Abstract
The drug combination of biapenem (BIPM) and xuebijing injection (XBJ) is commonly applied for the treatment of sepsis in China. However, the potential synergistic mechanism is still enigmatic. There have been no studies focused on the plasma metabolome alterations in sepsis after the intervention of this combination. In this work, an untargeted metabolomics approach was performed by liquid chromatography-mass spectrometry coupled with multivariate statistical analysis to provide new insights into the synergistic effect of BIPM in combination with XBJ. We characterized the metabolic phenotype of sepsis and described metabolic footprint changes in septic rats responding to XBJ and BIPM individually and in combination, in addition to histopathological and survival evaluation. A total of 91 potential biomarkers of sepsis were identified and 32 disturbed metabolic pathways were constructed. Among these biomarkers, 36 metabolites were reversely regulated by XBJ, mainly including glycerophospholipids, sphingolipids, free fatty acids (FFAs), bile acids and acylcarnitines; 42 metabolites were regulated by BIPM, mainly including amino acids, glycerophospholipids, and acylcarnitines; 72 metabolites were regulated after XBJ-BIPM combination treatment, including most of the 91 potential biomarkers. The results showed that the interaction between XBJ and BIPM indeed exhibited a synergistic effect by affecting some key endogenous metabolites, 15 metabolites of which could not be regulated when XBJ or BIPM was used alone. Compared with Model group, 13, 22, and 27 metabolic pathways were regulated by XBJ, BIPM, and XBJ-BIPM combination, respectively. It suggested that many more endogenous metabolites and metabolic pathways were significantly regulated after combination treatment compared with XBJ or BIPM monotherapy. Metabolisms of lipids, amino acids, acylcarnitines, and bile acids were common pathways involved in the synergistic action of XBJ and BIPM. This study was the first to employ metabolomics to elucidate the synergistic effect and decipher the underlying mechanisms of BIPM in combination with XBJ against sepsis. The results provide some support for clinical application of antibiotics in combination with traditional Chinese medicines and have important implications for the treatment of sepsis in clinic.
Keywords: metabolomics, drug combination, synergistic effect, xuebijing injection, biapenem
Introduction
Sepsis is considered as life-threatening organ dysfunction resulting from dysregulated host response to infection (Singer et al., 2016). Timely administration of antibiotics is the leading line of defense against infection and prescribed as one of the most common medications to treat sepsis. Biapenem (BIPM), a carbapenem antibiotic, exhibits broad spectrum antibacterial activity and has a good curative effect for severe infection (Ikawa et al., 2008). It is widely applied for the treatment of sepsis. However, simply acting on pathogenic microorganisms does not significantly improve survival in patients with sepsis. Antibiotic-mediated endotoxin release and overwhelming inflammatory mediator release play key roles in triggering systemic inflammation reaction syndrome (SIRS) and subsequent multiple organ dysfunction syndrome (MODS) (Lepper et al., 2002). Therefore, as a powerful complement of antibiotic medications, traditional Chinese medicines have been incorporated into routine therapy for the treatment of sepsis (Chinese Society of Critical Care Medicine, 2015).
Xuebijing injection (XBJ), an emerging antiseptic herbal medicine, is prepared from a combination of Carthamus tinctorius L., Paeonia lactiflora Pall., Conioselinum anthriscoides “Chuanxiong”, Salvia miltiorrhiza Bunge and Angelica sinensis (Oliv.) Diels (Supplementary Table S1). It is the only Chinese herb preparation approved by China Food and Drug Administration (CFDA) indicated for sepsis and MODS caused by infections. It has been clinically prescribed for septic patients with good effectiveness on toxic heat flourishing and blood stasis syndrome, as it is considered to possess the efficacies of activating circulation, removing blood stasis, and eliminating toxins (Yin and Li, 2014). In China, approximately 0.8 million patients received treatment with XBJ yearly, about 80% of which were sepsis or septic shock cases (Zhang et al., 2018). In fact, XBJ was most commonly used in combination with antibiotics (e.g., BIPM) for sepsis treatment in clinical practice, instead of being used alone. Numerous clinical studies have shown that addition of XBJ can effectively reduce the 28-day mortality, shorten the length of stay in intensive care units, and improve the physical condition and prognosis of patients with sepsis (Chen and Li, 2013; Gao et al., 2015; Song et al., 2016). The combination of antibiotics and XBJ is one of the medication characteristics of China in clinical treatment of sepsis, and has been showing its advantages in the therapeutic efficacy.
Despite high frequency of antibiotic-XBJ combination and promising clinical effects, the exact synergistic mechanism against sepsis is still enigmatic. Metabolomics provides a cutting-edge approach that measures metabolic alterations in responds to disease process or drug treatment. Characterization of metabolic phenotype facilitates a better understanding of the pathogenesis and interventional mechanism. Mass spectrometry plays an essential role in the metabolic profile analysis. Therefore, this work employs a global metabolomic approach to describe metabolic alterations of septic rats responding to XBJ and BIPM individually and in combination by ultra-high performance liquid chromatography coupled with a Q Exactive hybrid quadrupole-orbitrap high resolution mass spectrometry (UHPLC-Q-Orbitrap HRMS). This work will provide a novel insight into the mechanism of synergistic effect of BIPM in combination with XBJ against sepsis.
Materials and Methods
Drugs and Regents
Every 0.3g BIPM (Simcere Pharmaceutical Co., Ltd., Nanjing, China, 92-181004) was dissolved in 30 ml of 0.9% saline for injection. All the endogenous metabolite standards were purchased from Sigma-Aldrich Company (St Louis, MO, USA) and J&K Scientific Ltd. (Beijing, China). Ketoprofen (ESI- internal standard) was purchased from Sigma-Aldrich, and 2-chloro-L-phenylalanine (ESI+ internal standard) was purchased from J&K Scientific Ltd. The reference standards of Hydroxysafflor yellow A, Oxypaeoniflorin, Benzoylpaeoniflorin, Senkyunolide I, Succinic acid, Gallic acid, Rosmarinic acid, Caffeic acid, Protocatechuic aldehyde, Protocatechuic acid, Rutin, Salvianic acid A, Chlorogenic acid, and Naringenin were purchased from Chengdu Must Bio-technology Co., Ltd. (Sichuan, China), and the compounds' purities were over 98%. HPLC grade methanol and acetonitrile were obtained from Fisher Scientific (Fair Lawn, NJ, USA). HPLC grade formic acid was supplied by Aladdin Industrial Co., Ltd (Shanghai, China). Ultrapure water was prepared by a Milli-Q purification system (Millipore, Shanghai, China).
XBJ was provided by Tianjin Chase Sun Pharmaceutical Co., Ltd. (Tianjin, China, batch number: 1810221). Per milliliter of XBJ was prepared from the combination of each 0.1 g of Carthamus tinctorius L., Paeonia lactiflora Pall., Conioselinum anthriscoides “Chuanxiong”, Salvia miltiorrhiza Bunge and Angelica sinensis (Oliv.) Diels, yielding an herb-to-injection ratio of 1:2. The final product of XBJ for intravenous injection is a sterile and nonpyrogenic dosage form. Qualitative and quantitative analysis of XBJ was performed by UHPLC-Q-Orbitrap HRMS. The chromatographic separation was carried out on an ACQUITY UPLC BEH C18 column (50 mm × 2.1 mm, 1.7 μm, Waters, USA) maintained at 40°C. The mobile phase consisted of water containing 0.1% (v/v) formic acid (A) and acetonitrile (B). The gradient elution was set as follows at a flow rate of 0.25 ml/min: 5%–7.5% B at 0–6 min, 7.5%–40% B at 6–30 min, 40%–100% B at 30–50 min, 100% B at 50–55 min, 100%–5% B at 55–56 min, 5% B at 56–60 min. The total ion chromatograms of XBJ were shown in Supplementary Figure S1, and a total of 127 components were characterized (Supplementary Table S2). We also determined the concentrations of 14 dominating compounds contained in XBJ, the chemical structures of which were shown in Supplementary Figure S2. Satisfactory linearity and correlation coefficient were achieved with wide linear range (Supplementary Table S3). The precision, repeatability, stability and recovery met requirements (Supplementary Table S4).
Animal Model of Cecal Ligation and Puncture (CLP)-Induced Sepsis
Cecal ligation and puncture (CLP), the most widely used model for experimental sepsis, is considered as the gold standard in sepsis research (Buras et al., 2005; Hubbard et al., 2005). In this work, the rat model of CLP-induced sepsis was performed following previous established protocols (Rittirsch et al., 2009). In brief, rats were anesthetized with ketamine (80 mg/kg) and xylazine (15 mg/kg) by intraperitoneal injection. A midline laparotomy with the incision less than 3 cm was performed to expose the cecum. The cecum was then tightly ligated below the ileocecal valve, and the distal caecum was punctured with a sterile 18G needle. Then, the cecum was gently squeezed to extrude a small amount of intraluminal content. After that, the cecum was returned to abdominal cavity and the incision was sutured. The rats were resuscitated by injecting prewarmed normal saline (37°C, 10 ml/Kg) subcutaneously after the surgical operation. The sham-operated rats underwent the same procedures but the cecum was not subjected to ligation and puncture.
