Abstract
Alzheimer’s disease (AD) is a neurodegenerative condition, of which one of the cardinal pathological hallmarks is the extracellular accumulation of amyloid β (Aβ) peptides. These peptides are generated via proteolysis of the amyloid precursor protein (APP), in a manner dependent on the β-secretase, BACE1 and the multicomponent γ-secretase complex. Recent data also suggest a contributory role in AD of transactive response DNA binding protein 43 (TDP-43). There is little insight into a possible mechanism linking TDP-43 and APP processing. To this end, we used cultured human neuronal cells to investigate the ability of TDP-43 to interact with APP and modulate its proteolytic processing. Immunocytochemistry showed TDP-43 to be spatially segregated from both the extranuclear APP holoprotein and its nuclear C-terminal fragment. The latter (APP intracellular domain) was shown to predominantly localise to nucleoli, from which TDP-43 was excluded. Furthermore, neither overexpression of each of the APP isoforms nor siRNA-mediated knockdown of APP had any effect on TDP-43 expression. Doxycycline-stimulated overexpression of TDP-43 was explored in an inducible cell line. Overexpression of TDP-43 had no effect on expression of the APP holoprotein, nor any of the key proteins involved in its proteolysis. Furthermore, increased TDP-43 expression had no effect on BACE1 enzymatic activity or immunoreactivity of Aβ1-40, Aβ1-42 or the Aβ1-40:Aβ1-42 ratio. Also, siRNA-mediated knockdown of TDP-43 had no effect on BACE1 immunoreactivity. Taken together, these data indicate that TDP-43 function and/or dysfunction in AD is likely independent from dysregulation of APP expression and proteolytic processing and Aβ generation.
Keywords: Alzheimers disease, Amyloid precursor protein, BACE1, Gene regulation, proteolysis, TDP-43
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease and the most common form of dementia [1]. It is traditionally characterised by a bipartite pathology featuring plaques comprising amyloid β (Aβ) peptide and neurofibrillary tangles containing the microtubule-associated protein τ [1,2]. The former is produced by sequential proteolytic cleavage of the amyloid precursor protein (APP), of which there are three major isoforms (APP695, APP751 and APP770) [3,4].
There are two predominant proteolytic pathways of APP processing, though other pathways have been identified [5]. In neurons, amyloidogenic processing is the minor pathway and involves sequential cleavage by β-secretase, BACE1 and the γ-secretase complex (comprising presenilin 1 or 2, nicastrin, Aph1 and Pen2) [6–8]. This ultimately releases Aβ species (Aβ1-40 and Aβ1-42 are the most significant), which may play a role in the pathogenesis of AD [9]. The second, non-amyloidogenic, pathway involves α-secretase cleavage of APP, a function ascribed to the metalloprotease ADAM10 in neurons [10]. This cleavage occurs within the Aβ region [11,12], precluding generation and deposition of Aβ. A large, soluble, N-terminal ectodomain, the neuroprotective sAPPα, is shed from the cell surface as a product of this pathway [13]. The most C-terminal region of the APP holoprotein is referred to as the amyloid precursor protein intracellular domain (AICD), through which a number of signalling pathways and transcriptional regulatory events are mediated [14–17].
There is an emerging view that transactive response DNA binding protein 43 (TDP-43) plays a key role in AD pathogenesis. TDP-43 was first linked to neurodegenerative disease through its discovery as a component of classical ubiquitination inclusions in motor neuron disease and frontotemporal dementia [18]. Since then, several recent findings have indicated a key role in AD [19–21]. TDP-43 is a predominantly nuclear protein, although it does shuttle between the nucleus and cytosol [22]. It functions as a nucleic acid binding protein, regulating in excess of 6000 targets. It can modulate all aspects of RNA regulation: processing, splicing, transport and translation, targeting both coding and non-coding RNAs [23].
TDP-43 inclusions have been identified in AD brains in the form of high molecular weight [24] and phosphorylated [25] TDP-43 species. This has been subsequently validated by several neuropathological studies and developed into a staging scheme for TDP-43 pathology in AD [26–30]. In support for an active role for TDP-43 in AD progression, the presence of TDP-43 inclusions in AD is associated with a higher Braak stage and greater impairment in cognitive performance [20,21,31]. It is considered unlikely that there are any significant TDP-43 mutations in AD cases [32].
However, despite evidence showing the presence of TDP-43 inclusions in AD cases and an inverse correlation with cognitive performance, mechanistic insight is sparse. It is neither clear why TDP-43 forms inclusions in neurodegenerative disease generally, nor what specific effects this might have in AD. Overexpression of TDP-43 in AD transgenic mice has been shown to reduce Aβ plaque number, through mechanisms unclear [33], although TDP-43 depletion increased Aβ uptake by microglia [34], which corresponds to the decreased plaque formation in a mouse model [35]. In addition, TDP-43 regulates τ splicing [36] and suppresses its expression [37]. TDP-43 may modulate mitochondrial function [38,39], as evidenced by a recent report of small molecule inhibition of TDP-43 localisation to mitochondria having beneficial effects in an AD transgenic mouse [40].
Despite the centrality of APP, its proteolytic processing and metabolites in AD, there has only been one investigation, in mice, of the ability of TDP-43 to modulate APP processing [41]. To this end, we sought to elucidate the role of TDP-43 in modulating the expression and proteolytic processing of APP in human cells. The aim of the present study was to describe, for the first time, the possible dependence of APP processing on TDP-43 and the effects of possible dysfunction of this axis in the context of AD.
Experimental procedures
Materials
All chemicals were purchased from Fisher Scientific (Loughborough, Leicestershire, U.K.) unless otherwise stated. Actinomycin D was from Sigma–Aldrich (Gillingham, Dorset, U.K.).
Primary antibodies used: for APP (Y188; Abcam, Cambridge, U.K.; RRID:AB_2289606 and 22C11; Merck Millipore, Watford, Hertfordshire, U.K.; AB_94882), TDP-43 (Proteintech, Manchester, U.K.; AB_ 2200520), AICD (BioLegend, London, U.K.; AB_2564761, raised against a neo-epitope around the VMLK sequence at the ε-cleavage site), Tip60 (Novus Biologicals, Abingdon, Oxfordshire, U.K.; AB_1199339), Fe65 (Novus Biologicals; AB_789863), FLAG (Sigma–Aldrich; AB_439687), ADAM10 (Proteintech, Cat #25900-1-AP), BACE1 (Cell Signaling Technology, Leiden, The Netherlands; AB_1903900), nicastrin (Cell Signaling Technology; AB_10694688), Pen2 (ABcloncal, Woburn, MA, U.S.A.; AB_2771856), PS1 (Abcam; AB_1310605), PS2 (Cell Signaling Technology; AB_10831052), fibrillarin (Abcam; AB_1523617) and GAPDH (Proteintech, AB_2107436).
