Abstract
Background
Small heat shock proteins (sHSPs) are critical for plant response to biotic and abiotic stresses, especially heat stress. They have also been implicated in various aspects of plant development. However, the acting mechanisms of the sHSPs in plants, especially in perennial grass species, remain largely elusive.
Results
In this study, AsHSP26.8a, a novel chloroplast-localized sHSP gene from creeping bentgrass (Agrostis stolonifera L.) was cloned and its role in plant response to environmental stress was studied. AsHSP26.8a encodes a protein of 26.8 kDa. Its expression was strongly induced in both leaf and root tissues by heat stress. Transgenic Arabidopsis plants overexpressing AsHSP26.8a displayed reduced tolerance to heat stress. Furthermore, overexpression of AsHSP26.8a resulted in hypersensitivity to hormone ABA and salinity stress. Global gene expression analysis revealed AsHSP26.8a-modulated expression of heat-shock transcription factor gene, and the involvement of AsHSP26.8a in ABA-dependent and -independent as well as other stress signaling pathways.
Conclusions
Our results suggest that AsHSP26.8a may negatively regulate plant response to various abiotic stresses through modulating ABA and other stress signaling pathways.
Keywords: Creeping bentgrass, AsHSP26.8a, ABA, Abiotic stress, Small heat shock protein
Background
The heat shock proteins (HSPs) are virtually ubiquitous molecules in plants that are rapidly induced by heat stress [1, 2]. At least six types of HSPs, HSP100 (Clp), HSP90, HSP70 (DnaK), HSP60, HSP40 (DNAJ) and small HSPs (sHSPs) have been identified in higher plants, of which the sHSPs with a molecular mass of 12 to 42 kDa are found ubiquitously in all kingdoms of life [3]. The sHSPs function as molecular chaperones, both in vitro and in vivo, to protect cells from stress damage by preventing irreversible protein aggregation and maintaining denatured proteins in a folding-competent state [2, 4–8]. Based on sequence homology, immunological cross-reactivity and subcellular localization, the sHSPs can be further classified into different groups. A total of 12 subfamilies of sHSPs have been identified in various plant species, such as maize (Zea mays L.), Kashgar tamarisk (Tamarix hispida), creeping bentgrass (Agrostisc stolonifera L.), poplars (Populus), Pearl millet (Pennisetum glaucum L.), tall fescue (Festuca arundinacea Schreb.) [9–15]. They are localized in different places within the cell including cytosol or nucleus, mitochondria, plastids (P), endoplasmic reticulum (ER) and peroxisomes (Po) [15–17]. Most of the sHSP subfamily proteins are structurally similar with a highly variable N-terminal region, a conserved core domain of about 100 amino acids and a short C-terminal region [2, 18].
Apart from heat stress, sHSPs are induced in response to a number of other environmental adversities such as osmotic stress (e.g. salt, drought), oxidative stress, cold stress, heavy metal stress and phytohormone ABA [6, 11, 15, 16, 19], suggesting its involvement in plant response to general abiotic stresses. Additionally, their implication in various aspects of plant development have also been documented [16, 20–22].
The roles different sHSPs play in positively regulating plant responses to heat and other environmental stresses (drought, NaCl, mannitol and H2O2) have largely been reported in various plant species and most of them belong to cytosolic sHSPs [10, 21, 23–32]. Class I and II sHSPs have mainly contributed to the current model for sHSP chaperone activity. These two classes of sHSPs have both unique and overlapping functions and act in conjunction with HSP101 to either directly or indirectly protect specific translation factors in cytosolic stress granules [33]. Extensive evolutionary analysis indicates that organelle-targeted sHSPs (chloroplast, mitochondria, peroxisomes, and endoplasmic reticulum) are closely related to their class I counterparts [34–36]. Endoplasmic reticulum-located sHSP, sHSP22, together with ABA insensitive 1 (ABI1) protein phosphatase controls polar auxin transport and orchestrates ABA and auxin signaling crosstalk in Arabidopsis [37]. Peroxisome-located sHSPs activate catalase to regulate plant abiotic stress resistance [38]. A mitochondrial matrix-localized small heat shock protein in cotton, GhHSP24.7, positively controls seed germination via temperature-dependent ROS generation [39]. Overexpression of the chloroplast sHSP has been associated with cold, heat and oxidative stress tolerance [21, 26]. A typical chloroplast-localized sHSP, HSP21, has been identified in many plant species. Several studies have suggested that HSP21 protects the thermolabile photosystem II (PSII) against heat stress [9, 26, 40–42] and oxidative stress [26, 43]. HSP21 is activated by the GUN5-mediated retrograde signaling pathway and stabilizes PSII by directly binding to its subunits under heat stress [44]. HSP21 also interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development under heat stress by maintaining plastid-encoded RNA polymerase (PEP)-dependent transcription [22]. In addition, HSP21 is involved in extended thermomemory in Arabidopsis. Abundant HSP21 during the memory stage is negatively regulated by heat-induced plastid-localized metalloprotease FtsH6 [45]. Our recent study characterizing sHSPs in perennial grasses identified three creeping bentgrass sHSPs, AsHSP17 (previously named ApHSP16.5) (KT272405), AsHSP26.7 (previously named ApHSP26.7) (AY153761) and AsHSP26.8 (previously named ApHSP26.8) (AY153760) [9, 46–48]. AsHSP17 and AsHSP26.8 genes were significantly induced in transgenic creeping bentgrass overexpressing a rice SUMO E3 ligase gene, OsSIZ1 and exhibiting improved heat tolerance compared to wild type controls [46]. In transgenic creeping bentgrass ectopically expressing a cyanobacterial flavodoxin and exhibiting enhanced heat stress tolerance, AsHSP17, AsHSP26.7 and AsHSP26.8 were all significantly induced but differentially regulated in wild type (WT) and transgenic (TG) plants [47]. Additionally, overexpression of a rice miroRNA, OsmiR393 improves heat tolerance in transgenic creeping bentgrass that is associated with enhanced expression of sHSP genes, AsHSP17 and AsHSP26.7 [48]. Further analysis of AsHSP17, one of these turfgrass sHSPs revealed that AsHSP17 is a negative regulator attenuating plant response to abiotic stress through modulating plant photosynthesis and ABA-dependent and independent signaling [15].
To better understand how sHSPs function in perennial grasses to regulate plant stress response, we focused on AsHSP26.8, initially obtained from A. stolonifera cv Penncross as ApHSP26.8 [9], and cloned its ortholog gene, AsHSP26.8a from A. stolonifera cv Penn-A4 encoding a chloroplast-localized creeping bentgrass sHSP. The cloned AsHSP26.8a was introduced into Arabidopsis plants for characterization. Transgenic analysis revealed that constitutive expression of AsHSP26.8a led to significantly increased plant susceptibility to several abiotic stresses including heat and salt as well as treatment with hormone ABA. Our results suggested that similar to AsHSP17, AsHSP26.8a may also function as a protein chaperone to negatively regulate plant stress response.
Results
Cloning and sequence analysis of AsHSP26.8a and its protein subcellular localization
In our previous study manipulating sumoylation process in transgenic plants for enhanced tolerance to environmental adversities, it was revealed that overexpression of OsSIZ1, a rice E3 SUMO ligase in transgenic creeping bentgrass enhances plant tolerance to a number of abiotic stresses including heat stress associated with altered expression of two sHSP genes, AsHSP17 and AsHSP26.8 [46]. Further investigation of AsHSP17 uncovered its role in modulating plant photosynthesis and ABA-dependent and independent signaling to attenuate plant response to abiotic stress including heat and salt stress [15]. We were also curious about how AsHSP26.8 contributes to OsSIZ1-mediated plant stress response and therefore cloned its ortholog gene from the heat-stressed plants of creeping bentgrass cultivar, Penn A-4 and designated it as AsHSP26.8a. The AsHSP26.8a gene is 729 bp long encoding a protein of 242 amino acids. The predicted polypeptide has a molecular weight (MW) of 26.78 kD and an isoelectric point (pI) of approximately 6.03. Differences in nine nucleotides were identified between AsHSP26.8a and ApHSP26.8 DNA sequences, which led to changes in three amino acids (Fig. S1). Sequence alignment of AsHSP26.8a and the representative plant sHSPs allowed identification of a chloroplast transit peptide (I), a Met-rich region (II) and two consensus regions (III and IV) in AsHSP26.8a (Fig. S2), which was phylogenetically classified as a chloroplast-localized sHSP (Fig. S3).
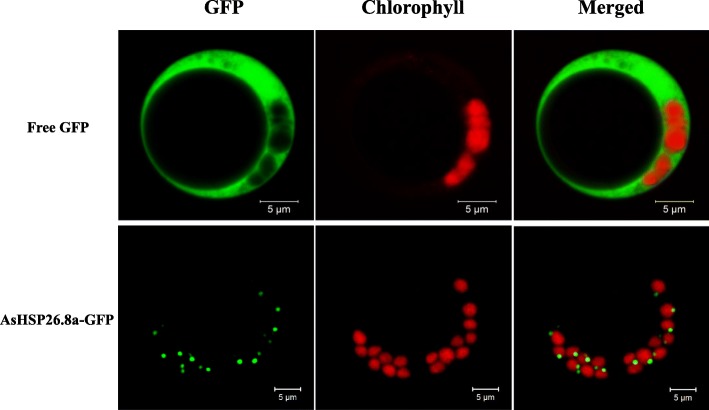
To further confirm its subcellular localization in the chloroplasts, AsHSP26.8a-green fluorescent protein (GFP) fusion gene was introduced into rice (Oryza sativa) protoplasts. The GFP fluorescence was found to be localized to the nucleoids inside chloroplasts, which is mostly associated with thylakoids (Fig. 1), similar to that of the AtHSP21 (AT4G27670), a well-characterized homolog sHSP of AsHSP26.8a in Arabidopsis thaliana [22]. This result indicates that AsHSP26.8a is specifically localized to the thylakoids membranes within chloroplasts.
Fig. 1.
