Abstract
A simple method for the identification of brush-border membrane α-glucosidases is described. The proteins were first solubilized and separated in a gel under native, non-denaturing, conditions. The gel was then incubated in substrate solutions (maltose or sucrose), and the product (glucose) exposed in situ by the oxidation of o-dianisidine, which yields a brown-orange color. Nano-liquid chromatography coupled to mass spectrometry analyses of proteins (nano LC-MS/MS) present in the colored bands excised from the gels, was used to confirm the presence of the enzymes. The stain is inexpensive and the procedure permits testing several substrates in the same gel. Once enzymes are identified, their abundance, relative to that of other proteins in the brush border, can be semi-quantified using nano LC-MS/MS.
Keywords: α-glucosidase, polyacrylamide gels, detection, proteomics, activity
1. Introduction
It is a common goal in physiology to assess the function of enzymes as well as to identify effector proteins in subcellular compartments. Numerous methods have been proposed based on split assays, one to measure the function of the enzyme and another to identify the protein that catalyzes the reaction. The latter step often requires an additional sample (not the one in which function was measured), and relies on the use of antibodies. However, antibodies typically are species-specific and do not cross-react when studying non-traditional model animals, or are not specific enough to distinguish between closely related enzymes. Specific antibodies can require time to be produced. We describe a method with the following two advantages: (1) It can perform both steps on the same sample, and (2) the enzyme identification is fast and accurate. We used the method to reveal the hydrolytic activity of α-glucosidases in brush border membrane (bbm) of intestinal epithelial cells (enterocytes), and their identification, by combining a gel electrophoresis for sample protein separation, a colored reaction to reveal the enzyme activity on the substrate, i.e., zymography [1], and a proteomic method, for enzyme identification-nano-liquid chromatography coupled to mass spectrometry (nano LC-MS/MS ) of the solubilized stained bands of the gels. If the method is followed by nano LC-MS/MS on the proteins from the bbm, the method also allows quantifying the relative abundance of the enzymes identified.
Our demonstration target enzymes hydrolyze the final products of the degradation of starch and glycogen (e.g., maltose and maltoligosaccharides) and disaccharides (e.g., maltose, sucrose, isomaltose). In mammals, the intestinal α-glucosidases are the complexes sucrase-isomaltase (SI) and maltase-glucoamylase (MGAM) of the bbm of enterocytes. SI and MGAM are orthologs that derive from an ancient duplication and are similar in structure and composition, sharing a 60% homology between complexes [2,3]. Both enzyme complexes include two subunits, an N-terminal subunit (Nt-MGAM and Nt-SI) and a C-terminal subunit (Ct-MGAM and Ct-SI) [4,5]. According to the classification system of the carbohydrate-active enzymes (CAZy, [6]), all of the subunits belong to the glycoside hydrolase 31 family (GH31) subgroup 1, characterized by the WIDMNE sequence in their catalytic centers [7]. All these similarities make it time consuming and costly to distinguish the different complexes. Moreover, because some of the subunits share substrate hydrolytic specificity [8,9], and in the reaction assay the activity gives similar colored products, distinguishing between the enzymes poses an even greater challenge. Therefore, we adapted a method using zymography to separate and detect activities plus nano LC-MS/MS to recognize the proteins in the gels slices where the reaction was apparent. We used a preparation of bbm of the small intestine of laboratory mice and two substrates hydrolyzed by the complexes, maltose and sucrose. According to the specificities of these enzymes, the proteomic analyses of the gel slices should reveal the presence of both proteins.
2. Material and Methods
2.1. Brush Border Membrane Preparation and Zymography
Dr. Sarah Newman from the University of Wisconsin—Madison Research Animal Resource Center (RARC) provided the study animals. The gastrointestinal tracts of two Swiss-Webster mice anesthetized with isoflurane were removed. The small intestine from the pyloric valve to the ileocecal junction was excised and placed in ice cold PBS solution for enterocyte isolation. Immediately after, animals were euthanized by anesthesia overdose. The isolation of enterocytes and bbm were performed according to protocols described by Mac Donal, et al. [10] and McConnell, et al. [11].
