Abstract
Purpose of review
Successful integration of artificial intelligence into extant clinical workflows is contingent upon a number of factors including clinician comprehension and interpretation of computer vision. This article discusses how image analysis and machine learning have enabled comprehensive characterization of kidney morphology for development of automated diagnostic and prognostic renal pathology applications.
Recent findings
The primordial digital pathology informatics work employed classical image analysis and machine learning to prognosticate renal disease. Although this classical approach demonstrated tremendous potential, subsequent advancements in hardware technology rendered artificial neural networks ‘(ANNs) the method of choice for machine vision in computational pathology’. Offering rapid and reproducible detection, characterization and classification of kidney morphology, ANNs have facilitated the development of diagnostic and prognostic applications. In addition, modern machine learning with ANNs has revealed novel biomarkers in kidney disease, demonstrating the potential for machine vision to elucidate novel pathologic mechanisms beyond extant clinical knowledge.
Summary
Despite the revolutionary developments potentiated by modern machine learning, several challenges remain, including data quality control and curation, image annotation and ontology, integration of multimodal data and interpretation of machine vision or ‘opening the black box’. Resolution of these challenges will not only revolutionize diagnostic pathology but also pave the way for precision medicine and integration of artificial intelligence in the process of care.
Keywords: artificial intelligence, digital pathology, image analysis, machine learning, renal pathology
INTRODUCTION
In the age of modern medicine and artificial intelligence integration, computational image analysis and machine learning are revolutionizing diagnostic pathology [1▪]. The advent of algorithms for automated classification of kidney morphology in digital histopathology has potential to enable computer-aided detection, diagnosis and prognosis of renal disease [2▪▪]. Integration of such machine learning applications with extant clinical practices will offer rapid and precise patient assessment on the part of the pathologist, with significant implications for patient outcome. Further expansion of computational and machine learning analysis to clinical biometrics and omics data will potentiate patient prognostication and personalized medicine. Beyond research and development of artificial intelligence tools, successful adoption of artificial intelligence in patient care requires physician comprehension and interpretation of computer vision’s methods and results, respectively. The goal of this review is to guide readers through the computational image analysis and machine learning pipelines applied to digital renal histopathology (Fig. 1), while also summarizing published, kidney-centric artificial intelligence tools (Table 1).
FIGURE 1.
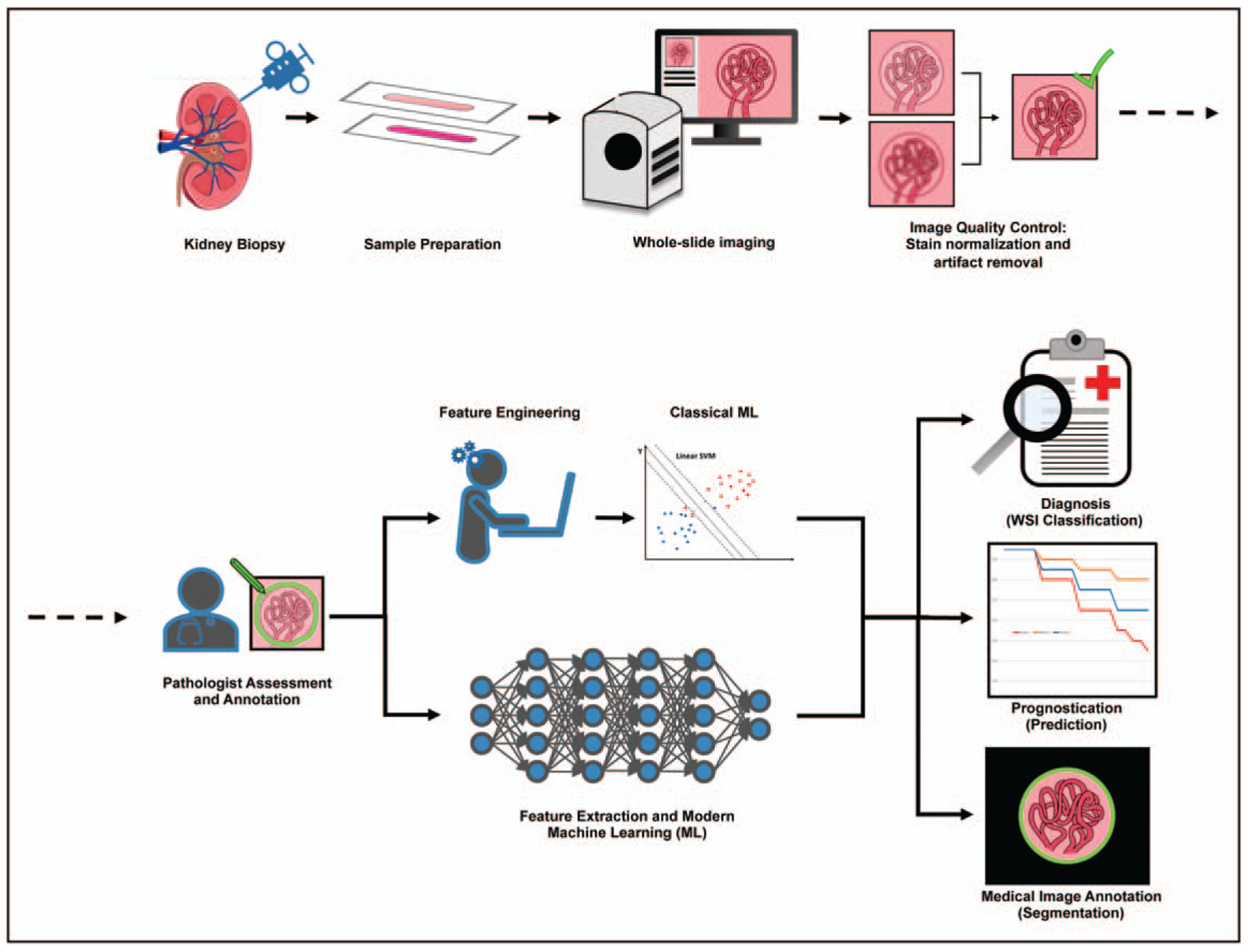
Clinical and computational workflow for artificial intelligence driven assessment of digital renal pathology. Following data acquisition and preparation, classical and modern artificial intelligence approaches provide novel insights for patient assessment and medical image analysis.