Subcellular localization of AsHSP26.8a protein. Free GFP, control with empty vector; AsHSP26.8a-GFP, AsHSP26.8a-GFP fusion. Bars = 5 μm
AsHSP26.8a expression in response to heat stress
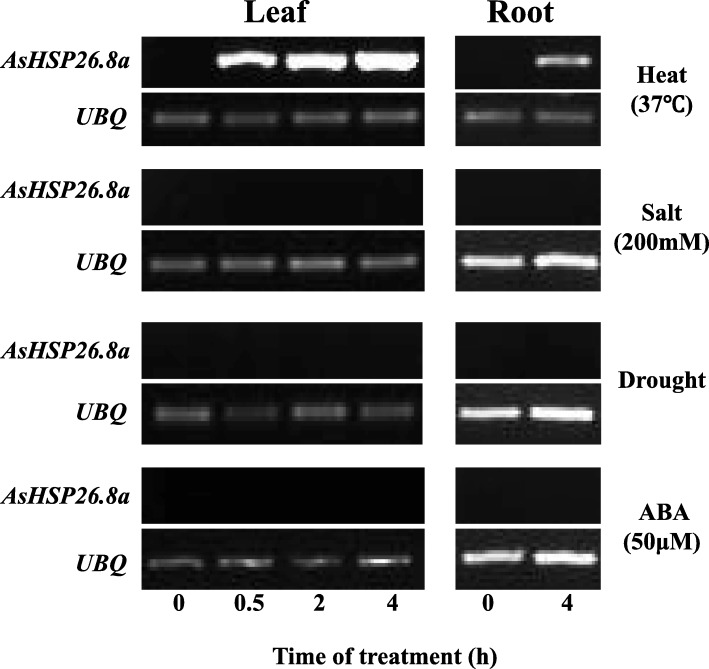
Our previous study showed that AsHSP26.8 is strongly induced by heat stress in creeping bentgrass leaf [46]. To investigate whether the AsHSP26.8a responds to heat and other abiotic stresses and ABA stimulus, we examined the expression pattern of the AsHSP26.8a in creeping bentgrass under heat, salinity and drought stress as well as exogenous ABA treatment. Total RNAs extracted from leaves and roots were subjected to semi-quantitative RT-PCR analysis. As shown in Fig. 2, the expression of AsHSP26.8a in leaves and roots was strongly induced by heat stress, confirming our previous observation [46]. However, no expression was detected in creeping bentgrass leaves or roots subjected to salinity and drought stress as well as ABA treatment (Fig. 2). It should be noted that the very low basal expression level of AsHSP26.8a may prevent its detection even if it does respond to salinity, drought and ABA. With the high sequence similarity between AsHSP26.8a and other sHSPs such as AsHSP26.2 (AY153578), AsHSP26.7 and AsHSP26.8, we were unable to perform quantitative-real-time reverse transcription PCR (qRT-PCR) analysis for AsHSP26.8a expression because of the difficulty in designing appropriate PCR primers.
Fig. 2.
Semi-quantitative RT-PCR analysis of AsHSP26.8a expression profile in leaf (left) and root (right) tissues under heat (37 °C), salt (200 mM NaCl), Drought, ABA (50 μM) treatment. Leaf samples were collected at 0, 0.5, 2 and 4 h after stress treatment and root samples were collected at 0 and 4 h after stress treatment. Total RNA was isolated using Trizol and 2 μg total RNA was used for reverse transcription by using reverse transcriptase II (NEB, USA). Creeping bentgrass ubiquitin gene AsUBQ was used as the endogenous control. The gel images were cropped to only retain PCR products. All original, full-length gel images were included as additional files in the supplementary materials. The PCR process: 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 65 °C for 30 s, 72 °C for 30 s, for AsHSP26.8a; 95 °C for 5 min, 25 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, for AsUBQ.
Generation of transgenic A. thaliana plants constitutively expressing AsHSP26.8a
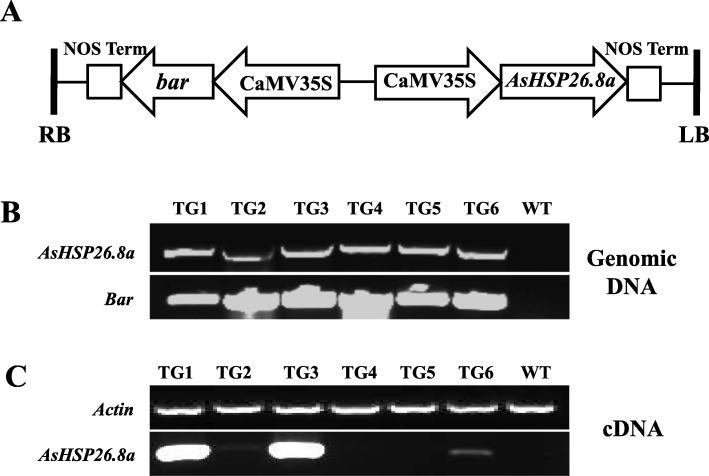
To further investigate the role of AsHSP26.8a in plant response to abiotic stress, we set to manipulate AsHSP26.8a expression in transgenic plants. To this end, an AsHSP26.8a overexpression vector was constructed (Fig. 3a) for transformation into A. thaliana ecotype Columbia. The six independent transgenic lines generated (Fig. 3b, c) did not appear different from wild type controls, of which four homozygous lines, TG1, TG2, TG3 and TG6 expressing different levels of AsHSP26.8a (Fig. 3b, c) were further analyzed. It should be noted that in this study, we chose to use wild type plants as the control in characterizing AsHSP26.8a transgenic plants based on the observation that their development and response to various stresses exhibited no significant difference from that of the transgenic lines harboring an empty vector or a gus reporter gene (data not shown).
Fig. 3.
Generation and molecular analysis of the AsHSP26.8a transgenic (TG) Arabidopsis thaliana.a, Schematic diagram of the AsHSP26.8a chimeric gene expression construct, p35S-AsHSP26.8a/p35S-bar. The AsHSP26.8a gene and an herbicide resistance gene, bar, are both under the control of the CaMV 35S promoter. RB, right border; LB, left border. b, PCR analysis of the AsHSP26.8a and bar gene in wild type (WT) and TG plants to detect transgene insertion in the Arabidopsis thaliana genome and transcription. Actin gene AtActin1 was used as the endogenous control. The gel images were cropped to only retain PCR products. All original, full-length gel images were included as additional files in the supplementary materials
AsHSP26.8a overexpression leads to increased heat susceptibility in transgenic plants
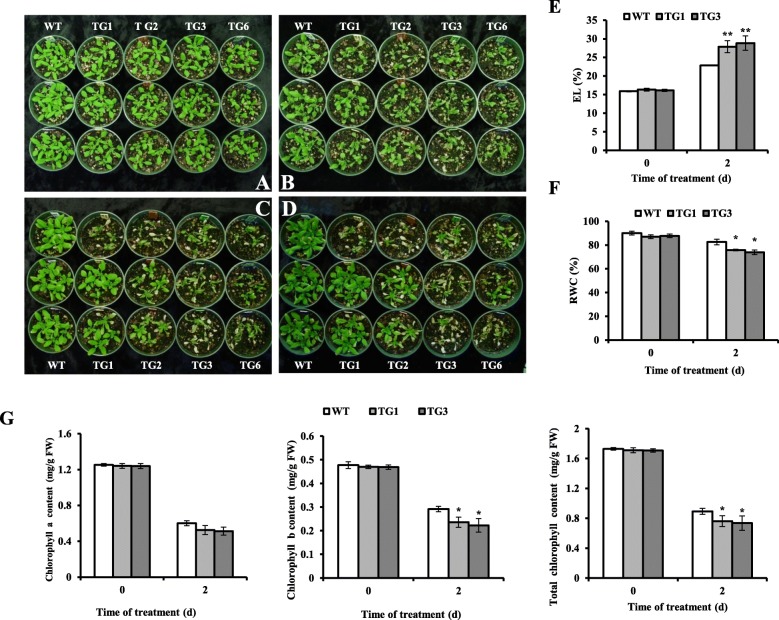
To evaluate how altered AsHSP26.8a expression impacts plant response to high temperature, we subject the well-developed 3 weeks old AsHSP26.8a-overexpressing transgenic plants to heat stress (40 °C) and evaluated their performance in comparison with non-transgenic plants. As shown in Fig. 4b, two-day heat stress led to non-recoverable severe damage in the majority of the transgenic plants, whereas the heat-elicited damage was barely observed in wild type controls, all of which recovered and survived the treatment (Fig. 4c, d). Plant relative water content (RWC) and electrolyte leakage (EL) were measured to exam their water status and cell membrane integrity. While similar RWC and EL were observed in both transgenic and wild type control plants under normal growth conditions, there was a significantly more water loss and a more severe heat-elicited cell membrane damage in the AsHSP26.8a-expressing transgenics than in the control plants 2 days after heat treatment (Fig. 4e, f), suggesting that AsHSP26.8a negatively impacts plant water retention capacity and cell membrane integrity. Similarly, although no significant difference in chlorophyll a, b and total chlorophyll contents was observed between wild type control and the AsHSP26.8a-containing plants under normal growth conditions, the AsHSP26.8a-expressing transgenic plants exhibited significantly lower leaf chlorophyll a, b and total chlorophyll contents than wild type control under heat-stress conditions (Fig. 4g) suggesting a reduced chlorophyll production in AsHSP26.8a transgenic plants under heat stress. It should be noted that the inconsistent phenotypes of the TG1 plants in the three pots used for heat stress response assessment (Fig. 4a-d) was most likely due to random experimental errors rather than the inherent difference between individual transgenic plants.
Fig. 4.
Responses of wild type (WT) and transgenic (TG) plants to heat stress. Two-week-old Arabidopsis thaliana WT and four AsHSP26.8a TG lines were grown under normal conditions in a growth chamber (a). Heat stress was applied by heating the plants to 40 °C for 2 d (b). The plants were then moved back to normal conditions, photographed 2 and 4 d after recovery (c and d). Leaf samples collected 2 d after heat stress were used for measuring electrolyte leakage (e), relative water content (f) and chlorophyll a content (G left), Chlorophyll b content (G middle) and total chlorophyll content (G right) (n = 3). Each column represents mean of three biological replicates. Error bars represent SE. ‘*’ or ‘**’ indicate significant differences between TG and WT plants at P < 0.05 or 0.01, respectively by Student’s t test
AsHSP26.8a transgenic plants exhibit increased salinity susceptibility
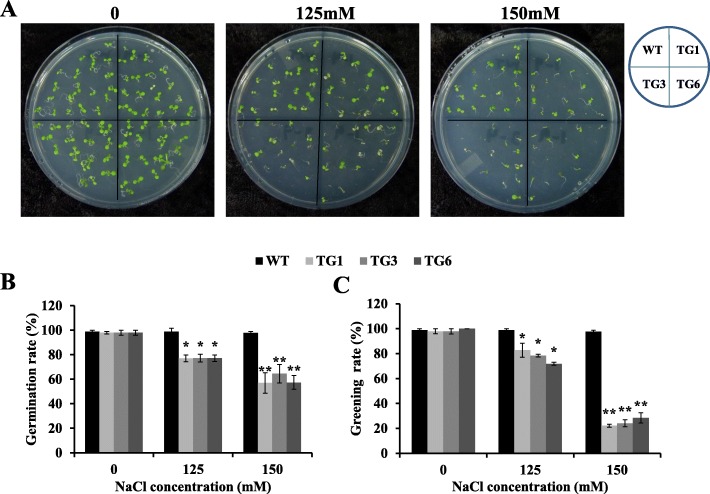
To investigate possible involvement of AsHSP26.8a in plant response to other abiotic stresses, we examined seed germination of AsHSP26.8a overexpressing plants subjected to salt stress (Fig. 5) and observed that transgenic plants exhibited significantly lower seed germination than the wild type controls when treated with 125 or 150 mM NaCl, under which the germination rate was only approximately 60–80% in the AsHSP26.8a transgenics versus 100% in wild type controls (Fig. 5a, b). Similarly, plant post-germination growth in AsHSP26.8a-expressing transgenic plants was also significantly impaired compared to wild type controls under the two different NaCl concentrations (Fig. 5a, c). These data indicate that besides heat stress, AsHSP26.8a also impacts how plants tackle other environmental adversities. More specifically, it negatively regulates plant salinity stress response.
Fig. 5.