Proteins from the bbm preparations were solubilized in loading buffer (60 mM Tris-HCl pH 6.8, 20% Glycerol, 1% Triton X 100 and 0.01% Bromophenol blue) and then incubated 10 min at 40 °C in a water bath, vortexed, and 10 µg of total protein was loaded per gel well. For each mouse, three gels were run; one to reveal maltose hydrolytic activity (identified as I in Table 1), one for sucrose activity (identified as II in Table 1), and the remaining gel was used to visualize the proteins by Coomassie Blue staining following Simpson [12]. All the gels were run with PageRuler Plus Prestained Protein ladder (ThermoFisher Scientific catalog number 26,619, Waltham, MA, USA) as a molecular marker. Gels I and II were run at room temperature in 4–12% Tris-Glycine Mini polyacrylamide gel (NovexTM TermoFisher Scientific) at 100 V constant and ~350 mA using Tris-glycine pH 8.3 as running buffer (Table 1). Coomassie blue gel running time was 2 h. Gels for disaccharidase activity were run for a total of 3 h to have better separation between these enzymes that have similar molecular weight and overlapping activity [8,9,13,14,15]. The two gels for disaccharidase activity were immersed in either maltose or sucrose 56 mM in maleate-OH buffer for 1 hour at 37 °C (Table 1). The buffered substrate solution was removed. Then the gels were soaked in assay reagent solution of the Glucose (GO) Assay Kit (Sigma-Aldrich GAGO20, Saint Louis, MO, USA) at 37 °C; for 1 h for the gel previously soaked in maltose, and for 5 h for the gel previously soaked in sucrose (Table 1, Figure 1). The reaction is based on the promotion of the oxidation of glucose produced by the hydrolytic enzymes tested by glucose oxidase and formation of H2O2 by a peroxidase, which in turn oxidizes the colorless-reduced o-dianisidine forming an orangish compound according to the manufacturer (Glucose (GO) Assay Kit, Sigma-Aldrich GAGO20). The reaction was stopped by rinsing the gel with deionized (DI) water 3 times. Finally, the bands were cut and submerged in DI water and stored at 4 °C for further analysis.
Table 1.
Summary of parameters used with each substrate tested in the zymography.
| Electrophoresis | ||
|---|---|---|
| Gel | I | II |
| Constituents 1 | 4–12% mini polyacrylamide gel 1 | |
| Temperature of the run | room temperature | |
| Total protein loaded per well | 10 µg | |
| Running buffer | Tris-glycine pH 8.3 | |
| Electric field applied | 100 V constant and ~350 mA | |
| Run time | 3 h | |
| Hydrolytic activity assay | ||
| Substrate solution and concentration | Maltose 56 mM | Sucrose 56 mM |
| Incubation time | 1 h | 1 h |
| Incubation temperature | 37 °C | |
| Assay reagent 2
Incubation time (reaction temperature) |
1 h (37 °C) |
5 h (37 °C) |
| Rinse the gel with deionized water | 3 × | |
| Gel storage for further analysis | 4 °C | |
References: 1 NovexTM 4–12% Tris-glycine mini polyacrylamide gel; 2 Glucose (GO) Assay Kit (Sigma GAGO-20).
Figure 1.
