Abstract
Innate immune perception is the first line of inducible defense against invading pathogens. Plants lack specialized circulating immune cells. Therefore, diverse cell types are able to recognize and respond to pathogens. Surface localized and intracellular plant innate immune receptors are capable of recognizing diverse pathogen components. Intracellular nucleotide-binding leucine-rich repeat (NLR) receptors recognize pathogen effectors delivered inside host cells. Recent advances shed light onto NLR activation, phosphorylation of defense signaling nodes and overlap in transcriptional responses between pathogen perception and abiotic stress.
Keywords: plant innate immunity, NLR, pathogen effector, resistosome
Introduction
With the exception of viruses and specialized insect-vectored bacteria, most pathogens do not replicate inside plant cells. To cause disease and modify their hosts, pathogens secrete proteins, called effectors, into the extracellular space or directly into host cells [1]. Plant innate immune receptors include surface localized pattern recognition receptors (PRRs) as well as intracellular NLR receptors [2,3]. PRRs can recognize conserved microbe- or pathogen-associated molecular patterns (M/PAMPs), damage-associated molecular patterns (DAMPs) and extracellular effector proteins [1,3]. NLRs detect the presence or activity of effectors delivered into host cells during infection [2]. Both PRR- and NLR-triggered immunity (PTI and NTI) lead to a suite of downstream defense responses including the generation of reactive oxygen species (ROS), an influx of extracellular calcium, kinase activation and global transcriptional reprogramming for defense [2,3] (Fig 1). After immune recognition, signals of pathogen perception are propagated from the initial infection site to distal tissues [4]. This systemic immune signaling primes naïve tissue against subsequent attack. Despite similarities, the timing, intensity and duration of defense can differ between PTI and NTI (Fig 1) [5]. NLR activation induces a quantitatively stronger, prolonged and robust response, frequently culminating in programmed cell death at the site of infection [2]. Here we will focus on recent advancements in NLR biology from receptor activation to downstream signaling.