Germination and greening rates of wild type (WT) and transgenic (TG1, TG3 and TG6) plants in response to salt treatment. a, seed germination of WT and AsHSP26.8a TG plants subjected to 0, 125 and 150 mM of NaCl 6 d after treatment. b, germination and c, greening percentages of WT and TG seeds subjected to 0, 125 and 150 mM NaCl 6 d after treatment (n = 3). Each column represents mean of three biological replicates. Error bars represent SE. ‘*’ or ‘**’ indicate significant differences between TG and WT plants at P < 0.05 or 0.01 respectively by Student’s t test. The layout of the plates is shown on the upper right-hand corner
AsHSP26.8a overexpression impacts ABA signaling in transgenic plants
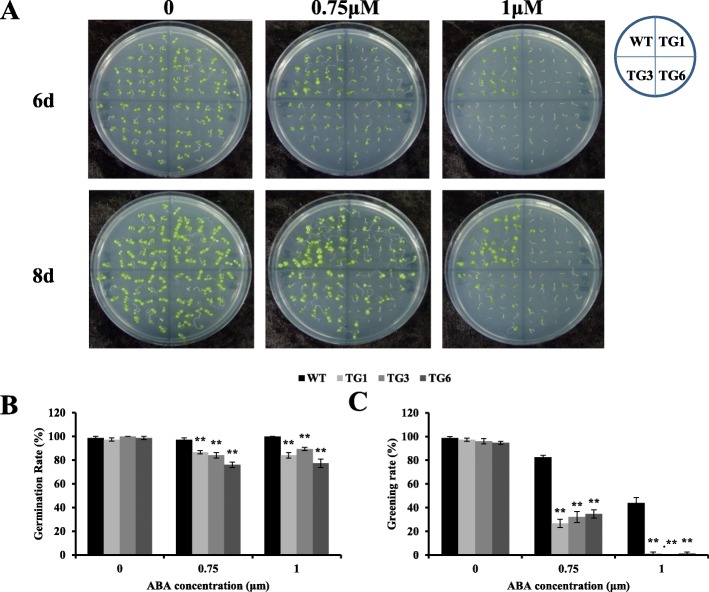
Among various phytohormones, ABA is the central regulator of abiotic stress resistance in plants and coordinates an array of functions [49–51]. We were curious about the possible involvement of AsHSP26.8a in plant ABA signaling. To this end, we conducted experiments assessing plant response to exogenous ABA treatment (0, 0.75 and 1 μM) and found that compared to wild type controls, seed germination and post germinative plant growth in AsHSP26.8a transgenics were both significantly reduced responding to ABA application (Fig. 6), suggesting that AsHSP26.8a may function as a positive regulator mediating ABA-associated plant development.
Fig. 6.
Germination and greening rates of wild type (WT) and transgenic (TG1, TG3 and TG6) plants in response to abscisic acid (ABA). a, seed germination of WT and AsHSP26.8a TG plants subjected to 0, 0.75 and 1 μM of ABA 6 and 8 d after treatment. b, germination percentages of WT and TG seeds subjected to 0, 0.75 and 1 μM of ABA 6 d after treatment (n = 3). C, greening percentages of WT and TG seeds (percentages of seeds with fully expanded cotyledons) subjected to 0, 0.75 and 1 μM of ABA 6 d after treatment (n = 3). Each column represents mean of three biological replicates. Error bars represent SE. ‘*’ or ‘**’ indicate significant differences between TG and WT plants at P < 0.05 or 0.01 respectively by Student’s t test. The layout of the plates is shown on the upper right-hand corner
Genome-wide gene expression analysis in AsHSP26.8a overexpression transgenic plants
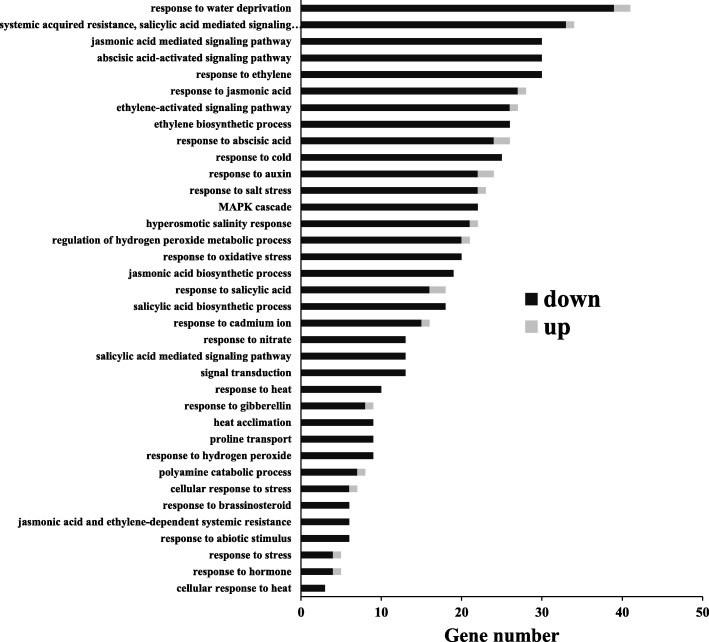
To better understand molecular mechanisms underlying AsHSP26.8a involvement in plant abiotic stress response, we conducted genome-wide gene expression analysis in wild type and AsHSP26.8a transgenic plants to screen for differentially expressed genes (DEGs) between the two genotypes (Table S1). Among a total of 269 DEGs identified, 20 (7%) were up-regulated and 249 (93%) down-regulated. Absolute log2 fold changes ranged from − 6.80 to 4.84 and adjusted P-value (FDR) ranged from 1.61 × 10− 77 to 0.99 × 10− 2. Allocated gene ontology (GO) term-based classification of the 269 DEGs resulted in 53 different groups. One of the most dominant GO terms was ‘Response to stimulus’ in the biological process category (Fig. S4). Additionally, many genes responsive to phytohormones (ABA, ethylene, jasmonic acid, auxin, salicylic acid, gibberellin) and abiotic stresses (drought, cold, salinity, oxidative stress, cadmium ion and heat) as well as those involved in various stress-related signaling pathways (salicylic acid mediated signaling pathway, jasmonic acid mediated signaling pathway, abscisic acid-activated signaling pathway, ethylene-activated signaling pathway) were also identified (Fig. 7).
Fig. 7.
Gene ontology (GO) classification for abiotic stress-related differentially expressed genes (DEGs) in wild type (WT) and AsHSP26.8a transgenic (TG) plants. The y-axis and x-axis indicate the names of clusters and the gene number of each cluster, respectively. Only the biological processes were used for GO term analysis
A total of 88 abiotic stress-related DEGs identified from the RNA-seq data were all functionally annotated that encode either regulatory or functional proteins (Table 1). The regulatory proteins consist of 30 transcription factors (e.g. AP2/ERF, DREB, HSF, MYB, NAC, WRKY and Zinc finger protein), 10 signaling proteins (e.g. calcium-binding proteins) and 17 kinases (e.g. cysteine-rich receptor-like protein kinase, leucine-rich repeat protein kinase, mitogen-activated protein kinase kinase kinase 14, S-locus lectin protein kinase family protein and wall-associated receptor kinase-like 2). The functional proteins include six cytochrome P450 family members, three LEA proteins, four carbohydrate metabolism-related proteins, thirteen nitrogen metabolism-related proteins, four proteins involved in oxidation-reduction process and one AAA-type ATPase family protein. The majority of these 88 abiotic stress-related DEGs were down-regulated except six encoding one MYB domain protein, one protein kinase, one cytochrome P450 family member, one alpha/beta-hydrolase family protein and two proteins involved in oxidation-reduction process). The differential expression patterns of the four representative abiotic stress responsive genes, DREB1B, ERF105, HSFB2a and HSFC1 between wild type control and the AsHSP26.8a transgenic plants were confirmed by qRT-PCR analysis (Table 2), further validating the RNA-seq data.
Table 1.
Annotation of genes up-regulated or down-regulated in AsHSP26.8a overexpressing transgenic (TG) Arabidopsis compared to wild type (WT) controls. Significant differences were corrected with FDR < 0.01 and expression ratio ≥ 2, (FC, fold change)
| Gene #ID | log2FC | nr_annotation |
|---|---|---|
| Signal proteins | ||
| Transcription factor | ||
| AT1G68840 | −1.12 | AP2-EREBP family, RAVE subfamily protein RAV2 |
| AT4G25490 | −6.80 | dehydration-responsive element-binding protein 1B |
| AT3G15210 | −2.12 | ethylene-responsive transcription factor 4 |
| AT5G47230 | −2.49 | ethylene-responsive transcription factor 5 |
| AT5G53290 | −2.27 | ethylene-responsive transcription factor CRF3 |
| AT3G50260 | −2.03 | ethylene-responsive transcription factor ERF011 |
| AT1G77640 | −3.41 | ethylene-responsive transcription factor ERF013 |
| AT1G22190 | −1.81 | ethylene-responsive transcription factor ERF058 |
| AT5G61600 | −2.52 | ethylene-responsive transcription factor ERF104 |
| AT5G51190 | −4.49 | ethylene-responsive transcription factor ERF105 |
| AT5G62020 | −1.50 | heat stress transcription factor B-2a |
| AT3G24520 | −1.68 | heat stress transcription factor C-1 |
| AT5G49330 | 1.39 | myb domain protein 111 |
| AT1G18570 | −1.31 | myb domain protein 51 |
| AT4G37260 | −1.35 | myb domain protein 73 |
| AT3G50060 | −2.70 | myb domain protein 77 |
| AT1G25550 | −1.63 | myb-like transcription factor-like protein |
| AT5G67300 | −1.39 | transcription factor MYB44 |
| AT1G69490 | −1.26 | NAC transcription factor protein family |
| AT4G31800 | −1.18 | WRKY DNA-binding protein 18 |
| AT1G62300 | −1.16 | WRKY transcription factor 6 |
| AT3G46620 | −3.92 | C3H4 type zinc finger protein |
| AT1G51700 | −1.19 | DOF zinc finger protein 1 |
| AT5G59820 | −4.87 | high light responsive zinc finger protein ZAT12 |
| AT3G52800 | −1.56 | zinc finger A20 and AN1 domain-containing stress-associated protein 6 |
| AT1G27730 | −3.74 | zinc finger protein STZ/ZAT10 |
| AT2G37430 | −5.33 | zinc finger protein ZAT11 |
| AT5G04340 | −2.28 | zinc finger protein ZAT6 |
| AT3G46090 | −4.18 | zinc finger protein ZAT7 |
| AT5G67450 | −3.06 | zinc-finger protein 1 |
| Signaling | ||
| AT5G49480 | −1.41 | Ca2 + −binding protein 1 |
| AT5G37770 | −2.55 | calcium-binding protein CML24 |
| AT1G76650 | −4.92 | calcium-binding protein CML38 |
| AT5G62570 | −1.17 | calmodulin binding protein-like protein |
| AT1G66400 | −3.17 | calmodulin like 23 |
| AT2G41100 | −1.02 | calmodulin-like protein 4 |
| AT3G25600 | −1.50 | putative calcium-binding protein CML16 |
| AT3G29000 | −4.55 | putative calcium-binding protein CML30 |
| AT5G39670 | −2.02 | putative calcium-binding protein CML45 |
| AT3G10300 | −1.02 | putative calcium-binding protein CML49 |
| Kinase | ||
| AT4G23190 | −1.43 | cysteine-rich receptor-like protein kinase 11 |
| AT2G19190 | −2.16 | FLG22-induced receptor-like kinase 1 |
| AT1G67470 | −1.37 | inactive serine/threonine-protein kinase |
| AT5G01540 | −1.60 | lectin receptor kinase A4.1 |
| AT1G33610 | −1.28 | leucine-rich repeat (LRR) family protein |
| AT1G51790 | −1.02 | leucine-rich repeat protein kinase-like protein |
| AT3G47090 | −1.02 | leucine-rich repeat protein kinase-like protein |
| AT2G30040 | −2.38 | mitogen-activated protein kinase kinase kinase 14 |
| AT1G51890 | −1.73 | probable LRR receptor-like protein kinase |
| AT2G44830 | 1.01 | protein kinase |
| AT3G57640 | −1.24 | protein kinase family protein |
| AT4G11521 | −1.26 | putative cysteine-rich receptor-like protein kinase 34 |
| AT4G04540 | −2.83 | putative cysteine-rich receptor-like protein kinase 39 |
| AT1G51800 | −1.51 | putative leucine-rich repeat protein kinase |
| AT1G74360 | −1.12 | putative LRR receptor-like serine/threonine-protein kinase |
| AT1G61370 | −1.21 | S-locus lectin protein kinase family protein |
| AT1G16130 | −1.09 | wall-associated receptor kinase-like 2 |
| Function protein | ||
| Cytochrome P450 | ||
| ATCG00730 | −1.68 | cytochrome b6/f complex subunit IV |
| AT4G31500 | −1.07 | cytochrome P450 83B1 |
| AT4G22710 | −2.44 | cytochrome P450, family 706, subfamily A, polypeptide 2 |
| AT4G12320 | 1.32 | cytochrome P450, family 706, subfamily A, polypeptide 6 |
| AT5G57220 | −1.43 | cytochrome P450, family 81, subfamily F, polypeptide 2 |
| AT2G27690 | −1.56 | cytochrome P450, family 94, subfamily C, polypeptide 1 |
| LEA | ||
| AT1G65690 | −1.51 | late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein |
| AT4G23610 | −2.52 | late embryogenesis abundant hydroxyproline-rich glycoprotein |
| AT3G54200 | −1.07 | late embryogenesis abundant hydroxyproline-rich glycoprotein |
