CDK12 and CDK13 regulate POLII elongation rate and processivity and influence the selection of transcription termination sites.
Abstract
The RNA polymerase II (POLII)–driven transcription cycle is tightly regulated at distinct checkpoints by cyclin-dependent kinases (CDKs) and their cognate cyclins. The molecular events underpinning transcriptional elongation, processivity, and the CDK-cyclin pair(s) involved remain poorly understood. Using CRISPR-Cas9 homology-directed repair, we generated analog-sensitive kinase variants of CDK12 and CDK13 to probe their individual and shared biological and molecular roles. Single inhibition of CDK12 or CDK13 induced transcriptional responses associated with cellular growth signaling pathways and/or DNA damage, with minimal effects on cell viability. In contrast, dual kinase inhibition potently induced cell death, which was associated with extensive genome-wide transcriptional changes including widespread use of alternative 3′ polyadenylation sites. At the molecular level, dual kinase inhibition resulted in the loss of POLII CTD phosphorylation and greatly reduced POLII elongation rates and processivity. These data define substantial redundancy between CDK12 and CDK13 and identify both as fundamental regulators of global POLII processivity and transcription elongation.
INTRODUCTION
RNA polymerase II (POLII)–driven transcription consists of discrete checkpoints at the initiation, pausing, elongation, and termination stages of the transcription cycle, each of which is regulated by a dedicated set of cyclin-dependent kinases (CDKs) and their cognate cyclin. The concerted action of transcriptional CDK-cyclin complexes tightly controls both POLII transcriptional activity and cotranscriptional processes, including splicing and polyadenylation, which are critical for normal development and homeostasis and can promote disease initiation and progression when disrupted (1–3).
The regulation of POLII initiation and pause-release checkpoints by CDK7/cyclin H and CDK9/cyclin T, respectively, has been relatively well characterized (4, 5) and has spurred the development of multiple small molecules targeting these CDKs to correct cancer-associated transcriptional dysregulation (6–8). In contrast, the molecular events controlling POLII transcriptional elongation and processivity after pause-release remain poorly understood. CDK12 has been reported to control transcriptional elongation (9); however, unlike the genomes of early metazoans, which only have a single-cell ancestral ortholog of CDK12, in vertebrates evolution has given rise to a closely related gene, CDK13, which shares the same cognate cyclin, cyclin K (9, 10). CDK12 and CDK13 are thus evolutionarily related and structurally similar kinases, and biochemical assays have demonstrated that both have POLII C-terminal domain (CTD) kinase activity and the ability to phosphorylate the Ser2 residue of the repetitive CTD heptad sequence (9). Recent studies have demonstrated that CDK12 regulates POLII elongation, processivity, and alternative polyadenylation in a gene-selective manner, controlling the expression of long genes such as BRCA1 and ATR involved in the DNA damage response, thus explaining the BRCA-like phenotype observed in CDK12 mutant cancers (9–16). Ovarian and prostate cancers with functional mutations in CDK12 have been demonstrated to have a BRCAness phenotype with associated hypersensitivity to poly(adenosine diphosphate–ribose) polymerase (PARP) inhibitors and DNA cross-linking agents (11, 12, 17). In contrast, the role of CDK13 in transcription remains poorly understood but has been suggested to be gene selective and distinct from CDK12, despite their conserved domain structure and interaction partners (15, 18). Unlike CDK12, CDK13 mutations have not been reported in cancer; however, amplification of CDK13 was reported in hepatocellular carcinoma (HCC), where CDK13 copy number was significantly associated with clinical onset of HCC (19). Heterozygous mutations within the kinase domain of CDK13 are linked with a syndromic form of intellectual disability, a disease not associated with CDK12 mutations (20). Recently, therapeutic targeting of both CDK12 and CDK13 by the covalent inhibitor THZ531 has demonstrated potent efficacy in a variety of cancers through the induction of cell death and down-regulation of DNA damage response genes (21). These studies have principally attributed the therapeutic activity of THZ531 to the inhibition of CDK12 activity and the associated impaired DNA damage response, a hypothesis that is supported by the therapeutic synergy demonstrated between THZ531 and PARP inhibitors (22). Together, these studies highlight gene-selective and nonoverlapping roles for both CDK12 and CDK13 in the regulation of POLII-driven transcription, thus leaving a molecular understanding of the global regulation of POLII elongation and processivity unexplored.
Here, we used CRISPR-Cas9 homology-directed repair (HDR) to generate a unique set of cell lines comprising analog-sensitive (AS) CDK12, CDK13, or dual-edited alleles to probe the independent and shared roles of CDK12 and CDK13 using phenotypic, phosphoproteomic, and genome-wide transcriptional assays. Our results highlight that while individual roles for both CDK12 and CDK13 in the regulation of POLII transcription exist, only dual inhibition of CDK12 and CDK13 results in genome-wide transcriptional changes, widespread alternative polyadenylation, and loss of POLII processivity and CTD phosphorylation, which is associated with the induction of cell death and decreased proliferation. Together, our results demonstrate the significant redundancy between CDK12 and CDK13 and fundamentally define both factors as key regulators of global POLII processivity.
RESULTS
CDK12 and CDK13 play functionally redundant roles in maintaining cell proliferation and viability
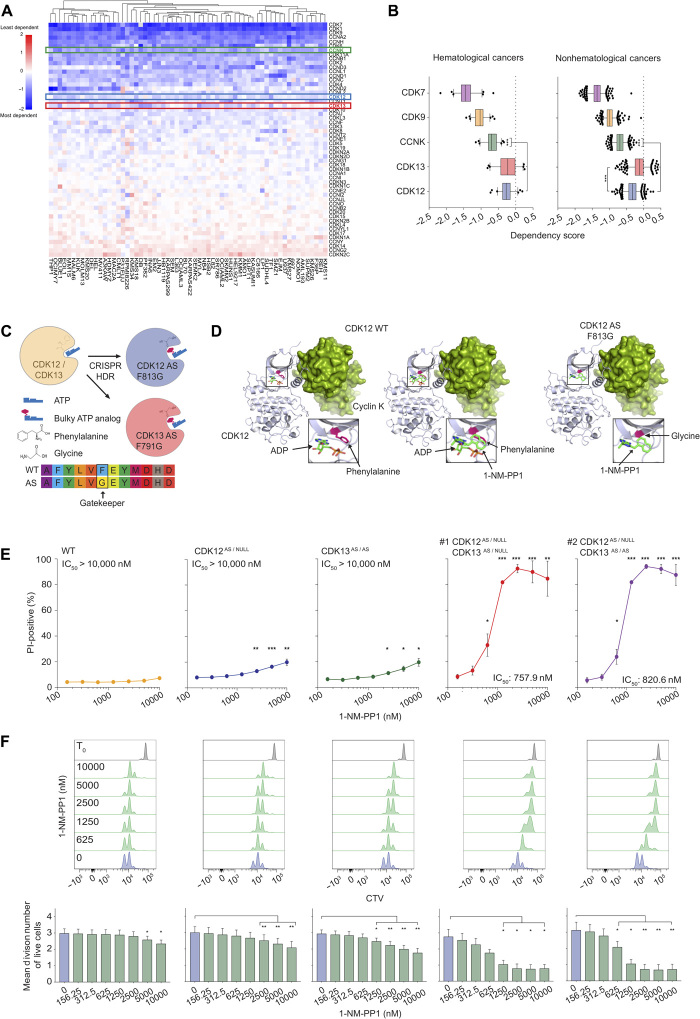
To determine the role of transcriptional CDKs and cyclins in maintaining the growth and survival of malignant cells of hematological origin, we mined the Cancer Dependency Map (DepMap) from the Broad Institute to identify essential CDKs, cyclins, and associated genes involved in transcription control across 62 hematological cancer cell lines from the Avana CRISPR public 19Q1 dataset. This analysis demonstrated the importance of CDK7 and CDK9 for the viability of all cell lines tested (Fig. 1, A and B). Targeted deletion of CDK12 or CDK13 revealed only a weak dependency on either kinase (Fig. 1A). In contrast cyclin K (CCNK), the cognate cyclin for CDK12 and CDK13, was identified as an essential gene across all 62 cells lines, with loss of cyclin K expression being significantly more detrimental than loss of either CDK12 or CDK13 (Fig. 1, A and B). The unfavorable effect of the loss of cyclin K expression was not restricted to hematological malignancies, with 496 solid cancer cell lines also demonstrating a reliance on this gene for their proliferation/survival (Fig. 1B and fig. S1, A and B). Similar to hematopoietic malignancies, deletion of CDK12 or CDK13 alone was not detrimental to many solid cancer cell lines tested (Fig. 1B and fig. S1A), and while the impact of CDK12 depletion was more potent in nonhematological tumor cells compared to CDK13 loss, ablation of cyclin K expression was once again significantly more detrimental to cell viability (Fig. 1B). This broader dependency of cancer cells on cyclin K expression over CDK12 or CDK13 alone is suggestive of potential redundant abilities of CDK12 and CDK13 to maintain cell viability.
