Abstract
rhTCRseq (RNase H-dependent PCR-enabled T cell receptor sequencing) is a procedure that can be used to determine paired alpha/beta T cell receptor (TCR) clonotypes in single cells or perform both alpha and beta TCR repertoire analysis in bulk RNA samples. Relying on the enhanced specificity of RNase H-dependent PCR (rhPCR), it achieves TCR-specific amplification and addition of dual index barcodes in a single PCR step. For single cells, the stages of the protocol are sorting single cells into 96- or 384-well plates, generation of full-length cDNA libraries, the single TCR-specific amplification step, a second PCR on a pooled sample to generate a sequencing library, and sequencing on the MiSeq platform. In the bulk RNA method, the sorting and cDNA library steps are replaced with a reverse transcriptase reaction that add a Unique Molecular Identifier (UMI) to each cDNA molecule in order to improve the accuracy of repertoire frequency measurements. Compared to other multiplex PCR methods for TCR sequencing, rhTCRseq has a streamlined workflow and the ability to analyze single cells in 384-well plates. Compared to TCR reconstruction from single-cell transcriptome sequencing data, it improves the success rate for obtaining paired alpha/beta information and ensures recovery of complete CDR3 sequences, which is a prerequisite for the cloning and expression of discovered TCRs. Although it does not have the throughput of droplet-based methods, rhTCRseq is well-suited to the analysis of small sorted populations, especially cases where analysis of 96 or 384 single cells is sufficient to identify predominant T cell clones. For single cells, sorting typically requires two to four hours and can be performed days, or even months, before library processing. The remainder of the single cell protocol takes on the order of four days, including data processing. For bulk RNA, the overall time is about three days, including data processing.
Keywords: T cell receptor repertoire, T cell receptor, paired TCRαβ single-cell sequencing, RNase H-dependent PCR, immunotherapy, neoantigen vaccination, rhPCR-enabled TCR sequencing, rhTCRseq
EDITORIAL SUMMARY
Here the authors describe rhTCRseq, RNase H-dependent PCR-enabled TCR sequencing, for repertoire analysis from bulk RNA samples or single-cell profiling.
Introduction
Characterization of the immune response to pathogens, cancer, and therapeutic interventions has long used T cell receptor (TCR) diversity/heterogeneity as a measure of clonal diversity and immune response because T cells are primary effector cells of antigen-specific immunity. A TCR interacts with a peptide antigen bound to a major histocompatibility complex (MHC) molecule mainly through the paired alpha- and beta-complementarity determining regions 3 (CDR3). Thus, characterizing the antigen specificity of a T cell involves determining the sequence of the CDR3 segments.
Recent single-cell sequencing technologies have facilitated the identification of which TCR interacts with a particular antigen through establishing the exact pairing of TCR-alpha and TCR-beta chains. The first methods described used multiplex PCR with a mixture of TRAV- and TRBV-specific forward primers for targeted TCR amplification from single-cell cDNA followed by sequencing of these low complexity libraries1–5. A second approach reconstructs TCR sequences from global single-cell RNA sequencing (scRNA-seq) data6–9. Because these computational methods require full-length sequence information, they cannot be applied to the end-counting scRNA-seq data generated by droplet-based procedures. Recently, 10x Genomics has commercialized a protocol that does enable determination of single-cell TCR sequences from their droplet-generated libraries and this product has begun generating published results10,11. Finally, pairSEQ is not a single-cell method, but it does generate paired alpha/beta information by computational imputation from TCR sequence data acquired by dividing a bulk sample into 96 subsets12.
Overview of the protocol
Here we detail the rhTCRseq (rhPCR-enabled TCR sequencing) protocol that was used in Keskin et al.13 to analyze TCR clonotypes in glioblastoma patients who were part of a neoantigen vaccination clinical trial. rhTCRseq uses highly specific PCR to reliably achieve targeted amplification of alpha and beta CDR3 segments from TCR transcripts in a streamlined workflow. In order to develop a method for amplifying TCR segments that is compatible with next-generation sequencing (NGS) library generation, rhTCRseq takes advantage of RNase H-dependent PCR (rhPCR), a technique that greatly enhances the specificity of PCR14. As diagrammed in Fig. 1a, rhPCR uses 3’-blocked oligonucleotides with a single ribo residue located approximately five nucleotides from the 3’ end. By including thermostable RNase H in the amplification reaction, these blocked oligonucleotides are cleaved at the RNA base if, and only if, the oligonucleotide is hybridized to an appropriate target. Cleavage generates a free 3’-hydroxyl that is extended by Taq DNA polymerase. Thus, functional primers are generated in situ during the PCR and accurate hybridization of the proto-primers is required during every round of PCR in order to achieve exponential amplification. This technique is very specific because the absence of free primers not hybridized to target essentially eliminates primer dimer formation, and the requirement of RNase H for high-fidelity base pairing severely reduces off-target amplification.
Fig. 1 |. Critical features and schemes of rhTCRseq.
a, Mechanism for the enhanced specificity of rhPCR. Instead of conventional primers, rhPCR uses 3’-blocked oligonucleotides each containing a single ribo base. Upon high fidelity hybridization to its target, each oligonucleotide is cleaved at the ribo base by thermostable RNase H2 to generate a primer with a 3’-hydroxyl that can be extended by DNA polymerase. b,c, Scheme for TCR-specific amplification from single cell cDNA libraries (b) and from bulk RNA (c). V, (D), J, and C indicate segments of the TCR transcript. Arrowheads indicate the 3’ end of primers. TSO refers to the NEBNext Template Switching Oligo. P1 refers to the NEBNext Single Cell cDNA PCR Primer. Segments specific for the variable (TRAV, TRBV) and constant (TRAC, TRBC) regions of alpha and beta TCR transcripts, and for the Illumina read 1 and read 2 sequences (Rd1, Rd2) are labelled. There are 69 rhPCR primers specific for TRAV and TRBV, 1 rhPCR primer specific for TRAC, and 1 rhPCR primer specific for TRBC. IDT*** indicates 8-nucleotide index sequence. P5 and P7 refer to the Illumina sequences required for flow cell attachment. P5.IDT***.Rd1 and P7.IDT***.Rd2 are rhPCR primers. UMI refers to Unique Molecular Identifier.
For the analysis of single cells, Fig. 1b diagrams the scheme for using rhPCR to re-amplify CDR3 segments from whole transcript single-cell cDNA libraries, such as those generated as an intermediate step in Smart-seq215,16. To amplify CDR3-containing segments, 69 rhPCR primers (38 alpha and 31 beta) were designed to hybridize to all functional alpha and beta TCR variable genes (Supplementary Table 1). These primers include a Rd1 (Read 1) sequence needed for Illumina-based sequencing. The two rhPCR primers specific for TRAC and TRBC (the constant segments of TCR alpha and beta loci, respectively) have an Illumina Rd2 (Read 2) sequence. In addition to the TCR-specific primers, the amplification reaction contains flanking rhPCR primers (Supplementary Table 2) that incorporate index sequences into the final amplification products and append the P5 and P7 sequences needed for Illumina-based sequencing. By using distinct index sequences for different single-cell libraries, a single PCR step specifically amplifies CDR3 segments and introduces barcodes for each sample. The 768 primers listed in Supplementary Table 2 enable dual indexing for pools of up to 384 samples. Dual indexing is used to minimize the complications of data interpretation caused by index switching17,18. After the first PCR step shown in Fig. 1b, samples are pooled for further processing and sequencing.
We observed the procedure of Fig. 1b to demonstrate high fidelity in determining TCR sequences in the Keskin et al. study13. In this case, TCR sequences in single tumor-infiltrating lymphocytes (TILs) from Patient 7 were determined both using TraCeR6 reconstruction from Smart-seq215 scRNA-seq data and using rhTCRseq on the same Smart-seq2 cDNA libraries. For the 277 cells from Patient 7 with paired alpha/beta-chain data by rhTCRseq, the predominant CDR3 sequences identified by TraCeR were identical to the most frequent CDR3 sequences detected by rhTCRseq. In addition, the TRAV and TRBV (the variable segments of TCR alpha and beta loci, respectively) gene assignments and the TRAJ and TRBJ (the joining segments of TCR alpha and beta loci, respectively) sequences matched between the two methods. There was no case of TraCeR analysis identifying a TRAV or TRBV allele that was not also detected by rhTCRseq, but it is difficult to assess the breadth of coverage achieved with the rhTCRseq primer set analyzing cells one at a time. Therefore, the rhTCRseq protocol was adapted for amplifying CDR3 segments in bulk RNA samples (Fig. 1c). In the first step, TRAC- and TRBC-specific primers are used in a reverse transcriptase (RT) reaction to synthesize TCR cDNA. These primers include a Rd2 sequence needed for Illumina-based sequencing and a Unique Molecular Identifier (UMI)19,20. There is an exonuclease treatment between the RT and PCR steps to digest the UMI-containing primers so that UMIs are only incorporated during the RT step. Use of the UMI enables counting of the original number of cDNA molecules prior to PCR, which improves the accuracy of repertoire frequency measurements21–26. The PCR1 step is the same as in the single-cell protocol except the rhPCR primers specific for TRAC and TRBC are not included. The steps after this point are the same for the two protocols. As detailed in the Supplementary Note under “Analysis of bulk RNA from five normal individuals,” determination of the TCR repertoire in peripheral blood mononuclear cell (PBMC) RNA samples (S. L., R. A., J. F., R. J., D. B. K., K. J. L., unpublished data) shows that the rhTCRseq primer set does have breadth of coverage across the functional alpha and beta TCR variable genes (Supplementary Fig. 1, Supplementary Table 3). Supplementary Fig. 2 and Supplementary Table 4 show the reproducibility of rhTCRseq when applied to the five normal bulk RNA samples.