Figure 1: Surface localized and intracellular plant innate immune receptors recognize diverse pathogens.
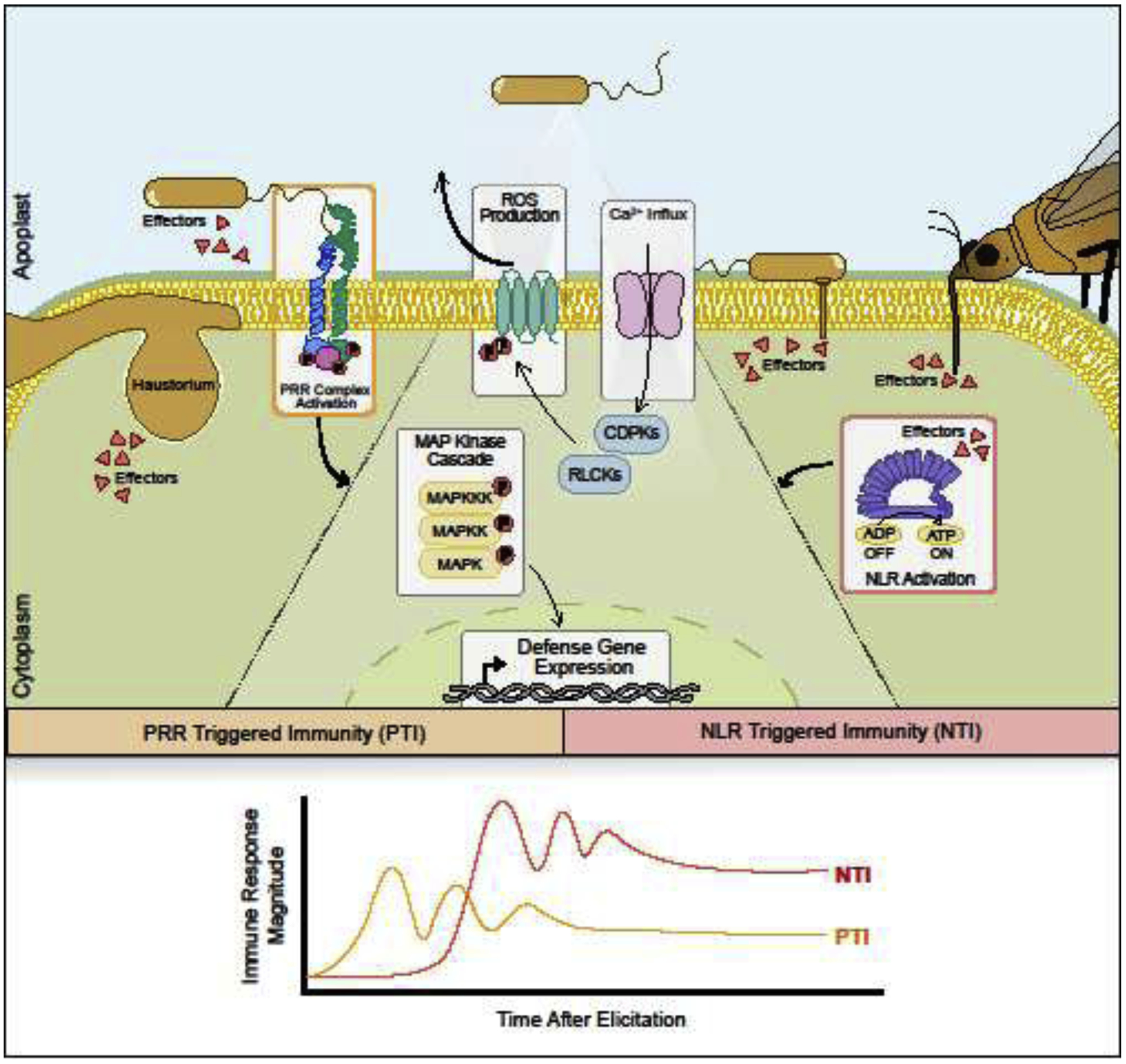
Plant immune receptors include surface localized pattern recognition receptors (PRRs) and intracellular nucleotide binding leucine rich repeat (NLR) receptors. PRRs can recognize microbial features, damage associated molecular patterns, and extracellular receptors from insects, bacteria and filamentous pathogens. NLRs perceive pathogen effectors directly or through effector-mediated perturbations. Both PTI and NTI induce downstream defense responses including an influx of extracellular calcium, reactive oxygen species (ROS) production, kinase activation and transcriptional reprogramming for defense. While downstream defense responses are similar between PTI and NTI, the timing, amplitude and duration of responses differ.
NLR architecture and diverse modes of effector recognition
Plant genomes possess diverse NLR repertoires, with many species possessing hundreds of distinct NLRs that can be used to control pathogen infection in crops [6]. NLRs are composed of a central nucleotide-binding site (NBS) and C-terminal leucine-rich repeats (LRRs). They can be divided into two broad classes based on their N-termini, with CNLs carrying a coiled-coil (CC) domain and TNLs carrying a Toll-interleukin 1 receptor (TIR) domain (Fig 2) [2]. Both pathogen effectors and NLRs can localize to diverse subcellular locations including the cytoplasm, nucleus, plasma membrane, tonoplast and endoplasmic reticulum [2]. The barley CNL MLA10 and the Arabidopsis TNL RPS4 reside in the nucleus and cytoplasm, with both locations required for full resistance [7,8]. The Arabidopsis CNL RPM1 constitutively associates with the plasma membrane and recognizes a membrane-targeted Pseudomonas effector protein [9,10]. How NLRs with diverse subcellular localizations are able to trigger similar defense responses remains unknown.
Figure 2: NLRs act as molecular switches to provide robust resistance against pathogens.
