi Arabia (n = 84); Tahiti, French Polynesia (n = 48); Cairns, Australia (n = 45); Nairobi, Kenya (n = 54); Cebu City, Philippines (n = 108); and Key West, Florida, USA (n = 52) (Gloria-Soria et al. 2016a,b; Kotsakiozi et al. 2018). Population structure was evaluated using twelve previously validated microsatellite markers (Slotman et al. 2007; Brown et al. 2011; Gloria-Soria et al. 2016b) via the Bayesian clustering method implemented in the STRUCTURE v. 2.3 software (Pritchard, Stephens, and Donnelly 2000). STRUCTURE identifies genetic clusters and assigns individuals to these clusters with no a priori information of sample location. Twenty independent runs were conducted for each predefined number of clusters (K = 1 to K = 6). Each run assumed an admixture model and correlated allele frequencies using a burn-in value of 100,000 iterations followed by 500,000 repetitions. Results were plotted with the program DISTRUCT v.1.1. (Rosenberg 2004). Pairwise genetic distances (Cavalli-Sforza and Edwards 1967) were calculated and used to generate a neighbor-joining tree with the adegenet package in R v. 3.4.0 (Jombart 2008). Bootstrap replicates were performed using the poppr R package (Kamvar, Tabima, and Grünwald 2014).
Results
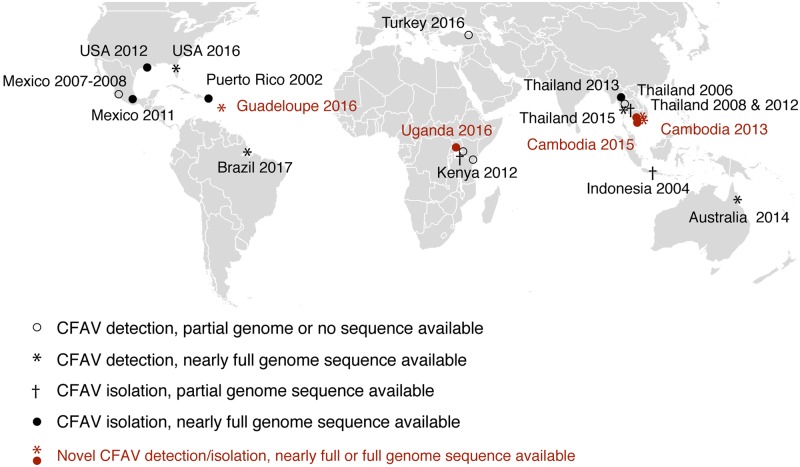
Our initial literature survey revealed that in the last two decades, CFAV has been detected and/or isolated throughout tropical and subtropical regions of the world, as summarized in Fig. 1. To shed light on the evolutionary history of CFAV, we retrieved all full or nearly full-genome sequences already available and produced six novel genome sequences to perform a global phylogenetic analysis of CFAV genetic diversity.
Figure 1.
Global distribution of CFAV. CFAV detection and isolation history are represented on the world map. Symbols represent detection or isolation events annotated with the country of mosquito origin and the year of the event. Novel isolations and detections from this study are shown in red symbols.
To generate novel CFAV genome sequences, we used two approaches. First, we sequenced the full genome of three CFAV strains that we recently isolated from early generations of A.aegypti laboratory colonies, named CFAV-KC (second laboratory generation, Kampong Cham, Cambodia), CFAV-PP (second laboratory generation, Phnom Penh, Cambodia), and CFAV-Zik (third laboratory generation, village near Zika forest, Uganda). CFAV was isolated by inoculation of mosquito homogenate onto C6/36 cell cultures and one to two amplification passages. The three virus genome sequences were obtained by deep sequencing of cDNA libraries generated from virus stocks, followed by de novo assembly and verification of the extremities by rapid amplification of cDNA ends. Second, we assembled three novel CFAV genome sequences by untargeted viral metagenomics. The first one (CFAV-mosq395) was obtained from single wild female A.aegypti caught in Kampong Cham, Cambodia. The second one (CFAV-mof2522) was obtained from a single female from the third laboratory generation of an A.aegypti colony originating from Kampong Cham, Cambodia. The third one (CFAV-Guadeloupe) was derived from a pool of wild A.aegypti mosquitoes caught in Guadeloupe, French West Indies.
In addition to the six novel CFAV sequences that we generated in this study, we assembled two nearly full genomes of CFAV (CFAV-FL-05-MOS and CFAV-FL-08-MOS) that had been previously detected in cDNA libraries from pools of wild-caught A.aegypti in FL, USA (Metsky et al. 2017). We also remapped two CFAV genome sequences from Thailand and Australia (named CFAV-Bangkok and CFAV-Cairns, respectively) that were previously obtained from RNA sequencing of wild-caught A.aegypti pools (Zakrzewski et al. 2018).
We first examined the patterns of minority genetic variants in all the CFAV deep-sequencing datasets available (Supplementary Fig. S1). With the exception of CFAV-PP, CFAV isolates (CFAV-Zik, CFAV-KC, and previously reported CFAV-KPP (Baidaliuk et al. 2019)) displayed a relatively homogeneous viral population with a majority of SNVs at frequencies <10 per cent. CFAV-PP exhibited multiple major SNVs at ∼25 per cent frequency across the genome, indicating a possible mixed infection. The majority of CFAV sequences obtained by untargeted viral metagenomics (CFAV_FL_05_MOS, CFAV_Cairns, CFAV_Bangkok, CFAV_mosq395, CFAV_Guadeloupe) was more genetically heterogeneous than those from virus stocks, which could reflect true genetic diversity or assembly errors. For instance, CFAV-mosq395 displayed multiple SNVs below 25 per cent in frequency across the whole genome as well as a few variants above 25 per cent. There were also multiple SNVs along the CFAV-Guadeloupe genome sequence. Despite a similar depth of coverage, CFAV-FL-05-MOS exhibited numerous SNVs at frequencies ranging from 1 per cent to 50 per cent across the genome, whereas CFAV-FL-08-MOS had localized clusters of low-frequency SNVs and only two SNVs at >25 per cent frequency. CFAV-Bangkok and CFAV-Cairns also displayed abundant high-frequency SNVs.