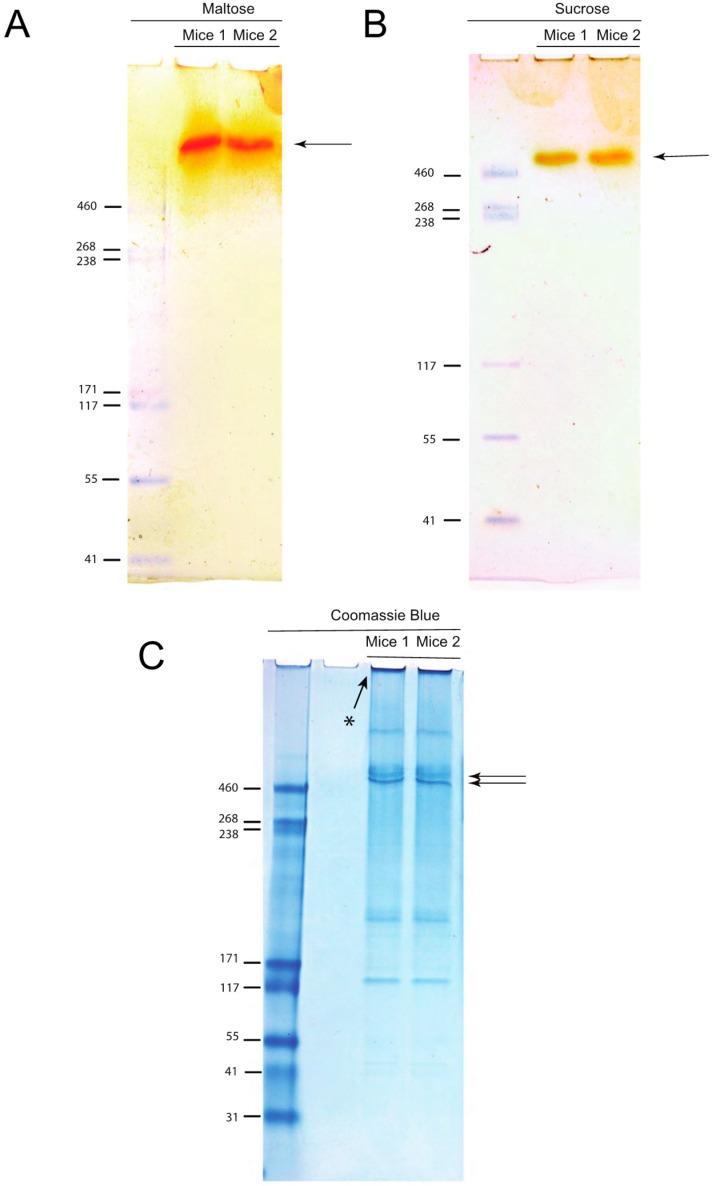
Bands developed indicating α-glucosidase activity by zymography. The polyacrylamide gels were incubated with either maltose (A) or sucrose (B) solution or Coomassie blue solution (C). Then for A and B only, the gel was immersed in assay reagent solution until a well-defined band appeared, as shown. Asterisk (*) on Figure 1C shows proteins that did not run through the gel.
2.2. Enzymatic “In Gel” Digestion
Stained gel slices from the native gel were excised based on the activity staining. Slices were placed for 5 min in MeOH/H2O/NH4HCO3 [50%:50%:25 mM], and the proteins entrapped in the gel were denatured for 10 min in SDS/DTT/Tris-HCl solution [2%/1 mM/50 mM pH 7] with subsequent 2 × 5 min washes in MeOH/H2O/NH4HCO3 [50%:50%:25 mM]. Gel fragments were washed twice for 5 min in MeOH/H2O/NH4HCO3 [50%:50%:100 mM], dehydrated for 5 min in acetonitrile (ACN)/H2O/NH4HCO3 [50%:50%:25 mM] then once more for 1 min in 100% ACN, dried in a vacuum concentrator (Speed-Vac, ThermoFisher Scientific) for 2 min, reduced in 25 mM DTT (dithiotreitol in 25 mM NH4HCO3) for 30 min at 52 °C, alkylated with 55 mM IAA (iodoacetamide in 25 mM NH4HCO3) in darkness at room temperature for 30 min, washed twice in H2O for 30 s equilibrated in 25 mM NH4HCO3 for 1 min, dehydrated for 5 min in ACN/H2O/NH4HCO3 [50%:50%:25 mM] then once more for 30 s in 100% ACN, dried again and rehydrated with 20 μL of trypsin solution [10 ng/μL trypsin (Promega, Madison, WI, USA) in 25 mM NH4HCO3/0.01% Protease MAX w/v (Promega)]. An additional 30 μL of digestion solution (25 mM NH4HCO3/0.01% Protease MAX w/v) was added to facilitate complete rehydration. The digestion was conducted for 3 h at 42 °C. The supernatant was removed, and gel pieces were extracted for peptides with 2 × gel volume of ACN/H2O/TFA solution [70/30/0.75%]. Extracted peptides were combined with the supernatant. Degraded Protease MAX was removed via centrifugation [max speed 16,000 g, 10 min] and the peptides solid phase extracted (ZipTipC18pipette tips Millipore, Burlington, VT, USA). The extracted peptides were subsequently submitted for nano-LC-MS/MS analysis. This protocol can be found at: https://www.biotech.wisc.edu/services/massspec/protocols/ingel.