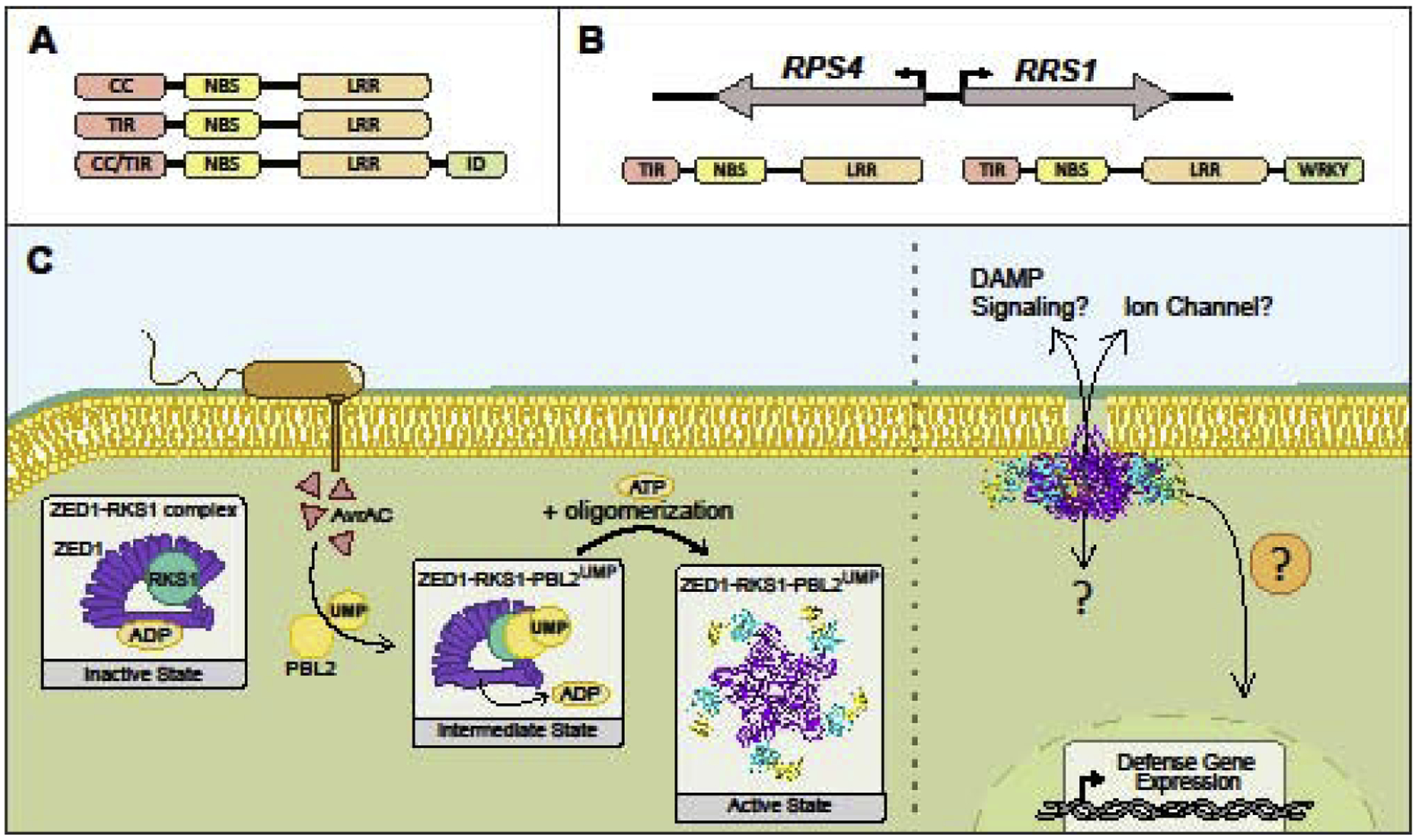
A. Plant NLR domain architecture includes an N-terminal coiled-coil (CC) or Toll/interleukin-1 receptor (TIR) domain, a central nucleotide binding site (NBS) and C-terminal leucine-rich repeats (LRRs). Some NLRs carry an integrated domain (ID) that can directly sense pathogen effectors. B. NLRs can act in pairs, such as Arabidopsis RPS4 and RRS1. Top: head-to-head genomic orientation, bottom: domain architecture from N- to C-termini. RRS1 is a sensor NLR with a WRKY ID. C. The Arabidopsis NLR ZAR1, pseudokinase RSK1, and kinase PBL2 can form a pentameric complex, or resistosome. Upon uridylation by the Xanthomonas effector AvrAC, PBL2 is recruited to the ZAR1-RKS1 complex (intermediate state). The active ZAR1 complex exhibits enhanced membrane affinity and the CC-domains of ZAR1 resemble a pore-like structure. Many questions remain regarding the activation of innate immune responses upon resistosome formation.