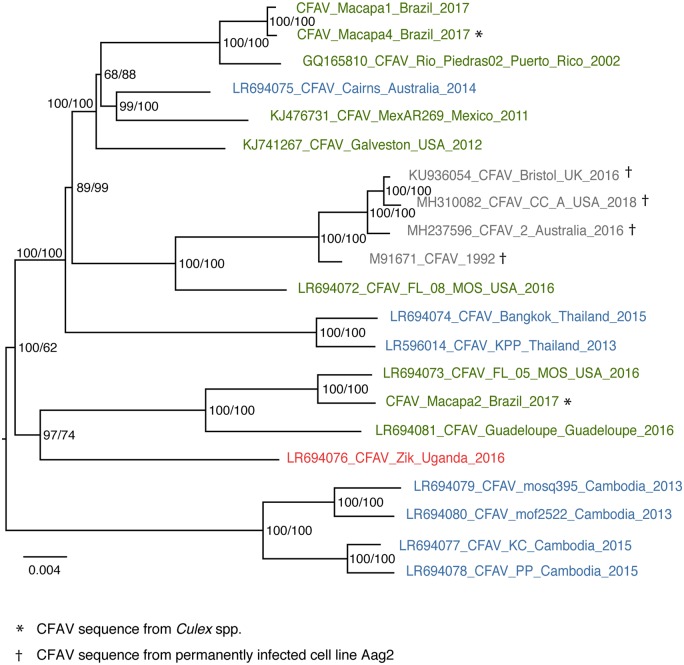
All the novel CFAV consensus sequences described above were combined with other published CFAV sequences, including four distinct CFAV genome sequences derived from the Aag2 cell line (https://github.com/artembaidaliuk/CFAV-cISF-phylogeny-2019.git), to perform phylogenetic analyses based on previously described annotations of single genes (Blitvich and Firth 2015). We first performed a phylogenetic analysis of all available nearly full-genome CFAV sequences to date (n = 21). The full polyprotein ORF or separate gene nucleotide alignments were constructed and concatenated into a full-ORF alignment. We initially performed seven recombination tests (RDP, GENECONV, BootScan, MaxChi, Chimera, SiScan, 3Seq) using the RDP4 software. Two recombination signals were detected with statistical significance in three or more recombination tests (Supplementary Fig. S2A). The first recombination signal (reported in its longest version, with confidence intervals of the breakpoint positions) was detected between ORF positions 3389 (99% CI 3,326–3,532) and 3927 (99% CI 3,726–4,082) by all seven recombination tests in the four CFAV sequences derived from the Aag2 cell line. Interestingly, this region corresponds to fifo, which is genetically divergent and disrupted by premature stop codons in all Aag2-derived CFAV strains (Firth et al. 2010). Recombination tests failed to assign a minor parent in the recombination triplet. The second recombination signal was detected in the NS4A-NS4B region between ORF positions 6053 (99% CI 5,792–7,164) and 7105 (99% CI 5,792–7,164) by only three recombination tests (RDP, BootScan, MaxChi). This recombination signal was detected in a subset of CFAV sequences (CFAV_Cairns, CFAV_Macapa1, CFAV_Macapa4, CFAV_Rio_Piedras02, CFAV_Galveston, and CFAV_MexAR269). The putative major parent was identified as Aag2-derived strain CFAV_CC_A, and the minor parent was identified as the CFAV_Guadeloupe strain. Given the passage history and known genetic divergence of the Aag2-derived CFAV strain in the fifo region, we considered these recombination signals as probable false positives (Firth et al. 2010).
The best-fitting substitution model for the full polyprotein ORF alignment (10,023 nucleotide sites in total, 9,441 for the shortest fully covered region) was GTR+F+G4 (log-likelihood = −29,929.5618, BIC = 60,301.3302). Overall, the global phylogenetic relationships of CFAV sequences showed some degree of geographical clustering and also notable discrepancies between geography and phylogeny (Fig. 2). All four Aag2-derived CFAV sequences clustered together and branched closest to a CFAV sequence from FL, USA. These five CFAV sequences formed a sister clade to a majority of the CFAV sequences from the Americas (Brazil, Puerto Rico, Mexico, and USA). However, the predominantly American clade also contained the CFAV sequence from Cairns, Australia. Moreover, several other CFAV sequences from the Americas (Brazil, Guadeloupe, and USA) formed another clade, which branched closest (albeit still rather distantly) to the only African full-genome sequence available (Uganda). The most striking discrepancy between geographical and phylogenetic distance was the case of Southeast Asian CFAV sequences. Whereas both CFAV sequences from Thailand branched in basal position of the predominantly American clade, all four CFAV sequences from Cambodia formed a separate clade that was highly divergent from all other sequences. The CFAV phylogeny based on all full and partial ORF sequences available was largely consistent with the full-ORF phylogeny (Supplementary Fig. S3). In particular, CFAV sequences from Cambodia remained phylogenetically distant from all CFAV sequences from Thailand in both full-ORF and partial gene trees.
Figure 2.
Limited geographical structuring of the CFAV phylogeny. Consensus tree of 1,000 ultrafast bootstrap replicate ML trees represents phylogenetic relationships between all CFAV sequences based on the nearly full ORF alignment. Tips are color-coded according to the geographic origin of the sequences (red: Africa; green: Americas; blue: Asia-Pacific). Tips in gray represent CFAV sequences derived from the Aag2 cell line. Tips indicate the GenBank or ENA accession number (if any), virus isolate/strain name, country of origin, and the year of detection/isolation. The tree is midpoint rooted. Node support values include both ultrafast bootstrap proportion (%, first value) and posterior probability derived from the Bayesian maximum clade credibility tree (%, second value). The scale bar represents branch length in substitutions per site.
The first explanation to the observed phylogenetic divergence between CFAV sequences from Thailand and Cambodia that we investigated was the existence of alternative tree topologies that we may have overlooked. We selected a subset of the CFAV full-genome sequences that included the four sequences from Cambodia, both sequences from Thailand, three sequences from the Americas (Puerto Rico, Mexico, and USA), and the single sequence from Africa (Uganda). This subset of sequences was chosen to exclude sequences from metagenomics datasets with potentially mixed infections, and to minimize the number of alternative topologies. We used this subset of wild-type CFAV sequences to build ORF phylogenetic trees that had a constrained topology between the clades and unconstrained topology within the clades. Using an AU tree topology test performed on unrooted trees, we found that alternative tree topologies (sequences from Cambodia and Thailand constrained to group together, or sequences from Cambodia and the Americas constrained to group together) were significantly less likely than the unconstrained topology (p = 0.0002). In addition, we explored the distinct tree topologies that were present in the Bayesian posterior tree distribution. To strictly compare the topologies and avoid rooting ambiguity, the tree root was artificially fixed between the African and non-African sequences. There were only two alternative tree topologies, which corresponded to rearrangements within the American clade. In both alternative topologies, the sequences from Thailand remained in the same clade as the sequences from the Americas and the sequences from Cambodia formed a sister clade to the clade comprising the sequences from Thailand and the Americas. Regardless of the tree rooting and the phylogenetic approach, these analyses confirmed that indeed the CFAV sequences from Thailand are phylogenetically closer to the sequences from the Americas than to the sequences from Cambodia.
