Abstract
Background
Paeoniae Radix Alba, the root of the plant Paeonia lactiflora Pall, is a common blood-enriching drug in traditional Chinese medicine. Its effectiveness in the clinical treatment of anaemia is remarkable, but its potential pharmacologic mechanism has not been clarified.
Methods
In this study, the potential pharmacologic mechanism of Paeoniae Radix Alba in the treatment of iron-deficiency anaemia was preliminarily elucidated through systematic and comprehensive network pharmacology.
Results
Specifically, we obtained 15 candidate active ingredients from among 146 chemical components in Paeoniae Radix Alba. The ingredients were predicted to target 77 genes associated with iron-deficiency anaemia. In-depth analyses of these targets revealed that they were mostly associated with energy metabolism, cell proliferation, and stress responses, suggesting that Paeoniae Radix Alba helps alleviate iron-deficiency anaemia by affecting these processes. In addition, we conducted a core target analysis and a cluster analysis of protein-protein interaction (PPI) networks. The results showed that four pathways, the p53 signalling pathway, the IL-17 signalling pathway, the TNF signalling pathway and the AGE-RAGE signalling pathway in diabetic complications, may be major pathways associated with the ameliorative effects of Paeoniae Radix Alba on iron-deficiency anaemia. Moreover, molecular docking verified the credibility of the network for molecular target prediction.
Conclusions
Overall, this study predicted the functional ingredients in Paeoniae Radix Alba and their targets and uncovered the mechanism of action of this drug, providing new insights for advanced research on Paeoniae Radix Alba and other traditional Chinese medicines.
Keywords: Paeoniae Radix Alba, Network pharmacology, Iron-deficiency anaemia
Background
Worldwide, 46% of children aged 5 to 14 years and 48% of pregnant women suffer from iron-deficiency anaemia (IDA) [1, 2]. IDA is one of the most widespread nutritional deficiency diseases [3] and can cause cognitive deficiency and irreversible auditory and visual system damage in infants [4, 5]. Pregnant women with anaemia may give birth to infants with foetal dysplasia and low birth weight [6, 7]. Iron supplements are widely used to treat IDA [8]. However, long-term use of supplements containing ferrous salts can cause side effects such as epigastric pain, diarrhoea and constipation [9, 10]. Thus, identification of a good alternative supplement with fewer side effects has become an important research objective.
In China, traditional Chinese medicine (TCM) is not simply a cultural practice, it also has a history of thousands of years of use for the treatment of various diseases. Under the guidance of the overall concepts and principles of syndrome differentiation and treatment, TCM has achieved satisfactory clinical results for anaemia treatment. Paeoniae Radix Alba (PRA), the root of the plant Paeonia lactiflora Pall (family Ranunculaceae), is a TCM with the functions of nourishing blood, astringing Yin, preventing perspiration, regulating menstruation, extinguishing liver wind and relieving pain [11, 12].
In recent years, an unconventional novel analytical technique called network pharmacology has been widely used in TCM research [13–16]. Combined with extensive data analysis, network pharmacology can systematically determine the effects and mechanisms of drugs employed to treat complex diseases at the molecular, cellular, tissue, and biological levels [17]. Although PRA is noteworthy in treating anaemia, for IDA, the active compositions, drug targets, and exact molecular mechanism are still unclear [12, 18–20].
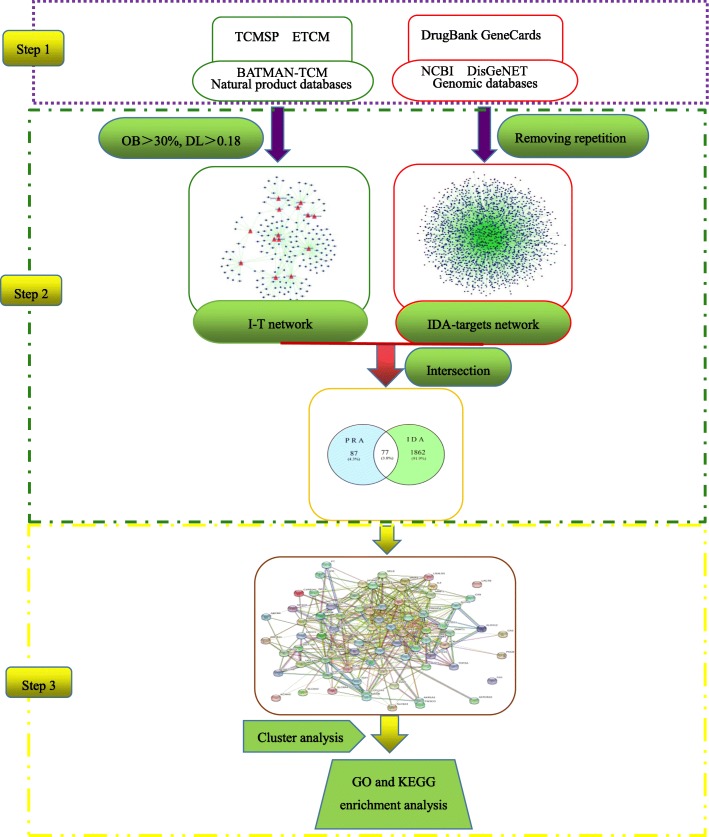
In this study, network pharmacology was utilized to analyse the active ingredients, drug targets and key pathways of PRA in the treatment of IDA, as shown in Fig. 1. This study provides a new perspective for studying the mechanisms of TCMs.
Fig. 1.
Diagram of the study design. Step 1: Gathered the chemical composition of PRA from three databases (TCMSP, ETCM, BATMAN-TCM), and collected IDA-related targets from four databases (DrugBank, GeneCards, NCBI, DisGeNET). Step 2: Select the candidate components and take the intersection of the component-target and the IDA-target. Step 3: KEGG and GO enrichment analysis
Methods
Data sources
Ingredients of PRA
Information on the chemical composition of PRA was gathered from three databases: the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, a unique pharmacologic platform for Chinese herbal medicine that can be used to search for the relationships among drugs, targets and diseases), the Encyclopedia of Traditional Chinese Medicine (ETCM, a database of commonly used herbs and herbal formulations that includes standardized information and ingredient information), and the Bioinformatics Analysis Tool for Molecular mechANisms of TCM (BATMAN-TCM, a biological online networking tool that provides users with basic information about herbs, such as their ingredients, targets, and disease relationships) [21–23]. In this experiment, “baishao” was invoked as the keyword, and the structures of the components were saved in MOL2 format. The structures of the components were verified with ChemSpider and SciFinder.
IDA-related targets
Targets of IDA were identified with four databases: GeneCards (a searchable, free, and comprehensive database that provides users with ample information for annotating and predicting human genes), DrugBank (a comprehensive, and freely accessible online database that includes information on drugs and drug targets), DisGeNET (a platform to explore the relationships between genes and diseases), and the NCBI database (National Center for Biotechnology Information, a platform integrating the PubMed, Bookshelf, Blast, Genome and other databases; users can search different targets through different databases of the platform) [24–26]. In this study, we used “iron-deficiency anaemia” as a keyword, searched the “gene” database type, and limited the species to “Homo sapiens” to identify IDA-related genes.
Data preprocessing
Screening of active ingredients of PRA
Screening of dynamic components can be conducted on the basis of the five rules of Lipinski: a molecular weight (MW) < 500, a hydrogen acceptor number ≤ 10, a hydrogen donor (HDon) number ≤ 5, a log P value of − 2 ~ 5, and a rotatable hydrogen bond number (RBN) ≤ 10. If a compound does not violate two or more of the above principles, it can be considered that the compound has soothing properties [27]. Functional components can also be screened according to oral bioavailability (OB; > 30%) and drug likeness (DL; > 0.18) [28]. The OB value is an important indicator for evaluation of the internal conversion of drugs [29]; a higher OB value of a drug is associated with a higher utilization rate of the drug after oral administration and a greater possibility of clinical application [30, 31]. In this experiment, an OB > 30% and a DL > 0.18 were utilized as the criteria for screening of functional components. A literature review was used to supplement the information on the active components.
Target prediction for the bioactive ingredients of PRA
At present, the methods and techniques of drug target prediction can be divided into four parts according to their principles: (1) ligand prediction based on chemical structure similarity and pharmacophore models; (2) machine intelligence learning and prediction, for which standardized names and clear molecular target correspondence are required; (3) molecular docking, in which receptors are used to make predictions; and (4) combined prediction [32]. Building on the current conditions and constraints, we selected ligand prediction as the main method and supplemented it with data from DrugBank.
First, the TCMSP, BATMAN-TCM, and ETCM were chosen as databases for the chemical components of PRA. The active components were screened with the criteria of an OB > 30% and a DL > 0.18, and the component targets were then rigorously predicted by SwissTargetPrediction (which compares the components to a library containing 28,000 compounds by two-dimensional and three-dimensional similarity and further predicts any applicable molecular targets from among more than 2000 targets in five different organisms) and Stitch (which can randomly select at least four predicted linked proteins based on a single protein name, multiple protein names, or amino acid sequences with moderate or better confidence) [33]. In addition, listed or laboratory-verified targets in DrugBank were identified as supplementary data. In this experiment, we converted the dynamic component into the “SMILES” format and selected Homo sapiens as the species. To ensure the accuracy of the results, we used P > 0.5 as the constraint condition for the predicted targets, yielding moderate credibility. After that, we obtained the drug active ingredient targets.
Second, to consolidate and standardize the data, we identified the gene names of the predicted target proteins with Universal Protein (UniProt), a comprehensive resource of protein sequences and annotation data. UniProt is a compilation of the UniProt Knowledgebase, the UniProt Reference Cluster, and the UniProt Archive [34]. We restricted the species to humans and created a protein-gene document.