Not only do NLRs localize to distinct cellular compartments, they also exhibit diversity in mechanisms of pathogen effector recognition. Some receptors can directly bind and recognize cognate pathogen effectors, while others monitor for effector-mediated perturbations of host targets [2]. For example, the Nicotiana TNL Roq1 confers resistance against Xanthomonas and is able to physically associate with the recognized effector XopQ [11]. In contrast, RPM1 recognizes effector-induced phosphorylation of the host protein RIN4 [12,13]. Animal NLRs recognize PAMPs as well as monitor for pathogen-mediated perturbations, such as the mouse NLR NOD1 that is activated by Salmonella SopE effector activity [14]. Plant NLRs can also act as pairs and can exhibit head-to-head chromosomal orientation to facilitate co-expression (Fig 2) [15,16]. These paired NLRs have been characterized from both monocots and dicots [17,18••]. Most NLR pairs consist of a canonical signaling NLR, such as RPS4, and a sensor NLR carrying an integrated domain that interacts with an effector target, such as RRS1-R with a WRKY domain (Fig 2) [15,16]. Finally, some receptors require downstream helper NLRs to form a functional unit for disease resistance [19••].
NLR activation and resistosome formation
NLR activity undergoes multilayered regulation, including self-inhibition, dimerization or oligomerization, epigenetic and transcriptional regulation, alternative splicing and proteasome‐ mediated degradation [20]. Their similar structure across diverse organisms, especially the presence of the NBS, indicates that nucleotide binding acts as a switch for receptor activity. Early work in plants identified the first NLRs, demonstrated conservation of NBS domains and determined that the NBS is essential to their functionality [21–23]. If any of the multiple ATP-binding motifs within the NBS are mutated, this renders the NLR either locked in an activated state (ATP bound) or nonfunctional (ADP bound or unbound) in terms of its ability to elicit a defense response [22,24]. A longstanding model within the field of plant immunology posited NLRs are tightly folded and bound to ADP in an inactive state and upon effector perception exhibit conformational changes enabling ATP binding and higher order complex formation (Fig 2) [25,26].
The first structure of a plant NLR complex in inactive, intermediate and activated state was recently elucidated using cryo–electron microscopy [27••,28••] (Fig 2). This was accomplished with the CNL ZAR1, which recognizes the Xanthomonas effector AvrAC indirectly, through effector-mediated uridylation of the host kinase PBL2 [29]. When inactive, ZAR1 self-associates through inter-domain interactions and interacts with the pseudokinase RKS1 through its LRR domain [28••]. Upon uridylation, PBL2 recruits and binds to RKS1 [29]. The allosteric binding of PLB2 to RSK1 induces conformational changes in ZAR1’s NBS domain, causing release of ADP and formation of a ZAR1-RSK1-PBL2 trimeric complex, likely representing a primed intermediate state [28••]. ZAR1 dATP or ATP binding induces conformational changes within the NBS domain, which in turn mediates oligomerization of the complex into a higher order wheel-like pentamer, called the resistosome [27••]. This multistep activation of ZAR1 may function to ensure appropriate activation of defense. When oligomerized, the N-terminal α‐helices of the ZAR1 CC domains form a protruding funnel-like structure with similarity to pore-forming toxins [27••,30] (Fig 3). The N-terminal α-helix is essential for enhanced membrane association and signaling upon ZAR1 activation [27••]. Animal NLRs undergo higher order complex formation upon pathogen perception, forming inflammasomes and apoptosomes that trigger cell death [31]. Thus, ATP binding, oligomerization and cell death induction appears to be a common feature of NLR activation.