Animals and Drug Administration
Fifty male Sprague-Dawley (SD) rats (weighing 250–300 g) were purchased from Henan experimental animal center (Zhengzhou, China). These animals were fed with normal diet and water under the controlled temperature (18–22°C) and humidity (55%–65%) to accommodate to the environment for seven days before the experiment. All animal studies were conducted in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experimental procedures were approved by the Animal Ethics Committee of the first affiliated hospital of Zhengzhou University.
All the rats were randomly divided into five groups. Sham group (n=10): Rats underwent sham surgery in which the cecum was not subjected to ligation and puncture, and intravenous injections of saline via tail vein were performed every 12 hours after the sham operation. Model group (n=10): The septic rat model was induced by CLP operation as described above, and saline solution was administrated via intravenous injection every 12 hours after the surgery. XBJ group (n=10): Rats received tail vein injection of XBJ (5.25 ml/Kg) every 12 h after CLP operation. BIPM group (n=10): Rats received tail vein injection of BIPM (31.5 mg/Kg) every 12 h after CLP operation. COM group (combined administration of XBJ and BIPM, n=10): Rats received tail vein injection of XBJ (5.25 ml/Kg) right after injection of BIPM (31.5 mg/Kg) every 12 h after CLP operation. The term of the experiment was three continuous days.
Sample Collection and Preparation
The rats were sacrificed at 72 h after surgical operation. Blood samples were collected from abdominal aorta, and plasma was separated by centrifugation at 3000 rpm for 10 min. The supernatants were stored at -80°C until subsequent analysis. The lung and liver tissues were dissected and fixed in 10% formalin for histopathological examination.
A 100 μl aliquot of plasma was added to a 1.5 ml Eppendorf tube followed by precipitation with 300 μl of methanol (containing 500 ng/ml ketoprofen and 50 ng/ml 2-chloro-L-phenylalanine as internal standard), and the mixture was vortexed for 1 min. The mixture was subsequently centrifuged at 13,000 rpm for 10 min at 4°C, and 200 μl of the supernatant was transferred to an autosampler vial for further UHPLC-Q-Orbitrap HRMS analysis. To ensure data quality of metabolomic profiling, pooled quality control (QC) samples were prepared by mixing 10 μl of plasma from each sample. The pretreatment of QC samples was conducted according to the procedure mentioned above, and one QC sample was injected after every eight samples throughout the run.
UHPLC-Q-Orbitrap HRMS Analysis
A 5 μl aliquot of the prepared sample was injected into ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm, Waters, USA) maintained at 40°C using a Thermo Scientific Dionex UltiMate 3000 UHPLC system for chromatographic separation. The mobile phase consisted of water containing 0.1% (v/v) formic acid (A) and acetonitrile (B). The gradient elution was set as follows at a flow rate of 0.35 ml/min: 5% B at 0–1min, 5%–100% B at 1–9 min, 100% B at 9–12 min, 100%–5% B at 12–12.1 min, 5% B at 12.1–15 min.
The mass spectrometry was performed on Q-Exactive orbitrap system (Thermo Fisher Scientific, San Jose, USA) equipped with a heated electrospray ionization source operated in both positive and negative ion modes. The MS parameters were optimized and set as follows: collision energy at 20, 40, and 60 eV, ion source temperature at 350°C, capillary temperature at 320°C, spray voltage at 3.50 kV for the positive ion mode and 2.8 kV for the negative ion mode, sheath gas flow rate at 40 arb for the positive ion mode and 38 arb for the negative ion mode, auxiliary gas flow rate at 10 arb. Metabolomic profiles were acquired with a mass range of 80–1200 m/z. The full scan spectra and MS/MS data were collected with the resolution of 70,000 and 17,500 FWHM respectively. The samples were injected in random order, and all the mass data were acquired and processed using Thermo Xcalibur 3.0 software.
Data Processing and Statistical Analysis
The pretreatment of LC-MS raw data was conducted by Thermo Scientific Compound Discoverer 3.0 software. The spectra were selected from input LC-MS data files and retention time alignment was accomplished based on mass tolerance and time shift criteria. Preliminary identification of metabolites was realized by searching databases including ChemSpider, Mass Lists, mzCloud, mzVault, and local database. Multiple nodes such as “Align Retention Times”, “Detect Unknown Compounds”, “Group Unknown Compounds”, “Search Database”, “Fill Gaps”, and “Normalize Areas” were combined to form an untargeted metabolomics workflow for raw data processing.
The data matrix obtained from Compound Discoverer were imported into SIMICA software (version 14.0, Umetrics, Sweden) for multivariate statistical analysis, including unsupervised principal component analysis (PCA), supervised partial least-squares discriminant analysis (PLS-DA) and orthogonal partial least-squares discriminant analysis (OPLS-DA). The PCA analysis was applied to assess the reproducibility and stability of QC samples, and the PLS-DA model was established to describe general separation of samples from five groups. The variables responsible for the discrimination between two groups were identified by OPLS-DA, and permutation test was performed 200 times to assess the risk of overfitting for the OPLS-DA model. In addition to the multivariate statistical method, Student's t test was also applied to measure the significance of each metabolite. The metabolites with variable importance in the projection (VIP) values >1.0 and p values <0.05 for Model versus Sham were considered as potential biomarkers of sepsis, the structures of which were further confirmed based on available reference standards.
To evaluate the effect of drug intervention, fold changes and p values of the potential biomarkers for comparisons of XBJ versus Model, BIPM versus Model, and COM versus Model were calculated respectively. The concentrations of metabolites detected in different groups were expressed as peak areas. Histograms were described to reflect the changes of potential biomarkers across the five groups using GraphPad Prism 8.0.2 software (GraphPad Software Inc., San Diego, USA). Correlation analysis, metabolic pathway analysis, heat map and hierarchical cluster analysis were conducted by MetaboAnalyst (http://www.metaboanalyst.ca/).
Results
Histopathological Examination and Survival Analysis
The effects of three different interventions on the histological morphology of lung and liver tissues in septic rats by hematoxylin- eosin (HE) staining were shown in Supplementary Figure S3. In the lung tissue of Sham group, the alveolar septum was normal and there was no inflammatory cell infiltration. In the Model group, the alveolar wall was thickened and the blood vessels were congested. In the XBJ and BIPM group, the alveolar walls were less thickened. The histological morphology of lung tissue in COM group was similar to that of Sham group. In the liver tissue sections of Sham group, the hepatic cords were arranged in a radial arrangement. There was no edema in the hepatocytes, and the structure of the hepatic lobule was clear. The Model group showed hepatic cord disorder and cellular swelling of the hepatocyte. Hepatic lobular structure was unclear, and there was a large number of inflammatory cell infiltration. After intervention by XBJ, BIPM, and XBJ-BIPM combination, the hepatic cord structures were acceptable, and the inflammatory cells were reduced. The hepatocytes were mildly edema in XBJ and BIPM groups, and the edema was significantly relieved in the COM group.
Survival analysis showed that the rats in Sham group all survived within 72 h. The survival rates of rats in Model, XBJ, BIPM, and COM groups were 60%, 60%, 70%, and 90%, respectively (Supplementary Figure S4). Survival time was prolonged after drug intervention, especially in the COM group. The results indicated that the combination of XBJ and BIPM could improve the survival rate of septic rats.
Reproducibility and Stability Assessment of the Analytical Strategy
Typical base peak chromatograms (BPCs) of representative samples from five groups were shown in Supplementary Figure S5. To characterize the general distribution of all the samples and assess the quality of QC data, unsupervised PCA analysis was performed. The tight cluster of QC samples both in positive and negative (Supplementary Figures S6A, B) ion modes demonstrated good reproducibility of analytical method and instrumental stability throughout the run. The reproducibility was also evaluated by the relative standard deviation (RSD) of peak areas across the QC samples, and over 90% of the peaks showed RSD values below 30% (Supplementary Figures S6C, D). Moreover, RSD values of ESI+ and ESI- internal standards across all the samples were 4.9% and 6.6% respectively. The relative abundance variabilities with respect to injection order for internal standards were presented in Supplementary Figures S6E, F. The results provided evidence that our analytical strategy possessed satisfactory reproducibility and stability.
Metabolic Disturbance Related to Sepsis and Identification of Potential Biomarkers
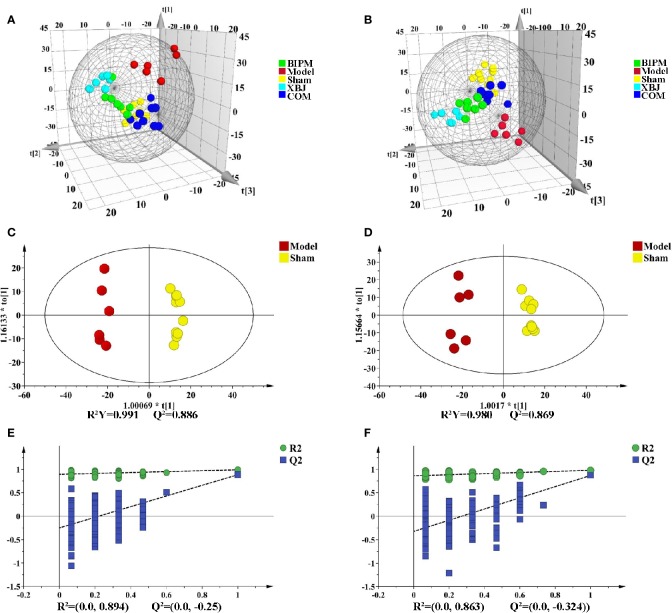
Supervised PLS-DA model was constructed to explore the distribution and tendency of five groups. As shown in Figures 1A, B, good separation trend between groups was observed in both positive and negative ion modes. It was obvious that metabolic change occurred in the Model group compared with Sham group. Three different ways of drug intervention exerted different effects by influencing endogenous metabolites, and the COM group was closer to the Sham group than XBJ and BIPM groups. The OPLS-DA model of comparison between Model and Sham group was next performed to characterize the metabolic disturbance related to sepsis. Significant distinctions were obtained with R2Y at 0.991 and Q2 at 0.886 for ESI+ mode (Figure 1C), and R2Y at 0.980 and Q2 at 0.869 for ESI- mode (Figure 1D). To assess the validity of the OPLS-DA model, permutation test with 200 measurements was performed and the result (R2 = 0.894, Q2 = -0.250 for ESI+ mode; R2 = 0.863, Q2 = -0.324 for ESI- mode) indicated that there was no overfitting of the model (Figures 1E, F).