Finally, to identify the universal targets between IDA and PRA, we uploaded the two target networks to the Venny 2.1 online server and obtained 77 common targets (Table 2) [35]. We then used the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) online tool to carry out Gene Ontology (GO) analysis on the shared targets for the Biological Process (BP), Cellular Component (CC), Molecular Function (MF) GO categories [36]. Enriched terms in the BP, CC, and MF categories were selected for display. Additionally, we utilized ClueGo in Cytoscape (v 3.6.1) for pathway analysis using data from the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Table 2.
Common targets between PRA and IDA
| NO. | Protein ID | Gene name | Protein name | Protein Class |
|---|---|---|---|---|
| 1 | P31645 | SLC6A4 | solute carrier family 6 member 4 | transporter |
| 2 | Q01959 | SLC6A3 | solute carrier family 6 member 3 | transporter |
| 3 | P23975 | SLC6A2 | solute carrier family 6 member 2 | transporter |
| 4 | P33527 | ABCC1 | ATP binding cassette subfamily C member 1 | transporter |
| 5 | P19793 | RXRA | retinoid X receptor alpha | nucleic acid binding; receptor; transcription factor |
| 6 | P03372 | ESR1 | estrogen receptor 1 | nucleic acid binding; receptor; transcription factor |
| 7 | P05412 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit | nucleic acid binding; transcription factor |
| 8 | P10275 | AR | androgen receptor | nucleic acid binding; receptor; transcription factor |
| 9 | P37231 | PPARG | peroxisome proliferator activated receptor gamma | nucleic acid binding; receptor; transcription factor |
| 10 | P27338 | MAOB | monoamine oxidase B | nucleic acid binding; oxidoreductase; transferase |
| 11 | P55055 | NR1H2 | nuclear receptor subfamily 1 group H member 2 | nucleic acid binding; receptor;transcription factor |
| 12 | O75469 | NR1I2 | nuclear receptor subfamily 1 group I member 2 | nucleic acid binding; receptor; transcription factor |
| 13 | P06746 | POLB | DNA polymerase beta | nucleic acid binding |
| 14 | O95342 | ABCB11 | ATP binding cassette subfamily B member 11 | hydrolase; protease |
| 15 | Q2M3G0 | ABCB5 | ATP binding cassettesubfamily B member 5 | hydrolase; protease |
| 16 | P22303 | ACHE | Acetylcholinesterase | hydrolase; protease |
| 17 | P03956 | MMP1 | matrix metallopeptidase 1 | hydrolase; protease |
| 18 | P00734 | F2 | coagulation factor II, thrombin | hydrolase; protease |
| 19 | P08253 | MMP2 | matrix metallopeptidase 2 | hydrolase; protease |
| 20 | P08183 | ABCB1 | ATP binding cassette subfamily B member 1 | hydrolase; protease |
| 21 | P08709 | F7 | coagulation factor VII | hydrolase; protease |
| 22 | P45983 | MAPK8 | mitogen-activated protein kinase 8 | kinase; transferase |
| 23 | Q13315 | ATM | ATM serine/threonine kinase | kinase;nucleic acid binding; transferase |
| 24 | P06493 | CDK1 | cyclin dependent kinase 1 | kinase; transferase |
| 25 | P48736 | PIK3CG | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma | kinase; transferase |
| 26 | P47989 | XDH | xanthine dehydrogenase | oxidoreductase |
| 27 | P04040 | CAT | catalase | oxidoreductase |
| 28 | P09917 | ALOX5 | arachidonate 5-lipoxygenase | oxidoreductase |
| 29 | P08684 | CYP3A4 | cytochrome P450 family 3 subfamily A member 4 | oxidoreductase |
| 30 | P16050 | ALOX15 | arachidonate 15-lipoxygenase | oxidoreductase |
| 31 | P23219 | PTGS1 | prostaglandin-endoperoxide synthase 1 | oxidoreductase |
| 32 | P05177 | CYP1A2 | cytochrome P450 family 1 subfamily A member 2 | oxidoreductase |
| 33 | P35354 | PTGS2 | prostaglandin-endoperoxide synthase 2 | oxidoreductase |
| 34 | Q16850 | CYP51A1 | cytochrome P450 family 51 subfamily A member 1 | oxidoreductase |
| 35 | P09601 | HMOX1 | heme oxygenase 1 | oxidoreductase |
| 36 | P18054 | ALOX12 | arachidonate 12-lipoxygenase, 12S type | oxidoreductase |
| 37 | P14550 | AKR1A1 | aldo-keto reductase family 1 member A1 | oxidoreductase |
| 38 | O60218 | AKR1B10 | aldo-keto reductase family 1 member B10 | oxidoreductase |
| 39 | P04141 | CSF2 | colony stimulating factor 2 | signaling molecule |
| 40 | P10415 | BCL2 | BCL2, apoptosis regulator | signaling molecule |
| 41 | P01375 | TNF | tumor necrosis facto | signaling molecule |
| 42 | P27487 | DPP4 | dipeptidyl peptidase 4 | enzyme modulator; hydrolase; protease |
| 43 | P05121 | SERPINE1 | serpin family E member 1 | enzyme modulator |
| 44 | P42574 | CASP3 | caspase 3 | enzyme modulator; hydrolase; protease |
| 45 | P07550 | ADRB2 | adrenoceptor beta 2 | receptor |
| 46 | P25105 | PTAFR | platelet activating factor receptor | receptor |
| 47 | P08238 | HSP90AB1 | heat shock protein 90 alpha family class B member 1 | chaperone |
| 48 | P04637 | TP53 | tumor protein p53 | transcription factor |
| 49 | P17931 | LGALS3 | galectin 3 | cell adhesion molecule; signaling molecule |
| 50 | P05231 | IL6 | interleukin 6 | None |
| 51 | P00918 | CA2 | carbonic anhydrase 2 | None |
| 52 | P00915 | CA1 | carbonic anhydrase 1 | None |
| 53 | P01130 | LDLR | low density lipoprotein receptor | None |
| 54 | P11388 | TOP2A | DNA topoisomerase II alpha | None |
| 55 | P35503 | UGT1A3 | UDP glucuronosyltransferase family 1 member A3 | None |
| 56 | O60656 | UGT1A9 | UDP glucuronosyltransferase family 1 member A9 | None |
| 57 | P04035 | HMGCR | 3-hydroxy-3-methylglutaryl -CoA reductase | None |
| 58 | P10636 | MAPT | microtubule associated protein tau | None |
| 59 | P26358 | DNMT1 | DNA methyltransferase 1 | None |
| 60 | P35228 | NOS2 | nitric oxide synthase 2 | None |
| 61 | P05093 | CYP17A1 | Cytochrome P450 family 17 subfamily A member 1 | None |
| 62 | P16581 | SELE | selectin E | None |
| 63 | P04114 | APOB | apolipoprotein B | None |
| 64 | P17612 | PRKACA | protein kinase cAMP-activated catalytic subunit alpha | None |
| 65 | O00255 | MEN1 | menin 1 | None |
| 66 | P08700 | IL3 | interleukin 3 | None |
| 67 | P19320 | VCAM1 | vascular cell adhesion molecule 1 | None |
| 68 | Q9HAW9 | UGT1A8 | UDP glucuronosyl transferase family 1member A8 | None |
| 69 | P29474 | NOS3 | nitric oxide synthase 3 | None |
| 70 | Q16790 | CA9 | carbonic anhydrase 9 | None |
| 71 | P83111 | LACTB | lactamase beta | None |
| 72 | P06276 | BCHE | butyrylcholinesterase | None |
| 73 | P09211 | GSTP1 | glutathione S-transferase pi 1 | None |
| 74 | P06213 | INSR | insulin receptor | None |
| 75 | P14679 | TYR | tyrosinase | None |
| 76 | Q12809 | KCNH2 | potassium voltage-gated channel subfamily H member 2 | None |
| 77 | P27169 | PON1 | paraoxonase 1 | None |
Protein-protein interaction (PPI) network construction for IDA and PRA
Physiological processes are not only affected by single signals; rather, the expression and function of a gene/protein are often impacted by multiple genes [37]. PPI networks are interaction networks between targets and proteins [38]. We uploaded the obtained targets to the tool of Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) Version 11.0 to develop a PPI network. STRING calculates comprehensive scores and probabilities based on distinct lines of evidence and corrects for the probabilities of random interactions. A minimum score of 0.4 indicates moderate credibility, a minimum score of 0.7 indicates high credibility, and a minimum score of 0.9 indicates the highest credibility [39]. In this study, we constructed a PPI network with a score of 0.4; thus, it was a moderate-credibility network.
Cluster analyses for the PPI network
Clustering refers to the identification of highly correlated groups of different compounds or objects with similar basic attributes [40]. Cluster analysis, an important classification method, indicates the effectiveness of the classification used for the PPI network. Many algorithms for Cytoscape PPI network clustering analysis have been reported, but previous studies have shown that the molecular complex detection (MCODE) algorithm for protein complex detection is the most reliable for gene network module analysis [41, 42]; thus, we choose MCODE for our PPI network cluster analysis.
GO function and KEGG pathway enrichment analyses
To determine the commonality among targets, the GO and KEGG pathways of clustered targets are commonly analysed [42]. WebGestalt is a feature-rich web analytics tool; as of 14 January 2019, it covers 354 databases that support 12 organisms and 321,251 functional classifications. It also enables analysis of genes that are not in the database and of data from experimental organisms on the platform [36]. In this study, we examined the enriched GO functions for each target classification and conducted KEGG pathway analysis of the targets with WebGestalt.
Drug-ingredient-target-pathway-disease (D-I-T-P-D) network construction
The network obtained from the above experiment was introduced into Cytoscape (v 3.6.1), and the “merge” tool was utilized to merge the network. The D-I-T-P-D network was obtained.
Molecular docking verification
The LibDock module of Discovery Studio 2016 was used to verify the molecular docking based on the functional components of PRA, and a heat map was constructed from the component-core target docking scores.