Resolving the resistosome structure is an important step in understanding mechanisms of NLR activation and opens new avenues for investigating downstream signaling. If activated CNLs are able to form pores in membranes, cell death may occur through disrupting selective membrane permeability in a similar manner to pore forming toxins. Membrane disruption could also induce DAMP signaling and be perceived by PRRs to amplify immune responses [3]. Alternatively, CNL resistosomes could form selective ion channels and transport signaling ions, such as Ca2+, but this would need additional layers of regulation to control ion selectivity (Fig 2). Given the diverse and dynamic NLR sub‐cellular localizations, it will be interesting to determine if other NLRs can form similar structures targeted to various membranes. CNL signaling may be two-fold, with pore/ion channel formation coupled to signaling initiated intracellularly through the resistosome complex.
Regulation of downstream signaling
Responses downstream of CNL receptors frequently require the NDR1 locus, while TNL receptors require a set of lipase-like proteins including EDS1 and SAG101 [2,32,33]. NDR1 is anchored to the plasma membrane, mediates plasma-membrane cell wall adhesions and possess similarity to plant proteins involved in abiotic stress responses and mammalian integrins [34]. With the recent discovery that CNLs may form pore-like structures, it will be important to address the role of NDR1 and other immune signaling nodes for effects in plasma membrane integrity. While TNLs lack CC domains, they frequently require downstream helper NLRs of the CNL class, including ADR1, NRG1 and NRCs [19••,35,36]. For example, the TNLs Roq1 and RPP1 require the helper NLR NRG1 [37]. Furthermore, multiple CNLs in Solanaceous plants that require NRC helpers possess extended N-terminal regions before their CC domain, including the tomato CNLs Prf and Mi [19••,38]. Deletion of the N-terminal 13aa from the CC-domain of the NRG1 helper blocks its cell death inducing activity [27••]. It is possible that primary NLRs with diverse subcellular localizations or N-terminal domains unable to form pore-like structures partner with helper NLRs to achieve a robust NTI response.
Accumulation of extracellular ROS by NAPDH oxidases is a hallmark of both PTI and NTI responses [2,3]. Extracellular ROS also physically strengthens the plant cell wall, induces cell wall depositions and functions as a secondary signal required for both local and systemic innate immune responses [39]. In Arabidopsis, the primary NADPH oxidase required for ROS production, RBOHD, is activated by conformational changes induced by Ca2+ binding and N-terminal phosphorylation of conserved residues [40–43]. Phosphorylation of RBOHD S343 and S347 occurs during both PTI and NTI, but through distinct kinases [44••]. Downstream MAPK cascades are also similarly induced, but it is unknown if the upstream activating kinases are similar for both receptor types [5]. Thus, distinct kinases may converge upon critical signaling nodes with varying intensity to regulate the duration and magnitude of responses during PTI and NTI (Fig 1).
Transcriptional regulation of immunity: Overlap with general stress response
NLR recognition of pathogen effectors induces massive transcriptional reprogramming towards defense. Genetic studies have demonstrated that several transcription factor families play critical roles in innate immune and abiotic stress responses, including those in the AP2/ERF, bHLH, MYB, NAC, WRKY, bZIP and CAMTA families [45,46••]. Transcriptional profiling after activation of the barley CNL MLA1, the Arabidopsis TNL RPS4 and various PRRs recognizing bacterial and fungal PAMPs revealed significant overlap in early response genes [46••]. Early response genes are enriched in loci encoding signaling components, such as transcription factors, with CAMTA binding motifs enriched in their promoters [46••]. CAMTAs are a group of calmodulin binding transcription factors involved in both positive and negative regulation of various Ca2+ dependent stress responses [47]. Upon abiotic and biotic stress, CAMTAs rapidly and transiently induce gene expression by binding the Rapid Stress Response Element (RSRE). RSREs are overrepresented in the promoters of general stress response-associated genes [48]. The enrichment of CAMTA binding motifs in early response genes support the notion that both innate immune activation and abiotic stress induce a similar and rapid general stress response, with differential transcriptional outputs at later time points depending on pathogen or abiotic stimulus.