| Carbohydrate metabolism-related proteins | ||
| AT3G09020 | −1.37 | alpha 1,4-glycosyltransferase family protein |
| AT5G26340 | −1.37 | sugar transport protein 13 |
| AT5G18840 | −2.66 | sugar transporter ERD6-like 16 |
| AT3G49790 | −1.18 | Carbohydrate-binding protein |
| Nitrogen metabolism-related proteins | ||
| AT1G02920 | −1.26 | glutathione S-transferase 7/11 |
| AT2G02930 | −1.48 | glutathione S-transferase F3 |
| AT5G62480 | −2.05 | glutathione S-transferase tau 9 |
| AT5G02780 | −1.78 | glutathione transferase lambda 1 |
| AT1G77760 | −1.38 | nitrate reductase [NADH] |
| AT1G66180 | −1.61 | aspartyl protease family protein |
| AT5G19120 | −1.14 | aspartyl protease family protein |
| AT2G38860 | −1.03 | protease I (pfpI)-like protein YLS5 |
| AT4G22470 | −1.24 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein |
| AT4G14290 | 4.42 | alpha/beta-hydrolase family protein |
| AT1G02660 | −1.37 | alpha/beta-Hydrolases superfamily protein |
| AT3G05200 | −1.06 | E3 ubiquitin-protein ligase ATL6 |
| AT3G52450 | −2.75 | E3 ubiquitin-protein ligase PUB22 |
| Oxidation-reduction process | ||
| AT3G09940 | −1.38 | monodehydroascorbate reductase (NADH) |
| AT4G37925 | 1.01 | NAD(P)H-quinone oxidoreductase subunit M |
| AT4G11290 | −1.27 | peroxidase 39 |
| AT3G47430 | 1.25 | peroxisomal membrane protein 11B |
| Energy | ||
| AT3G28510 | −2.42 | AAA-type ATPase family protein |
Table 2.
Differentially expressed genes in AsHSP26.8a overexpression transgenic (TG) and wild type (WT) Arabidopsis plants by qRT-PCR analysis
| Gene | TG/WT (log2 FC) | |
|---|---|---|
| RNA-seq | qRT-PCR | |
| DREB1B | −6.80 | −6.35 |
| ERF105 | −4.49 | −4.82 |
| HSFB2a | −1.68 | −2.53 |
| HSFC1 | −1.50 | −2.17 |
Discussion
The sHSPs, especially those in plants, are a large and complex family of proteins [35]. The plant sHSPs are divided into 12 subfamilies, of which chloroplast-localized sHSPs family has been identified in diverse higher plant species [52, 53]. This family of sHSPs have a Met-rich domain and a unique amphipathic domain at their N-terminus, not present in other sHSPs. AsHSP26.8a possesses both a Met-rich domain and a chloroplast transit peptide at its N-terminus (Fig. S2). Phylogenetic analysis and subcellular localization further support the classification of the AsHSP26.8a protein as a member of the chloroplast-localized sHSPs family (Fig. S3 and Fig. 1).
AsHSP26.8a-mediated HSP/HSF pathway and plant abiotic stress tolerance
The major abiotic stresses such as drought, high salinity, extreme temperature, negatively influence plant survival, growth and productivity. As sessile organisms, plants are unable to change their sites to escape from the unfavorable environmental adversities but have developed a great degree of resilience to conditions that would be considered harmful to many other organisms. A network of interconnected cellular stress response systems is essential for plant survival and productivity [54]. Within the complex stress response network, transcription factors (TFs) play a core role in the conversion of stress signal perception to stress-responsive gene expression by interacting with the promoter regions of various target stress-responsive genes, thus activating the whole network of genes to act together in enhancing plant tolerance to the harsh environmental conditions [55]. Heat shock transcription factors (HSFs) are the central regulators in plant cellular response to various abiotic stresses, especially to heat stress [56, 57]. Class B HSF and Class C HSF have been implicated in plant response to heat stress. HSFB1 and HSFB2b repress the expression of HSFs, but positively impact the acquired thermotolerance [58]. Capsicum annuum HSFB2a forms a transcriptional cascade with CaWRKY6 and CaWRKY40 to positively regulate the response to high temperature and high humidity [59]. Guan et al. [60] found that regulation of heat stress-responsive genes including HSFC1 and other HSFs by RCF2 and its interacting partner NAC transcription factor NAC019 is critical for thermotolerance in Arabidopsis. A recent study showed that HSFC1b from tall fescue plays a positive role in plant tolerance to heat stress in association with the induction and upregulation of heat-protective genes [61]. Our results showed that overexpression of AsHSP26.8a alters plant heat stress response (Fig. 4) and results in down-regulated expression of the two HSFs, HSFB2a and HSFC1 in transgenic plants (Table 1), suggesting that AsHSP26.8a may function to repress HSF gene expression, modulating heat-responsive genes and thus attenuating plant response to heat stress.
AsHSP26.8a modulates ABA-dependent stress signaling and plant abiotic stress response
ABA, commonly known as the “stress hormone”, responds to an array of biotic and abiotic stresses [62]. Under osmotic stress condition such as drought and high salinity stress, a number of genes functioning in stress response and tolerance are induced, and ABA is accumulated [49, 63, 64]. The expression of stress-responsive genes is regulated by ABA-dependent and ABA-independent pathways [64]. These genes encode the late embryogenesis abundant (LEA) proteins, enzymes, transcription factors, protein kinases et al. LEA accumulation is a functional adaptation of plants in gaining tolerance against osmotic as well as oxidative stresses [65]. Overexpression of genes encoding LEA proteins can improve the stress tolerance of transgenic plants [66–69]. MYB transcription factors are a large group of proteins identified in eukaryotes and widely distributed in plants [70–72]. Some of the MYB protein family members are involved in ABA-dependent signaling pathways regulating stress adaption and conferring plant stress tolerance [71, 73–75]. In this study, overexpression of AsHSP26.8a alters plant development and plant response to ABA and salt stress (Figs. 5 and 6) and leads to significantly reduced expression of several genes encoding LEA and MYB proteins (Table 1) as well as some stress-responsive transcription factors, such as WRKY transcription factors, involved in ABA-dependent signaling [76]. Abiotic stresses such as heat, high salinity and drought also induce the WRKY genes and trigger a cascade of signaling pathways for improved plant stress tolerance [77, 78]. Many studies showed that overexpression of a WRKY family gene confers abiotic stress tolerance in transgenic plants. For example, AtWRKY25 and AtWRKY26 overexpression enhanced plant heat tolerance in transgenic Arabidopsis [79]. Transgenic Arabidopsis overexpressing a wheat WRKY transcription factor, TaWRKY33 exhibited enhanced heat tolerance [80]. In cotton, GhWRKY17 overexpression increased plant sensitivity to drought and salt stress as well as ABA-mediated seed germination and root growth by reducing the levels of ABA and transcripts of ABA-inducible genes including NbAREB1 (ABA-responsive element binding), NbDREB (dehydration-responsive element binding), NbNCED (9-cis-epoxycarotenoid dioxygenase), NbERD (early responsive to dehydration) and LEA protein, NbLEA [81]. These data suggest that AsHSP26.8a may function as a chaperone protein, contributing to ABA-dependent signaling in plant abiotic stress responses.
AsHSP26.8a modulates ABA-independent stress signaling and plant abiotic stress response
Our results also showed that AsHSP26.8a modulates stress-related transcription factor gene expression in the ABA-independent signaling pathways. The ABA-independent stress-responsive gene expression is regulated by DREB proteins. DREBs belong to Ethylene Response Factor (ERF)/AP2 family and consist of two subclasses, DREB1/CBF and DREB2 induced by cold and dehydration/high salinity, respectively [64, 82–89]. Arabidopsis DREB1A overexpression was reported to enhance LEA protein levels and therefore abiotic stress tolerance in Arabidopsis [90, 91] and various crops including rice, soybean, peanut and wheat [92–95]. Heterologous expression of AtDREB1B in Salvia miltiorrhiza enhanced plant drought tolerance by activating different downstream DREB/CBF genes [96]. Moreover, AtHSFA3 is a transcription factor that is transcriptionally induced during heat stress by DREB2A and in turn regulates the expression of HSP-encoding genes [97]. Overexpressing AtDREB2A in Arabidopsis plants induces not only drought- and salt-responsive genes but also heat-shock-related genes. Thermotolerance was significantly increased in plants overexpressing DREB2A and decreased in DREB2A knockout plants [85]. The ERF (ethylene-responsive element binding factor) is another subfamily of the AP2/ERF family of TFs and plays vital roles in the regulation of biotic and abiotic stress responses [98–100]. Overexpression of a tomato ERF transcription factor, SlERF84 in Arabidopsis endows transgenic plants with ABA hypersensitivity and enhanced tolerance to drought and salt stress [101]. Overexpression of CmERF053 of chrysanthemum could enhance drought tolerance [102]. In Tamarix hispida, constitutive expression of an ERF transcription factor, ThCRF1, increased biosynthesis of trehalose and proline and the activities of SOD and POD, resulting in an altered osmotic potential and an enhanced reactive oxygen species (ROS) scavenging, and therefore significantly improved salt tolerance in transgenic plants. On the contrary, suppression of ThCRF1 led to decreased plant salt tolerance [103]. In this study, transgenic plants overexpressing AsHSP26.8a displayed significant expression changes in ten ERF/AP2 family genes (Table 1), for example, compared to the wild type, DREB1B/CBF1 and ERF105 expression in AsHSP26.8a transgenic plants was down-regulated over sixty-fold and thirty-fold, respectively (Table 1). These results suggest that AsHSP26.8a-modulated expression of the genes in the ABA-independent signaling pathways may also contribute to plant response to various abiotic stresses.
