Abstract
The tuberal hypothalamus is comprised of the dorsomedial, ventromedial, and arcuate nuclei, as well as parts of the lateral hypothalamic area, and it governs a wide range of physiologies. During neurogenesis, tuberal hypothalamic neurons are thought to be born in a dorsal-to-ventral and outside-in pattern, although the accuracy of this description has been questioned over the years. Moreover, the intrinsic factors that control the timing of neurogenesis in this region are poorly characterized. Proneural genes, including Achate-scute-like 1 (Ascl1) and Neurogenin 3 (Neurog3) are widely expressed in hypothalamic progenitors and contribute to lineage commitment and subtype-specific neuronal identifies, but the potential role of Neurogenin 2 (Neurog2) remains unexplored. Birthdating in male and female mice showed that tuberal hypothalamic neurogenesis begins as early as E9.5 in the lateral hypothalamic and arcuate and rapidly expands to dorsomedial and ventromedial neurons by E10.5, peaking throughout the region by E11.5. We confirmed an outside-in trend, except for neurons born at E9.5, and uncovered a rostrocaudal progression but did not confirm a dorsal-ventral patterning to tuberal hypothalamic neuronal birth. In the absence of Neurog2, neurogenesis stalls, with a significant reduction in early-born BrdU+ cells but no change at later time points. Further, the loss of Ascl1 yielded a similar delay in neuronal birth, suggesting that Ascl1 cannot rescue the loss of Neurog2 and that these proneural genes act independently in the tuberal hypothalamus. Together, our findings show that Neurog2 functions as a classical proneural gene to regulate the temporal progression of tuberal hypothalamic neurogenesis.
SIGNIFICANCE STATEMENT Here, we investigated the general timing and pattern of neurogenesis within the tuberal hypothalamus. Our results confirmed an outside-in trend of neurogenesis and uncovered a rostrocaudal progression. We also showed that Neurog2 acts as a classical proneural gene and is responsible for regulating the birth of early-born neurons within the ventromedial hypothalamus, acting independently of Ascl1. In addition, we revealed a role for Neurog2 in cell fate specification and differentiation of ventromedial -specific neurons. Last, Neurog2 does not have cross-inhibitory effects on Neurog1, Neurog3, and Ascl1. These findings are the first to reveal a role for Neurog2 in hypothalamic development.
Keywords: Neurog2, neurogenesis, proneural genes, specification, VMH
Introduction
The hypothalamus is a multinucleated structure that is highly conserved across species (Bedont et al., 2015; Nesan and Kurrasch, 2016; Xie and Dorsky, 2017), likely due to its important role in regulating homeostasis (Chrousos, 2007; Kurrasch et al., 2007; Alvarez-Bolado et al., 2015; Nesan and Kurrasch, 2016). The hypothalamus is divided into three regions across the rostral-caudal plane: anterior, tuberal, and mammillary hypothalamus (Bedont et al., 2015; Nesan and Kurrasch, 2016; Xie and Dorsky, 2017). Each region also is divided into three mediolateral zones: the periventricular (closest to third ventricle), medial (adjacent to periventricular zone), and lateral (farthest from third ventricle) zones (Nesan and Kurrasch, 2016). Extrinsic factors, such as Shh (ventralization) (Szabo et al., 2009; Alvarez-Bolado et al., 2012) and Wnt (caudalization) (Newman et al., 2018; Alvarez-Bolado, 2019), work in combination with intrinsic factors, including the transcription factors Sox2/3, Tbx1/2, Rax, and Lhx2 (Braun et al., 2003; Zhao et al., 2012; Lu et al., 2013; Trowe et al., 2013; Orquera et al., 2016), to pattern the emerging hypothalamus. In addition, Shh is required for proper regionalization of anterior and tuberal structures within the hypothalamus (Shimogori et al., 2010). After patterning, neural progenitor cells undergo rapid proliferative cell divisions to expand the progenitor pool, followed by asymmetric divisions to produce a daughter neural progenitor cell and a neuron, a process referred to as neurogenesis (Xie and Dorsky, 2017). The cues that drive the onset and propagation of hypothalamic neurogenesis remain poorly understood.
The tuberal hypothalamus is a medial hypothalamic region that includes the dorsomedial hypothalamus (DMH), ventromedial hypothalamus (VMH), arcuate nucleus (ARC), and a loosely defined region of the lateral hypothalamus (LH) (Nesan and Kurrasch, 2016). Neurons clustered into tuberal hypothalamic nuclei play important roles in regulating satiety and energy balance, aggression, sexual behavior, and stress responses (Dhillon et al., 2006; King, 2006; Joly-Amado et al., 2014; Cheung et al., 2015; Yang et al., 2017). Work in the 1970s showed that hypothalamic neurons are established in a so-called outside-in pattern, whereby neurons born at early developmental stages are localized in lateral zones, whereas neurons born later occupy the medial and periventricular zones (Shimada and Nakamura, 1973). However, this model has been challenged, with more recent studies showing later-born neurons also located in lateral regions of the tuberal hypothalamus and early-born neurons located in the periventricular zone (Padilla et al., 2010; Alvarez-Bolado et al., 2012). In addition, an early study suggested that neurogenesis advances along a dorsal-to-ventral gradient in the third ventricle, with DMH-residing neurons born before ARC neurons (Shimada and Nakamura, 1973), although other studies have data inconsistent with this regional pattern of neurogenesis but lack a full characterization (Markakis and Swanson, 1997; Padilla et al., 2010; Alvarez-Bolado et al., 2012), demonstrating the need to revisit tuberal hypothalamic neurogenesis in a detailed manner.
Alongside questions regarding the patterning of neuronal birth in the tuberal hypothalamus, the intrinsic signals governing the onset and duration of neurogenesis are still emerging. Proneural genes are basic-helix-loop-helix (bHLH) transcription factors that control multiple functions in the developing brain, including neurogenesis (Bertrand et al., 2002; Schuurmans and Guillemot, 2002; Wilkinson et al., 2013). Neurogenins are a proneural gene family consisting of three members: Neurog1, Neurog2, and Neurog3 (Bertrand et al., 2002), with each gene playing well-known roles in a variety of developmental processes (Fode et al., 2000; Akagi et al., 2004; Florio et al., 2012; Dennis et al., 2017; Chouchane and Costa, 2019). Neurog3 is the only neurogenin family member that has been studied in the VMH and ARC nucleus, with reports showing that it is required for proper differentiation of a subset of ARC and VMH neurons (Pelling et al., 2011) that go on to control feeding (Anthwal et al., 2013). Achaete-Scute homolog 1 (Ascl1) is another bHLH family transcription factor that controls neurogenesis, progenitor maintenance, neural cell fate specification, neuronal differentiation, and migration in many regions of the CNS (Casarosa et al., 1999; Schuurmans et al., 2000; Nieto et al., 2001; Parras et al., 2002; Sugimori et al., 2007; Pacary et al., 2011), including the hypothalamus (McNay et al., 2006; Pelling et al., 2011). In the retina (Hufnagel et al., 2010) and forebrain (Fode et al., 2000; Parras et al., 2002; Schuurmans et al., 2004; Anthwal et al., 2013), Ascl1 expression is upregulated and can rescue neurogenesis in the absence of Neurog2, demonstrating the remarkable compensatory potential of one proneural gene for another.
Here, we asked whether Neurog2, which promotes cell cycle exit, neurogenesis, neuronal fate specification, differentiation, and migration in several CNS domains, including the neocortex and retina (Parras et al., 2002; Akagi et al., 2004; Helms et al., 2005; Heng et al., 2008; Florio et al., 2012; Dixit et al., 2014), also regulates neurogenesis in the hypothalamus, acting in a cross-regulatory manner with Ascl1. Using Neurog2 and Ascl1 loss-of-function animals, we found that Neurog2 functions as a classical proneural gene to drive early neurogenesis in the hypothalamus in an Ascl1-independent fashion.
Materials and Methods
Animals and genotyping
Animal protocols were approved by the University of Calgary Animal Care Committee (AC17-0191) and were housed according to the Guidelines of the Canadian Council of Animal Care. In this study, Neurog2GFPKI (Britz et al., 2006) and Ascl1GFPKI (Leung et al., 2007) heterozygous animals were maintained on a CD1 (Charles River) background. Vaginal plugs were checked each morning shortly after lights on at 8:00 A.M., and the plug dates were considered as embryonic day (E) 0.5. Both male and female embryos were used in our study. PCR was used to establish genotype using the following primers: Neurog2GFPKI: mutant forward 5′-GGACATTCCCGGACACACAC-3′, mutant reverse 5′-GCATCACCTTCACCCTCTCC-3′, WT forward 5′-TAGACGCAGTGACTTCTGTGACCG-3′ and WT reverse 5′-ACCTCCTCTTCCTCCTTCAACTCC-3′. Ascl1GFPKI: mutant forward 5′-AACTTTCCTCCGGGGCTCGTTTC-3′, mutant-reverse 5′-TGGCTGTTGTAGTTGTACTCCAGC-3′, WT forward 5′-TCCAACGACTTGAACTCTATGG-3′, WT reverse 5′-CCAGGACTCAATACGCAGGG-3′.
Tissue preparation
Pregnant dams were killed using cervical dislocation and embryos were collected at E12.5, E15.5, and E19.5. Brains were dissected out of the skull, and tissue fixation and preparation were conducted as previously described (Marsters et al., 2016; Rosin and Kurrasch, 2018).
BrdU labeling
Pregnant females were injected with 100 µg/g body weight BrdU (Sigma Millipore) at embryonic stages E9.5, E10.5, E11.5, E12.5, E13.5, and E14.5. Embryos were harvested at E19.5 and fixed as described (Marsters et al., 2016; Rosin and Kurrasch, 2018). For BrdU staining, we used a modified antigen retrieval protocol as follows: 1 h in 50% formamide/2× SSC at 65°C, followed by 15 min in 2× SSC wash, 30 min in 2N HCl at 37°C, 10 min in borate buffer, pH 8.5, and 5× wash with PBS following by regular immunostaining procedure for anti-BrdU (Lai et al., 2008).
Immunostaining
Using a cryostat, fixed brains were coronally sectioned (10 μm), with tissue collection starting at trigeminal ganglion nerve for E10.5–14.5 and anterior commissure for E15.5–E19.5 before immunostaining. The immunostaining procedure has been previously described (Marsters et al., 2016; Rosin and Kurrasch, 2018). Primary antibodies included the following: rabbit anti-Fezf1 (Fitzgerald; 1:100); rabbit anti-TTF-1 (alternatively Nkx2.1; Santa Cruz Biotechnology; 1:500); and rat anti-BrdU (Abcam; 1:300). Secondary antibodies used were goat anti-IgG AlexaFluor-488 or -546-conjugated (Thermo Fisher Scientific; 1:500). In addition, we applied Hoechst nuclear stain on all samples (Thermo Fisher Scientific; 1:2000).