Methods
Cell culture
SH-SY5Y cells (RRID:CVCL_0019) were cultured in 1:1 Dulbecco’s modified Eagle’s medium: Ham’s F12 with UltraGlutamine (DMEM F12, Cat #BE04-687F/U1, Lonza, Slough, Berkshire, U.K.), supplemented with 10% foetal bovine serum (FBS) (Life Technologies, Thermo Fisher Scientific, Paisley, U.K.), 100 U/ml penicillin, 100 µg/ml streptomycin (both Life Technologies) and 1% non-essential amino acids (Sigma–Aldrich). Cells were incubated at 37°C and 5% CO2 in a humidified atmosphere. For differentiation, cells were seeded and cultured in full growth medium (above) for 24 h, then differentiated for 7 days in DMEM F12 supplemented with 1% FBS (Cat #631107, Takara, Saint-Germain-en-Laye, France), 100 U/ml penicillin, 100 µg/ml streptomycin, 1% non-essential amino acids and 1 µM retinoic acid. SH-SY5Y cells overexpressing APP isoforms were generated as previously described [42].
The iPSC line, OX1-19 (obtained from S. Cowley, University of Oxford) was maintained on Matrigel (BD Biosciences, Wokingham, Berkshire, U.K.) in mTeSR1 medium (StemCell Technologies, Cambridge, U.K.) containing 50 U/ml penicillin and 50 µg/ml streptomycin (Life Technologies) in a humidified incubator at 37°C in a 5% CO2, 95% air atmosphere. iPSC pluripotency and successful cortical neuron differentiation were confirmed using immunofluorescence microscopy with appropriate markers [43]. The iPSCs were differentiated into cortical neurons as described previously [44,45], using dual-SMAD inhibition by 1 µM dorsomorphin and 10 mM SB431452 (Bio-Techne, Abingdon, Oxfordshire, U.K.). Following successful differentiation, neural progenitor cells (NPCs) were re-plated on day 35 post-induction at 300000 cells/well on to poly-ornithine and laminin-coated (Sigma–Aldrich) six-well tissue culture plates and cultured until between days 75 and 90 post-induction with media changes every 2–3 days. Post-induction culture medium was 1:1 DMEM F12: neurobasal medium containing B27 and N2 supplements, 2 mM l-glutamine, 100 µM 2-mercaptoethanol, 25 µM insulin, 100 U/ml penicillin, 100 µg/ml streptomycin (all Life Technologies) and 0.5% non-essential amino acids.
Inducible cell line
For the generation of the inducible cell line, the TRE-x system was used (Thermo Fisher Scientfic). Two plasmid vectors were used: the regulatory pcDNA6/TR and pT-Rex-DEST30 (expressing FLAG-TDP-43) (GeneArt, Thermo Fisher Scientific). Both vectors were transfected into SH-SY5Y cells (2 µg total DNA, 6:1 ratio) using the Amaxa 4D-Nucleofector on programme CA-137 (Lonza). After 48 h, cells stably expressing both plasmids were selected using blasticidin (5 µg/ml) and G418 (500 µg/ml), selecting for the regulatory and expression vectors, respectively. TDP-43 expression was induced using 500 ng/ml doxyxlycline hyclate (Sigma–Aldrich) for 72 h. Expression of FLAG-TDP-43 was verified using immunoblotting.
siRNA knockdown
SH-SY5Y cells were differentiated for 7 days as described and then nucleofected with siRNA duplexes targeting either APP (100 nM; Ambion Silencer Select, Thermo Fisher Scientific; ID s229520, siRNA #1 or Dharmacon ON-TARGETplus SMARTpool, Horizon Discovery Cambridge, U.K.; Cat #L-003731-00-005, siRNA #2), TDP-43 (120 nM; Ambion Silencer Select, Thermo Fisher Scientific; ID s23879, siRNA #1 or Dharmacon ON-TARGETplus SMARTpool, Horizon Discovery Cambridge, U.K.; Cat #L-012394-00-005, siRNA #2) or a non-targeting control (Negative Control siRNA; Qiagen, Manchester, U.K.) according to the manufacturer’s instructions (Amaxa 4D-Nucleofector; programme CA-137). Cells were further cultured for 72 h and target knockdown verified using immunoblotting.
Immunocytochemistry
Cells were cultured on gelatin-coated glass coverslips (BioReagent, Sigma–Aldrich). After treatment, cells were fixed in 4% paraformaldehyde for 10 min then washed in phosphate-buffered saline (PBS). Cells were subsequently permeabilised in 0.5% Triton X-100, washed in PBS and incubated with blocking buffer (0.5% fish skin gelatin (FSG, Sigma–Aldrich) in PBS) for 1 h. Coverslips were incubated with primary antibody for 1 h (in 0.5% FSG, 0.5% Triton X-100 in PBS), washed in PBS and incubated for 1 h in secondary antibody (0.5% FSG, 0.5% Triton X-100 in PBS and either Alexa Fluor 488 (Thermo Fisher Scientific; AB_2610666), Alexa Fluor 555 (Abcam; AB_2801638) or CF 647 (Sigma–Aldrich, Cat #SAB4600183). After washing, coverslips were incubated with 1 µg/ml DAPI (emp Biotech, Berlin, Germany) for 5 min and mounted on microscope slides with Prolong Diamond mounting medium (Thermo Fisher Scientific). Images were acquired on an Olympus IX83 inverted microscope using Lumencor LED excitation, a 40× or 60× objective and the Sedat QUAD filter set (Cat #89000, Chroma Technology, Bellows Falls, VT, U.S.A.). The images were collected using a Retiga R6 Q-Imaging CCD camera and Metamorph v7.8.4.0 (Molecular Devices, San Jose, CA, U.S.A.). Images were then processed and analysed using ImageJ (NIH, U.S.A.).
Cell lysis
Cells were washed twice in ice-cold PBS and harvested in PBS. Cells were pelleted at 3000×g for 5 min (4°C) and re-suspended in 6× volume of lysis buffer (RIPA buffer: 50 mM Tris/HCl (pH 8.0), 150 mM sodium chloride, 1% Igepal CA-630 (Sigma–Aldrich), 0.5% sodium deoxycholate, 0.1% SDS, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and Complete Protease Inhibitor cocktail (Roche Diagnostics, Burgess Hill, West Sussex, U.K.)). Lysis was performed for 30 min on ice, followed by centrifugation at 3000×g for 5 min (4°C) to yield the RIPA-soluble fraction as the supernatant, which was used for immunoblotting.