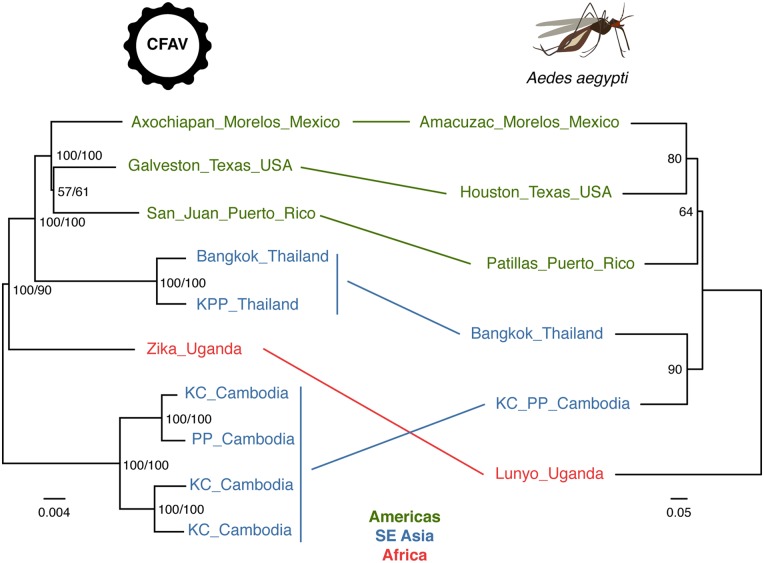
The alternative hypothesis that we investigated to explain the phylogenetic divergence between CFAV sequences from Thailand and Cambodia was that their main mosquito host A.aegypti was phylogenetically divergent too. Assuming that vertical transmission from mother to offspring is the main mode of ISF transmission, CFAV phylogeny is expected to match its host phylogeny. Thus, phylogenetic divergence of CFAV sequences in Thailand and Cambodia could reflect a large genetic distance between A.aegypti populations at these locations. We used microsatellite markers to genotype newly field-collected A.aegypti specimens from Cambodia, which were analyzed against the backdrop of known A.aegypti genotypes based on the same set of microsatellite markers (Gloria-Soria et al. 2016b; Kotsakiozi et al. 2018). A distance tree derived from the microsatellite data showed that A.aegypti from Thailand and Cambodia clustered together, in disagreement with the CFAV phylogeny (Fig. 3). This phylogenetic discrepancy was also supported by admixture analyses, which confirmed that A.aegypti populations from Thailand and Cambodia were genetically homogeneous (Supplementary Fig. S4).
Figure 3.
Lack of phylogenetic congruence between CFAV and A.aegypti in Southeast Asia. Left: consensus tree of 1,000 ultrafast bootstrap replicate ML trees represents phylogenetic relationships between CFAV sequences based on the nearly full ORF alignment. The tree is midpoint rooted. Node support values include both ultrafast bootstrap proportion (%, first value) and posterior probability derived from the Bayesian maximum clade credibility tree (%, second value). The scale bar represents branch length in substitutions per site. Right: midpoint rooted consensus tree of 1,000 bootstrap replicates trees based on the Edwards’ genetic distances calculated from microsatellite allele frequencies among A.aegypti populations. Node support values represent bootstrap proportions (%). Lines between the trees connect the tips sharing the same geographic origin.
To further probe the apparent discrepancy between CFAV and A.aegypti phylogenies, we built a microsatellite-based distance tree with nine additional A.aegypti populations included in a previous study that used a genome-wide panel of ∼19,000 single-nucleotide polymorphisms (SNPs) to produce a well-resolved A.aegypti phylogeny (Kotsakiozi et al. 2018) (Supplementary Fig. S5). A relatively high bootstrap value of 88 per cent indicates that mosquito specimens from Cambodia are genetically closest to mosquitoes from Ho Chi Minh City, Vietnam. In 73 per cent of the bootstrap replicates, they belong to a larger clade that also includes mosquitoes from Thailand and Hanoi, Vietnam. Importantly, considering the best-supported nodes and overall topology, our distance tree is consistent with the SNP-based A.aegypti phylogeny (Kotsakiozi et al. 2018), in which mosquitoes from Vietnam are closely related to mosquitoes from Thailand. Based on the accumulated evidence of the position of Cambodian mosquitoes in the A.aegypti phylogeny, the following evolutionary scenario can be inferred. African populations diverged from the clade encompassing American and Asian-Pacific populations, in which the Northern American clade is basal to the Asian-Pacific clade. Among the three distinct topologies of CFAV trees artificially rooted with the Uganda sequence present in the Bayesian posterior tree distribution, none of the topologies were strictly consistent with the A.aegypti divergence scenario. All three topologies showed the Thai-American clade as a sister clade to the Cambodian clade, and only the relationships inside the American clade alternated. This analysis provides additional evidence of the unlikely phylogenetic congruence between CFAV and A.aegypti in Southeast Asia.