Single cell applications of rhTCRseq
A key finding of Keskin et al.13 is the evidence provided by single-cell TCR analysis that neoantigen-specific T cells generated in response to peptide vaccination can migrate from the peripheral blood to a glioblastoma in the brain. For Patient 7, TraCeR reconstruction identified paired alpha and beta information for 160 of 384 cells (41.7%), but predominantly only partial CDR3 sequences were recovered. Using rhTCRseq on the Smart-seq2 cDNA libraries in 96-well plates, the yield of paired alpha/beta sequences was improved to 277 of 384 cells (72.1%) and, for all 277 cells, complete CDR3 sequences were obtained. Having complete CDR3 sequences proved to be critical for the cloning and expression of discovered TCRs27, and the subsequent testing to demonstrate neoantigen specificity of individual TCRs.
As detailed in the Supplementary Note under “Single-cell analysis of tumor-specific T cells in 384-well format,” rhTCRseq has been applied to single T cells sorted into a 384-well plate (Supplementary Figure 3) and used to determine the clonality of peripheral T cells that are specific for tumor tissue (S. L., R. A., G. O., J. F., R. J., K. J. L., unpublished data). The paired alpha/beta clonotype for each single cell are reported in Supplementary Table 5. As shown in Supplementary Fig. 4, 49 putative tumor-specific clones were detected, ranging in size from 2–30 cells per clone. For this experiment, a workflow was developed for processing single cells in 384-well plates, starting with preparation of full-length cDNA libraries followed by re-amplification of CDR3 segments as depicted in Fig. 1b. Compared to the Smart-seq2 procedure for preparing full-length cDNA libraries, streamlining and reduced cost were achieved by minimizing the number of purifications and lowering reaction volumes.
The pairing of TCR sequence information and RNA expression data is vital for understanding how a particular clonotype relates to the functional state of the T cell. rhTCRseq amplifies TCR segments from single-cell cDNA libraries, which can be further processed to generate single-cell sequencing libraries to obtain whole-transcript scRNA-seq data. By following this strategy, TCR clonotype can be linked to the most complete method currently available for determining single-cell function. Expense precludes very deep sequencing of single-cell libraries which means it can be difficult to detect specific mutations (or splice variants) in scRNA-seq data. However, because the cDNA libraries are whole-transcript libraries, rhPCR primers can be designed that flank known alterations and adding these primers to the initial PCR step of rhTCRseq should enable very sensitive detection of known mutations and splice variants.
Bulk RNA applications of rhTCRseq
For Patient 8 in the Keskin et al. study13, single TILs were not available for analysis, so a different strategy was adopted to associate TCR clonotypes found in the peripheral blood with those in the tumor. T cells from a post-vaccination peripheral blood sample were stimulated with individual neoantigen peptides used in the vaccine. Single-cell TCR sequencing of these stimulated T cells using rhTCRseq in 96-well plates identified putative peptide-specific clonotypes. For this analysis, single-cell cDNA libraries were made as described in this protocol. Peripheral blood and tumor RNA were then analyzed using the rhTCRseq protocol for bulk RNA. Peptide-specific alpha- and beta-chain sequences were detected in a post-vaccination blood sample and in a post-vaccination tumor sample obtained when the patient relapsed. These sequences were not detected in pre-vaccination blood or tumor samples.
Related to cancer, studies28–30 have investigated how changes in TCR-beta chain diversity and clonality in peripheral blood are associated with response to treatment with immune checkpoint inhibitors. These analyses would have been enhanced by using methods, such as rhTCRseq and others23,31, that add information about TCR-alpha chain usage and provide improved UMI-based frequency measurement. These methods would provide the same advantages to the TCR analysis of bulk tumor RNA samples. Li et al.32 were able to extract beta CDR3 information from bulk RNA-seq data deposited in The Cancer Genome Atlas (TCGA). From the subsequent analysis of 9,142 RNA-seq samples across 29 cancer types, they obtained an average of 65 clonotypes per tumor sample. In comparison, targeted methods like rhTCRseq can identify many more TCR clonotypes in a tumor RNA sample. As reported in Supplementary Table 10 of Keskin et al.13, rhTCRseq identified 1716–2559 unique TCR-alpha clonotypes and 2197–4084 unique TCR-beta clonotypes in two glioblastoma samples. Although interpretation can be confounded by variability in tumor purity, studies33–36 have used targeted TCR sequencing of bulk tumor RNA samples to show that differences in TCR-beta chain diversity are associated with response to immunotherapy. Beausang et al.37 used TCR sequencing of bulk DNA to show that TILs in breast cancer have a higher TCR-beta clonality compared to adjacent normal breast tissue.
Strengths and limitations of rhTCRseq
Many protocols have been devised that employ multiplex PCR with a mixture of TRAV- and TRBV-specific forward primers for targeted TCR amplification from bulk genomic DNA or bulk transcript RNA38–43. This approach has been extended to the determination of paired TCR alpha/beta clonotypes in single cells1–5. These existing methods use nested primers to achieve specific amplification of CDR3 segments. As described here, the major advantage of rhTCRseq is its high level of specificity, which enables specific TCR amplification and addition of barcodes in a single PCR step. After this barcoding, samples can be pooled so there is only one sample for the subsequent steps of library generation. This greatly simplifies the workflow and reduces cost compared to previous multiplex PCR methods for targeted TCR amplification.
For repertoire analysis, rhTCRseq uses transcript RNA as a substrate. This has pluses and minuses compared to using genomic DNA. Because each cell has multiple TCR transcripts compared to just one genomic DNA copy, RNA analysis can be performed with limited amounts of input material. This also means that clonotype frequencies derived from RNA data are skewed by differential TCR expression levels. This emphasis on more highly expressed TCRs can be advantageous for certain analyses, but makes it difficult, if not impossible, to quantify clonal expansions on a cell count basis. The dependence on expression levels also means that direct comparison of RNA- and DNA-based frequency values is problematic.
One limitation of rhTCRseq is that it does not have the throughput of droplet-based methods. To date, though, droplet-based methods can only generate end-counting scRNA-seq data. Thus, rhTCRseq coupled with whole-transcript scRNA-seq balances the moderate throughput of 384 cells per processing run with the sensitive detection of paired complete CDR3 segments and with the information gained from whole-transcript, single-cell analysis. rhTCRseq is well-suited to the analysis of small sorted populations where the economics of droplet-based methods are less favorable. It has the added flexibility of decoupling isolating single cells from library processing, enabling archiving of single cell plates.
Experimental Design
Cell sorting and lysis
For sorting (Steps 1A(i-iii)), we have found that we can dry sort single cells into 384-well plates. This simplifies the sorting process somewhat because it obviates the need to dispense lysis buffer into the wells prior to sorting. Thorough lysis is important for maximizing the yield of cDNA in single-cell protocols. Trombetta et al.15 used a guanidine thiocyanate buffer to ensure complete lysis, but this required a bead cleanup of the RNA to remove the guanidine thiocyanate prior to the RT reaction. Performing this bead cleanup, especially in a 384-well plate, is cumbersome and can lead to loss of RNA. The rhTCRseq protocol achieves robust liberation of cellular RNA by incubating the cells with Proteinase K at 50 °C (Steps 1A(iv-ix)). Inactivation of the Proteinase K is achieved by a combination of heat inactivation at 72 °C and the inclusion of a protease inhibitor in the RT reaction. Thus, there is no need for a post-lysis purification step.
Single-cell cDNA library construction
This protocol generates single-cell cDNA libraries in 96- or 384-well plates using reagents from the NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (New England BioLabs E6421L). Steps 1A(x-xvii) are essentially the same as for Smart-seq215,16: a RT reaction with an oligo dT primer and a template-switching oligo followed by PCR amplification of full-length cDNA using a single primer. In order to make the volumes compatible with a 384-well plate and to reduce cost, reaction volumes have been reduced by a factor of ten compared to those recommended in the kit manual. For 96-well plates, the reduction factor is five. Following PCR, the individual single-cell cDNA libraries are purified using bead cleanup (Steps 1A(xviii-xxv)) and normalized to equal concentrations (Steps 1A(xxvi-xxviii)). The normalized libraries are ready for TCR-specific amplification as described below. The cDNA libraries can also be used for making sequencing libraries to perform whole-transcript scRNA-seq. Library construction can be done using the Nextera XT system15 or the more conventional route of fragmentation followed by ligation of adaptors with Illumina sequences44–46.
TCR-specific amplification of single-cell cDNA libraries
Targeted TCR amplification (Steps 1A(xxix-xxxv)) is performed by first transferring an aliquot of each normalized single-cell cDNA library to a 96- or 384-well plate. Then, a pair of barcode rhPCR primers with the structures P5.IDTxxx.Rd1x and P7.IDTyyy.Rd2x are added to each well. P5 and P7 refer to the Illumina sequences required for attachment to the flow cell; IDTxxx and IDTyyy refer to 8-nucleotide index sequences; and Rd1x and Rd2x refer to the 5’ portions of the Illumina TruSeq Read1 and Read 2 primers. The designation IDTxxx and IDTyyy indicates that the two index sequences added to any particular well are not the same sequence. Using the pipetting scheme outlined in Table 1 enables transferring barcode primers stored in 96-well plates to a 384-well plate such that each well gets a unique dual index. Next, a PCR mix containing rhPCR primers specific for TCR is added to each well. The structures of these primers are Rd1.AVxx, Rd1.BVxx, Rd2.AC, and Rd2.BC. Rd1 and Rd2 refer to the sequences corresponding to the Illumina TruSeq Read1 and Read 2 primers; AVxx refer to sequences specific for all functional TRAV genes; BVxx refer to sequences specific for all functional TRBV genes; and AC and BC refer to TRAC- and TRBC-specific sequences, respectively. In the PCR, the concentrations of the TCR-specific primers are low (50 nM) and the barcode primers are high (2 μM). This is done to favor the formation of full-length P5/P7-containing amplicons, but this strategy is not 100% effective. Therefore, after pooling and purification (Steps 2–16) of the 96 or 384 reactions from the TCR-specific amplification, a second PCR is performed using the generic primers P5 and P7 (Steps 17–18). The products of this second PCR are purified and qualified for sequencing by BioAnalyzer analysis (Steps 19–29). Standard paired-end MiSeq sequencing is performed using the 300-cycle kit and reads of 248 nt read 1, 48 nt read 2, 8 nt index 1, and 8 nt index 2 (Steps 30–35).