Determination of protein concentration
Protein concentration in the RIPA-soluble fraction was determined using the bicinchoninic acid (BCA) method [46], using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Absorbance at 562 nm was measured using a plate reader (ELx800, BioTek, Swindon, U.K.). Sample concentration was determined using bovine serum albumin (BSA) as a standard at concentrations from 0 to 1 mg/ml.
SDS/PAGE and immunoblotting
Protein samples were separated by electrophoresis at 120 V for 90 min on a polyacrylamide gel. After SDS/PAGE, proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hemel Hempstead, Hertfordshire, U.K.). Blots were incubated for 2 h in blocking solution (5% (w/v) milk power, 2% (w/v) BSA in TBS + 1% (v/v) Tween-20 (TBST)). The blots were then incubated overnight in primary antibody (5% (w/v) milk powder in TBS). Blots were washed 4 × 10 min with TBST before the addition of secondary antibody (HRP–conjugated anti-IgG; 5% (w/v) milk powder in TBST, 1:5000 (Thermo Fisher Scientific)) for 1 h, followed by 4 × 10 min washes with TBST. Protein bands were visualised by chemiluminescence (Clarity Western ECL Blotting Substrate, Bio-Rad) using a G:BOX and GeneTools software (Syngene, Cambridge, U.K.).
Quantitative PCR
RNA was isolated from differentiated SH-SY5Y cells using the RNeasy Mini Kit according to the manufacturers’ instructions (Qiagen). cDNA was subsequently prepared using the Applied Biosystems High Capacity cDNA Synthesis Kit after which quantitative PCRs (qPCRs) were prepared as follows (total 20 µl): 1 µl cDNA, 500 nM each of forward and reverse primers with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). Thermal cycler (QuantStudio 3, Applied Biosystems, Thermo Fisher Scientific) parameters were set as follows: 2 min @ 50°C, 2 min @ 95°C and 40 cycles of 15s @ 95°C, 15s @ 53°C and 60s @ 72°C and data analysed using the ΔΔCT method with succinate dehydrogenase complex, subunit A (SDHA) as a reference gene. Primer sequences were as follows: BACE1 (F: GTAAAGCAGACCCACGTTCC; R: CAATGATCATGCTCCCTCCG) and SDHA (F: CAGCATGCAGAAGTCAATGC; R: ACGTCTTCAGGTGCTTTAGG).
BACE1 activity assay
Differentiated SH-SY5Y cells were lysed in assay buffer (10 mM sodium acetate, 1.5 mM NaCl, 0.1% Triton X-100, 0.32 M sucrose, pH 5.0) for 30 min on ice, before clarification by centrifugation (5000×g, 5 min) and protein quantitation of the supernatant. Subsequently, 2 µl of BACE1 substrate (Abcam) was added to 50 µl supernatant (containing 50 µg total protein) followed by kinetic output measurement on a fluorescent microplate reader at Ex/Em = 335/495 nm (Synergy HT, Biotek). Rate (ΔRFU) was calculated in the linear reaction phase.
Amyloid-β enzyme-linked immunosorbent assay
Conditioned media samples (48 h) were isolated from differentiated SH-SY5Y cells and centrifuged at low speed to remove cell debris (500×g, 5 min). The resultant supernatant was concentrated ten-fold using a Vivaspin 2 (2000 MWCO) and the protein concentration assessed. Conditioned media samples were then added to enzyme-linked immunosorbent assay (ELISA) plates to quantify Aβ1-40 (5 µg total protein, Cat #KHB3481) and Aβ1-42 (30 µg total protein, Cat #KHB3544) (both Thermo Fisher) ELISAs were performed according to the manufacturer’s instructions, including incubation of condition medium with biotinylated detection antibody, followed by HRP–conjugated secondary antibodies, chromagen and finally the stop solution, resulting in a colorimetric response proportional to target abundance, the absorbance of which was measured on a microplate reader at 450 nm (ELx800, Biotek).
Statistical analysis
All experiments are n=3 unless otherwise indicated, where n indicates independent experiments on independent cell cultures. Statistical tests were either Mann–Whitney U test, Student’s t test (ELISA and BACE1 activity data only) or Kruskal–Wallis with Dunn’s post hoc test as indicated; P<0.05 (*), P<0.01 (**), P<0.001 (***) or P<0.0001 (****). Error bars indicate standard deviation. All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, U.S.A.).
Results and discussion
APP and TDP-43 have distinct intracellular locations in cultured neuronal cells
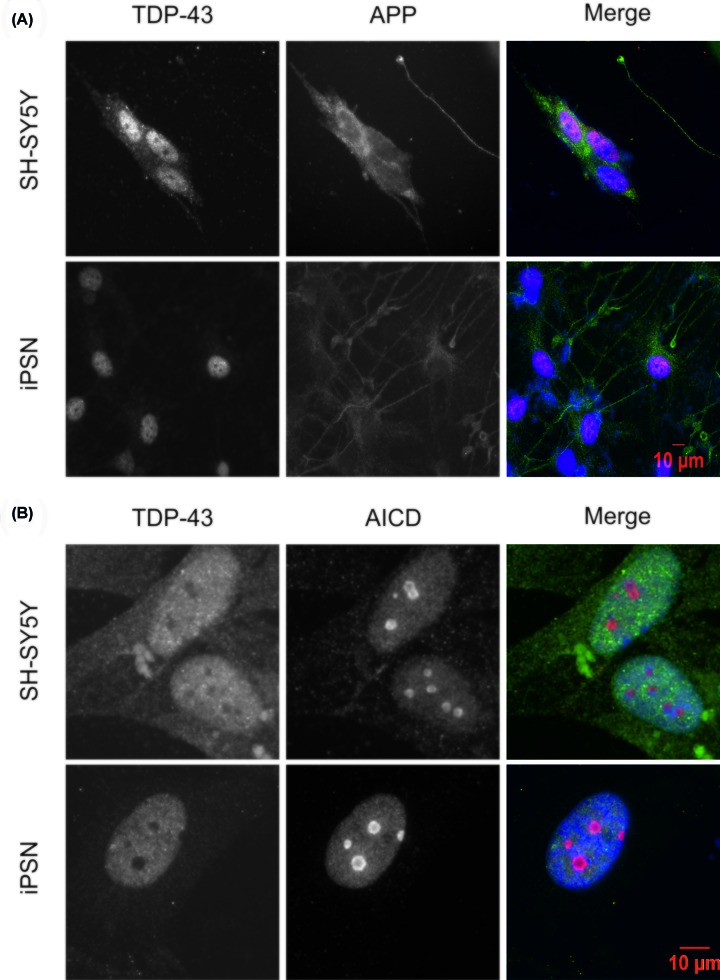
In order to investigate a possible direct relationship between APP and TDP-43, putative co-localisation was assessed using immunofluorescence microscopy. Using differentiated SH-SY5Y cells and, separately, OX1-19 iPSC-derived neurons (iPSNs), the localisation of the APP holoprotein and TDP-43 was investigated. As expected, TDP-43 was localised exclusively in the nucleus, whereas the APP holoprotein was excluded from the nucleus (Figure 1A). AICD translocates to the nucleus after proteolysis of the APP holoprotein [15,17]. Using an AICD-specific antibody (targeting a neo-epitope) [47], we probed the subcellular localisation of AICD in comparison with TDP-43. Though there was some evidence of AICD immunoreactivity throughout the cell, AICD was most strongly localised to the nucleus. More specifically, AICD was present as a component of several large subnuclear structures. In contrast, TDP-43 was completely excluded from these structures, seen as voids in the TDP-43 immunostaining. This was recapitulated in SH-SY5Y cells and iPSNs (Figure 1B).