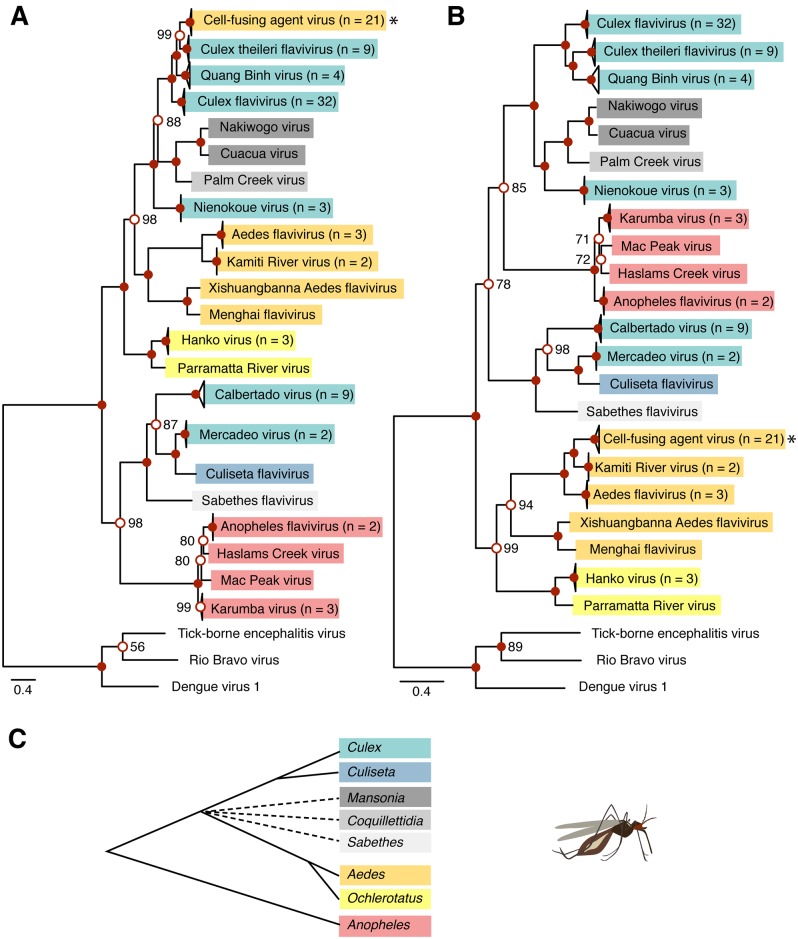
Finally, we examined whether CFAV had recombined with other cISFs. We aligned all publicly available cISF full or nearly full ORF sequences (https://github.com/artembaidaliuk/CFAV-cISF-phylogeny-2019.git), by splitting them into individual genes and concatenating the amino-acid-guided nucleotide alignments into the full-ORF alignment. Recombination tests detected 10 putative recombination events (Supplementary Fig. S2B), of which none could explain CFAV divergence in Southeast Asia. However, the full-ORF cISF recombination analysis provided additional insights into the evolutionary history of cISFs. We detected one outstanding recombination signal in all Culex flavivirus sequences with CFAV as the potential minor parent and Culiseta flavivirus as the potential major parent (Supplementary Fig. S2B). The recombinant region encompassed all three structural genes from position 44 (99% CI 0–84) to position 1900 (99% CI 1,850–1,939) in the final alignment. Recombination of the E region had been previously suggested for cISFs (Cook et al. 2012). This recombination event might provide an explanation to the topological discrepancy observed in the current study between phylogenetic trees based on structural vs. nonstructural genomic regions of cISFs (Fig. 4). This observation is consistent with previous studies that reported topological incongruence between cISF phylogenies based on E or based on NS3, NS5, and the full ORF (Cook et al. 2012; Yamanaka et al. 2013). Our phylogeny based on the nonstructural part of the polyprotein strongly supports the existence of a clade including the Aedes- and Ochlerotatus-associated cISFs (Fig. 4B). However, when the phylogeny is based on the structural part of the polyprotein, this clade is interspersed among Culex- and Culiseta-associated cISFs (Fig. 4A). Phylogenies based on both structural and nonstructural parts of the polyprotein show a well-supported Anopheles-associated cISF clade (Fig. 4). This clade groups together with a clade composed of two Culex-associated cISFs (Mercadeo and Calbertado viruses), Sabethes flavivirus, and Culiseta flavivirus, when the phylogeny is based on structural genes. Relationships of this clade with other cISFs are not clearly resolved when the phylogeny is based on nonstructural genes.
Figure 4.
Differing topology of cISF phylogenetic trees based on structural vs. nonstructural ORF regions. Consensus trees of 1,000 ultrafast bootstrap replicate ML trees (A, B) represent phylogenetic relationships among all cISFs based on the concatenated amino-acid alignment of C, prM, and E (A) and NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 (B). Both consensus trees are rooted with an outgroup composed of three non-cISF sequences shown at the bottom of the tree. Node support values represent ultrafast bootstrap proportions (%) and are only shown if <100% next to the open red circles. All nodes represented by filled red circles have 100 per cent support values. The scale bar represents branch length in substitutions per site. Tree tips indicate cISF species and number of sequences per species (n) if n > 1. The star symbol points to CFAV. Color highlighting of the tree leaves indicates the predominant mosquito host (red = Anopheles-associated cISFs; yellow = Aedes- and Ochlerotatus-associated cISFs; cyan =Culex- and Culiseta-associated cISFs; gray = Mansonia-, Coquillettidia-, and Sabethes-associated cISFs). The same color-coding is used in the schematic tree representation of evolutionary relationships between mosquito genera (C) based on limited phylogenetic studies (Shepard, Andreadis, and Vossbrinck 2006; Harbach 2007; Reidenbach et al. 2009; Chu et al. 2018; Aragao et al. 2019). In (C), the multifurcating dashed branches indicate uncertain clade assignment and the branch length is arbitrary.
Discussion
In this study, we generated six novel full-genome CFAV sequences by sequencing three new virus isolates and subjecting two wild-caught mosquito samples and one recently colonized mosquito to untargeted viral metagenomics. In addition, we de novo assembled two full-genome CFAV sequences from deep-sequencing datasets in which CFAV had been previously detected. We combined these newly obtained CFAV genome sequences with published ones to perform the first phylogenetic analysis of CFAV genetic diversity at a global scale. Overall, we found that despite some degree of geographical clustering among CFAV sequences, there were also notable discrepancies between geographical and phylogenetic distances.
For instance, two CFAV sequences initially detected in Miami, Florida, USA within the same study (Metsky et al. 2017) fell into two distinct clades (Fig. 2; Supplementary Fig. S3). Likewise, two of the three CFAV sequences originating from the same city in Brazil clustered together with most other American sequences, whereas the third one was phylogenetically distant (Fig. 2; Supplementary Fig. S3), as was previously noticed (Fernandes et al. 2018). CFAV sequences originating from the Americas, which were the most represented in our dataset, were divided into two major clades. One clade grouped with the only CFAV genome sequence from Uganda, and the other clade included sequences from Thailand, Indonesia, and Australia (Fig. 2; Supplementary Fig. S3). Such examples of high phylogenetic divergence observed for CFAV strains originating from the same locality indicate that the genetic structure of CFAV genetic diversity does not simply reflect geographical distance and point to a high degree of genetic mixing.
It is worth noting that several of the CFAV sequences that we used in our phylogenetic analysis were derived from untargeted viral metagenomics of wild-caught mosquitoes or an early-generation laboratory colony. We observed that these CFAV sequences were typically associated with larger amounts of minority variants than CFAV deep-sequencing datasets generated from cell-culture isolates (Supplementary Fig. S1). We cannot exclude that some of these sequences may reflect chimeric assemblies from mixed CFAV infections or pooled single infections. Moreover, the well-known presence of CFAV-derived endogenous viral elements in mosquito genomes (Crochu 2004; Whitfield et al. 2017) may also contribute to produce chimeric sequences. Although such methodological issues are unlikely to explain all the phylogenetic discrepancies, the high phylogenetic divergence of some CFAV strains should be considered with caution.
