CONSPECTUS
α-Helices constitute the largest class of protein secondary structures and play a major role in mediating protein-protein interactions. Development of stable mimics of short α-helices would be invaluable for inhibition of protein—protein interactions. This Account describes our efforts in developing a general approach for constraining short peptides in α-helical conformations by a main-chain hydrogen bond surrogate (HBS) strategy. The HBS α-helices feature a carbon-carbon bond derived from a ring-closing metathesis reaction in place of an N-terminal intramolecular hydrogen bond between the peptide i and i + 4 residues. Our approach is centered on the helix-coil transition theory in peptides, which suggests that the energetically demanding organization of three consecutive amino acids into the helical orientation inherently limits the stability of short α-helices. The HBS method affords preorganized α-turns to overcome this intrinsic nucleation barrier and initiate helix formation.
The HBS approach is an attractive strategy for generation of ligands for protein receptors because placement of the cross-link on the inside of the helix does not block solvent-exposed molecular recognition surfaces of the molecule. Our metathesis-based synthetic strategy utilizes standard Fmoc solid phase peptide synthesis methodology, resins, and reagents and provides HBS helices in sufficient amounts for subsequent biophysical and biological analyses. Extensive conformational analysis of HBS α-helices with 2D NMR, circular dichroism spectroscopies and X-ray crystallography confirms the α-helical structure in these compounds. The crystal structure indicates that all i and i + 4 C=O and NH hydrogen-bonding partners fall within distances and angles expected for a fully hydrogen-bonded α-helix. The backbone conformation of HBS α-helix in the crystal structure superimposes with an rms difference of 0.75 Å onto the backbone conformation of a model α-helix. Significantly, the backbone torsion angles for the HBS helix residues fall within the range expected for a canonical α-helix.
Thermal and chemical denaturation studies suggest that the HBS approach provides exceptionally stable α-helices from a variety of short sequences, which retain their helical conformation in aqueous buffers at exceptionally high temperatures. The high degree of thermal stability observed for HBS helices is consistent with the theoretical predictions for a nucleated helix.
The HBS approach was devised to afford internally constrained helices so that the molecular recognition surface of the helix and its protein binding properties are not compromised by the constraining moiety. Notably, our preliminary studies illustrate that HBS helices can target their expected protein receptors with high affinity.
Graphical Abstract
Introduction
Selective modulation of protein—protein interactions by small molecules is a fundamental challenge for bioorganic and medicinal chemists.1–4 Protein interfaces often feature large shallow surfaces, which are difficult for small molecules to target with high affinity and selectivity. Natural products are frequently used as templates by organic chemists for the design of more potent and selective agents, but the selection of natural products that target protein receptors with high specificity is limited. Examination of complexes of proteins with other biomolecules reveals that proteins tend to interact with partners via folded subdomains or protein secondary structures.5–7 α-Helices constitute the largest class of protein secondary structures and play a major role in mediating protein-protein interactions.5–7 Importantly, the average length of helical domains in proteins is rather small and spans two to three helical turns (or eight to twelve residues).8 Figure 1 shows a selection of complexes in which a short α-helical domain targets the biomolecule. These complexes suggest that it may be possible to develop low molecular weight helix mimetics that potentially participate in selective interactions with biomolecules.9,10
FIGURE 1.
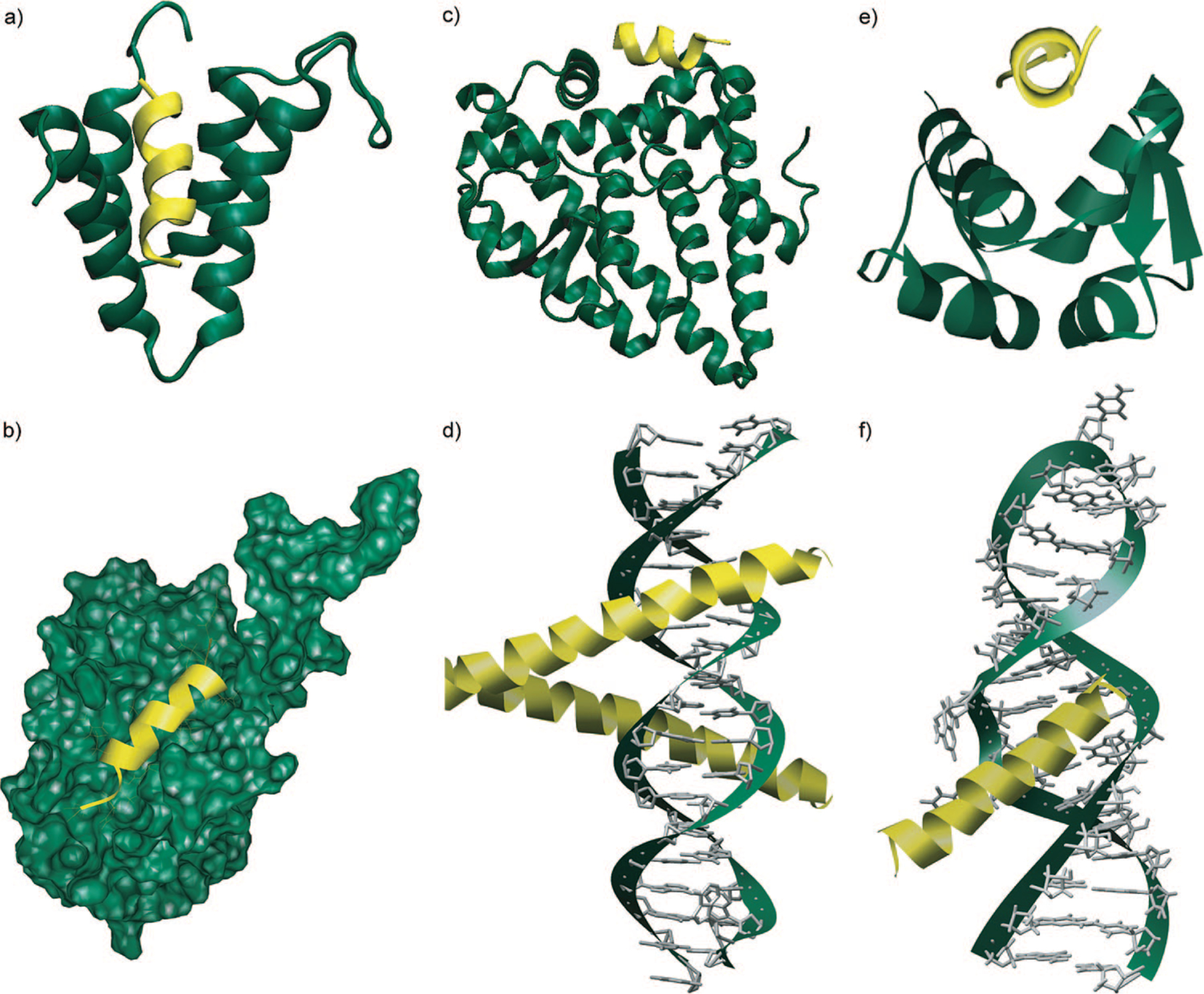
Biomolecular recognition with short α-helices: (a) corepressor Sin3B bound with transcription factor Mad (PDB code 1E91); (b) recognition between Bcl-xL-Bak regulators of apoptosis (PDB code 1BXL); (c) subunit of human estrogen receptor R ligand-binding domain in complex with glucocorticoid receptor interacting protein (PDB code 3ERD); (d) GCN4 region of leucine zipper bound to DNA (PDB code 1YSA); (e) MDM2 oncoprotein complexed with the p53 tumor suppressor-transactivation domain (PDB code 1YCR); (f) α-helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex (PDB code 1ETF).
Given the importance of the α-helical domain in biomolecular recognition, the chemical biology community has been developing several approaches to either stabilize this conformation in peptides or mimic this domain with nonnatural scaffolds. Figure 2 provides a summary of the most advanced strategies including β-peptide helices, terphenyl helix mimetics, miniproteins, peptoid helices, and side-chain cross-linked helices. Excellent reviews of these and other approaches are available in the literature;9,11–15 in this Account, we discuss our efforts in developing a new constraining strategy that affords stable α-helices from very short peptide sequences.
FIGURE 2.