Table 1 |.
Scheme for pipetting from four 96-well plates to one 384-well plate.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| B | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| C | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| E | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| F | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| G | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| H | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| I | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| J | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| K | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| L | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| M | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| N | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
| O | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| P | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D | C | D |
The numbers in the first row and the letters in the leftmost column indicate the coordinates of a 384-well plate. The remaining letters A, B, C, and D refer to four 96-well plates. Using an 8-channel pipette, primers in each column of 96-well plate A are pipetted to a column with wells marked A in this diagram of a 384-well plate. This requires 12 transfers. The process is repeated for 96-well plates B, C, and D. The overall operation requires 48 transfers, with 8 wells being pipetted per transfer.
Adaptation of single cell protocol for bulk RNA
For bulk RNA, the TRAC- and TRBC-specific primers are included as part of the RT reaction. To do this, conventional primers (not rhPCR primers) with structure Rd2.UM7.AC and Rd2.UM7.BC are mixed with total RNA, NP-40, and dNTPs, then incubated at 65 °C to relax RNA secondary structure and allow the primers to hybridize to the RNA (Steps 1B(i-v)). Rd2 refers to the sequences corresponding to the Illumina TruSeq Read 2 primer; UM7 is a 7-nucleotide random sequence that is the UMI; and AC and BC refer to TRAC- and TRBC-specific sequences, respectively. A standard RT reaction is performed (Steps 1B(vi-ix)) without a template-switching oligo. Then, an exonuclease reaction is performed (Steps 1B(x-xiii)) to digest the RT primers. This prevents the UMI-containing primers from being incorporated during subsequent PCR steps. TCR-specific PCR amplification (Steps 1B(xiv-xix)) is similar to the single cell protocol except there are no TRAC- and TRBC-specific primers in the reaction. Following pooling and purification of the PCR products (Steps 2–16), a second PCR is performed using the generic primers P5 and P7 (Steps 17–18). After this, the bulk RNA protocol is the same as the single cell protocol (Steps 19–35). For tumor samples, RNA isolated from fresh frozen tissue always works better than formalin-fixed paraffin-embedded (FFPE) material. RNA should have an RNA Integrity Number (RIN) of at least 4.
Data analysis
After preparing the fastq files from the sequencing run and installing the required software (Step 36), a pipeline written in Python and Bash shell scripts takes sequencing data from single cell or bulk RNA sequencing and analyzes it to report information about the TCR alpha and beta clonotypes present in each sample (Steps 37–39). As detailed in Box 1, the pipeline first determines which TCR locus each sequencing read came from and separates the reads by gene (TRA locus or TRB locus). Then the program MiXCR47 aligns reads to reference variants of V, D, J, and C regions and assembles clonotypes. For single cell data, clonotypes are filtered and collapsed based on sequence similarity. For bulk RNA data, clonotypes are filtered and collapsed based on UMIs, first within each well and then between replicates. The single cell outputs (Step 39) are used to determine the paired alpha/beta TCR clonotype of each single cell and to identify any clones in the collection of single cells analyzed. The bulk RNA outputs (Step 39) report the frequencies of alpha and beta clonotypes separately. These frequencies can be used to calculate a variety of parameters to characterize TCR repertoire diversity.
Box 1 |. Steps in the pipeline used to analyze rhTCRseq data.
Validate demultiplexing results of the fastq files from the Illumina MiSeq and count the total number of reads per well for initial quality assessment.
Separate reads into alpha- and beta-chain files by using BLAST48 to align to the chain-specific primers used in the amplification.
Assemble and annotate the reads using MiXCR47 with default parameters for alignment to known V, D and J regions, assembly of each TCR, and identification of the CDR3 region.
Merge TRBV genes that are indistinguishable based on the locations of the primers for those genes.
- Additional steps were performed to adjust for overestimations of clonotypes and UMIs introduced by errors in the PCR or sequencing steps:
- For single cell results, collapse clonotypes if they share identical V- and J-segments and the CDR3 nucleotide sequences have 95% or greater similarity. The clonotype with higher frequency is retained, and the read counts of the subsumed clonotype are merged with the higher frequency clonotype.
- For bulk RNA results, delete UMIs that are supported by only one read, then delete clonotypes that have no remaining UMIs.
- For bulk RNA results, collapse clonotypes to the majority clonotype if they share identical V- and J-segments, the CDR3 nucleotide sequences have 95% or greater similarity, and the CDR3 amino acid sequences have the same length.
- For bulk RNA results, collect clonotypes from all the replicates of a given sample, sort in groups with the same J segment and CDR3 sequence, and collapse to the clonotype within each group that has the highest UMI count.
- For bulk RNA results, remove nonproductive clonotypes.
Controls
Negative controls with no cells or RNA added should be included with each plate run. Typically, positive controls are not used. For sequencing, the spike-in library PhiX Control v3 is included at 5% as a quality control to ensure robust cluster identification.
Materials
T cells from sample of interest for single cell protocol ! CAUTION For work with human subjects, informed consent and appropriate approvals from user’s institution are required. Informed consent was obtained from all study subjects as part of DF/HCC IRB approved protocols.
Total RNA from blood or tumor for bulk RNA protocol ! CAUTION For work with human subjects, informed consent and appropriate approvals from user’s institution are required. Informed consent was obtained from all study subjects as part of DF/HCC IRB approved protocols.
Reagents
NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (New England BioLabs, cat. no. E6421L). This kit contains NEBNext Cell Lysis Buffer (10×), Murine RNase Inhibitor, NEBNext Single Cell RT Primer Mix, NEBNext Single Cell RT Buffer (4×), NEBNext Template Switching Oligo, NEBNext Single Cell RT Enzyme Mix, NEBNext Single Cell cDNA PCR Master Mix (2×), and NEBNext Single Cell cDNA PCR Primer.
Proteinase K (800 units/mL, New England BioLabs, cat. no. P8107S)
Prionex (MilliporeSigma, cat. no. G0411–100ML)
Elastase Inhibitor III (AAPV, MilliporeSigma, cat. no. 324745–5MG)
▲CRITICAL This is the only inhibitor that has been tested in the rhTCRseq protocol. Use and concentration of a different Proteinase K inhibitor would need to be validated.
ProNex Size-Selective Purification System (125 mL, Promega, cat. no. NG2002)
Quant-iT High-Sensitivity dsDNA Assay Kit (Thermo Fisher, cat. no. Q33120)
Water, PCR certified (Teknova, cat. no. W3331)
TE Buffer (10 mM Tris, pH 8.0; 1 mM EDTA, Teknova, cat. no. T0224)
DNA Suspension Buffer (10 mM Tris, pH 8.0; 0.1 mM EDTA, Teknova, cat. no. T0221)
P5.IDTxxx.Rd1x.x1 and P7.IDTyyy.Rd2x.x1 oligonucleotides (Supplementary Table 2, Integrated DNA Technologies, custom product)
▲CRITICAL All oligonucleotides are listed in the 5′ to 3′ direction.
RNase H2 Enzyme Kit (includes RNase H2 Enzyme at 2 units/μL and RNase H2 Dilution Buffer, Integrated DNA Technologies, cat. no. 11–02-12–01)
1 M Tris-HCl, pH 8.4 (Teknova, cat. no. T1084)
1 M KCl (Teknova, cat. no. P0325)
1 M MgCl2 (Teknova, cat. no. M0302)
dNTP Mix, 25 mM each (Thermo Fisher, cat. no. FERR1121)
Rd1.AVxx.x1 and Rd1.BVxx.x1 oligonucleotides (Supplementary Table 1, Integrated DNA Technologies, custom product)
Rd2.AC.x1and Rd2.BC.x1 oligonucleotides (Supplementary Table 1, Integrated DNA Technologies, custom product)
Hot Start Taq DNA Polymerase (New England BioLabs, cat. no. M0495L)
AMPure XP beads (Beckman Coulter, cat. no. A63880)
200 Proof Ethanol (Decon Labs, cat. no. 2716) ! CAUTION Ethanol is flammable, keep away from open flame.
Beads Buffer: 20% polyethylene glycol (PEG 8000), 2.5M sodium chloride (Teknova, cat. no. P4137)
P5 primer, AATGATACGGCGACCACCGAGATCTACAC (Integrated DNA Technologies, custom product)
P7 primer, CAAGCAGAAGACGGCATACGAGAT (Integrated DNA Technologies, custom product)
Q5 Hot Start HiFi PCR Master Mix (New England BioLabs, cat. no. M0543S)
1 N NaOH (MilliporeSigma, cat. no. 109137) ! CAUTION Causes severe skin burns and eye damage.
PhiX spike-in library (Control v3, Illumina, cat. no. FC-110–3001)
MiSeq 300-cycle Reagent Kit v2 (Illumina, cat. no. MS-102–2002)
10% NP-40 (Thermo Fisher, cat. no. 28324)
Rd2.UM7.AC primer, gtgactggagttcagacgtgtgctcttccgatctNHNNNHVTCAGCTGGTACACGGCA (Integrated DNA Technologies, custom product)
Rd2.UM7.BC primer, gtgactggagttcagacgtgtgctcttccgatctNHNNNHVTCTCTGCTTCTGATGGCTCAA (Integrated DNA Technologies, custom product)
50% glycerol (Teknova, cat. no. G1796)
1 M dithiothreitol (Teknova, cat. no. D9750) ! CAUTION Dithiothreitol is harmful if swallowed. It causes skin irritation and serious eye irritation.