Results
Component-target networks of PRA
We collected data on the chemical components of PRA from three databases, namely, the TCMSP, ETCM, and BATMAN-TCM. The numbers of chemical components derived from these three databases were 85, 59, and 35, respectively. After removing duplicates, we obtained 146 chemical components. Through screening of OB values and DL values, 13 qualified chemical components were obtained (Table 1). The literature shows that albiflorin and paeoniflorin are the active ingredients of PRA responsible for its ameliorative effects on anaemia [20], and gallic acid has anti-inflammatory, antioxidant and antitumour effects [43]. Therefore, these chemical components were also considered candidate components, and their structures were verified with SciFinder and ChemSpider.
Table 1.
Candidate active ingredients of PRA
| NO. | ID | Name | OB% | DL |
|---|---|---|---|---|
| 1 | MOL001910 | 11alpha,12alpha-epoxy-3beta-23-dihydroxy-30-norolean-20-en-28,12beta-olide | 64.77 | 0.38 |
| 2 | MOL001919 | (3S,5R,8R,9R,10S,14S)-3,17-dihydroxy-4,4,8,10,14-pentamethyl-2,3,5,6,7,9-hexahydro-1H-cyclopenta [a]phenanthrene-15,16-dione | 43.56 | 0.53 |
| 3 | MOL001918 | paeoniflorigenone | 87.59 | 0.37 |
| 4 | MOL001921 | Lactiflorin | 49.12 | 0.8 |
| 5 | MOL001924 | paeoniflorin | 53.87 | 0.79 |
| 6 | MOL001925 | paeoniflorin_qt | 68.18 | 0.4 |
| 7 | MOL001928 | albiflorin_qt | 66.64 | 0.33 |
| 8 | MOL001930 | benzoylpaeoniflorin | 31.27 | 0.75 |
| 9 | MOL000211 | Mairin | 55.38 | 0.78 |
| 10 | MOL000358 | beta-sitosterol | 36.91 | 0.75 |
| 11 | MOL000359 | sitosterol | 36.91 | 0.75 |
| 12 | MOL000422 | kaempferol | 41.88 | 0.24 |
| 13 | MOL000492 | (+)-catechin | 54.83 | 0.24 |
| 14 | MOL001927 | Albiflorin | 12.09 | 0.77 |
| 15 | MOL000513 | gallic acid | 31.69 | 0.04 |
To more intuitively indicate the relationships between components and targets, we constructed a component-target network diagram with Cytoscape (v 3.6.1) [44] that contained 178 nodes and 264 edges. In this network diagram, we found that the median degree of connectivity among 12 components was greater than 6; specifically, kaempferol, beta-sitosterol, (+)-catechin and gallic acid exhibited 61, 38, 32 and 32 degrees of connectivity, respectively, indicating that these four components are important active ingredients in PRA.
Target networks associated with IDA
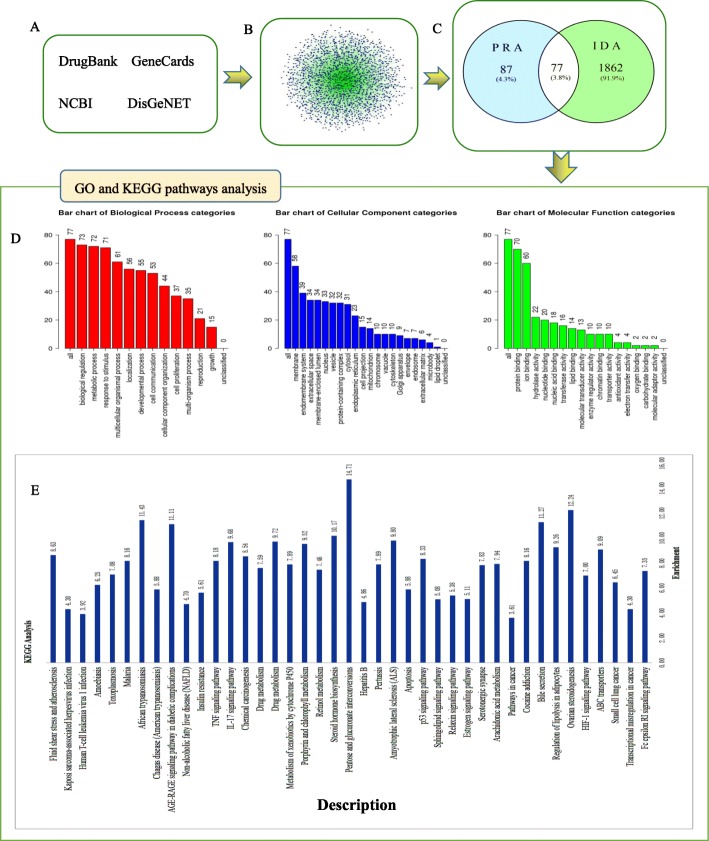
The development of a disease is usually associated with multiple genes or proteins, as is the case for IDA. In this study, we identified 1923, 60, 25 and 29 IDA-related genes from the GeneCards, DisGeNET, NCBI and DrugBank databases, respectively. Duplicates were removed, and 1939 related genes were obtained. A total of 77 genes were shared between PRA targets and IDA-related genes (Table 2). To investigate the relationships between the 77 common targets and IDA, we conducted GO and KEGG analyses of the shared targets (Fig. 2). Ultimately, we obtained 12 enriched BP terms, 19 enriched CC terms, 16 enriched MF terms and 40 enriched KEGG pathways. The BP category results mainly indicated enrichment for the biological regulation (73/77), metabolic process (72/77), response to stimulus (71/77), multicellular organismal process (76/77), localization (56/77), developmental process (55/77), and cell communication (53/77) terms. The membrane (58/77), endomembrane system (39/77) and membrane-enclosed lumen (34/77) terms were significantly enriched in the CC category. The protein binding (70/77) and ion binding (60/77) terms were the primary enriched MF terms identified in our study. In addition, nitrogen metabolism was the most significantly enriched pathway. This suggests that nitrogen metabolism may be the core process affecting IDA. IDA development has been found to play roles in a variety of diseases, such as hepatitis B, amoebiasis, toxoplasmosis, malaria, African trypanosomiasis, and prostate cancer, suggesting that IDA may be affected by one or more diseases. The NF-kappa B signalling pathway, the HIF-1 signalling pathway, the AGE-RAGE signalling pathway in diabetic complications, the pentose and glucuronate interconversion pathways, and the IL-17 signalling pathway were also identified in this study. The results show that the TCM PRA affects multiple pathways and processes in the context of IDA treatment.
Fig. 2.
IDA-related target network. a Four disease-related gene target databases. b IDA target network containing 1940 nodes and 1939 edges. c Seventy-seven common targets between IDA and PRA. d GO and e KEGG pathway enrichment analysis results for PRA-targeted genes associated with IDA
PRA-IDA PPI networks
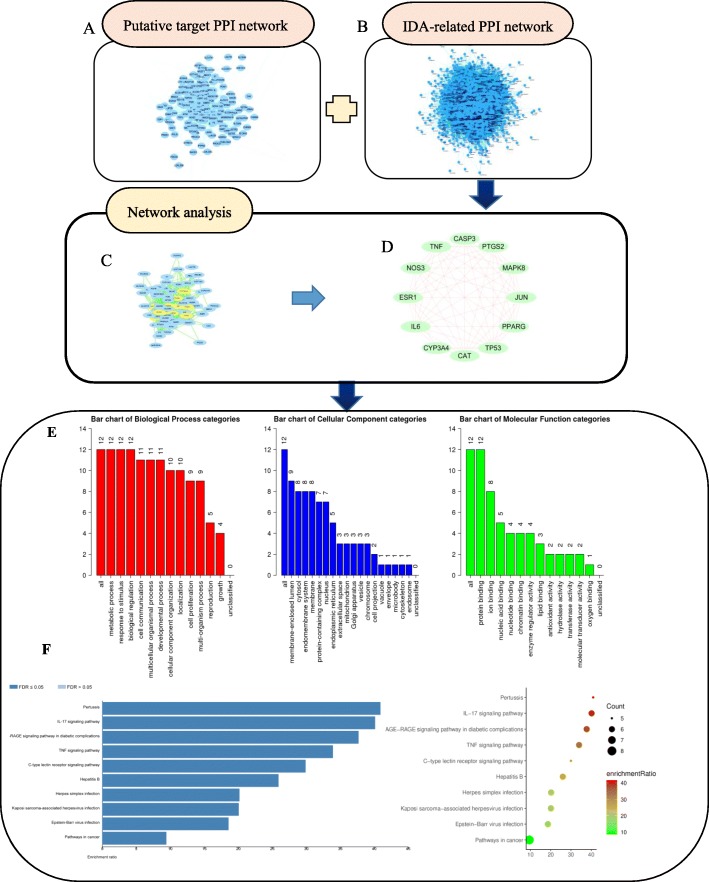
To develop a better understanding of the association between PRA and IDA, we analysed the relationships between them through assessment of their core targets. The screening condition of a degree centrality (DC) > 2× the average degree for the core targets yielded 12 strategic targets. The results of the GO function and KEGG pathway enrichment analyses were very similar to the enrichment results for the 77 targets (Fig. 3). The top eight functional terms were the biological regulation (12/12), metabolic process (12/12), response to stimulus (12/12), membrane-enclosed lumen (9/12), cytosol (8/12), endomembrane system (8/12), protein binding (12/12), and ion binding (8/12) terms. These enriched terms were highly correlated with anti-inflammatory activity, especially in the context of chronic or allergic rhinitis.
Fig. 3.