2.3. Nano LC-MS/MS
Peptides were analyzed by nano LC-MS/MS using the Agilent 1100 nanoflow system (Agilent Technologies, Santa Clara, CA, USA) connected to a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap Elite™, ThermoFisher Scientific) equipped with an EASY-Spray™ electrospray source. Chromatography of peptides prior to mass spectral analysis was accomplished using capillary column with integrated emitter (PepMap® C18, 3 µM, 100 A, 150 × 0.075 mm, ThermoFisher Scientific) onto which 3 µL of extracted peptides was automatically loaded. The nanoHPLC system delivered solvents A: 0.1% (v/v) formic acid, and B: 99.9% (v/v) acetonitrile, 0.1% (v/v) formic acid at 0.50 µL/min to load the peptides (over a 30 min period) and 0.3 µL/min to elute peptides directly into the electrospray source using a gradual gradient from 3% (v/v) B to 30% (v/v) B over 77 min and concluded with 5 min fast gradient from 30% (v/v) B to 50% (v/v) B at which time a 5 min flush-out from 50–95% (v/v) B took place. As peptides eluted from the HPLC-column/electrospray source survey, MS1 scans were acquired in the Orbitrap over the mass range 300 to 2000 m/z at resolving power of 120,000 followed by MS2 fragmentation in the linear ion trap of the 20 most intense peptides detected in the MS1 scan. Precursor redundancy was limited by dynamic exclusion.
3. Data Analysis and Protein Identification
Raw MS/MS data were converted to mgf file format using MSConvert (ProteoWizard: Open Source Software for Rapid Proteomics Tools Development). The resulting mgf files were used to search against the M. musculus (87,156 entries) Uniprot amino acid sequence database which also contained a list of common protein contaminants, plus sequence-reversed decoy entries to establish false discovery rates using the University of Wisconsin’s Mass Spectrometry and Proteomics Facility’s Mascot search engine 2.2.07 (Matrix Science) with variable methionine oxidation, variable asparagine and glutamine deamidation, plus fixed cysteine carbamidomethylation. Peptide precursor mass tolerance was set at 15 ppm and fragment mass tolerance at 0.6 Da. Protein annotations, significance of identification and spectral counting based quantification were done using the Scaffold software package (version 4.3.2, Proteome Software Inc., Portland, OR, USA). Protein identifications were accepted if they could be established at greater than 99.0% probability within 1% false discovery rate and contained at least 5 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [16]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
4. Results and Discussion
Orange-colored stains were apparent in the native gels where the reaction with either substrate, i.e., maltose (Figure 1A) or sucrose (Figure 1B), took place. The intensity of the bands could be increased by extending incubation times in the assay reagent. Maltase activity was apparently greater because the develop reagent required less time to be visualized for the staining than for sucrase (Table 1). The locations of the orange-colored stains on the sucrose and maltose gels are above the molecular mass (Mr) marker 460, which is also the case for the two most intense bands of proteins that ran through the Coomassie blue stained gel (intense bands marked with arrows in Figure 1C). Considering this proximity, and that α-glucosidases are the two most abundant proteins in the orange colored bands (see below), we suspect that the bands indicated by the arrows on the Coomassie blue stained gel correspond to SI and MGAM, but additional tests (e.g., Western blots) would be necessary to confirm this. Because electrophoretic mobility of native proteins results from size and charge as well as the overall bulk or cross-sectional area of the protein, Mr markers do not allow the exact determination of the Mr of test proteins [17]. Additionally, the differences in migration over the polyacrylamide gel between the weight marker and the band that contains the α-glucosidases might also be due to the level of glycosylation of these two proteins [4,18].