RNaseOUT (40 units/μL, Thermo Fisher, cat. no. 10777019)
SuperScript II Reverse Transcriptase (200 units/μL, Thermo Fisher, cat. no. 18064014)
Exonuclease I (20 units/μL, New England BioLabs, cat. no. M0293S)
RNase H2 Dilution Buffer (Integrated DNA Technologies, cat. no. 11–01-02–12)
NEBNext Ultra II FS DNA Library Prep Kit (New England BioLabs, cat. no. E7805L). The single-cell cDNA libraries prepared using the rhTCRseq protocol can be processed to make sequencing libraries using this kit. E6421L for cDNA library construction and E7805L for sequencing library construction can be purchased together as the NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina (New England BioLabs, cat. no. E6420L)
Equipment
Twin.tec PCR plate (384 wells, LoBind, skirted, PCR clean, colorless, Eppendorf, cat. no. 0030129547)
Twin.tec PCR plate (96 wells, LoBind, skirted, PCR clean, colorless, Eppendorf, cat. no. 0030129512)
0.2-mL PCR 8-tube FLEX-FREE strip (attached clear flat caps, natural, USA Scientific, cat. no. 1402–4700)
20-μL 8-channel pipette (Pipet-Lite Multi Pipette L8–20XLS+, Rainin, cat. no. 17013803)
200-μL 8-channel pipette (Pipet-Lite Multi Pipette L8–200XLS+, Rainin, cat. no. 17013805)
2-μL pipette (Pipet-Lite LTS Pipette L-2XLS+, Rainin, cat. no. 17014393)
20-μL pipette (Pipet-Lite LTS Pipette L-20XLS+, Rainin, cat. no. 17014392)
200-μL pipette (Pipet-Lite LTS Pipette L-200XLS+, Rainin, cat. no. 17014391)
1000-μL pipette (Pipet-Lite LTS Pipette L-1000XLS+, Rainin, cat. no. 17014382)
20-μL pipette tips (RT-LTS-A-10μL-/F/L-960/10, Rainin, cat. no. 30389226)
200-μL pipette tips (RT-LTS-A-200μL-/F/L-960/10, Rainin, cat. no. 30389240)
1000-μL pipette tips (RT-LTS-A-1000μL-/F/L-768/8, Rainin, cat. no. 30389213)
MagnaBot 384 magnetic separation stand (Promega, cat. no. V8241)
10x magnetic separation stand for 8-tube strip (10x Genomics, cat. no. 230003)
DynaMag-96 side skirted magnetic separation stand (Thermo Fisher, cat. no. 12027)
MagneSphere magnetic separation stand (12-hole, 1.5 mL vial, Promega, cat. no. Z5342)
CoolRack XT cooling block for 384-well PCR plate (Corning, cat. no. 432055) for keeping plate cool on the deck of the Mantis liquid dispenser. When not in use, block is stored at −20 °C.
Cooling block for 96-well PCR plate (Eppendorf, cat. no. 022510509) for keeping plate cool on the deck of the Mantis liquid dispenser. When not in use, block is stored at −20 °C.
DNA LoBind microcentrifuge tubes (1.5 mL, PCR clean, colorless, Eppendorf, cat. no. 022431021)
LSE vortex mixer (Corning, cat. no. 6775)
3-inch head for vortex mixer (Corning, cat. no. 480100)
Mantis liquid dispenser (Formulatrix, cat. no. MANTV3.2)
1250-μL pipette tips (Sterile Non-Filtered Extra Long Pipet Tip, Thomas Scientific, cat. no. 1158U40). For use as reservoir on Mantis liquid dispenser.
1000-μL pipette (PIPETMAN Classic P1000, Gilson, cat. no. F123602). Required for using the 1250-μL pipette tips.
C1000 Touch Thermal Cycler (with 96 Deep Well Reaction Module, Bio-Rad, cat. no. 1851197)
C1000 TOUCH 384 Well Reaction Module (Bio-Rad, cat. no. 1840138)
PX1 PCR Plate Sealer (Bio-Rad, cat. no. 1814000)
Peelable foil heat seal (Bio-Rad, cat. no. 1814045)
5430 microcentrifuge (with 2-place microplate swing bucket rotor, Eppendorf, cat. no. 022620572)
Synergy H1 hybrid multi-mode microplate reader (BioTek, cat. no. H1MF)
2100 Bioanalyzer (Agilent, cat. no. G2939BA)
High Sensitivity DNA Kit (Agilent, cat. no. 5067–4626)
MiSeq system (Illumina, cat. no. SY-410–1003)
Software
BLAST48 version NCBI-BLAST-2.2.30+ (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.2.30/)
MiXCR47 version 2.1.5 (https://github.com/milaboratory/mixcr/releases/tag/v2.1.5)
Java JDK version 8 (https://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html)
GNU parallel version 20180722 (http://git.savannah.gnu.org/cgit/parallel.git/) [Tange, O. GNU Parallel 2018 at <https://zenodo.org/record/1146014#.W_1SJhNKjmV>]
Scripts and reference files from the Github repository at https://github.com/julietforman/rhTCRseq
Reagent setup
25 mM AAPV Proteinase K inhibitor
Dissolve 5 mg Elastase Inhibitor III in 400 μL DMSO, dispense 20 μL aliquots, and store at −80 °C for up to six months.
P5.IDTxxx.Rd1x.x1 / P7.IDTyyy.Rd2x.x1 barcode primer plates
There are 768 barcode primers in Supplementary Table 2. These were obtained in eight 96-well plates with each primer at a concentration of 200 μM. As shown in Supplementary Table 2, these plates are designated: P5.1, P5.2, P5.3, P5.4, P7.1, P7.2, P7.3, and P7.4. The table includes the well position of each primer and its corresponding barcode, with the barcodes designated IDT001-IDT384. The 8-nucleotide barcode is underlined in each of the primer sequences. Four Quadrant Plates at concentration 6 μM each primer were made by mixing 3 μL 200 μM P5 barcode primer, 3 μL 200 μM P7 barcode primer, and 94 μL TE Buffer following this scheme:
Quadrant Plate A: P5.1 + P7.2
Quadrant Plate B: P5.2 + P7.3
Quadrant Plate C: P5.3 + P7.4
Quadrant Plate D: P5.4 + P7.1
Table 1 shows the pipetting pattern used to transfer primers from the Quadrant Plates to a 384-well plate. The last tab in Supplementary Table 2 is a MiSeq sample sheet showing the correspondence of index sequences to individual samples. The Sample_ID column in the sample sheet shows the well position in the 384-well plate. These barcode primer plates are stored at −20 °C for up to one year.
Rd1.AV.x1 / Rd1.BV.x1 primer mix
Combine 69 rhPCR primers at a concentration of 5 μM each by mixing 5 μL 500 μM each primer with 155 μL TE Buffer. These primers are specific for the V segments of the human alpha and beta TCR genes and are designed to amplify all productive alpha and beta alleles. The sequences are in Supplementary Table 1. Store at −20 °C for up to one year.
Rd2.AC.x1 / Rd2.BC.x1 primer mix
Combine the two rhPCR primers at a concentration of 5 μM each prepared by mixing 5 μL 500 μM each primer with 490 μL TE Buffer. These primers are specific for the C segments of the human alpha and beta TCR genes. The sequences are in Supplementary Table 1. Store at −20 °C for up to one year.
Ethanol, 80% (vol/vol)
Prepare 80% (vol/vol) ethanol just before use by mixing 4 mL 200 proof ethanol with 1 mL water.
5× RT Buffer (75 mM Tris-HCl, pH 8.4; 375 mM KCl; 50 mM MgCl2; 25% glycerol)
Mix 15 μL1 M Tris-HCl, pH 8.4, 75 μL 1 M KCl, 10 μL 1 M MgCl2, 100 μL 50% glycerol. Store at room temperature for up to one year.
NaOH, 0.2 N
Prepare just before use by mixing 4 μL 1 N NaOH with 16 μL water.
Procedure
CRITICAL: The workflow of this protocol is diagrammed in Fig. 2.
Fig. 2 |. Schema of the single cell and bulk RNA rhTCRseq workflows.
Diagram outlining the parts of the protocol that are described in detail in steps 1–35.
-
1
Choose Option A for processing single cells and Option B for processing bulk RNA.
Option A: rhTCRseq for single cells
Single cell sorting ● Timing 30–60 min sorter set-up, 10–45 min per plate
-
i
Dry sort single cells into a 96- or 384-well PCR plate. In order to have a negative control, do not sort cells into 2–4 wells.
▲CRITICAL Ensure the sorter is properly aligned so that each cell is deposited to the center of its well.
? TROUBLESHOOTING
-
ii
Immediately centrifuge the plate at 800× g for 1 min at 4 °C and place on dry ice.
-
iii
Store the plate at −80 °C.
■PAUSE POINT Single-cell plates can be stored for at least one year at −80 °C.
cDNA library construction ● Timing 8 h
-
iv
Dilute Proteinase K to 80 units/mL by mixing 5 μL 800 units/mL Proteinase K with 45 μL Prionex and keep on ice.
-
vPrepare the following lysis mix and keep on ice:
Component Volume for 96-well plate with overage (μL) Volume for 384-well plate with overage (μL) Final concentration NEBNext Cell Lysis Buffer (10×) 12 20.8 0.5× Murine RNase Inhibitor 6 10.4 - Proteinase K (80 units/mL) 24 41.5 8 units/mL NEBNext Single Cell RT Primer Mix 24 41.5 - Water 174 301 - Total volume 240 415.2 -
vi
Remove single-cell plate from −80 °C freezer (Step 1Aiii), place on ice for 2 min, centrifuge briefly, and place plate in cooling block on the deck of the Mantis.