Enrichment analysis of 12 core targets. a Putative target PPI network of PRA. b IDA-related PPI network. c and d Analysis network; targets with DC values > 27.789 were considered core targets. e GO and f KEGG pathway analysis results
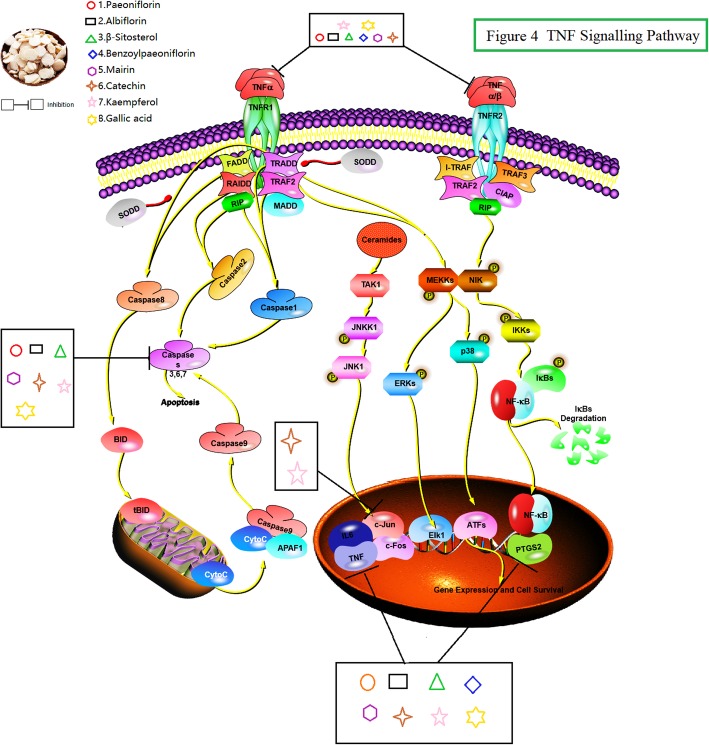
The 12 targets were enriched in 10 KEGG pathways with significant false discovery rate (FDR)-adjusted P-values, including pertussis and the TNF signalling pathway (Fig. 4). The details of the KEGG pathways are outlined in Additional file 3.
Fig. 4.
TNF signalling pathway. As shown in the figure, PRA may treat IDA by inhibiting the TNF signalling pathway
Enrichment analyses of 75 core targets
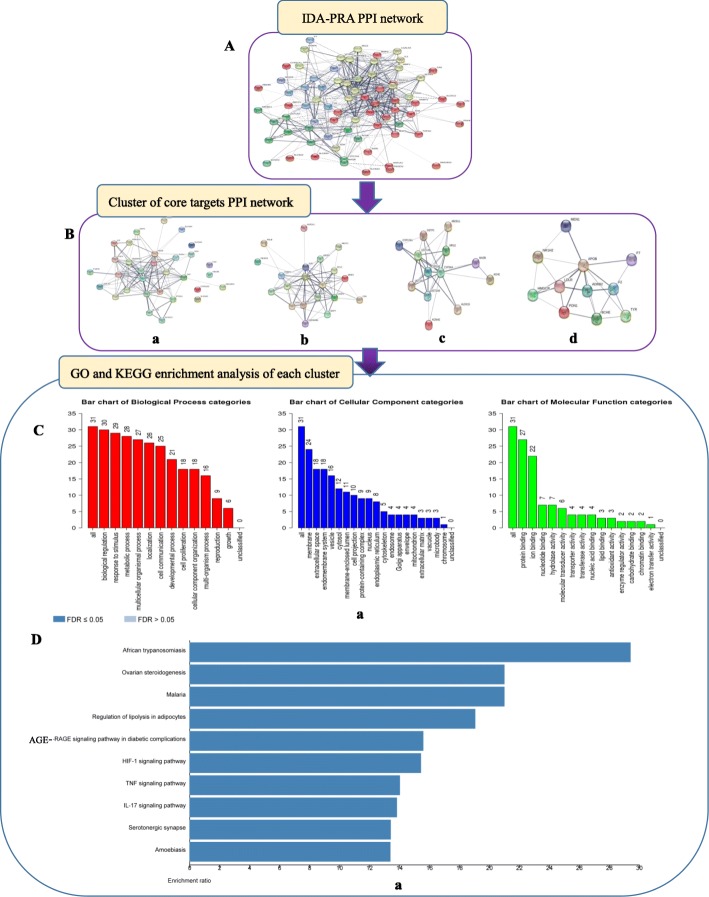
The PPI network comparison of IDA and PRA revealed 75 core targets. To elucidate the biological functions of these targets, we divided the 75 targets into four clusters and subjected them to GO and KEGG pathway analyses (Fig. 5). Based on the GO term results, we found that biological regulation-related processes, such as gene expression, smooth muscle cell proliferation, and nitric oxide biosynthesis; metabolic processes, such as aerobic metabolism and steroid metabolism; responses to stimuli such as hypoxia, oestradiol, and lipopolysaccharide; and other processes, such as enzyme binding, protein homodimerization activity, iron ion binding, RNA polymerase II transcription factor activity, and ligand-activated sequence-specific DNA binding, were enriched for our clusters, suggesting that PRA may help alleviate IDA by affecting enzymes, iron ion binding, stress responses and nitric oxide biosynthesis.
Fig. 5.
Enrichment analysis of 75 core targets. A IDA-PRA PPI network. B Clusters of the PPI network. C GO and D KEGG pathway analysis results for each cluster. Due to space constraints, other Cluster analysis (b, c, d) is shown in file 3
The occurrence and development of diseases can be affected by other diseases and processes. In our study, we found that African trypanosomiasis, malaria, amoebiasis, colorectal cancer, pertussis, hepatitis B, serotonergic synapse-related processes, ovarian steroidogenesis, apoptosis, and cell proliferation could also indirectly affect the development of IDA, suggesting that PRA alleviates IDA by affecting cell-, nerve-, and inflammation-related processes.
In addition, we believe that the top four KEGG pathways identified for these clusters, namely, the HIF-1 signalling pathway, the AGE-RAGE signalling pathway in diabetic complications, the TNF signalling pathway, and the IL-17 signalling pathway, might play significant roles in IDA treatment.
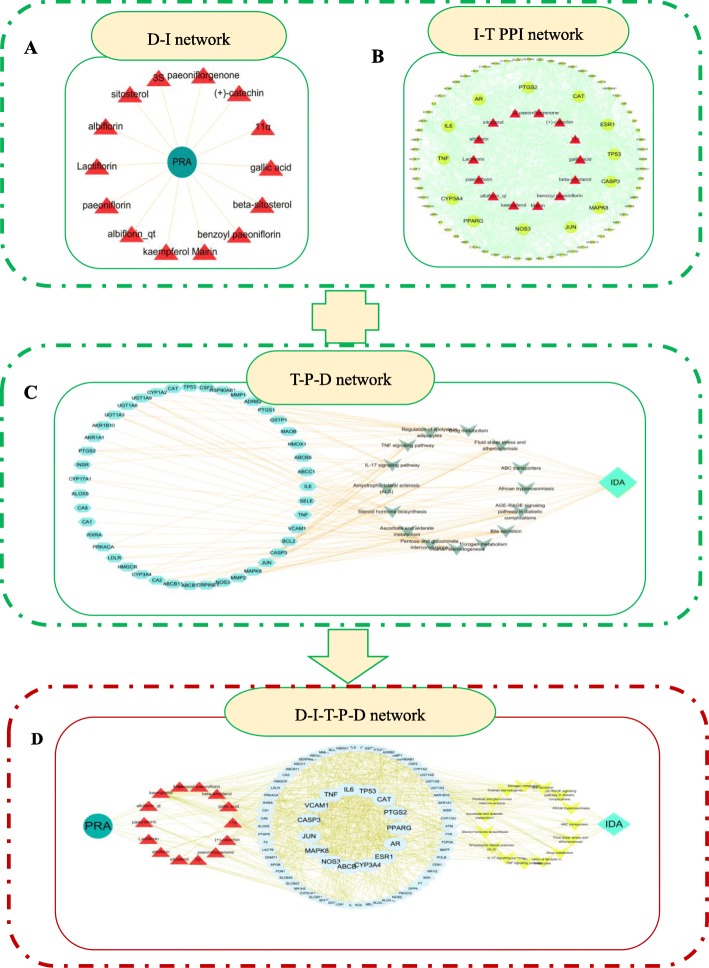
D-I-T-P-D network construction
On the basis of the PPI targets and pathway analyses, a D-I-T-P-D network was constructed using Cytoscape (v 3.6.1). As illustrated in Fig. 6, this D-I-T-P-D network had 108 nodes and 785 edges. The dark cyan circles, red triangles, celadon ellipses, yellow inverted triangles, and cyan diamond represent PRA, PRA ingredients, target genes, pathways, and IDA, respectively.
Fig. 6.
Construction of the D-I-T-P-D network. a Ingredient-PRA (ingredient-drug [I-D]) network. b Ingredient-target (I-T) network. c Target-pathway-disease (T-P-D) network. d D-I-T-P-D network containing 108 nodes and 785 edges
Component-core target docking scores
The docking score diagram is given in Fig. 7a. Notably, each active component and core target had good docking ability. PPARG, CYP3A4, and TNF may be the most important targets. Paeoniflorin, benzoylpaeoniflorin, and albiflorin may be the components associated with the ameliorative effects of PRA on IDA, which further confirms the reliability of the network pharmacology. MOL001910 and MOL001919 were replaced by 11α and 3S, respectively. The ligands paeoniflorigenone and lactiflorin could not be constructed, so docking verification was not carried out. Example of molecular docking: benzoylpaeoniflorin-TNF, Fig. 7b.
Fig. 7.