RNA ISH
ISH was performed on coronally sectioned brains at different embryonic stages from E10.5 to P0. ISH performed in this study had been described previously (Kurrasch et al., 2007). The riboprobes used in this study were as follows: Neurog1 (Blader et al., 2004), Neurog2 (Gradwohl et al., 1996), Neurog3 (Gradwohl et al., 2000), and Ascl1 (Cau et al., 1997).
RNAScope
RNAScope Multiplex Fluorescent Detection Kit v2 (catalog #323110) was used on E12.5 fixed brains that were coronally sectioned (10 μm). Three RNA probe mixtures were applied for 2 h at 40°C: RNAscope 3-plex Negative Control Probe (catalog #320871), RNAscope 3-plex Positive Control Probe-Mm (catalog #320881), and a mixture of Ascl1 (catalog #313291) and Neurog2-C2 (catalog #417291-C2). Amplification and staining steps were performed as described by the manufacturer. Opal 570 reagent (FP1488001KT) (red, 1:1500) was used for Channel 1, and Opal 520 reagent (FP1487001KT) (green, 1:1500) was used for Channel 2 according to the procedure described by the manufacturer. After staining, samples were imaged using Axiovert 200 M confocal microscope (Carl Zeiss).
Quantification and statistical analysis
An Axioplan 2 manual compound microscope (Carl Zeiss) with an Axiocam HRc camera (Carl Zeiss) was used to capture the images. ImageJ software was used for quantification of cell numbers in producing binary images and plots. The whole image was used to make the plots. At least three brain sections across the rostral to caudal plane (∼30 µm apart) for each embryo were analyzed (focusing on the tuberal hypothalamus). A Fezf1 immunolabel was used to mark the beginning and end of the VMH nucleus. For each experimental group, three or four embryonic samples from at least two pregnant dams were analyzed. GraphPad Prism 7 and unpaired Student's t test were used to assess statistical differences between controls and Neurog2 mutants. Results are displayed as mean ± SEM.
Results
Neurogenesis within tuberal hypothalamus follows an outside-in pattern
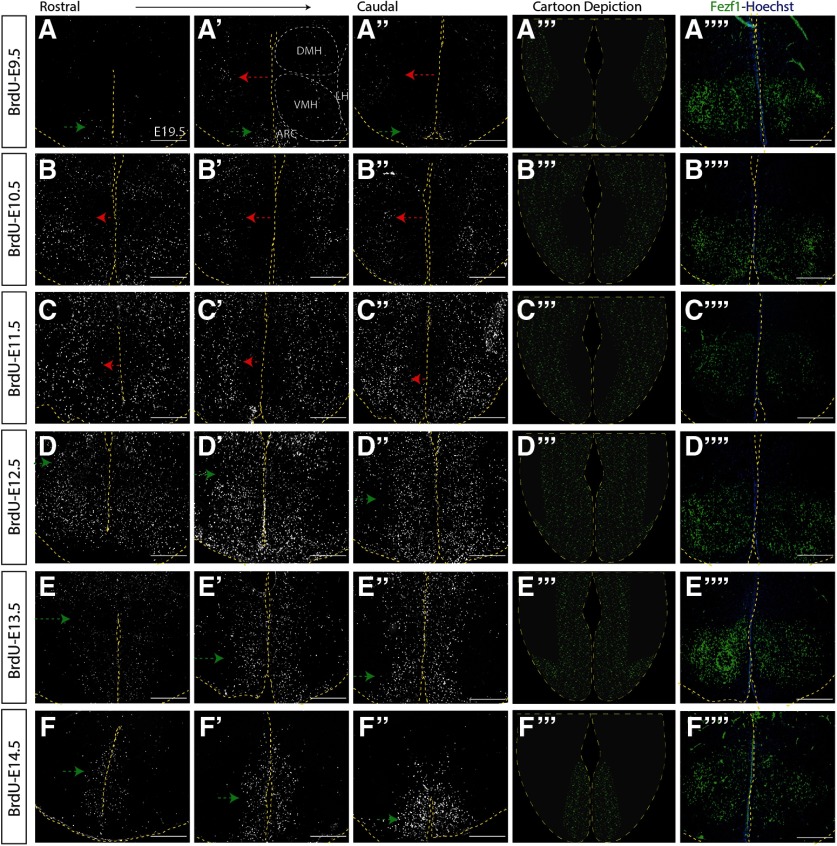
Given the conflicting evidence both supporting (Shimada and Nakamura, 1973) and contradicting (Markakis and Swanson, 1997; Padilla et al., 2010; Alvarez-Bolado et al., 2012) an outside-in model for hypothalamic neurogenesis, we first conducted detailed birthdating analyses in the developing tuberal hypothalamus of WT embryos. To do so, we crossed CD1 mice and injected pregnant dams at E9.5, E10.5, E11.5, E12.5, E13.5, or E14.5 and then collected all embryos at E19.5, a time point when neurogenesis is complete and newborn neurons have coalesced into their mature nucleus (Altman and Bayer, 1986; Kurrasch et al., 2007). We stained for BrdU+ cells (Lai et al., 2008) at this E19.5 time point and imaged multiple sections from the anterior to posterior border of the tuberal hypothalamus, as defined by the pan-VMH marker Fezf1. Unlike in other brain regions, such as the neocortex, where a division into six layers is consistent, hypothalamic nuclear morphology changes considerably in coronal sections at different axial levels (e.g., picture an American football cross-sectioned at either the tip or in the middle of the ball). Furthermore, also in contrast to neocortical cells that are evenly distributed (more-or-less) within their designated layer, phenotypically similar hypothalamic neurons often cluster into discrete subdomains within a single nucleus, further complicating the 3D composition of the nucleus. Combined, nuclear formation is distinct across three axes (rostrocaudal, dorsoventral, and mediolateral), suggesting that hypothalamic neurogenesis and neuronal migration are tightly regulated across these three planes. To capture an accurate distribution pattern of BrdU+ newborn neurons across the entire tuberal hypothalamus, we aligned three sections (20 μm apart) spanning this region (Fig. 1). In addition, to show the VMH location within the tuberal hypothalamus, we immunostained for Fezf1, a pan-VMH marker (Fig. 1A′′′′–F′′′′).
Figure 1.
Neurogenesis within the tuberal hypothalamus has an outside-in trend. Immunostaining showing BrdU expression in the sectioned E19.5 mouse brain injected with BrdU at the following: E9.5 (A-A′′), E10.5 (B-B′′), E11.5 (C-C′′), E12.5 (D-D′′), E13.5 (E-E′′), and E14.5 (F-F′′). A′′′–F′′′, Schematic figure of tuberal hypothalamic neurogenesis at E9.5 (A′′′), E10.5 (B′′′), E11.5 (C′′′), E12.5 (D′′′), E13.5 (E′′′), and E14.5 (F′′′). A′′′′–F′′′′, E14.5 immunostaining showing Fezf1 expression in the sectioned E19.5 mouse brain injected with BrdU at the following: E9.5 (A′′′′), E10.5 (B′′′′), E11.5 (C′′′′), E12.5 (D′′′′), E13.5 (E′′′′), and E14.5 (F′′′′). Scale bars, 100 μm.
As predicted, few tuberal hypothalamic neurons were born at E9.5, with nearly all BrdU+ cells born at this time restricted to the ARC and LH (Fig. 1A-A′′). Significantly more neurons were born at E10.5, indicative of a sharp rise in neurogenesis at this time point (Fig. 1B-B′′). A rostrocaudal gradient in the timing of neuronal birth was also evident, with rostral-most sections populated by overall higher numbers of E10.5-labeled BrdU+ cells, resembling BrdU+ cell density in more caudal sections of brains exposed to BrdU at E11.5 (Fig. 1C-C′′). In contrast, in more caudal sections of E10.5-injected brains, BrdU+ cells were more abundant in lateral regions than in the periventricular zone (Fig. 1B′,B′′). Peak neurogenesis occurred between E11.5 and E12.5, with a noticeable switch in the outside-in patterning. Specifically, in brains injected with BrdU at E11.5, BrdU+ cells were still primarily positioned in the lateral and medial zone with a paucity of BrdU+ cells around the third ventricle (Fig. 1C-C′′). In contrast, cells labeled with BrdU at E12.5 displayed a complementary pattern whereby neurons born at this stage were located primarily in the medial and periventricular zones and to a lesser extent in lateral areas (Fig. 1D-D′′).
We next examined the late window of tuberal hypothalamic neurogenesis. Injection of BrdU at E13.5 revealed a noticeable decrease in the number of cells born at this time point, with BrdU+ cells primarily restricted to medial and periventricular zones (Fig. 1E-E′′). Fewer rostral tuberal hypothalamic neurons were also born at E13.5 (Fig. 1E) compared with the medial and caudal hypothalamic sections (Fig. 1E′,E′′). Finally, labeling newborn neurons at E14.5 revealed a further decrease in BrdU+ cells (Fig. 1F-F′′), suggesting that neurogenesis was nearly complete. Additionally, nearly all BrdU+ neurons in E14.5-injected brains resided in the periventricular zone, with very few cells located in the medial or lateral regions of the tuberal hypothalamus (Fig. 1F-F′′). Together, these data confirm a neurogenic window from E9.5–E14.5 that spreads in an outside-in (with the exception of neurons born at E9.5) and rostrocaudal pattern (Fig. 1, diagrams; see Fig. 9A).
Figure 9.
Schematic figure depicting neurogenesis within the tuberal hypothalamus and VMH. A, Schematic curve for the timing of neurogenesis within the tuberal hypothalamus, in control compared with Neurog2−/− and Ascl1−/− brains. B, Diagram summarizing the pattern of neurogenesis within the VMH nucleus.