-
vii
Use Mantis to dispense 1 μL of lysis mix to each of the wells if using a 384-well plate, or 2 μL per well for a 96-well plate.
-
viii
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
ix
Centrifuge briefly, then transfer plate to thermal cycler and run the protocol: 50 °C for 30 min, 72 °C for 5 min, hold at 4 °C.
-
xPrepare the following reverse transcriptase (RT) mix and keep on ice:
Component Volume for 96-well plate with overage (μL) Volume for 384-well plate with overage (μL) Final concentration in RT reaction NEBNext Single Cell RT Buffer (4×) 120 207.5 1× AAPV Proteinase K inhibitor (25 mM) 9.6 16.6 0.5 mM NEBNext Template Switching Oligo 24 41.5 - NEBNext Single Cell RT Enzyme Mix 48 83 - Water 38.4 66.4 - Total volume 240 415 -
xi
With the plate in cooling block on the deck of the Mantis, use Mantis to dispense 1 μL of RT mix to each of the wells if using a 384-well plate, or 2 μL per well for a 96-well plate.
-
xii
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
xiii
Vortex and centrifuge briefly, then transfer plate to thermal cycler and run the protocol: 42 °C for 90 min, 70 °C for 10 min, hold at 4 °C.
-
xivPrepare the following PCR mix and keep on ice:
Component Volume for 96-well plate with overage (μL) Volume for 384-well plate with overage (μL) Final concentration in PCR reaction NEBNext Single Cell cDNA PCR Master Mix (2×) 1200 2065 1× NEBNext Single Cell cDNA PCR Primer 48 82.6 - Water 672 1156.4 - Total volume 1920 3304 -
xv
With the sample plate in cooling block on the deck of the Mantis, use Mantis to dispense 8 μL of PCR mix to each of the wells if using a 384-well plate, or 16 μL per well for a 96-well plate, using a 1250-μL pipette tip.
-
xvi
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
xviiVortex and centrifuge briefly. Transfer plate to thermal cycler and run the following PCR protocol:
Cycle number Denature Anneal Extend 1 98 °C, 45 s 2–23 98 °C, 10 s 62 °C, 15 s 72 °C, 3 min 24 72 °C, 5 min -
xviii
Warm ProNex beads to room temperature.
-
xix
With the sample plate at room temperature, use Mantis to dispense 11 μL of ProNex beads to each of the wells if using a 384-well plate, or 21 μL per well for a 96-well plate, using a 1250-μL pipette tip.
-
xx
Vortex briefly and incubate at room temperature for 10 min.
-
xxi
Place the plate on a magnetic stand, wait until solution clears (approximately 2 min for beads to collect), then discard supernatants using 8-channel pipette.
-
xxii
With plate remaining in the magnetic stand, wash the beads by adding 30 μL of Promega Wash Buffer to each of the wells if using a 384-well plate, or 100 μL per well for a 96-well plate, using an 8-channel pipette, waiting 30 s, and discarding the supernatants.
-
xxiii
Repeat Step 1Axxii one more time. Remove as much residual Wash Buffer as possible.
-
xxiv
After air drying the beads for 10 min, use Mantis to add 17 μL Promega Elution Buffer to each well, using a 1250-μL pipette tip.
-
xxv
Vortex briefly, incubate at room temperature for 5 min, place the plate on the magnetic stand, wait for solution to clear (approximately 2 min for beads to collect), then transfer 15 μL supernatant from each well to a fresh plate.
■PAUSE POINT Amplified libraries can be stored for at least one year at −20 °C.
-
xxvi
Determine the concentrations of the cDNA libraries using 1-μL aliquots and the Quant-iT High-Sensitivity dsDNA Assay Kit. Measure fluorescence on the Synergy H1MF microplate reader.
-
xxvii
Normalize DNA concentration to 0.2 ng/μL by combining 1.5 μL of each library with the appropriate volume of TE in wells of a fresh plate. If concentration is less than 0.2 ng/μL, combine 1.5 μL of library with 0.6 μL TE. Dispense variable volumes of TE using the Mantis.
-
xxviii
Store both the original and normalized cDNA library plates at −20 °C.
■PAUSE POINT Original and normalized cDNA library plates can be stored for at least one year at −20 °C.
TCR-specific amplification ● Timing 3 h
-
xxix
Transfer 2 μL (400 pg) of each normalized cDNA library to a fresh 96- or 384-well plate on ice. CRITICAL STEP: cDNA libraries prepared by another method can alternatively be used at this point as long as the other method uses a template-switch reverse transcriptase step and full-length cDNA amplification. For example, for Patient 7 in Keskin et al.13, cDNA libraries prepared using a modified version of Smart-seq215 were used as starting material for Step 1Axxix.
-
xxx
From the four Quadrant Plates A-D of barcode primers, use an 8-channel pipette to add 2 μL 6 μM each P5.IDTxxx.Rd1x.x1 / P7.IDTyyy.Rd2x.x1 to each of the wells following the pipetting scheme in Table 1. CRITICAL STEP: For a 96-well plate, only one of the Quadrant Plates is used.
-
xxxi
Dilute RNase H2 to 20 mU/μL by mixing 1 μL 2 units/μL RNase H2 Enzyme with 99 μL RNase H2 Dilution Buffer and keep on ice.
-
xxxiiPrepare the following PCR mix (PCR1) and keep on ice:
Component Volume for 96-well plate with overage (μL) Volume for 384-well plate with overage (μL) Final concentration in PCR reaction Tris-HCl, pH 8.4 (1 M) 10.8 36.9 15 mM KCl (1 M) 18 61.5 25 mM MgCl2 (1 M) 2.9 9.84 4 mM dNTPs (25 mM each) 11.5 39.36 0.4 mM each Rd1.AV.x1 / Rd1.BV.x1 primer mix (5 μM each) 7.2 24.6 50 nM each Rd2.AC.x1 / Rd2.BC.x1 primer mix (5 μM each) 7.2 24.6 50 nM each RNase H2 (20 mU/μL) 18 61.5 0.5 mU/μL Hot Start Taq DNA Polymerase (5 units/μL) 28.8 98.4 0.2 units/μL Water 135.6 463.3 - Total volume 240 820 -
xxxiii
With sample plate in cooling block on the deck of the Mantis, use Mantis to dispense 2 μL PCR mix to each of the wells.
-
xxxiv
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
xxxvTransfer plate to thermal cycler and run the following protocol:
Cycle number Denature Anneal Extend 1 95 °C, 5 min 2–19 96 °C, 20 s 60 °C, 6 min
Option B: rhTCRseq for bulk RNA
CRITICAL: For bulk RNA rhTCRseq, we recommend using 1–50 ng total RNA per reaction. For any particular study, RNA samples should be diluted to the same concentration. RNA can be isolated by any of the standard methods for purifying total RNA. For each sample, we recommend running four to eight replicates. The protocol is written for running 96 reactions. This could be 24 samples with 4 replicates, 12 samples with 8 replicates, or other combinations. Twelve samples with 8 replicates will be used as an example here. One of the samples should be a negative control with no RNA added.
cDNA synthesis ● Timing 3 h
-
iPrepare the following mix and keep on ice:
Component Volume per tube (μL) Volume for 12 tubes with overage (μL) Final concentration in RT reaction NP-40 (10%) 1 15 0.25% dNTPs (25 mM each) 0.8 12 0.5 mM Rd2.UM7.AC primer (10 μM) 0.84 12.6 210 nM Rd2.UM7.BC primer (10 μM) 0.28 4.2 70 nM Water 7.08 106.2 - Total volume 10 150 -
ii
Dispense 10 μL of this mix to each of 12 1.5-mL microcentrifuge tubes.
-
iii
Add 10 μL 1–50 ng/μL RNA to each tube.
-
iv
From each tube, transfer 2 μL to each of 8 wells in a 96-well PCR plate.
-
v
Transfer plate to thermal cycler and run the protocol: 65 °C for 5 min, hold at 4 °C.
-
viPrepare the following RT mix and keep on ice:
Component Volume per reaction (μL) Volume for 96 reactions with overage (μL) Final concentration in RT reaction RT Buffer (5×) 0.8 96 1× Dithiothreitol (1 M) 0.04 4.8 10 mM RNaseOUT (40 units/μL) 0.08 9.6 0.8 units/μL Superscript II Reverse Transcriptase (200 units/μL) 0.02 2.4 1 units/μL Water 1.06 127.2 - Total volume 2 240 -
vii
With a 96-well sample plate on the cooling block on the deck of the Mantis, use Mantis to dispense 2 μL RT mix to each of the 96 wells.
-
viii
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
ix
Transfer plate to thermal cycler and run the protocol: 42 °C for 90 min, 85 °C for 5 min, hold at 4°C.
-
xPrepare diluted exonuclease as follows and keep on ice:
Component Volume per reaction (μL) Volume for 96 reactions with overage (μL) Final concentration in exonuclease reaction Exonuclease I (20 units/μL) 0.025 3 0.083 units/μL RNase H2 Dilution Buffer 1.975 237 - Total volume 2 240 -
xi
With the 96-well sample plate on the cooling block on the deck of the Mantis, use Mantis to dispense 2 μL diluted exonuclease to each of the 96 wells.
-
xii
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
xiii
Transfer the plate to the thermal cycler and run the protocol: 37 °C for 15 min, 85 °C for 15 min, hold at 4 °C.
TCR-specific amplification ● Timing 3 h
-
xiv
From one of the four Quadrant Plates A-D of barcode primers, use an 8-channel pipette to add 4 μL 6 μM each P5.IDTxxx.Rd1x.x1 / P7.IDTyyy.Rd2x.x1 to each of the 96 wells.
-
xv
Dilute RNase H2 to 20 mU/μL by mixing 1 μL 2 units/μL RNase H2 Enzyme with 99 μL RNase H2 Dilution Buffer and keep on ice.