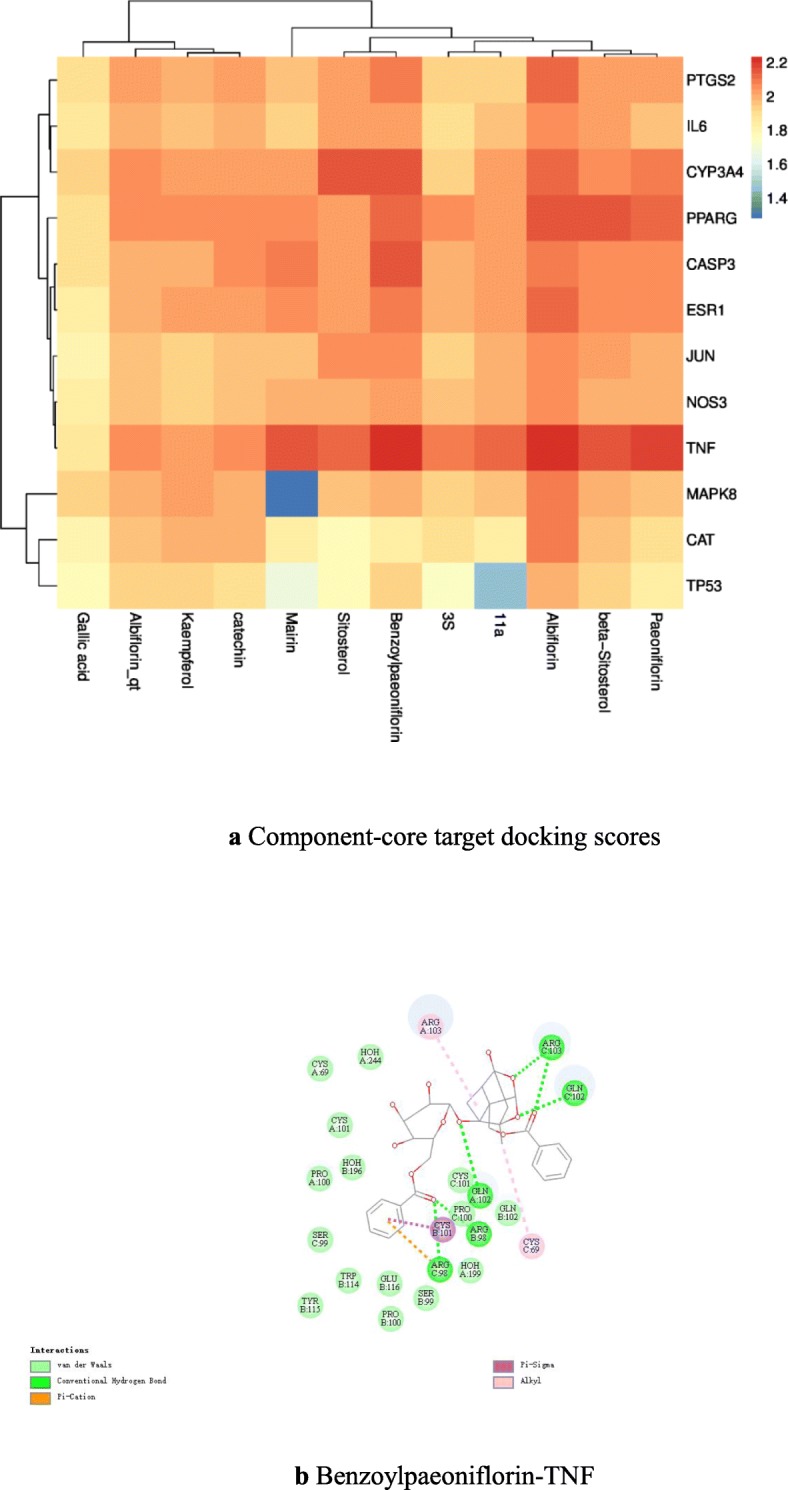
a Component-core target docking scores. In this heat map, the docking score between the component and the target is converted by Log10, and the color is closer to red, indicating that the docking score between the component and the target is high and the docking strength is high. b benzoylpaeoniflorin-TNF. A two-dimensional diagram of the interaction between benzoylpaeoniflorin and TNF. Most of the oxygen atoms in benzoylpaeoniflorin form hydrogen bonds with ARG98, ARG103, GLN102, and some benzene rings form Pi-Sigma with CYS101
Discussion
IDA is a condition that occurs among children in both developing and developed countries, leading to impaired development, activity intolerance, behavioural changes, irritability, and reduced learning ability; women and elderly individuals are also affected [45, 46]. The iron deficiency associated with IDA can be classified as either absolute or functional iron deficiency: absolute iron deficiency is defined as a severe reduction in or loss of iron reserves in the bone marrow, liver, and spleen, while functional iron deficiency is defined as deficiency due to inadequate intake, malabsorption, or metabolic disorders [47–49]. Iron is an important component of human metabolism that plays crucial roles in cellular respiration, DNA synthesis, cell proliferation and oxygen storage [50]. Disorders in iron absorption and metabolism result in severe oxidative stress and tissue damage [51]. PRA is a blood tonic drug that regulates menstruation, but its precise mechanism of action is not yet clear [11]. Therefore, it is imperative to explore the mechanism of PRA in IDA treatment by using network pharmacology methods combined with functional ingredient screening, drug target prediction, and network and pathway analyses.
In this study, we identified 77 common targets between PRA and IDA. These targets were mainly enriched for energy metabolism-, cell proliferation-, and apoptosis-related terms. The main properties of these targets were associated with nucleic acid binding, receptors, and transcription factors (9/77); hydrolases and proteases (8/77); and oxidoreductases (13/77). Many other targets had unknown or other attributes. We reviewed several literature sources and found that most of the components downregulate TNF, IL6, PTGS2, CYP3A4, and CASP3 and upregulate PPARG and CAT, further indicating the reliability of the target prediction, as detailed in Table 3. In addition, we found that IL6 can induce the growth of myeloma and plasma cell tumours and induce the differentiation of nerve cells and that its overexpression in inflammation sites is a main cause of anaemia and chronic inflammation [88, 89]. TP53, which is involved in a variety of cell death pathways and can be used as a marker of neuronal injury, has been widely studied in the context of cancer treatment [90, 91]. TNF and IL6 are involved in the occurrence and development of chronic anaemia/IDA, and the level of TNF expression can reflect the degree of disease of patients with aplastic anaemia [92–94]. IL3 and TNF participate in relieving blood deficiencies in mice caused by cyclophosphamide [19].
Table 3.
Correlations between components and targets
| Components | Downregulate | Upregulate | References |
|---|---|---|---|
| Paeoniflorin | TNF,IL6,PTGS2,CYP3A4,CASP3 | PPARG,CAT | [52–58] |
| Albiflorin | TNF,IL6,PTGS2,CYP3A4,CASP3 | NOS3 | [52, 54, 57, 59] |
| β-Sitosterol | IL6,PTGS2,TNF,CYP3A4,CASP3 | PPARG,CAT,NOS3 | [60–64] |
| Mairin | PTGS2,TNF,CYP3A4,IL6,CASP3, | PPARG,CAT | [65–69] |
| (+)-Catechin | TNF,PTGS2,CYP3A4,IL6,CASP3,JUN | PPARG,ESR1,TP53 | [70–76] |
| Kaempferol | PTGS2,CYP3A4,TNF,IL6,CASP3,JUN | PPARG,CAT | [77–82] |
| Gallic acid | PTGS2,CYP3A4,IL6,TNF,CASP3 | PPARG,CAT,NOS3 | [61, 83–87] |
| Benzoylpaeoniflorin | PTGS2,IL6,TNF | [53, 84] |
In addition, we compared the KEGG and GO results for the 12 core targets, the 75 targets revealed in the cluster analyses, and the common targets, and the results showed an IDA correlation. We believe that the processes of nitric oxide biosynthesis, arachidonic acid metabolism, and Th17 cell differentiation play important roles in IDA [95–97]. Effects on gene expression, steroid metabolism, hypoxia responses, and protein homodimerization activity may be responsible for a large proportion of the treatment effects.
Subsequently, we searched for the top four KEGG pathways and identified the p53 signalling pathway, the IL-17 signalling pathway, the TNF signalling pathway, and the AGE-RAGE signalling pathway in diabetic complications. We concluded that these pathways are involved in the treatment of anaemia [98–101]. Interestingly, toxoplasmosis, malaria, African trypanosomiasis, hepatitis B and prostate cancer are all associated with anaemia [102–106], demonstrating that the progression of this disease is influenced by other diseases. To some extent, these results also indicate that PRA contains multiple active components and can achieve multiple objectives through multiple pathways. The findings also serve as a basis for investigation into the collaboration among multiple components in follow-up research.
Conclusions
In general, we elucidated the multi-component, multi-target, and multi-pathways characteristics of PRA in the context of IDA treatment through network pharmacology. Four pathways, the p53 signalling pathway, the IL-17 signalling pathway, the TNF signalling pathway, and the AGE-RAGE signalling pathway in diabetic complications, were identified, providing new directions for the development of drugs for IDA.
Supplementary information
Additional file 1: Table 1. Candidate active ingredients of PRA; Table 2. Common targets between PRA and IDA; Table 3. Correlations between components and targets.
Additional file 2: File 2a: Table 1 Known therapeutic targets for IDA; File 2b: Table 2 Targets for PRA.
Additional file 3. File for all pictures.
Additional file 4. File for partial targets with components docking.
Acknowledgements
Special thanks to Jiangxi University of traditional Chinese medicine for the support of this research.
Abbreviations
- IDA
Iron-deficiency anaemia
- TCM
Traditional Chinese medicine
- PRA
Paeoniae Radix Alba
- TCMSP
Traditional Chinese Medicine Systems Pharmacology database and analysis platform
- PPI
Protein-protein interaction network
- ETCM
Encyclopedia of Traditional Chinese Medicine
- BATMAN-TCM
Bioinformatics Analysis Tool for Molecular mechANisms of TCM
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- OB
Oral bioavailability
- DL
Drug likeness
- UniProt
Universal Protein
- NCBI
National Center for Biotechnology Information
- DC
Degree centrality
- GO
Gene ontology
- BP
Biological Process
- CC
Cellular Component
- MF
Molecular Function
- KEEG
Kyoto encyclopedia of genes and genome
- MCODE
Molecular complex detection
- WebGestalt
WEB-based Gene SeT AnaLysis Toolkit
Authors’ contributions
YXW and ZJL conceived and designed the experiments; YXW, DYL, and XLT performed the experiments and wrote the paper; RHM analyzed the data. All authors have read and approved the final manuscript.