Figure 1.
PLS-DA and OPLS-DA score plots in positive and negative ion modes. PLS-DA score plots of different groups in positive (A) and negative (B) ion modes. OPLS-DA score plots for Model group vs. Sham group in positive (C) and negative (D) ion modes. Permutation tests obtained from Model group vs. Sham group in positive (E) and negative (F) ion modes. PLS-DA, partial least-squares discriminant analysis; OPLS-DA, orthogonal partial least-squares discriminant analysis.
With the criteria of VIP values >1.0 and p values <0.05, 459 and 426 differential ions were selected for Model vs. Sham in ESI+ and ESI- mode, respectively. The volcano plot was subsequently performed on the basis of p values and fold change values between the two groups. As shown in Supplementary Figure S7, the red dots represented metabolites with p values <0.05 and fold change values >1.20 (or <0.83). According to accurate m/z and fragmentation information, a total of 91 differential endogenous metabolites were identified, 22 metabolites of which were confirmed by standard references. The MS/MS spectra and fragment ions for some of these metabolites in certain samples compared with standard references were provided in Supplementary Figure S8.
As listed in Table 1, 63 metabolites elevated and 28 metabolites decreased in Model group. Compared with Sham group, the lipids metabolic disturbance was observed in sepsis including declined lysophosphatidylcholines (LysoPCs) and lysophosphatidylethanolamines (LysoPEs), increased phosphatidylcholines (PCs), elevated free fatty acids (FFAs), and decreased sphingolipids. Altered lipid levels reflected systemic changes resulted from inflammatory response and oxidative stress in sepsis (Sevastou et al., 2013). Significant decreases in LysoPCs and LysoPEs levels in septic rats were consistent with a previous study (Xu et al., 2018). The elevated amino acids, especially the aromatic and sulfur containing amino acids (phenylalanine, tyrosine, and methionine), suggested the imbalance between the hepatic protein synthesis and muscle protein breakdown in energy metabolism of sepsis (Freund et al., 1979). The plasma amino acid pattern was considered as an indication of the severity of metabolic disturbance in sepsis and might be predictive of mortality (Freund et al., 1979). Significantly increased levels of acyl carnitines (hexanoylcarnitine, stearoylcarnitine, oleoylcarnitine, etc.) in Model group indicated enhanced fatty acid β-oxidation in sepsis. In addition, the increased level of N-Acetylneuraminic (Neu5Ac) acid was observed as one of the most significantly changed metabolic markers for sepsis. Neu5Ac was produced by the catalysis of neuraminidase, which played a central role in NF-κB activation and pro-inflammatory responses (Abdulkhalek and Szewczuk, 2013). Multiple organ dysfunction has been implicated in the pathogenesis of sepsis, as indicated by elevated level of creatinine, the biomarker for kidney damage. The increased plasma bile acids might be a consequence of liver injury and dysfunction in the course of sepsis. These disregulated endogenous metabolites were considered as potential biomarkers of sepsis.
Table 1.
Metabolomic-based potential biomarkers of sepsis and the levels change of metabolites responding to three different interventions.
| No. | Metabolites | Theoretical mass (m/z) | Measured mass (m/z) | Delta (ppm) | RT (min) | Ion Mode | VIP | Fold Change (Model/Sham) | Fold Change (XBJ/Model) | Fold Change (BIPM/Model) |
Fold Change (COM/Model) | Classification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lactic acid# | 89.02441 | 89.02436 | -0.644 | 1.15 | n | 1.63 | 1.83↑** | 0.55↓** | 0.55↓** | 0.59↓** | Organic acids |
| 2 | Choline | 104.10699 | 104.10700 | 0.090 | 0.88 | p | 1.42 | 2.15↑* | – | 0.53↓* | 0.51↓* | Organonitrogen compounds |
| 3 | Glyceric acid | 105.01933 | 105.01932 | -0.114 | 1.14 | n | 1.12 | 1.57↑* | – | – | – | Organic acids |
| 4 | Cytosine | 112.05053 | 112.05052 | -0.164 | 0.88 | p | 1.47 | 1.81↑** | – | 0.71↓* | 0.74↓* | Pyrimidines |
| 5 | Uracil | 113.03455 | 113.03449 | -0.566 | 1.14 | p | 1.37 | 3.41↑* | – | – | – | Pyrimidines |
| 6 | Creatinine# | 114.06618 | 114.06629 | 0.890 | 0.86 | p | 1.36 | 1.48↑** | – | – | 0.65↓** | other |
| 7 | 2-Oxovaleric acid | 115.04006 | 115.03997 | -0.847 | 2.48 | n | 1.24 | 1.34↑** | – | – | – | Organic acids |
| 8 | Betaine# | 118.08625 | 118.08626 | 0.041 | 0.89 | p | 1.18 | 0.44↓** | – | – | – | other |
| 9 | Nicotinamide# | 123.05528 | 123.05530 | 0.086 | 1.14 | p | 1.14 | 0.54↓* | – | – | 1.78↑* | other |
| 10 | Pyroglutamic acid† | 130.04986 | 130.05000 | 1.002 | 0.89 | p | 1.60 | 1.47↑** | 0.72↓** | 0.71↓** | 0.75↓* | Amino acids |
| 11 | 4-Oxoproline† | 128.03531 | 128.03531 | -0.050 | 1.15 | n | 1.56 | 1.74↑** | 0.67↓* | 0.59↓** | 0.63↓** | Amino acids |
| 12 | Pipecolic acid | 130.08625 | 130.08636 | 0.806 | 0.88 | p | 1.35 | 1.55↑** | – | – | – | Amino acids |
| 13 | Creatine# | 132.07675 | 132.07684 | 0.658 | 0.87 | p | 1.72 | 2.45↑** | – | 0.64↓* | 0.49↓** | Organic acids |
| 14 | 2-hydroxycaproic acid | 131.07136 | 131.07121 | -1.202 | 3.98 | n | 1.16 | 1.85↑* | – | 0.57↓* | 0.58↓* | Organic acids |
| 15 | Threonic acid | 135.02989 | 135.02983 | -0.493 | 0.90 | n | 1.71 | 1.77↑** | – | 0.58↓** | 0.60↓** | Organic acids |
| 16 | 3-Dehydroxycarnitine | 146.11755 | 146.11763 | 0.511 | 0.90 | p | 1.52 | 1.58↑** | – | – | 0.69↓** | other |
| 17 | Glutamine#† | 147.07641 | 147.07649 | 0.485 | 0.89 | p | 1.58 | 1.49↑** | 0.71↓** | 0.72↓** | 0.70↓** | Amino acids |
| 18 | β-Ureidoisobutyric acid† | 147.07641 | 147.07652 | 0.689 | 0.89 | p | 1.50 | 1.46↑** | 0.73↓* | 0.72↓** | 0.72↓** | Organic acids |
| 19 | Lysine# | 147.11280 | 147.11284 | 0.243 | 0.75 | p | 1.40 | 1.58↑** | – | – | 0.68↓** | Amino acids |
| 20 | Glutamate#† | 146.04588 | 146.04597 | 0.609 | 0.90 | n | 1.33 | 1.81↑* | 0.57↓* | 0.55↓* | 0.64↓* | Amino acids |