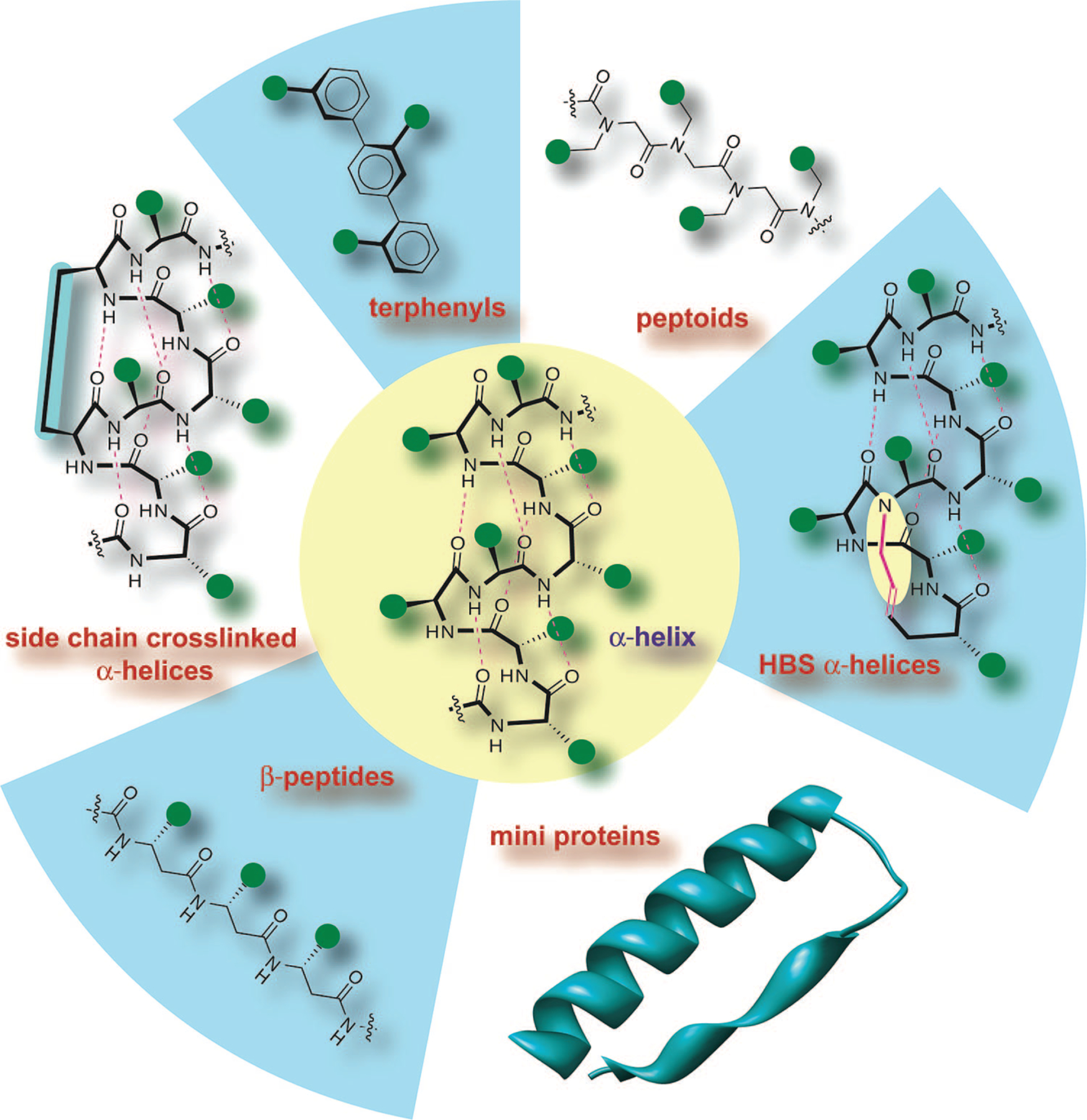
Stabilized helices and nonnatural helix mimetics: Several strategies that stabilize the α-helical conformation in peptides or mimic this domain with nonnatural scaffolds have been described. The most advanced efforts include β-peptide helices, terphenyl helix mimetics, miniproteins, peptoid helices, and side-chain cross-linked α-helices. This Account reviews our efforts to develop the hydrogen bond surrogate (HBS) derived α-helices. Green circles represent amino acid side chains.
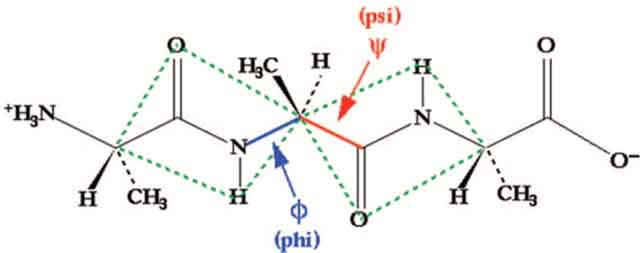
Stabilization of short peptides in α-helical conformation is a challenging endeavor. The helix-coil transition theory in peptides suggests that the energetically demanding organization of three consecutive amino acids into the helical orientation inherently limits the stability of short α-helices.16–18 According to this theory, α-helices composed of ten or fewer amino acids are expected to be essentially unstable due to a low nucleation probability.19,20 Preorganization of amino acid residues in an α-turn is expected to overwhelm the intrinsic nucleation propensities and initiate helix formation.21,22 In an α-helix, a hydrogen bond between the C=O of the ith amino acid residue and the NH of the (i + 4)th amino acid residue stabilizes and nucleates the helical structure (Figure 3). Our strategy for the preparation of artificial α-helices involves replacement of one of the main chain hydrogen bonds with a covalent linkage.23 To mimic the C=O⋯H–N hydrogen bond as closely as possible, we envisioned a covalent bond of the type C=X–Y–N, where X and Y would be part of the i and the i + 4 residues, respectively. The exceptional functional group tolerance displayed by the olefin metathesis catalysts for the facile introduction of nonnative carbon–carbon constraints in the preparation of peptidomimetics suggested that X and Y could be two carbon atoms connected through a ring-closing metathesis (RCM) reaction (Figure 3).24,25
FIGURE 3.
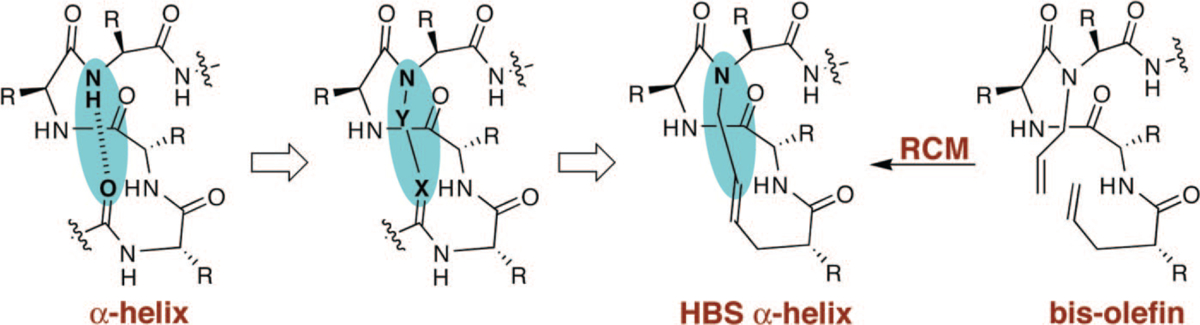
Nucleation of short α-helices by replacement of an N-terminal i and i + 4 hydrogen bond (C=O ⋯ H–N) with a covalent link (C=X–Y–N). The hydrogen bond surrogate-based (HBS) α-helices contain a carbon-carbon bond derived from a ring-closing metathesis reaction.
The main chain hydrogen bond surrogate (HBS) strategy is attractive because placement of the cross-link on the inside of the helix does not block solvent-exposed molecular recognition surfaces of the molecule. Traditional α-helix stabilization methods have relied predominantly on side-chain constraints.13,25–27 These methods either block solvent-exposed surfaces of the target α-helices or remove important side-chain functionalities. Our strategy allows strict preservation of the helix surfaces.
Synthesis of HBS α-Helices
The macrocycle that nucleates the HBS helices may be formed by several synthetic approaches. We concentrated our efforts on the design of a RCM-mediated synthetic strategy that (1) is compatible with standard resins and commercially available amino acid derivatives, (2) allows incorporation of any amino acid residue at any position in the helix, (3) is adaptable to solid phase, and (4) provides the final product on resin such that no extra modifications are required except cleavage and purification. We chose these criteria to potentially access libraries of stabilized helices from short peptide sequences. Scheme 1 depicts our optimized synthetic strategy.28,29 We utilize the standard Fmoc solid phase peptide synthesis methodology and Rink amide (or Wang) resin to prepare resin-bound bis-olefins (1). The synthesis of bis-olefin peptides utilizes three sets of monomers, which include commercially available Fmoc amino acids (3) and 4-pentenoic acid (5), and Fmoc-N-allyl dipeptides (4) prepared by the Fukuyama alkylation procedure.30,31 The resin-bound bis-olefins are converted to the corresponding HBS α-helices by treatment with the metathesis catalyst.
SCHEME 1.
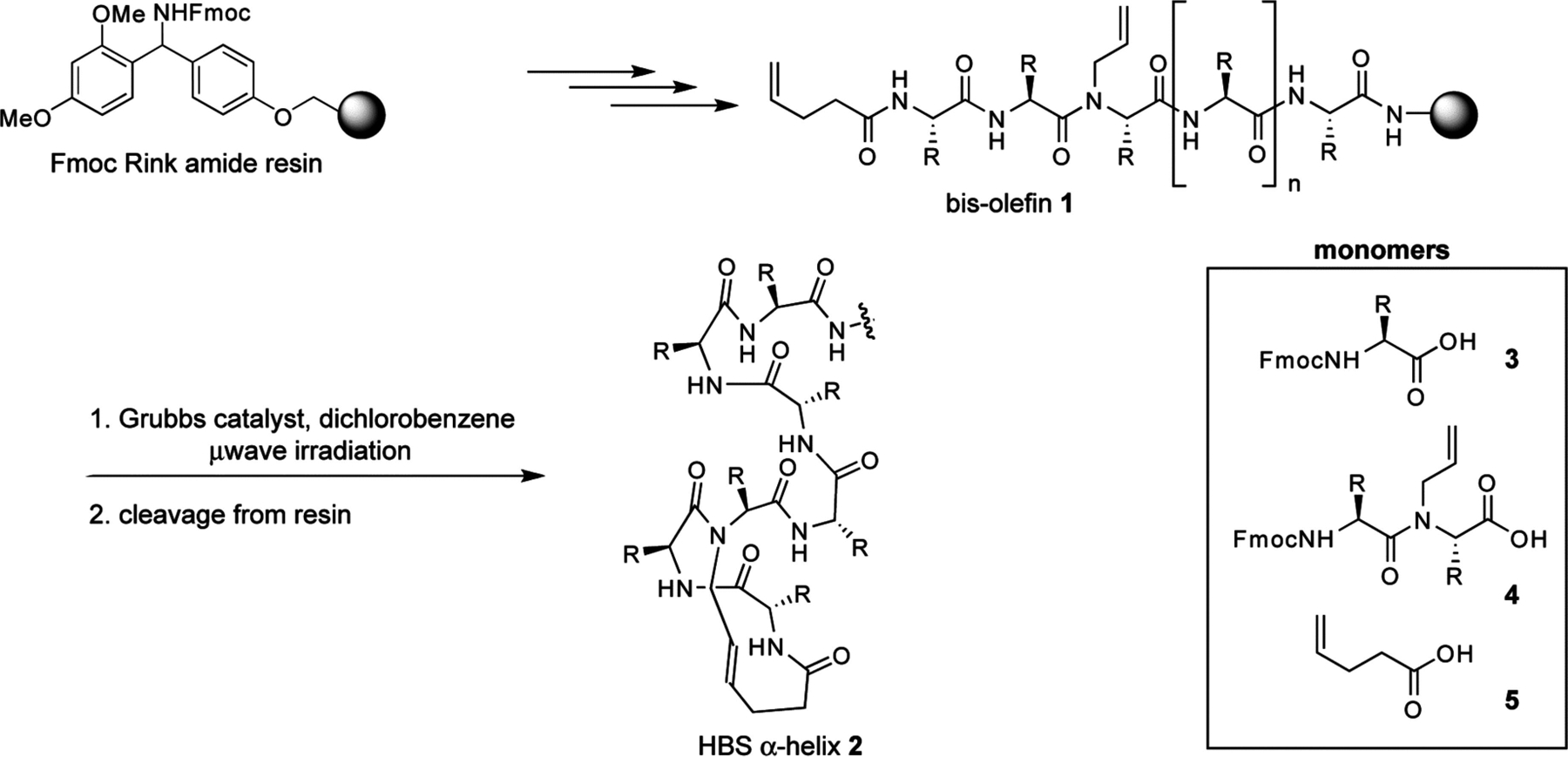
Solid Phase Synthesis of HBS Helicesa
a R = amino acid side chain.