Figure 1. TDP-43 does not co-localise with either the APP holoprotein or its intracellular domain.
SH-SY5Y or OX1-19 iPSCs were cultured and differentiated as described, followed by fixation and immunocytochemistry using primary antibodies against TDP-43 and (A) APP holoprotein (antibody 22C11) and (B) AICD. iPSN denotes iPSC-derived neurons.
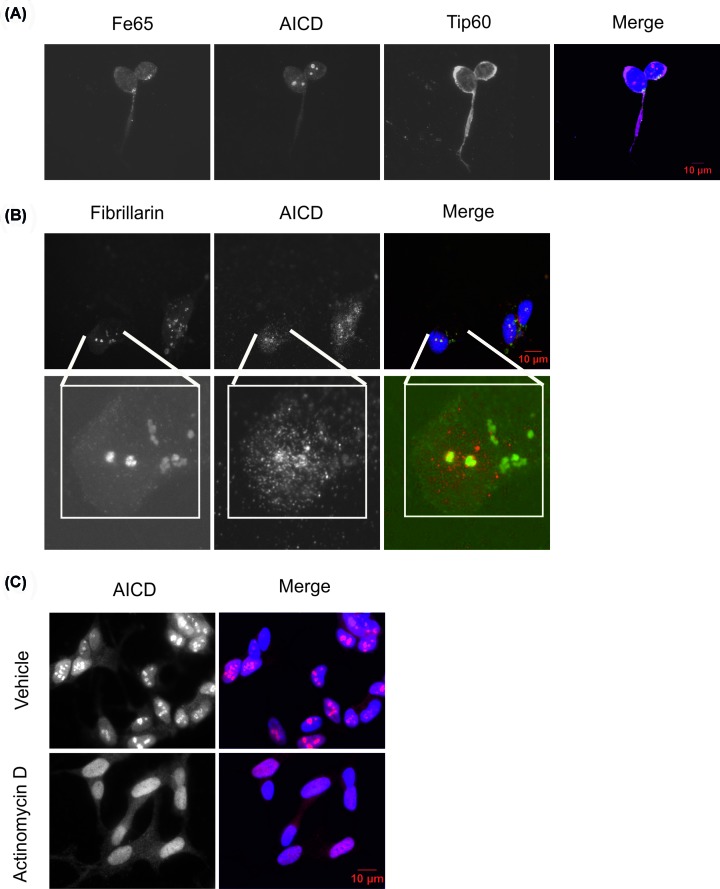
In order to investigate the identity of these AICD-positive, TDP-43-negative subnuclear structures, we assessed whether they represented the AFT (AICD-Fe65-Tip60) transcriptional regulatory complex. However, there was no evidence of Fe65 or Tip60 co-staining with these structures. Moreover, Tip60 immunostaining is absent from these AICD-positive puncta in a similar manner to TDP-43 (Figure 2A). It is unclear exactly how the endogenous AFT complex manifests. Several publications report the use of fluorescently tagged AFT complex components [48–51], but Tip60 in these studies shows a markedly different distribution to those reports using smaller tags, or untagged Tip60 [52–54]. Further differences may arise from the use of non-neuronal [51,52] or neuronal-like [53] cell lines. Altogether, these data give a contradictory picture of the exact nature of the endogenous AFT complex in neurons.
Figure 2. AICD is present in nucleoli.
SH-SY5Y cells were cultured and differentiated as described, followed by fixation and immunocytochemistry using primary antibodies against (A) FE65, AICD and Tip60 (B) AICD and fibrillarin. (C) Cells were treated with actinomycin D (0.02 µg/ml, 3 h) and immunocytochemistry performed to assess the effect on AICD localisation.
Given the large size of the AICD-positive structures, it was hypothesised that they may represent nucleoli. Indeed, the nucleolar marker fibrillarin showed strong co-localisation with AICD, indicating that AICD may be localised to the nucleolus (Figure 2B). Localisation of AICD to nucleoli has not previously been reported in the literature. Pharmacological treatment with actinomycin D is known to disrupt nucleoli and treatment of SH-SY5Y cells resulted in a homogenous redistribution of AICD throughout the nucleus (Figure 2C). Taken together, these data indicate that nuclear AICD may be localised to nucleoli, from which TDP-43 is excluded, potentially suggesting that there is no direct interaction between TDP-43 and APP/AICD.
Modulation of APP expression does not affect TDP-43 expression
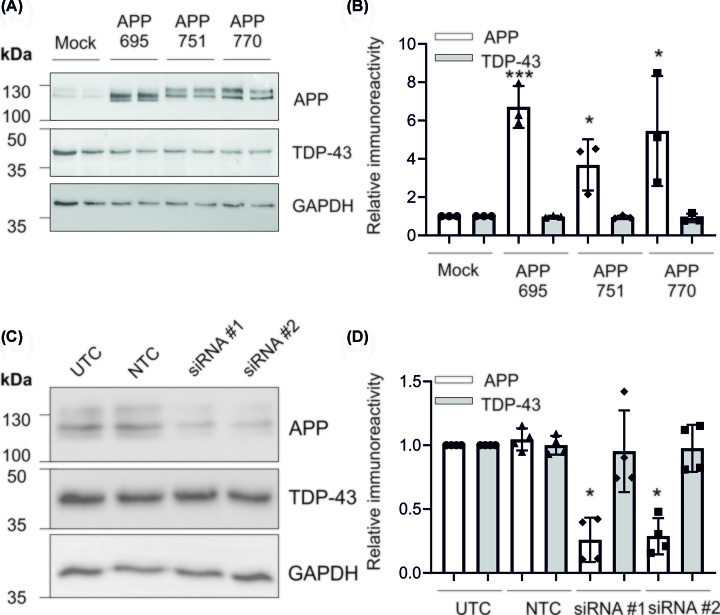
APP has been shown to regulate the expression of numerous genes, both directly through AICD and indirectly via its binding partners [17,55,56]. In order to assess whether APP could modulate the expression of TDP-43, each APP isoform (695, 751, 770) was expressed separately in SH-SY5Y cells. Individual isoforms were expressed to assess any differential effects of APP, as AICD is preferentially generated from APP695, suggesting isoform-specific processing pathways [42]. There were no substantial isoform differences in the level of APP expression, with all isoforms expressed at an approximately eight-fold higher level than in the mock control (Figure 3A,B). None of the APP isoforms had an effect on TDP-43 immunoreactivity (Figure 3A,B). Similarly, siRNA-mediated knockdown of APP had no effect on TDP-43 expression (Figure 3C,D).