| 21 | Methionine#† | 150.05832 | 150.05843 | 0.694 | 1.14 | p | 1.70 | 1.64↑** | – | 0.73↓** | 0.69↓** | Amino acids |
| 22 | Allantoin† | 157.03671 | 157.03656 | -0.977 | 0.90 | n | 1.12 | 1.58↑* | 0.58↓* | – | 0.62↓* | other |
| 23 | 3-Dehydrocarnitine | 158.08226 | 158.08223 | -0.231 | 3.89 | n | 1.07 | 0.30↓* | – | – | – | other |
| 24 | 2-Hydroxycinnamic acid# | 165.05462 | 165.05475 | 0.784 | 1.16 | p | 1.04 | 1.36↑* | – | 0.71↓* | 0.76↓* | Organic acids |
| 25 | Phenylalanine#† | 166.08625 | 166.08633 | 0.451 | 1.95 | p | 1.07 | 1.34↑* | – | 0.66↓* | 0.70↓* | Amino acids |
| 26 | Uric acid#† | 169.03561 | 169.03566 | 0.257 | 1.14 | p | 1.42 | 6.08↑* | 0.30↓* | – | 0.26↓* | Organic acids |
| 27 | 1-Methylhistidine | 170.09240 | 170.09254 | 0.805 | 0.77 | p | 1.55 | 2.54↑* | – | 0.59↓* | 0.44↓** | Amino acids |
| 28 | Hippuric acid#† | 178.05096 | 178.05099 | 0.132 | 3.73 | n | 1.32 | 0.19↓** | – | – | 2.45↑* | Organic acids |
| 29 | Inositol | 179.05611 | 179.05617 | 0.328 | 0.94 | n | 1.15 | 0.63↓* | – | – | 1.56↑* | other |
| 30 | Tyrosine#† | 182.08116 | 182.08131 | 0.770 | 1.15 | p | 1.04 | 1.36↑* | – | 0.71↓* | 0.76↓* | Amino acids |
| 31 | Phosphorylcholine | 184.07332 | 184.07318 | -0.766 | 7.96 | p | 1.45 | 0.71↓** | – | 1.31↑* | 1.41↑** | Organonitrogen compounds |
| 32 | 5-Hydroxyisourate | 183.01597 | 183.01587 | -0.590 | 1.14 | n | 1.53 | 4.99↑* | 0.42↓* | 0.39↓* | 0.36↓** | Organic acids |
| 33 | Indoleacrylic acid | 186.05605 | 186.05608 | 0.151 | 5.18 | n | 1.19 | 0.15↓** | 2.62↑* | – | 4.56↑* | Indole derivatives |
| 34 | N6,N6,N6-Trimethyl-L-lysine | 189.15975 | 189.15982 | 0.347 | 0.80 | p | 1.58 | 2.29↑** | – | 0.45↓** | 0.52↓** | Amino acids |
| 35 | Citric acid# | 191.01972 | 191.01987 | 0.755 | 1.12 | n | 1.15 | 0.47↓* | – | 1.81↑* | 1.90↑* | Organic acids |
| 36 | Acetyl-L-carnitine# | 204.12303 | 204.12308 | 0.223 | 1.13 | p | 1.35 | 1.45↑** | – | – | 0.60↓** | Acyl carnitine |
| 37 | Lipoic acid | 207.05079 | 207.05067 | -0.617 | 0.79 | p | 1.71 | 2.39↑** | 0.69↓* | 0.70↓* | 0.54↓** | Organic acids |
| 38 | N-Acetyl-L-phenylalanine† | 206.08226 | 206.08238 | 0.550 | 4.40 | n | 1.08 | 0.61↓* | – | – | – | Amino acids |
| 39 | 3-Oxododecanoic acid | 213.14961 | 213.14978 | 0.761 | 6.98 | n | 1.44 | 4.16↑* | 0.38↓* | – | 0.40↓* | Free fatty acids |
| 40 | 12-Hydroxydodecanoic acid | 215.16526 | 215.16531 | 0.196 | 7.49 | n | 1.06 | 1.86↑* | – | – | – | Free fatty acids |
| 41 | Prolylleucine | 229.15466 | 229.15454 | -0.563 | 0.90 | p | 1.26 | 2.75↑* | – | – | – | Amino acids |
| 42 | Pseudouridine† | 243.06225 | 243.06239 | 0.538 | 0.90 | n | 1.43 | 1.86↑** | – | 0.65↓* | 0.59↓* | other |
| 43 | 3-hydroxybutyrylcarnitine | 248.14924 | 248.14923 | -0.078 | 1.15 | p | 1.13 | 1.53↑* | – | – | 0.56↓** | Acyl carnitine |
| 44 | Hexanoylcarnitine | 260.18563 | 260.18576 | 0.481 | 4.37 | p | 1.68 | 3.01↑** | 0.73↓* | 0.54↓* | 0.40↓** | Acyl carnitine |
| 45 | Neuraminic acid† | 266.08813 | 266.08823 | 0.339 | 0.88 | n | 1.71 | 3.03↑** | – | 0.54↓* | 0.34↓** | Organic acids |
| 46 | His-asp | 269.08914 | 269.08789 | -4.656 | 0.89 | n | 1.63 | 2.37↑** | 0.55↓* | 0.45↓** | 0.44↓** | Amino acids |
| 47 | Juniperic acid | 271.22786 | 271.22806 | 0.707 | 9.67 | n | 1.48 | 1.54↑** | 0.82↓* | – | 0.74↓* | Free fatty acids |
| 48 | Methyl aspartylphenylalaninate† | 295.12884 | 295.12888 | 0.108 | 3.53 | p | 1.40 | 1.79↑* | – | 0.59↓* | 0.55↓* | Amino acids |
| 49 | Sphingosine# | 300.28970 | 300.28952 | -0.619 | 7.32 | p | 1.21 | 0.32↓** | 2.16↑** | – | 2.77↑* | Sphingolipids |
| 50 | Sphinganine | 302.30535 | 302.30524 | -0.384 | 7.08 | p | 1.30 | 0.75↓** | 1.16↑* | – | 1.24↑* | Sphingolipids |
| 51 | Arachidonic acid# | 303.23295 | 303.23315 | 0.648 | 9.78 | n | 1.21 | 1.30↑** | 0.81↓* | – | 0.83↓* | Free fatty acids |
| 52 | N-Acetylneuraminic acid | 308.09870 | 308.09891 | 0.668 | 0.95 | n | 1.70 | 7.06↑** | – | 0.37↓* | 0.20↓** | Organic acids |
| 53 | Docosahexaenoic acid# | 327.23295 | 327.23325 | 0.906 | 9.62 | n | 1.50 | 1.94↑** | 0.74↓* | – | 0.64↓* | Free fatty acids |
| 54 | Trihydroxy octadecenoic acid | 329.23334 | 329.23315 | -0.600 | 5.90 | n | 1.36 | 2.41↑* | 0.44↓* | 0.41↓* | 0.56↓* | Free fatty acids |
| 55 | Docosapentaenoic acid# | 329.24860 | 329.24881 | 0.627 | 10.02 | n | 1.08 | 1.99↑* | 0.53↓* | – | 0.58↓* | Free fatty acids |
| 56 | 3-Indole carboxylic acid glucuronide† | 336.07248 | 336.07285 | 1.072 | 3.61 | n | 1.05 | 0.08↓* | – | – | 5.13↑* | other |
| 57 | 5-Hydroxy-6-methoxyindole glucuronide† | 338.08813 | 338.08853 | 1.154 | 3.86 | n | 1.28 | 0.07↓** | – | – | – | other |
| 58 | N-Acetyl-4-O-Acetylneuraminate† | 350.10926 | 350.10959 | 0.917 | 1.15 | n | 1.52 | 11.90↑* | – | – | 0.11↓* | Organic acids |
| 59 | Tetradecenoylcarnitine | 370.29518 | 370.29489 | -0.797 | 7.05 | p | 1.23 | 2.04↑** | 1.60↑* | – | – | Acyl carnitine |
| 60 | Ursodeoxycholic acid† | 391.28538 | 391.28583 | 1.142 | 7.02 | n | 1.37 | 3.05↑** | 0.41↓* | – | 0.36↓** | Bile acids |
| 61 | Deoxycholic acid#† | 391.28538 | 391.28580 | 1.066 | 7.65 | n | 1.21 | 3.21↑** | 0.31↓* | – | 0.31↓* | Bile acids |
| 62 | Hexadecenoylcarnitine | 398.32648 | 398.32642 | -0.164 | 7.54 | p | 1.42 | 2.15↑** | 0.72↓* | – | 0.64↓* | Acyl carnitine |
| 63 | Palmitoylcarnitine# | 400.34213 | 400.34189 | -0.613 | 7.94 | p | 1.45 | 2.23↑** | 0.74↓* | – | 0.60↓* | Acyl carnitine |
| 64 | Linoleyl carnitine | 424.34213 | 424.34198 | -0.366 | 7.70 | p | 1.37 | 1.87↑** | 0.76↓* | 0.68↓* | 0.66↓* | Acyl carnitine |
| 65 | Oleoylcarnitine | 426.35778 | 426.35764 | -0.341 | 8.04 | p | 1.37 | 2.28↑** | – | – | – | Acyl carnitine |
| 66 | Stearoylcarnitine | 428.37343 | 428.37335 | -0.200 | 8.40 | p | 1.48 | 2.63↑** | – | – | – | Acyl carnitine |
| 67 | Chenodeoxyglycocholic acid† | 448.30684 | 448.30734 | 1.100 | 6.83 | n | 1.44 | 4.79↑** | 0.37↓* | – | 0.31↓** | Bile acids |
| 68 | LysoPE(18:0)† | 482.32411 | 482.32391 | -0.427 | 8.05 | p | 1.22 | 0.31↓** | – | 1.89↑* | 2.85↑** | Glycerophospholipids |