Neurog1, Neurog2, Neurog3, and Ascl1 are expressed within tuberal hypothalamic progenitors throughout embryonic development
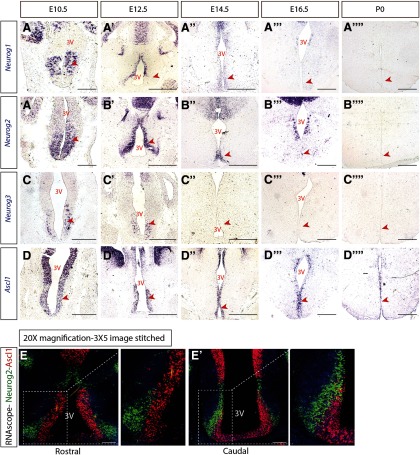
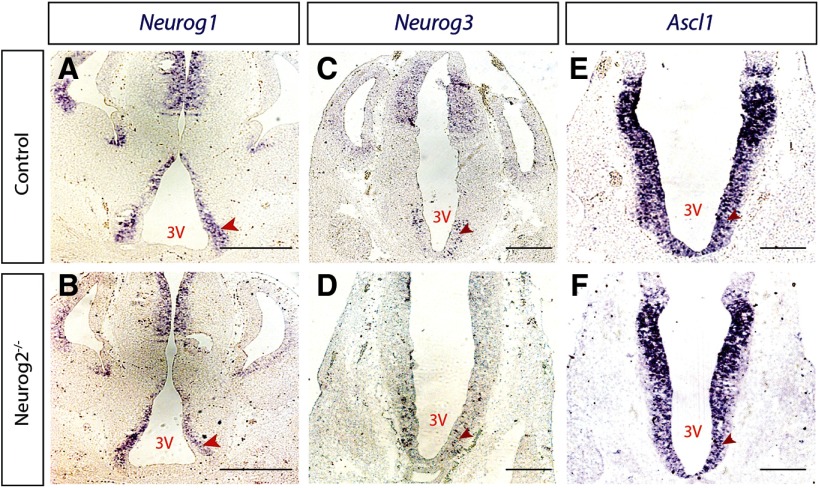
To begin to identify the intrinsic factors that drive tuberal hypothalamic neurogenesis, we first investigated the expression of neurogenin proneural genes (Neurog1, Neurog2, Neurog3) and Ascl1 within the developing tuberal hypothalamus. We conducted ISH for Neurog1, Neurog2, Neurog3, and Ascl1 at different embryonic stages (E10.5 to P0) on CD1 WT brains. The three neurogenin genes were expressed in tuberal hypothalamic progenitor cells starting at E10.5, reached their peak expression at E12.5; Neruog1 and 2 transcripts were largely reduced by E14.5, and Neurog3 transcript was absent by E14.5 (Fig. 2A-A′′,B-B′′,C-C′′, arrowheads). Additionally, at E12.5, Neurog1 and Neurog2 expression was absent in the ventral progenitors that give rise to ARC and median eminence neurons (Fig. 2A′,B′). By E16.5, weak expression of Neurog1 and Neurog2, but not Neurog3, was detected (Fig. 2A′′′, B′′′, C′′′, arrowheads); and by P0, no transcript of any neurogenin gene was observed within tuberal hypothalamic progenitors (Fig. 2A′′′′, B′′′′, C′′′′, arrowheads). In addition, we confirmed Ascl1 expression in tuberal hypothalamic progenitors and across all embryonic time points, as we reported previously (Aslanpour et al., 2020) (Fig. 2D-D′′′). Among the three neurogenin genes, Neurog2 displayed the strongest expression levels (Fig. 2B-B′′′′), causing us to focuson the role of this proneural gene in tuberal hypothalamic development.
Figure 2.
Proneural genes are expressed within VMH progenitors across embryonic development. ISH results demonstrating Neurog1 (A-A′′′′), Neurog2 (B-B′′′′), Neurog3 (C-C′′′′), and Ascl1 (D-D′′′′) expression at E10.5, E12.5, E14.5, E16.5, and P0. E, E′, RNAScope results for Ascl1 (red) and Neurog2 (green) on a rostral (E) and caudal (E′) section of E12.5 hypothalamus. Scale bars, 100 μm.
In addition, to determine the spatial relationship of Ascl1 and Neurog2 in tuberal hypothalamic progenitors, we used RNAScope to detect their transcript in WT E12.5 brains (a time point whereby both genes display strong expression). We showed that Ascl1 and Neurog2 are expressed in distinct progenitor populations in both the rostral and caudal tuberal hypothalamus, with no colocalization observed (Fig. 2E,E′).
Neurog2 is required for proper neurogenesis of early-born but not later-born neurons within the embryonic tuberal hypothalamus
To determine whether Neurog2 was required for the onset and/or propagation of neurogenesis within the tuberal hypothalamus, we used Neurog2GFPKI mice that carry a null allele. To monitor the timing of neurogenesis in the absence of Neurog2, we crossed Neurog2GFPKI/+ animals, and pregnant dams were injected with BrdU at 24 h intervals between E9.5-E14.5. All brains were harvested at E19.5, and Neurog2GFPKI/+ (e.g., WT) embryos were compared with Neurog2GFP/GFP (hereafter referred to as Neurog2−/−) embryos.
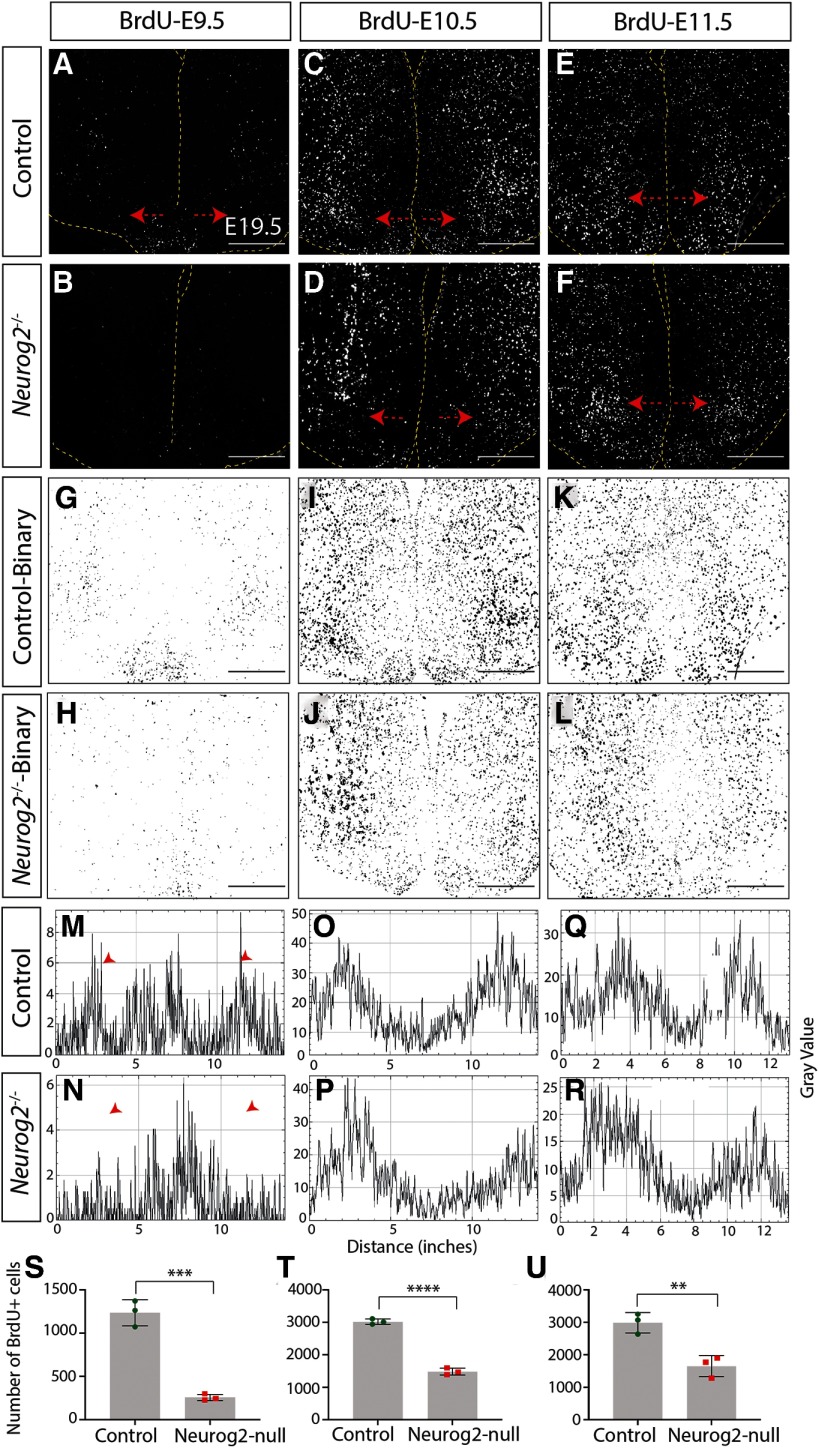
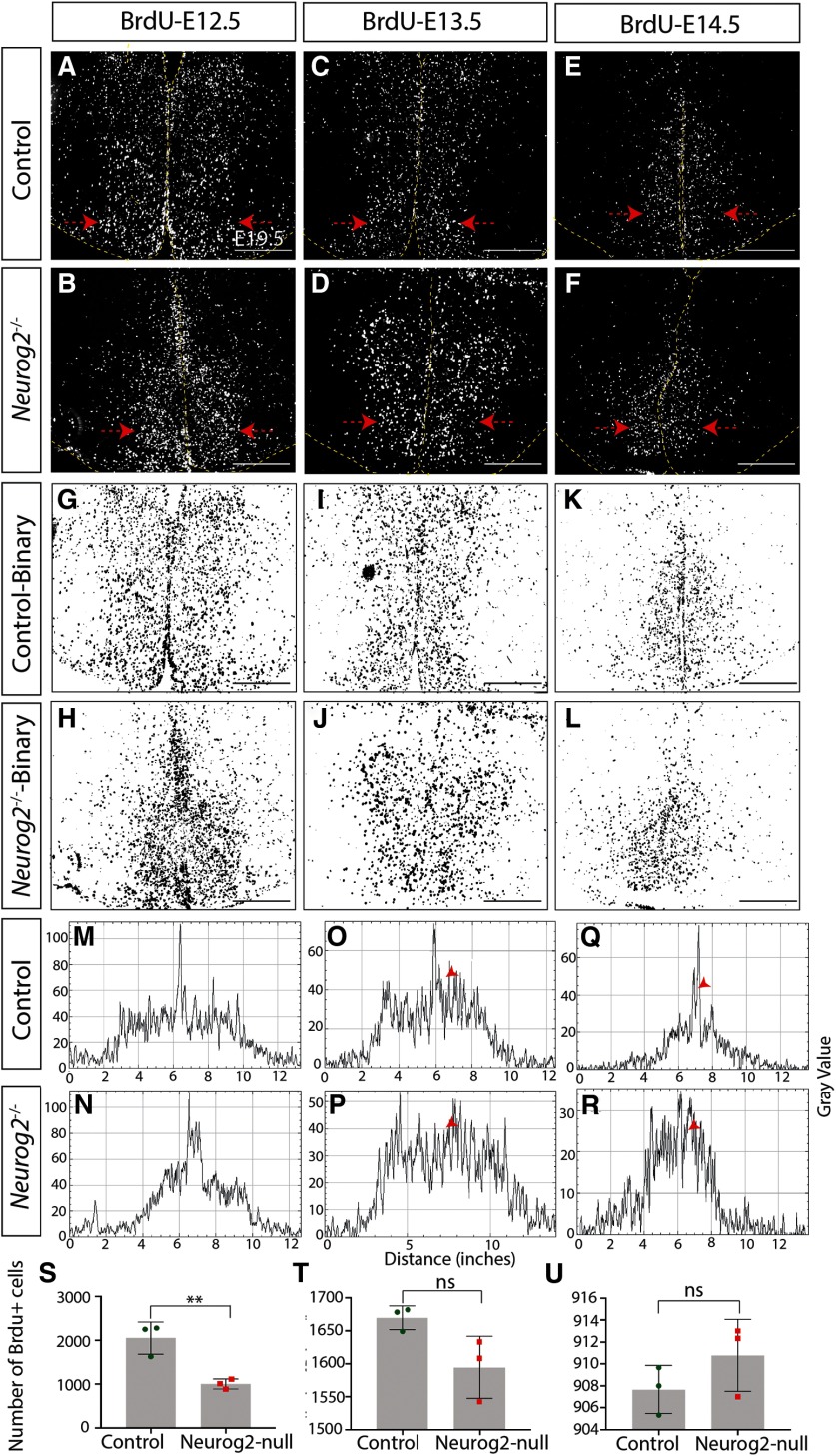
We first assessed overall numbers of newborn neurons and observed a significant decrease in the number of neurons born at E9.5 in the absence of Neurog2 compared with WT controls (Control: 1234 ± 87.61 cells, n = 3; Neurog2−/−: 255.6 ± 21.62 cells, n = 3; p = 0.0004, unpaired t test; Fig. 3A,B,S). This decrease in neurogenesis continued with fewer neurons born at E10.5 (Control: 2931 ± 49.97 cells, n = 3; Neurog2−/−: 1483 ± 61.64 cells, n = 3; p < 0.0001, unpaired t test; Fig. 3C,D,T), E11.5 (Control: 3242 ± 77.21 cells, n = 3; Neurog2−/−: 1650 ± 188.1 cells, n = 3; p = 0.0014, unpaired t test; Fig. 3E,F,U), and E12.5 (Control: 2053 ± 212.7 cells, n = 3; Neurog2−/−: 1004 ± 65.78 cells, n = 3; p = 0.0092, unpaired t test; Fig. 4A,B,S) in Neurog2−/− brains compared with WT controls. After this peak period of neurogenesis, however, neuronal birth in the Neurog2−/− brains continued at equivalent levels to that observed in WT embryos, with no change in the number of BrdU+ cells born at E13.5 (Control: 1670 ± 10.38 cells, n = 3; Neurog2−/−: 1594 ± 27.4 cells, n = 3; p = 0.06, unpaired t test; Fig. 4C,D,T) or E14.5 (Control: 907 ± 1.26 cells, n = 3; Neurog2−/−: 910.8 ± 1.89 cells, n = 3; p = 0.24, unpaired t test; Fig. 4E,F,U) in Neurog2−/− brains compared with WT embryos (Fig. 9A).
Figure 3.