Other stress signaling pathways mediated by AsHSP26.8a and plant abiotic stress response
Calcium (Ca2+) is the most widely accepted second massager and involved in plant stress responses and cytoplasmic Ca2+ signal is recognized by Ca2+ sensors including calmodulins (CaM), calmodulin-like proteins (CMLs), calcium dependent protein kinases (CDPKs) and calcineurin B-like proteins (CBLs) [104–111]. Overexpression of AtCML24 enhances transgenic Arabidopsis tolerance to various ions including Co2+, Zn2+ and Mg2+ [112]. CML18 directly interacts with Na+/H+ antiporter NHX1 to regulate plant salinity tolerance [113]. CML9 is suggested to negatively regulate ABA-dependent salinity tolerance [114]. OsANN1, a calcium-binding protein of rice modulates antioxidant accumulation under abiotic stress to confer abiotic stress tolerance. OsANN1-knockdown led to increased plant sensitivity to heat and drought stresses, whereas OsANN1 overexpression resulted in improved plant growth with higher expression of OsANN1 under abiotic stress [115]. Receptor-like protein kinases (RLKs), a class of single-pass transmembrane proteins located in the plasma membrane, sense and transmit a variety of signals to regulate plant growth and development [116, 117]. Many RLKs have been implicated in abiotic stress responses, including the abscisic acid response, calcium signaling and antioxidant defense. Upon drought stress, the Arabidopsis LRK10L1.2 responds to directly or indirectly regulate stomata closure via ABA-mediated signaling [118], while a Glycine soja ABA-responsive receptor-like cytoplasmic kinase (RLCK), GsRLCK, responds to modulate ABA sensitivity in plants by regulating the expression of ABA-responsive genes [119]. In Pisum sativum, salinity induced lectin receptor-like kinase (PsLecRLK) gene expression and overexpression of PsLecRLKs led to improved plants salt tolerance due to enhanced ROS-scavenging [120]. In Medicago spp., the LRR-RLK gene, SRLK has been shown to regulate the root response to salt stress [121]. Large families of zinc finger transcription factors are abundant in plants and have diverse functions including DNA binding and transcriptional regulation [122]. Cys2/His2-type (C2H2) zinc finger proteins are implicated in plant response to a variety of adversities including low-temperatures, salt, drought, oxidative stress, excessive light and silique shattering [123–125]. One example is ZAT6 in Arabidopsis that positively regulates cadmium tolerance via the glutathione-dependent pathway [126]. Another example is the C2H2 zinc finger protein gene, Zat7 whose constitutive expression suppressed growth and enhanced salt tolerance in transgenic Arabidopsis plants [127]. Moreover, transgenic analysis in Arabidopsis points to the involvement of ZAT10 or STZ (salt tolerance zinc finger) in determining plant tolerance to drought, salt, osmotic, heat, photo-inhibitory light and oxidative stresses [128–131]. In this study, AsHSP26.8a overexpression led to significantly reduced expression of the genes encoding four CMLs, fifteen RLKs and nine zinc finger proteins in transgenic Arabidopsis (Table 1). For example, ZAT11 expression in AsHSP26.8a TG Arabidopsis plants was down-regulated over thirty-fold and ZAT12, ZAT7, CML38 and CML30 were down-regulated over sixteen-fold compared to wild type controls (Table 1). These results suggest that other than the ABA-dependent and -independent signaling pathways, AsHSP26.8a may also participate in other signaling pathways responding to abiotic stresses.
A few studies about the negative effect of HSPs on plant response to abiotic stress have previously been reported. Song et al. [132] found that transgenic Arabidopsis overexpressing cytosolic and organellar AtHSP90s exhibited suppressed expression of stress-responsive genes and consequently reduced salt and drought tolerance. Our previous study also showed that overexpression of AsHSP17, a creeping bentgrass sHSP, attenuates plant response to abiotic stress by modulating plant photosynthesis and ABA-dependent and -independent signaling [15]. Moreover, Wang and Luthe [9] were unable to amplify ApHSP26.8 (renamed as AsHSP26.8 in this study) from the heat-tolerant variant selected bentgrass, which was regenerated from callus that survived selection at 40 °C for 1 week, but they were able to amplify the gene from the heat-sensitive variant non-selected bentgrass, which was not subjected to heat stress. Taken together, these data imply that similar to AsHSP17, as a chaperone protein, AsHSP26.8a may be regulated to maintain an appropriate level in protecting a stressed plant. The excessive amount of AsHSP26.8a in transgenic plants may negatively impact the stress response regulatory network, compromising on plant stress tolerance. Indeed, our results assessing plant performance under stressful conditions showed that the plant response to heat stress in different transgenic lines appeared quite consistent regardless of the level of the elevated AsHSP26.8a expression (Fig. 3c and Fig. 4b-d). It is therefore tempting to speculate that any change in AsHSP26.8a expression above the basal level would have significant impact on plant response to environmental stress. Alternatively, AsHSP26.8a may need to stay inactive under normal condition not to become a stress itself. Using transgenic approach, we are currently investigating the impact of the regulated expression of AsHSP26.8a in creeping bentgrass itself in order to better understand molecular mechanisms of AsHSP26.8a-mediated plant development and stress response.
Conclusions
We have cloned and characterized a chloroplast localized sHSP gene, AsHSP26.8a, whose expression is induced by heat stress. Overexpression of AsHSP26.8a in transgenics attenuates plant response to various abiotic stresses including heat, salt and ABA. AsHSP26.8a may be involved in several aspects of plant stress response including ABA-dependent and -independent signaling and some other stress response pathways. Although the molecular mechanisms of the possible chaperone role AsHSP26.8a may play in plant stress response remain to be unraveled, the results obtained from the current study allow a better understanding of sHSPs involvement in plant abiotic stress response providing information for prospecting studies towards the development of novel molecular strategies for enhancing crop performance under adverse environments.
Methods
Plant materials and abiotic stress treatment
The creeping bentgrass cultivar ‘Penn-A4’ originally provided by HybriGene (Hubbard, OR) was clonally propagated from stolon and maintained as described previously [133]. Plants were maintained in the growth room and subjected to heat, salt, drought and phytohormone treatment as described previously [15]. The leaf samples were collected at 0, 0.5, 2 and 4 h and roots were collected at 0 and 4 h after stress treatment for RNA extraction to clone the AsHSP26.8a gene and analyze AsHSP26.8a gene expression.
The wild type A. thaliana plants (ecotype Columbia) and four transgenic lines (TG1, TG2, TG3 and TG6) generated by Xinbo Sun were cultured in a growth chamber and subjected to heat stress as described previously [15]. The seeds of the transgenic lines obtained are maintained and available in Hong Luo’s lab at Clemson University (Clemson, SC).
Plant genomic DNA, RNA isolation and gene expression analysis
Plant genomic DNA was extracted as described previously [134]. Plant total RNA was extracted from 100 mg of fresh leaves/roots with Trizol reagent (Invitrogen, Carlsbad, CA) following the manufacturers’ protocol. First-stand cDNA was synthesized from 2 μg RNA with SuperScript III System (Invitrogen) and oligo (dT) or gene specific primers.
Semi-quantitative RT-PCR was conducted on 25–30 cycles based on its exponential phase. PCR products were analyzed by electrophoresis using a 0.8% or 1.5% (w/v) agarose gel and photographed with the BioDoc-It imaging system (Ultra -Violet Products). The creeping bentgrass ubiquitin gene, AsUBQ (JX570760) and A. thaliana Actin1 (AT2G37620) were used as reference genes.
Using the Bio-Rad iQ5 real-time detection system with 12.5 μL of iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), quantitative real-time PCR was conducted to verify the expression of four representative DEGs (DREB1B, ERF105, HSFB2a and HSFC1) in WT and TG Arabidopsis plants identified by RNA-seq analysis. The reaction mix was preincubated at 95 °C for 3 min followed by 40 cycles of denaturing at 95 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 20 s. The data were collected by using iQ5 Optical System Software version 2.0 (Bio-Rad Laboratories) with AtActin1 and AtTub6 (AT5G12250), the two reference genes as endogenous controls for A. thaliana analysis. The ΔΔCt method was used for real-time PCR analysis with three biological replicates [133].
Subcellular localization of GFP protein in rice protoplasts
For subcellular localization, the full-length of AsHSP26.8a without a stop codon was subcloned into the pUC19/35S-EGFP vector with GFP at C terminus. The resulting fusion construct and empty vector were transformed into rice protoplasts by PEG (polyethylene glycol)-mediated transformation method, respectively [135]. After incubation in the dark for 16 h, the GFP fluorescence of transiently transformed rice protoplasts was examined and photographed under a confocal scanning microscopy (LSM510 Meta; Zeiss). For GFP, we used 488 and 519 nm for excitation and emission, respectively. For chlorophyll autofluorescence, we used 488 and 650–750 nm for excitation and emission, respectively.
Plasmid construction and plant transformation
The AsHSP26.8a overexpression construct, p35S-AsHSP26.8a/35S-bar contains the open reading frame of the turfgrass sHSP gene, AsHSP26.8a, driven by the cauliflower mosaic virus 35S (CaMV35S) promoter and linked to a CaMV35S-driven bar gene for herbicide resistance. The construct was introduced into wild type A. thaliana (Col-0) by Agrobaterium tumefaciens (LBA4404)-mediated plant transformation using floral dip. Individual transgenic plants were selected by herbicide screening.
RNA-seq analysis
The 4-week-old wild type and AsHSP26.8a TG3 seedlings were harvested for total RNA isolation with Trizol reagent (Invitrogen, Carlsbad, CA). RNA concentration, purity and integrity were monitored by Nanodrop, Qubit 2.0 and Agilent 2100 Bioanalyzer. The mRNAs were isolated using oligo (dT) magnetic beads and randomly broken into 150- to 250- pieces in fragmentation reagent. The first strand cDNA was generated by employing random hexamer-primers and reverse transcriptase, followed by the synthesis of the second-strand cDNA in the presence of dNTPs, RNase H and DNA polymerase I. The purified cDNA products after end-reparation, adaptor ligation and size selection by AMPure XP beads were PCR-amplified and then sequenced on an Illumina HiSeq2500 (Biomaker Technologies, Beijing, China). The clean reads obtained after removal of low-quality reads and adaptor sequences from the raw reads of each library were mapped to the reference Arabidopsis genome (TAIR 10, ftp://ftp.ensemblgenomes.org/pub/ plants/release-25/fasta/arabidopsis_thaliana/) using TopHat2 [136]. The FPKM (fragments per kilobase of transcript per million fragments mapped) values were used to measure Gene expression levels [137]. The FDR (false discovery rate) values of ≤0.01 and the FC (fold change) value of ≥2 (the absolute value of log2 ratio ≥ 1) were used to determine differentially expressed genes (DEGs) in the four libraries from wild type and AsHSP26.8a TG plants (two libraries from two biological replicates for each sample). Gene Ontology (GO) enrichment analysis was conducted by mapping all the DEGs to GO terms in the GO database (http://www.geneontology.org/) and the unigene number in each term was also determined.
Pearson’s correlation coefficients [138] between each pair of the biological replicates in WT and TG samples calculated were both close to 1 (Fig. S5), indicating a high RNA-seq data reliability.