| 69 | LysoPC(O-16:0) | 482.36050 | 482.36060 | 0.205 | 8.73 | p | 1.10 | 0.80↓* | – | 1.34↑* | 1.32↑* | Glycerophospholipids |
| 70 | LysoPC(16:0) | 496.33976 | 496.33972 | -0.092 | 8.49 | p | 1.21 | 0.79↓** | – | – | 1.16↑* | Glycerophospholipids |
| 71 | LysoPE(20:4)† | 502.29281 | 502.29251 | -0.609 | 8.07 | p | 1.31 | 1.57↑** | – | – | 0.69↓* | Glycerophospholipids |
| 72 | LysoPE(20:3)† | 504.30846 | 504.30826 | -0.408 | 8.38 | p | 1.17 | 0.21↓* | – | – | 3.74↑** | Glycerophospholipids |
| 73 | LysoPC(17:0) | 510.35541 | 510.35541 | -0.012 | 8.93 | p | 1.48 | 0.45↓** | 1.17↑* | 1.55↑* | 2.13↑** | Glycerophospholipids |
| 74 | LysoPC(18:3) | 518.32411 | 518.32416 | 0.085 | 7.73 | p | 1.34 | 0.75↓** | – | – | – | Glycerophospholipids |
| 75 | LysoPC(18:2) | 520.33976 | 520.33984 | 0.143 | 8.11 | p | 1.36 | 0.68↓** | – | – | 1.38↑* | Glycerophospholipids |
| 76 | LysoPC(18:1) | 522.35541 | 522.35553 | 0.218 | 8.68 | p | 1.16 | 0.66↓* | 1.27↑* | 1.28↑* | 1.50↑* | Glycerophospholipids |
| 77 | LysoPE(22:6)† | 526.29281 | 526.29272 | -0.182 | 8.02 | p | 1.13 | 0.75↓* | 1.98↑** | 1.64↑* | 1.74↑** | Glycerophospholipids |
| 78 | LysoPC(20:5) | 542.32411 | 542.32410 | -0.029 | 7.64 | p | 1.41 | 0.68↓** | – | 1.27↑* | 1.31↑* | Glycerophospholipids |
| 79 | LysoPC(20:4) | 544.33976 | 544.33936 | -0.746 | 8.09 | p | 1.68 | 0.65↓** | – | 1.35↑** | 1.43↑** | Glycerophospholipids |
| 80 | LysoPC(20:2) | 548.37106 | 548.37091 | -0.284 | 8.86 | p | 1.21 | 0.42↓** | 1.34↑* | 1.63↑* | 2.25↑* | Glycerophospholipids |
| 81 | Taurolithocholic acid-3-sulfate | 562.25138 | 562.25158 | 0.356 | 13.50 | n | 1.74 | 13.29↑** | – | – | – | Bile acids |
| 82 | LysoPC(22:4) | 572.37106 | 572.37103 | -0.063 | 8.70 | p | 1.23 | 0.60↓** | 1.27↑* | 1.34↑* | 1.52↑** | Glycerophospholipids |
| 83 | LysoPI(20:4) | 619.28888 | 619.28912 | 0.377 | 13.50 | n | 1.66 | 0.52↓** | 1.61↑** | 1.51↑* | 1.62↑* | Glycerophospholipids |
| 84 | PC(16:0/18:2) | 758.56943 | 758.56940 | -0.041 | 13.42 | p | 1.29 | 1.51↑** | – | – | – | Glycerophospholipids |
| 85 | PC(16:0/18:1) | 760.58508 | 760.58337 | -2.250 | 13.48 | p | 1.50 | 0.38↓** | – | – | – | Glycerophospholipids |
| 86 | PC(18:3/18:2) | 780.55378 | 780.55139 | -3.064 | 13.42 | p | 1.39 | 2.38↑* | – | – | 0.63↓* | Glycerophospholipids |
| 87 | PC(18:1/18:2) | 784.58508 | 784.58270 | -3.035 | 13.52 | p | 1.62 | 4.31↑** | 0.54↓* | 0.61↓* | 0.61↓* | Glycerophospholipids |
| 88 | PC(18:0/18:2) | 786.60073 | 786.60071 | -0.027 | 13.44 | p | 1.25 | 1.56↑** | 0.81↓* | 0.75↓* | 0.82↓* | Glycerophospholipids |
| 89 | PC(16:0/22:6) | 806.56943 | 806.56696 | -3.064 | 13.42 | p | 1.08 | 1.21↑* | – | – | – | Glycerophospholipids |
| 90 | PC(16:0/22:5) | 808.58508 | 808.58246 | -3.242 | 13.41 | p | 1.24 | 1.91↑** | 0.69↓* | 0.71↓* | 0.67↓* | Glycerophospholipids |
| 91 | PC(18:0/22:6) | 834.60073 | 834.60034 | -0.469 | 13.53 | p | 1.39 | 3.12↑* | – | – | – | Glycerophospholipids |
#metabolites were identified by reference standards; †metabolites were detected in both negative and positive ion modes; *p values < 0.05; **p values < 0.01; RT, retention time; VIP, variable importance in the projection obtained from Model group vs. Sham group; XBJ, xuebijing injection; BIPM, biapenem; COM, combination of xuebijing injection and biapenem; LysoPE, lysophosphatidylethanolamine; LysoPC, lysophosphatidylcholine; LysoPI, lysophosphatidylinositol; PC, phosphatidylcholine; p, positive ion mode; n, negative ion mode.
Metabolomic Insights Into the Interventional Effects of XBJ, BIPM, and XBJ-BIPM Combination
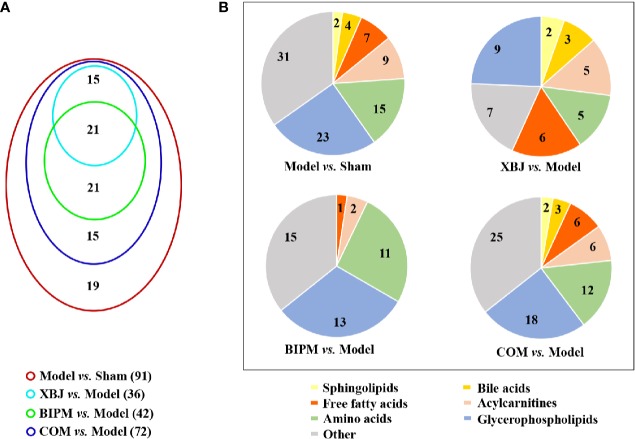
The OPLS-DA models for XBJ vs. Model, BIPM vs. Model, and COM vs. Model were constructed respectively, and distinct discrimination was obtained for each comparison (Supplementary Figure S9). To further investigate the different interventional effects of XBJ and BIPM in alone or in combination, we explored the metabolic phenotypes of different groups focused on the level changes of identified potential biomarkers. In total, 36 metabolites were reversely regulated by XBJ when compared with Model group, mainly including glycerophospholipids, sphingolipids, FFAs, bile acids and acylcarnitines; 42 metabolites were regulated by BIPM, mainly including amino acids, glycerophospholipids, and acylcarnitines; 72 metabolites were regulated after XBJ-BIPM combination treatment, including most of the 91 potential biomarkers. Detailed information of the metabolites was presented in Table 1. It was shown that the combination intervention induced more altered metabolites, 15 metabolites of which were not affected when XBJ or BIPM was administrated alone. A venn diagram was described to display the overlapping relationship of regulated metabolites for different comparisons (Figure 2A), and the classification of regulated metabolites for different comparisons was shown in Figure 2B.
Figure 2.
(A) Venn diagram showing the number of regulated metabolites for different comparisons. (B) Pie charts showing the classification of regulated metabolites for different comparisons. XBJ, xuebijing injection; BIPM, biapenem; COM, combination of xuebijing injection and biapenem.
Compared with Model group, there were no significant changes in bile acids profile (ursodeoxycholic acid, deoxycholic acid, and chenodeoxyglycocholic acid), creatinine, uric acid, and allantoin in BIPM group, but the levels of these metabolites were apparently down-regulated by the combination of BIPM and XBJ. The results evidently showed that treatment with this combination conferred protection against liver and kidney dysfunctions caused by infection in sepsis. Similarly, the decreased levels of FFAs in COM group compared with Model group might be the defensive consequence of XBJ against inflammation and oxidative stress. Beyond this point, more amino acids, glycerophospholipids, and acylcarnitines were regulated after combination treatment compared with XBJ or BIPM monotherapy, exhibiting a synergistic performance of XBJ and BIPM as well.