Figure 3. APP overexpression or knockdown does not affect TDP-43 expression.
SH-SY5Y cells were cultured as described and (A) stably transfected with each of the three APP isoforms. Expression was confirmed by immunoblot, compared with TDP-43 expression and (B) changes in protein expression quantified. (C) Cells were nucleofected with two independent siRNA targeting APP and protein expression of APP and TDP-43 assessed by immunoblot and (D) quantified; n=4. *P<0.05 and ***P<0.001.
Inducible expression of TDP-43 does not affect expression of proteins involved in APP proteolytic processing
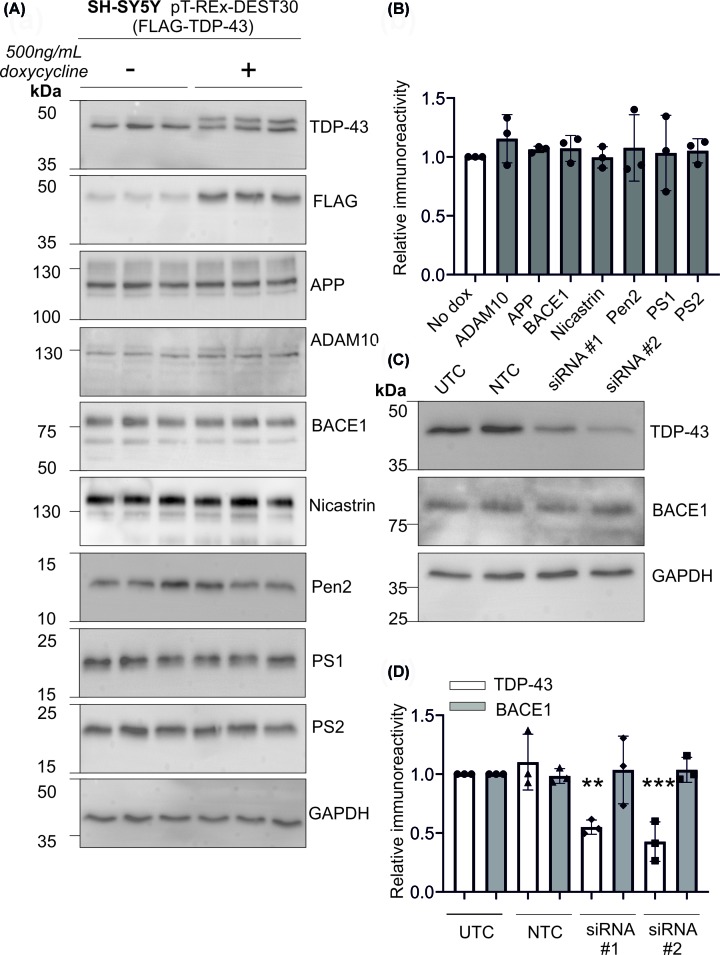
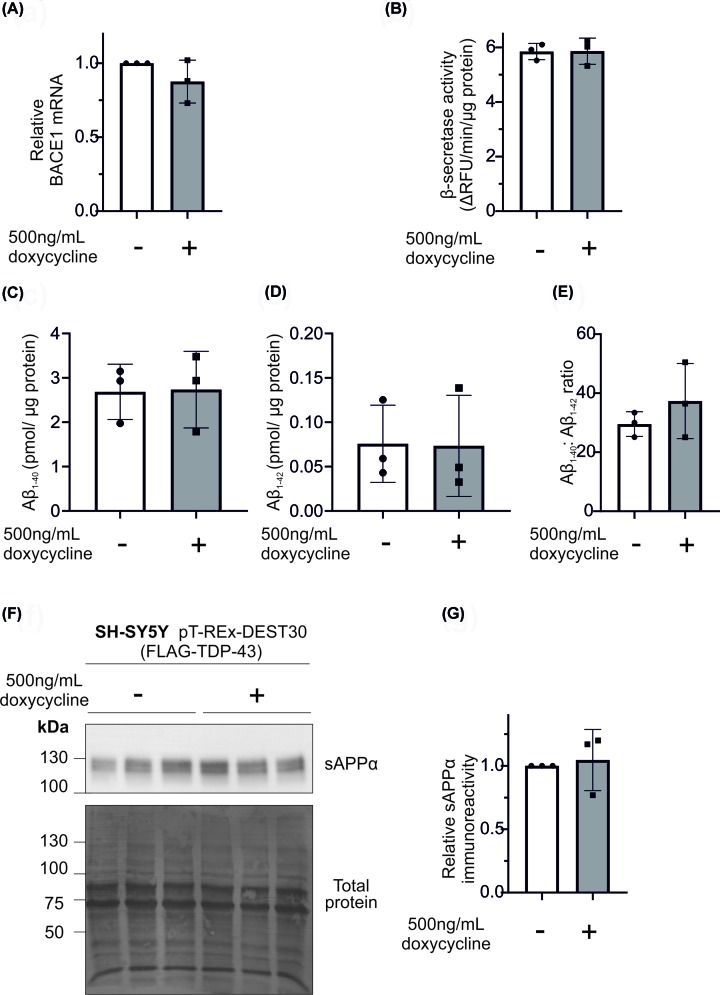
An inducible cell line was generated, whereby TDP-43 was only expressed when activated by doxycycline. Incubation of cells with doxycycline resulted in increased expression of TDP-43, combined with FLAG immunoreactivity (Figure 4A), indicating high expression of FLAG-TDP-43 when induced with doxycycline. Using this cell model, TDP-43 expression was induced and the expression of APP and key APP-related proteins assessed. The protein panel comprised the APP holoprotein, the α-secretase ADAM10, the β-secretase BACE1 and the γ-secretase components (PS1, PS2, nicastrin and Pen2). Overall, the expression of TDP-43 did not have any significant effect on the expression of any of these proteins (Figure 4A,B). Given a previous report linking TDP-43 to BACE1 [41], we further focused on these proteins. Using siRNA knockdown, we assessed the effect of a reduction in TDP-43 on the expression of BACE1. Despite significant reductions in TDP-43 expression, this did not alter the immunoreactivity of BACE1 (Figure 4C,D). Our data show a lack of change in BACE1 protein expression in response to TDP-43 overexpression or knockdown. To confirm that TDP-43 did not alter BACE1 protein expression, qPCR was used to measure the level of BACE1 mRNA. After doxycycline-induced expression of TDP-43, there was no significant change in BACE1 mRNA (Figure 5A). Overall, these data show that TDP-43 does not affect the expression of key proteins involved in APP proteolytic processing.