Mass spectrometry results from the excised orange stained bands revealed the presence of 32 proteins (Table 2; all proteins detected are listed in Supplementary Table S1), which was <5% of 650 proteins that were identified in crude bbm without electrophoresis (unpublished data). Of the 32 proteins, 51 ± 2% (S.D.; n = 2 mice) of those were hydrolases of which 75 ± 1% were SI and MGAM (Table 2). SI and MGAM overlap in their hydrolysis activity for some substrates, while for others they are unique. For instance, α-1,4 bonds characteristic of the disaccharide maltose are hydrolyzed by both subunits, Nt and Ct, of SI and Nt-MGAM [8,9]. Thus, the orange spot on the gel incubated with sucrose accounts for the activity of the α-1, 2 action of Ct-SI (Figure 1B) [8,9]. Alternatively, the band that is developed in the gel incubated with maltose accounts for the summed hydrolysis of both enzyme complexes, retrieving the total maltase activity for that given sample (Figure 1A). The fact that we do not see two bands for both complexes is because we maximized the color reaction to develop all the potential glucose formed in the gel. Early in the time course of the color reaction development for glucose, we observed the apparent two incipient bands until they merged (not shown).
Table 2.
Hydrolases identified by nano-liquid chromatography coupled to mass spectrometry in the slices cut from gels incubated with maltose and sucrose. Quantitative value and coverage of enzymes in the analyzed sample was determined using Scaffold software package (version 4.3.2, Proteome Software Inc., Portland, OR, USA). The % coverage refers to the percentage of the protein sequence covered by identified peptides. Protein identifications were accepted if they could be established at greater than 99.0% probability with a 1% false discovery rate and containing at least 5 identified peptides.
| Enzyme | Uniprot Accesion Number | Molecular Mass (kDa) | Maltose Substrate | Sucrose Substrate | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse 1 | Mouse 2 | Mouse 1 | Mouse 2 | |||||||
| Quantitative Value (Normalized Total Spectra) | Coverage % | Quantitative Value (Normalized Total Spectra) | Coverage % | Quantitative Value (Normalized Total Spectra) | Coverage % | Quantitative Value (Normalized Total Spectra) | Coverage % | |||
| SI (sucrase-isomaltase) | F8VQM5 | 209 | 104 | 23 | 42 | 3 | 100 | 35 | 81 | 6 |
| MGAM (maltase-glucoamylase) | B5THE2 | 209 | 56 | 15 | 94 | 6 | 83 | 31 | 38 | 4 |
| ANPEP (aminopeptidase-N) | P97449 | 110 | 25 | 11 | 42 | 2 | 49 | 39 | 49 | 5 |
| ENPEP (Glutamyl Aminopeptidase) | P16406 | 108 | 9 | 8 | - | - | 12 | 21 | - | - |
| DPP4 (Dipeptidyl peptidase 4) | P28843 | 87 | 3 | 4 | - | - | 7 | 16 | - | - |
| MEP1B (Meprin A Subunit Beta) | Q61847 | 80 | 3 | 10 | - | - | 5 | 12 | - | - |
| All other non-hydrolase proteins (details in Supplementary Table S1) |
194 | 147 | 210 | 211 | ||||||
| Hydrolases as % of total proteins | 51% | 55% | 55% | 44% | ||||||
| α-glucosidases as % of total hydrolases | 80% | 76% | 71% | 71% | ||||||
Our method description here relied on technical replicates of proteins run on gels (e.g., gels I and II) that were derived from bbm of two mice (two biological replicates). We had similar success applying the method to laboratory rats, chickens, and three species of wild birds (n = 2 individuals/species) [19], so we have become confident in the method’s applicability and replicability. The method can identify the activity of α-glucosidases, but we consider that it can be broadly applied to any enzyme that releases D-glucose or even other products of breakdown that can be identified in gel runs. Furthermore, an indication of the relative ratios of activity can be obtained by comparing the relative staining intensities. After identifying the proteins and assessing their function, their relative abundance relative to other proteins in the bbm components (Table 2) can be semi-quantified using nano LC-MS/MS loading equal amounts of total protein per sample.