Table 1.
Kidney-centric artificial intelligence tools
| Publication | Journal | Authors | Year | Tools | Applied Task |
|---|---|---|---|---|---|
| Computational pathology analysis of tissue microarrays predicts survival of renal clear cell carcinoma patients | Lect. Notes Comput. Sc., MICCAI | Fuchs et al. [3] | 2008 | Local Binary Patterns (LBP) Random Forest Survival Analysis | Tissue Microarray (TMA) Analysis Nuclei Detection Prognostication |
| Weakly supervised cell nuclei detection and segmentation on tissue microarrays of renal clear cell carcinoma | Lect. Notes Comput. Sc., DAGM | Fuchs et al. [3] | 2008 | Edge Detection Morphological Image Processing Support Vector Machine (SVM) |
TMA Analysis Nuclei Detection |
| Computational TMA analysis and cell nucleus classification of renal cell carcinoma | Lect. Notes Comput. Sc., DAGM | Schuffler et al. [4] | 2010 | Graph-Cut Optimization Feature Engineering SVM |
TMA Analysis Nuclei Segmentation Nuclei Classification |
| Segmental HOG: new descriptor for glomerulus detection in kidney microscopy image | BMC Bioinformatics | Kato et al. [5] | 2015 | Segmental Histogram of Oriented Gradients SVM |
Glomerulus Detection |
| Unsupervised labelling of glomerular boundaries using Gabor filters and statistical testing in renal histology | J. Med. Imaging | Ginley et al. [6] | 2017 | Gabor Filterbank Statistical Testing |
Glomerulus Segmentation |
| Multiradial LBP features as a tool for rapid glomerular detection and assessment in whole slide histopathology images | Sci. Rep. | Simon et al. [7] | 2018 | Multiradial Colour LBP SVM Convolutional Neural Network (CNN) |
Glomerulus detection Glomerulus subclassification |
| Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. | J. Am. Soc. Nephrol. | Bukowy et al. [8] | 2018 | Region-based CNN | Glomerulus detection |
| Association of pathological fibrosis with renal survival using deep neural networks | Kidney Int. Reports | Kolachalama et al. [9▪▪] | 2018 | CNN | Prediction of clinical phenotype from trichrome image features |
| Glomerulus classification and detection based on convolutional neural networks | J. Imaging | Gallego et al. [10] | 2018 | CNN | Glomerulus detection |
| Segmentation of glomeruli within trichrome images using deep learning | Kidney Int. Reports | Kannan et al. [11▪▪] | 2019 | CNN | Glomerulus detection Glomerulus classification |
| An integrated iterative annotation technique for easing neural network training in medical image analysis | Nat. Mach. Intell. | Lutnick et al. [12▪] | 2019 | CNN - Semantic segmentation | Glomerulus segmentation Multiclass segmentation of renal morphology |
| Computational segmentation and classification of diabetic glomerulosclerosis | J. Am. Soc. Nephrol. | Ginley et al. [13▪] | 2019 | Feature engineering Recurrent Neural Network (RNN) |
Glomerulus Classification Diabetic nephropathy classification |
| Deep learning-based histopathologic assessment of kidney tissue | J. Am. Soc. Nephrol. | Hermsen et al. [2▪▪] | 2019 | CNN - Semantic segmentation | Multiclass segmentation of renal morphology |
In recent years, the field of renal pathology has seen a surge in the volume of works applying computational image analysis and machine learning for analysis of benign renal histomorphometry.