Figure 4. Inducible TDP-43 overexpression does not affect APP processing.
A stable cell line with doxycycline-inducible TDP-43 expression (pT-REx-DEST30-TDP-43-FLAG) was generated and (A) TDP-43 overexpression confirmed by immunoblot. APP and its proteolytic enzymes were also probed by immunoblot and (B) changes quantified by densitometry. (C) Cells were nucleofected with two independent siRNA targeting TDP-43 and protein expression of TDP-43 and BACE1 assessed by immunoblot and (D) quantified. **P<0.01 and ***P<0.001.
Figure 5. TDP-43 overexpression does not modulate BACE1 mRNA, activity or the generation of Aβ peptides.
TDP-43 expression was induced in SH-SY5Y (pT-REx-DEST30-TDP-43-FLAG) cells and cells were analysed for (A) BACE1 mRNA levels, (B) β-secretase activity and culture medium assessed for (C) Aβ1-40 and (D) Aβ1-42 followed by (E) calculation of the Aβ1-40: Aβ1-42 ratio (F) sAPPα immunoreactivity (using antibody 22C11). Total protein was assessed using Amido Black stain to ensure equal loading and (G) protein changes were quantified by densitometry.
TDP-43 expression does not alter APP proteolysis or BACE1 activity
APP processing is an enzymatic process and, as such, may be modulated by changes in secretase activity which are independent of protein expression. In order to assess this, β-secretase activity was assessed using a fluorescent substrate. Doxycycline-induced expression of TDP-43 had no effect on β-secretase activity (Figure 5B). Increased expression of TDP-43 as induced by doxycycline did not have any effect on levels of Aβ1-40 or Aβ1-42, nor did it affect the ratio between the two peptides (Figure 5C–E). The mean abundance in the absence of doxycycline treatment was 2.68 pg/µg protein (Aβ1-40) or 0.076 pg/µg protein (Aβ1-42) and in the presence of doxycycline was 2.73 pg/µg protein (Aβ1-40) or 0.073 pg/µg protein (Aβ1-42). In addition, there was no effect on non-amyloidogenic processing of APP, as immunoblotting conditioned medium for sAPPα showed no change upon TDP-43 induction (Figure 5F,G). Together these data show that TDP-43 does not modulate either amyloidogenic or non-amyloidogenic proteolytic processing of APP. Our data conflicts with Herman et al. who reported TDP-43 mediated up-regulation of BACE1 in a mouse model [41]. However, their lentiviral overexpression of TDP-43 also caused significant increases in TNFα and IL-6, both of which can increase BACE1 expression and activity [41]. Due to the confounding inflammation, it is not possible to determine whether TDP-43 directly increases BACE1 activity or merely promotes an inflammatory phenotype which drives BACE1 activity. However, as we observed no direct effect of TDP-43 on BACE1 activity, it is more likely that the increased BACE1 activity in the mice was due to the inflammation.
Conclusion
In the present study, we have focused on the interaction of TDP-43 with APP proteolytic processing. TDP-43 did not influence APP expression, nor did APP modulate TDP-43 expression. TDP-43 had no effect on the non-amyloidogenic cleavage of APP and did not alter the production of either Aβ1-40 or Aβ1-42. In addition, TDP-43 did not directly affect BACE1 activity. Our data strongly suggest that TDP-43 is not directly involved in any AD-linked alterations in APP expression or proteolytic cleavage. Given these findings and the notable regional differences between TDP-43 and Aβ pathology in AD [26], it is highly likely that AD-linked alterations in Aβ are independent of TDP-43 dysfunction.
Acknowledgements
Special thanks goes to Steven Marsden for help with the microscopy.
Abbreviations
- AD
Alzheimer’s disease
- ADAM10
a disintegrin and metalloprotease 10
- AICD
amyloid precursor protein intracellular domain
- APP
amyloid precursor protein
- Aβ
amyloid β
- BACE1
β-site APP cleaving enzyme 1
- BCA
bicinchoninic acid
- BSA
bovine serum albumin
- ELISA
enzyme-linked immunosorbent assay
- FBS
foetal bovine serum
- FSG
fish skin gelatin
- iPSC
induced pluripotent stem cell
- iPSN
iPSC-derived neuron
- PBS
phosphate-buffered saline
- qPCR
quantitative PCR
- sAPPα
soluble amyloid precursor protein α
- SDHA
succinate dehydrogenase complex, subunit A
- TBST
TBS + 1% (v/v) Tween-20
- TDP-43
transactive response DNA binding protein 43
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Alzheimer’s Society Fellowship [grant number 285 (AS-JF-15b-006) (to D.A.H)]; iPSC culture by the Medical Research Council (MRC) [grant numbers MR/M024997/1, MR/N013255/1]; the Donald Dean Fund for Dementia Research (to N.H.); the Bioimaging Facility microscopes used in the present study were purchased with grants from BBSRC; Wellcome; and the University of Manchester Strategic Fund.
Author Contribution
D.A.H., S.M.P.-B. and N.H. designed the study. D.A.H., S.M.P.-B. and N.H. acquired the funding. D.A.H. and A.C.J. performed experiments and analysed the data. D.A.H. wrote the manuscript. D.A.H. and N.H. edited the manuscript.