Acknowledgments
We thank Grzegorz Sabat, member of the Mass Spectrometry and Proteomics Facility team of the University of Wisconsin-Madison.
Supplementary Materials
The following are available online at https://www.mdpi.com/2409-9279/3/1/15/s1, Table S1: Proteins identified by nano-liquid chromatography coupled to mass spectrometry in the slices cut from gels incubated with maltose and sucrose.
Author Contributions
A.B., M.E.M., C.M.d.R., G.A.B.-W., W.H.K. and E.C.-V. participated in project conception; A.B. and M.E.M. performed zymography and contributed equally; G.A.B.-W. run proteomic assays; A.B., M.E.M., C.M.d.R., G.A.B.-W., W.H.K. and E.C.-V. analyzed, discussed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Science Foundation (IOS-1354893 to W.H.K.), the Universidad Nacional de San Luis (Ciencia y Técnica 2-0814 to E.C.-V.), the Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 2015-834 to E.C.-V.) and the Department of Forest and Wildlife Ecology, UW-Madison.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Morales R.L.C., Zazueta-Novoa V., Leal-Morales C.A., Martínez A.F., Noyola P.P., Zazueta-Sandoval R. Polyacrylamide gel electrophoresis an important tool for the detection and analysis of enzymatic activities by electrophoretic zymograms. In: Magdelin S., editor. Gel Electrophoresis—Advances Techniques. InTech; Rijeka, Croatia: 2012. p. 500. [Google Scholar]
- 2.Sim L., Quezada-Calvillo R., Sterchi E.E., Nichols B.L., Rose D.R. Human Intestinal Maltase–Glucoamylase: Crystal Structure of the N-Terminal Catalytic Subunit and Basis of Inhibition and Substrate Specificity. J. Mol. Biol. 2008;375:782–792. doi: 10.1016/j.jmb.2007.10.069. [DOI] [PubMed] [Google Scholar]
- 3.Jones K., Sim L., Mohan S., Kumarasamy J., Liu H., Avery S., Naim H.Y., Quezada-Calvillo R., Nichols B.L., Mario Pinto B., et al. Mapping the intestinal alpha-glucogenic enzyme specificities of starch digesting maltase-glucoamylase and sucrase-isomaltase. Bioorg. Med. Chem. 2011;19:3929–3934. doi: 10.1016/j.bmc.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 4.Nichols B.L., Eldering J., Avery S., Hahn D., Quaroni A., Sterchi E. Human small intestinal maltase-glucoamylase cDNA cloning: Homology to sucrase-isomaltase. J. Biol. Chem. 1998;273:3076–3081. doi: 10.1074/jbc.273.5.3076. [DOI] [PubMed] [Google Scholar]
- 5.Nichols B.L., Avery S., Sen P., Swallow D.M., Hahn D., Sterchi E. The maltase-glucoamylase gene: common ancestry to sucrase-isomaltase with complementary starch digestion activities. Proc. Natl. Acad. Sci. USA. 2003;100:1432–1437. doi: 10.1073/pnas.0237170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst H.A., Lo Leggio L., Willemoës M., Leonard G., Blum P., Larsen S. Structure of the Sulfolobus solfataricus α-Glucosidase: Implications for domain conservation and substrate recognition in GH31. J. Mol. Biol. 2006;358:1106–1124. doi: 10.1016/j.jmb.