The most striking example of inconsistency between geography and phylogeny was observed for CFAV sequences in Southeast Asia. The four CFAV sequences from Cambodia originated from two locations ∼70 km apart (Phnom Penh and Kampong Cham) and the two full-genome CFAV sequences from Thailand originated from two locations ∼300 km apart (Bangkok and Kamphaeng Phet). Although the distance between the closest locations between the two countries was as short as ∼500 km, CFAV sequences from each country were highly divergent phylogenetically. The two sequences from Thailand clustered with the majority of CFAV sequences from the Americas, whereas the four sequences from Cambodia formed a separate clade (Fig. 2; Supplementary Fig. S3).
The high phylogenetic divergence of CFAV between Thailand and Cambodia was unlikely to result from methodological artifacts because multiple CFAV sequences from both countries had been independently obtained by different methods. CFAV sequences from Cambodia were derived from two cell-culture isolates, one field-collected female mosquito, and one recently colonized female mosquito. CFAV sequences from Thailand were derived from one cell-culture isolate and from one field-collected mosquito pool. The subset of CFAV sequences from the Americas and from Uganda that was used to investigate CFAV phylogenetic divergence in Southeast Asia was obtained from cell-culture isolates. Therefore, it is unlikely that any chimeric assembly of multiple distinct strains could have caused the high phylogenetic divergence observed between CFAV sequences from Thailand and Cambodia.
We further explored the hypothesis that phylogenetic divergence between geographically close CFAV sequences could reflect the underlying population genetic structure of the mosquito host A.aegypti. We assumed that A.aegypti was the primary host of CFAV because the majority of detections and isolations were associated with wild or colonized A.aegypti. Accordingly, the subset of CFAV sequences that we used to investigate phylogenetic divergence in Southeast Asia was exclusively obtained from A.aegypti samples. Aedes aegypti as a species originated in Africa and spread throughout the rest of the tropical and subtropical during the last few centuries (Kotsakiozi et al. 2018; Powell, Gloria-Soria, and Kotsakiozi 2018). Recent population genetic studies suggested that A.aegypti most likely spread from Africa to the Americas via the slave trade and was subsequently brought to Asia either via the Suez Canal and the Indian Ocean or through the Black Sea (Kotsakiozi et al. 2018; Powell, Gloria-Soria, and Kotsakiozi 2018). Under this scenario, A.aegypti populations in Thailand and Cambodia were expected to be genetically similar, as was observed for A.aegypti populations in Vietnam and Thailand (Gloria-Soria et al. 2016b); however, specimens from Cambodia had not been included in these earlier population genetic studies. We, therefore, obtained wild-caught A.aegypti samples from the two locations where CFAV sequences had been isolated in Cambodia and analyzed their genetic relationships with A.aegypti populations worldwide (Gloria-Soria et al. 2016b; Kotsakiozi et al. 2018). Based on microsatellite allele frequencies, we found that A.aegypti from Cambodia were genetically closely related to A.aegypti from Vietnam and Thailand (Fig. 3, Supplementary Fig. S5). Interestingly, mosquitoes from Cambodia were genetically closer to mosquitoes collected in Ho Chi Minh City, Vietnam than from Hanoi, Vietnam, indicating a tight link between genetic and geographic distance in this area. Moreover, a previously published SNP-based phylogeny of A.aegypti (Kotsakiozi et al. 2018) showed that A.aegypti specimens from Thailand were most closely related to mosquitoes sampled in Ho Chi Minh City, providing additional evidence for the close genetic relatedness between A.aegypti in Thailand and in Cambodia. This result ruled out the hypothesis that CFAV phylogenetic divergence between Thailand and Cambodia reflected the genetic differentiation of its host A.aegypti. It is worth noting that the relatively small number of A.aegypti samples used for population genetic analyses were not identical to the samples from which CFAV was isolated or sequenced. Thus, our analysis assumed that the CFAV-infected mosquitoes were adequately represented by the newly collected samples at the same locations.
Our analyses showed that the phylogeny of CFAV was not entirely consistent with that of A.aegypti, especially in Southeast Asia. However, this result should be interpreted with caution for two reasons. First, the A.aegypti genetic distance trees based on the microsatellite loci (Fig. 3, Supplementary Fig. S5) are not fully resolved and a SNP-based phylogeny including samples from Cambodia would be desirable to further strengthen our confidence that mosquitoes in Cambodia and Thailand are indeed closely genetically related. Second, using the closest related viruses to CFAV, such as Aedes flavivirus, Kamiti River virus or all cISFs combined could not fully resolve the root position among CFAV sequences. Therefore, we used a midpoint-rooting method that placed the root of the CFAV phylogenies between the clade of sequences from Cambodia and the rest of the tree (Figs 2 and 3). To assess the robustness of our conclusions based on this rooting assumption, we compared tree topologies for different scenarios based on unrooted trees and by artificially fixing the root between African and non-African CFAV sequences (i.e., in line with the A.aegypti phylogeny). Even though all these analyses supported our conclusion that CFAV and A.aegypti phylogenies are inconsistent in Southeast Asia, additional full CFAV genomes from Africa and Southeast Asia (e.g., Vietnam, Laos) are required to further improve our understanding of CFAV evolutionary history globally and locally in Southeast Asia.
We did not directly address the possibility of CFAV acquisition from other mosquito species in this study. Nevertheless, several previous findings suggested that such horizontal transmission of CFAV between mosquito species might indeed occur in nature. For example, CFAV was isolated from various mosquito species, including A.aegypti, A.albopictus, and Culex spp. in Puerto Rico (Cook 2006). At least three CFAV isolates showed identical sequences and, based on the primer pairs used, the viruses were genetically similar in another forty-one isolates. Likewise, two partial CFAV genome sequences from Indonesia (Hoshino et al. 2009), one from A.aegypti (CFAV-Surabaya10) and another from A.albopictus (CFAV-Surabaya12), were closely related (Supplementary Fig. S3). Another study detected one CFAV sequence from A.aegypti and two sequences from Culex spp. in Brazil (Fernandes et al. 2018). Interestingly, one of the Culex-derived CFAV sequences (CFAV-Macapa4) was remarkably similar to the Aedes-derived CFAV sequence (CFAV-Macapa1); however, the other Culex-derived sequence (CFAV-Macapa2) was genetically distant from the other two (Fig. 2). Finally, another study detected two CFAV sequences from A.albopictus as well as two sequences from A.aegypti in Turkey (Akiner et al. 2019). We included one sequence from each mosquito species in our supplementary phylogenetic analysis, and despite the short size of the genomic region, one of the sequences from A.albopictus was phylogenetically distant from the rest of CFAV sequences and the sequence from A.aegypti was even more distant (Supplementary Fig. S3). Further experimental and observational evidence is required to elucidate CFAV host range and cross-species transmission patterns.