References
- 1.Masters C.L., Bateman R., Blennow K., Rowe C.C., Sperling R.A. and Cummings J.L. (2015) Alzheimer’s disease. Nat. Rev. Dis. Primers 1, 15056 10.1038/nrdp.2015.56 [DOI] [PubMed] [Google Scholar]
- 2.Querfurth H.W. and LaFerla F.M. (2010) Alzheimer’s disease. N. Engl. J. Med. 362, 329–344 10.1056/NEJMra0909142 [DOI] [PubMed] [Google Scholar]
- 3.Tanzi R.E., Gusella J.F., Watkins P.C., Bruns G.A., St George-Hyslop P., Van Keuren M.L. et al. (1987) Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science 235, 880–884 10.1126/science.2949367 [DOI] [PubMed] [Google Scholar]
- 4.Sandbrink R., Masters C.L. and Beyreuther K. (1996) APP gene family. Alternative splicing generates functionally related isoforms. Ann. N.Y. Acad. Sci. 777, 281–287 10.1111/j.1749-6632.1996.tb34433.x [DOI] [PubMed] [Google Scholar]
- 5.Andrew R.J., Kellett K.A., Thinakaran G. and Hooper N.M. (2016) A Greek tragedy: the growing complexity of alzheimer amyloid precursor protein proteolysis. J. Biol. Chem. 291, 19235–19244 10.1074/jbc.R116.746032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson J.P., Chen Y., Kim K.S. and Robakis N.K. (1992) An alternative secretase cleavage produces soluble Alzheimer amyloid precursor protein containing a potentially amyloidogenic sequence. J. Neurochem. 59, 2328–2331 10.1111/j.1471-4159.1992.tb10128.x [DOI] [PubMed] [Google Scholar]
- 7.Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P. et al. (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 10.1126/science.286.5440.735 [DOI] [PubMed] [Google Scholar]
- 8.Bergmans B.A. and De Strooper B. (2010) Gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol. 9, 215–226 10.1016/S1474-4422(09)70332-1 [DOI] [PubMed] [Google Scholar]
- 9.Hardy J.A. and Higgins G.A. (1992) Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185 10.1126/science.1566067 [DOI] [PubMed] [Google Scholar]
- 10.Kuhn P.H., Wang H., Dislich B., Colombo A., Zeitschel U., Ellwart J.W. et al. (2010) ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 29, 3020–3032 10.1038/emboj.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allinson T.M., Parkin E.T., Turner A.J. and Hooper N.M. (2003) ADAMs family members as amyloid precursor protein α-secretases. J. Neurosci. Res. 74, 342–352 10.1002/jnr.10737 [DOI] [PubMed] [Google Scholar]
- 12.Esch F.S., Keim P.S., Beattie E.C., Blacher R.W., Culwell A.R., Oltersdorf T. et al. (1990) Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science 248, 1122–1124 10.1126/science.2111583 [DOI] [PubMed] [Google Scholar]
- 13.Allinson T.M., Parkin E.T., Condon T.P., Schwager S.L., Sturrock E.D., Turner A.J. et al. (2004) The role of ADAM10 and ADAM17 in the ectodomain shedding of angiotensin converting enzyme and the amyloid precursor protein. Eur. J. Biochem. 271, 2539–2547 10.1111/j.1432-1033.2004.04184.x [DOI] [PubMed] [Google Scholar]
- 14.Chang K.A. and Suh Y.H. (2010) Possible roles of amyloid intracellular domain of amyloid precursor protein. BMB Rep. 43, 656–663 10.5483/BMBRep.2010.43.10.656 [DOI] [PubMed] [Google Scholar]
- 15.Beckett C., Nalivaeva N.N., Belyaev N.D. and Turner A.J. (2012) Nuclear signalling by membrane protein intracellular domains: the AICD enigma. Cell. Signal. 24, 402–409 10.1016/j.cellsig.2011.10.007 [DOI] [PubMed] [Google Scholar]
- 16.Pardossi-Piquard R. and Checler F. (2012) The physiology of the beta-amyloid precursor protein intracellular domain AICD. J. Neurochem. 120, 109–124 10.1111/j.1471-4159.2011.07475.x [DOI] [PubMed] [Google Scholar]
- 17.Leissring M.A., Murphy M.P., Mead T.R., Akbari Y., Sugarman M.C., Jannatipour M. et al. (2002) A physiologic signaling role for the gamma -secretase-derived intracellular fragment of APP. Proc. Natl. Acad. Sci. U.S.A. 99, 4697–4702 10.1073/pnas.072033799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 19.Josephs K.A., Dickson D.W., Tosakulwong N., Weigand S.D., Murray M.E., Petrucelli L. et al. (2017) Rates of hippocampal atrophy and presence of post-mortem TDP-43 in patients with Alzheimer’s disease: a longitudinal retrospective study. Lancet Neurol. 16, 917–924 10.1016/S1474-4422(17)30284-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josephs K.A., Murray M.E., Whitwell J.L., Parisi J.E., Petrucelli L., Jack C.R. et al. (2014) Staging TDP-43 pathology in Alzheimer’s disease. Acta Neuropathol. (Berl) 127, 441–450 10.1007/s00401-013-1211-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josephs K.A., Whitwell J.L., Weigand S.D., Murray M.E., Tosakulwong N., Liesinger A.M. et al. (2014) TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta Neuropathol. (Berl.) 127, 811–824 10.1007/s00401-014-1269-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buratti E. and Baralle F.E. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 10.1074/jbc.M104236200 [DOI] [PubMed] [Google Scholar]
- 23.Sun Y. and Chakrabartty A. (2017) Phase to phase with TDP-43. Biochemistry 56, 809–823 10.1021/acs.biochem.6b01088 [DOI] [PubMed] [Google Scholar]
- 24.Amador-Ortiz C., Lin W.L., Ahmed Z., Personett D., Davies P., Duara R. et al. (2007) TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann. Neurol. 61, 435–445 10.1002/ana.21154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai T., Mackenzie I.R., Hasegawa M., Nonoka T., Niizato K., Tsuchiya K. et al. (2009) Phosphorylated TDP-43 in Alzheimer’s disease and dementia with Lewy bodies. Acta Neuropathol. (Berl.) 117, 125–136 10.1007/s00401-008-0480-1 [DOI] [PubMed] [Google Scholar]
- 26.Josephs K.A., Murray M.E., Whitwell J.L., Tosakulwong N., Weigand S.D., Petrucelli L. et al. (2016) Updated TDP-43 in Alzheimer’s disease staging scheme. Acta Neuropathol. (Berl) 131, 571–585 10.1007/s00401-016-1537-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson Y., Kelley T., Mackenzie I.R., Pickering-Brown S., Du Plessis D., Neary D. et al. (2007) Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 113, 521–533 10.1007/s00401-006-0189-y [DOI] [PubMed] [Google Scholar]