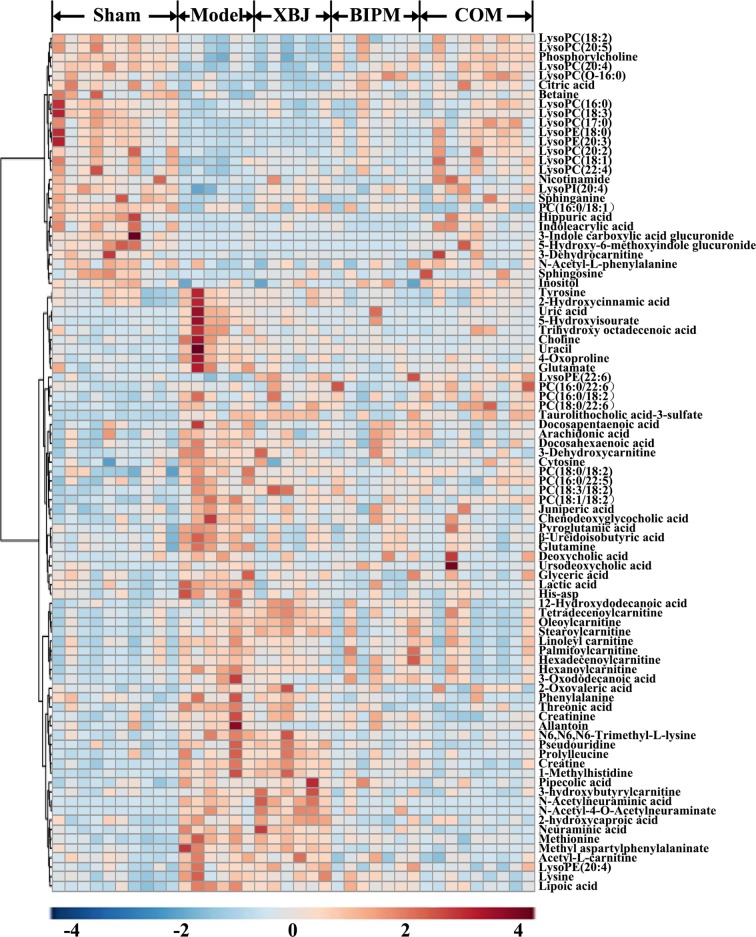
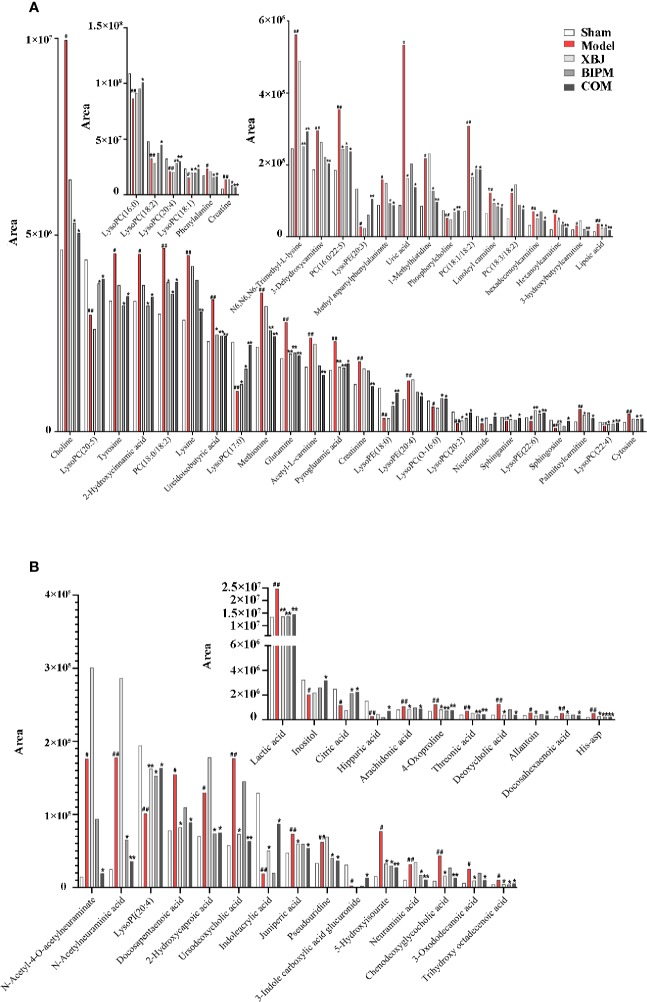
Furthermore, a heat map was depicted to intuitively show the content changing trend of 91 identified potential biomarkers among different groups. As shown in Figure 3, an obvious color discrimination was observed between Model group and Sham group, and different metabolites were reversibly regulated in XBJ, BIPM, and COM group respectively. The COM and Sham group shared similar color distribution, indicating an excellent synergistic effect of XBJ-BIPM combination for sepsis. Among these 91 metabolites, we focused on 72 metabolites which could be reversely regulated by XBJ-BIPM combination, and the average peak area of each metabolite in each group was shown in Figure 4. The histograms directly exhibited levels of 72 metabolites across the five groups.
Figure 3.
Heat map showing the level changing trend of 91 differential metabolites across the five groups. The colors from blue to red indicate higher levels of metabolites. XBJ, xuebijing injection; BIPM, biapenem; COM, combination of xuebijing injection and biapenem.
Figure 4.
The average peak areas of each metabolite in each group for the 72 metabolites detected in positive (A) and negative (B) ion modes. #p < 0.05, ##p < 0.01 versus the Sham group; *p < 0.05, **p < 0.01 versus the Model group. XBJ, xuebijing injection; BIPM, biapenem; COM, combination of xuebijing injection and biapenem.
To explore the latent relationships of potential biomarkers, a heat map was produced according to the Pearson correlation coefficients between metabolites (Supplementary Figure S10). The levels of LysoPCs and LysoPEs showed an obvious negative correlations with FFAs, bile acids, and acylcarnitines, while acylcarnitines had significant positive correlation with part of FFAs and amino acids.
Metabolic Pathway Analysis
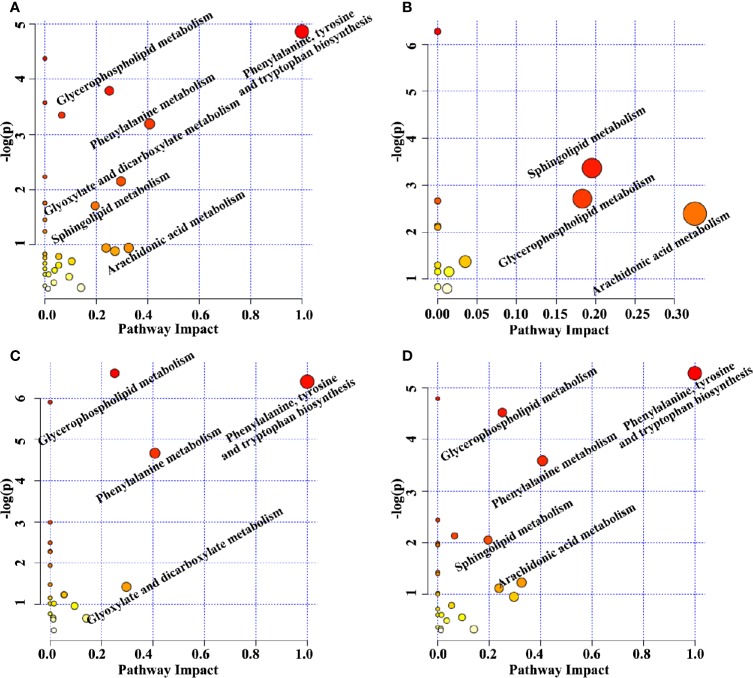
The perturbed metabolic pathways were enriched on the basis of altered metabolites. The principle disturbed metabolic pathways for Model group included phenylalanine, tyrosine and tryptophan biosynthesis, glycerophospholipid metabolism, phenylalanine metabolism, glyoxylate and dicarboxylate metabolism, sphingolipid metabolism, and arachidonic acid metabolism (Figure 5A), which were valuable for elucidating pathogenic mechanism of sepsis. Compared with Model group, 13, 22, and 27 metabolic pathways were regulated by XBJ, BIPM, and XBJ-BIPM combination, respectively (Figures 5B–D). The results suggested that many more metabolic pathways were affected by the synergistic treatment of XBJ and BIPM. A metabolic correlation network (Figure 6) was further established by searching online database KEGG (https://www.kegg.jp/) and HMDB (http://www.hmdb.ca/).
Figure 5.
Topology maps of the altered metabolic pathways analyzed by metaboanalyst between Model group vs. Sham group (A), XBJ group vs. Model group (B), BIPM group vs. Model group (C), COM group vs. Model group (D). XBJ, xuebijing injection; BIPM, biapenem; COM, combination of xuebijing injection and biapenem.
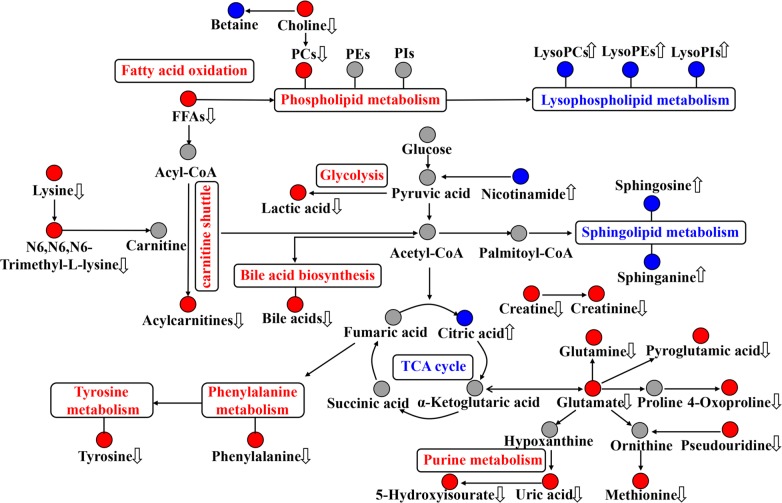
Figure 6.
Disordered metabolic pathway networks in sepsis and the interventional effects of biapenem (BIPM) in combination with xuebijing injection (XBJ). The red dots represent increased metabolites in septic rats, the blue dots represent decreased metabolites in septic rats, and the gray dots represent undetected metabolites. The disturbed metabolic pathways in septic rats are marked in red (up-regulated) and blue (down-regulated). The metabolites regulated by the combination of XBJ and BIPM are marked with up and down arrows.