Fig. 1. CDK12 and CDK13 are functionally redundant.
(A) Heat map of dependency scores for 55 CDKs and cyclins in 62 hematological cancer cell lines from the Avana CRISPR public 19Q1 dataset. (B) Box plot comparisons of dependency scores for cyclin K and CDK7, CDK9, CDK12, and CDK13 in 62 hematological and 496 nonhematological cancer cell lines. (C) Overview of CDK12 and CDK13 ATP AS mutant kinase generation through mutation of the gatekeeper residue. (D) Crystal structures of WT and AS CDK12 with 1-NM-PP1 interaction. (E) Propidium iodide (PI) incorporation assay of MV4;11 WT and AS clones treated with 1-NM-PP1 as indicated for 48 hours. (F) Representative CellTrace Violet dye (CTV) profiles and mean division number of PI-negative MV4;11 WT and AS clones treated with 1-NM-PP1 as indicated for 48 hours. (B) represents 62 hematological and 496 nonhematological cell lines. (E) and (F) represent the mean ± SEM of three independent experiments, and Student’s t tests were performed for (B), (E), and (F) (*P < 0.05, **P < 0.001, and ***P < 0.0001).
To determine whether pharmacological inhibition of CDK12 and/or CDK13 could phenocopy genetic depletion of these genes, we used CRISPR-mediated gene editing to develop a novel biological system expressing “analog-sensitive” mutant versions of CDK12 and CDK13 in MV4;11 mixed lineage leukemia (MLL)–rearranged acute myeloid leukemia (AML) cells (Fig. 1C and fig. S1, C and D). Mutation of the “gatekeeper” phenylalanine residue to a glycine expands the adenosine triphosphate (ATP)–binding pocket of CDK12 or CDK13, allowing binding of the inhibitory ATP analog 1-NM-PP1 (Fig. 1, C and D). Editing of CDK12 and CDK13 alleles did not affect their expression at the mRNA or protein levels (fig. S1, E and F). Wild-type (WT) MV4;11 cells and single-cell clones edited to only express mutant CDK12 (CDK12AS/NULL), mutant CDK13 (CDK13AS/AS), and two independent clones that express mutant alleles of both CDK12 and CDK13 (#1 CDK12AS/NULL;CDK13AS/NULL and #2 CDK12AS/NULL;CDK13AS/AS) were tested for sensitivity to the ATP analog 1-NM-PP1 (Fig. 1E and fig. S1G). WT clones showed little sensitivity to 1-NM-PP1, with concentrations of 5 μM and above exhibiting a minor impact on cell proliferation. The selective inhibition of CDK12 or CDK13 had only a marginal impact on cell survival even at relatively high concentrations of 1-NM-PP1 (Fig. 1E and fig. S1G). However, 1-NM-PP1 treatment of CDK12AS/NULL and CDK13AS/AS cells significantly impaired cell cycle progression, with CDK13 inhibition appearing to have a more robust effect on proliferation than CDK12 inhibition (Fig. 1, E and F, and fig. S1G). In contrast, combined inhibition of both CDK12 and CDK13 in two independent clones treated with 1-NM-PP1 resulted in a dose-dependent induction of cell death and inhibition of proliferation, with submicromolar IC50 (median inhibitory concentration) values observed for cell death (Fig. 1, E and F). These results showing the cell death–inducing effects of the dual, but not individual, inhibition of CDK12 and CDK13 were concordant with experiments using THZ531, an irreversible small-molecule inhibitor of CDK12 and CDK13 (21). THZ531 treatment of parental MV4;11 (fig. S1H) and THP1 cells (fig. S1I) resulted in a dose-dependent induction of cell death. Further analysis of MV4;11 cells treated with low nanomolar concentrations of THZ531 showed that this dual CDK12/CDK13 inhibitor could reduce proliferation of surviving cells (fig. S1J). Together, these data indicate that CDK12 and CDK13 regulate the survival and proliferation of MLL-rearranged AML cells, and using our novel series of isogenic cell lines expressing AS versions of CDK12 and/or CDK13, we unequivocally demonstrate that these enzymes exhibit significant functional redundancy for the maintenance of cell viability.
CDK12 and CDK13 coordinately regulate gene expression and proximal polyadenylation site usage
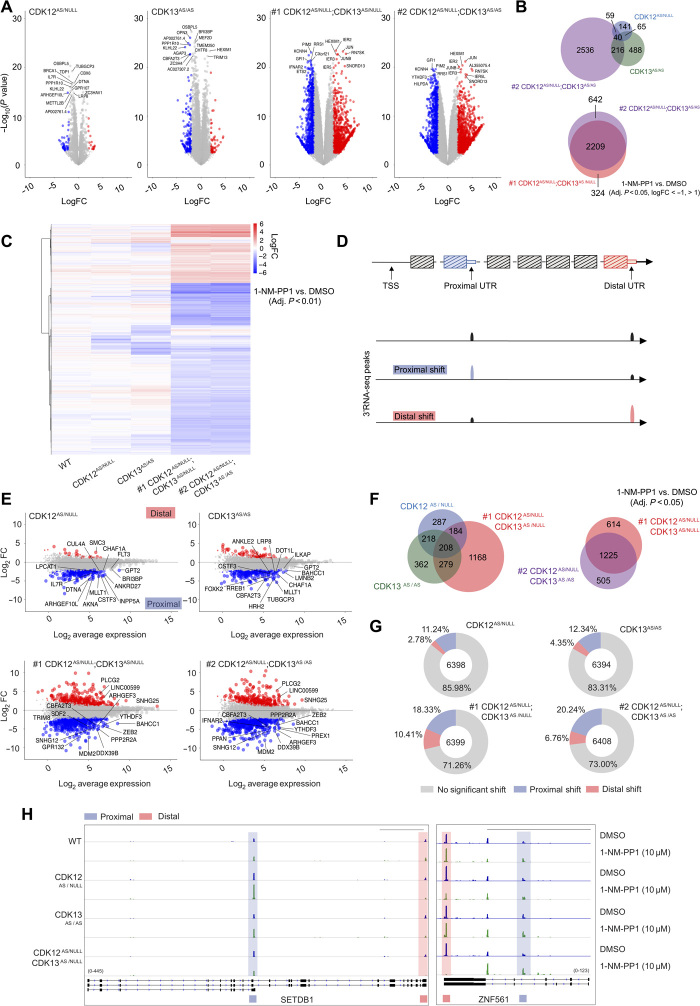
To determine whether the functional redundancy between CDK12 and CDK13 observed at the phenotypic level was reflected on the transcriptome level, we performed 3′ RNA sequencing (3′RNA-seq) (QuantSeq) on WT, CDK12AS/NULL, CDK13AS/AS, #1 CDK12AS/NULL;CDK13AS/NULL, and #2 CDK12AS/NULL;CDK13AS/AS MV4;11 clones treated with 1-NM-PP1 or vehicle for 4 hours. Differential gene expression analysis revealed that while 1-NM-PP1 had minimal effects on the WT MV4;11 clone (fig. S2A), variable transcriptional changes occurred following inhibition of CDK12 or CDK13 alone, with 305 and 809 genes detected as differentially expressed, respectively (Fig. 2, A to C). Hypergeometric analysis indicated that there was a significant overlap between differentially expressed genes following selective CDK12 and CDK13 inhibition, with the observed number of overlapping genes determined to be 6.6-fold higher than would be expected if CDK12 and CDK13 gene targets were completely independent (hypergeometric P value of 2.03 × 10−58). A global effect on approximately 15% of all expressed transcripts was observed following treatment of #1 CDK12AS/NULL;CDK13AS/NULL and #2 CDK12AS/NULL;CDK13AS/AS cells with 1-NM-PP1, with more than 2500 genes differentially expressed, the overwhelming majority of which (2209 genes) were shared by the two independent clones (Fig. 2, A to C). Consistent with previous studies (10, 15), inhibition of CDK12 resulted in altered expression of DNA damage response genes including BRCA1 as one of top significantly down-regulated genes (Fig. 2A), and analysis of differentially expressed protein-coding genes exclusive to selective inhibition of CDK12 demonstrated a strong DNA repair Gene Ontology (GO) signature (fig. S2B). Furthermore, GO analysis revealed that the gene set responsive exclusively to CDK13 inhibition was enriched for DNA repair–related terms, while genes associated with target of rapamycin (TOR)–dependent growth signaling were also differentially expressed (fig. S2B). Consistent with the 3′RNA-seq data, quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis revealed time-dependent down-regulation of BRCA1 following selective inhibition of CDK12 or CDK13, with greater down-regulation of BRCA1 expression observed following dual inhibition of CDK12 and CDK13 (fig. S2C). In line with the different GO signatures obtained following selective inhibition of CDK12 and CDK13, comparison of the genes differentially regulated following concomitant inhibition of the two kinases showed only fractional overlap with 315 overlapping genes from a total of 1114 (Fig. 2, B and C). GO analysis of the protein-coding genes responsive to 1-NM-PP1 in the CDK12AS/NULL;CDK13AS/NULL and CDK12AS/NULL;CDK13AS/AS cells showed that chromatin modification, DNA repair, and cell division pathways were the major biological programs affected (fig. S2B). In addition, gene set enrichment analysis following dual CDK12/CDK13 inhibition identified the ultraviolet (UV) radiation response signature, which has been previously linked to the regulation of transcriptional elongation by POLII (fig. S2D) (23). Treatment of parental MV4;11 cells with THZ531 resulted in altered gene expression, with unsupervised hierarchical clustering of the top 200 most variable genes demonstrating clustering of THZ531-treated parental MV4;11 cells with 1-NM-PP1–treated #1 CDK12AS/NULL;CDK13AS/NULL and #2 CDK12AS/NULL;CDK13AS/AS MV4;11 clones (fig. S2E).