THE DATA: DIGITAL RENAL HISTOPATHOLOGY
Terminology
Computational image analysis and machine learning utilize a wealth of technical jargon. To achieve the aims of this review, several terms are bolded and defined throughout. For additional terminology, we recommend Roeder et al.’s Computational Image Analysis Glossary for Biologists [14] and Deo’s Machine Learning in Medicine [15].
The advent of digital pathology
The implementation of telepathology in the 1960s demonstrated the power of data sharing and communication for diagnostic assessment [16]. Later development of whole-slide imaging technologies enabled digitization of pathology slides, establishment of digital pathology platforms and curation of whole-slide image (WSI) repositories. Modern digital pathology capitalizes upon these developments, providing a WSI-based, data-driven environment for pathology research, education and clinical practice [17,18]. Primordial digital pathology informatics efforts employed computational image analysis, feature engineering and classical machine learning to prognosticate survival in patients with renal cellcarcinoma (RCC) [3]. More recent works focus on the extension of these methods to benign processes of the kidney.
Data sources and challenges
Benign renal histopathology data are primarily derived from human kidney biopsies performed in patients with primary renal or allograft dysfunction. To date, exemplary renal pathology repositories featuring comprehensive human data include the Nephrotic Syndrome Network (NEPTUNE) [19] and CureGN [20]. Enlistment of these vast data for computational purposes is ideal. However, these datasets lack ground-truth annotations. As a direct result, all projects investigating kidney morphology for classification or predictive purposes must invest significant time and manpower to generate annotation sets. Depending on the kidney structure of interest, machine learning algorithms require anywhere from hundreds to thousands of examples [21] to achieve optimal performance. This annotation burden [12▪] is but one of many data-related challenges facing digital pathology research.
Large-scale analysis of digital pathology is further complicated by data heterogeneity. The lack of standardized protocols for sample preparation and digitization results in heterogeneous, multiinstitutional datasets, which feature significant variation in stain and image quality. To mitigate this issue, de Bel et al. [22▪] recently proposed an innovative solution to the shortage and heterogeneity of kidney data: cycle generative adversarial networks (cycleGANs) for stain transformation and structural segmentation of kidney histopathology. GANs are deep neural network architectures [23], which learn to generate synthetic images resembling authentic input images. In the aforementioned work, the cycleGAN is trained to take input histology images of disparate stain quality and generate images featuring an identical colourspace. The ability to generate synthetic images representative of renal histopathology navigates disparities in tissue preparation. An additional computational tool for quality control of digital pathology data is HistoQC [24▪▪]. Designed to automatically detect image artefacts and normalize sample stain variation, HistoQC offers a rapid and reproducible alternative to the manual review of WSIs.
DETECTION AND CHARACTERIZATION OF KIDNEY STRUCTURES IN WHOLE-SLIDE IMAGES
Traditionally, diagnostic renal pathology involves examination of kidney biopsies for known structural abnormalities indicative of disease [13▪]. On the basis of the type and severity of abnormality, pathologists classify biopsies and provide a patient diagnosis. Automation of this process requires the development of artificial intelligence tools, which automatically detect kidney structures in biopsy WSIs. To achieve this end, engineers employ computer vision: the scientific field that explores how computers can achieve high-level understanding from digital images [25]. When provided and trained with digital pathology data, computers have the potential to detect, analyse and classify kidney structural morphology, effectively emulating pathologist visual assessment. Computers accomplish these tasks by conducting pattern recognition via image analysis [18].