Discussion
In this work, we characterized the metabolic phenotype of sepsis and described metabolic footprints change of septic rats responding to XBJ and BIPM individually and in combination, in addition to histopathological and survival evaluation. This study was the first to employ metabolomics to elucidate the synergistic effect and decipher the underlying mechanisms of BIPM in combination with XBJ against sepsis. The major findings on the synergistic effect of this combination in our study include: 1) BIPM monotherapy regulated metabolism levels of amino acids, glycerophospholipids, and acylcarnitines in sepsis, but had no effect on sphingolipids, bile acids and FFAs. 2) Many more endogenous metabolites and metabolic pathways were significantly regulated after combination treatment compared with XBJ or BIPM monotherapy. 3) Metabolisms of lipids, amino acids, acylcarnitines, and bile acids were common pathways involved in the synergistic action of XBJ and BIPM.
Lipid Metabolism
The perturbed lipid metabolism including reduced lysophospholipids (LPLs) metabolism, increased PCs metabolism, elevated FFAs metabolism, and decreased sphingolipids metabolism as reflective of systemic changes resulted from inflammatory response and oxidative stress in sepsis (Sevastou et al., 2013). Our results were consistent with previous studies (Mecatti et al., 2018). Decreased LysoPCs might enhance the production of lysophosphatidic acids (Tokumura, 2002), which induced an immune response by activating various immune cells (Gräler and Goetzl, 2002). The LysoPCs have been considered useful for discriminating septic patients from normal control (Drobnik et al., 2003; Cho et al., 2012), and another research demonstrated that LysoPCs increased after administration with appropriate antibiotics in septic patients (Park et al., 2014), which was also agreement with our results. Elevated PCs and reduced LysoPCs might be related to a lack of phospholipase A2 (PLA2), which could hydrolyze PCs to LysoPCs. After intervention by XBJ, BIPM and XBJ-BIPM combination, a total of 8, 12, and 17 glycerophospholipids were reversely regulated respectively, and PLAs probably acted as the therapeutic targets. In addition, lipolysis, an adaptive response to inflammation (Fong et al., 1990), together with increased biosynthesis and decreased oxidation in liver, leading to the accumulation of FFAs in plasma of septic rats (Kamisoglu et al., 2013). The increased arachidonic acid, a precursor of pro-inflammation eicosanoid mediators (Sanak, 2016), might be closely related to the systemic inflammation reaction in sepsis. Sphingolipids are important building blocks of cell membranes, and a large of evidences have demonstrated that sphingolipid metabolites participate in many cell regulation processes as key signal molecules in immunity and inflammation (Michael and Sarah, 2014). Previous studies had shown that immune response in sepsis was associated with disordered sphingolipid metabolism (Winkler et al., 2015). In our study, FFAs and sphingolipids could barely be affected by BIPM monotherapy, but they were well regulated when BIPM combined with XBJ, indicating a synergistic effect of this combination. In short, BIPM in combination with XBJ resulted in enhanced regulating effect on perturbed lipid metabolism in sepsis.
Amino Acid Metabolism
The alteration of amino acid metabolism was another metabolic characteristic of sepsis. Our results showed that both BIPM used alone and combined with XBJ displayed excellent effect on the regulation of amino acids in septic rats, but XBJ monotherapy had a weaker action on amino acids. It was observed that most amino acids elevated in sepsis compared with Sham group, and this results seemed contrary with our previous report (Zuo et al., 2018). In fact, it should be noticed that the time of blood sampling in this work was much later than that of our previous work. In the early stage of sepsis, the organism was in a state of hypermetabolism, and peripheral protein catabolism was activated (Su et al., 2015). Augmented metabolic activity in the liver region promoted the shift of amino acids from peripheral tissues to liver for the hepatic protein synthesis (Druml et al., 2001). However, as the disease progressed, this protein synthesis was reduced in liver because of the hepatic dysfunction (Vaidyanath et al., 1976). Increased muscle protein breakdown and relative hepatic incompetence resulted in accumulation of amino acids (Freund et al., 1979). These reasons might explain the opposite trend of plasma amino acids in our previous study and this work. In this aspect, the plasma accumulation of aromatic and sulfur containing amino acids (phenylalanine, tyrosine, and methionine), which must be metabolized by the liver, was likely to be an important indicator of the combined effect of muscle breakdown and hepatic dysfunction in sepsis.
Energy Metabolism
Elevated levels of acylcarnitines suggested activated fatty acids oxidation (FAO) and energy metabolism in sepsis. The carnitine shuttle, as an essential pathway to transport fatty acids from cytoplasm to mitochondria, played a pivotal role in β-oxidation (Adeva-Andany et al., 2017). Long-chain acyl-CoAs were converted to acylcarnitines by carnitine acyl transferase in the process (Calvani et al., 2000). As one of the major ways of energy production, FAO enhanced in septic rats to maintain the energy supplement, resulting in the increased acylcarnitines. In this work, more acylcarnitines were down-regulated after combination treatment compared with XBJ or BIPM monotherapy. The results implied that energy metabolism was alleviated under the synergistic effect of XBJ and BIPM.
Bile Acid Metabolism
Septic patients usually exhibit cholestasis, and plasma bile acid levels have been viewed as related to mortality (Thomas, 2017). In the present study, elevated bile acids were observed in septic rats, which implied a liver dysfunction induced by infection. It has been suggested that bile acids accumulation promoted sepsis-associated inflammation via NLRP3 inflammasome activation (Hao et al., 2017). The bile acids were decreased after administration of XBJ, but not regulated by BIPM. This phenomenon might attribute to multiple components with hepatoprotective activities existed in XBJ. Therefore, the combination would have more advantages against sepsis in clinic.
In addition, plasma lactic acid levels are traditionally interpreted as a marker of tissue hypoxia and usually employed as an indicator of severity and outcome of sepsis in clinical practice (Nolt et al., 2018). A significant increase of lactic acid in septic rats was also observed in this work, and it was reversely regulated by BIPM in combination with XBJ. The results indicated that administration of this combination could improve hypoxia and energy metabolism in sepsis. Uric acid is a product of purine metabolism and considered as an independent risk factor for weakness in kidney function (Pilemann-Lyberg et al., 2019). In most mammals, uric acid is further oxidized by the enzyme uricase into allantoin (Andries et al., 2018). Creatinine is a biomarker for renal function in clinic. Therefore, the elevated levels of uric acid, allantoin, and creatinine demonstrated kidney dysfunction in sepsis. These metabolites were down-regulated by XBJ-BIPM combination, suggesting a renal protection of this combination.
To sum up, we described a UHPLC-Q-Orbitrap HRMS method in both positive and negative modes for metabolomic evaluation of synergistic effect of BIPM in combination with XBJ against sepsis. The two drugs may interact each other to regulate the plasma levels of amino acids, glycerophospholipids, sphingolipids, FFAs, acylcarnitines, and bile acids in sepsis. The specific effects of these endogenous metabolites in sepsis therapy and metabolomics-based key pathways remain to be further researched. Nevertheless, this work provided a better understanding on the synergistic mechanisms of XBJ and BIPM from metabolomic insights. The results provide some support for clinical application of antibiotics in combination with XBJ and have important implications for the treatment of sepsis in clinic.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of the first affiliated hospital of Zhengzhou University.