Fig. 2. Dual CDK12 and CDK13 inhibition drives global disruption of transcriptional landscapes.
(A) Log2 fold change (FC) gene expression volcano plots for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours [relative to dimethyl sulfoxide (DMSO) treatment]. The top 15 differentially expressed genes [ranked by adjusted P value] are labeled. Genes exhibiting a log2FC of >2 (red) or <−2 (blue) with an adjusted P value of <0.01 are highlighted. (B) Venn diagram of the number of differentially expressed genes for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours (logFC < −1 and logFC > 1 and adjusted P < 0.05 relative to DMSO treatment). (C) Heat map of differentially expressed genes for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours (log2FC < −1 and log2FC > 1 and adjusted P < 0.001 relative to DMSO treatment). (D) Schematic of proximal versus distal UTR/polyadenylation sites. (E) MA plots representing proximal and distal 3′UTR peak shifts for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours. (F) Venn diagram of significant proximal and distal 3′UTR peak shift events for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours (adjusted P < 0.05 relative to DMSO treatment). (G) Proportion of genes with a significant change in proximal or distal 3′UTR peak signal for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours (relative to DMSO treatment). (H) Representative IGV profiles of differential 3′UTR peak usage for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours. Scale bars, 5 kb (SETDB1) and 10 kb (ZNF561). Data are representative of two biologically independent replicates.
The variable gene expression changes observed following inhibition of only CDK12 or CDK13 compared to the extensive changes seen following dual inhibition of both kinases demonstrates the redundant molecular activities of these proteins. This is concordant with the phenotypic changes observed following single and dual inhibition of CDK12 and CDK13 demonstrated in Fig. 1. These results also demonstrate that phenotypically and transcriptionally CDK13 is of comparable importance to CDK12. Furthermore, CDK13 also regulated growth signaling pathways when inhibited selectively, consistent with a previously reported role of CDK13 (15, 18), and demonstrating additional roles for CDK13 beyond shared regulation of DNA damage–associated pathways with CDK12 (10, 13, 14).
Given the reported interaction between CDK12 and CDK13 with RNA splicing factors (24), we treated parental MV4;11 cells with 100 and 200 nM THZ531 for 6 hours and performed total RNA-seq. Using Mixture Isoform (MISO) analysis (25) to detect differential splicing events, 711 and 1084 alternative splicing events were identified in parental MV4;11 cells treated with 100 and 200 nM THZ531, respectively (fig. S3A). Most of the altered mRNA processing events identified following treatment with THZ531 were classified as alternative last exon (ALE) events, a phenomenon that can be caused by alternative polyadenylation (APA) (26). As an example, short transcript variants of ASCC3 were detected (fig. S3B), indicating that proximal polyadenylation sites were being used preferentially after THZ531 treatment. To confirm this finding, we performed 3′RNA-seq (fig. S3B) and RT-PCR (fig. S3C) using cells treated for 6 hours with 100 and 200 nM THZ531. Consistent with the total RNA-seq analysis, THZ531 again demonstrated increased proximal polyadenylation site usage in the ASCC3 gene (fig. S3, B and C).
Having devised a 3′RNA-seq–based method to identify alternative polyadenylation usage, we treated WT, CDK12AS/NULL, CDK13AS/AS, and CDK12AS/NULL;CDK13AS/AS cells with 1-NM-PP-1 for 4 hours and used pairwise peak analysis of our 3′RNA-seq data to analyze both proximal and distal shifts in polyadenylation signals (Fig. 2, D and E, and fig. S3D). Two main peaks were defined for each gene with multiple 3′RNA-seq peaks, and the ratio between the distal and proximal peaks was calculated. A negative logFC (fold change) value indicated that the proximal peak was preferentially used in comparison to the vehicle-treated control, while a positive logFC value was indicative of preferential utilization of a distal polyadenylation site (Fig. 2D). As a specificity control, very few peak shifts were observed following treatment of the WT MV4;11 clone with 1-NM-PP1 (fig. S3D). Following treatment of CDK12AS/NULL and CDK13AS/AS cells with 1-NM-PP1, 897 and 1067 significant peak shifts were detected, respectively (Fig. 2, E and F). A greater number of APA events were detected in CDK12AS/NULL;CDK13AS/NULL and CDK12AS/NULL;CDK13AS/AS cells treated with 1-NM-PP1, with more than 1700 peak shifts detected in each individual clone (Fig. 2, E and F). Similar to the large degree of overlap for differentially expressed genes (Fig. 2, B and C), there was very strong concordance of genes with peak shift changes between the two dual CDK12/CDK13 AS clones (Fig. 2F). While there was a considerable number of shared peak shifts following selective inhibition of CDK12 or CDK13 compared to dual inhibition, there were a large number of unique events detected following single inhibition indicating that CDK12 and, to a greater extent, CDK13 have independent roles for the regulation of polyadenylation site usage (Fig. 2F). When assessing the overall effect of single or dual inhibition of CDK12 and CDK13, it was apparent that the dominant effect was a proximal peak shift consistent with alternative polyadenylation site usage and early termination of POLII (Fig. 2G). The dominance of proximal peak shifts was also observed in parental MV4;11 cells treated with THZ531 (fig. S3, E and F). This is exemplified by analysis of SETDB1 and ZNF561 that both demonstrate increased proximal peak and decreased distal peak changes following dual inhibition of CDK12 and CDK13 (Fig. 2H). Proximal peak shifts observed in NACC2 and MTHFSD are also examples of APA events induced by selective inhibition of CDK12 or CDK13, respectively, as well as dual inhibition of both kinases (fig. S3G). Together, these results demonstrate that both CDK12 and CDK13 influence 3′ polyadenylation site usage, with inhibition of CDK12 and CDK13 resulting in ALE events and preferential usage of more transcription start site (TSS)–proximal polyadenylation sites. While both CDK12 and CDK13 were capable of regulating this process, the profound effect of dual inhibition of both CDK12 and CDK13 demonstrated substantial functional redundancy for these kinases, suggesting that, under homeostatic conditions, both kinases act in a cooperative manner to prevent early termination at TSS-proximal polyadenylation sites.