We took advantage of the newly generated CFAV sequences to investigate the possibility of recombination among CFAV strains and between CFAV and other cISFs. We detected two recombination signals among CFAV strains (Supplementary Fig. S2A); however, both of them involved CFAV strains derived from the Aag2 cell line and are likely false positives. Since CFAV infection has persisted in Aag2 cells for at least four decades, it is reasonable to assume the inability of this virus to recombine with any of the wild-type CFAV strains during this time period. The first recombination signal corresponds to the fifo genomic region, which is divergent in Aag2-derived CFAV strains, and recombination tests failed to assign a minor parent in the recombination triplet. This recombination signal was also detected in the cISF alignment (Supplementary Fig. S2B), but again the minor parent was unassigned. The cISF recombination analysis identified multiple putative recombinants both within and between cISF species (Supplementary Fig. S2B). Interestingly, we detected recombination between CFAV, Culex flavivirus, and Culiseta flavivirus, in line with conclusions from a previous study (Cook et al. 2012). In particular, we found that Culex flavivirus recombined with CFAV over the entire genomic region encoding structural genes. This recombination signal is corroborated by the topological discrepancy we observed between phylogenetic trees based on structural vs. nonstructural parts of the cISF polyprotein (Fig. 4), which was previously observed by comparing phylogenies based on E vs. NS3, NS5, and the full ORF (Cook et al. 2012; Yamanaka et al. 2013). The assignment of the putative recombinant and its parents has to be considered with caution because CFAV is only distantly related to Culex flavivirus even in the structural part of the polyprotein (∼60–70% nucleotide identity in structural vs. ∼50% in nonstructural ORF regions). This could reflect the ancient nature of the recombination event, which would have occurred before the separation of the Culicini and Aedini tribes. Alternatively, the recombination event could be more recent but the true members of the recombination triplet would not have been present in the dataset tested. Recent CFAV detection in Culex mosquitoes (Fernandes et al. 2018) provided circumstantial evidence that coinfection and potential recombination with Culex-associated flaviviruses is possible, but the likelihood of such event is yet to be investigated.
Although the evolutionary history of CFAV in Southeast Asia remains obscure, our results revealed that the patterns of CFAV genetic diversity in Southeast Asia, and presumably in other parts of the world, do not merely reflect the population genetic structure of A.aegypti. It is possible that CFAV was introduced multiple times in Southeast Asia through reintroductions of A.aegypti after it was already established in the region. The virus could have persisted by vertical transmission even if the introgression events did not leave a clear signal of admixed ancestry. Alternatively, the virus may have been horizontally transferred to A.aegypti from other mosquito species. Addressing these hypotheses will require further studies on CFAV host range, transmission routes, and genetic diversity.
The rapid accumulation of full-genome sequences for cISFs in publicly available databases, such as the ones provided in this study, shed new light on the evolutionary history of cISFs and flaviviruses in general. For instance, we detected phylogenetic evidence of recombination between CFAV and Culex-associated cISFs with a breakpoint between structural and nonstructural parts of the polyprotein ORF. We accounted for this probable recombination event to update the phylogeny of all cISFs. More generally, additional knowledge about understudied members of the Flavivirus genus contributes to improve our understanding of the evolutionary history of the genus, and more reliably anticipate the public health threats associated with some of its members.
Data availability
All novel sequencing data are available from the SRA database under accession number PRJNA556544 and in the ENA database under accession numbers PRJEB33801 (LR694072, LR694073, LR694074, LR694075) and PRJEB33690 (LR694076, LR694077, LR694078, LR694079, LR694080, LR694081).
Acknowledgements
We thank Catherine Lallemand for assistance with mosquito rearing, and Jeff Powell, Carla Saleh, and Marie Flamand for their insights. We are grateful to Martin Mayanja and Julius Lutwama for providing the original mosquito colony from Uganda. We thank Gordana Rašić, Lícia Natal Fernandes, Bronwyn MacInnis, and Hayden Metsky for providing CFAV sequences. We thank Oliver Pybus, Peter Simmonds, Xavier de Lamballerie, and one anonymous reviewer for their insights and constructive suggestions on earlier versions of the manuscript. This work was supported by Agence Nationale de la Recherche (grant numbers ANR-16-CE35-0004-01 and ANR-17-ERC2-0016-01), the French Government’s Investissement d’Avenir program Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant number ANR-10-LABX-62-IBEID), the INCEPTION program (Investissements d’Avenir grant number ANR-16-CONV-0005), the City of Paris Emergence(s) program in Biomedical Research, and the European Union Seventh Framework Programme (FP7/2007/2011) under Grant Agreement 282378. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest: None declared.