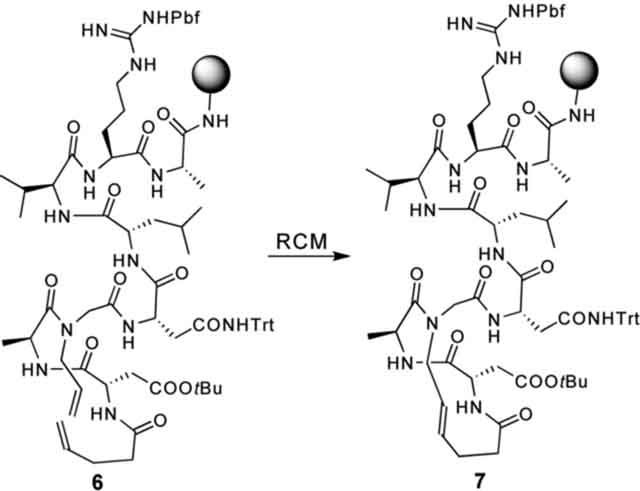
The RCM reaction is the key step in the preparation of HBS helices. In preliminary experiments, we found that the standard ring-closing metathesis reaction conditions did not yield significant amounts of the metathesized product from the bis-olefins shown in Scheme 1. This initial result prompted us to systematically investigate several of the commonly used RCM catalysts and reaction conditions, including microwave irradiation. A typical set of results obtained for the metathesis of bis-olefin peptides to form HBS helices is illustrated in Table 1 with macrocyclization of 6 to form 7.28 We discovered that with conventional heating, metathesis of resin-bound bis-olefins required the second-generation Hoveyda–Grubbs catalyst because the more common Grubbs catalysts failed to provide the desired macrocycle.29 Both Grubbs II and Hoveyda–Grubbs II catalysts provided efficient macrocyclization of various bis-olefin peptides under microwave irradiation conditions.28 Our optimized RCM conditions utilize microwave irradiation and provide access to a variety of HBS macrocycles in shorter reaction times and higher yields compared with conventional heating. In general, the RCM reaction predominantly affords the trans alkene product.29,32,33
TABLE 1.
Typical Results Obtained for the Metathesis of Bio-olefin Peptides
 | ||||
|---|---|---|---|---|
| Grubbs II, oil bath | Grubbs II, microwave | HG II, oil bath | HG II, microwave | |
| % conversion of 6 to 7 | <5% | 80% | 40% | 80% |
Structure of HBS α-Helices
We have extensively analyzed conformation adopted by HBS α-helices with 2D NMR and circular dichroism spectroscopies.32–34 Thermal and chemical denaturation studies suggest that the HBS approach provides exceptionally stable α-helices from a variety of short sequences. The NMR-derived solution structure of an HBS α-helix has been reported.33 Recently, we were also able to obtain a high-resolution (1.15 Å) crystal structure of a short HBS α-helix, which unequivocally confirms our helix design principles.35
The helix-coil transition theory in peptides suggests that α-helices composed of ten or fewer amino acids are expected to be essentially unstable due to a low nucleation probability.16,17,20 Two 10-mer peptide sequences derived from the Bak BH3 and the c-Jun coiled-coil domains were selected to examine the HBS strategy.36,37 These peptides were chosen because they represent two different α-helices involved in protein-protein interactions allowing us to gauge the potential of our method for the preparation of biologically important compounds from very short peptide sequences. Below we outline a summary of results obtained with the Bak BH3 10-mer sequence, AcQVARQLAEIY (8), and its HBS α-helix analog 9 (Figure 4a).
FIGURE 4.
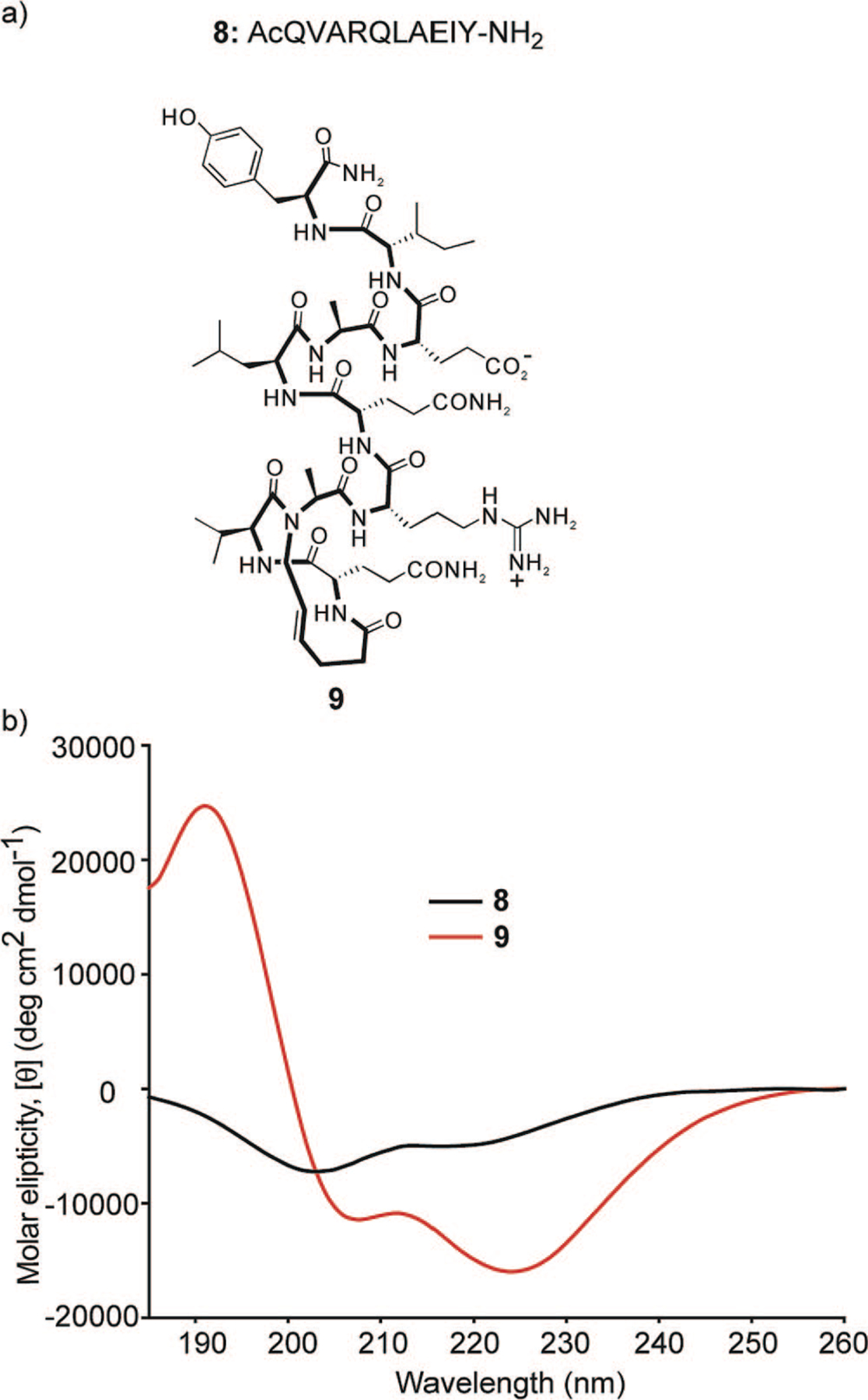
(a) Unconstrained peptide and HBS α-helix used to evaluate the hydrogen bond surrogate strategy and (b) circular dichroism spectra of unconstrained peptide 8 and HBS α-helix 9. The CD spectra were obtained in 10% TFE/PBS.
Characterization by Circular Dichroism.33
The circular dichroism spectra of 8 and 9 are shown in Figure 4b. As expected, 8 displays spectra typical of unstructured or slightly helical peptides. HBS α-helix 9 displays double minima at 208 and 222 nm and a maximum near 190 nm consistent with those observed for canonical α-helices. The relative percent helicity of peptides can be estimated by the mean residue ellipticity at 222 nm, although these estimates are typically not accurate for short helices.33,38,39 HBS helix 9 was calculated to be roughly 70% helical in 10% trifluoroethanol (TFE) in phosphate-buffered saline (PBS), while the unconstrained peptide 8 remained essentially unstructured.
Characterization by 2D NMR Spectroscopy.33
A combination of 2D total correlation spectroscopy (TOCSY), double-quantum-filtered correlation spectroscopy (DQF-COSY), and nuclear Overhauser effect spectroscopy (NOESY) was used to assign 1H NMR resonances for the HBS helices. A peptide folded in the α-helical conformation would be expected to provide sequential NN (i and i + 1) NOEs and medium-range NOEs, including, dαN(i, i + 3), dαN(i, i + 4), and dαβ(i, i + 3) (Figure 5a). All of these major cross-peaks expected from a stable α-helix were observed from the spectrum of 9 as shown in the NOE correlation chart (Figure 5b), although, spectral overlap prevented assignment of some key cross-peaks. The fact that we can detect NOEs involving the last residues in 9 indicates that the helix has not started fraying at the C-terminus.
FIGURE 5.