We deposited the raw sequence reads into the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) repository with the accession numbers SRP065638 and SRA308886.
Seed germination assays
Seed germination assays under salt stress conditions and ABA treatment were performed as described previously [15]. The numbers of germinated seeds and green seedlings were counted on the fourth day. Each assay was repeated three times.
Leaf electrolyte leakage, chlorophyll, relative water contents and photosynthesis parameters
Measurements of Leaf electrolyte leakage (EL), chlorophyll and relative water content (RWC) were conducted as described previously [133, 139]. Each measurement was conducted in three replicates.
Statistical analysis
The data were analyzed by Microsoft excel 2010 (Microsoft, USA) and significance of differences between data sets was evaluated by SAS software 9.2 (SAS institute Inc. USA)
Supplementary information
Additional file 1: Table S1. List of assembled transcripts up-regulated or down-regulated (log2FC > 1 or < − 1, FDR < 0.01) in AsHSP26.8a transgenic (TG) Arabidopsis, relative to wild type (WT) plants.
Additional file 2: Table S2. Primers used in this study.
Additional file 3: Figure S1. AsHSP26.8a nucleic acid sequence (A) and amino acid sequence (B) alignment with AsHSP26.8. Figure S2. Amino acid sequence alignment of AsHSP26.8a and other plant chloroplast localized (CP) sHSPs. Accession umbers of sHSPs are: Agrostis stolonifera HSP26.8a (KU353578); Aegilops kotschyi HSP26.8 (CAI96511); Triticum dicoccoides HSP26.4 (CAI96512); Hordeum vulgare HSP26 (AAB28590); Brachypodium distachyon sHSP (XP_003558381.1); Spartina alterniflora HSP27.1 (AFP96756.1); Zea mays HSP18 (CAM12752.1); Oryza sativa HSP26 (BAA78385.1). Figure S3. The phylogenetic relationship of the AsHSP26.8a with different classes of sHSPs from other plant species. Accession numbers of the sHSPs are: Arabidopsis thaliana HSP17.4 (X17293); Arabidopsis thaliana HSP17.6 (X16076); Oryza sativa HSP17.4 (D12635); Daucus carota HSP18.0 (X53852); Helianthus annuus HSP17.6 (X59701); Agrostis stolonifera HSP17 (KT272405); Triticum aestivum HSP16.9A (X13431); Oryza sativa HSP16.9A (X60820); Zea mays HSP16.9 (X65725); Solanum lycopersicum HSP17.8 (X56138); Glycine max HSP17.5 (M11318); Glycine max HSP17.6 (MI1317); Medicago sativa HSP18.1 (X58710); Pisum sativum HSP18.1 (M33899); Arabidopsis thaliana HSP22 (U11501); Glycine max HSP22 (X63198); Pisum sativum HSP22 (M33898); Arabidopsis thaliana HSP18.5 (816448); Oryza sativa HSP17.6 (113595340); Arabidopsis thaliana HSP15.7 (833746); Arabidopsis thaliana HSP26.5 (841687); Oxybasis rubra HSP23 (X15333); Arabidopsis thaliana HSP23.6 (828623); Oryza sativa HSP22 (4330786); Agrostis stolonifera HSP26.8a (KU353578); Triticum aestivum HSP26.6 (X58280); Zea mays HSP26 (L28712); Petunia x hybrida HSP21 (X54103); Arabidopsis thaliana HSP21 (X54102); Glycine max HSP22 (X07188); Pisum sativium HSP21 (X07187); Arabidopsis thaliana HSP15.4 (828276); Oryza sativa HSP18.8 (4343386); Arabidopsis thaliana HSP17.4 (841843); Oryza sativa HSP17.6B (4330933); Lilium longiflorum HSP17.6 (D21816); Zea mays HSP17.5 (X54076); Zea mays HSP17.8 (X54075); Triticum aestivum HSP17.3 (X58279); Arabidopsis thaliana HSP17.6 (X63443); Ipomoea nil HSP18.8 (M99430); Ipomoea nil HSP17.2 (M99429); Glycine max HSP17.9 (X07159); Pisum sativum HSP17.7 (M33901); Arabidopsis thaliana HSP21.7 (835555); Oryza sativa HSP22.2 (4339231). Figure S4. Gene ontology (GO) classification of the annotated unigenes or differentially expressed genes (DEGs). Unigenes with best BLAST hits were aligned to GO database. A total of 26,326 unigenes (red) and 261 DEGs (blue) were assigned to at least one GO term and grouped into three main GO categories (cellular component, molecular function and biological process) and 53 GO terms. Left Y-axis represents the percentages of unigenes or DEGs in each main category. Right Y-axis indicates the numbers of unigenes or DEGs in each GO term. Figure S5. Correlation assessment of a pair of biological replicates in wild type (WT) and AsHSP26.8a transgenic (TG) plants, respectively, used for RNA-seq. A, Pearson’s correlation coefficient between the two biological replicates of the wild type (E01 and E02) or AsHSP26.8a transgenic plants (E05 and E06). B and C, Correlation plots of a pair of biological replicates from wild type (E01 and E02), (B) and AsHSP26.8a transgenic line TG3 (E05 and E06) (C), respectively. Each dot in the plot represents a gene, denoting the log10 of the gene expression (FPKM) in the two replicate samples (x-axis and y-axis). The closer is the dot to the diagonal, the smaller is the difference in expression for the corresponding gene in the two replicates.
Acknowledgements
We wish to thank Dr. Dayong Li for his help in AsHSP26.8a subcellular localization experiment.
Abbreviations
- CaM
Calmodulins
- CaMV35S
Cauliflower mosaic virus 35S;
- CBLs
Calcineurin B-like proteins (CBLs)
- CDPKs
Calcium dependent protein kinases
- CMLs
Calmodulin-like proteins
- DEGs
Differentially expressed genes
- EL
Electrolyte leakage
- FDR
False discovery rate
- GFP
Green florescent protein
- GO
Gene Ontology
- HSFs
Heat shock transcription factors
- HSPs
Heat shock proteins
- LEA
Late embryogenesis abundant
- MW
Molecular weight
- RNA-seq
RNA sequencing
- RWC
Relative water content
- TF
Transcription factor
Authors’ contribution
XS, LH and HL conceived and designed the experiments. XS, JZ, XL, and ZL conducted experiments. XS and HL analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by the National Science Foundation for Young Scientists of China (31701955), the Natural Science Foundation of Hebei Province (C2018204056), Young Talents Support Program of Hebei Province and in part by the Biotechnology Risk Assessment Grant Program competitive grant no. 2019–33522-30102 from the USDA National Institute of Food and Agriculture and the United States Golf Association, Inc. These funding bodies did not play any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All relevant data are included in this article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liebao Han, Email: hanliebao@163.com.
Hong Luo, Email: hluo@clemson.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02369-5.
References
- 1.Howarth CJ. Molecular responses of plants to an increased incidence of heat shock. Plant Cell Environ. 1991;14:831–841. doi: 10.1111/j.1365-3040.1991.tb01446.x. [DOI] [Google Scholar]
- 2.Basha E, O’Neill H, Vierling E. Small heat shock proteins and a-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 4.Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun W, Bernard C, van de Cotte B, van Montagu M, Verbruggen N. AtHSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhanceosmotolerance upon overexpression. Plant J. 2001;27:407–415. doi: 10.1046/j.1365-313X.2001.01107.x. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McHaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry. 2009;48:3828–3837. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyles SJ. Gierasch LM Nature’s molecular sponges: small heat shock proteins grow into their chaperone roles. Proc Natl Acad Sci U S A. 2010;107:2727–2728. doi: 10.1073/pnas.0915160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Luthe DS. Heat sensitivity in a bentgrass variant. Failure to accumulate a chloroplast heat shock protein isoform implicated in heat tolerance. Plant Physiol. 2003;133:319–327. doi: 10.1104/pp.102.018309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun L, Liu Y, Kong X, Zhang D, Pan J, Zhou Y, et al. ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 2012;31:1473–1484. doi: 10.1007/s00299-012-1262-8. [DOI] [PubMed] [Google Scholar]
- 11.Yang G, Wang Y, Zhang K, Gao C. Expression analysis of nine small heat shock protein genes from Tamarix hispida in response to different abiotic stresses and abscisic acid treatment. Mol Biol Rep. 2014;41:1279–1289. doi: 10.1007/s11033-013-2973-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Liu B, Li J, Zhang L, Wang Y, Zheng H, et al. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress response. BMC Genomics. 2015;16:181. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu T, Liu S, Amombo E. Fu J. Stress memory induced rearrangements of HSP transcription, photosystem II photochemistry and metabolism of tall fescue (Festuca arundinacea Schreb.) in response to high-temperature stress. Front. Plant Sci. 2015; 6:403. [DOI] [PMC free article] [PubMed]
- 14.Nitnavare RB, Yeshvekar RK, Sharma KK, Vadez V, Reddy MK, Reddy PS. Molecular cloning, characterization and expression analysis of a heat shock protein 10 (Hsp10) from Pennisetum glaucum (L.), a C4 cereal plant from the semi-arid tropics. Mol Biol Rep. 2016;43:861–870. doi: 10.1007/s11033-016-4012-0. [DOI] [PubMed] [Google Scholar]
- 15.Sun X, Sun C, Li Z, Hu Q, Han L, Luo H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 2016;39:1320–1337. doi: 10.1111/pce.12683. [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta. 2002;1577:1–9. doi: 10.1016/S0167-4781(02)00417-7. [DOI] [PubMed] [Google Scholar]
- 17.Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 19.Park H, Ko E, Jang E, Park S, Lee J, Ahn Y. Expression of DcHSP17.7, a small heat shock protein gene in carrot (Daucus carota L.) Hortic Environ Biote. 2013;54:121–127. doi: 10.1007/s13580-013-0027-9. [DOI] [Google Scholar]
- 20.Volkov RA, Panchuk II, Schöffl F. Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol Biol. 2005;57:487–502. doi: 10.1007/s11103-005-0339-y. [DOI] [PubMed] [Google Scholar]
- 21.Chauhan H, Khurana N, Nijhavan A, Khurana JP, Khurana P. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 2012;35:1912–1931. doi: 10.1111/j.1365-3040.2012.02525.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, et al. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell. 2013;25:2925–2943. doi: 10.1105/tpc.113.111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami T, Matsuba S, Funatsuki H, Kawaguchi K, Saruyama H, Tanida M, Sato Y. Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol Breed. 2004;13:165–175. doi: 10.1023/B:MOLB.0000018764.30795.c1. [DOI] [Google Scholar]
- 24.Sato Y, Yokoya S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep. 2008;27:329–334. doi: 10.1007/s00299-007-0470-0. [DOI] [PubMed] [Google Scholar]
- 25.Jiang C, Xu J, Zhang H, Zhang X, Shi J, Li M, Ming F. A cytosolic class I small heat shock protein, RcHSP17.8, of Rosa chinensis confers resistance to a variety of stresses to Escherichia coli, yeast and Arabidopsis thaliana. Plant Cell Environ. 2009;32:1046–1059. doi: 10.1111/j.1365-3040.2009.01987.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim KH, Alam I, Kim YG, Sharmin SA, Lee KW, Lee SH, Lee BH. Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol Lett. 2012;34:371–377. doi: 10.1007/s10529-011-0769-3. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, Xu Z, Hwang I. AtHSP17.8 overexpression in transgenic lettuce gives rise to dehydration and salt stress resistance phynotypes through modulation of ABA-mediated signaling. Plant Cell Rep. 2013;32:1953–1963. doi: 10.1007/s00299-013-1506-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Chen H, Chu P, Li Y, Tan B, Ding Y, et al. NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 2012;31:379–389. doi: 10.1007/s00299-011-1173-0. [DOI] [PubMed] [Google Scholar]
- 29.Ham DJ, Moon JC, Hwang SG, Jang CS. Molecular characterization of two small heat shock protein genes in rice: their expression patterns, localizations, networks, and heterogeneous overexpressions. Mol Biol Rep. 2013;40:6709–6720. doi: 10.1007/s11033-013-2786-x. [DOI] [PubMed] [Google Scholar]