Image analysis is the extraction of meaningful information from digitized images via image processing techniques [25]. Given that images are composed of pixels, or a matrix of numbers, these processing techniques involve mathematical calculations to detect and characterize specific patterns in image pixels [25]. To selectively analyse image regions of interest, we apply segmentation: the partitioning of an image into multiple, distinct segments [25] of select pixels. For example, when analysing kidney morphology, WSIs must be segmented or partitioned into regions featuring anatomical structures of interest such as glomeruli, tubules, vessels or inflammatory cells. Once segmented, kidney structures may be quantified – enumerated and measured – to provide a set of descriptive features for subsequent classification by type and morphological variation [26,27].
Classical feature extraction and classification from renal biopsy whole-slide images
A myriad of classical image analysis algorithms has been developed for exploration of kidney morphology in WSIs [27]. These algorithms capitalize upon textural and morphological image processing techniques [28] to detect regions wherein disease may manifest. In image processing, morphology describes the shapes of image regions, while texture describes the arrangement and heterogeneity of colour or pixel intensities. Object edges in images are described as discrete regions wherein an abrupt change in intensity is observed for a stretch of contiguous pixels. Textural operations may be employed to detect object edges or distinct textures in images. For example, the texture of glomeruli is very different from the surrounding tubulo-interstitium. This observation, among many others, facilitated the application of textural analysis techniques for glomerulus detection and feature engineering, including Gabor filtering [6], local binary pattern (LBP) [7] and histogram of oriented gradients [5].
Morphological processing techniques have been used for detection of kidney microstructures that have characteristic morphologies. For example, the glomerular unit is typically elliptical in histology images, and therefore may be detected via minimization of the variance between the glomerulus boundary and a predefined shape (e.g. ellipse) [29]. Unfortunately, despite reproducible detection of glomerulus boundaries, these shape fitting methods have a substantial false-positive rate. More specifically, the similarity of the glomerular boundary to large, transversely cut tubules and arterioles renders each candidate for detection [29]. Algorithms based upon textural analysis also have drawbacks. In pathologic states, such as renal parenchymal scarring, there is a decrease in textural contrast between globally sclerosed glomeruli and surrounding cortical fibrosis that may hinder texture-based segmentation [30]. Therefore, algorithms may perform poorly when analysing disease tissue. To date, some of the most successful algorithms use morphological and textural analysis in tandem to provide for robust detection, segmentation and characterization of glomeruli, intraglomerular compartments and tubules [13▪,31].
Once segmented, image regions are further processed to derive discriminative features for subsequent classification tasks. Features descriptive of image texture can be as simple as the standard deviation of pixel intensities in a given region [7], or as complex as the amplitude response of a Gabor filter [6]. Similarly, morphological features may be more apparent, such as image object area defined as the sum of resident pixels, or the computation of nuclear shape signatures that compare object radii at incremental polar angles [32]. Given the ability to selectively apply techniques for image analysis, derived features are often referred to as handcrafted or engineered. Similar to segmentation tasks, handcrafted feature sets derived via a combination of textural and morphological processes provide for optimal image classification. Although classical classification algorithms in machine learning are numerous, methods commonly used for classification of kidney microstructures using handcrafted image features include support vector machines (SVMs) [7,31,33] and decision trees [29]. Strong classifier performance has also been reported when features are extracted using artificial neural networks (ANNs), and subsequently classified using a SVM [33].
Neural networks for feature extraction and classification from renal biopsy whole-slide images
In recent years, ANNs have taken the field of image analysis by storm, exploiting their tremendous depth of vision and comprehension for a myriad of image-based applications. With respect to computational renal pathology, ANNs have emerged as the ‘panacea’ for existing challenges, enabling not only detection and classification of kidney morphology [2▪▪,8,11▪▪,12▪,26,33,34,35] but also prediction of disease status and outcome [13▪,9▪▪,35,36,37,38▪▪,39]. ANNs achieve high-level understanding through their biologically inspired architecture, wherein interconnected artificial neurons simulate the neuronal pathways of the human brain [40]. Input data (observations) are passed through a series of layers, or mathematical processes, which transform the input into a desired output. As data are passed from layer to layer, the network gains increasingly abstract insight and learns to recognize trends in the input data. Similar to the brain, ANNs also feature weighted connections between their artificial neurons [41]. These connections enable neurons to selectively influence each other, with higher number weights corresponding to greater influence. This fully connected, weighted design facilitates ANN learning or training.
During training, ANNs are provided with vast input data and ground truth labels defining each input’s expected output. Comparison of transformed output with the ground-truth defines the error of fitting, which the machine optimizes using an operation called back propagation [41]. Back propagation involves adjustment of learning parameters, such as the weights of neuronal connections, to produce updated transformed outputs. When the actual and expected outputs converge, error is minimized, and the network achieves optimal performance.