Author Contributions
L-WL and Y-YS contributed equally to this work. ZS and X-JZ designed the research. L-WL, Y-YS, Z-LL, and L-HZ performed the experiments. L-WL, Y-YS, MT, and Z-WJ analyzed data. L-WL and H-YZ wrote the manuscript. PX, LZ, and Q-ZD revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81873188, 81703666), the Key Research and Promotion Project of Henan Province (182102310243), the Key Scientific Research Project of Colleges and Universities in Henan Province (20A350018, 18A360022), and Hong Ri Medical Research Foundation (HRJJ18002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.00502/full#supplementary-material
References
- Abdulkhalek S., Szewczuk M. R. (2013). Neu1 sialidase and matrix metalloproteinase-9 cross-talk regulates nucleic acid-induced endosomal TOLL-like receptor-7 and -9 activation, cellular signaling and pro-inflammatory responses. Cell Signal. 25, 2093–2105. 10.1016/j.cellsig.2013.06.010 [DOI] [PubMed] [Google Scholar]
- Adeva-Andany M. M., Calvo-Castro I., Fernández-Fernández C., Donapetry-García C., Pedre-Piñeiro A. M. (2017). Significance of l-carnitine for human health. IUBMB Life. 69, 578–594. 10.1002/iub.1646 [DOI] [PubMed] [Google Scholar]
- Andries A., De Rechter S., Janssens P., Mekahli D., Van Schepdael A. (2018). Simultaneous determination of allantoin and adenosine in human urine using liquid chromatography – UV detection. J. Chromatogr. B. Anal. Technol. BioMed. Life Sci. 1096, 201–207. 10.1016/j.jchromb.2018.08.026 [DOI] [PubMed] [Google Scholar]
- Buras J. A., Holzmann B., Sitkovsky M. (2005). Animal models of sepsis: setting the stage. Nat. Rev. Drug Discovery 4, 854–865. 10.1038/nrd1854 [DOI] [PubMed] [Google Scholar]
- Calvani M., Reda E., Arrigoni-Martelli E. (2000). Regulation by carnitine of myocardial fatty acid and carbohydrate metabolism under normal and pathological conditions. Basic Res. Cardiol. 95, 75–83. 10.1007/s003950050167 [DOI] [PubMed] [Google Scholar]
- Chen Y., Li C. S. (2013). The effectiveness of XueBiJing injection in therapy of sepsis: a multicenter clinical study. Chin. J. Emerg. Med. 22, 130–135. 10.3760/cma.j.issa1671-0282.2013.02.006 [DOI] [Google Scholar]
- Chinese Society of Critical Care Medicine (2015). Chinese guidelines for management of severe sepsis and septic shock 2014. Chin. Crit. Care Med. 27, 401–426. 10.3760/cma.j.issn.2095-4352.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Cho W. H., Park T., Park Y. Y., Huh J. W., Lim C. M., Koh Y., et al. (2012). Clinical significance of enzymatic lysophosphatidylcholine (LPC) assay data in patients with sepsis. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1805–1810. 10.1007/s10096-011-1505-6 [DOI] [PubMed] [Google Scholar]
- Drobnik W., Liebisch G., Audebert F. X., Frohlich D., Gluck T., Vogel P., et al. (2003). Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 44, 754–761. 10.1194/jlr.M200401-JLR200 [DOI] [PubMed] [Google Scholar]
- Druml W., Heinzel G., Kleinberger G. (2001). Amino acid kinetics in patients with sepsis. Am. J. Clin. Nutr. 73, 908–913. 10.1093/ajcn/73.5.908 [DOI] [PubMed] [Google Scholar]
- Fong Y. M., Marano M. A., Moldawer L. L., Wei H., Calvano S. E., Kenney J. S., et al. (1990). The acute splanchnic and peripheral tissue metabolic response to endotoxin in humans. J. Clin. Invest. 85, 1896–1904. 10.1172/JCI114651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund H., Atamian S., Holroyde J., Fischer J. E. (1979). Plasma amino acids as predictors of the severity and outcome of sepsis. Ann. Surg. 190, 571–576. 10.1097/00000658-197911000-00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Kong L. B., Liu S., Feng Z. Q., Shen H., Liu Q. Q. (2015). A prospective multicenter clinical study of XueBiJing injection in the treatment of sepsis and multiple organ dysfunction syndrome. Chin. Crit. Care Med. 27, 465–470. 10.3760/cma.j.issn.2095-4352.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Gräler M. H., Goetzl E. J. (2002). Lysophospholipids and their G protein-coupled receptors in inflammation and immunity. Biochim. Biophys. Acta 1582, 168–174. 10.1016/s1388-1981(02)00152-x [DOI] [PubMed] [Google Scholar]
- Hao H. P., Cao L. J., Jiang C. T., Che Y., Zhang S. Y., Takahashi S., et al. (2017). Farnesoid X Receptor Regulation of the NLRP3 Inflammasome Underlies Cholestasis-Associated Sepsis. Cell Metab. 25, 856–867.e5. 10.1016/j.cmet.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard W. J., Choudhry M., Schwacha M. G., Kerby J. D., Rue L. W., Bland K. I., et al. (2005). Cecal ligation and puncture. Shock 24, 52–57. 10.1097/01.shk.0000191414.94461.7e [DOI] [PubMed] [Google Scholar]
- Ikawa K., Morikawa N., Ikeda K., Ohge H., Sueda T. (2008). Pharmacodynamic evaluation of biapenem in peritoneal fluid using population pharmacokinetic modelling and Monte Carlo simulation. Int. J. Antimicrob. Ag. 32, 339–343. 10.1016/j.ijantimicag.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Kamisoglu K., Sleight K. E., Calvano S. E., Coyle S. M., Corbett S. A., Androulakis I. P. (2013). Temporal metabolic profiling of plasma during endotoxemia in humans. Shock 40, 519–526. 10.1097/SHK.0000000000000063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper P., Held T., Schneider E., Bölke E., Gerlach H., Trautmann M. (2002). Clinical implications of antibiotic-induced endotoxin release in septic shock. Intens Care Med. 28, 824–833. 10.1007/s00134-002-1330-6 [DOI] [PubMed] [Google Scholar]
- Mecatti G. C., Fernandes Messias M. C., Sant'Anna Paiola R. M., Figueiredo Angolini C. F., Barbosa D. S. C. I., Eberlin M. N., et al. (2018). Lipidomic profiling of plasma and erythrocytes from septic patients reveals potential biomarker candidates. Biomarker Insights 13, 1–13. 10.1177/1177271918765137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael M., Sarah S. (2014). Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67. 10.1038/nature13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolt B., Tu F., Wang X. H., Ha T. Z., Winter R., Williams D. L., et al. (2018). Lactate and immunosuppression in sepsis. Shock 49, 120–125. 10.1097/SHK.0000000000000958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. W., Kwak D. S., Park Y. Y., Chang Y., Huh J. W., Lim C. M., et al. (2014). Impact of serial measurements of lysophosphatidylcholine on 28-day mortality prediction in patients admitted to the intensive care unit with severe sepsis or septic shock. J. Crit. Care 29, 882.e5–882.e11. 10.1016/j.jcrc.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Pilemann-Lyberg S., Hansen T. W., Tofte N., Winther S. A., Theilade S., Ahluwalia T. S., et al. (2019). Uric acid is an independent risk factor for decline in kidney function, cardiovascular events, and mortality in patients with type 1 diabetes. Diabetes Care 42, 1088–1094. 10.2337/dc18-2173 [DOI] [PubMed] [Google Scholar]
- Rittirsch D., Huber-Lang M. S., Flierl M. A., Ward P. A. (2009). Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 4, 31–36. 10.1038/nprot.2008.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanak M. (2016). Eicosanoid mediators in the airway inflammation of asthmatic patients: what is new? Allergy Asthma Immunol. Res. 8, 481–490. 10.4168/aair.2016.8.6.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevastou I., Kaffe E., Mouratis M. A., Aidinis V. (2013). Lysoglycerophospholipids in chronic inflammatory disorders: the PLA(2)/LPC and ATX/LPA axes. Biochim. Biophys. Acta 1831, 42–60. 10.1016/j.bbalip.2012.07.019 [DOI] [PubMed] [Google Scholar]
- Singer M., Deutschman C. S., Seymour C. W., Shankar-Hari M., Annane D., Bauer M., et al. (2016). The third international consensus definitions for sepsis and septic shock (sepsis-3). J. Am. Med. Assoc. 315, 801–810. 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. L., Yao C., Shang H. C., Yao X. Q., Bai C. X. (2016). Late-breaking abstract: intravenous infusion of Chinese medicine XueBiJing significantly improved clinical outcome in severe pneumonia patients in a multiple center randomized controlled clinical trials. Eur. Respir. J. 48, OA3323. 10.1183/13993003.congress-2016.OA3323 [DOI] [Google Scholar]
- Su L. X., Li H., Xie A. M., Liu D., Rao W. Q., Lan L. P., et al. (2015). Dynamic Changes in Amino Acid Concentration Profiles in Patients with Sepsis. PloS One 10, e0121933. 10.1371/journal.pone.0121933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2017). Bile acids promote inflammation in cholestasis-associated sepsis. Nat. Rev. Gastroenterol. Hepatol. 14, 324–325. 10.1038/nrgastro.2017.55 [DOI] [PubMed] [Google Scholar]
- Tokumura A. (2002). Physiological and pathophysiological roles of lysophosphatidic acids produced by secretory lysophospholipase D in body fluids. Biochim. Biophys. Acta 1582, 18–25. 10.1016/s1388-1981(02)00133-6 [DOI] [PubMed] [Google Scholar]
- Vaidyanath N., Oswald G., Trietley G., Weissenhofer W., Moritz E., McMenamy R. H., et al. (1976). Turnover of amino acids in sepsis and starvation: effect of glucose infusion. J. Trauma 16, 125–135. 10.1097/00005373-197602000-00008 [DOI] [PubMed] [Google Scholar]
- Winkler M. S., Nierhaus A., Holzmann M., Mudersbach E., Bauer A., Robbe L., et al. (2015). Decreased serum concentrations of sphingosine-1-phosphate in sepsis. Crit. Care 19, 372. 10.1186/s13054-015-1089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T. Y., Zhou L., Shi Y. Y., Liu L. W., Zuo L. H., Jia Q. Q., et al. (2018). Metabolomics approach in lung tissue of septic rats and the interventional effects of Xuebijing injection using UHPLC-QOrbitrap-HRMS. J. Biochem. 164, 427–435. 10.1093/jb/mvy070 [DOI] [PubMed] [Google Scholar]
- Yin Q., Li C. (2014). Treatment effects of xuebijing injection in severe septic patients with disseminated Intravascular Coagulation. Evid. Based. Complement Alternat. Med. 2014, 949254. 10.1155/2014/949254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. T., Cheng C., Olaleye O. E., Sun Y., Li L., Huang Y. H., et al. (2018). Pharmacokinetics-based identification of potential therapeutic phthalides from XueBiJing, a Chinese herbal injection used in sepsis management. Drug Metab. Dispos. 46, dmd.117.079673. 10.1124/dmd.117.079673 [DOI] [PubMed] [Google Scholar]
- Zuo L. H., Zhou L., Xu T. Y., Li Z. L., Liu L. W., Shi Y. Y., et al. (2018). Antiseptic activity of ethnomedicinal Xuebijing revealed by the metabolomics analysis using UHPLC-Q-Orbitrap HRMS. Front. Pharmacol. 9, 300. 10.3389/fphar.2018.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.