- 30.Mu C, Zhang S, Yu G, Chen N, Li X, Liu H. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS One. 2013;8:e82264. doi: 10.1371/journal.pone.0082264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merino I, Contreras A, Jing ZP, Gallardo F, Cánovas FM, Gómez L. Plantation forestry under global warming: hybrid poplars with improved thermotolerance provide new insights on the in vivo function of small heat shock protein chaperones. Plant Physiol. 2014;164:978–991. doi: 10.1104/pp.113.225730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santhanagopalan I, Basha E, Ballard KN, Bopp NE, Vierling E. Model chaperones: small heat shock proteins from plants. In: Tanguay RM, Hightower LE, editors. The big book on small heat shock proteins. Cham, Switzerland: Springer International Publishing; 2015. pp. 119–153. [Google Scholar]
- 33.McLoughlin F, Basha E, Fowler ME, Kim M, Bordowitz J, Katiyar-Agarwal S, Vierling E. Class I and II small heat shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiol. 2016;172:1221–1236. doi: 10.1104/pp.16.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters ER, Vierling E. The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol Biol Evol. 1999;16:127–139. doi: 10.1093/oxfordjournals.molbev.a026033. [DOI] [PubMed] [Google Scholar]
- 35.Waters ER, Rioflorido I. Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J Mol Evol. 2007;65:162–174. doi: 10.1007/s00239-006-0223-7. [DOI] [PubMed] [Google Scholar]
- 36.Waters ER. The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Li Y, Liu Y, Wu Y, Xie Q. The sHSP22 heat shock protein requires the ABI1 protein phosphatase to modulate polar auxin transport and downstream responses. Plant Physiol. 2018;176:2406–2425. doi: 10.1104/pp.17.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Li J, Hao R, Guo Y. Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6II. J Genet Genomics. 2017;44:395–404. doi: 10.1016/j.jgg.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Ma W, Guan X, Li J, Pan R, Wang L, Liu F, et al. Mitochondrial small heat shock protein mediates seed germination via thermal sensing. Proc Natl Acad Sci U S A. 2019;116:4716–4721. doi: 10.1073/pnas.1815790116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckathorn SA, Downs CA, Sharkey TD, Coleman JS. The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol. 1998;116:439–444. doi: 10.1104/pp.116.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heckathorn SA, Ryan SL, Baylis JA, Wang DF, Hamilton EW, Cundiff L, et al. In vivo evidence from an Agrostis stolonifera selection genotype that chloroplast small heat-shock proteins can protect photosystem II during heat stress. Funct Plant Biol. 2002;29:933–944. doi: 10.1071/PP01191. [DOI] [PubMed] [Google Scholar]
- 42.Shakeel S, Ul Haq N, Heckathorn SA, Hamilton EW, Luthe DS. Ecotypic variation in chloroplast small heat-shock proteins and related thermotolerance in Chenopodium album. Plant Physiol Biochem. 2011;49:898–908. doi: 10.1016/j.plaphy.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Harndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF. Sundby C. The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones 1999;4: 129–138. [DOI] [PMC free article] [PubMed]
- 44.Chen S, He N, Chen J, Guo F. Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5-mediated retrograde pathway in Arabidopsis. Plant J. 2017;89:1106–1118. doi: 10.1111/tpj.13447. [DOI] [PubMed] [Google Scholar]
- 45.Sedaghatmehr M, Mueller-Roeber B, Balazadeh S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat Commun. 2016;7:12439. doi: 10.1038/ncomms12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Hu Q, Zhou M, Vandenbrink J, Li D, Menchyk N, et al. Heterologous expression of OsSIZ1, a rice SUMO E3 ligase, enhances broad abiotic stress tolerance in transgenic creeping bentgrass. Plant Biotechnol J. 2013;11:432–445. doi: 10.1111/pbi.12030. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Yuan S, Jia H, Gao F, Zhou M, Yuan N, et al. Ectopic expression of a cyanobacterial flavodoxin in creeping bentgrass impacts plant development and confers broad abiotic stress tolerance. Plant Biotechnol J. 2016;15:433–446. doi: 10.1111/pbi.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Yuan S, Zhou M, Yuan N, Li Z, Hu Q, et al. Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol J. 2018;17:233–251. doi: 10.1111/pbi.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finkelstein R: Abscisic acid synthesis and response. Arabidopsis Book 2013, 11:e0166 http://dx.doi.org/10.1199/tab.%200166. [DOI] [PMC free article] [PubMed]
- 50.Wani SH, Kumar V. Plant stress tolerance: engineering ABA: a potent phytohormone. Transcriptomics: Open Access. 2015;3:1000113. doi: 10.4172/2329-8936.1000113. [DOI] [Google Scholar]
- 51.Sah SK, Reddy KR, Li J. Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci. 2016;7:571. doi: 10.3389/fpls.2016.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vierling E, Harris LM, Chen Q. The major low-molecularweight heat-shock protein in chloroplasts shows antigenic conservation among diverse higher-plant species. Mol Cell Biol. 1989;9:461–468. doi: 10.1128/MCB.9.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vierling E. The roles of heat-shock proteins in plants. Annu. Rev. plant Physiol. Plant Mol. Biol. 1991;42:579–620. [Google Scholar]
- 54.Scharf KD, Berberich T, Ebersberbger I, Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta. 1819;2012:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Akhtar M, Jaiswal A, Taj G, Jaiswal JP, Qureshi MI, Singh NK. DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J Genet. 2012;91:385–395. doi: 10.1007/s12041-012-0201-3. [DOI] [PubMed] [Google Scholar]
- 56.Fragkostefanakis S, Röth S, Schleiff E, Scharf KD. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 2015;38:1881–1895. doi: 10.1111/pce.12396. [DOI] [PubMed] [Google Scholar]
- 57.Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol J. 2017;15:405–414. doi: 10.1111/pbi.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011;157:1243–1254. doi: 10.1104/pp.111.179036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashraf M, Yang S, Wu R, Wang Y, Hussain A, Noman A, et al. Capsicum annuum HsfB2a positively regulates the response to Ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with CaWRKY6 and CaWRKY40. Plant Cell Physiol. 2018;59:2608–2623. doi: 10.1093/pcp/pcy181. [DOI] [PubMed] [Google Scholar]
- 60.Guan Q, Yue X, Zeng H, Zhu J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell. 2014;26:438–453. doi: 10.1105/tpc.113.118927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhuang L, Cao W, Wang J, Yang Z, Huang B. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis. Int J Mol Sci. 2018;19:2702. doi: 10.3390/ijms19092702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang D. Abscisic acid: metabolism, transport and signaling. New York, NY: Springer; 2014. [Google Scholar]
- 63.Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- 64.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 65.Shao HB, Liang ZS, Shao MA. LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids Surf B Biointerfaces. 2005;45:131–135. doi: 10.1016/j.colsurfb.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 66.Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- 67.Tang N, Ma S, Zong W, Yang N, Lv Y, Yan C, et al. MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell. 2016;28:2161–2177. doi: 10.1105/tpc.16.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang L, Zhang M, Jia J, Zhao X, Huang X, Ji E, et al. An atypical late embryogenesis abundant protein OsLEA5 plays a positive role in ABA-induced antioxidant defense in Oryza Sativa L. Plant Cell Physiol. 2018;59:916–929. doi: 10.1093/pcp/pcy035. [DOI] [PubMed] [Google Scholar]
- 69.Magwanga RO, Lu P, Kirungu JN, Dong Q, Hu Y, Zhou Z, et al. Cotton late embryogenesis abundant (LEA2) genes promote root growth and confers drought stress tolerance in transgenic Arabidopsis thaliana. G3-genes genomes. Genetics. 2018;8:2781–2803. doi: 10.1534/g3.118.200423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C, Ng CKY, Fan LM. MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot. 2015;114:80–91. doi: 10.1016/j.envexpbot.2014.06.014. [DOI] [Google Scholar]
- 73.Yu Y, Ni Z, Chen Q, Qu Y. The wheat salinity-induced R2R3-MYB transcription factor TaSIM confers salt stress tolerance in Arabidopsis thaliana. Biochem Biophys Res Commun. 2017;491:642–648. doi: 10.1016/j.bbrc.2017.07.150. [DOI] [PubMed] [Google Scholar]
- 74.Wei Q, Luo Q, Wang R, Zhang F, He Y, Zhang Y, et al. A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front Plant Sci. 2017;8:1374. doi: 10.3389/fpls.2017.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao F, Zhou J, Deng R, Zhao H, Li C, Chen H, et al. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J Plant Physiol. 2017;214:81–90. doi: 10.1016/j.jplph.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 76.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- 77.Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 78.Schluttenhofer C, Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167:295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 80.He GH, Xu JY, Wang YX, Liu JM, Li PS, Chen M, et al. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016;16:116. doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yan H, Jia H, Chen X, Hao L, An H, Guo X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014;55:2060–2076. doi: 10.1093/pcp/pcu133. [DOI] [PubMed] [Google Scholar]
- 82.Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura SK, Yamaguchi-Shinozaki K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/A:1006321900483. [DOI] [PubMed] [Google Scholar]
- 83.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 84.Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA. 2006;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen H, Hwang JE, Lim CJ, Kim DY, Lee SY, Lim CO. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem Biophys Res Commun. 2010;401:238–244. doi: 10.1016/j.bbrc.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 87.Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol Plant. 2010;54:201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- 88.Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011;62:4731–4738. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 89.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 90.Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, et al. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 91.Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009;150:1972–1980. doi: 10.1104/pp.109.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, et al. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- 93.Suo H, Ma Q, Ye K, Yang C, Tang Y, Hao J, et al. Overexpression of AtDREB1A causes a severe dwarf phenotype by decreasing endogenous gibberellin levels in soybean [Glycine max (L.) Merr] PLoS One. 2012;7:e45568. doi: 10.1371/journal.pone.0045568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhatnagar-Mathur P, Rao JS, Vadez V, Dumbala SR, Rathore A, Yamaguchi-Shinozaki K, et al. Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol Breed. 2014;33:327–340. doi: 10.1007/s11032-013-9952-7. [DOI] [Google Scholar]