-
xviPrepare the following PCR mix (PCR1) and keep on ice:
Component Volume per reaction (μL) Volume for 96 reactions with overage (μL) Final concentration in PCR reaction Tris-HCl, pH 8.4 (1 M) 0.18 21.6 15 mM KCl (1 M) 0.3 36 25 mM MgCl2 (1 M) 0.048 5.76 4 mM dNTPs (25 mM each) 0.192 23.04 0.4 mM each Rd1.AV.x1 / Rd1.BV.x1 primer mix (5 μM each) 0.12 14.4 50 nM each RNase H2 (20 mU/μL) 0.3 36 0.5 mU/μL Hot Start Taq DNA Polymerase (5 units/μL) 0.48 57.6 0.2 units/μL Water 0.38 45.6 - Total volume 2 240 -
xvii
With the 96-well sample plate on the cooling block on the deck of the Mantis, use Mantis to dispense 2 μL PCR mix to each of the 96 wells.
-
xviii
Cover the plate with foil and heat seal using plate sealer at 168 °C for 3 s.
-
xixTransfer plate to thermal cycler and run the protocol:
Cycle number Denature Anneal Extend 1 95 °C, 5 min 2–21 96 °C, 20 s 60 °C, 6 min
Pooling, purification, and final PCR. ● Timing 3.5 h
-
2
Pool all the reactions by mixing 2 μL of each sample. Pooling is done by using an 8-channel pipette to transfer samples from all wells of each column repeatedly into the same wells of one column of a new 96-well PCR plate. This requires 12 transfers for a 96-well plate and 48 transfers for a 384-well plate. The eight pooled samples are then combined by transferring to a 1.5-mL microcentrifuge tube to generate a 192-μL pooled sample for a 96-well plate and a 768-μL pooled sample for a 384-well plate. Store remainder of unpooled samples at −20 °C for up to six months.
-
3
Warm AMPure XP beads to room temperature.
-
4To the pooled sample, add:
Component Volume for pool from 96-well plate (μL) Volume for pool from 384-well plate (μL) AMPure XP beads 19.2 76.8 Beads Buffer 96 384 -
5
Pipet up and down, then incubate at room temperature for 5 min.
-
6
Place the tube on the magnetic stand and wait 5 min or until solution clears for beads to collect.
-
7Transfer supernatant to fresh tube and add:
Component Volume for pool from 96-well plate (μL) Volume for pool from 384-well plate (μL) AMPure XP beads 6.4 25.6 Beads Buffer 32 128 -
8
Pipet up and down, then incubate at room temperature for 5 min.
-
9
Place the tube on the magnetic stand, wait until solution clears (approximately 5 min for beads to collect), and discard supernatant.
-
10
With tube remaining on the magnetic stand, wash the beads by adding 1 mL freshly prepared 80% ethanol, waiting 30 s, and discarding the supernatant.
-
11
Repeat Step 10 one more time. Remove as much residual 80% ethanol as possible.
-
12
After air drying the beads for 5 min, add 21 μL DNA Suspension Buffer.
-
13
Pipet up and down, place the tube on the magnetic stand, wait 2 min for beads to collect, then transfer 20 μL supernatant to one of the unused wells in the 96-well PCR plate used to pool samples (Step 2).
-
14
Add 16 μL AMPure XP beads.
-
15
Repeat Steps 8–13 using the 96-well magnetic stand and 100 μL 80% ethanol for Steps 10 and 11, then transferring 20 μL supernatant to another unused well in the 96-well PCR plate.
-
16
In an adjacent unused well in the 96-well PCR plate, prepare 1:10 dilution by mixing 2 μL purified pooled sample and 18 μL DNA Suspension Buffer.
■PAUSE POINT Samples can be stored for up to one week at −20 °C.
-
17In 8-tube PCR strip, prepare two PCR reactions (PCR2) on ice. Each reaction contains:
Component Volume per sample (μL) Final concentration in PCR reaction Purified pooled sample from Step 15 (original) or Step 16 (1:10 dilution) 5 - P5 primer (10 μM) 2.5 0.5 μM P7 primer (10 μM) 2.5 0.5 μM Q5 Hot Start HiFi PCR Master Mix (2×) 25 1× Water 15 - Total volume 50 -
18Transfer strip to thermal cycler and run one of these protocols:
- Protocol for single cell procedure (Option A):
Cycle number Denature Anneal Extend 1 98 °C, 30 s 2–13 98 °C, 10 s 62 °C, 2 min 14 75 °C, 2 min - Protocol for bulk RNA procedure (Option B):
Cycle number Denature Anneal Extend 1 98 °C, 30 s 2–18 98 °C, 10 s 62 °C, 2 min 19 75 °C, 2 min
-
19
Warm ProNex beads to room temperature.
-
20
To each of the two PCR samples from Step 18, add 55 μL ProNex beads.
-
21
Pipet up and down, then incubate at room temperature for 10 min.
-
22
Place the strip on the 10x magnetic stand in the High orientation, wait until solution clears (approximately 2 min for beads to collect), then transfer supernatants to two unused tubes in the strip.
-
23
Add 15 μL ProNex beads to each, pipet up and down, and incubate at room temperature for 10 min.
-
24
Place the strip on the 10x magnetic stand in the High orientation, wait until solution clears (approximately 2 min for beads to collect), then discard supernatants.
-
25
With strip remaining in the magnetic stand, wash the beads by adding 100 μL Promega Wash Buffer per well, waiting 30 s, and discarding the supernatants.
-
26
Repeat wash step 25 one time. Remove as much residual Wash Buffer as possible.
-
27
After air drying the beads for 10 min, add 20 μL Promega Elution Buffer to each sample.
-
28
Pipet up and down, place in the 10x magnetic stand in the Low orientation, wait until solution clears (approximately 2 min for beads to collect), then transfer the two supernatants to unused wells in the 96-well plate used to store the samples after purification of the pooled TCR-specific PCR (Step16).
-
29
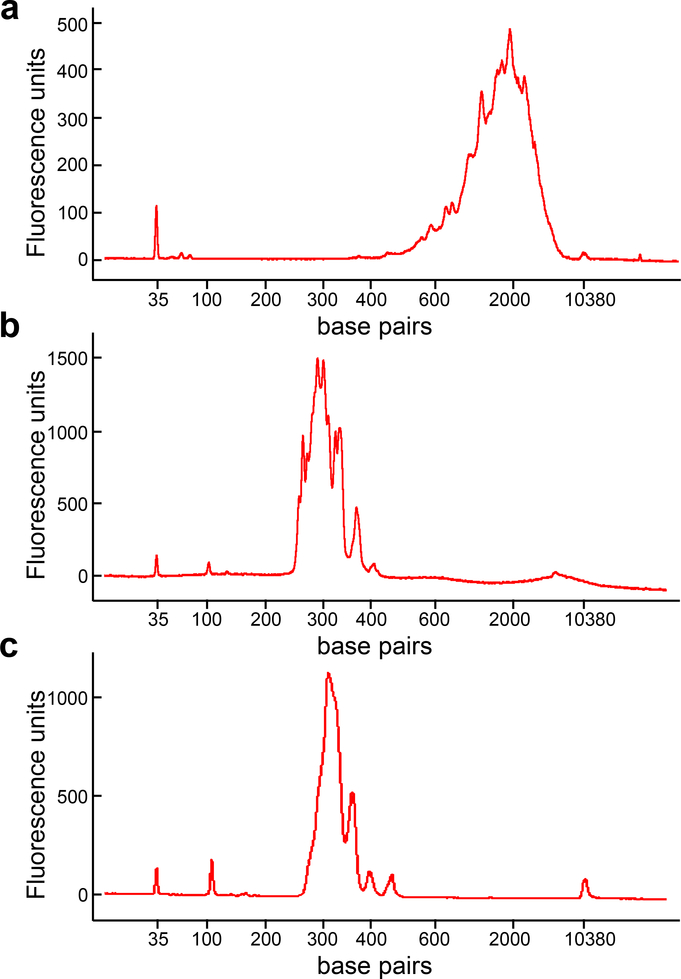
Use 1 μL of each of the two final PCR products from Step 28 to measure the fragment size distribution and estimate concentration using the 2100 BioAnalyzer and High Sensitivity DNA Kit. Fragment size should be a set of peaks predominantly in the range 260–400 bp (see Fig. 3b,c). Concentration is determined setting the size range to 200–1000 bp. Of the two libraries, the one with the flattest baseline is used for sequencing.
Fig. 3 |. Expected results for library constructions using the rhTCRseq protocol for single cells.
a, Bioanalyzer trace for single-cell, full-length cDNA library from Step 1Axxv. The distribution of fragments looks very similar to that reported in Figure 3 of Trombetta et al.15 for a Smart-Seq2 single-cell cDNA library. b,c, Bioanalyzer trace for final rhTCRseq libraries prepared from 384 single cells (b) and bulk RNA (c) from Step 29.
■PAUSE POINT Amplified libraries can be stored for at least one year at −20 °C.
Sequencing ● Timing 28 h
-
30
Perform sequencing using the 300-cycle Reagent Kit v2 on the Illumina MiSeq according to the manufacturer’s protocol. As part of the set-up, upload a sample sheet like the one found in the last tab in Supplementary Table 2 to the instrument. This sample sheet specifies 248 nt read 1, 48 nt read 2, 8 nt index 1, and 8 nt index 2. We have found that the sequencing depth using the MiSeq has been adequate for analyzing 384 single cells or 96 bulk RNA reactions at 50 ng RNA per reaction.
? TROUBLESHOOTING
-
31
Dilute the selected (see Step 29) final library from Step 28 to 4 nM using water.
-
32
Mix 4.5 μL 4 nM library, 0.5 μL PhiX spike-in, and 5 μL freshly prepared 0.2 N NaOH, then incubate at room temperature for 5 min.