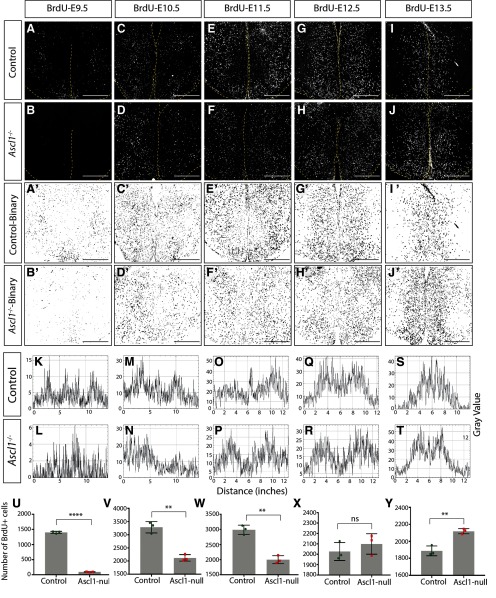
Neurog2 is required for proper neurogenesis within the embryonic tuberal hypothalamus. Immunostaining results for anti-BrdU on E19.5.5 coronal sections of control and Neurog2−/− brains injected with BrdU at E9.5 (A,B), E10.5 (C,D), or E11.5 (E,F). G–L, Binary images of the data presented in A–F. Histogram plots of these binary images demonstrate the location of BrdU+ cells following injection at E9.5 (M,N), E10.5 (O,P), or E11.5 (Q,R). Cell counts of BrdU+ cells within control and Neurog2−/− tuberal hypothalamus injected with BrdU at E19.5 (S), E10.5 (T), or E11.5 (U). Bar graphs represent mean ± SEM (n = 3 embryos per group; 3 brain sections per embryo). **p < 0.007; ***p < 0.0004; ****p < 0.0001; unpaired t test. Scale bars, 100 μm.
Figure 4.
Neurog2 is required for proper neurogenesis within the embryonic tuberal hypothalamus. Immunostaining results for anti-BrdU on E19.5.5 coronal sections of control and Neurog2−/− brains injected with BrdU at E12.5 (A,B), EC13.5 (C,D), or E14.5 (E,F). G–L, Binary images of the data presented in A–F. Histogram plots of these binary images demonstrate the location of BrdU+ cells following injection at E12.5 (M,N), E13.5 (O,P), or E14.5 (Q,R). Cell counts of BrdU+ cells within control and Neurog2−/− tuberal hypothalamus injected with BrdU at E12.5 (S), E13.5 (T), or E14.5 (U). Bar graphs represent mean ± SEM (n = 3 embryos per group; 3 brain sections per embryo). **p < 0.009 (unpaired t test), ns - not significant. Scale bars, 100 μm.
We next questioned whether the overall pattern of neurogenesis was perturbed, first examining mediolateral patterning. For this purpose, we generated binary images of BrdU+ newborn cells within the tuberal hypothalamus (Figs. 3G–L, 4G–L) and used histograms to plot their distribution (Figs. 3M–R, 4M–R). BrdU intensity was plotted from the leftmost to rightmost edge of the brain, with the third ventricle in the middle of each histogram. At E9.5, the earliest time point analyzed, we observed a specific reduction in laterally located BrdU+ newborn cells in the presumptive LH, but no change in the number of more medially located ARC neurons born at this time point in Neurog2−/− brains compared with WT controls (Fig. 3M,N, arrowhead). In contrast, the reduction in neurogenesis observed at E10.5 and E11.5 in the Neurog2−/− tuberal hypothalamus was widespread throughout, as there was no change in the distribution of BrdU+ newborn neurons (E10.5 and E11.5; Fig. 3O–R, E12.5; Fig. 4M,N). At later time points, although neurogenic differences were not detected in Neurog2−/− hypothalami (E13.5 and E14.5), a wider distribution of BrdU+ newborn cells was observed around the third ventricle in Neurog2−/− brains (Fig. 4O–R, arrowheads).
Finally, in contrast to the altered pattern of neurogenesis in the mediolateral plane, the distribution of BrdU+ cells did not change across the rostrocaudal axis between Neurog2−/− versus WT control brains (data not shown). The outside-to-inside pattern of neurogenesis was also maintained in Neurog2−/− brains, with the shape of the histograms sharply changed from a biphasic curve with a dip in BrdU+ signal at the third ventricle in neurons born at E9.5–E11.5 to an inverted U shape with peak intensity measured around the third ventricle in cells born at E12.5–E14.5 (compare Fig. 3M with Fig. 4R), as observed in Figure 1.
Neurog2 is required for specification and positioning of VMH-specific neurons
Given that Neurog2 is required for early neurogenesis in the tuberal hypothalamus (Fig. 3M–O), and thatneurogenesis follows an outside-in pattern, with lateral-residing neurons born before medially residing (Fig. 1), we next asked whether lateral-residing neurons were specifically lost in Neurog2-null brains. We focused on the VMH as it is subdivided into three domains in the mediolateral axis: the VMH-dorsomedial (VMHDM), VMH-central (VMHC), and VMH-ventrolateral (VMHVL) domains (for general marking of these subdomains, seeFig. 5D). Each subdomain expresses unique markers that define eachcompartment: Fezf1 is a pan-VMH marker, whereas Vgll2 labels the VMHDM, Satb2 the VMHC-VL, and Nkx2.1 the VMHVL (Kurrasch et al., 2007). We were particularly interested in whether the loss of Neurog2 would affect VMHVL-residing neurons more than those clustered in the VMHDM, for example. We assayed marker expression at E12.5 (peak neurogenesis) and E15.5 (neurons coalescing into a nuclear structure).
Figure 5.