Funding
This study was supported by National Key R&D Program - Special Topics for Modernization of Traditional Chinese Medicine (No. 2018YFC1707206), Key R & D Program of Jiangxi Province (No. 20192BBG70073), these funds provide project funding for PRA research. Doctoral Research start-up Fund (No. 2018WBZR009), and Jiangxi Province’s double first-class discipline (Traditional Chinese Medicine) construction project (NO. JXSYLXK-ZHYAO039/141), these funds provide financial support for the study of network pharmacology, expert consulting fees, and article publishing fees.
Availability of data and materials
Datasets supporting the results of this article have been included in the additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12906-020-02925-4.
References
- 1.Jia N, Qiao H, Zhu W, Zhu M, Meng Q, Lu Q, Zu Y. Antioxidant, immunomodulatory, oxidative stress inhibitory and iron supplementation effect of Astragalus membranaceus polysaccharide-iron (III) complex on iron-deficiency anemia mouse model. Int J Biol Macromol. 2019;132:213–221. doi: 10.1016/j.ijbiomac.2019.03.196. [DOI] [PubMed] [Google Scholar]
- 2.Cottin SC, Gambling L, Hayes HE, Stevens VJ, McArdle HJ. Pregnancy and maternal iron deficiency stimulate hepatic CRBPII expression in rats. J Nutr Biochem. 2016;32:55–63. doi: 10.1016/j.jnutbio.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Lynch SR. The impact of iron fortification on nutritional anaemia. Best Pract Res Clin Haematol. 2005;18(2):333–346. doi: 10.1016/j.beha.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Collard KJ. Iron homeostasis in the neonate. Pediatrics. 2009;123(4):1208–1216. doi: 10.1542/peds.2008-1047. [DOI] [PubMed] [Google Scholar]
- 5.Algarin C, Peirano P, Garrido M, Pizarro F, Lozoff B. Iron deficiency anemia in infancy: long-lasting effects on auditory and visual system functioning. Pediatr Res. 2003;53(2):217–223. doi: 10.1203/01.PDR.0000047657.23156.55. [DOI] [PubMed] [Google Scholar]
- 6.Haider BA, Olofin I, Wang M, Spiegelman D, Ezzati M, Fawzi WW. Anaemia, prenatal iron use, and risk of adverse pregnancy outcomes: systematic review and meta-analysis. BMJ. 2013;346:f3443. doi: 10.1136/bmj.f3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahajan SD, Singh S, Shah P, Gupta N, Kochupillai N. Effect of maternal malnutrition and anemia on the endocrine regulation of fetal growth. Endocr Res. 2004;30(2):189–203. doi: 10.1081/ERC-200027380. [DOI] [PubMed] [Google Scholar]
- 8.Al Hassan NN. The prevalence of iron deficiency anemia in a Saudi University female students. J Microsc Ultrastruct. 2015;3(1):25–28. doi: 10.1016/j.jmau.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren F, Qian XH, Qian XL. Astragalus polysaccharide upregulates hepcidin and reduces iron overload in mice via activation of p38 mitogen-activated protein kinase. Biochem Biophys Res Commun. 2016;472(1):163–168. doi: 10.1016/j.bbrc.2016.02.088. [DOI] [PubMed] [Google Scholar]
- 10.Dias AM, Hussain A, Marcos AS, Roque AC. A biotechnological perspective on the application of iron oxide magnetic colloids modified with polysaccharides. Biotechnol Adv. 2011;29(1):142–155. doi: 10.1016/j.biotechadv.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Chinese Pharmacopoeia Commission . Pharmacopoeia of People's Republic of China. Beijing: Chemical Industry Press; 2015. [Google Scholar]
- 12.Qu SS, Zhang JJ, Huang YF, Li W, Zhu YL, Yang Q. Study on blood enriching effects of γ-ray radiation of paeoniflorin and albiflorin on mouse model of blood deficiency. Zhongguo Zhong Yao Za Zhi. 2014;39(15):2952–2955. [PubMed] [Google Scholar]
- 13.Ren G, Zhong Y, Ke G, Liu X, Li H, Li X, Zheng Q, Yang M. The mechanism of compound Anshen essential oil in the treatment of insomnia was examined by network pharmacology. Evid Based Complement Alternat Med. 2019;2019:9241403. doi: 10.1155/2019/9241403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G, Luo Z, Zhou Y, Zhang L, Wu Y, Ding L, Shi Y. Uncovering the pharmacological mechanism of Carthamus tinctorius L on cardiovascular disease by a systems pharmacology approach. Biomed Pharmacother. 2019;117:109094. doi: 10.1016/j.biopha.2019.109094. [DOI] [PubMed] [Google Scholar]
- 15.Yang SH, Zhang JL, Yan YQ, Yang M, Li C, Li JM, Zhong LY, Gong QF, Yu H. Network Pharmacology-Based Strategy to Investigate the Pharmacologic Mechanisms of Atractylodes macrocephala Koidz. for the Treatment of Chronic Gastritis. Front Pharmacol. 2020;10:1629. doi: 10.3389/fphar.2019.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang YL, Zhang XF, Zou JB, Shi YJ, Wang Y, Tai J, et al. Pharmacology mechanism of Flos magnoliae and Centipeda minima for treating allergic rhinitis based on pharmacology network. Drug Dev Ind Pharm. 2019;45(9):1547–1555. doi: 10.1080/03639045.2019.1635150. [DOI] [PubMed] [Google Scholar]
- 17.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 18.Zhu YL, Wang LY, Wang JX, Wang C, Wang CL, Zhao DP, Wang ZC, Zhang JJ. Protective effects of paeoniflorin and albiflorin on chemotherapy-induced myelosuppression in mice. Chin J Nat Med. 2016;14(8):599–606. doi: 10.1016/S1875-5364(16)30070-X. [DOI] [PubMed] [Google Scholar]
- 19.Zhu YL, Zhang JJ, Wang JX, Yang ZH, Huang YF, Qu SS, Zhou TT. Comparative study on effects of blood enriching on mouse model of blood deficiency syndrome induced by cyclophosphamide of albiflorin, paeoniflorin on levels of GM-CSF, IL-3 and TNF-α. Zhongguo Zhong Yao Za Zhi. 2015;40(2):330–333. [PubMed] [Google Scholar]
- 20.Zhang JJ, Huang YF, Wang LL, Li W, Wang JX, Wang C, Qu SS. Comparative study on effects of blood enriching on mouse model of blood deficiency syndrome induced by compound method of bleeding, starved feeding and exhausting of Paeoniae Radix Alba and Paeoniae Radix Rubra, paeoniflorin and albiflorin. Zhongguo Zhong Yao Za Zhi. 2013;38(19):3358–3362. [PubMed] [Google Scholar]
- 21.Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, Zhang XB, Zhang W, Li ZY, Zhou RR, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1):D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, Diao L, Gu J, Wang W, Li D, et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci Rep. 2016;6:21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzer G, Rosen R, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Iny Stein T, et al. The GeneCards suite: from gene data mining to disease genome sequence analysis. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 25.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, Centeno E, Garcia-Garcia J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Zhao P, Li Y, Tian Y, Wang Y. Systems pharmacology-based dissection of mechanism of Chinese medicinal formula Bufei Yishen as an effective treatment for chronic obstructive pulmonary disease. Sci Rep. 2015;5:15290. doi: 10.1038/srep15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, Duan J, Ling Y. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Wang J, Zhou W, Wang Y, Yang L. Systems approaches and polypharmacology for drug discovery from herbal medicines: an example using licorice. J Ethnopharmacol. 2013;146(3):773–793. doi: 10.1016/j.jep.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Alam MA, Al-Jenoobi FI, Al-Mohizea AM, Ali R. Understanding and managing oral bioavailability: physiological concepts and patents. Recent Pat Anticancer Drug Discov. 2015;10(1):87–96. doi: 10.2174/1574892809666140917103834. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Chen J, Xu X, Li Y, Zhao H, Fang Y, Li X, Zhou W, Wang W, Wang Y. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PLoS One. 2012;7(5):e37608. doi: 10.1371/journal.pone.0037608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. 2007-2015. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 36.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45(W1):W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosque G, Folch-Fortuny A, Pico J, Ferrer A, Elena SF. Topology analysis and visualization of Potyvirus protein-protein interaction network. BMC Syst Biol. 2014;8:129. doi: 10.1186/s12918-014-0129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Las RJ, Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6(6):e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu NQ, Hou JY, Ma GY, Liu JX. Network pharmacology identifies the mechanisms of action of Shaoyao Gancao decoction in the treatment of osteoarthritis. Med Sci Monit. 2019;25:6051–6073. doi: 10.12659/MSM.915821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Li C, Zhu Y, Sun L, Sun H, Liu Y, Zhang Z, Wang C. Integrating GO and KEGG terms to characterize and predict acute myeloid leukemia-related genes. Hematology. 2015;20(6):336–342. doi: 10.1179/1607845414Y.0000000209. [DOI] [PubMed] [Google Scholar]