- 95.Wei T, Deng K, Gao Y, Liu Y, Yang M, Zhang L, et al. Arabidopsis DREB1B in transgenic Salvia miltiorrhiza increased tolerance to drought stress without stunting growth. Plant Physiol Biochem. 2016;104:17–28. doi: 10.1016/j.plaphy.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 96.Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- 97.Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, et al. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53:264–274. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- 98.Büttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci U S A. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zarei A, Korbes AP, Younessi P, Montiel G, Champion A, Memelink J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol. 2011;75:321–331. doi: 10.1007/s11103-010-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Licausi F, Ohme-Takagi M, Perata P. APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199:639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- 101.Li Z, Tian Y, Xu J, Fu X, Gao J, Wang B, et al. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. Tomato DC3000. Plant Physiol Biochem. 2018;132:683–695. doi: 10.1016/j.plaphy.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 102.Nie J, Wen C, Xi L, Lv S, Zhao Q, Kou Y, et al. The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 2018;37:1049–1060. doi: 10.1007/s00299-018-2290-9. [DOI] [PubMed] [Google Scholar]
- 103.Qin L, Wang L, Guo Y, Li Y, Ümüt H, Wang Y. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant Sci. 2017;265:154–166. doi: 10.1016/j.plantsci.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 104.Kim KN, Cheong YH, Gupta R, Luan S. Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 2000;124:1844–1853. doi: 10.1104/pp.124.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene encoding a Ca2+−binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol. 2000;41:898–903. doi: 10.1093/pcp/pcd010. [DOI] [PubMed] [Google Scholar]
- 106.Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C. Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol. 2004;134:898–908. doi: 10.1104/pp.103.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gong D, Guo Y, Schumaker KS, Zhu JK. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004;134:919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hashimoto K, Kudla J. Calcium decoding mechanisms in plants. Biochimie. 2011;93:2054–2059. doi: 10.1016/j.biochi.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 110.Reddy AS. Calcium: silver bullet in signaling. Plant Sci. 2001;160:381–404. doi: 10.1016/S0168-9452(00)00386-1. [DOI] [PubMed] [Google Scholar]
- 111.Jiang Z, Zhu S, Ye R, Xue Y, Chen A, An L, et al. Relationship between NaCl- and H2O2-induced cytosolic Ca2+ increases in response to stress in Arabidopsis. PLoS One. 2013;8:e76130. doi: 10.1371/journal.pone.0076130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Delk NA, Johnson KA, Chowdhury NI, Braam J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 2005;139:240–253. doi: 10.1104/pp.105.062612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamaguchi T, Aharon GS, Sottosanto JB. Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+− and pH-dependent manner. Proc Natl Acad Sci U S A 2005;102: 16107–16112. [DOI] [PMC free article] [PubMed]
- 114.Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008;56:575–589. doi: 10.1111/j.1365-313X.2008.03622.x. [DOI] [PubMed] [Google Scholar]
- 115.Qiao B, Zhang Q, Liu D, Wang H, Yin J, Wang R, et al. A calcium-binding protein, rice annexin OsANN1, enhances heat stress tolerance by modulating the production of H2O2. J Exp Bot. 2015;66:5853–5866. doi: 10.1093/jxb/erv294. [DOI] [PubMed] [Google Scholar]
- 116.Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci STKE. 2001;(113):re22. [DOI] [PubMed]
- 117.Wei Z, Li J. Receptor-like protein kinases: key regulators controlling root hair development in Arabidopsis thaliana. J Integr Plant Biol. 2018;60:841–850. doi: 10.1111/jipb.12663. [DOI] [PubMed] [Google Scholar]
- 118.Lim CW, Yang SH, Shin KH, Lee SC, Kim SH. The AtLRK10L1.2, Arabidopsis, ortholog of wheat LRK10, is involved in ABA-mediated signaling and drought resistance. Plant Cell Rep. 2015;34:447–455. doi: 10.1007/s00299-014-1724-2. [DOI] [PubMed] [Google Scholar]
- 119.Sun XL, Sun M, Luo X, Ding XD, Cai H, Bai X, et al. Erratum to: a Glycine soja, ABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses. Planta. 2013;237:1527–1545. doi: 10.1007/s00425-013-1864-6. [DOI] [PubMed] [Google Scholar]
- 120.Vaid N, Pandey P, Srivastava VK, Tuteja N. Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol Biol. 2015;88:1–14. doi: 10.1007/s11103-015-0319-9. [DOI] [PubMed] [Google Scholar]
- 121.de Lorenzo L, Merchan F, Laporte P, Thompson R, Clarke J, Sousa C, et al. A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell. 2009;21:668–680. doi: 10.1105/tpc.108.059576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol. 1999;39:1073–1078. doi: 10.1023/A:1006184519697. [DOI] [PubMed] [Google Scholar]
- 123.Kim SH, Ahn YO, Ahn M, Jeong JC, Lee H, Kwak S. Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol Biochem. 2013;70:445–454. doi: 10.1016/j.plaphy.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 124.Muthamilarasan M, Bonthala VS, Mishra AK, Khandelwal R, Khan Y, Roy R, et al. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct Integr Genomic. 2014;14:531–543. doi: 10.1007/s10142-014-0383-2. [DOI] [PubMed] [Google Scholar]
- 125.Yue X, Que Y, Xu L, Deng S, Peng Y, Talbot NJ, et al. ZNF1 encodes a putative C2H2 zinc-finger protein essential for appressorium differentiation by the rice blast fungus Magnaporthe oryzae. Mol Plant-Microbe Interact. 2016;29:22–35. doi: 10.1094/MPMI-09-15-0201-R. [DOI] [PubMed] [Google Scholar]
- 126.Chen J, Yang L, Yan X, Liu Y, Wang R, Fan T, et al. Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol. 2016;171:707–719. doi: 10.1104/pp.15.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, et al. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem. 2007;282:9260–9268. doi: 10.1074/jbc.M611093200. [DOI] [PubMed] [Google Scholar]
- 128.Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, et al. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, et al. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell. 2007;19:4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nguyen XC, Kim SH, Hussain S, An J, Yoo Y, Han HJ, et al. A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. J Plant Biol. 2016;59:55–61. doi: 10.1007/s12374-016-0442-4. [DOI] [Google Scholar]
- 132.Song H, Zhao R, Fan P, Wang X, Chen X, Li Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta. 2009;229:955–964. doi: 10.1007/s00425-008-0886-y. [DOI] [PubMed] [Google Scholar]
- 133.Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013;161:1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luo H, Kausch AP, Hu Q, Nelson K, Wipff JK, Fricker CCR, et al. Controlling transgene escape in GM creeping bentgrass. Mol Breed. 2005;16:185–188. doi: 10.1007/s11032-005-4784-8. [DOI] [Google Scholar]
- 135.Bart R, Chern M, Park CJ, Battley L, Ronald PC. A novel system for gene slienceing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods. 2006;2:13. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schulze SK, Kanwar R, Gölzenleuchter M, Therneau TM, Beutler AS. SERE: single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics. 2012;13:524. doi: 10.1186/1471-2164-13-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Z, Baldwin CM, Hu Q, Liu H, Luo H. Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.) Plant Cell Environ. 2010;33:272–289. doi: 10.1111/j.1365-3040.2009.02080.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of assembled transcripts up-regulated or down-regulated (log2FC > 1 or < − 1, FDR < 0.01) in AsHSP26.8a transgenic (TG) Arabidopsis, relative to wild type (WT) plants.
Additional file 2: Table S2. Primers used in this study.
Additional file 3: Figure S1. AsHSP26.8a nucleic acid sequence (A) and amino acid sequence (B) alignment with AsHSP26.8. Figure S2. Amino acid sequence alignment of AsHSP26.8a and other plant chloroplast localized (CP) sHSPs. Accession umbers of sHSPs are: Agrostis stolonifera HSP26.8a (KU353578); Aegilops kotschyi HSP26.8 (CAI96511); Triticum dicoccoides HSP26.4 (CAI96512); Hordeum vulgare HSP26 (AAB28590); Brachypodium distachyon sHSP (XP_003558381.1); Spartina alterniflora HSP27.1 (AFP96756.1); Zea mays HSP18 (CAM12752.1); Oryza sativa HSP26 (BAA78385.1). Figure S3. The phylogenetic relationship of the AsHSP26.8a with different classes of sHSPs from other plant species. Accession numbers of the sHSPs are: Arabidopsis thaliana HSP17.4 (X17293); Arabidopsis thaliana HSP17.6 (X16076); Oryza sativa HSP17.4 (D12635); Daucus carota HSP18.0 (X53852); Helianthus annuus HSP17.6 (X59701); Agrostis stolonifera HSP17 (KT272405); Triticum aestivum HSP16.9A (X13431); Oryza sativa HSP16.9A (X60820); Zea mays HSP16.9 (X65725); Solanum lycopersicum HSP17.8 (X56138); Glycine max HSP17.5 (M11318); Glycine max HSP17.6 (MI1317); Medicago sativa HSP18.1 (X58710); Pisum sativum HSP18.1 (M33899); Arabidopsis thaliana HSP22 (U11501); Glycine max HSP22 (X63198); Pisum sativum HSP22 (M33898); Arabidopsis thaliana HSP18.5 (816448); Oryza sativa HSP17.6 (113595340); Arabidopsis thaliana HSP15.7 (833746); Arabidopsis thaliana HSP26.5 (841687); Oxybasis rubra HSP23 (X15333); Arabidopsis thaliana HSP23.6 (828623); Oryza sativa HSP22 (4330786); Agrostis stolonifera HSP26.8a (KU353578); Triticum aestivum HSP26.6 (X58280); Zea mays HSP26 (L28712); Petunia x hybrida HSP21 (X54103); Arabidopsis thaliana HSP21 (X54102); Glycine max HSP22 (X07188); Pisum sativium HSP21 (X07187); Arabidopsis thaliana HSP15.4 (828276); Oryza sativa HSP18.8 (4343386); Arabidopsis thaliana HSP17.4 (841843); Oryza sativa HSP17.6B (4330933); Lilium longiflorum HSP17.6 (D21816); Zea mays HSP17.5 (X54076); Zea mays HSP17.8 (X54075); Triticum aestivum HSP17.3 (X58279); Arabidopsis thaliana HSP17.6 (X63443); Ipomoea nil HSP18.8 (M99430); Ipomoea nil HSP17.2 (M99429); Glycine max HSP17.9 (X07159); Pisum sativum HSP17.7 (M33901); Arabidopsis thaliana HSP21.7 (835555); Oryza sativa HSP22.2 (4339231). Figure S4. Gene ontology (GO) classification of the annotated unigenes or differentially expressed genes (DEGs). Unigenes with best BLAST hits were aligned to GO database. A total of 26,326 unigenes (red) and 261 DEGs (blue) were assigned to at least one GO term and grouped into three main GO categories (cellular component, molecular function and biological process) and 53 GO terms. Left Y-axis represents the percentages of unigenes or DEGs in each main category. Right Y-axis indicates the numbers of unigenes or DEGs in each GO term. Figure S5. Correlation assessment of a pair of biological replicates in wild type (WT) and AsHSP26.8a transgenic (TG) plants, respectively, used for RNA-seq. A, Pearson’s correlation coefficient between the two biological replicates of the wild type (E01 and E02) or AsHSP26.8a transgenic plants (E05 and E06). B and C, Correlation plots of a pair of biological replicates from wild type (E01 and E02), (B) and AsHSP26.8a transgenic line TG3 (E05 and E06) (C), respectively. Each dot in the plot represents a gene, denoting the log10 of the gene expression (FPKM) in the two replicate samples (x-axis and y-axis). The closer is the dot to the diagonal, the smaller is the difference in expression for the corresponding gene in the two replicates.
Data Availability Statement
All relevant data are included in this article and its supplementary information files.