Systemic immunity and transgenerational resistance
After pathogen perception, immune signals are subsequently propagated within a tissue, systemically move to distal tissues, and prime the plant for heightened resistance against subsequent attack. Local immune priming can be established by NLR activation as well as crosstalk between PRRs and their co-receptors after MAMP perception [49]. After bacterial challenge, the flagellin co-receptor, BAK1, phosphorylates the receptor-like kinase for chitin perception, CERK1, which primes the plant and enhances defense activation upon subsequent fungal attack [49]. The plant hormone salicylic acid (SA) is required for defense in local and distal tissues. Systemic immunity in distal tissues induces transcriptional and metabolic reprogramming leading to heightened resistance against biotrophic pathogens that typically lasts for several weeks [50,51]. SA dependent immune priming can be propagated between individual Arabidopsis plants through monoterpene emissions [52,53]. This monoterpene-associated response depends on signals associated with systemic resistance, potentially mediating propagation of resistance at a population level [53].
SA application induces chromatin modifications, including acetylation and methylation, on the promoters of defense genes, which correlate with stronger and more robust expression upon pathogen challenge [54–56]. Progeny of Arabidopsis infected with P. syringae or treated with an SA analog displayed stronger induction of SA defense genes and enhanced resistance to P. syringae and the oomycete pathogen Hyaloperonospora arabidopsidis [54,55]. Furthermore, repeated pathogen challenge within a single generation increased the longevity of transgenerational resistance [54]. A greater mechanistic understanding of the interplay between defense priming, post-translational modifications, epigenetic changes and plant growth can be used to enhance disease resistance and minimize the growth penalty.
Conclusions
Plants represent excellent model systems to study NLR innate immune receptors. Recent evidence has revealed the structure of an NLR complex in various states of activation, demonstrating the formation of the first plant resistosome. Despite differences in defense timing and amplitude between innate immune receptor types, there is overlap in protein phosphorylation and early transcriptional responses. Future research focusing on how diverse NLR receptors induce cell death and resistance upon activation will significantly advance our understanding of this common protein family. Furthermore, given the impact of disease for agricultural production, a comprehensive understanding of NLR biology has significant translational applications for crop improvement.
Highlights:
Plant NLRs recognize intracellular effectors, inducing cell death and resistance
NLRs exhibit diverse localization and modes of effector recognition
The Arabidopsis ZAR1 NLR forms a pentameric resistosome complex upon activation
Abiotic stress and innate immunity induce overlapping transcriptional reprogramming
Acknowledgements
We thank members of the Coaker laboratory for fruitful discussions and critical reading of the manuscript.
Funding: This work was supported by the National Institutes of Health [grant number RO1 GM092772] and the United States Department of Agriculture [grant numbers 2019-70016-29796, 2016-70016-24833] to GC. S.L. was supported by the Independent research fund Denmark [grant number 7026-00053B].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and recommended reading
Papers of paritcular interest, published within the period of review, have been highlighted as:
•• of outstanding interest
- 1.Toruno TY, Stergiopoulos I, Coaker G: Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu Rev Phytopathol 2016, 54:419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang Y, Coaker G: Effector Triggered Immnity: NLR Immune Perception and Downstream Defense Responses In The Arabidopsis Book 11. Edited by: The American Society of Plant Biologists; 2015. [Google Scholar]
- 3.Couto D, Zipfel C: Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 2016, 16:537–552. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann M, Zeier J: N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr Opin Plant Biol 2019, 50:44–57. [DOI] [PubMed] [Google Scholar]
- 5.Peng Y, van Wersch R, Zhang Y: Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol Plant Microbe Interact 2018, 31:403–409. [DOI] [PubMed] [Google Scholar]