CDK12 and CDK13 cooperate to modulate POLII CTD phosphorylation and POLII activity/processivity
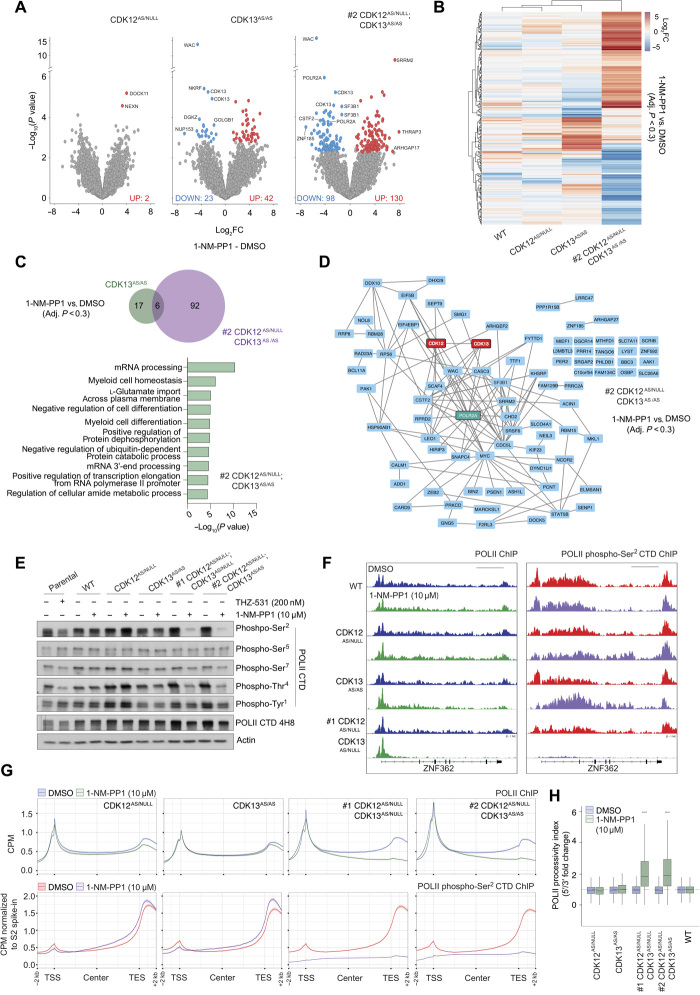
To investigate whether the phenotypic and transcriptional redundancy is reflected on the biochemical level and to identify putative direct or indirect phosphotargets for CDK12 and CDK13, we performed global phosphopeptide mass spectrometry (MS) on CDK12AS/NULL, CDK13AS/AS, and CDK12AS/NULL;CDK13AS/AS cells treated with 1-NM-PP1 for 2 hours. The abundance of only a limited number of phosphorylation sites exhibited changes following selective inhibition of CDK12, while a greater number of phosphorylation sites exhibited increased (42 phosphosites) or decreased (23 phosphosites) abundance in response to CDK13 inhibition, including CDK13 itself as a potential autosubstrate (Fig. 3A and table S1). Similar to the increased impact of dual compared to single kinase inhibition at the transcriptomic level (Fig. 2), dual CDK12 and CDK13 inhibition identified 228 phosphorylation sites that exhibited altered abundance including 98 sites with decreased and 130 sites with increased phosphorylation (Fig. 3, A and B). There was limited overlap of proteins exhibiting sites with differential phosphorylation as a result of selective CDK12 or CDK13 inhibition compared to dual inhibition, with only 6 of 98 proteins exhibiting decreased phosphorylation shared between CDK13AS/AS and CDK12AS/NULL;CDK13AS/AS cells (Fig. 3, B and C). These data suggest that dual inhibition of CDK12 and CDK13 is required to reveal the full impact of the loss of cyclin K–containing complexes on the phosphoproteome, indicating that, under selective inhibition of CDK12 or CDK13, there is significant compensation by the uninhibited kinase, resulting in a minimal impact on the phosphoproteome under these conditions. More differentially phosphorylated peptides were identified following selective inhibition of CDK13 compared to CDK12. GO and STRING analysis of proteins displaying decreased phosphorylation following dual CDK12 and CDK13 inhibition identified modulators of POLII elongation and multiple regulators of pre-mRNA processing (Fig. 3, C and D), which is consistent with the demonstrated ability of CDK12 and CDK13 activity to modulate polyadenylation site usage (Fig. 2). POLII (POLR2A) was identified as a putative phosphosubstrate for CDK12 and CDK13, with the Ser2 and Ser5 residues within the POLII CTD heptad repeat identified as candidate phosphosites (Fig. 3, A and D). Western blot analysis using selective antibodies for phosphorylated Tyr1, Ser2, Thr4, Ser5, and Ser7 within the POLII CTD demonstrated that selective inhibition of CDK12 or CDK13 following treatment with 1-NM-PP1 for 4 hours did not alter the phosphorylation status at any of these residues (Fig. 3E and fig. S4A). In contrast, dual inhibition of CDK12 and CDK13 in #1 CDK12AS/NULL;CDK13AS/NULL and #2 CDK12AS/NULL;CDK13AS/AS cells significantly decreased the abundance of phospho-Ser2 POLII and phospho-Thr4 POLII (Fig. 3E and fig. S4A). Reduced phosphorylation of the Tyr1 and Ser7 residues was also observed following concomitant inhibition of CDK12 and CDK13, while the phosphorylation of Ser5 was relatively unaffected (Fig. 3E and fig. S4A). The impact of dual CDK12 and CDK13 inhibition on POLII CTD phosphorylation mirrored the effect of THZ531 treatment of parental MV4;11 cells, with the most marked effects observed on Ser2 and Thr4 phosphorylation (Fig. 3E and fig. S4A). The impact of dual CDK12 and CDK13 inhibition appeared to be most profound against the Ser2 and Thr4 residues, with only minimal effects observed using a total phospho-CTD antibody (4H8; Fig. 3E). These results demonstrate that CDK12 and CDK13 share most of their substrates including critical regulators of RNA processing and POLII, suggesting that the loss of phosphorylation of these proteins is responsible for the observed transcriptional and phenotypic consequences observed upon dual CDK12 and CDK13 inhibition.
Fig. 3. CDK12 and CDK13 redundantly control POLII CTD phosphorylation to control processivity.