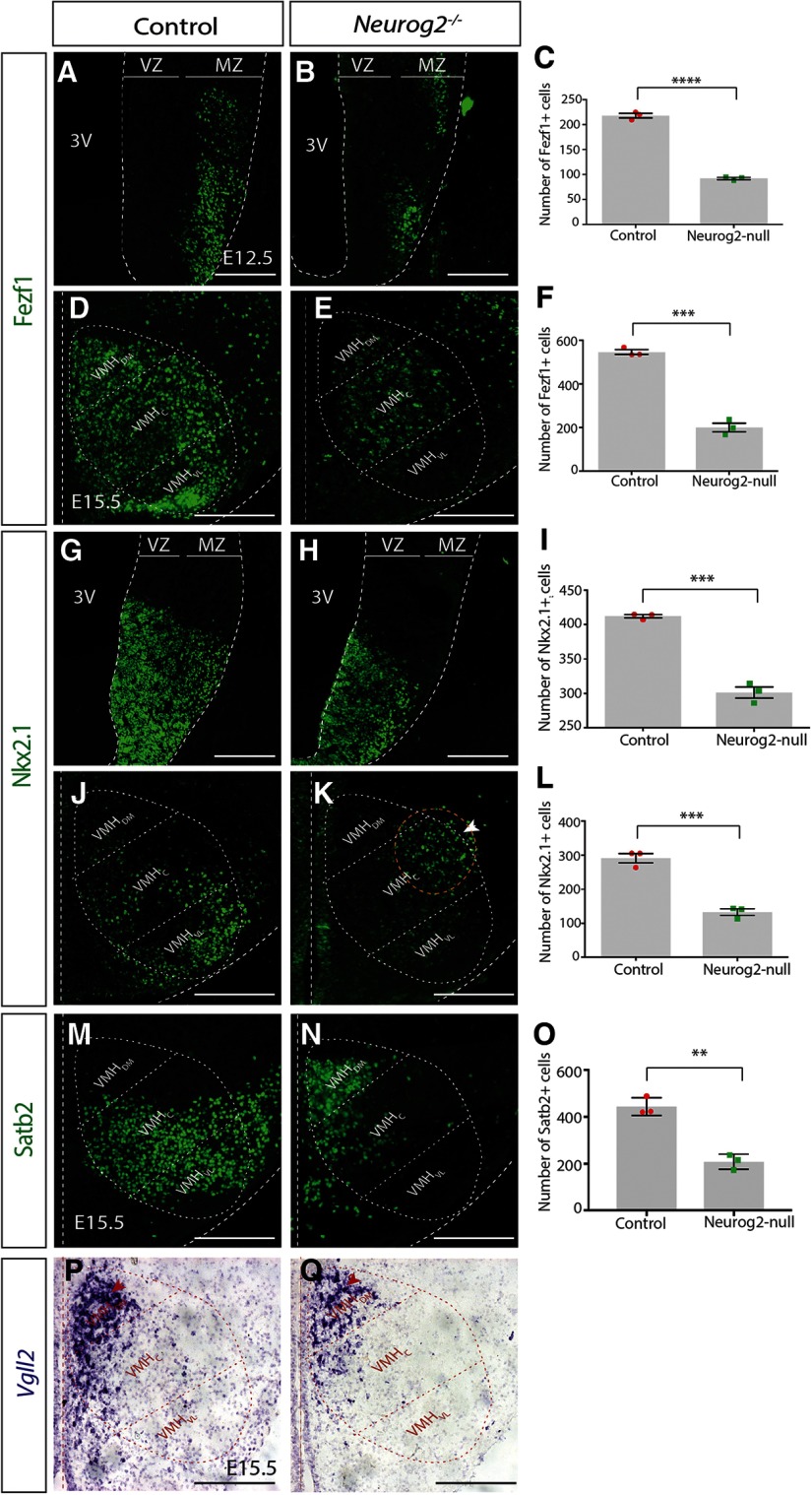
Neurog2 is required for specification of VMH neurons. A, B, Immunostaining results for anti-Fezf1 on E12.5 mouse coronal sections on control and Neurog2−/−. C, Fezf1+ cell counts for whole VMH at E12.5 on both control and Neurog2−/− brains. D, E, Immunostaining results for anti-Fezf1 on E15.5 mouse coronal sections on control and Neurog2−/−. F, Fezf1+ cell counts for whole VMH at E15.5 on both control and Neurog2−/− brains. G, H, Immunostaining results for anti-Nkx2.1 on E12.5 mouse coronal sections on control and Neurog2−/−. I, Nkx2.1+ cell counts for whole VMH at E12.5 on both control and Neurog2−/− brains. J, K, Immunostaining results for anti-Nkx2.1 on E15.5 mouse coronal sections on control and Neurog2−/−. L, Nkx2.1+ cell counts for whole VMH at E15.5 on both control and Neurog2−/− brains. M, N, Immunostaining results for anti-Stab2 on E15.5 mouse coronal sections on control and Neurog2−/−. O, Satb2+ cell counts for whole VMH at E15.5 on both control and Neurog2−/− brains. P, Q, ISH results for Vgll2 ribo-probe on E15.5 mouse coronal sections on control and Neurog2−/−. Bar graphs represent mean ± SEM (n = 3 embryos per group; 3 brain sections per embryo). *p < 0. 01; **p < 0.005; ***p < 0.0001; unpaired t test. Dashed oval represents VMH nucleus and its three subdomains. Scale bars, 50 μm.
At E12.5, we observed a significant reduction in the number of Fezf1+ pan-VMH neurons in Neurog2−/− compared with WT controls (Control: 218.1 ± 4.62 cells, n = 3; Neurog2−/−: 92.11 ± 2.11 cells, n = 3; p < 0.0001, unpaired t test; Fig. 5A–C), which persisted at E15.5 (Control: 545.8 ± 10.78 cells, n = 3; Neurog2−/−: 200 ± 19.74 cells, n = 3; p = 0.0001, unpaired t test; Fig. 5D–F). The positioning of Fezf1+ cells at E15.5 was notable, with an overall reduction in Fezf1+ neurons observed in the VMHDM and VMHC subdomains but a nearly complete loss of Fezf1+ neurons in the VMHVL (Fig. 5D,E). Next, we examined Nkx2.1+ cells, first largely as a general marker of VMH progenitors in the ventricular zone, although some postmitotic neurons in the mantle zone are also labeled at E12.5, and later, as a VMHVL-specific neuronal marker at E15.5. We detected a significant reduction in the number of Nkx2.1+ cells in the Neurog2−/− VMH compared with controls at E12.5 (Control: 412 ± 2.39 cells, n = 3; Neurog2−/−: 301 ± 8.60 cells, n = 3; p = 0.0002, unpaired t test; Fig. 5G–I) and E15.5 (Control: 291 ± 13.83 cells, n = 3; Neurog2−/−: 133.2 ± 9.46 cells, n = 3; p = 0.0007, unpaired t test; Fig. 5J–L). Moreover, we observed a reproducible mispositioning of Nkx2.1+ cells in the E15.5 Neurog2−/− hypothalamus (Fig. 5K, white arrowhead), whereby the remaining Nkx2.1+ cells continue to cluster into a discreet subdomain but positioned outside the VMH core, suggesting perhaps an additional migration phenotype in the absence of Neurog2.
In addition, to more fully investigate whether Neurog2 was required for the generation of distinct populations of VMH neurons, we assayed for Satb2, a VMHVL-C marker. A significant reduction in the number of Satb2+ cells was observed in hypothalami lacking Neurog2 compared with WT controls at E15.5 (Control: 444 ± 22.18 cells, n = 3; Neurog2−/−: 208.1 ± 18.79 cells, n = 3; p = 0.0013, unpaired t test; Fig. 5M–O). Notably, the remaining Satb2+ neurons were clustered in the VMHDM and not in the VMHVL-C, where they are normally located again suggesting a migration phenotype in the absence of Neurog2.
Finally, we examined the localization of cells expressing Vgll2 transcripts, a VMHDM marker, and likewise showed an apparent overall reduction in Vgll2+ cells (although ISH data are not quantitative) with no changes in the positioning of these cells in Neurog2−/− compared with control VMH at E15.5 (Fig. 5P,Q). Together, these studies are consistent with a role for Neurog2 in VMH neurogenesis, leading to overall decreases in VMH neuronal numbers and mis-positioning of the remaining neurons away from the VMHVL.
VMHVL-residing neurons are particularly affected in Neurog2-null brains
To better characterize the influence of Neurog2 on VMH neuronal birth, we examined in the mature nucleus at E19.5 the distribution of BrdU+/Fezf1+ cells (pan-VMH) and BrdU+/Nkx2.1+ cells (VMHVL) born at different time points. In WT embryos, we observed few (∼5%) Fezf1+ cells born at E9.5 (Fig. 6A), high levels of Fezf1+ neuronal birth between E10.5 and E11.5 (∼15% and ∼20%, respectively), and a sizable decrease in Fezf1+ cell birth between E12.5 and E14.5 (∼5% each time point; Fig. 6C,E,G,I,K). Consistent with an outside-in neurogenic pattern, Fezf1+ neurons born at E10.5 primarily resided in the VMHVL (Fig. 6C, white arrowhead), whereas Fezf1+ cells born at E11.5 were localized to the VMHC (Fig. 6E, white arrowhead), with some Fezf1+ neurons dispersed across the lateral and periventricular regions (Fig. 6E, yellow arrows) (Fig. 9B). In brains injected with BrdU at E12.5–E14.5, BrdU+/Fezf1+ cells were positioned primarily in the periventricular zone (Fig. 6G,I,K, white arrowhead). In the absence of Neurog2, we likewise observed a severe reduction in the overall numbers of Fezf1+ neurons in the mature nucleus (i.e., E19.5), with fewer BrdU+/Fezf1+ cells born at each embryonic day (Fig. 6A–L). Interestingly, independent of the day on which the Fezf1+ neurons were born in the Neurog2−/− brain, most Fezf1+ cells localized to the dorsomedial VMH (Fig. 6D,F,H,J,L, white arrowhead), including those born at E10.5 and E11.5 that normally reside in the VMHVL in WT brains (Fig. 6C–F, arrowheads). Furthermore, when the percentage of BrdU+/Fezf1+ neurons was quantified for each day during neurogenesis (E9.5–E14.5), we observed a shift in the neurogenic curve whereby fewer Fezf1+ neurons were born in the early phase and more neurons were generated in the later phase in Neurog2-null brains compared with WT controls, suggesting that a secondary wave of neurogenesis partially compensates for the loss of Fezf1+ cells in the first wave (BrdU-E9.5: Control: 17.67 ± 2.33 cells, n = 3; Neurog2−/−: 0.77 ± 0.11 cells, n = 3; p = 0.0019, unpaired t test; BrdU-E10.5: Control: 212.7 ± 7.21 cells, n = 3; Neurog2−/−: 24 ± 2.51 cells, n = 3; p < 0.0001, unpaired t test; BrdU-E11.5: Control: 252.7 ± 6.47 cells, n = 3; Neurog2−/−: 57.67 ± 2.33 cells, n = 3; p < 0.0001, unpaired t test; BrdU-E12.5: Control: 133 ± 3.60 cells, n = 3; Neurog2−/−: 27.33 ± 1.85 cells, n = 3; p < 0.0001, unpaired t test; BrdU-E13.5: Control: 82 ± 2.3 cells, n = 3; Neurog2−/−: 25.33 ± 0.88 cells, n = 3; p < 0.0001, unpaired t test; BrdU-E14.5: Control: 45.67 ± 4.37 cells, n = 3; Neurog2−/−: 19.33 ± 0.88 cells, n = 3; p = 0.0042, unpaired t test; Fig. 6Y). In addition, in the Neurog2-null background, many singly labeled BrdU+ cells were observed at peak neurogenesis (Fig. 6F, red cells), in contrast to WT brains, in which these cells mainly acquired a Fezf1+ fate (Fig. 6E, yellow cells). Thus, in the absence of Neurog2, neurons born at E11.5 still exit the cell cycle and move into the brain parenchyma but fail to express a VMH-specific marker.
Figure 6.