-
33
Add 990 μL Hyb Buffer (from MiSeq kit), mix by inversion, and place on ice. The library concentration is now 20 pM.
-
34
To prepare final loading library at 8 pM, mix 400 μL 20 pM library with 600 μL Hyb Buffer.
-
35
Load 600 μL 8 pM library into MiSeq cartridge then follow the Illumina instructions to run the sequencer.
Data analysis ● Timing 8 h
CRITICAL: The required input for the rhTCRseq pipeline includes the sequencing reads in fastq.gz files separated by well, with four files per well. Each filename for each well should begin with the fastq_basename prefix for that well, and the four filenames should end in _L001_R1_001, _L001_R2_001, _L001_I1_001, and _L001_I2_001. Also required is the SampleSheet.csv file for the run.
-
36
Install MiXCR-2.1.5, NCBI Blast, Java, and GNU Parallel.
-
37Set up directory structure as follows:
-
choose a ROOT_DIR in which all requirements for the pipeline will go
-
create a directory <ROOT_DIR>/scripts (add to this folder all of the files from the Github repository folder rhTCRseq/scripts)
-
create a directory <ROOT_DIR>/data
-
create a directory <ROOT_DIR>/out
-
create a directory <ROOT_DIR>/blast_database (add to this folder all of the blast database files from the Github repository folder rhTCRseq/blast_database)
-
-
38
Prepare to analyze a particular run:
open the config file <ROOT_DIR>/scripts/config.py and edit the variables in the file to match your run
create a directory in <ROOT_DIR>/data/ with the same name as RUN_NAME in config.py (place the fastq.gz files from the sequencing run in this folder)
create a directory in <ROOT_DIR>/out/ with the same name as RUN_NAME in config.py (place SampleSheet.csv for the run into this folder)
After these steps are complete, the directory structure should look like this:
ROOT_DIR blast_database TRV_primer.fasta TRV_primer.fasta.nhr TRV_primer.fasta.nin TRV_primer.fasta.nog TRV_primer.fasta.nsd TRV_primer.fasta.nsi TRV_primer.fasta.nsq TRC_primer.fasta TRC_primer.fasta.nhr TRC_primer.fasta.nin TRC_primer.fasta.nog TRC_primer.fasta.nsd TRC_primer.fasta.nsi TRC_primer.fasta.nsq target_gene_primer_forward.fasta target_gene_primer_forward.fasta.nhr target_gene_primer_forward.fasta.nin target_gene_primer_forward.fasta.nog target_gene_primer_forward.fasta.nsd target_gene_primer_forward.fasta.nsi target_gene_primer_forward.fasta.nsq target_gene_primer_reverse.fasta target_gene_primer_reverse.fasta.nhr target_gene_primer_reverse.fasta.nin target_gene_primer_reverse.fasta.nog target_gene_primer_reverse.fasta.nsd target_gene_primer_reverse.fasta.nsi target_gene_primer_reverse.fasta.nsq target_gene_reverse.fasta target_gene_reverse.fasta.nhr target_gene_reverse.fasta.nin target_gene_reverse.fasta.nog target_gene_reverse.fasta.nsd target_gene_reverse.fasta.nsi target_gene_reverse.fasta.nsq target_gene_forward.fasta target_gene_forward.fasta.nhr target_gene_forward.fasta.nin target_gene_forward.fasta.nog target_gene_forward.fasta.nsd target_gene_forward.fasta.nsi target_gene_forward.fasta.nsq data <RUN_NAME> <fastq.gz files> out <RUN_NAME> SampleSheet.csv scripts blast.sh count_umi.py separate_fastq.py collapse_rules.txt parse_blast_results.py analyze_tcr.py compare_clonotype.py merge_TRBV.py mixcr.sh get_parallel_range.py print_description.py config.py run_pipeline.sh make_index_list.py
-
6
Run the pipeline:
cd <ROOT_DIR>/scripts ./run_pipeline.sh
The steps in the pipeline are detailed in Box 1. The output files are shown in Table 2 for the single cell protocol and in Table 3 for the bulk RNA protocol.
Table 2 |.
Data analysis output files for single cell protocol
| Results folder | Content |
|---|---|
| clone_stat_all.txt | The final number of clonotypes and the percentage collapsed for each locus in each well |
| collapse_stat_all.txt | Information about clonotypes that were collapsed |
| mixcr_clonotype_per_well.list | Information about the top clonotypes in each well |
| mixcr_clonotype_recurrence.list | Information about which clonotypes are found in which wells |
| mixcr_clonotype_TRA_across_well.table | With one row per well, records the clone fraction of the top TRA clonotype in that well and its occurrence in the other wells |
| mixcr_clonotype_TRB_across_well.table | With one row per well, records the clone fraction of the top TRB clonotype in that well and its occurrence in the other wells |
Table 3 |.
Data analysis output files for bulk RNA protocol
| Results folder | Content |
|---|---|
| [well]_clonotype_count_TRA.csv | For each well, reports all unique TRA clonotypes with V identity, J identity, CDR3 nucleic acid sequence, CDR3 amino acid sequence, read count, and UMI count |
| [well]_clonotype_count_TRB.csv | For each well, reports all unique TRB clonotypes with V identity, J identity, CDR3 nucleic acid sequence, CDR3 amino acid sequence, read count, and UMI count |
| [sample]_clones_count.csv | Read counts for combined replicates |
| [sample]_clones_umi_count.csv | UMI counts for combined replicates |
| [sample]_observation_across_replicates.csv | Number of replicates in which each clonotype appears |
| [sample]_clonotype_count.csv | Number of clonotypes with each V gene for combined replicates |
| [sample]_clonotype_count.png | Bar plot of clonotypes per V gene for combined replicates |
Troubleshooting
Troubleshooting advice can be found in Table 4.
Table 4 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 1Ai | >25% of cells have no productive clonotype | Misalignment of the sorter so that cells are not being sorted to the center of the wells | Ensure the sorter is properly aligned and that the plate is put squarely in place. Alignment can be checked using fluorescent beads or by placing parafilm across a plate and seeing that sorted droplets are in the center of each well. Rodrigues and Monard49 describe a method using microspheres that can be used to validate sorter alignment. |
| 1Ai | >25% of cells have no productive clonotype plus >25% have alpha only or beta only | Dormant or dying cells | If thawing frozen cells, let cells recover longer before sorting. Be sure to include a viability marker in the sorting. |
| 1A(vii-xxxiv) | >10% of cells have multiple clonotypes | Cross-well contamination prior to pooling | Scrupulous attention to detail during pipetting. A particularly vulnerable point is the washing of the beads during purification of the cDNA libraries. Cross-contamination of barcode primer plates will also produce multiple clonotypes. In this case, the contaminated P5.IDTxxx.Rd1x.x1 / P7.IDTyyy.Rd2x.x1 barcode primer plate needs to be re-made from stocks. |
| 1Biii or 30 | For the bulk RNA protocol, a low number of clonotypes are detected. A possible cutoff is <500 clonotypes (alpha + beta) detected per sample. | This could indicate the fraction of T cells is low in the starting material used to prepare RNA | The read depth can be increased by using a platform other than MiSeq or the amount of input RNA can be increased. |
| This could be due to poor RNA quality | Re-extract RNA to obtain a sample with a RIN of at least 4 |
Timing
Sorting for single cell protocol
Steps 1Ai-1Aiii, single cell sorting: 40 min to 2 h.
Cells can be sorted when the cells are ready and sorting time is available. Plates with sorted cells can be stored at −80 °C for at least one year and the rhTCRseq protocol can be commenced at Step 1Aiv whenever it is convenient.
Day 1
Steps 1Aiv-ix, lysis mix preparation and cell lysis: 50 min
Steps 1Bi-v, RNA sample preparation and primer annealing: 15 min
Steps 1Ax-xiii or 1Bvi-ix, RT mix preparation and RT reaction: 2 h
Steps 1Axiv-xvii, cDNA PCR mix preparation, Mantis dispense, and cDNA PCR: 2 h 30 min
Steps 1Axviii-xxv, cDNA PCR product purification: 1 h
Steps 1Axxvi-xxviii, cDNA quantification and normalization: 1 h 30 min
Steps 1Bx-xiii, Exonuclease I treatment: 45 min
Steps 1Bxiv-xix, PCR1 mix preparation and PCR1: 3 h
Day 2
Steps 1Axxix-xxxv, PCR1 mix preparation and PCR1: 3 h
Steps 2–16, PCR1 pooling and purification: 1 h (Day 1 for Option B)
Steps 17–18, PCR2 amplification: 45 min (started on Day 1 for Option B)
Steps 19–28, PCR2 purification: 45 min
Step 29, library quality control: 1 h
Day 3
Steps 30–35, sequencing: 28 h (Day 2 for Option B)
Day 4
Steps 36–39, data processing: 8 h (Day 3 for Option B)
Anticipated results
Fig. 3a shows the Bioanalyzer trace for the cDNA library from one cell. We have observed the average concentration per run of single-cell cDNA libraries to be in the range 0.5 to 6.1 ng/μL. Fig. 3b shows the expected Bioanalyzer trace of the final rhTCRseq library from a pool of 384 single cells. For single-cell TCR sequencing, the range of average reads per cell has been 5760–13919 reads for TRA and 6767–17619 reads for TRB. Fig 3c shows the expected Bioanalyzer trace of the final rhTCRseq library for the bulk RNA protocol. For both single cells and bulk RNA, predominantly fragments in the range 260–400 nt should be observed. The difference is that the pattern for single-cell libraries tends to be spikier than for bulk RNA libraries.