(A) Log2FC volcano plots of differentially phosphorylated peptides in MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 2 hours (relative to DMSO treatment). Highlighted points represent proteins with an adjusted P value of <0.3 relative to DMSO treatment, and the number of increased (UP) and decreased (DOWN) phosphopeptides is highlighted. (B) Heat map of differentially phosphorylated peptides for MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 2 hours (adjusted P < 0.3 relative to DMSO treatment). (C) Venn diagram, GO analysis, and (D) STRING interaction network for proteins with significantly decreased phosphopeptides in MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 2 hours (adjusted P < 0.3 relative to DMSO condition). (E) Western blot analysis of MV4;11 parental cells and MV4;11 WT and AS clones treated as indicated with 200 nM THZ531 (6 hours) or 10 μM 1-NM-PP1 (4 hours). (F) Representative IGV images and (G) average gene profiles of 9552 expressed genes for total and phospho-Ser2 POLII ChIP-seq analysis of MV4;11 AS clones treated with 10 μM 1-NM-PP1 for 4 hours. Scale bars, 10 kb. (H) POLII processivity index (FC ratio of the 5′/3′ POLII signal ratio in the gene body) for MV4;11 WT and AS clones treated with 10 μM 1-NM-PP1 for 4 hours. Western blots are representative of three independent experiments. (H) is representative of 9552 expressed genes, and Student t test was performed (****P < 0.001).
To assess the relationship between the transcriptional roles of CDK12 and CDK13 and the potential regulation of POLII function by CDK12-CDK13–dependent phosphorylation, we performed total and phospho-Ser2 POLII chromatin immunoprecipitation sequencing (ChIP-seq) using the AS CDK12/CDK13 system. Representative Integrative Genomics Viewer (IGV) profiles of the ZNF362 locus (Fig. 3F) and metagene profiles of 9552 expressed genes (Fig. 3G and fig. S4B) demonstrate a minimal impact of 1-NM-PP1 treatment on total or phospho-POLII occupancy in WT, CDK12AS/NULL, or CDK13AS/AS cells (Fig. 3G and fig. S4B), while 1-NM-PP1 treatment of #1 CDK12AS/NULL;CDK13AS/NULL and #2 CDK12AS/NULL;CDK13AS/AS cells resulted in reduced total POLII occupancy toward the 3′ end of the gene body and the transcription end site (TES), as well as the near-complete loss of phospho-POLII occupancy (Fig. 3, F and G, and fig. S4C). The differential impact of selective versus dual CDK12 or CDK13 inhibition is further illustrated by the calculation of a POLII processivity index based on the fold-change ratio of the total POLII ChIP signal in the 5′ versus the 3′ regions of the gene body, excluding the TSS and the TES (Fig. 3H and fig. S4D). Inhibition of CDK12 or CDK13 had no substantial effect on the POLII processivity index. In contrast, concomitant inhibition of both kinases resulted in a significant change in the POLII processivity index (Fig. 3H). The impact of CDK12 inhibition on POLII activity and genome occupancy has been demonstrated previously to be more potent at longer (>64.5 kb) genes (14). By subsetting genes based on gene-length quartiles, analysis of the total POLII ChIP-seq data generated in Fig. 3G revealed that there was an increased impact of dual, but not single, CDK12 and CDK13 inhibition on POLII occupancy at longer genes compared to shorter genes (fig. S4E). Specifically, there was a robust change in the calculated POLII processivity index for genes in length quartiles 2, 3, and 4, but not quartile 1 (shortest genes), in #1 CDK12AS/NULL;CDK13AS/NULL and #2 CDK12AS/NULL;CDK13AS/AS cells treated with 1-NM-PP1 (fig. S4E). The impact of dual, but not selective, CDK12 and CDK13 inhibition on total and phospho-POLII occupancy across the genome mirrored that observed following treatment of parental MV4;11 cells with THZ531, with a significant dose-dependent loss of total POLII occupancy across the 3′ and TES regions of gene bodies, as well as a dose-dependent loss of phospho-POLII signal across the genome (fig. S4, F to H). Precision nuclear run-on sequencing (PRO-seq) analysis was also performed to assess the impact of THZ531 treatment on actively transcribing POLII in parental MV4;11 cells, the results demonstrating a significant dose-dependent reduction of active POLII signal from 3′ gene body regions (fig. S4, H and I). Together, these studies demonstrate the important, yet largely redundant, role for CDK12 and CDK13 for the regulation of POLII CTD phosphorylation at multiple residues, as well as the global role for both of these kinases in regulating POLII occupancy across the genome.
Perturbation of POLII transcription cycle at the elongation checkpoint by dual CDK12 and CDK13 inhibition
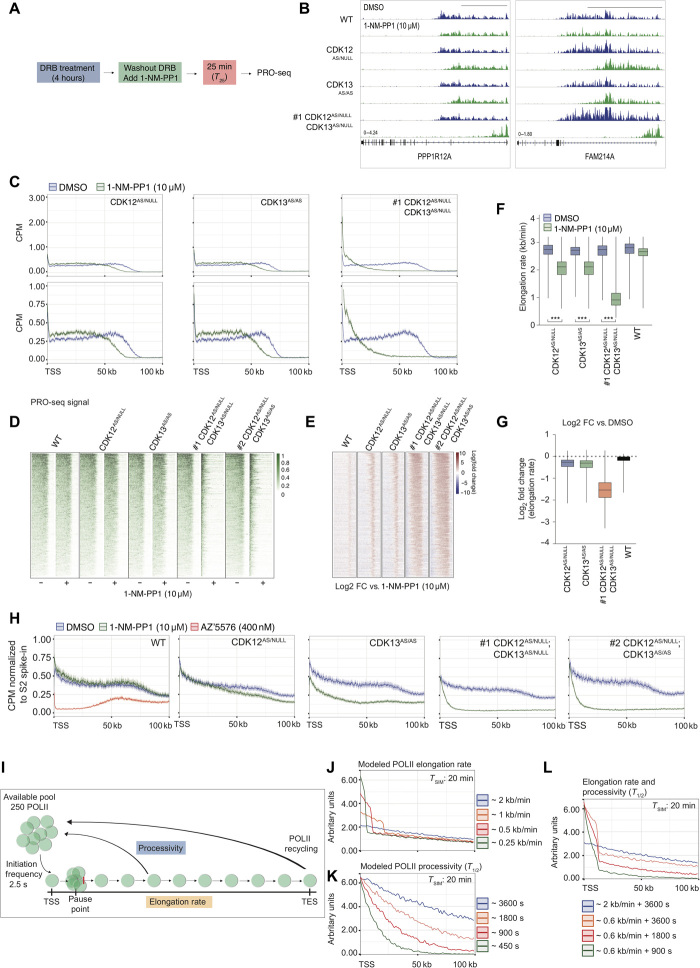
The gradual loss of POLII occupancy toward the 3′ end of the gene body following dual inhibition of CDK12 and CDK13 could be indicative of a global processivity or elongation defect. To investigate whether CDK12 and/or CDK13 affect POLII elongation rates directly, we performed 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) release assays coupled with PRO-seq. In these assays, DRB was used to induce global, synchronized pausing of POLII at the promoter-proximal region across all cells (Fig. 4A and fig. S5, A and B). Following the washout of DRB, the ability of newly released POLII to actively elongate in the presence or absence of 1-NM-PP1 or THZ531 was measured using PRO-seq in WT, CDK12AS/NULL, CDK13AS/AS, # 1CDK12AS/NULL;CDK13AS/NULL, and # 2CDK12AS/NULL;CDK13AS/AS MV4;11 clones or parental MV4;11 cells, respectively. IGV profiles of the PRO-seq signal, which represents the distance traveled by POLII following DRB washout, across the PPP1R12A and FAM214A loci demonstrate that while 1-NM-PP1 treatment had minimal effect on POLII elongation in WT clones, individual and combined inhibition of CDK12 and CDK13 impaired POLII elongation (Fig. 4B and fig. S5B). Metagene analysis of 368 “long” expressed genes (>100 kb) demonstrated that CDK12 (CDK12AS/NULL) or CDK13 (CDK13AS/AS) inhibition impaired POLII elongation, with a robust decrease in the PRO-seq signal observed beyond 50 kb of the TSS, while dual CDK12/13 inhibition resulted in a profound inhibition of POLII elongation, with decreased PRO-seq signal observed within 25 kb of the TSS (Fig. 4C and fig. S5C). This demonstrated the reduced capacity of POLII to actively transcribe through TSS-proximal regions following release from DRB-induced pausing in the absence of CDK12 and CDK13 kinase activity. Occupancy heat maps depicting the normalized and log fold-change [dimethyl sulfoxide (DMSO) versus 1-NM-PP1] PRO-seq signal demonstrate that single and combined inhibition of CDK12 and CDK13 had a global effect on POLII elongation; however, there was some variability at the individual gene level (Fig. 4, D and E). The negative impact of combined CDK12 and CDK13 inhibition on POLII transcription was further demonstrated in parental MV4;11 cells, where THZ531 treatment induced a dose- and time-dependent decrease in POLII elongation following the removal of DRB (fig. S5, A, B, and D to F). To quantify the effect of CDK12/CDK13 inhibition on POLII elongation, the PRO-seq signal wavefront at 25-min post-DRB washout was measured to estimate the elongation rate (kb/min) for each gene. Single inhibition of CDK12 or CDK13 significantly decreased the POLII elongation rate; however, a much greater inhibition of the POLII elongation rate was observed in response to dual CDK12 and CDK13 inhibition, which was reduced by more than 60% of the normal rate [DMSO (2.96 kb/min) versus 1-NM-PP1 (1.11 kb/min)] (Fig. 4, F and G, and fig. S5, G and H). THZ531 also resulted in a dose-dependent decrease in the calculated POLII elongation rate, which was reduced by approximately 50% following treatment with 200 nM THZ531 (fig. S5, I and J). These results demonstrate that CDK12 and CDK13 both have independent capacities to regulate POLII elongation, yet consistent with results observed at the phenotypic, transcriptomic, and phosphoproteomic levels, CDK12 and CDK13 maintain some redundancy for the control of POLII elongation.