Initially, the computationally intensive nature of network parameter (weights) estimation and optimization deterred use of ANNs for image-based applications. Later discovery of convolutional neural networks (CNNs) – a more efficient network design – rendered ANNs ideal for image analysis tasks. Digital pathology has since seen an explosion in the application of CNNs for morphological and textural analysis of tissue structure in disease [1▪,39,42,43]. CNNs pass input images through a series of convolutional filters (layers) and extract image features via convolution, a signal processing operation. The progressive complexity of convolutional filters at increasing network depth results in the extraction of a hierarchy of image features, branching from simple (edges and textures) to abstract (objects). These hierarchical features, along with the input image’s ground truth label, provide for subsequent image segmentation and classification tasks.
To date, CNNs have been applied for the detection and classification of kidney morphological structures, including glomeruli, tubules, interstitium, arteries and even empty Bowman’s spaces [2▪▪,8,11▪▪,12▪,26,33,34]. In addition, CNNs have enabled discrimination of healthy and pathologic kidney morphology [2▪▪,13▪,34]. Many of these methods capitalize upon select histological stains for multiclass structural segmentation. For example, WSIs stained with Masson’s trichrome enabled detection and quantification of glomeruli and renal scarring. In addition, PAS-stained WSIs provided for comprehensive compartmentalization of both healthy and pathologic kidney morphology [2▪▪]. These methods brought to light challenges facing multiclass segmentation of renal tissue, including reproducible detection and subclassification of tubules and sclerosed glomeruli, and overestimation of renal pathologies in distinct stains. Despite significant training data, composed of both pathologist manual segmentations and ground truth labels, networks struggle to reproducibly detect tubules (esp. atrophied) and sclerosed glomeruli. In addition, studies have shown that quantification of trichrome stain tends towards overestimation of renal fibrosis [44]. Taken together, these two challenges limit network performance for detection and classification of pathologic kidney morphologies essential to disease assessment.
Lutnick et al. [12▪] recently developed Human-Artificial Intelligence-Loop (H-AI-L), a unique, iterative technique for multiclass semantic segmentation of kidney structures from WSIs. Rather than providing a single image label, semantic segmentation involves linking a class label to each pixel in an image. Pixel class labels are assigned based on ground truth annotations defining distinct image regions of interest. H-AI-L allows humans to interact with the machine through manual correction of structural boundary predictions for additional rounds of network training [12▪]. The ability to iteratively train the network and guide the machine to identify kidney structures, as pathologists do, offers a solution to the aforementioned challenges in multiclass image segmentation. In addition, H-AI-L’s ability to automatically generate structural boundary predictions will ease the annotation burden in medical image analysis.
Recent works have demonstrated the successful application of an alternative ANN architecture – Recurrent Neural Networks (RNNs) [45] – for classification of digital pathology [13▪,46]. Traditional ANNs process inputs independently, providing an output for each input. In comparison, RNNs inter-relate all inputs, using the output of a previous input to provide context for the next. As a result, RNNs have the ability to recognize the sequential characteristics of a dataset, and use learned patterns in data to make informed predictions and effectively exhibit memory. Ginley et al. [13▪] capitalized upon this network ‘memory’ to predict the stage of diabetic nephropathy from WSIs of kidney biopsies. Given that current diagnostic schemes involve holistic evaluation of a biopsy’s glomerular pathologies, machine learning applications designed for automated diagnosis must analyse and classify all glomeruli in a WSI. To achieve this, Ginley et al. [13▪] strategically trained a RNN with a sequence of labelled feature sets for each glomerulus in a biopsy, and trained the network to predict an ‘average’ stage for the entire input dataset (kidney biopsy) based on learned data trends. For the input data, glomeruli were staged by an expert pathologist, and glomerular features were engineered via computational image analysis. This work’s successful prediction of disease stage through a combination of computational image analysis, feature engineering and modern machine learning demonstrates the tremendous potential for artificial intelligence in diagnostic pathology.
Correlation of image features with clinical biometrics and progression data
ANNs are not limited to image data. The ability of ANNs to extract features and classify image, numerical and categorical data, renders them ideal for correlation of histopathology with clinical phenotypes. For example, multiple CNN models have been trained to correlate and predict CKD stage, proteinuria and other measures of kidney function at time of biopsy based on input histopathology images derived from renal biopsies [9▪▪,35]. Additional CNN models predict 1, 3 and 5-year renal survival based on input biopsy images [9▪▪]. At this time, few kidney-centric works feature such predictive capacity. However, a variety of ANN models have been developed for other types of histopathology data. Landmark discoveries in the field of cancer pathology have been made as a product of artificial intelligence methods for evaluation of histopathology data [38▪▪,39,47,48]. The predictive capacity of these models for patient biometrics and survival rates demonstrates artificial intelligence’s potential in clinical diagnostics, prognostics and theragnostics.