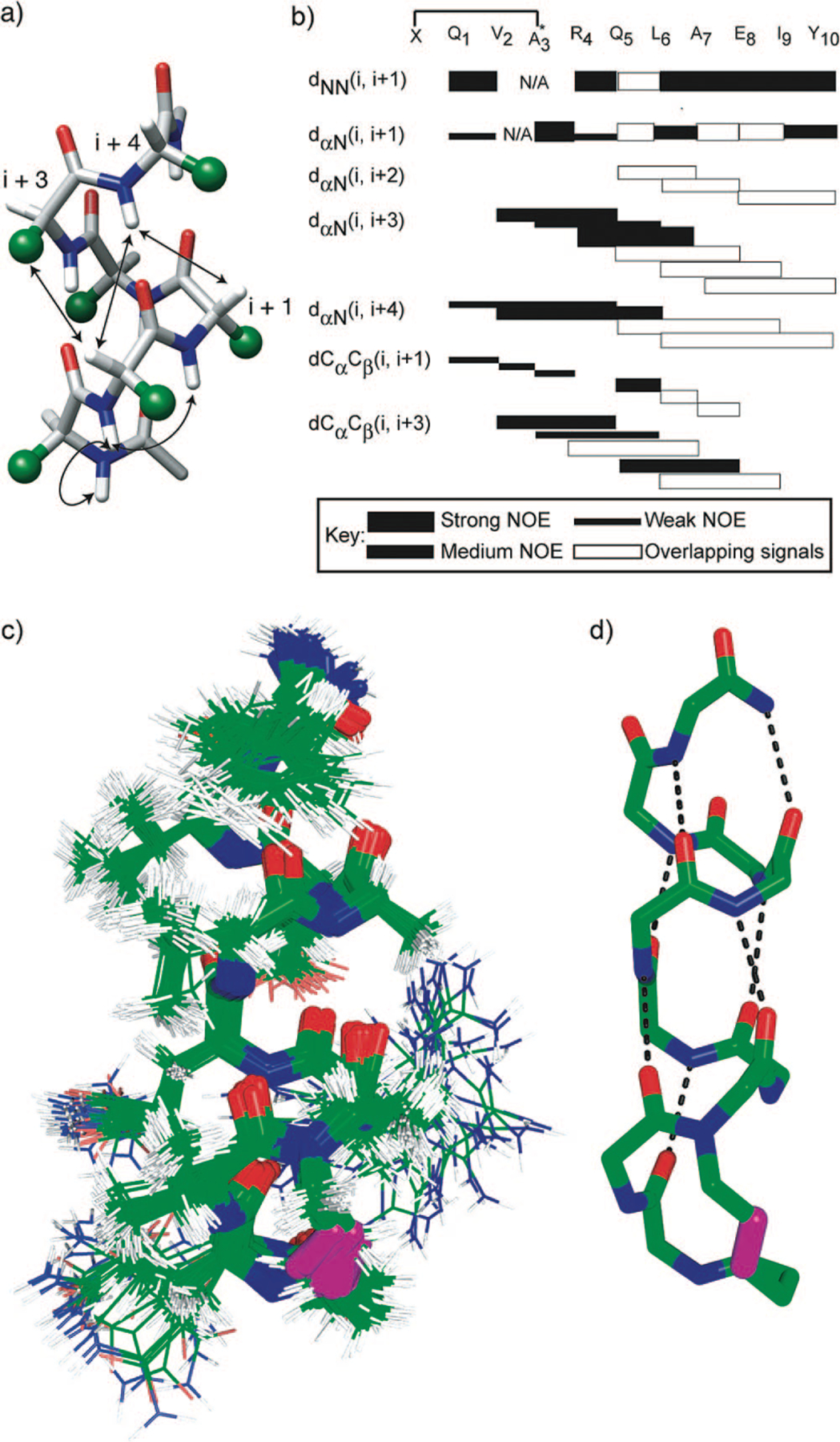
(a) Medium-range NOEs expected from an idealized α-helix–amino acid side chains are represented as green spheres; (b) NOESY correlation chart for HBS α-helix 9–the alanine-3 residues in both artificial helices are alkylated; filled rectangles indicate relative intensity of the NOE cross-peaks, while empty rectangles indicate NOE that could not be unambiguously assigned because of overlapping signals; (c, d) NMR-derived structures of HBS α-helix 9, views of (c) 20 lowest energy structures and (d) lowest energy structure showing hydrogen-bonding pattern within the artificial α-helix. All carbon, nitrogen, and oxygen atoms are shown in green, blue, and red, respectively, with the exception of the trans alkene group, which is shown in magenta.
The solution structure of 9 was determined from NOESY cross-peaks and 3JNHCHα coupling constants (Table 2) using simulated annealing and energy minimization protocol. The final 20 lowest energy structures obtained for the peptide show a backbone root mean squared deviation (rmsd) of 0.58 ± 0.10 Å and all heavy atom rmsd of 1.15 ± 0.30 Å (Figure 5c).33 The final structures exhibit a hydrogen bonding network along the backbone in an i and i + 4 configuration consistent with a well-defined α-helix (Figure 5d).
TABLE 2.
Summary of NMR Data for HBS α-Helix 9
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| peptide | residues | ||||||||
| Q1 | V2 | R4 | Q5 | L6 | A7 | E8 | I9 | Y10 | |
| 3JNH-CαH (ppm) | 8.4 | 4.0 | 4.4 | 4.8 | 4.4 | 3.3 | 4.8 | 6.0 | 6.9 |
| calculated φ (deg)a | −95 | −58 | −62 | −65 | −62 | −52 | −65 | −74 | −81 |
| temperature coefficient (ppb/K) | −4.45 | −8.88 | 2.70 | −2.64 | −5.69 | −4.50 | −2.98 | −3.26 | −5.85 |
| H/D exchange rate (log k) | −4.10 | −3.96 | −5.19 | −4.66 | −4.79 | −4.44 | −3.78 | −4.49 | −3.96 |
| protection factor (log) | 1.64 | 1.13 | 2.94 | 2.62 | 2.09 | 1.90 | 1.60 | 1.94 | 1.13 |
| −ΔG (kcal/mol) | 2.18 | 1.47 | 3.94 | 3.51 | 2.80 | 2.54 | 2.12 | 2.33 | 1.48 |
Characterization by X-ray Crystallography.35
We were recently successful in obtaining an X-ray crystal structure of an HBS α-helix at 1.15 Å resolution (Figure 6a), the coordinates of which have been deposited into the Cambridge Structural Database. The structure of the helix superimposes well onto a model of an idealized α-helix, supporting our hypothesis that stable short helices can be accessed by the HBS strategy. Because crystal structures of short α-helices have been typically reported with the α-helix in complex with a protein partner,40–44 we believe that the solid-state structure of an isolated short helix suggests a high degree of conformational stability being conferred by the HBS strategy. All i and i + 4 C=O and NH hydrogen bonding partners in the HBS helix fall within distances and angles expected for a fully hydrogen-bonded short α-helix (Figure 6b). The backbone conformation of the HBS α-helix in the crystal structure superimposes with an rms difference of 0.75 Å onto the backbone conformation of a model α-helix of sequence AcQVARQLAEIY-NH2 (Figure 6c).
FIGURE 6.
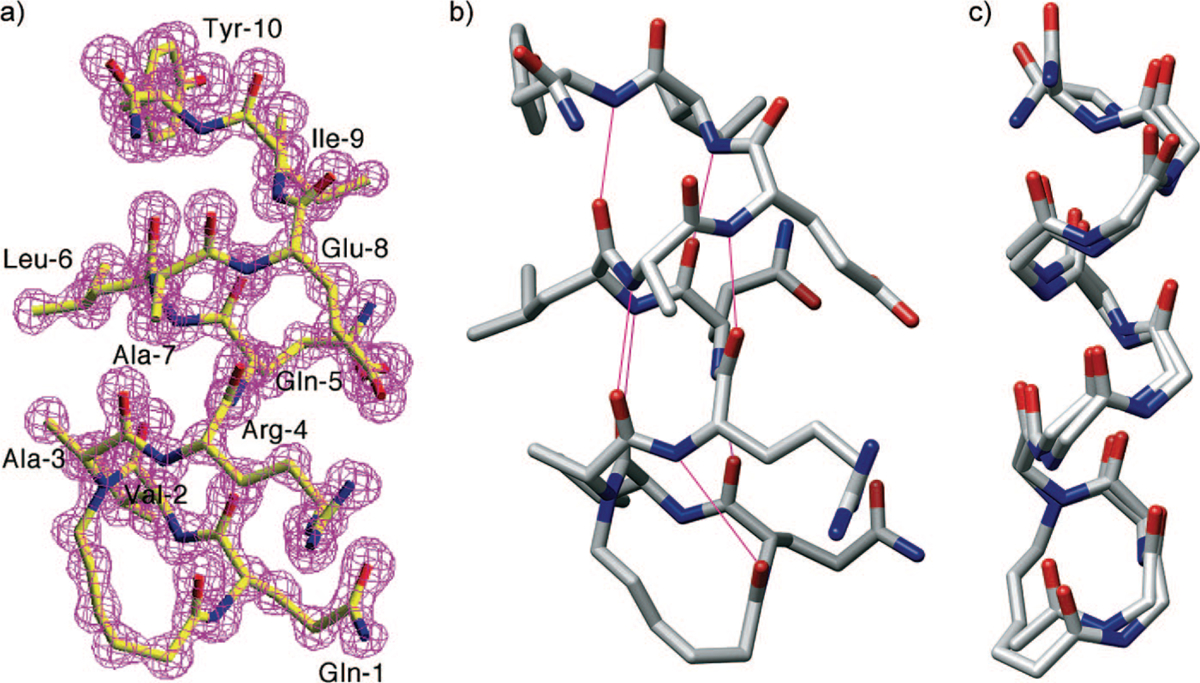
(a) Crystal structure of the HBS α-helix with electron density map superimposed onto the refined molecular model, (b) putative i and i + 4 hydrogen bonds (magenta lines) in crystal structure-derived molecular model of HBS helix, and (c) overlay of crystal structure and a model of an idealized α-helix.