Fig. 4. Perturbation of POLII transcription cycle at the elongation checkpoint by dual CDK12 and CDK13 inhibition.
(A) Schematic overview of DRB-release PRO-seq assay for (B) to (G). (B) Representative IGV images for DRB-release PRO-seq of MV4;11 WT and AS clones treated with 10 μM 1-NM-PP1. Scale bars, 50 kb. (C) Average gene profiles for DRB-release PRO-seq analysis of 368 genes in MV4;11 AS clones treated with 10 μM 1-NM-PP1. Heat map analysis of (D) PRO-seq signal and (E) log2FC PRO-seq signal (DMSO relative to 1-NM-PP1) in MV4;11 WT and AS clones treated with 10 μM 1-NM-PP1. (F) POLII elongation rate and (G) log2FC in POLII elongation rate (relative to DMSO) in MV4;11 WT and AS clones treated with 10 μM 1-NM-PP1. (H) Average gene profiles for TT-seq analysis of 368 genes normalized to Drosophila S2 RNA spike-in in MV4;11 WT and AS clones treated as indicated for 25 min. (I) Schematic overview of POLII elongation rate and processivity computational modeling parameters. Computational modeling of simulated defects in POLII (J) elongation rates, (K) processivity half-life (T1/2), and (L) combined elongation and processivity. (F) and (G) are representative of 368 genes, and Student’s t test was performed for (F) (***P < 0.001).
To assess whether the reduced elongation rate alone could be causative for the widespread alternative polyadenylation detected upon dual CDK12 and CDK13 inhibition, we studied the impact of decreased POLII elongation rate on the utilization of proximal polyadenylation sites. To this end, we used human embryonic kidney (HEK) 293 (HEK293) cells expressing two independent doxycycline-inducible, α-amanitin–resistant (Amr) Rbp1 mutant alleles (R749H and H1108Y) that result in slow rates of POLII elongation (27). Using 3′RNA-seq analysis to detect APA events (as detailed in Fig. 2), we measured the number of genes exhibiting a significant change in either proximal or distal 3′ untranslated region (3′UTR) peak signal following induction of slow elongation mutant compared to WT Amr POLII in α-amanitin–treated HEK293 cells (fig. S5K). Comparison of these datasets to HEK293T cells treated with THZ531 showed that cells expressing Rbp1 mutant alleles demonstrated a comparable number of significant 3′UTR peak signal shifts indicative of APA (R749H, 2409; H1108Y, 2437) to that observed with THZ531 (2197 events) (fig. S5K). Approximately half (1047 of 2197; 47.7%) of the genes, where APA site usage was detected following THZ531 treatment of HEK293T cells, overlapped with genes exhibiting APA in R489H- and H1108Y-Rpb1 mutant cells, suggesting that inhibition of CDK12 and CDK13 may modulate polyadenylation site usage via other phosphosubstrates (Fig. 3D and fig. S5K). Consistent with the impact of CDK12 and CDK13 inhibition in MV4;11 cells (Fig. 2D), a predominance of proximal rather than distal APA events was observed in HEK293 cells expressing Rbp1 mutant alleles and THZ531-treated HEK293T cells (fig. S5L). These results indicate that reductions in the POLII elongation rate alone are sufficient to promote APA events and demonstrate the importance of regulators of POLII elongation rates for determining polyadenylation site usage.
Although DRB-release PRO-seq experiments revealed a reduction in the POLII elongation rate, the decreased travel of POLII after pause-release could also be the result of a global loss of POLII processivity. To further dissect functional links between altered transcriptional elongation and processivity, we performed transient transcriptome sequencing (TT-seq) to dynamically measure nascent RNA production and compare this directly with an in silico simulation-based analysis of POLII dynamics upon either perturbed elongation or processivity (Fig. 4I and fig. S5M). TT-seq was performed on WT, CDK12AS/NULL, CDK13AS/AS, CDK12AS/NULL;CDK13AS/NULL, and CDK12AS/NULL;CDK13AS/AS MV4;11 cells treated with 1-NM-PP1 for 25 min. As a control for transcriptional pausing at the pause-release checkpoint, the WT MV4;11 clone was also treated with the small-molecule CDK9 inhibitor, AZ′5576 (28). Metagene analysis of 368 long genes demonstrated that 1-NM-PP1 treatment did not affect nascent RNA production in the WT MV4;11 clone, while single inhibition of CDK12 or CDK13 resulted in a decrease in nascent RNA production, with a more profound effect observed following inhibition of CDK13 compared to loss of CDK12 kinase activity (Fig. 4H). Dual inhibition of CDK12 and CDK13 resulted in a substantial reduction in RNA production after the TSS, with a far greater magnitude of change observed following concomitant inhibition of CDK12 and CDK13 compared to single inhibition of either kinase, once again highlighting a level of functional redundancy that exists between these transcriptional kinases (Fig. 4H). In comparison to AZ′5576-mediated CDK9 inhibition in WT MV4;11 cells, disruption of CDK12 or CDK13 activity did not impair transcription at TSS, suggesting that theses kinases regulate POLII elongation or processivity downstream of the pause-release checkpoint (Fig. 4H). The impact of CDK12 and CDK13 inhibition on nascent RNA synthesis (TT-seq) and POLII elongation rates (DRB–PRO-seq) is consistent with the observed loss of POLII occupancy across 3′ regions of gene bodies (Fig. 3, G and H), with these results suggesting that both POLII elongation rate and processivity are regulated by CDK12 and CDK13 kinase activity.