CONCLUSION
Despite the successful application of ANNs in digital pathology, our interpretation of machine vision and the metaphorical ‘black box’ remains unclear [49]. Elucidation of the mechanisms governing machine decision making is essential to exploiting the maximal potential of these algorithms and gaining insight beyond extant clinical knowledge. Such insights will facilitate streamlined ANN training for recognition of rare, yet highly informative image features (biomarkers) indicative of distinct clinical phenotypes. Reproducible detection of clinical phenotypes from digital pathology will further the development of diagnostic and prognostic applications requiring analysis of multimodal data. In truth, the greatest obstacle facing the precision medicine initiative is integration of omic – genomic, proteomic – and image data [50]. Although select biomarkers have been identified in digital pathology, a method for comprehensive correlation of image biomarkers with patient omic data has yet to be developed. Additional challenges facing digital pathology include expansion of computational pipelines from two-dimensional tissue sections to three-dimensional reconstructions, and establishment of image ontology protocols [51–53] for curation of optimal machine learning datasets. Resolution of these challenges will revolutionize our understanding of renal pathology and will actualize future integration of artificial intelligence in the process of care.
KEY POINTS.
The ability of ANNs to not only extract image features but also classify image, numerical and categorical data potentiates the integration of omic and image data in pursuit of precision medicine.
Given that machine learning algorithms require a high volume of annotated examples to learn to reproducibly detect and classify distinct kidney structures, computational tools (e.g. H-AI-L) offer a unique solution to ease pathologists’ annotation burden and augment machine learning data sets.
Due to the lack of standardized sample preparation and imaging protocols, robust algorithms must be developed to perform data quality control and navigate disparities in digital pathology data.
Although modern machine learning with ANNs provides rapid, reproducible results, the handcrafted or engineered features of classical machine learning are more biologically and clinically interpretable.
RNNs have enabled the evaluation of whole kidney biopsies, taking into account all pathologically relevant regions of interest, and strong approximation of pathologist visual assessment.
Acknowledgements
We would like to thank Brendon R. Lutnick for discussion pertaining to neural networks while preparing the manuscript.
Financial support and sponsorship
The project was supported by the faculty startup funds from the Jacobs School of Medicine and Biomedical Sciences, University at Buffalo, NIDDK Diabetic Complications Consortium grant U24 DK076169, NIDDK grant R01 DK114485 and NIDDK CKD Biomarker Consortium grant U01 DK103225.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.▪.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019; 20:e253–e261. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides an overview of artificial intelligence approaches in digital pathology with a focus on cancer diagnosis.
- 2.▪▪.Hermsen M, de Bel T, den Boer M, et al. Deep learning–based histopathologic assessment of kidney tissue. J Am Soc Nephrol 2019; 30:1968–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates the application of CNNs for multiclass segmentation of renal morphology in digital pathology images.
- 3.Fuchs TJ, Wild PJ, Moch H, Buhmann JM. (2008). Computational Pathology Analysis of Tissue Microarrays Predicts Survival of Renal Clear Cell Carcinoma Patients. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008 Lecture Notes in Computer Science, p. 1–8. doi: 10.1007/978-3-540-85990-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Schüffler PJ, Fuchs TJ, Ong CS, Roth V, and Buhmann JM. (2010). Computational TMA Analysis and Cell Nucleus Classification of Renal Cell Carcinoma Lecture Notes in Computer Science Pattern Recognition, Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 202–211. [Google Scholar]
- 5.Kato T, Relator R, Ngouv H, et al. Segmental HOG: new descriptor for glomerulus detection in kidney microscopy image. BMC Bioinformatics 2015; 16:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginley B, Tomaszewski JE, Yacoub R, et al. Unsupervised labeling of glomerular boundaries using Gabor filters and statistical testing in renal histology. J Med Imaging (Bellingham) 2017; 4:021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon O, Yacoub R, Jain S, et al. Multi-radial LBP features as a tool for rapid glomerular detection and assessment in whole slide histopathology images. Scientific Reports 2018; 8:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukowy JD, Dayton A, Cloutier D, et al. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol 2018; 29:2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.▪▪.Kolachalama VB, Martini S, Barisoni L, et al. Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep 2018;3:464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this landmark work, the authors describe successful prediction of clinical phenotypes, including 1, 3 and 5- year survival, from patient-specific histopathology images via deep learning.