The X-ray crystal structure provides explicit support for our hypothesis that replacement of the hydrogen bond between the i and i + 4 residues at the N-terminus of a short peptide with a carbon–carbon bond results in a highly stable constrained α-helix. Importantly, the crystal structure shows that the alkene-based macrocycle faithfully reproduces the conformation of a prenucleated α-turn.
Effect of Nucleation Macrocycles on Overall Helicity.34
In the HBS approach, the macrocycle mimicking the N-terminal α-turn nucleates the α-helix; in initial studies, we replaced the 13-membered hydrogen-bonded ring that characterize α-helices with a 13-membered macrocycle, based on the conjecture that this ring size may be best at nucleating the helix. In subsequent studies, we also characterized analogs that contain 14-membered rings in which the hydrogen bond acceptor (carbonyl groups) functionality is offset by one atom (Figure 7a).34 To fully determine the effects of the macrocycle on nucleation, we also prepared the hydrogenated HBS analogs containing 13- and 14-membered rings. We hypothesized that a minor change in the macrocycle may have a large impact on the stability of the helix, thus supporting our original design principles that a precise α-turn mimic is critical for helix nucleation.
FIGURE 7.
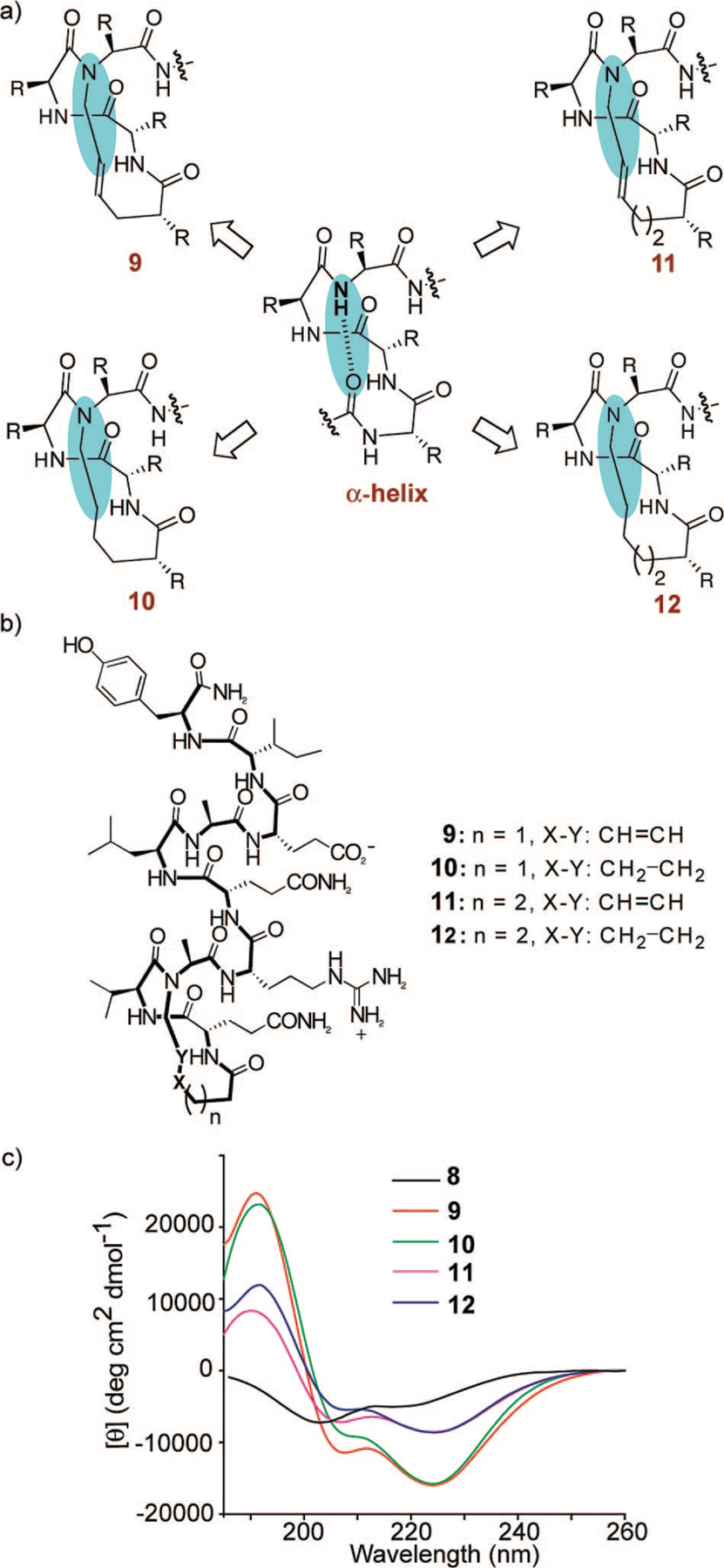
(a) Determination of the optimum nucleation macrocycle in HBS helices, (b) control peptide and HBS helices designed to determine the effect of nucleation macrocycle on the overall helicity, and (c) circular dichroism spectra of unconstrained peptide 8 (black) and HBS analogs 9 (red), 10 (green), 11 (pink), and 12 (blue). The CD spectra were obtained in 10% TFE-PBS.
The CD studies show that HBS analog 11 containing the 14-membered macrocycle is significantly less helical than the 13-membered ring analog 9. These results are consistent with the hypothesis that the macrocycle that most closely mimics the 13-membered hydrogen-bonded α-turn in canonical α-helices should afford the most stable artificial α-helix. Interestingly, the CD studies suggest that the hydrogenated and transalkene HBS helices are similar in overall helicity (Figure 7c). The CD spectra are superimposable except for the 208 nm bands, which are less negative in the hydrogenated series. This result is significant because it indicates that a saturated hydrocarbon link may be sufficient to nucleate an α-helix.
Stability of HBS α-Helices
The conformational stability of HBS helices was examined by thermal and chemical denaturation studies using CD and NMR spectroscopies. These experiments indicate that the HBS strategy confers highly stable helical conformation in short peptides.
Thermal Stability of HBS Helices.32–34
The thermal stability of 9 was investigated by monitoring the temperature-dependent change in the intensity of the 222 nm bands in the CD spectra (Figure 8a). We observe a gradual increase in the signal at 222 nm with temperature, which indicates helix unwinding at high temperatures. Nevertheless, we find that 9 shows a remarkable degree of thermal stability because the peptide retains 60–70% of its room temperature helicity at 85 °C. Although HBS helices are conformationally stable at high temperatures, they can be denatured with concentrated guanidinium chloride.32 CD spectroscopy indicates that 9 retains 25% of its maximum helicity in 8 M GuHCl at 85 °C (Figure 8a). We attribute the CD signal at 8 M GuHCl ([θ]222 = −4500) to the contribution of the nucleation macrocycle to the overall CD spectra.
FIGURE 8.
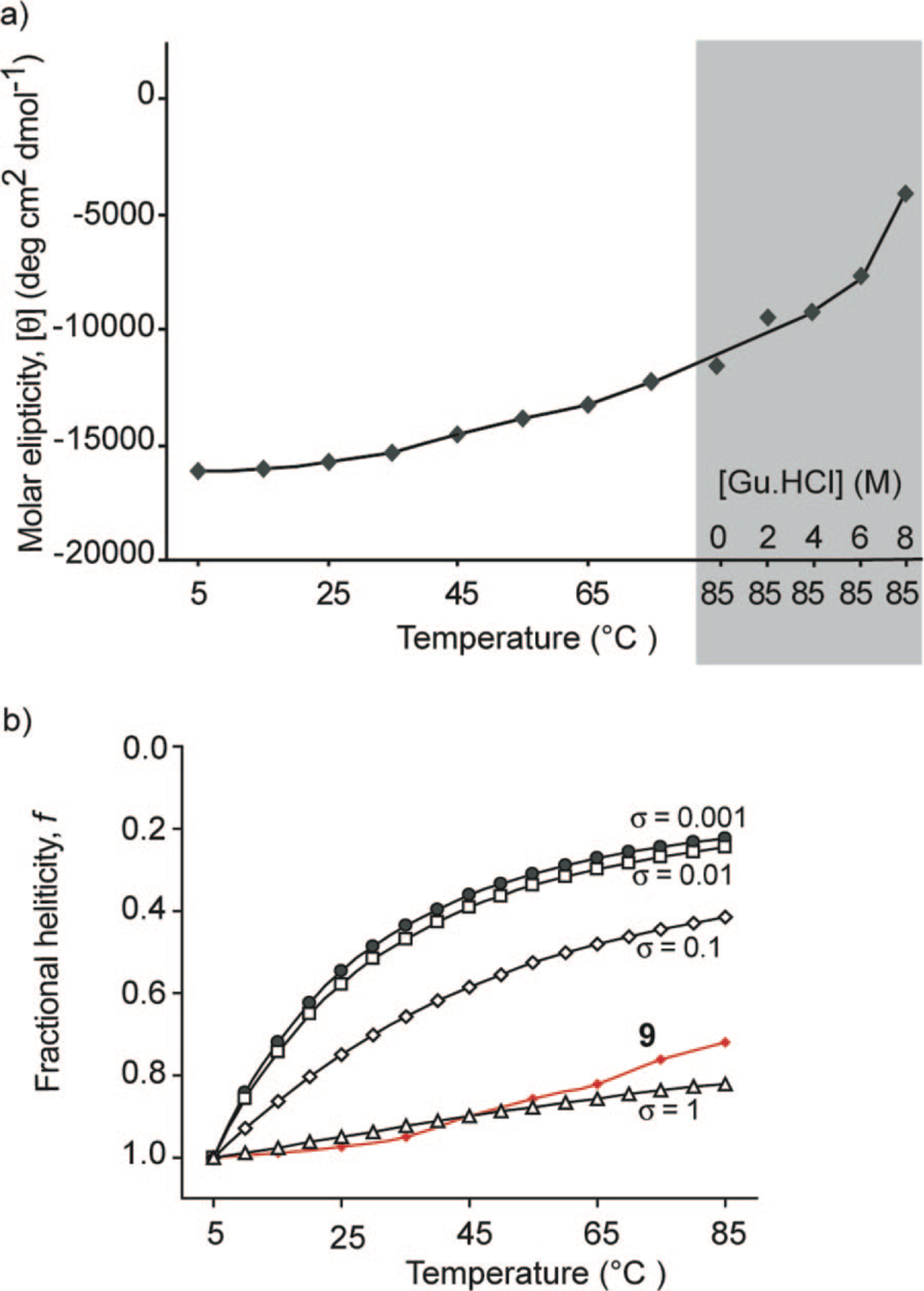
(a) Effect of temperature and guanidinium chloride denaturant (at 85 °C) on HBS α-helix 9 and (b) comparison of theoretical denaturation curves as a function of different nucleation constant, σ, values with the normalized experimental denaturation curve.