To test whether the TT-seq profiles obtained following dual inhibition of CDK12 and CDK13 are a result of impaired POLII elongation and/or processivity, we used mathematical modeling (Fig. 4I). Active transcription profiles equivalent to TT-seq were simulated in three scenarios: (i) decreased POLII elongation rate (kb/min; Fig. 4, I and J), (ii) decreased POLII processivity [half-life (T1/2) of POLII occupancy across the transcriptional unit; Fig. 4K], and (iii) a combined decrease in POLII elongation rate and processivity (Fig. 4L). In silico profiles were then captured in steady-state (“untreated”) conditions and 20 min after the infliction of the corresponding parameter change to closely match experimental TT-seq data (1-NM-PP1 treatment for 25 min; Fig. 4H). Modeling of decreased elongation rate alone results in a sharp, TSS-proximal peak of active transcription, with only a minor reduction of active transcription in the remaining TSS-distal part of the transcription unit (Fig. 4J). In comparison, reducing POLII processivity alone gave rise to a gradual loss of active transcription signal across the entire gene (Fig. 4K). Experimental TT-seq data derived from individual inhibition of CDK12 or CDK13 (Fig. 4H) qualitatively resembled simulated defects in both POLII elongation and processivity (Fig. 4, J and K), with dual inhibition of CDK12 and CDK13 resulting in TT-seq profiles that most closely mimicked the simulation of a combined reduction of both POLII elongation rate and processivity (Fig. 4, H and L). In response to dual inhibition, transcription was substantially affected throughout the entire transcriptional unit with a sharp decrease in the TSS-proximal transcription signal followed by a reduction of active transcription throughout the remainder of the gene body. Together, these observations suggest that CDK12 and CDK13 not only have an important role in the maintenance of POLII elongation rate but also control POLII processivity, with the combined inhibition of these kinases resulting in the inability of POLII to transcribe through to the 3′TES site, as observed in the POLII ChIP-seq (Fig. 3G), PRO-seq (Fig. 4, B and C), and TT-seq data (Fig. 4H). In combination with the observation that dual CDK12 and CDK13 inhibition results in widespread proximal APA events, our data demonstrate that, under normal conditions, CDK12 and CDK13 control the phosphorylation of the POLII CTD and RNA processing factors to control elongation and prevent early termination at proximal polyadenylation signals. This study thus places CDK12 and CDK13 downstream from CDK7 and CDK9 in the global POLII transcription cycle and fundamentally demonstrates that the activity of both factors is required for proper transcription after pause-release.
DISCUSSION
POLII-driven transcription is tightly controlled by CDK-cyclin pairs at distinct checkpoints along the gene body. The initiation and pause-release checkpoints regulated by CDK7/cyclin H and CDK9/cyclin T, respectively, have been relatively well characterized; however, the regulation of POLII-dependent transcriptional processes beyond these initial stages remains poorly understood. Here, we demonstrate that CDK12 and CDK13 regulate POLII elongation rate and processivity and influence the selection of transcription termination sites. Dual, but not single, inhibition of CDK12 and CDK13 resulted in the loss of POLII CTD phosphorylation, concurrent with global POLII elongation and processivity defects and the widespread induction of alternative polyadenylation events. These findings uncover that, under homeostatic conditions, CDK12 and CDK13 activity is fundamentally required to prevent TSS-proximal early termination as POLII elongates along the gene body. A number of recent studies have described gene-selective and nonoverlapping roles for CDK12 and CDK13 in transcription, with genetic CDK12 depletion or selective CDK12 kinase inhibition resulting in aberrant transcription of DNA damage response genes and CDK13 regulating a distinct set of genes (13–15, 29). Transcriptional defects induced by CDK12 perturbation were associated with premature termination at intronic polyadenylation sites and an apparent decrease in POLII elongation at DNA damage response genes (13, 14). Our data are concordant with this, as transcriptional responses following targeted CDK12 inhibition were associated with altered expression of DNA damage response genes. We observed that selective inhibition of AS CDK12 or AS CDK13 moderately affected global POLII elongation and processivity. However, complete redundancy between CDK12 and CDK13 was not apparent, as aberrant transcriptional responses including alternative polyadenylation events reduced POLII elongation rates and impaired processivity did arise upon selective inhibition of CDK12 or CDK13. The magnitude of the phenotypic, transcriptional, and phosphoregulatory aberrations induced by selective CDK13 inhibition was more profound than the response achieved by targeting CDK12 alone, highlighting an important independent role for CDK13 kinase activity for cancer cell viability and regulation of gene expression. The molecular underpinnings of CDK12- and CDK13-specific transcriptional responses remain to be determined but may be the result of selective recruitment of either kinase to particular genomic loci, thus skewing the stoichiometry of cyclin K containing complexes in a locus-specific manner. Although CDK12 and CDK13 domain composition is highly conserved and both kinases share a large degree of their interactomes (9), it is possible that selective recruitment may be mediated by CDK12- or CDK13-specific interaction partners.
The gene-selective effects of CDK12 and CDK13 may also arise due to the activity of nonoverlapping kinase substrates of CDK12 and CDK13. Our results demonstrate that CDK12 and CDK13 are regulators of POLII CTD phosphorylation and thus strongly suggest that the loss of CTD phosphorylation upon their inhibition is, at least in part, causative of the POLII elongation and processivity defects observed in this study. This is supported by the observation that demonstrates that the accumulation of Ser2 CTD phosphorylation along the gene body is associated with the POLII elongation rate, phase separation, and cotranscriptional effects (30–32). In addition to their direct effect on POLII, the importance of distinct CDK12 and CDK13 kinase substrates, such as SF3B1 and SRRM2, for regulation of POLII processivity and elongation remains to be determined. Chronic lymphocytic leukemias with mutant SF3B1 exhibit a significantly elevated number of intronic polyadenylation events compared to SF3B1 WT counterparts, suggesting that CDK12/CDK13-dependent phosphorylation of SF3B1 may be involved in the regulation of alternative polyadenylation in addition to its normal role in splicing (33).
Although our work defines the molecular roles of CDK12 and CDK13 in POLII transcription, it remains to be determined how the activity of CDK12 and CDK13 is controlled under normal homeostasis or in circumstances of cellular stress. Saccharomyces cerevisiae CAK1 has the ability to phosphorylate human CDK12 on a regulatory site within the T-loop (34), suggesting that human CDK7/CAK activity may control CDK12 and CDK13 activity and highlighting putative molecular interplay between CDKs that regulate different stages of the transcription cycle. Intriguingly, the transcriptional consequences of dual CDK12/CDK13 inhibition are highly reminiscent of those induced upon cellular exposure to UV light. Similar to CDK12/CDK13 inhibition, UV exposure induces widespread loss of POLII elongation/processivity associated with the induction of alternative polyadenylation, affecting genes such as HERC4 and ASCC3, which was highlighted in our study as a THZ531-sensitive gene (23). In addition, decreased POLII elongation rates in response to UV damage have been suggested to promote increased utilization of proximal polyadenylation sites, further linking POLII elongation and processivity to the regulation of transcriptional termination (23, 35). In agreement with this, we observed that the induced expression of slow elongation POLII mutants was sufficient to induce proximal alternative polyadenylation events, demonstrating the importance of maintained POLII elongation rates for the prevention of premature transcription termination and indicating that the concordance between observed CDK12-, CDK13-, and UV-dependent responses may be the result of their similar effects on POLII elongation rates.
Our findings have important implications in the context of tumorigenesis and cancer therapy. In contrast to CDK12, few genetic aberrations in CDK13 have been documented in the context of cancer. Down-regulation of DNA damage response gene expression as a key feature of CDK12 loss has been suggested to be fundamental to its tumor-suppressive capacity (36). The essentiality of cyclin K and the functional redundancy existing between CDK12 and CDK13 for cell survival, demonstrated by our study and pan-cancer cell line dependency data, suggests that while biallelic loss-of-function mutations in cyclin K are unlikely, it is possible that partial loss of cyclin K through either monoallelic loss-of-function or dominant-negative mutations may phenocopy CDK12 loss and impact DNA damage responses. The important function of CDK12 in the DNA damage response has spurred the development of CDK12 targeting agents such as THZ531, which has demonstrated therapeutic potential by potently inducing cell death in a variety of tumor types. THZ531 targets both CDK12 and CDK13 with similar efficacy, and we demonstrate that the therapeutic activities of this compound can therefore not be solely attributed to CDK12 inhibition. Our results demonstrate that selective inhibition of both CDK12 and CDK13 is required to potently induce cell cycle arrest and cell death in MV4;11 cells and phenocopies the effect of THZ531. We therefore posit that the therapeutic effects of THZ531 are reliant on its ability to inhibit CDK13 as much as CDK12, and the reported synergy observed with PARP inhibitors (22) as CDK12 inhibition predominantly affects DNA damage response gene expression. In this context, specific targeting of CDK12 or CDK13 in combination with PARP inhibitors may reduce unwanted side effects associated with global disruption of POLII-dependent transcription while maintaining the synergistic effect of this strategic combination therapeutic approach.