- 42.Song W, Ni S, Fu Y, Wang Y. Uncovering the mechanism of maxing Ganshi decoction on asthma from a systematic perspective: a network pharmacology study. Sci Rep. 2018;8(1):17362. doi: 10.1038/s41598-018-35791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehraban Z, Ghaffari Novin M, Golmohammadi MG, Sagha M, Pouriran K, Nazarian H. Protective effect of gallic acid on apoptosis of sperm and in vitro fertilization in adult male mice treated with cyclophosphamide. J Cell Biochem. 2019;120(10):17250–17257. doi: 10.1002/jcb.28987. [DOI] [PubMed] [Google Scholar]
- 44.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18(2):623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nazari M, Mohammadnejad E, Dalvand S, Ghanei GR. Prevalence of iron deficiency anemia in Iranian children under 6 years of age: a systematic review and meta-analysis. J Blood Med. 2019;10:111–117. doi: 10.2147/JBM.S196102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Askar S, Deveboynu SN, Er H, Askar TK, Hismiogullari AA. Changes in pro-inflammatory cytokines and antimicrobial proteins in elderly women with iron deficiency anemia. Pak J Med Sci. 2019;35(2):298–301. doi: 10.12669/pjms.35.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gafter-Gvili A, Schechter A, Rozen-Zvi B. Iron deficiency Anemia in chronic kidney disease. Acta Haematol. 2019;142(1):44–50. doi: 10.1159/000496492. [DOI] [PubMed] [Google Scholar]
- 48.Auerbach M, Adamson JW. How we diagnose and treat iron deficiency anemia. Am J Hematol. 2016;91(1):31–38. doi: 10.1002/ajh.24201. [DOI] [PubMed] [Google Scholar]
- 49.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 50.Zager RA. Parenteral iron compounds: potent oxidants but mainstays of anemia management in chronic renal disease. Clin J Am Soc Nephrol. 2006;1(Suppl 1):S24–S31. doi: 10.2215/CJN.01410406. [DOI] [PubMed] [Google Scholar]
- 51.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of mammalian iron metabolism. Cell. 2010;142(1):24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 52.Wang QS, Gao T, Cui YL, Gao LN, Jiang HL. Comparative studies of paeoniflorin and albiflorin from Paeonia lactiflora on anti-inflammatory activities. Pharm Biol. 2014;52(9):1189–1195. doi: 10.3109/13880209.2014.880490. [DOI] [PubMed] [Google Scholar]
- 53.Zhu X, Fang ZH. New monoterpene glycosides from the root cortex of Paeonia suffruticosa and their potential anti-inflammatory activity. Nat Prod Res. 2014;28(5):301–305. doi: 10.1080/14786419.2013.858340. [DOI] [PubMed] [Google Scholar]
- 54.Gao LN, Zhang Y, Cui YL, Akinyi OM. Comparison of Paeoniflorin and Albiflorin on human CYP3A4 and CYP2D6. Evid Based Complement Alternat Med. 2015;2015:470219. doi: 10.1155/2015/470219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong P, Chi R, Zhang L, Wang N, Lu Y. Effects of paeoniflorin on tumor necrosis factor-α-induced insulin resistance and changes of adipokines in 3T3-L1 adipocytes. Fitoterapia. 2013;91:44–50. doi: 10.1016/j.fitote.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Hu PF, Chen WP, Bao JP, Wu LD. Paeoniflorin inhibits IL-1β-induced chondrocyte apoptosis by regulating the Bax/Bcl-2/caspase-3 signaling pathway. Mol Med Rep. 2018;17(4):6194–6200. doi: 10.3892/mmr.2018.8631. [DOI] [PubMed] [Google Scholar]
- 57.Xie P, Cui L, Shan Y, Kang WY. Antithrombotic effect and mechanism of Radix Paeoniae Rubra. Biomed Res Int. 2017;2017:9475074. doi: 10.1155/2017/9475074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai Z, Zhang X, Zhang Y, Li X, Xu J, Li X. The impact of Paeoniflorin on α-synuclein degradation pathway. Evid Based Complement Alternat Med. 2015;2015:182495. doi: 10.1155/2015/182495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu YJ, Mei Y, Shi XQ, Zhang YF, Wang XY, Guan L, Wang Q, Pan HF. Albiflorin ameliorates memory deficits in APP/PS1 transgenic mice via ameliorating mitochondrial dysfunction. Brain Res. 2019;1719:113–123. doi: 10.1016/j.brainres.2019.05.037. [DOI] [PubMed] [Google Scholar]
- 60.Lee IA, Kim EJ, Kim DH. Inhibitory effect of β-Sitosterol on TNBS-induced colitis in mice. Planta Med. 2012;78(9):896–898. doi: 10.1055/s-0031-1298486. [DOI] [PubMed] [Google Scholar]
- 61.Vijayakumar TM, Kumar RM, Agrawal A, Dubey GP, Ilango K. Comparative inhibitory potential of selected dietary bioactive polyphenols, phytosterols on CYP3A4 and CYP2D6 with fluorometric high-throughput screening. J Food Sci Technol. 2015;52(7):4537–4543. doi: 10.1007/s13197-014-1472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moustafa EM, Thabet NM. Beta-sitosterol up-regulated Paraoxonase-1 via peroxisome proliferator-activated receptor-γ in irradiated rats. Can J Physiol Pharmacol. 2017;95(6):661–666. doi: 10.1139/cjpp-2016-0397. [DOI] [PubMed] [Google Scholar]
- 63.Park C, Moon DO, Rhu CH, Choi BT, Lee WH, Kim GY. β-Sitosterol induces anti-proliferation and apoptosis in human leukemic U937 cells through activation of Caspase-3 and induction of Bax/Bcl-2 ratio. Biol Pharm Bull. 2007;30(7):1317–1323. doi: 10.1248/bpb.30.1317. [DOI] [PubMed] [Google Scholar]
- 64.Yin Y, Liu X, Liu J, Cai E, Zhu H, Li H, Zhang L, Li P, Zhao Y. Beta-Sitosterol and its derivatives repress lipopolysaccharide / D-galactosamine-induced acute hepatic injury by inhibiting the oxidation and inflammation in mice. Bioorg Med Chem Lett. 2018;28(29):1525–1533. doi: 10.1016/j.bmcl.2018.03.073. [DOI] [PubMed] [Google Scholar]
- 65.Takada Y, Aggarwal BB. Betulinic acid suppresses carcinogen-induced NF-kappa B activation through inhibition of I kappa B alpha kinase and p65 phosphorylation: abrogation of cyclooxygenase-2 and matrix metalloprotease-9. J Immunol. 2003;171(6):3278–3286. doi: 10.4049/jimmunol.171.6.3278. [DOI] [PubMed] [Google Scholar]
- 66.Liu R, Dobson CC, Foster BC, Durst T, Sanchez P, Arnason JT, Harris CS. Effect of an anxiolytic botanical containing Souroubea sympetala and Platanus occidentalis on in-vitro diazepam human cytochrome P450-mediated metabolism. J Pharm Pharmacol. 2018;71(3):429–437. doi: 10.1111/jphp.13045. [DOI] [PubMed] [Google Scholar]
- 67.Laavola M, Haavikko R, Hämäläinen M, Leppänen T, Nieminen R, Alakurtti S, Moreira VM, Yli-Kauhaluoma J, Moilanen E. Betulin derivatives effectively suppress inflammation in vitro and in vivo. J Nat Prod. 2016;79(2):274–280. doi: 10.1021/acs.jnatprod.5b00709. [DOI] [PubMed] [Google Scholar]
- 68.Dash S, Chattopadhyay S, Ghosh T, Dash SS, Tripathy S, Das B, Bag BG, Das D, Roy S. Self-assembled betulinic acid protects doxorubicin induced apoptosis followed by reduction of ROS-TNF-alpha-caspase-3 activity. Biomed Pharmacother. 2015;72:144–157. doi: 10.1016/j.biopha.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 69.Ou Z, Zhao J, Zhu L, Huang L, Ma Y, Ma C, Luo C, Zhu Z, et al. Anti-inflammatory effect and potential mechanism of betulinic acid on λ-carrageenan-induced paw edema in mice. Biomed Pharmacother. 2019;118:109347. doi: 10.1016/j.biopha.2019.109347. [DOI] [PubMed] [Google Scholar]
- 70.Li T, Li F, Liu X, Liu J, Li D. Synergistic anti-inflammatory effects of quercetin and catechin via inhibiting activation of TLR4-MyD88-mediated NF-κB and MAPK signaling pathways. Phytother Res. 2019;33(3):756–767. doi: 10.1002/ptr.6268. [DOI] [PubMed] [Google Scholar]
- 71.Chow HH, Hakim IA, Vining DR, Crowell JA, Cordova CA, Chew WM, Xu MJ, Hsu CH, Ranger-Moore J, Alberts DS. Effects of repeated green tea Catechin administration on human cytochrome P450 activity. Cancer Epidemiol Biomark Prev. 2006;15(12):2473–2476. doi: 10.1158/1055-9965.EPI-06-0365. [DOI] [PubMed] [Google Scholar]
- 72.Shin DW, Kim SN, Lee SM, Lee W, Song MJ, Park SM, Lee TR, Baik JH, Kim HK, Hong JH, Noh M. (−)-Catechin promotes adipocyte differentiation in human bone marrow mesenchymal stem cells through PPAR gamma transactivation. Biochem Pharmacol. 2009;77(1):125–133. doi: 10.1016/j.bcp.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 73.Zhai W, Zheng J, Yao X, Peng B, Liu M, Huang J, Wang G, Xu Y. Catechin prevents the calcium oxalate monohydrate induced renal calcium crystallization in NRK-52E cells and the ethylene glycol induced renal stone formation in rat. BMC Complement Altern Med. 2013;13:228. doi: 10.1186/1472-6882-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shalan NA, Mustapha NM, Mohamed S. Noni leaf and black tea enhance bone regeneration in estrogen-deficient rats. Nutrition. 2017;33:42–51. doi: 10.1016/j.nut.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Limtrakul P, Yodkeeree S, Pitchakarn P, Punfa W. Anti-inflammatory effects of proanthocyanidin-rich red rice extract via suppression of MAPK, AP-1 and NF-κB pathways in raw 264.7 macrophages. Nutr Res Pract. 2016;10(3):251–258. doi: 10.4162/nrp.2016.10.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alshatwi AA. Catechin hydrate suppresses MCF-7 proliferation throughTP53 / Caspase-mediated apoptosis. J Exp Clin Cancer Res. 2010;29(1):167. doi: 10.1186/1756-9966-29-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang DR, Belal SA, Choe HS, Shin DK, Shim KS. Effect of Kaempferol on cyclooxygenase 2 (Cox2) and cytosolic phospholipase A2 (cPLA2) protein expression in BALB/c mice. Iran J Allergy Asthma Immunol. 2018;17(5):428–435. doi: 10.18502/ijaai.v17i5.301. [DOI] [PubMed] [Google Scholar]