- 28.McAleese K.E., Walker L., Erskine D., Thomas A.J., McKeith I.G. and Attems J. (2017) TDP-43 pathology in Alzheimer’s disease, dementia with Lewy bodies and ageing. Brain Pathol. 27, 472–479 10.1111/bpa.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foulds P., McAuley E., Gibbons L., Davidson Y., Pickering-Brown S.M., Neary D. et al. (2008) TDP-43 protein in plasma may index TDP-43 brain pathology in Alzheimer's disease and frontotemporal lobar degeneration. Acta Neuropathol. (Berl.) 116, 141–146 10.1007/s00401-008-0389-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tremblay C., St-Amour I., Schneider J., Bennett D.A. and Calon F. (2011) Accumulation of transactive response DNA binding protein 43 in mild cognitive impairment and Alzheimer disease. J. Neuropathol. Exp. Neurol. 70, 788–798 10.1097/NEN.0b013e31822c62cf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohn T.T. (2008) Caspase-cleaved TAR DNA-binding protein-43 is a major pathological finding in Alzheimer's disease. Brain Res. 1228, 189–198 10.1016/j.brainres.2008.06.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ticozzi N., LeClerc A.L., van Blitterswijk M., Keagle P., McKenna-Yasek D.M., Sapp P.C. et al. (2011) Mutational analysis of TARDBP in neurodegenerative diseases. Neurobiol. Aging 32, 2096–2099 10.1016/j.neurobiolaging.2009.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis S.A., Gan K.A., Dowell J.A., Cairns N.J. and Gitcho M.A. (2017) TDP-43 expression influences amyloidbeta plaque deposition and tau aggregation. Neurobiol. Dis. 103, 154–162 10.1016/j.nbd.2017.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paolicelli R.C., Jawaid A., Henstridge C.M., Valeri A., Merlini M., Robinson J.L. et al. (2017) TDP-43 depletion in microglia promotes amyloid clearance but also induces synapse loss. Neuron 95, 297e6–308e6 10.1016/j.neuron.2017.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaClair K.D., Donde A., Ling J.P., Jeong Y.H., Chhabra R., Martin L.J. et al. (2016) Depletion of TDP-43 decreases fibril and plaque beta-amyloid and exacerbates neurodegeneration in an Alzheimer’s mouse model. Acta Neuropathol. (Berl) 132, 859–873 10.1007/s00401-016-1637-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu J., Chen F., Iqbal K., Gong C.X., Wang X. and Liu F. (2017) Transactive response DNA-binding protein 43 (TDP-43) regulates alternative splicing of tau exon 10: implications for the pathogenesis of tauopathies. J. Biol. Chem. 292, 10600–10612 10.1074/jbc.M117.783498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu J., Wu F., Xu W., Shi J., Hu W., Jin N. et al. (2017) TDP-43 suppresses tau expression via promoting its mRNA instability. Nucleic Acids Res. 45, 6177–6193 10.1093/nar/gkx175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis S.A., Itaman S., Khalid-Janney C.M., Sherard J.A., Dowell J.A., Cairns N.J. et al. (2018) TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci. Lett. 678, 8–15 10.1016/j.neulet.2018.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J., Wang L., Yan T., Perry G. and Wang X. (2019) TDP-43 proteinopathy and mitochondrial abnormalities in neurodegeneration. Mol. Cell. Neurosci. 100, 103396 10.1016/j.mcn.2019.103396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J., Wang L., Gao C., Arakawa H., Perry G. and Wang X. (2020) TDP-43 inhibitory peptide alleviates neurodegeneration and memory loss in an APP transgenic mouse model for Alzheimer's disease. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165580 10.1016/j.bbadis.2019.165580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herman A.M., Khandelwal P.J., Rebeck G.W. and Moussa C.E. (2012) Wild type TDP-43 induces neuro-inflammation and alters APP metabolism in lentiviral gene transfer models. Exp. Neurol. 235, 297–305 10.1016/j.expneurol.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belyaev N.D., Kellett K.A., Beckett C., Makova N.Z., Revett T.J., Nalivaeva N.N. et al. (2010) The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J. Biol. Chem. 285, 41443–41454 10.1074/jbc.M110.141390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Wilgenburg B., Browne C., Vowles J. and Cowley S.A. (2013) Efficient, long term production of monocyte-derived macrophages from human pluripotent stem cells under partly-defined and fully-defined conditions. PLoS ONE 8, e71098 10.1371/journal.pone.0071098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Y., Kirwan P., Smith J., Robinson H.P. and Livesey F.J. (2012) Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 15, 477–486, 10.1038/nn.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarosz-Griffiths H.H., Corbett N.J., Rowland H.A., Fisher K., Jones A.C., Baron J. et al. (2019) Proteolytic shedding of the prion protein via activation of metallopeptidase ADAM10 reduces cellular binding and toxicity of amyloid-beta oligomers. J. Biol. Chem. 294, 7085–7097 10.1074/jbc.RA118.005364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D. et al. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- 47.Stieren E.S., El Ayadi A., Xiao Y., Siller E., Landsverk M.L., Oberhauser A.F. et al. (2011) Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J. Biol. Chem. 286, 35689–35698 10.1074/jbc.M111.243147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakshi K., Ranjitha B., Dubey S., Jagannadham J., Jaiswal B. and Gupta A. (2017) Novel complex of HAT protein TIP60 and nuclear receptor PXR promotes cell migration and adhesion. Sci. Rep. 7, 3635 10.1038/s41598-017-03783-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu N., Wang J., Wang J., Wang R., Liu Z., Yu Y. et al. (2013) ING5 is a Tip60 cofactor that acetylates p53 in response to DNA damage. Cancer Res. 73, 3749–3760 10.1158/0008-5472.CAN-12-3684 [DOI] [PubMed] [Google Scholar]
- 50.Gersbacher M.T., Goodger Z.V., Trutzel A., Bundschuh D., Nitsch R.M. and Konietzko U. (2013) Turnover of amyloid precursor protein family members determines their nuclear signaling capability. PLoS ONE 8, e69363 10.1371/journal.pone.0069363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Rotz R.C., Kohli B.M., Bosset J., Meier M., Suzuki T., Nitsch R.M. et al. (2004) The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J. Cell Sci. 117, 4435–4448 10.1242/jcs.01323 [DOI] [PubMed] [Google Scholar]
- 52.Cao X. and Sudhof T.C. (2001) A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 10.1126/science.1058783 [DOI] [PubMed] [Google Scholar]
- 53.Hass M.R. and Yankner B.A. (2005) A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J. Biol. Chem. 280, 36895–36904 10.1074/jbc.M502861200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y., Jiang X., Chen S., Fernandes N. and Price B.D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 10.1073/pnas.0504211102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hicks D.A., Makova N.Z., Gough M., Parkin E.T., Nalivaeva N.N. and Turner A.J. (2013) The amyloid precursor protein represses expression of acetylcholinesterase in neuronal cell lines. J. Biol. Chem. 288, 26039–26051 10.1074/jbc.M113.461269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pierrot N., Tyteca D., D’Auria L., Dewachter I., Gailly P., Hendrickx A. et al. (2013) Amyloid precursor protein controls cholesterol turnover needed for neuronal activity. EMBO Mol. Med. 5, 608–625 10.1002/emmm.201202215 [DOI] [PMC free article] [PubMed] [Google Scholar]