Supplementary Material
References
- Ajamma Y. U. et al. (2018) ‘Vertical Transmission of Naturally Occurring Bunyamwera and Insect-Specific Flavivirus Infections in Mosquitoes from Islands and Mainland Shores of Lakes Victoria and Baringo in Kenya’, PLoS Neglected Tropical Diseases, 12: e0006949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiner M. M. et al. (2019) ‘Arboviral Screening of Invasive Aedes Species in Northeastern Turkey: West Nile Virus Circulation and Detection of Insect-Only Viruses’, PLOS Neglected Tropical Diseases, 13: e0007334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. (1997) ‘Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs’, Nucleic Acids Research, 25: 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragao A. O. et al. (2019) ‘Description and Phylogeny of the Mitochondrial Genome of Sabethes chloropterus, Sabethes glaucodaemon and Sabethes belisarioi (Diptera: Culicidae)’, Genomics, 111: 607–11. [DOI] [PubMed] [Google Scholar]
- Baidaliuk A. et al. (2019) ‘Cell-Fusing Agent Virus Reduces Arbovirus Dissemination in Aedes aegypti Mosquitoes In Vivo’, Journal of Virology, 93: e00705–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A. et al. (2012) ‘SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing’, Journal of Computational Biology, 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S. et al. (2013) ‘The Global Distribution and Burden of Dengue’, Nature, 496: 504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitvich B. J., Firth A. E. (2015) ‘Insect-Specific Flaviviruses: A Systematic Review of Their Discovery, Host Range, Mode of Transmission, Superinfection Exclusion Potential and Genomic Organization’, Viruses, 7: 1927–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitvich B. J., Firth A. E. (2017) ‘A Review of Flaviviruses That Have No Known Arthropod Vector’, Viruses, 9: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S., Laviolette F., Corbeil J. (2010) ‘Ray: Simultaneous Assembly of Reads from a Mix of High-Throughput Sequencing Technologies’, Journal of Computational Biology, 17: 1519–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014) ‘Trimmomatic: A Flexible Trimmer for Illumina Sequence Data’, Bioinformatics, 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling B. G. et al. (2012) ‘Transmission Dynamics of an Insect-Specific Flavivirus in a Naturally Infected Culex pipiens Laboratory Colony and Effects of Co-Infection on Vector Competence for West Nile Virus’, Virology, 427: 90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling B. G. et al. (2015) ‘Insect-Specific Viruses Detected in Laboratory Mosquito Colonies and Their Potential Implications for Experiments Evaluating Arbovirus Vector Competence’, The American Journal of Tropical Medicine and Hygiene, 92: 422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. E. et al. (2011) ‘Worldwide Patterns of Genetic Differentiation Imply Multiple ‘Domestications’ of Aedes aegypti, a Major Vector of Human Diseases’, Proceedings of the Royal Society B: Biological Sciences, 278: 2446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Cammisa-Parks H. et al. (1992) ‘The Complete Nucleotide Sequence of Cell Fusing Agent (CFA): Homology between the Nonstructural Proteins Encoded by CFA and the Nonstructural Proteins Encoded by Arthropod-Borne Flaviviruses’, Virology, 189: 511–24. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. (1967) ‘Phylogenetic Analysis. Models and Estimation Procedures’, American Journal of Human Genetics, 19: 233–57. [PMC free article] [PubMed] [Google Scholar]
- Chu H. et al. (2018) ‘The Phylogenetic Relationships of Known Mosquito (Diptera: Culicidae) Mitogenomes’, Mitochondrial DNA Part A, 29: 31–5. [DOI] [PubMed] [Google Scholar]
- Cock P. J. et al. (2009) ‘Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics’, Bioinformatics, 25: 1422–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição-Neto N. et al. (2015) ‘Modular Approach to Customise Sample Preparation Procedures for Viral Metagenomics: A Reproducible Protocol for Virome Analysis’, Scientific Reports, 5:16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Gutierrez M. A. et al. (2017) ‘Experimental Infection with and Maintenance of Cell Fusing Agent Virus (Flavivirus) in Aedes aegypti’, The American Journal of Tropical Medicine and Hygiene, 97: 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S. (2006) ‘Isolation of a New Strain of the Flavivirus Cell Fusing Agent Virus in a Natural Mosquito Population from Puerto Rico’, Journal of General Virology, 87: 735–48. [DOI] [PubMed] [Google Scholar]
- Cook S. et al. (2009) ‘Isolation of a Novel Species of Flavivirus and a New Strain of Culex Flavivirus (Flaviviridae) from a Natural Mosquito Population in Uganda’, Journal of General Virology, 90: 2669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S. et al. (2012) ‘Molecular Evolution of the Insect-Specific Flaviviruses’, Journal of General Virology, 93: 223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A., Gribaldo S. (2010) ‘BMGE (Block Mapping and Gathering with Entropy): A New Software for Selection of Phylogenetic Informative Regions from Multiple Sequence Alignments’, BMC Evolutionary Biology, 10: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochu S. (2004) ‘Sequences of Flavivirus-Related RNA Viruses Persist in DNA Form Integrated in the Genome of Aedes spp.’, Journal of General Virology, 85: 1971–80. [DOI] [PubMed] [Google Scholar]
- Di Giallonardo F. et al. (2018) ‘Complete Genome of Aedes aegypti Anphevirus in the Aag2 Mosquito Cell Line’, Journal of General Virology, 99: 832–6. [DOI] [PubMed] [Google Scholar]
- Espinoza-Gomez F. et al. (2011) ‘Detection of Sequences from a Potentially Novel Strain of Cell Fusing Agent Virus in Mexican Stegomyia (Aedes) Aegypti Mosquitoes’, Archives of Virology, 156: 1263–7. [DOI] [PubMed] [Google Scholar]
- Fernandes L. N. et al. (2018) ‘A Novel Highly Divergent Strain of Cell Fusing Agent Virus (CFAV) in Mosquitoes from the Brazilian Amazon Region’, Viruses, 10: 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A. E. et al. (2010) ‘Evidence for Ribosomal Frameshifting and a Novel Overlapping Gene in the Genomes of Insect-Specific Flaviviruses’, Virology, 399: 153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloria-Soria A. et al. (2016a) ‘Temporal Genetic Stability of Stegomyia Aegypti (= Aedes aegypti) Populations’, Medical and Veterinary Entomology, 30: 235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloria-Soria A. et al. (2016b) ‘Global Genetic Diversity of Aedes aegypti’, Molecular Ecology, 25: 5377–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenaga S. et al. (2015) ‘Potential for Co-Infection of a Mosquito-Specific Flavivirus, Nhumirim Virus, to Block West Nile Virus Transmission in Mosquitoes’, Viruses, 7: 5801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E. A., Solomon T. (2008) ‘Pathogenic Flaviviruses’, The Lancet, 371: 500–9. [DOI] [PubMed] [Google Scholar]
- Guzman H. et al. (2018) ‘Characterization of Three New Insect-Specific Flaviviruses: Their Relationship to the Mosquito-Borne Flavivirus Pathogens’, The American Journal of Tropical Medicine and Hygiene, 98: 410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S. et al. (2016) ‘The Insect-Specific Palm Creek Virus Modulates West Nile Virus Infection in and Transmission by Australian Mosquitoes’, Parasites & Vectors, 9: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach R. E. (2007) ‘The Culicidae (Diptera): a Review of Taxonomy, Classification and Phylogeny’, Zootaxa, 1668: 591–638. [Google Scholar]