For single cells, clonotypes are assembled based on the CDR3 sequence results in output file mixcr_clonotype_per_well.list (Table 2) and are classified as containing: (1) one alpha and one beta; (2) two alpha and one beta; (3) one alpha and two beta; (4) one or two alpha only; (5) one or two beta only (6) multiple alpha and beta; and (7) lacking a productive alpha and beta. Table 5 shows the distribution of these classes for the single-cell rhTCRseq experiment described in the Supplementary Note (384 single cells). These results indicate a success rate of 90% (344/384) in obtaining useful paired alpha/beta information (categories 1–3). The success rate depends predominantly on the quality of the input cells and the precision of sorting. For high quality cells, we have observed success rates of 65–90% of single cells analyzed per plate. For bulk RNA results, the number of unique clonotypes observed depends entirely on the amount of RNA analyzed and the percent T cells in the original sample.
Table 5 |.
Enumeration of TCR-alpha and -beta TCR clonotypes detected in 384 putative tumor-specific T cells.
| 1 Alpha + 1 Beta | 2 Alpha + 1 Beta | 1 Alpha + 2 Beta | 1 or 2 Alpha Only | 1 or 2 Beta Only | Multiple Alpha and Beta | No Alpha and Beta | |
|---|---|---|---|---|---|---|---|
| Number of cells | 289 | 49 | 6 | 5 | 17 | 1 | 17 |
| Total cells | 344 | 40 | |||||
In each single cell, and separately for alpha and beta, detected clonotypes are ranked by number of CDR3 reads. Nonproductive CDR3 sequences with stop codon or frame shift are ignored. If less than 100 CDR3 reads are counted for the predominant sequence, then no alpha/beta clonotype is reported for that cell. If the CDR3 reads for the predominant sequence are 20× or greater than the second sequence, one alpha/beta clonotype is reported for that cell. If the sum of the CDR3 reads for the first and second sequences is 20× or greater than the third sequence, two alpha/beta clonotypes are reported for that cell. Any remaining cells are reported as multiple. If a cell has two alpha clonotypes and two beta clonotypes, it is also reported as multiple.
Code availability statement
Code and reference files used for data analysis are publicly available from the Github repository at https://github.com/julietforman/rhTCRseq
Data availability statement
All TCR clonotype data for the results presented in Supplementary Figures 1, 2, and 4 are provided in Supplementary Tables 3 and 5. All other data are available from the corresponding author upon reasonable request. Example data used for data analysis are publicly available from the Github repository at https://github.com/julietforman/rhTCRseq
Supplementary Material
Supplementary Table 1: TCR Primers, Excel file
Supplementary Table 2: Barcode Primers, Excel file
Supplementary Table 4: Number of unique TCR clonotypes detected in five PBMC RNA samples from healthy adult volunteers, in Supplementary Note
Supplementary Table 5: Single Cell Clonotypes, Excel file
Supplementary Table 3: Bulk RNA Clonotypes, Excel file
Supplementary Fig. 2: Reproducibility of the bulk RNA rhTCRseq protocol
Supplementary Fig. 3: Gating strategy for isolating tumor-specific T cells
Supplementary Fig. 4: Distribution of clones identified by single-cell TCR sequencing of tumor-stimulated T cells
Supplementary Fig. 1: TCR repertoire analysis for five individuals
Acknowledgements
We thank Wandi Zhang for preparation of total RNA from PBMC samples. S.L., J.F., D.B.K., S.A.S., and K.J.L are supported by NIH/NCI U24 CA224331. G.O. receives generous support from the Mathers Family Foundation. J.F. is a participant in the Broad Cancer Genomics Scholars program at the Broad Institute. D.B.K, is supported by NIH/NCI R21 CA216772–01A1 and NCI-SPORE-2P50CA101942–11A1. S.A.S. is supported by NIH R50CA211482. C.J.W. is supported by NCI 1R01 CA155010–01A1, Leukemia Lymphoma Translational Research Program Award 6460–15, and is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Competing interests
J.S. is currently an employee of Moderna Therapeutics. K.D. is an employee of IDT; Y.B. was an employee of IDT during the time of this work; K.J.L. was a paid consultant of IDT during a portion of this work; D.B.K. has previously advised Neon Therapeutics, and owns equity in Aduro Biotech, Agenus Inc., Ampliphi BioSciences Corp., Biomarin Pharmaceutical Inc., Bristol Myers Squibb Com., Celldex Therapeutics Inc., Editas Medicine Inc., Exelixis Inc., Gilead Sciences Inc., IMV Inc., Lexicon Pharmaceuticals Inc., Sangamo Therapeutics, and Stemline Therapeutics Inc.; C.J.W. is a founder of Neon Therapeutics and a member of its scientific advisory board. The remaining authors declare no competing financial interests. C.J.W. is subject to a conflict of interest management plan for the reported studies because of her competing financial interests in Neon Therapeutics. Under this plan, C.J.W. may not access identifiable human subjects data nor otherwise participate directly in the IRB-approved protocol reported herein. C.J.W.’s contributions to the overall program strategy and data analyses occurred on a de-identified basis.
TWEET rhTCRseq for Highly Specific and Efficient Targeted Sequencing of T Cell Receptor mRNA for Single-Cell and Repertoire Analysis
Related links
Key reference using this protocol
1. Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019). doi: 10.1038/s41586-018-0792-9.
References
- 1.Dash P et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J. Clin. Invest. 121, 288–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang GC, Dash P, McCullers JA, Doherty PC & Thomas PG T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci. Transl. Med. 4, 128ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han A, Glanville J, Hansmann L & Davis MM Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dash P, Wang GC & Thomas PG Single-Cell Analysis of T-Cell Receptor αβ Repertoire. Methods Mol. Biol. 1343, 181–197 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Dash P et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stubbington MJT et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 13, 329–332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redmond D, Poran A & Elemento O Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Med. 8, 80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eltahla AA et al. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol. Cell Biol. 94, 604–611 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Afik S et al. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 45, e148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neal JT et al. Organoid modeling of the tumor immune microenvironment. Cell 175, 1972–1988.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howie B et al. High-throughput pairing of T cell receptor α and β sequences. Sci. Transl. Med. 7, 301ra131 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Keskin DB et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobosy JR et al. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol. 11, 80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trombetta JJ et al. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Kircher M, Sawyer S & Meyer M Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacConaill LE et al. Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics 19, 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hug H & Schuler R Measurement of the number of molecules of a single mRNA species in a complex mRNA preparation. J. Theor. Biol. 221, 615–624 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Kivioja T et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Britanova OV et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 192, 2689–2698 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Shugay M et al. Towards error-free profiling of immune repertoires. Nat. Methods 11, 653–655 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Egorov ES et al. Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. J. Immunol. 194, 6155–6163 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Turchaninova MA et al. High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat. Protoc 11, 1599–1616 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Ma K-Y et al. Immune repertoire sequencing using molecular identifiers enables accurate clonality discovery and clone size quantification. Front. Immunol. 9, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heather JM, Ismail M, Oakes T & Chain B High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Brief. Bioinformatics 19, 554–565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z et al. A cloning and expression system to probe T-cell receptor specificity and assess functional avidity to neoantigens. Blood 132, 1911–1921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert L et al. Distinct immunological mechanisms of CTLA-4 and PD-1 blockade revealed by analyzing TCR usage in blood lymphocytes. Oncoimmunology 3, e29244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cha E et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci. Transl. Med. 6, 238ra70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postow MA et al. Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J. Immunother. Cancer 3, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heather JM et al. Dynamic Perturbations of the T-Cell Receptor Repertoire in Chronic HIV Infection and following Antiretroviral Therapy. Front. Immunol. 6, 644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet. 48, 725–732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue H et al. Intratumoral expression levels of PD-L1, GZMA, and HLA-A along with oligoclonal T cell expansion associate with response to nivolumab in metastatic melanoma. Oncoimmunology 5, e1204507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riaz N et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 171, 934–949.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roh W et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beausang JF et al. T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc. Natl. Acad. Sci. USA 114, E10409–E10417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Dongen JJM et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 17, 2257–2317 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Boyd SD et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 1, 12ra23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robins HS et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2, 47ra64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mariani S et al. Comprehensive assessment of the TCRBV repertoire in small T-cell samples by means of an improved and convenient multiplex PCR method. Exp. Hematol. 37, 728–738 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Wang C et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc. Natl. Acad. Sci. USA 107, 1518–1523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherwood AM et al. Deep sequencing of the human TCRγ and TCRβ repertoires suggests that TCRβ rearranges after αβ and γδ T cell commitment. Sci. Transl. Med. 3, 90ra61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quail MA et al. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods 5, 1005–1010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knierim E, Lucke B, Schwarz JM, Schuelke M & Seelow D Systematic comparison of three methods for fragmentation of long-range PCR products for next generation sequencing. PLoS One 6, e28240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Head SR et al. Library construction for next-generation sequencing: overviews and challenges. BioTechniques 56, 61–4, 66, 68, passim (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolotin DA et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Camacho C et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodrigues OR & Monard S A rapid method to verify single-cell deposition setup for cell sorters. Cytometry A 89, 594–600 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: TCR Primers, Excel file
Supplementary Table 2: Barcode Primers, Excel file
Supplementary Table 4: Number of unique TCR clonotypes detected in five PBMC RNA samples from healthy adult volunteers, in Supplementary Note
Supplementary Table 5: Single Cell Clonotypes, Excel file
Supplementary Table 3: Bulk RNA Clonotypes, Excel file
Supplementary Fig. 2: Reproducibility of the bulk RNA rhTCRseq protocol
Supplementary Fig. 3: Gating strategy for isolating tumor-specific T cells
Supplementary Fig. 4: Distribution of clones identified by single-cell TCR sequencing of tumor-stimulated T cells
Supplementary Fig. 1: TCR repertoire analysis for five individuals
Data Availability Statement
All TCR clonotype data for the results presented in Supplementary Figures 1, 2, and 4 are provided in Supplementary Tables 3 and 5. All other data are available from the corresponding author upon reasonable request. Example data used for data analysis are publicly available from the Github repository at https://github.com/julietforman/rhTCRseq