- 78.Zhang ZJ, Xia ZY, Wang JM, Song XT, Wei JF, Kang WY. Effects of flavonoids in Lysimachia clethroides Duby on the activities of cytochrome P450 CYP2E1 and CYP3A4 in rat liver Microsomes. Molecules. 2016;21(6):E738. doi: 10.3390/molecules21060738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beekmann K, Rubió L, de Haan LH, Actis-Goretta L, van der Burg B, van Bladeren PJ, Rietjens IM. The effect of quercetin and kaempferol aglycones and glucuronides on peroxisome proliferator-activated receptor-gamma (PPAR-γ) Food Funct. 2015;6(4):1098–1107. doi: 10.1039/C5FO00076A. [DOI] [PubMed] [Google Scholar]
- 80.Suchal K, Malik S, Khan SI, Malhotra RK, Goyal SN, Bhatia J, Ojha S, Arya DS. Molecular pathways involved in the amelioration of myocardial injury in diabetic rats by Kaempferol. Int J Mol Sci. 2017;18(5):E1001. doi: 10.3390/ijms18051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen HJ, Lin CM, Lee CY, Shih NC, Peng SF, Tsuzuki M, Amagaya S, Huang WW, Yang JS. Kaempferol suppresses cell metastasis via inhibition of the ERK-p38-JNK and AP-1 signaling pathways in U-2 OS human osteosarcoma cells. Oncol Rep. 2013;30(2):925–932. doi: 10.3892/or.2013.2490. [DOI] [PubMed] [Google Scholar]
- 82.Liao W, Chen L, Ma X, Jiao R, Li X, Wang Y. Protective effects of kaempferol against reactive oxygen species-induced hemolysis and its antiproliferative activity on human cancer cells. Eur J Med Chem. 2016;114:24–32. doi: 10.1016/j.ejmech.2016.02.045. [DOI] [PubMed] [Google Scholar]
- 83.Yang YH, Wang Z, Zheng J, Wang R. Protective effects of gallic acid against spinal cord injury-induced oxidative stress. Mol Med Rep. 2015;12(2):3017–3024. doi: 10.3892/mmr.2015.3738. [DOI] [PubMed] [Google Scholar]
- 84.Jiang M, Zhou M, Han Y, Xing L, Zhao H, Dong L, Bai G, Luo G. Identification of NF-κB inhibitors in Xuebijing injection for sepsis treatment based on bioactivity-integrated UPLC-Q/TOF. J Ethnopharmacol. 2013;147(2):426–433. doi: 10.1016/j.jep.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 85.Variya BC, Bakrania AK, Patel SS. Antidiabetic potential of gallic acid from Emblica officinalis: improved glucose transporters and insulin sensitivity through PPAR-γ and Akt signaling. Phytomedicine. 2019. 10.1016/j.phymed.2019.152906. [DOI] [PubMed]
- 86.Jin L, Piao ZH, Liu CP, Sun S, Liu B, Kim GR, Choi SY, Ryu Y, Kee HJ, Jeong MH. Gallic acid attenuates calcium calmodulin-dependent kinase II-induced apoptosis in spontaneously hypertensive rats. J Cell Mol Med. 2018;22(3):1517–1526. doi: 10.1111/jcmm.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang N, Lee JH, Lee W, Ko JY, Kim EA, Kim JS, Heu MS, Kim GH, Jeon YJ. Gallic acid isolated from Spirogyra sp. improves cardiovascular disease through a vasorelaxant and antihypertensive effect. Environ Toxicol Pharmacol. 2015;39(2):764–772. doi: 10.1016/j.etap.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 88.Ali ET, Jabbar AS, Mohammed AN. A comparative study of interleukin 6, inflammatory markers, ferritin, and hematological profile in rheumatoid arthritis patients with Anemia of chronic disease and Iron deficiency Anemia. Anemia. 2019;2019:3457347. doi: 10.1155/2019/3457347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raj DS. Role of interleukin-6 in the anemia of chronic disease. Semin Arthritis Rheum. 2009;38(5):382–388. doi: 10.1016/j.semarthrit.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 90.Volodko N, Salla M, Eksteen B, Fedorak RN, Huynh HQ, Baksh S. TP53 codon 72 Arg/Arg polymorphism is associated with a higher risk for inflammatory bowel disease development. World J Gastroenterol. 2015;21(36):10358–10366. doi: 10.3748/wjg.v21.i36.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu T, Kim P, Luo Y. Tp53 gene mediates distinct dopaminergic neuronal damage in different dopaminergic neurotoxicant models. Neural Regen Res. 2017;12(9):1413–1417. doi: 10.4103/1673-5374.215243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fraenkel PG. Understanding anemia of chronic disease. Hematology Am Soc Hematol Educ Program. 2015;2015:14–18. doi: 10.1182/asheducation-2015.1.14. [DOI] [PubMed] [Google Scholar]
- 93.Birgegard G, Samuelsson J, Ahlstrand E, Ejerblad E, Enevold C, Ghanima W, Hasselbalch H, Nielsen CH, et al. Inflammatory functional iron deficiency common in myelofibrosis, contributes to anaemia and impairs quality of life. From the Nordic MPN study group. Eur J Haematol. 2019;102(3):235–240. doi: 10.1111/ejh.13198. [DOI] [PubMed] [Google Scholar]
- 94.Weiler HA, Jean-Philippe S, Cohen TR, Vanstone CA, Agellon S. Depleted iron stores and iron deficiency anemia associated with reduced ferritin and hepcidin and elevated soluble transferrin receptors in a multiethnic group of preschool-age children. Appl Physiol Nutr Metab. 2015;40(9):887–894. doi: 10.1139/apnm-2014-0328. [DOI] [PubMed] [Google Scholar]
- 95.Chung IJ, Lee JJ, Nam CE, Kim HN, Kim YK, Park MR, Cho SH, Kim HJ. Increased inducible nitric oxide synthase expression and nitric oxide concentration in patients with aplastic anemia. Ann Hematol. 2003;82(2):104–108. doi: 10.1007/s00277-002-0602-0. [DOI] [PubMed] [Google Scholar]
- 96.Hua YL, Ma Q, Yuan ZW, Zhang XS, Yao WL, Ji P, Hu JJ, Wei YM. A novel approach based on metabolomics coupled with network pharmacology to explain the effect mechanisms of Danggui Buxue Tang in anaemia. Chin J Nat Med. 2019;17(4):275–290. doi: 10.1016/S1875-5364(19)30031-7. [DOI] [PubMed] [Google Scholar]
- 97.Li Y, Wang F, Guo R, Zhang Y, Chen D, Li X, Tian W, Xie X, Jiang Z. Exosomal sphingosine 1-phosphate secreted by mesenchymal stem cells regulated Treg/Th17 balance in aplastic anemia. IUBMB Life. 2019;71(9):1284–1292. doi: 10.1002/iub.2035. [DOI] [PubMed] [Google Scholar]
- 98.Semba RD, Ferrucci L, Sun K, Patel KV, Guralnik JM, Fried LP. Elevated serum advanced glycation end products and their circulating receptors are associated with anaemia in older community-dwelling women. Age Ageing. 2009;38(3):283–289. doi: 10.1093/ageing/afp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X, Wilson AF, Du W, Pang Q. Cell-cycle-specific function of p53 in Fanconi Anemia hematopoietic stem and progenitor cell proliferation. Stem Cell Reports. 2018;10(2):339–346. doi: 10.1016/j.stemcr.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Du W, Erden O, Pang Q. TNF-α signaling in Fanconi anemia. Blood Cells Mol Dis. 2014;52(1):2–11. doi: 10.1016/j.bcmd.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, Burk RD, Kunisaki Y, et al. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525(7570):528–532. doi: 10.1038/nature15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oz HS, Tobin T. Diclazuril protects against maternal gastrointestinal syndrome and congenital toxoplasmosis. Int J Clin Med. 2014;5(3):93–101. doi: 10.4236/ijcm.2014.53017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Totino PRR, de Souza HADS, Correa EHC, Daniel-Ribeiro CT, Ferreira-da-Cruz MF. Eryptosis of non-parasitized erythrocytes is related to anemia in Plasmodium berghei low parasitema malaria of Wistar rats. Parasitol Res. 2019;118(1):377–382. doi: 10.1007/s00436-018-6167-1. [DOI] [PubMed] [Google Scholar]
- 104.Stijlemans B, Vankrunkelsven A, Brys L, Raes G, Magez S, De Baetselier P. Scrutinizing the mechanism underlying the induction of anemia of inflammation through GPI-mediated modulation of macrophage activation in a model of African trypanosomiasis. Microbes Infect. 2010;12(5):389–399. doi: 10.1016/j.micinf.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 105.Ocak S, Kaya H, Cetin M, Gali E, Ozturk M. Seroprevalence of hepatitis B and hepatitis C in patients with thalassemia and sickle cell anemia in a long-term follow-up. Arch Med Res. 2006;37(7):895–898. doi: 10.1016/j.arcmed.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 106.Hicks BM, Klil-Drori AJ, Yin H, Campeau L, Azoulay L. Androgen deprivation therapy and the risk of Anemia in men with prostate Cancer. Epidemiology. 2017;28(5):712–718. doi: 10.1097/EDE.0000000000000678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table 1. Candidate active ingredients of PRA; Table 2. Common targets between PRA and IDA; Table 3. Correlations between components and targets.
Additional file 2: File 2a: Table 1 Known therapeutic targets for IDA; File 2b: Table 2 Targets for PRA.
Additional file 3. File for all pictures.
Additional file 4. File for partial targets with components docking.
Data Availability Statement
Datasets supporting the results of this article have been included in the additional files.