- 10.Gallego J, Pedraza A, Lopez S, et al. Glomerulus classification and detection based on convolutional neural networks. Journal of Imaging 2018; 4:20. doi: 10.3390/jimaging4010020. [DOI] [Google Scholar]
- 11.▪▪.Kannan S, Morgan LA, Liang B, et al. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep 2019; 4:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this work, the authors describe the development of a deep learning framework for detection and segmentation of glomeruli in digital pathology. The developed model demonstrates the power of artificial intelligence for detecting and assessing complex histologic structures from digital renal pathology.
- 12.▪.Lutnick B, Ginley B, Govind D, et al. An integrated iterative annotation technique for easing neural network training in medical image analysis. Nat Mach Intell 2019; 1:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work proposes an innovative Human-AI-Loop (H-AI-L) approach for segmentation of anatomical structures of interest in digital pathology. The authors describe how strategic application of the developed H-AI-L tool will ease the existing medical image annotation burden.
- 13.▪.Ginley B, Lutnick B, Jen K-Y, et al. Computational segmentation and classification of diabetic glomerulosclerosis. Journal of the American Society of Nephrology 2019; 30:1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this landmark work, the authors describe the development of a ‘digital pipeline’ for automated classification of diabetic glomerulosclerosis.
- 14.Roeder AH, Cunha A, Burl MC, Meyerowitz EM. A computational image analysis glossary for biologists. Development 2012; 139:3071–3080. [DOI] [PubMed] [Google Scholar]
- 15.Deo RC. Machine learning in medicine. Circulation 2015; 132: 1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross S, Dennis T, Start RJH. Telepathology: current status and future prospects in diagnostic histopathology. Histopathology 2002; 41:91–109. [DOI] [PubMed] [Google Scholar]
- 17.Pantanowitz L, Valenstein PN, Evans AJ, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform 2011; 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madabhushi A. Digital pathology image analysis: opportunities and challenges. Imaging Med 2009; 1:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gadegbeku CA, Gipson DS, Holzman LB, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nphropathy by a multidisciplinary approach. Kidney Int 2013; 83:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariani LH, Bomback AS, Canetta PA, et al. CureGN study rationale, design, and methods: establishing a large prospective observational study of glomerular disease. Am J Kidney Dis 2019; 73:218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Computational and Structural Biotechnology Journal 2018; 16:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.▪.de Bel T, Hermsen M, Kers J, et al. Stain-transforming cycle-consistent generative adversarial networks for improved segmentation of renal histopathology. Proceedings of the 2nd International Conference on Medical Imaging with Deep Learning 2019; 102:151–163. [Google Scholar]; In this work, the authors propose an innovative solution to navigate the heterogeneous nature of kidney data when segmenting digital pathology images.
- 23.Zhu J-Y, Park T, Isola P, Efros AA. (2017). Unpaired image-to-image translation using cycle-consistent adversarial networks, 2017 IEEE International Conference on Computer Vision (ICCV), 18 pages. doi: 10.1109/iccv.2017.244. [DOI] [Google Scholar]
- 24.▪▪.Janowczyk A, Zuo R, Gilmore H, et al. HistoQC: an open-source quality control tool for digital pathology slides 2019; 3:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work features Histo-QC, an innovative tool for quality control of digital pathology data that detects sample preparation and image artefacts. Application of Histo-QC will expedite diagnostic workflows by eliminating the laborious task of manual slide review (glass/digital) review.