Estimates of the Nucleation Constant in HBS Helices.34
The nucleation constant, σ, refers to the organization of three consecutive amino acid residues in an α-turn and is typically very low (10−3–10−4) in unconstrained peptides, disfavoring helix formation.20,45 The intent behind our hydrogen bond surrogate approach was to afford prenucleated helices such that σ would be ~1. We obtained estimates of σ in HBS helices by comparing theoretical denaturation curves, obtained from the Zimm–Bragg model, as a function of different nucleation constants with the experimental denaturation curve for 9 (Figure 8b). This analysis suggests that the nucleation constant is close to unity in HBS helices.34
Amide Proton Temperature Coefficients.33,34
The amide proton chemical shift is temperature sensitive, and the magnitude of this shift is indicative of the extent to which the particular amide proton is hydrogen-bonded.46 If an amide proton exchanges slowly with a temperature coefficient more positive than −4.5 ppb/K, it is considered to be hydrogen-bonded. Figure 9a,b shows the temperature-dependent chemical shifts and NMR spectra for main-chain amide protons in 9, respectively. The temperature coefficients for 9 are listed in Table 2. We find that most amide temperature coefficients in 9 fall in the range considered indicative of hydrogen-bonded amides. The major exception is valine-2 in 9 with coefficients between 8 and 10 ppb/K indicating that the amide proton is not forming intramolecular hydrogen bonds. This is expected because this residue resides at the N-terminus of the helix and does not have intramolecular hydrogen-bonding partners.
FIGURE 9.
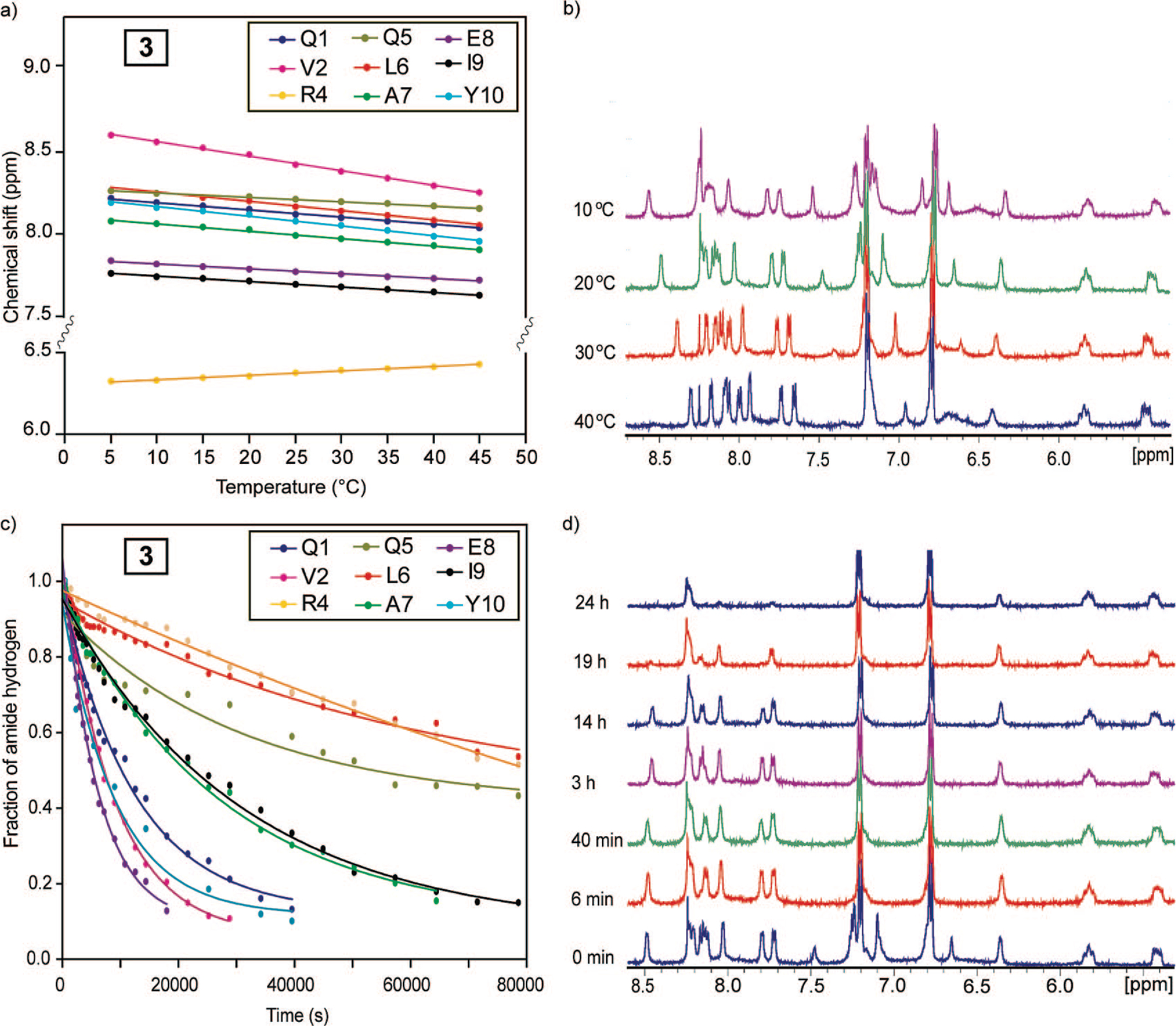
Temperature-dependent chemical shift plot (a) and spectra (b) for backbone amide hydrogens in HBS α-helix 9 and hydrogen-deuterium exchange plot (c) and spectra (d) for backbone amide hydrogens in 9.
Amide Proton Exchange Rates.33,34
Backbone amide hydrogen–deuterium exchange offers a sensitive tool for examining protein structure and dynamics.47,48 The amide exchange rates for unstructured peptides in aqueous solutions are often too fast to measure; however if the amide hydrogen is protected from exchange, that is, through hydrogen bonding, the exchange rates can slow by several orders of magnitude. Relative rate constants for the H/D exchange provide important insights into the involvement of individual amino acid residues in intramolecular hydrogen bonds. Figure 9c,d shows a H/D exchange curve and NMR spectra of 9. The data in Figure 9 shows that the individual hydrogen-ex change rates in this helix can be determined precisely.49 The measured exchange rates, kex, can be compared with the predicted intrinsic chemical exchange rate, kch, for an unstructured peptide of the same sequence to assess individual protection factors (log kch/kex) and the corresponding free energies of protection (ΔG). The predicted intrinsic chemical exchange rates, protection factors, and free energy of protection were calculated using the spreadsheet at http://hx2.med.upenn.edu/download.html and are shown in Table 2 for 9. The data indicates that 9 contains a highly stable hydrogen-bonded network with significant protection factors and associated free energies of protection (1.5–3.9 kcal/mol). Such a degree of stabilization is typically observed for buried amide protons in proteins but not in short peptides.26,50 As expected, we observed rapid exchange rates for 8 (k ≈ 10−3 s−1, data not shown).
Biological Potential of HBS α-Helices
We developed the hydrogen bond surrogate derived α-helices because of our interests in regulating biomolecular inter actions with small molecular weight protein secondary structure mimetics. Our early efforts, as outlined above, have focused on developing an efficient synthetic strategy and examining the stability of short helices derived from biologically relevant (i.e., non-alanine-rich) peptide sequences. Our studies have highlighted the considerable promise of the HBS approach in affording stable two- to three-turn α-helices. In our ongoing efforts, we are examining the potential of these compounds to inhibit chosen protein–protein interactions. We expected HBS α-helices to offer three significant advantages over their unconstrained peptide counterparts. (1) HBS helices are preorganized and do not need to pay the entropic cost of folding; thus these compounds would be expected to bind the target receptor with higher affinities than the unconstrained peptide analogs.53 (2) The biological potential of peptides is limited due to their proteolytic instability. Proteases are known to bind and cleave their target substrate in β-strand conformation.54 Stabilization of peptides in helical conformation should enhance their resistance to proteolytic degradation because the helix must be unfolded before cleavage occurs. In preliminary studies, we have found that HBS helices show enhanced metabolic stability compared with their unconstrained counterparts.55 (3) The cellular uptake of noncationic peptides is considered to be inefficient because of the hydration of amide bonds and the potential need for desolvation before transport through the membrane. Intramolecular hydrogen bonding, as featured in helical conformations, would be expected to reduce hydration of amide bonds and potentially enhance membrane permeability of HBS helices.56
To probe the potential of HBS helices to selectively target a protein receptor, we chose the interaction of Bak BH3 domain with Bcl-xL.37 Bcl-xL is an antiapoptotic protein that regulates cell death by binding the α-helical BH3 domain of a family of proapoptotic proteins (including Bak, Bad, Bid, and Bax).57,58 NMR studies by Fesik and co-workers have shown that a 16-mer peptide derived from the Bak BH3 domain adopts an α-helical conformation upon binding to Bcl-xL (Figure 10).37 Circular dichroism (CD) studies demonstrate that this peptide is unstructured at physiological conditions in the absence of the protein partner and only slightly helical in trifluoroethanol (TFE), a helix-promoting solvent.55
FIGURE 10.