Neurog2 is required for proper neurogenesis of VMH-specific neurons. Double-immunostaining results for anti-BrdU and anti-Fezf1 on E19.5.5 mouse coronal sections injected with BrdU on control and Neurog2−/− at E9.5 (A,B), E10.5 (C,D), E11.5 (E,F), 12.5 (G,H), E13.5 (I,J), and E14.5 (K,L). Double-immunostaining results for anti-BrdU and anti-Nkx2.1 on E19.5.5 mouse coronal sections injected with BrdU on control and Neurog2−/− at E9.5 (M,N), E10.5 (O,P), E11.5 (Q,R), E12.5 on control (S,T), E13.5 (U,V), and E14.5 (W,X). Y, Number of Fezf1+ cells colabeled with BrdU in control compared with Neurog2−/− backgrounds. Z, Number of Nkx2.1+ cells colabeled with BrdU in control compared with Neurog2−/− backgrounds. Bar graphs represent mean ± SEM (n = 3 embryos per group; 3 brain sections per embryo). **p < 0.001; ***p < 0.0002; ****p < 0.0001, ns - not significant; unpaired t test. Dashed oval represents VMH nucleus and its three subdomains. Scale bars, 50 μm.
We next investigated whether the loss of a VMHVL-residing population of cells can be compensated for by a secondary wave of neurogenesis in the absence of Neurog2. To do so, we conducted a similar experiment as per above and coimmunostained for BrdU+/Nkx2.1+ cells in E19.5 brains injected with BrdU at 24 h intervals from E9.5 to E14.5. In the E19.5 WT brain, Nkx2.1+ cells were primarily born at the early stages (E10.5 and E11.5) and populated the VMHVL (Fig. 6M,O,Q,S,U,W), consistent with an outside-in pattern of neurogenesis within the tuberal hypothalamus. We also observed a significant decrease of Nkx2.1+ neurons in Neurog2−/− brains (Fig. 6K–V), as we showed previously at E15.5 (Fig. 5J,K). Concomitantly, although we observed fewer BrdU+/Nkx2.1+ cells in Neurog2−/− brains compared with WT (Fig. 7M), the overall neurogenic curve aligned with the WT pattern, with no second wave of neurogenesis compensating for the decrease in Nkx2.1+ neurons born during the early phase (BrdU-E9.5: Control: 15.33 ± 1.20 cells, n = 3; Neurog2−/−: 0.00 ± 0.00 cells, n = 3; p = 0.0002, unpaired t test; BrdU-E10.5: Control: 46 ± 2.64 cells, n = 3; Neurog2−/−: 23.33 ± 1.20 cells, n = 3; p = 0.0015, unpaired t test; BrdU-E11.5: Control: 58.33 ± 1.85 cells, n = 3; Neurog2−/−: 19 ± 0.57 cells, n = 3; p < 0.0001, unpaired t test; BrdU-E12.5: Control: 10 ± 1 cells, n = 3; Neurog2−/−: 3 ± 0.57 cells, n = 3; p = 0.0037, unpaired t test; BrdU-E13.5: Control: 10.67 ± 2.33 cells, n = 3; Neurog2−/−: 6 ± 1.15 cells, n = 3; p = 0.14, unpaired t test; BrdU-E14.5: Control: 1.66 ± 0.66 cells, n = 3; Neurog2−/−: 1.33 ± 0.33 cells, n = 3; p = 0.67, unpaired t test; Fig. 6Z). These results suggest that Neurog2 is important for early neurogenesis and might particularly affect the specification of neurons that reside in the VMHVL.
Figure 7.
Ascl1 is required for proper neurogenesis within embryonic tuberal hypothalamus. Immunostaining results for anti-BrdU on E19.5.5 mouse coronal sections injected with BrdU on control and Ascl1−/− at E9.5 (A,B), E10.5 (C,D), E11.5 (E,F), E12.5 (G,H), and E13.5 (I,J). A′–J′, Binary images of the data presented in A–J. Histogram plots of these binary images demonstrate the location of BrdU+ cells following injection at E9.5 (K,L), E10.5 (M,N), E11.5 (O,P), E12.5 (Q,R), and E13.5 (S,T). BrdU+ cell counts for tuberal hypothalamus on E19.5 mouse brains injected with BrdU at E9.5 (U), E10.5 (V), E11.5 (W), E12.5 (X), and E13.5 (Y) for both control and Ascl1−/−. Bar graphs represent mean ± SEM (n = 3 embryos per group; 3 brain sections per embryo). **p < 0.005; ****p < 0.0001, ns - not significant; unpaired t test. Scale bars, 100 μm.
Ascl1 similarly drives neurogenesis of early-born neurons in the tuberal hypothalamus
Mechanistically, in the retina (Hufnagel et al., 2010) and neocortex (Fode et al., 2000; Parras et al., 2002; Schuurmans et al., 2004), Ascl1 becomes upregulated and rescues the effects of Neurog2 loss-of-function in the first neurogenic phase, as well as independently driving a second wave of neurogenesis. However, in the tuberal hypothalamus, Ascl1 expression overlaps with Neurog2 at the early neurogenic time points (Fig. 2D-D′′), suggesting that Ascl1 and Neurog2 might play complementary roles in driving the same early neurogenic wave. To determine whether the Neurog2-Ascl1 interplay is similar to the cortex or unique to the hypothalamus (not mutually exclusive), we used Ascl1GFPKI mice and crossed adult heterozygotes to generate both Ascl1GFP/+ controls and Ascl1GFP/GFP (hereafter referred to as Ascl1−/−) mutant brains. Pregnant dams were injected with BrdU at 24 h intervals from E9.5–E13.5, and embryos were collected at E19.5. In the absence of Ascl1, we observed a significant reduction in the number of tuberal hypothalamic neurons born at early stages (e.g., E9.5 to E11.5) compared with WT controls (BrdU-E9.5: Control: 1404 ± 20 cells, n = 3; Ascl1−/−: 92.89 ± 5.20 cells, n = 3; p < 0.0001, unpaired t test; BrdU-E10.5: Control: 3282 ± 125.5 cells, n = 3; Ascl1−/−: 2118 ± 71.3 cells, n = 3; p = 0.0013, unpaired t test; BrdU-E11.5: Control: 2985 ± 89.16 cells, n = 3; Ascl1−/−: 1999 ± 75.82 cells, n = 3; p < 0.0011, unpaired t test; Fig. 7A–F,U–W). In contrast, we did not measure any significant change in the number of neurons born at E12.5 in Ascl1−/− compared with control (BrdU-E12.5: Control: 2026 ± 50.16 cells, n = 3; Ascl1−/−: 2099 ± 56.78 cells, n = 3; p < 0.39, unpaired t test; Fig. 7G,H,X) and observed a significant increase in thenumber of neurons born at E13.5 in Ascl1−/− compared with control (BrdU-E13.5: Control: 1888 ± 33.11 cells, n = 3; Ascl1−/−: 2119 ± 17.39 cells, n = 3; p < 0.003, unpaired t test; Fig. 7I,J,Y), suggesting that Ascl1 is required for early neurogenesis but not a later secondary wave. In addition, the histogram plots do not reveal any obvious change in the outside-in pattern of neurogenesis in the absence of Ascl1 compared with control (Fig. 8A′–J′,K–T), although the shift from laterally born to medially born neurogenesis occurs later in the Ascl1−/− (E12.5–E13.5) versus Neurog2−/− (E11.5–E12.5) brains. Combined, these results suggest that Ascl1 drives a complementary, and perhaps slightly delayed, early phase of neurogenesis within the tuberal hypothalamus and does not compensate for the loss of Neurog2 (Fig. 9).
Figure 8.
Neurog2 does not regulate expression of Neurog1, Neurog3, or Ascl1 within the tuberal hypothalamus. Neurog1(A,B), Neurog3 (C,D), and Ascl1 (E,F) expression levels in E12.5 mouse brain sections in control and Neurog2−/− backgrounds. Scale bar, 100 μm.
Proneural genes are not upregulated in the absence of Neurog2 in tuberal hypothalamus
If Ascl1 cannot compensate for Neurog2, we next examined the expression of other proneural genes in the absence of Neurog2 to determine whether other neurogenins might be upregulated. We chose E12.5 as the best time point to test proneural gene expression since all neurogenin family members and Ascl1 genes are highly expressed within the tuberal hypothalamic progenitor zone at this stage and peak neurogenesis is occurring. We conducted ISH on Neurog2−/− and control embryos (n = 4) for Neurog1, Neurog3, and Ascl1 riboprobes. We detected no obvious change in the transcription levels of Ascl1 in the Neurog2−/− background compared with controls (Fig. 8E,F), consistent with our data that Ascl1 might be functioning independent from Neurog2 to drive an early wave of neurogenesis. Additionally, neither Neurog1 nor Neuro3 was upregulated in the Neurog2−/− hypothalamus, suggesting that these neurogenin family members do not compensate for Neurog2 in hypothalamic neurogenesis.
Discussion
The hypothalamus is a neuronally diverse and morphologically complex brain region (Chrousos, 2007; Alvarez-Bolado, 2019). The identification of programs that drive its development is an active area of research (Kurrasch et al., 2007; Szarek et al., 2010; Lu et al., 2013; Burbridge et al., 2016; Yoo and Blackshaw, 2018; Kim et al., 2019). Here, we focused on neurogenesis within the tuberal hypothalamus given its importance in energy balance and reproduction. Our data support the outside-in pattern of neurogenesis within this region proposed by some groups (Shimada and Nakamura, 1973; Padilla et al., 2010; Alvarez-Bolado et al., 2012) and introduced a new rostrocaudal gradient for neuronal birth within the tuberal hypothalamus. In addition, we demonstrated a novel role for Neurog2 in hypothalamic development, joining studies for other proneural genes, including Ascl1 (McNay et al., 2006; Marsters et al., 2016) and Neurog3 (Pelling et al., 2011; Anthwal et al., 2013). Finally, we showed that the loss of Neurog2−/− particularly affects early born neurons and cannot be compensated for by Ascl1 as is true in other brain regions, suggesting that Neurog2 functions as a classical proneural gene in the developing VMH.