- 25.Gonzalez RC, Wintz PJR. Digital image processing. MA: Addison-Wesley Publishing Co., Inc; 1977:451. [Google Scholar]
- 26.Zee J, Hodgin JB, Mariani LH, et al. Reproducibility and feasibility of strategies for morphologic assessment of renal biopsies using the nephrotic syndrome study network digital pathology scoring system. Arch Pathol Lab Med 2018; 142:613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangan GK, Tesch GH. Quantification of renal pathology by image analysis. Nephrology (Carlton) 2007; 12:553–558. [DOI] [PubMed] [Google Scholar]
- 28.Sarder P, Ginley B, Tomaszewski JE. Automated renal histopathology: digital extraction and quantification of renal pathology BT Medical imaging 2016: digital pathology. Proceedings of SPIE (SPIE Medical Imaging 2016: Digital Pathology) 2016; 9791:pp. 97910F: 1–12, doi: 10.1117/12.2217329. [DOI] [Google Scholar]
- 29.Marée R, Dallongeville S, Olivo-Marin J and Meas-Yedid V, An approach for detection of glomeruli in multisite digital pathology, 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, 2016, pp. 1033–1036. doi: 10.1109/isbi.2016.7493442. [DOI] [Google Scholar]
- 30.Barisoni L, Gimpel C, Kain R, et al. Digital pathology imaging as a novel platform for standardization and globalization of quantitative nephropathology. Clin Kidney J 2017; 10:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tey WK, Kuang YC, Ooi MP, Khoo JJ. Automated quantification of renal interstitial fibrosis for computer-aided diagnosis: a comprehensive tissue structure segmentation method. Comput Methods Programs Biomed 2018; 155:109–120. [DOI] [PubMed] [Google Scholar]
- 32.Tang JR, Mat Isa NA, Ch’ng ES. Evaluating nuclear membrane irregularity for the classification of cervical squamous epithelial cells. PLoS One 2016; 11:e0164389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chagas P, Souza L, Araújo I, et al. Classification of glomerular hypercellularity using convolutional features and support vector machine. arXiv e-prints 2019; p. arXiv: 1907.00028. [DOI] [PubMed] [Google Scholar]
- 34.Marsh JN, Matlock MK, Kudose S, et al. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging 2018; 37:2718–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ledbetter D, Ho L, Lemley KV. Prediction of kidney function from biopsy images using convolutional neural networks. arXiv preprint 2017; p. arXiv: 1702.01816. [Google Scholar]
- 36.Mariani LH, Martini S, Barisoni L, et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant 2017; 33: 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair V, Komorowsky CV, Weil EJ, et al. A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int 2018; 93:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.▪▪.Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology: new tools for diagnosis and precision oncology. Nature Reviews Clinical Oncology 2019; 16:703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article proposes how artificial intelligence and machine learning tools for analysis of digital pathology may be incorporated into clinical oncology. The authors further discuss anticipated challenges and innovative solutions, as well as potential opportunities for precision oncology.
- 39.Bauer S, Carion N, Schüffler S, et al. Multiorgan cancer classification and survival analysis. arXiv preprint 2016; p. arXiv: 1606.00897. [Google Scholar]
- 40.Zou J, Han Y, So S-S. Overview of artificial neural networks, in artificial neural networks. Springer; 2008; 14–22. [Google Scholar]
- 41.Hastie T, Friedman J, Tibshirani R. The elements of statistical learning: data mining, inference, and prediction. 2 ed New York: Springer; 2017. [Google Scholar]
- 42.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform 2016; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tizhoosh HR, Pantanowitz L. Artificial intelligence and digital pathology: challenges and opportunities. J Pathol Infrom 2018; 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brodsky SV, Albawardi A, Satoskar AA, et al. When one plus one equals more than two: a novel stain for renal biopsies is a combination of two classical stains. Histol Histopathol 2010; 25:1379–1383. [DOI] [PubMed] [Google Scholar]
- 45.Zuo Z, Shuai B, Wang G, et al. Convolutional recurrent neural networks: learning spatial dependencies for image representation, 2015 IEEE conference on computer vision and pattern recognition workshops (CVPRW), Boston, MA, 2015, pp. 18–26. doi: 10.1109/cvprw.2015.7301268. [DOI] [Google Scholar]
- 46.Zhang Z, Chen P, Mcgough M, et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nature Machine Intelligence 2019; 1:236–245. [Google Scholar]
- 47.Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019; 25:1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cima I, Schiess R, Wild P, et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci U S A 2011; 108:3342–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castelvecchi D. Can we open the black box of AI? Nature 2016; 538:20. [DOI] [PubMed] [Google Scholar]
- 50.Barsoum I, Tawedrous E, Faragalla H, Yousef GM. Histo-genomics: digital pathology at the forefront of precision medicine. Diagnosis (Berl) 2019; 6:203–212. [DOI] [PubMed] [Google Scholar]
- 51.Lindman K, Rose JF, Lindvall M, et al. Annotations, ontologies, and whole slide images: development of an annotated ontology-driven whole slide image library of normal and abnormal human tissue. J Pathol Inform 2019; 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith B, Arabandi S, Brochhausen M, et al. Biomedical imaging ontologies: a survey and proposal for future work. J Pathol Inform 2015; 6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurcan MN, Tomaszewski J, Overton JA, et al. Developing the Quantitative Histopathology Image Ontology (QHIO): a case study using the hot spot detection problem 2017; 66:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]