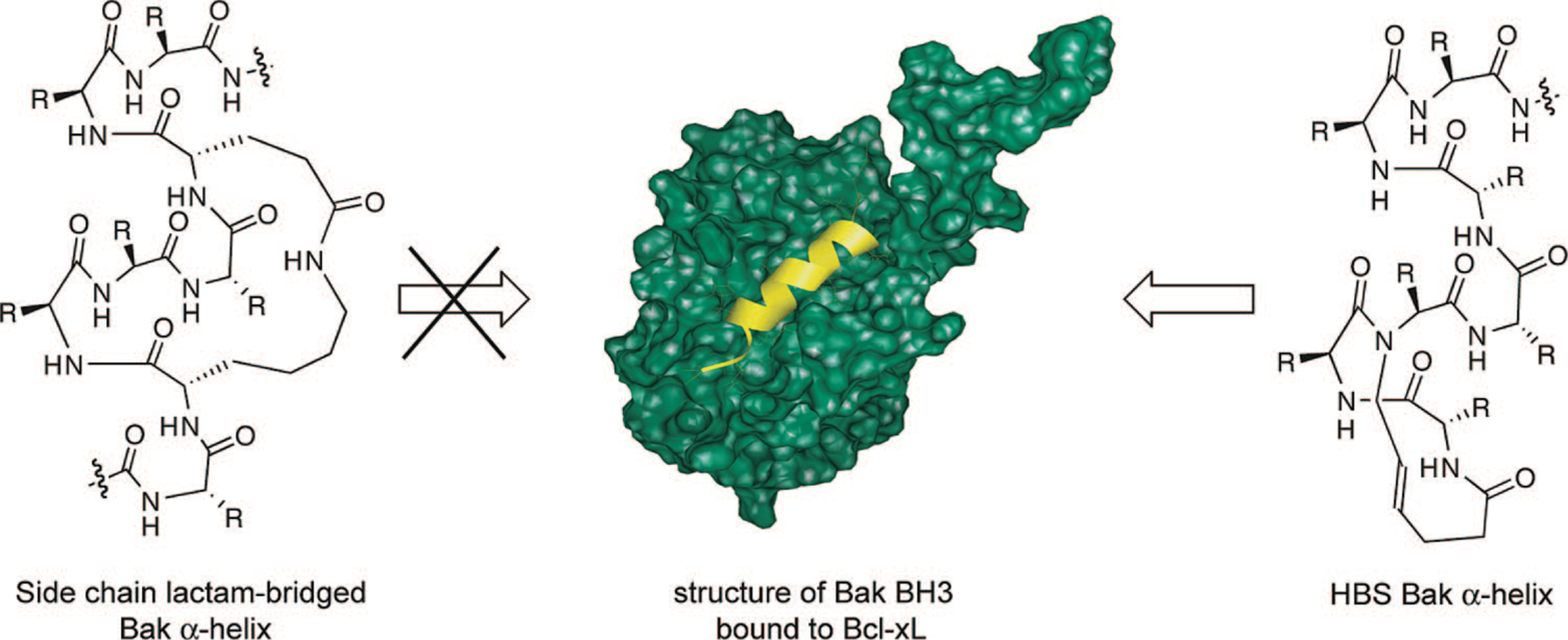
Recognition of Bcl-xL with side-chain cross-linked and HBS Bak BH3 α-helices (Bak72–87: GQVGRQLAIIGDDINR). The lactam-bridged helices fail to target Bcl-xL,59 whereas HBS Bak α-helix binds the same protein target with high affinity.55 The structure of Bak BH3/Bcl-xL was solved by Fesik and co-workers (PDB code 1BXL).37
Several methods that afford stabilized α-helices or helix mimetics have already been used to target Bcl-xL allowing us to directly compare the performance of our artificial α-helices (Figure 10).10,60–64 Significantly, Huang and co-workers had reported that Bak BH3 α-helices stabilized by a lactam-based side-chain cross-linking strategy were unable to bind Bcl-xL.59 The authors speculated that the lack of binding might be due to steric clashes between the cross-link and the narrow binding pocket of Bcl-xL. [Verdine, Korsmeyer, and co-workers found that side-chain bridged α-helices corresponding to the BH3 domain of a different proapoptotic protein, Bid, can target Bcl-xL and suppress the growth of leukemia cells in mice.10] The protein binding studies from the Huang laboratory illustrate potential problems with the side-chain bridging strategy. The fundamental advantage of the hydrogen bond surrogate approach over the side-chain bridging strategy for stabilizing helices is that the helix surfaces are not encumbered by the constraining element. The hydrogen bond surrogate approach should therefore greatly simplify the helix design process. The key question we asked was whether the HBS-derived Bak α-helix can bind Bcl-xL when the side-chain constrained (lactam-bridge) Bak helix is unable to target this same protein receptor. Fluorescence anisotropy studies indicated that the HBS Bak α-helix bound Bcl-xL with high affinity.55 This data validates our helix design principle because we found that the internally constrained HBS α-helices indeed access the deep hydrophobic cleft presented by Bcl-xL, whereas α-helices prepared by the side-chain bridging method showed no affinity for the same target. In continuing studies in the laboratory, we are exploring the efficacy of HBS helices to target Bak/Bcl-xL and other protein-protein interactions in cell culture.
In summary, the hydrogen bond surrogate (HBS) approach affords stable α-helices from a variety of short peptide sequences. Solution NMR and crystal structures of the HBS helices unequivocally confirm our fundamental helix design principles. Preliminary (and unpublished) studies indicate that HBS helices can target their expected protein partners with high affinity. Importantly, analyses of the helix-coil parameters suggest that the HBS strategy fixes the nucleation constant for helix formation near unity. Control over this fundamental parameter of helix folding will allow us, and members of the biophysical community, to test basic tenets of protein folding in minimal models.
Acknowledgments
This work was supported by the NIH (Grant GM073943), the donors of the American Chemical Society Petroleum Research Fund, Research Corporation (Cottrell Scholar Award), NYU (Whitehead Fellowship), and the New York State Office of Science, Technology and Academic Research (James D. Watson Investigator Award).
BIOGRAPHICAL INFORMATION
Anupam Patgiri received his B.Sc. in Chemistry from Gauhati University, India, in 2002 and an M.Sc. in Chemistry from Indian Institute of Technology, Guwahati, India, in 2005. He is currently a second-year graduate student in the Ph.D. program at NYU and is investigating the synthesis and molecular recognition properties of helix mimetics.
Andrea L. Jochim is a graduate of Cal Poly, San Luis Obispo, where she received her B.S. in Chemistry and Biochemistry in 2003. She is currently a fourth-year graduate student in the Ph.D. program at NYU; her research interests span computational and experimental aspects of chemical biology.
Paramjit S. Arora obtained his B.S. in Chemistry from UC Berkeley and his earliest research experience in the research group of Prof. Richard Mathies. He received his Ph.D. degree in Organic Chemistry under the guidance of Prof. James Nowick at UC Irvine and pursued a postdoctoral fellowship with Prof. Peter Dervan at Caltech before joining the faculty of NYU as an Assistant Professor of Chemistry.
REFERENCES
- 1.Arkin MR; Wells JA Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discovery 2004, 3, 301–317. [DOI] [PubMed] [Google Scholar]
- 2.Yin H; Hamilton AD Strategies for targeting protein-protein interactions with synthetic agents. Angew. Chem., Int. Ed 2005, 44, 4130–4163. [DOI] [PubMed] [Google Scholar]
- 3.Cochran AG Antagonists of protein-protein interactions. Chem. Biol 2000, 7, R85–R94. [DOI] [PubMed] [Google Scholar]
- 4.Cochran AG Protein-protein interfaces: Mimics and inhibitors. Curr. Opin. Chem. Biol 2001, 5, 654–659. [DOI] [PubMed] [Google Scholar]
- 5.Guharoy M; Chakrabarti P Secondary structure based analysis and classification of biological interfaces: identification of binding motifs in protein-protein interactions. Bioinformatics 2007, 23, 1909–1918. [DOI] [PubMed] [Google Scholar]
- 6.Jones S; Thornton JM Principles of protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A 1996, 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S; Thornton JM Protein-protein interactions - a review of protein dimer structures. Prog. Biophys. Mol. Biol 1995, 63, 31–65. [DOI] [PubMed] [Google Scholar]
- 8.Barlow DJ; Thornton JM Helix geometry in proteins. J. Mol. Biol 1988, 201, 601–619. [DOI] [PubMed] [Google Scholar]
- 9.Davis JM; Tsou LK; Hamilton AD Synthetic non-peptide mimetics of alpha-helices. Chem. Soc. Rev 2007, 36, 326–334. [DOI] [PubMed] [Google Scholar]
- 10.Walensky LD; Kung AL; Escher I; Malia TJ; Barbuto S; Wright RD; Wagner G; Verdine GL; Korsmeyer SJ Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 2004, 305, 1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng RP; Gellman SH; DeGrado WF β-Peptides: From structure to function. Chem. Rev 2001, 101, 3219–3232. [DOI] [PubMed] [Google Scholar]
- 12.Bautista AD; Craig CJ; Harker EA; Schepartz A Sophistication of foldamer form and function in vitro and in vivo. Curr. Opin. Chem. Biol 2007, 11, 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garner J; Harding MM Design and synthesis of α-helical peptides and mimetics. Org. Biomol. Chem 2007, 5, 3577–3585. [DOI] [PubMed] [Google Scholar]
- 14.Goodman CM; Choi S; Shandler S; DeGrado WF Foldamers as versatile frameworks for the design and evolution of function. Nat. Chem. Biol 2007, 3, 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray JK; Gellman SH Targeting protein-protein interactions: Lessons from p53/MDM2. Biopolymers 2007, 88, 657–686. [DOI] [PubMed] [Google Scholar]
- 16.Lifson S; Roig A On the theory of helix-coil transitions in polypeptides. J. Chem. Phys 1961, 34, 1963–1974. [Google Scholar]
- 17.Zimm BH; Bragg JK Theory of the phase transition between helix and random coil in polypeptide chains. J. Chem. Phys 1959, 31, 526–535. [Google Scholar]
- 18.Qian H; Schellman JA Helix-coil theories: A comparative study for finite length preferences. J. Phys. Chem 1992, 96, 3987–3994. [Google Scholar]
- 19.Siedlecka M; Goch G; Ejchart A; Sticht H; Bierzyski A Alpha-helix nucleation by a calcium-binding peptide loop. Proc. Natl. Acad. Sci. U.S.A 1999, 96, 903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JX; Zhao K; Gong YX; Vologodskii A; Kallenbach NR α-Helix nucleation constant in copolypeptides of alanine and ornithine or lysine. J. Am. Chem. Soc 1998, 120, 10646–10652. [Google Scholar]
- 21.Austin RE; Maplestone RA; Sefler AM; Liu K; Hruzewicz WN; Liu CW; Cho HS; Wemmer DE; Bartlett PA Template for stabilization of a peptide R helix: Synthesis and evaluation of conformational effects by circular dichroism and NMR. J. Am. Chem. Soc 1997, 119, 6461–6472. [Google Scholar]