An early study using tritiated thymidine radiography reported that neurogenesis occurs in an outside-in and dorsal-ventral pattern within the tuberal hypothalamus (Shimada and Nakamura, 1973). However, since they did not show any histologic data and instead presented diagrams of their results, it is hard to determine the preciseness of this pattern. Moreover, newer studies contradict Shimada's proposed outside-in pattern by showing either that neurons born at later stages (e.g., E13.5) localize to the far lateral region of the hypothalamus or neurons born at early time points (e.g., E12.5 in rat/∼E10.5 mouse) reside in the periventricular zones (Markakis and Swanson, 1997; Padilla et al., 2010; Alvarez-Bolado et al., 2012). Moreover, our study showed that, while most of the neurons born in the tuberal hypothalamus follow an outside-in pattern, ARC neurons born at E9.5 and LH neurons born at E13.5 and E14.5 do not. These findings are opposite to the expected outside-in strategy, whereby early-born neurons are positioned in the lateral regions and later-born neurons near the ventricular zone. Here, we demonstrated that, while an outside-in pattern was supported, it was not fully definitive, thereby providing support for the original paper (Shimada and Nakamura, 1973) and also the newer findings (Markakis and Swanson, 1997; Padilla et al., 2010; Alvarez-Bolado et al., 2012). Consistent with the outside-in model, we showed that Nkx2.1+ neurons primarily located in the VMHVL (far lateral region) were born as early as E10.5 and E11.5, whereas Fezf1+ cells that were distributed throughout the whole VMH were born across several embryonic stages, with early-born Fezf1+ cells preferentially located in the VMHVL and lateral region of the VMHC, and the later-born neurons occupying VMHDM and the medial part of VMHC. Moreover, we described a rostrocaudal trend to neurogenesis across the entire tuberal hypothalamus, which had previously only been reported for a few restricted neuronal subtypes in the ARC nucleus (Altman and Bayer, 1978), suprachiasmatic, and tuberomammillary nuclei (Altman and Bayer, 1978; Reiner et al., 1988). In contrast, we did not observe a dorsal-ventral gradient to tuberal hypothalamic neurogenesis,as reported previously (Shimada and Nakamura, 1973; Altman and Bayer, 1978), suggesting that hypothalamic neurons are born around the same time along the dorsal-to-ventral axis, with the exception of E9.5.
Proneural genes are well characterized for their many roles in regulating neurodevelopment across various brain regions, especially the neocortex and retina (Bertrand et al., 2002; Helms and Johnson, 2003; Akagi et al., 2004; Huang et al., 2014; Dennis et al., 2018; Chouchane and Costa, 2019). Ascl1 and Neurog3 are the only proneural genes studied for a functional role in the developing tuberal hypothalamus (McNay et al., 2006; Pelling et al., 2011; Anthwal et al., 2013; Marsters et al., 2016). In particular, Ascl1 is required for genomic screen homeobox 1 (Gsh1) and steroidogenic factor 1 (SF1) expression in the ARC and VMH, respectively, and suppresses neuropeptide Y (NPY) and TH transcripts in the ARC (McNay et al., 2006). Interestingly, ectopic Neurog2 expression under the control of the Ascl1 promoter rescues general neurogenesis but cannot restore normal differentiation of ARC and VMH neurons (McNay et al., 2006), suggesting a potential compensatory role for Neurog2 in Ascl1-mediated neurogenesis. Moreover, although Neurog3 does not play a role in neurogenesis per se, it contributes to the promotion of POMC+ and SF1+ neurons as well as the inhibition of NPY+ and TH+ neurons within ARC nucleus (Pelling et al., 2011). The contribution of these proneural genes to the myriad of other tuberal hypothalamic cell types remains unknown, as does the interaction of these factors to govern development of the VMH, a key brain region. Here, we introduced Neurog2 as another proneural gene important for regulating neurodevelopment within the tuberal hypothalamus. Specifically, we propose that Neurog2 acts as a classical proneural within the VMH, acting alone to influence the timing of early neurogenesis and in its absence causing a significant reduction in the number of neurons born in this nucleus. These findings make Neurog2 in the VMH one of the few examples of a vertebrate proneural family member that has kept its classical proneural activity from its homologous family ato in Drosophila (Fode et al., 1998; Lo et al., 2002; Guillemot et al., 2006; Huang et al., 2014). Moreover, Neurog2 plays a critical role in regulating cortical neuronal migration, and the absence of Neurog2 causes a significant increase in the number of neurons mispositioned in the intermediate zone and a concomitant reduction in the number of neurons reaching the cortical plate (Heng et al., 2008; Pacary et al., 2011). Thus, despite differences in the phenotype of the mispositioned Nkx2.1+ cells in the absence of Neurog2 versus what is reported in the cortex, it is possible the Neurog2, in addition to its role in neurogenesis, might also act to regulate migration within the VMH.
In the CNS and PNS, there are several examples in which other proneural genes compensate for the loss of Neurog2 and act to restore neurogenesis. Specifically, in the retina, Neurog2 leads the early wave of neurogenesis, with Ascl1 inducing a second neurogenic phase at later time points that can compensate for the neuronal deficits found in the Neurog2-null retina (Hufnagel et al., 2010). In the DRG, Neurog2 drives neurogenesis of early-born neurons and Neurog1 initiates the second neurogenic wave, with Neurog1 also able to compensate for the deficit in the number of early-born neurons in the Neurog2-null background (Ma et al., 1999). And finally, in the dorsal telencephalon, Ascl1 upregulation in the Neurog2-null compensates for neurogenic defects, as does the maintenance of Neurog1 expression in lateral domains (Fode et al., 2000; Schuurmans et al., 2004). In contrast to these findings, here we showed that the loss of Neurog2 does not positively or negatively affect transcription of Neurog1, Neurog3, or Ascl1 in the tuberal hypothalamus, suggesting that these proneural genes do not compensate for the elimination of Neurog2 in this region. Moreover, we demonstrated that Ascl1 itself can influence the neurogenesis of early-born neurons, unlike the above reports showing that Neurog2 and Ascl1 lead different waves of neurogenesis. Together, our findings suggest that Neurog2 might function independently from the other proneural genes to guide neurogenesis and neuronal specification within the VMH.
In the Neurog2-null hypothalamus, a decrease in neurons born between E9.5-E12.5 was observed, although neurons born at E13.5 or later were not affected, raising the intriguing question as to what factor gives rise to this second wave of neurogenesis. Moreover, in the absence of Ascl1, an increase in later-born neurons (e.g., E13.5) was demonstrated, suggesting that Ascl1 might actually play a cross-inhibitory role with another factor to drive this second wave of neurogenesis. In the dorsal spinal cord, Ascl1 and Neurog2 are epistatic to each other; however, Ascl1, Neurog1, and Math1 are responsible for neurogenesis of specific interneurons, with Neurog2 only cooperating with them to modulate the number of neurons born at each neuronal population (Helms et al., 2005). It is possible that a combination of Ascl1, Neurog1, Neurog2, Neurog3, and/or Math1 all work in concert to drive the waves of neurogenesis within the tuberal hypothalamus. Alternatively, the satiety signaling molecule leptin can function as a neurotrophic factor, with its elimination causing a neurogenesis defect of later born neurons (e.g., E14) within VMH and ARC nucleus (Garris, 1989). Thus, leptin is an unexpected and interesting candidate to consider for driving the recovery of neuronal birth observed at later time points in the absence of Neurog2 or Ascl1.
In conclusion, our findings add Neurog2 to the list of proneural genes controlling neurogenesis and specification of ventromedial hypothalamic neurons, and raises new questions as to how these proneural genes interact with other factors to specify discreet neurons with the VMH.
Footnotes
This work was supported by Canadian Institutes of Health Research MOP-275053 to D.M.K., Alberta Children's Hospital Research Institute Training Fellowship to S.A., and Vision Science Research Program Fellowship to S.H. C.S. holds the Dixon Family Chair in Ophthalmology Research. We thank Natalia Klenin for technical assistance; and the D.M.K. is co-founder of an epilepsy biotech company, Path Therapeutics. The other authors have nothing to declare.
The authors declare no competing financial interests.
References
- Aslanpour SA, Rosin JM, Balakrishnan A, Klenin N, Blot F, Gradwhol G, Schuurmans C, Kurrasch DM (2020) Ascl1 is required to specify a subset of ventromedial hypothalamic neurons. Development. Advance online publication. Retrieved April 6, 2020. doi: 10.1242/dev.180067. [DOI] [PubMed] [Google Scholar]
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee J, Guillemot F, Kageyama R (2004) Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem 279:28492–28498. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA (1978) Development of the diencephalon in the rat: I. Autoradiographic study of the time of origin and settling patterns of neurons of the hypothalamus. J Comp Neurol 182:945–971. 10.1002/cne.901820511 [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer S (1986) The development of the rat hypothalamus. Adv Anat Embryol Cell Biol 100:1–178. [PubMed] [Google Scholar]
- Alvarez-Bolado G. (2019) Development of neuroendocrine neurons in the mammalian hypothalamus. Cell Tissue Res 375:23–39. 10.1007/s00441-018-2859-1 [DOI] [PubMed] [Google Scholar]
- Alvarez-Bolado G, Paul FA, Blaess S (2012) Sonic hedgehog lineage in the mouse hypothalamus: from progenitor domains to hypothalamic regions. Neural Dev 7:4 10.1186/1749-8104-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Bolado G, Grinevich V, Puelles L (2015) Editorial: development of the hypothalamus. Front Neuroanat 9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthwal N, Pelling M, Claxton S, Mellitzer G, Collin C, Kessaris N, Richardson W, Gradwohl G, Ang SL (2013) Conditional deletion of neurogenin-3 using Nkx2.1iCre results in a mouse model for the central control of feeding, activity and obesity. Dis Model Mech 6:1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, Newman EA, Blackshaw S (2015) Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip Rev Dev Biol 4:445–468. 10.1002/wdev.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F (2002) Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3:517–530. 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- Blader P, Sok Lam C, Rastegar S, Scardigli R, Nicod JC, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, Strähle U (2004) Conserved and acquired features of neurogenin1 regulation. Development 131:5627–5637. [DOI] [PubMed] [Google Scholar]
- Braun MM, Etheridge A, Bernard A, Robertson CP, Roelink H (2003) Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 130:5579–5587. 10.1242/dev.00685 [DOI] [PubMed] [Google Scholar]
- Britz O, Mattar P, Nguyen L, Langevin LM, Zimmer C, Alam S, Guillemot F, Schuurmans C (2006) A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex 16:i138–i151. 10.1093/cercor/bhj168 [DOI] [PubMed] [Google Scholar]
- Burbridge S, Stewart I, Placzek M (2016) Development of the neuroendocrine hypothalamus. Compr Physiol 6:623–643. 10.1002/cphy.c150023 [DOI] [PubMed] [Google Scholar]