- 6.Giolai M, Paajanen P, Verweij W, Witek K, Jones JDG, Clark MD: Comparative analysis of targeted long read sequencing approaches for characterization of a plant’s immune receptor repertoire. BMC Genomics 2017, 18:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FL, et al. : Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 2012, 8:e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P: Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315:1098–1103. [DOI] [PubMed] [Google Scholar]
- 9.Gao Z, Chung EH, Eitas TK, Dangl JL: Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci U S A 2011, 108:7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL: Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 2000, 101:353–363. [DOI] [PubMed] [Google Scholar]
- 11.Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B: Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J 2017, 92:787–795. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Elmore JM, Lin ZJ, Coaker G: A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 2011, 9:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL: Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 2011, 9:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN, Winter SE, Kim A, Poon V, Ravesloot MM, Waldenmaier JF, et al. : Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature 2013, 496:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. : A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161:1089–1100. [DOI] [PubMed] [Google Scholar]
- 16.Le Roux C, Huet G, Jauneau A, Camborde L, Tremousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. : A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 2015, 161:1074–1088. [DOI] [PubMed] [Google Scholar]
- 17.Kroj T, Chanclud E, Michel-Romiti C, Grand X, Morel JB: Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol 2016, 210:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cesari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T: The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 2014, 33:1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study identified two rice NLRs in head-to-head genomic orientation that work together to recognize and respond to a fungal pathogen effector protein. The RGA5 NLR acts as a senstor, exhibits an integrated domain and directly binds the percieved AVR-Pia effector. The RGA4 NLR exhibits canonical domain architecture and is required for innate immune signaling.
- 19.Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S: NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci U S A 2017, 114:8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study identifies the NRC superclade that arose over 100 million years ago. Members in the NRC superclade, found in asterids and caryophyllales, act as helper NLRs that are required for the function of multiple sensor NLRs. Both the NRC family and NRC-dependent NLRs cluster phylogenetically and enable predictions of which NLRs require downstream helpers.
- 20.Li X, Kapos P, Zhang Y: NLRs in plants. Curr Opin Immunol 2015, 32:114–121. [DOI] [PubMed] [Google Scholar]
- 21.Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND: Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 1999, 20:317–332. [DOI] [PubMed] [Google Scholar]
- 22.Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL: Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 2006, 140:1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ: RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 1994, 265:1856–1860. [DOI] [PubMed] [Google Scholar]
- 24.Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ: The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 2002, 14:2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffett P, Farnham G, Peart J, Baulcombe DC: Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 2002, 21:4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takken FL, Albrecht M, Tameling WI: Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 2006, 9:383–390. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J: Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364: 6435 eaav5870. [DOI] [PubMed] [Google Scholar]; •• This study reconstituted an active plant NLR innate immune complex. The complex oligomerizes upon activation and cryo-electron microscopy revealed a pentameric resistasome complex. The N-terminal alpha-helix of the NLR ZAR1 exhibits a funnel-shaped structure upon resistasome formation.
- 28.Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, et al. : Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364: 6435 eaav5868. [DOI] [PubMed] [Google Scholar]; •• This study reports cryo-electron microscopy structures of a plant NLR innate immune complex in an inactive and intermediate state, revealing differences in domain partitioning between plant and animal NLRs. Pathogen effector modification of the PBL2 kinase recruits this kinase to the NLR complex, which initiates receptor activation.