- 22.Kemp DS; Curran TP; Davis WM; Boyd JG; Muendel C Studies of N-terminal templates for α-helix formation. Synthesis and conformational-analysis of (2S,5S,8S,11S)-1-acetyl-1,4-diaza-3-keto-5-carboxy-10-thiatricyclo[2.8.1.04,8]tridecane (Ac-Hel1-OH). J. Org. Chem 1991, 56, 6672–6682. [Google Scholar]
- 23.Cabezas E; Satterthwait AC The hydrogen bond mimic approach: Solid-phase synthesis of a peptide stabilized as an R helix with a hydrazone link. J. Am. Chem. Soc 1999, 121, 3862–3875. [Google Scholar]
- 24.Trnka TM; Grubbs RH The development of L2X2RudCHR olefin metathesis catalysts: An organometallic success story. Acc. Chem. Res 2001, 34, 18–29. [DOI] [PubMed] [Google Scholar]
- 25.Blackwell HE; Grubbs RH Highly efficient synthesis of covalently cross-linked peptide helices by ring-closing metathesis. Angew. Chem., Int. Ed. Engl 1998, 37, 3281–3284. [DOI] [PubMed] [Google Scholar]
- 26.Phelan JC; Skelton NJ; Braisted AC; McDowell RS A general method for constraining short peptides to an α-helical conformation. J. Am. Chem. Soc 1997, 119, 455–460. [Google Scholar]
- 27.Schafmeister CE; Po J; Verdine GL An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J. Am. Chem. Soc 2000, 122, 5891–5892. [Google Scholar]
- 28.Chapman RN; Arora PS Optimized synthesis of hydrogen-bond surrogate helices: Surprising effects of microwave heating on the activity of grubbs catalysts. Org. Lett 2006, 8, 5825–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimartino G; Wang D; Chapman RN; Arora PS Solid-phase synthesis of hydrogen-bond surrogate-derived α-helices. Org. Lett 2005, 7, 2389–2392. [DOI] [PubMed] [Google Scholar]
- 30.Miller SC; Scanlan TS oNBS-SPPS: A new method for solid-phase peptide synthesis. J. Am. Chem. Soc 1998, 120, 2690–2691. [Google Scholar]
- 31.Reichwein JF; Liskamp RMJ Site-specific N-alkylation of peptides on the solid phase. Tetrahedron Lett 1998, 39, 1243–1246. [Google Scholar]
- 32.Chapman RN; Dimartino G; Arora PS A highly stable short R helix constrained by a main-chain hydrogen-bond surrogate. J. Am. Chem. Soc 2004, 126, 12252–12253. [DOI] [PubMed] [Google Scholar]
- 33.Wang D; Chen K; Kulp JL III; Arora PS Evaluation of biologically relevant short α-helices stabilized by a main-chain hydrogen-bond surrogate. J. Am. Chem. Soc 2006, 128, 9248–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D; Chen K; Dimartino G; Arora PS Nucleation and stability of hydrogen-bond surrogate-based alpha-helices. Org. Biomol. Chem 2006, 4, 4074–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J; Wang D; Zheng Q; Lu M; Arora PS Atomic structure of a short α-helix stabilized by a main chain hydrogen bond surrogate. J. Am. Chem. Soc 2008, 130, 4334–4337. [DOI] [PubMed] [Google Scholar]
- 36.O’Shea EK; Rutkowski R; Stafford WF 3rd; Kim PS Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science 1989, 245, 646–648. [DOI] [PubMed] [Google Scholar]
- 37.Sattler M; Liang H; Nettesheim D; Meadows RP; Harlan JE; Eberstadt M; Yoon HS; Shuker SB; Chang BS; Minn AJ; Thompson CB; Fesik SW Structure of Bcl-x(L)-Bak peptide complex: Recognition between regulators of apoptosis. Science 1997, 275, 983–986. [DOI] [PubMed] [Google Scholar]
- 38.Chin DH; Woody RW; Rohl CA; Baldwin RL Circular dichroism spectra of short, fixed-nucleus alanine helices. Proc. Natl. Acad. Sci. U.S.A 2002, 99, 15416–15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shepherd NE; Hoang HN; Abbenante G; Fairlie DP Single turn peptide R helices with exceptional stability in water. J. Am. Chem. Soc 2005, 127, 2974–2983. [DOI] [PubMed] [Google Scholar]
- 40.Kim EE; Varadarajan R; Wyckoff HW; Richards FM Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures. Biochemistry 1992, 31, 12304–12314. [DOI] [PubMed] [Google Scholar]
- 41.Kussie PH; Gorina S; Marechal V; Elenbaas B; Moreau J; Levine AJ; Pavletich NP Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996, 274, 948–953. [DOI] [PubMed] [Google Scholar]
- 42.Sia SK; Carr PA; Cochran AG; Malashkevich VN; Kim PS Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. U.S.A 2002, 99, 14664–14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karle IL; Flippenanderson JL; Uma K; Balaram H; Balaram P Alpha-helix and mixed 310/alpha-helix in cocrystallized conformers of Boc-Aib-Val-Aib-Aib-Val-Val-Val-Aib-Val-Aib-Ome. Proc. Natl. Acad. Sci. U.S.A 1989, 86, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang DSC; Sax M; Chakrabartty A; Hew CL Crystal-structure of an antifreeze polypeptide and its mechanistic implications. Nature 1988, 333, 232–237. [DOI] [PubMed] [Google Scholar]
- 45.Matheson RR; Scheraga HA Calculation of the Zimm-Bragg cooperativity parameter σ from a simple model of the nucleation process. Macromolecules 1983, 16, 1037–1043. [Google Scholar]
- 46.Baxter NJ; Williamson MP Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR 1997, 9, 359–369. [DOI] [PubMed] [Google Scholar]
- 47.Bai Y; Milne JS; Mayne L; Englander SW Primary structure effects on peptide group hydrogen exchange. Proteins 1993, 17, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connelly GP; Bai Y; Jeng MF; Englander SW Isotope effects in peptide group hydrogen exchange. Proteins 1993, 17, 87–92. [DOI] [PubMed] [Google Scholar]
- 49.Englander SW; Kallenbach NR Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys 1983, 16, 521–655. [DOI] [PubMed] [Google Scholar]
- 50.Zhou HXX; Hull LA; Kallenbach NR; Mayne L; Bai YW; Englander SW Quantitative evaluation of stabilizing interactions in a prenucleated α-helix by hydrogen exchange. J. Am. Chem. Soc 1994, 116, 6482–6483. [Google Scholar]
- 51.Karplus M Contact electron-spin coupling of nuclear magnetic moments. J. Chem. Phys 1959, 30, 11–15. [Google Scholar]
- 52.Wuthrich K NMR of Proteins and Nucleic Acids; Wiley: New York, 1986. [Google Scholar]
- 53.Williams DH; Stephens E; O’Brien DP; Zhou M Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew. Chem., Int. Ed 2004, 43, 6596–6616. [DOI] [PubMed] [Google Scholar]
- 54.Tyndall JD; Nall T; Fairlie DP Proteases universally recognize β strands in their active sites. Chem. Rev 2005, 105, 973–999. [DOI] [PubMed] [Google Scholar]
- 55.Wang D; Liao W; Arora PS Enhanced metabolic stability and protein-binding properties of artificial alpha-helices derived from a hydrogen-bond surrogate: Application to Bcl-xL. Angew. Chem., Int. Ed 2005, 44, 6525–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rezai T; Yu B; Millhauser GL; Jacobson MP; Lokey RS Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc 2006, 128, 2510–2511. [DOI] [PubMed] [Google Scholar]
- 57.Cory S; Huang DC; Adams JM The Bcl-2 family: Roles in cell survival and oncogenesis. Oncogene 2003, 22, 8590–8607. [DOI] [PubMed] [Google Scholar]
- 58.Letai A; Bassik MC; Walensky LD; Sorcinelli MD; Weiler S; Korsmeyer SJ Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002, 2, 183–192. [DOI] [PubMed] [Google Scholar]
- 59.Yang B; Liu D; Huang Z Synthesis and helical structure of lactam bridged BH3 peptides derived from pro-apoptotic Bcl-2 family proteins. Bioorg. Med. Chem. Lett 2004, 14, 1403–6. [DOI] [PubMed] [Google Scholar]
- 60.Sadowsky JD; Murray JK; Tomita Y; Gellman SH Exploration of backbone space in foldamers containing alpha- and beta-amino acid residues: Developing protease-resistant oligomers that bind tightly to the BH3-recognition cleft of Bcl-x(L). ChemBioChem 2007, 8, 903–916. [DOI] [PubMed] [Google Scholar]
- 61.Chin JW; Schepartz A Design and evolution of a miniature bcl-2 binding protein. Angew. Chem., Int. Ed 2001, 40, 3806–3809. [DOI] [PubMed] [Google Scholar]
- 62.Degterev A; Lugovskoy A; Cardone M; Mulley B; Wagner G; Mitchison T; Yuan J Identification of small-molecule inhibitors of interaction between the BH3 domain and Bcl-xL. Nat. Cell Biol 2001, 3, 173–82. [DOI] [PubMed] [Google Scholar]
- 63.Kutzki O; Park HS; Ernst JT; Orner BP; Yin H; Hamilton AD Development of a potent Bcl-x(L) antagonist based on R helix mimicry. J. Am. Chem. Soc 2002, 124, 11838–11839. [DOI] [PubMed] [Google Scholar]
- 64.Gemperli AC; Rutledge SE; Maranda A; Schepartz A Paralog-selective ligands for bcl-2 proteins. J. Am. Chem. Soc 2005, 127, 1596–1597. [DOI] [PubMed] [Google Scholar]