- Hoshino K. et al. (2009) ‘Isolation and Characterization of a New Insect Flavivirus from Aedes albopictus and Aedes Flavopictus Mosquitoes in Japan’, Virology, 391: 119–29. [DOI] [PubMed] [Google Scholar]
- Hunt M. et al. (2015) ‘IVA: Accurate De Novo Assembly of RNA Virus Genomes’, Bioinformatics, 31: 2374–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita H. et al. (2018) ‘Mosquito Arbovirus Survey in Selected Areas of Kenya: Detection of Insect-Specific Virus’, Tropical Medicine and Health, 46: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. (2008) ‘Adegenet: A R Package for the Multivariate Analysis of Genetic Markers’, Bioinformatics, 24: 1403–5. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S. et al. (2017) ‘ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates’, Nature Methods, 14: 587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar Z. N., Tabima J. F., Grünwald N. J. (2014) ‘Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction’, PeerJ, 2: e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. (2002) ‘MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform’, Nucleic Acids Research, 30: 3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara Y. et al. (2007) ‘Rapid Determination of Viral RNA Sequences in Mosquitoes Collected in the Field’, Journal of Virological Methods, 146: 372–4. [DOI] [PubMed] [Google Scholar]
- Kotsakiozi P. et al. (2018) ‘Aedes aegypti in the Black Sea: Recent Introduction or Ancient Remnant?’, Parasites & Vectors, 11: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequime S. et al. (2017) ‘Full-Genome Dengue Virus Sequencing in Mosquito Saliva Shows Lack of Convergent Positive Selection during Transmission by Aedes aegypti’, Virus Evolution, 3: vex031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009) ‘Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform’, Bioinformatics, 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutomiah J. J. et al. (2007) ‘Infection and Vertical Transmission of Kamiti River Virus in Laboratory Bred Aedes aegypti Mosquitoes’, Journal of Insect Science, 7: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringer K. et al. (2017) ‘Proteomics Informed by Transcriptomics for Characterising Active Transposable Elements and Genome Annotation in Aedes aegypti’, BMC Genomics, 18: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C. B. et al. (2014) ‘Enhanced Methods for Unbiased Deep Sequencing of Lassa and Ebola RNA Viruses from Clinical and Biological Samples’, Genome Biology, 15: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsky H. C. et al. (2017) ‘Zika Virus Evolution and Spread in the Americas’, Nature, 546: 411–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman C. M. et al. (2011) ‘Culex Flavivirus and West Nile Virus Mosquito Coinfection and Positive Ecological Association in Chicago, United States’, Vector-Borne and Zoonotic Diseases, 11: 1099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S. et al. (2017) ‘metaSPAdes: A New Versatile Metagenomic Assembler’, Genome Research, 27: 824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004) ‘APE: Analyses of Phylogenetics and Evolution in R Language’, Bioinformatics, 20: 289–90. [DOI] [PubMed] [Google Scholar]
- Parry R., Asgari S. (2019) ‘Discovery of Novel Crustacean and Cephalopod Flaviviruses: Insights into Evolution and Circulation of Flaviviruses between Marine Invertebrate and Vertebrate Hosts’, Journal of Virology, 93: e00432–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R., Gloria-Soria A., Kotsakiozi P. (2018) ‘Recent History of Aedes aegypti: Vector Genomics and Epidemiology Records’, BioScience, 68: 854–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. (2000) ‘Inference of Population Structure Using Multilocus Genotype Data’, Genetics, 155: 945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010) ‘BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features’, Bioinformatics, 26: 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2013) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
- Rambaut A. et al. (2018) ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1’, Systematic Biology, 67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach K. R. et al. (2009) ‘Phylogenetic Analysis and Temporal Diversification of Mosquitoes (Diptera: Culicidae) Based on Nuclear Genes and Morphology’, BMC Evolutionary Biology, 9: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A. (2004) ‘DISTRUCT: A Program for the Graphical Display of Population Structure’, Molecular Ecology Notes, 4: 137–8. [Google Scholar]
- Shepard J. J., Andreadis T. G., Vossbrinck C. R. (2006) ‘Molecular Phylogeny and Evolutionary Relationships among Mosquitoes (Diptera: Culicidae) from the Northeastern United States Based on Small Subunit Ribosomal DNA (18S rDNA) Sequences’, Journal of Medical Entomology, 43: 443–54. [DOI] [PubMed] [Google Scholar]
- Shi M. et al. (2018) ‘The Evolutionary History of Vertebrate RNA Viruses’, Nature, 556: 197–202. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. (2002) ‘An Approximately Unbiased Test of Phylogenetic Tree Selection’, Systematic Biology, 51: 492–508. [DOI] [PubMed] [Google Scholar]
- Skoge R. H. et al. (2018) ‘New Virus of the Family Flaviviridae Detected in Lumpfish (Cyclopterus Lumpus)’, Archives of Virology, 163: 679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M. A. et al. (2007) ‘Polymorphic Microsatellite Markers for Studies of Aedes aegypti (Diptera: Culicidae), the Vector of Dengue and Yellow Fever’, Molecular Ecology Notes, 7: 168–71. [Google Scholar]
- Stollar V., Thomas V. L. (1975) ‘An Agent in the Aedes aegypti Cell Line (Peleg) Which Causes Fusion of Aedes albopictus Cells’, Virology, 64: 367–77. [DOI] [PubMed] [Google Scholar]
- Suchard M. A. et al. (2018) ‘Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10’, Virus Evolution, 4: vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkin S. E., Sims R. A., Allen R. (1966) ‘Isolation of St. Louis Encephalitis Virus from Bats (Tadarida B. Mexicana) in Texas’, Science, 152: 223–5. [DOI] [PubMed] [Google Scholar]
- Weger-Lucarelli J. et al. (2018) ‘Adventitious Viruses Persistently Infect Three Commonly Used Mosquito Cell Lines’, Virology, 521: 175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield Z. J. et al. (2017) ‘The Diversity, Structure, and Function of Heritable Adaptive Immunity Sequences in the Aedes aegypti Genome’, Current Biology, 27: 3511–9.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Wilke C. O. (2019) cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2 <https://wilkelab.org/cowplot/>.
- Wilm A. et al. (2012) ‘LoFreq: A Sequence-Quality Aware, Ultra-Sensitive Variant Caller for Uncovering Cell-Population Heterogeneity from High-Throughput Sequencing Datasets’, Nucleic Acids Research, 40: 11189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A. et al. (2013) ‘Genetic and Evolutionary Analysis of Cell-Fusing Agent Virus Based on Thai Strains Isolated in 2008 and ’, Infection, Genetics and Evolution, 19: 188–94. [DOI] [PubMed] [Google Scholar]
- Zakrzewski M. et al. (2018) ‘Mapping the Virome in Wild-Caught Aedes aegypti from Cairns and Bangkok’, Scientific Reports, 8:4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All novel sequencing data are available from the SRA database under accession number PRJNA556544 and in the ENA database under accession numbers PRJEB33801 (LR694072, LR694073, LR694074, LR694075) and PRJEB33690 (LR694076, LR694077, LR694078, LR694079, LR694080, LR694081).