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 8.Chaudet M.M., Amiri M., Marth N., Naim H.Y., Rose D.R. Phylogenetic analysis reveals key residues in substrate hydrolysis in the isomaltase domain of sucrase-isomaltase and its role in starch digestion. Biochim. Biophys. Acta Gen. Subj. 2019;1863:1410–1416. doi: 10.1016/j.bbagen.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Rose D.R., Chaudet M.M., Jones K. Structural studies of the intestinal α-glucosidases, maltase-glucoamylase and sucrase-isomaltase. J. Pediatr. Gastroenterol. Nutr. 2018;66:S11–S13. doi: 10.1097/mpg.0000000000001953. [DOI] [PubMed] [Google Scholar]
- 10.Mac Donal O., Chediack J.G., Caviedes-Vidal E. Isolation of epithelial cells, villi and crypts from small intestine of pigeons (Columba livia) Biocell. 2008;32:219–227. [PubMed] [Google Scholar]
- 11.McConnell R.E., Benesh A.E., Mao S., Tabb D.L., Tyska M.J. Proteomic analysis of the enterocyte brush border. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G914–G926. doi: 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson R.J. Staining proteins in gels with coomassie blue. CSH Protoc. 2007;2007:pdb.prot4719. doi: 10.1101/pdb.prot4719. [DOI] [PubMed] [Google Scholar]
- 13.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 14.Wang G., de Jong R.N., van den Bremer E.T.J., Parren P.W.H.I., Heck A.J.R. Enhancing accuracy in molecular weight determination of highly heterogeneously glycosylated proteins by native tandem mass spectrometry. Anal. Chem. 2017;89:4793–4797. doi: 10.1021/acs.analchem.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gericke B., Amiri M., Naim H.Y. The multiple roles of sucrase-isomaltase in the intestinal physiology. Mol. Cell. Pediatr. 2016;3:2. doi: 10.1186/s40348-016-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biviano A.B., Del Río C.M., Phillips D.L. Ontogenesis of intestine morphology and intestinal disaccharidases in chickens (Gallus gallus) fed contrasting purified diets. J. Comp. Physiol. B. 1993;163:508–518. doi: 10.1007/BF00346936. [DOI] [PubMed] [Google Scholar]
- 17.Lee B.H., Rose D.R., Lin A.H.M., Quezada-Calvillo R., Nichols B.L., Hamaker B.R. Contribution of the individual small intestinal α-glucosidases to digestion of unusual α-linked glycemic disaccharides. J. Agric. Food Chem. 2016;64:6487–6494. doi: 10.1021/acs.jafc.6b01816. [DOI] [PubMed] [Google Scholar]
- 18.Semenza G., Auricchio S., Mantei N. Small-Intestinal Disaccharidases. In: Valle D., Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., Gibson K.M., Mitchell G., editors. The Online Metabolic and Molecular Bases of Inherited Disease. The McGraw-Hill Companies, Inc.; New York, NY, USA: 2014. [Google Scholar]
- 19.Brun A., Mendez-Aranda D., Magallanes M.E., Karasov W.H., Martinez Del Rio C., Baldwin M., Caviedes-Vidal E. Evolution of intestinal α-glucosidases in vertebrates: Genomic and proteomic data upend previous hypotheses. Integr. Comp. Biol. 2019;59:E25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.