- 29.Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud MF, Chabannes M, et al. : The Decoy Substrate of a Pathogen Effector and a Pseudokinase Specify Pathogen-Induced Modified-Self Recognition and Immunity in Plants. Cell Host Microbe 2015, 18:285–295. [DOI] [PubMed] [Google Scholar]
- 30.Los FC, Randis TM, Aroian RV, Ratner AJ: Role of pore-forming toxins in bacterial infectious diseases. Microbiol Mol Biol Rev 2013, 77:173–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai J, Shi Y: Apoptosome and inflammasome: conserved machineries for caspase activation. National Science Review 2014, 1:101–118. [Google Scholar]
- 32.Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE: Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 2005, 17:2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Century KS, Holub EB, Staskawicz BJ: NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci U S A 1995, 92:6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knepper C, Savory EA, Day B: Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol 2011, 156:286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC: NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 2005, 15:968–973. [DOI] [PubMed] [Google Scholar]
- 36.Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL: Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S 2011, 108:16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ: NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci U S A 2018, 115:E10979–e10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu CH, Kamoun W: Tomato Prf requires NLR helpers NRC2 and NRC3 to confer resistance against the bacterial speck pathogen Pseudomonas syringae pv. tomato. bioRxiv 2019. 595744 [Google Scholar]
- 39.Kadota Y, Shirasu K, Zipfel C: Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol 2015, 56:1472–1480. [DOI] [PubMed] [Google Scholar]
- 40.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. : Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 2014, 54:43–55. [DOI] [PubMed] [Google Scholar]
- 41.Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T: Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci U S A 2013, 110:8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Chiang YH, Toruno TY, Lee D, Ma M, Liang X, Lal NK, Lemos M, Lu YJ, Ma S, et al. : The MAP4 Kinase SIK1 Ensures Robust Extracellular ROS Burst and Antibacterial Immunity in Plants. Cell Host Microbe 2018, 24:379–391.e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. : The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 2014, 15:329–338. [DOI] [PubMed] [Google Scholar]
- 44.Kadota Y, Liebrand TWH, Goto Y, Sklenar J, Derbyshire P, Menke FLH, Torres MA, Molina A, Zipfel C, Coaker G, et al. : Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol 2019, 221:2160–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study investigated protein phosphorylation dynamics upon NLR activation, revealing novel phosphorylated targets and overlap in phosphorylated proteins during PRR activation. While phosphorylation occurred on similar residues of the NADPH oxidase, different kinases are involved based on receptor type.
- 45.Ng DW, Abeysinghe JK, Kamali M: Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int J Mol Sci 2018, 19: E3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacob F, Kracher B, Mine A, Seyfferth C, Blanvillain-Baufume S, Parker JE, Tsuda K, Schulze-Lefert P, Maekawa T: A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol 2018, 217:1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study compared the transcriptional response upon activation of plant innate immunity and abiotic stress responses from over 40 datasets. Abiotic stress and innate immune activation trigger a similar and rapid general stress response, whose transcriptional outputs then diverge at later timepoints depending on the initial stimulus.
- 47.Bjornson M, Benn G, Song X, Comai L, Franz AK, Dandekar AM, Drakakaki G, Dehesh K: Distinct roles for mitogen-activated protein kinase signaling and CALMODULIN-BINDING TRANSCRIPTIONAL ACTIVATOR3 in regulating the peak time and amplitude of the plant general stress response. Plant Physiol 2014, 166:988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benn G, Wang CQ, Hicks DR, Stein J, Guthrie C, Dehesh K: A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J 2014, 80:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong BQ, Guo J, Zhang N, Yao X, Wang HB, Li JF: Cross-Microbial Protection via Priming a Conserved Immune Co-Receptor through Juxtamembrane Phosphorylation in Plants. Cell Host Microbe 2019, 26:810–822 [DOI] [PubMed] [Google Scholar]
- 50.Ross AF: Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology 1961, 14:329–339. [DOI] [PubMed] [Google Scholar]
- 51.Bernsdorff F, Doring AC, Gruner K, Schuck S, Brautigam A, Zeier J: Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28:102–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riedlmeier M, Ghirardo A, Wenig M, Knappe C, Koch K, Georgii E, Dey S, Parker JE, Schnitzler JP, Vlot AC: Monoterpenes Support Systemic Acquired Resistance within and between Plants. Plant Cell 2017, 29:1440–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenig M, Ghirardo A, Sales JH, Pabst ES, Breitenbach HH, Antritter F, Weber B, Lange B, Lenk M, Cameron RK, et al. : Systemic acquired resistance networks amplify airborne defense cues. Nat Commun 2019, 10:3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luna E, Bruce TJ, Roberts MR, Flors V, Ton J: Next-generation systemic acquired resistance. Plant Physiol 2012, 158:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B: Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol 2012, 158:835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaskiewicz M, Conrath U, Peterhansel C: Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 2011, 12:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]