- Casarosa S, Schuurmans C, Guillemot F (1999) Mash1 regulates neurogenesis in the ventral telencephalon. Development 126:525–534. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F (1997) Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124:1611–1624. [DOI] [PubMed] [Google Scholar]
- Cheung CC, Krause WC, Edwards RH, Yang CF, Shah NM, Hnasko TS, Ingraham HA (2015) Sex-dependent changes in metabolism and behavior, as well as reduced anxiety after eliminating ventromedial hypothalamus excitatory output. Mol Metab 4:857–866. 10.1016/j.molmet.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchane M, Costa MR (2019) Instructing neuronal identity during CNS development and astroglial-lineage reprogramming: roles of NEUROG2 and ASCL1. Brain Res 1705:66–74. 10.1016/j.brainres.2018.02.045 [DOI] [PubMed] [Google Scholar]
- Chrousos GP. (2007) Organization and integration of the endocrine system. Sleep Med Clin 2:125–145. 10.1016/j.jsmc.2007.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DJ, Wilkinson G, Li S, Dixit R, Adnani L, Balakrishnan A, Han S, Kovach C, Gruenig N, Kurrasch DM, Dyck RH, Schurmans C (2017) Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc Natl Acad Sci USA 114:E4934–E4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DJ, Han S, Schuurmans C (2018) bHLH transcription factors in neural development, disease, and reprogramming. Brain Res 1705:48–65. [DOI] [PubMed] [Google Scholar]
- Dhillon H, Zigman JM, Ye C, Lee CE, McGovern RA, Tang V, Kenny CD, Christiansen LM, White RD, Edelstein EA, Coppari R, Balthasar N, Cowley MA, Chua S Jr, Elmquist JK, Lowell BB (2006) Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron 49:191–203. 10.1016/j.neuron.2005.12.021 [DOI] [PubMed] [Google Scholar]
- Dixit R, Wilkinson G, Cancino GI, Shaker T, Adnani L, Li S, Dennis D, Kurrasch D, Chan JA, Olson EC, Kaplan DR, Zimmer C, Schuurmans C (2014) Neurog1 and Neurog2 control two waves of neuronal differentiation in the piriform cortex. J Neurosci 34:539–553. 10.1523/JNEUROSCI.0614-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, Leto K, Muzio L, Tinterri A, Badaloni A, Croci L, Zordan P, Barili V, Albieri I, Guillemot F, Rossi F, Consalez GG (2012) Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development 139:2308–2320. 10.1242/dev.075861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F (1998) The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode–derived sensory neurons. Neuron 20:483–494. 10.1016/S0896-6273(00)80989-7 [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F (2000) A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Garris DR. (1989) Morphometric analysis of obesity (ob/ob)- and diabetes (db/db)-associated hypothalamic neuronal degeneration in C57BL/KsJ mice. Brain Res 501:162–170. 10.1016/0006-8993(89)91037-8 [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, Guillemot F (1996) Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol 180:227–241. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F (2000) neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA 97:1607–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Molnár Z, Tarabykin V, Stoykova A (2006) Molecular mechanisms of cortical differentiation. Eur J Neurosci 23:857–868. 10.1111/j.1460-9568.2006.04626.x [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE (2003) Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol 13:42–49. 10.1016/S0959-4388(03)00010-2 [DOI] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE (2005) Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development 132:2709–2719. 10.1242/dev.01859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, Guillemot F (2008) Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455:114–118. 10.1038/nature07198 [DOI] [PubMed] [Google Scholar]
- Huang C, Chan JA, Schuurmans C (2014) Proneural bHLH genes in development and disease. Curr Top Dev Biol 110:75–127. 10.1016/B978-0-12-405943-6.00002-6 [DOI] [PubMed] [Google Scholar]
- Hufnagel RB, Le TT, Riesenberg AL, Brown NL (2010) Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev Biol 340:490–503. 10.1016/j.ydbio.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly-Amado A, Cansell C, Denis RG, Delbes AS, Castel J, Martinez S, Luquet S (2014) The hypothalamic arcuate nucleus and the control of peripheral substrates. Best Pract Res Clin Endocrinol Metab 28:725–737. 10.1016/j.beem.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Kim DW, Washington PW, Wang ZQ, Lin S, Sun C, Jiang L, Blackshaw S (2019) Single cell RNA-Seq analysis identifies molecular mechanisms controlling hypothalamic patterning and differentiation. bioRxiv 657148. doi: 10.1101/657148. [DOI] [Google Scholar]
- King BM. (2006) The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight. Physiol Behav 87:221–244. 10.1016/j.physbeh.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Cheung CC, Lee FY, Tran PV, Hata K, Ingraham HA (2007) The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J Neurosci 27:13624–13634. 10.1523/JNEUROSCI.2858-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD (2008) SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron 57:232–247. 10.1016/j.neuron.2007.12.023 [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR (2007) Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 10:720–726. 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Lo L, Dormand E, Greenwood A, Anderson DJ (2002) Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development 129:1553–1567. [DOI] [PubMed] [Google Scholar]
- Lu F, Kar D, Gruenig N, Zhang ZW, Cousins N, Rodgers HM, Swindell EC, Jamrich M, Schuurmans C, Mathers PH, Kurrasch DM (2013) Rax is a selector gene for mediobasal hypothalamic cell types. J Neurosci 33:259–272. 10.1523/JNEUROSCI.0913-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ (1999) NEUROGENIN1 and NEUROGENIN2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev 13:1717–1728. 10.1101/gad.13.13.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis EA, Swanson LW (1997) Spatiotemporal patterns of secretomotor neuron generation in the parvicellular neuroendocrine system. Brain Res Rev 24:255–291. 10.1016/S0165-0173(97)00006-4 [DOI] [PubMed] [Google Scholar]
- Marsters CM, Rosin JM, Thornton HF, Aslanpour S, Klenin N, Wilkinson G, Schuurmans C, Pittman QJ, Kurrasch DM (2016) Oligodendrocyte development in the embryonic tuberal hypothalamus and the influence of Ascl1. Neural Dev 11:20 10.1186/s13064-016-0075-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNay DE, Pelling M, Claxton S, Guillemot F, Ang SL (2006) Mash1 is required for generic and subtype differentiation of hypothalamic neuroendocrine cells. Mol Endocrinol 20:1623–1632. 10.1210/me.2005-0518 [DOI] [PubMed] [Google Scholar]
- Nesan D, Kurrasch DM (2016) Genetic programs of the developing tuberal hypothalamus and potential mechanisms of their disruption by environmental factors. Mol Cell Endocrinol 438:3–17. 10.1016/j.mce.2016.09.031 [DOI] [PubMed] [Google Scholar]
- Newman EA, Wu D, Taketo MM, Zhang J, Blackshaw S (2018) Canonical Wnt signaling regulates patterning, differentiation and nucleogenesis in mouse hypothalamus and prethalamus. Dev Biol 442:236–248. 10.1016/j.ydbio.2018.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F (2001) Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29:401–413. 10.1016/S0896-6273(01)00214-8 [DOI] [PubMed] [Google Scholar]
- Orquera DP, Nasif S, Low MJ, Rubinstein M, de Souza FS (2016) Essential function of the transcription factor Rax in the early patterning of the mammalian hypothalamus. Dev Biol 416:212–224. 10.1016/j.ydbio.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E, Heng J, Azzarelli R, Riou P, Castro D, Lebel-Potter M, Parras C, Bell DM, Ridley AJ, Parsons M, Guillemot F (2011) Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron 69:1069–1084. 10.1016/j.neuron.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla SL, Carmody JS, Zeltser LM (2010) Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med 16:403–405. 10.1038/nm.2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras C, Schuurmans C, Scardigli R, Kim J, Anderson D, Guillemot F (2002) Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev 16:324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelling M, Anthwal N, McNay D, Gradwohl G, Leiter AB, Guillemot F, Ang SL (2011) Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev Biol 349:406–416. 10.1016/j.ydbio.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Reiner PB, Semba K, Fibiger HC, McGeer EG (1988) Ontogeny of histidine-decarboxylase-immunoreactive neurons in the tuberomammillary nucleus of the rat hypothalamus: time of origin and development of transmitter phenotype. J Comp Neurol 276:304–311. 10.1002/cne.902760212 [DOI] [PubMed] [Google Scholar]
- Rosin JM, Kurrasch DM (2018) In utero electroporation induces cell death and alters embryonic microglia morphology and expression signatures in the developing hypothalamus. J Neuroinflammation 15:181 10.1186/s12974-018-1213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Guillemot F (2002) Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol 12:26–34. 10.1016/S0959-4388(02)00286-6 [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Ma Q, Casarosa S, Ang SL, Anderson D, Guillemot F (2000) A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Curr Opin Neurobiol 14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F (2004) Sequential phases of cortical specification involve Neurogenin‐dependent and ‐independent pathways. EMBO J 23:2892–2902. 10.1038/sj.emboj.7600278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Nakamura T (1973) Time of neuron origin in mouse hypothalamic nuclei. Exp Neurol 41:163–173. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S (2010) A genomic atlas of mouse hypothalamic development. Nat Neurosci 13:767–775. 10.1038/nn.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M (2007) Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development 134:1617–1629. 10.1242/dev.001255 [DOI] [PubMed] [Google Scholar]
- Szabo NE, Zhao T, Cankaya M, Theil T, Zhou X, Alvarez-Bolado G (2009) Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J Neurosci 29:6989–7002. 10.1523/JNEUROSCI.1089-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarek E, Cheah PS, Schwartz J, Thomas P (2010) Molecular genetics of the developing neuroendocrine hypothalamus. Mol Cell Endocrinol 323:115–123. 10.1016/j.mce.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Trowe MO, Zhao L, Weiss AC, Christoffels V, Epstein DJ, Kispert A (2013) Inhibition of Sox2-dependent activation of Shh in the ventral diencephalon by Tbx3 is required for formation of the neurohypophysis. Development 140:2299–2309. 10.1242/dev.094524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G, Dennis D, Schuurmans C (2013) Proneural genes in neocortical development. Neuroscience 253:256–273. 10.1016/j.neuroscience.2013.08.029 [DOI] [PubMed] [Google Scholar]
- Xie Y, Dorsky RI (2017) Development of the hypothalamus: conservation, modification and innovation. Development 144:1588–1599. 10.1242/dev.139055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Yang CF, Chizari MD, Maheswaranathan N, Burke KJ Jr, Borius M, Inoue S, Chiang MC, Bender KJ, Ganguli S, Shah NM (2017) Social control of hypothalamus-mediated male aggression. Neuron 95:955–970.e54. 10.1016/j.neuron.2017.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Blackshaw S (2018) Regulation and function of neurogenesis in the adult mammalian hypothalamus. Prog Neurobiol 170:53–66. 10.1016/j.pneurobio.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zevallos Solsire E, Rizzoti K, Jeong Y, Lovell-Badge R, Epstein DJ (2012) Disruption of SoxB1-dependent sonic hedgehog expression in the hypothalamus causes septo-optic dysplasia. Dev Cell 22:585–596. 10.1016/j.devcel.2011.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]