Together, this study characterizes both CDK12 and CDK13 as critical regulators of global POLII processivity and elongation and defines premature termination at intragenic polyadenylation sites as a universal CDK12/CDK13 controlled checkpoint along the transcription cycle.
MATERIALS AND METHODS
Cell culture and reagents
MV4;11 and THP1 AML cells (parental cells and HDR-edited clones) were cultured in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA, USA, 11875093) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (HI-FBS; Thermo Fisher Scientific, 10099), penicillin (100 U/ml), streptomycin (100 μg/ml) (Thermo Fisher Scientific, 15140122), 2 mM GlutaMAX (Thermo Fisher Scientific, 35050061), and 1 mM sodium pyruvate (Thermo Fisher Scientific, 11360070) at 37°C and 5% carbon dioxide. HEK293Ts were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, 11965) supplemented with 10% (v/v) HI-FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C and 10% carbon dioxide. Flp-In-HEK293 pcDNA5/FRT/TO-Rpb1 (WT Amr, R749H Amr, and H1108Y Amr) cells were a gift from D. Bentley [University of Colorado (UC) Denver] and were cultured as above with hygromycin (200 μg/ml; Sigma-Aldrich, St. Louis, MO, USA, 10843555001) and blasticidin (6.5 μg/ml; Thermo Fisher Scientific, A1113903). Experiments performed with Flp-In-HEK293 pcDNA5/FRT/TO-Rpb1 cells involved 16-hour incubation with doxycycline (2 μg/ml; Sigma-Aldrich, D1822) to induce mutant Rpb1 expression followed by 42-hour incubation with α-amanitin (2.5 μg/ml; Sigma-Aldrich, A2263) to inactivate endogenous POLII. Drosophila melanogaster S2 cells were cultured in Schneider’s Drosophila medium (Thermo Fisher Scientific, 21720) supplemented with 10% HI-FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM GlutaMAX at room temperature and atmospheric carbon dioxide. THZ531 (AOBIOUS, Gloucester, MA, USA, AOB8107), DRB (Sigma-Aldrich, D1916), 1-NM-PP1 (Merck Millipore, 529581), and AZ′5576 (provided by AstraZeneca) were prepared in DMSO (Sigma-Aldrich, 472301) and stored at −20°C.
CRISPR HDR generation of AS mutant clones
AS CDK12 mutations were generated at the previously identified gatekeeper residue [F813; (37)] using an HDR strategy adapted from (38). The corresponding gatekeeper residue for CDK13 (F791) was identified by aligning the CDK13 (NP_003709.3) and CDK12 (NP_057591.2) sequences. Parental MV4;11 cells (5 × 105) were washed twice in phosphate-buffered saline (PBS) and resuspended in 20 μl of nucleofector solution (16.4 μl of SF nucleofector solution + 3.6 μl of supplement 1) from the SF Cell Line 4D-Nucleofector X Kit (Lonza, Basel, Switzerland, V4XC-2032). Short guide RNAs (sgRNAs) targeting CDK12 or CDK13 (300 pmol; table S1) were incubated with 100 pmol of Alt-R SpCas9 nuclease (Integrated DNA Technologies, San Jose, CA, USA, 1074182) at room temperature for 20 min before being placed on ice. HDR donor template DNA (100 pmol) encoding mutated CDK12 or CDK13 (table S2) was added to SpCas9-sgRNA complexes for a final volume of 5 μl. Ribonucleoprotein complex (5 μl) was added to 20 μl of cell suspension and transferred to a 16-well Nucleocuvette strip before electroporation using the 4D-Nucleofector X Unit (program DJ100, Lonza, AAF-1002X). Prewarmed culture medium (100 μl) was added to the cell suspension, and cells were incubated for 10 min at 37°C and 5% CO2. Cells were then transferred to 24-well cell culture plates containing 500 μl of additional culture medium and were cultured at 37°C and 5% CO2. After expansion, single cells were isolated into 96-well cell culture plates using a Becton Dickinson (BD) FACSAria sorter. Mutant clones were identified by quantitative PCR (qPCR) analysis (see below; table S2) of genomic DNA (gDNA) isolated using DNeasy Blood & Tissue Kits (Qiagen, catalog no. 69506). Confirmation of CDK12 and/or CDK13 mutation status was confirmed by total RNA-seq analysis (see below) and Sanger sequencing of the CDK12 or CDK13 locus amplified from gDNA using KAPA-HIFI (Roche, 7958935001) at the Australian Genome Research Facility (sequencing primers detailed in table S2). A nonedited MV4;11 clone was isolated following HDR and used as a WT control.
Crystal structures of WT and AS CDK12
Native [Protein Data Bank (PDB) accession code 4NST] and AS mutant CDK12/cyclin K complexes were modeled. 1-NM-PP1 interaction with the gatekeeper residue [F813 (WT) and G813 (AS)] at the nucleotide binding site of CDK12/cyclin K was modeled using an overlay with the crystal structure of TgCDPK1 (PDB accession code 3I7B). All pictures and protein models were generated using PyMOL (v2.3.2).
Propidium iodide exclusion and CTV labeling
Cells were resuspended in PBS (1 × 107 cells/ml) and incubated with 5 μM CellTrace Violet dye (CTV; Thermo Fisher Scientific, C34557) in a 37°C water bath for 10 min, mixing every 5 min. Unbound dye was quenched through the addition of five volumes of ice-cold cell culture medium. CTV-positive cells were isolated using the BD Fusion FACS sorter and treated with DMSO, THZ531, or 1-NM-PP1 for 48 hours. Cells were incubated with propidium iodide (PI) (1 μg/ml; Sigma-Aldrich, P4170) for 15 min and analyzed using the BD LSRII. Flow cytometric data were analyzed using FlowJo v10 software (Ashland, OR, USA).
SDS–polyacrylamide gel electrophoresis and Western blotting
Cells were washed once in cold PBS before whole-cell lysis with Laemmli buffer [60 mM tris-HCl (pH 6.8), 10% (v/v) glycerol, and 2% (w/v) SDS] and incubated at 95°C for 10 min. Protein concentration was determined with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225), and samples equivalent to equal protein concentration were prepared with sample loading buffer [120 mM tris-HCl (pH 6.8), 20% (v/v) glycerol, 4% (w/v) SDS, 71.5 mM β-mercaptoethanol, and bromophenol blue]. Whole-cell lysates were separated using Mini-PROTEAN TGX 4 to 15% gradient gels [25 mM tris, 192 mM glycine, and 0.1% (w/v) SDS; Bio-Rad, Hercules, CA, USA, 4561086] and transferred at 4°C to Immobilon-P (IPVH00010) or Immobilon-FL (IPFL00010) polyvinylidene fluoride membranes (Merck, Kenilworth, NJ, USA) [2 hours; 200 mA, 25 mM tris, 192 mM glycine, and 5% (v/v) methanol]. Immobilon-FL membranes were blocked with Odyssey blocking buffer (Li-COR, Lincoln, NE, USA, P/N 927-40000), and Immobilon-P membranes were blocked with tris-buffered saline (TBS) supplemented with 0.1% (v/v) Tween 20 (Sigma-Aldrich, P9416) and 5% (w/v) skim milk powder. Membranes were incubated with primary antibodies (table S3) overnight at 4°C and incubated with horseradish peroxidase–conjugated secondary antibodies (Immobilon-P; table S3) or IRDye-conjugated secondary antibodies (Immobilon-FL; table S3) at room temperature for 1 hour. Membranes were washed three to five times with TBS and 0.1% (v/v) Tween 20. Protein was visualized using Amersham ECL Plus (GE Healthcare, Chicago, IL, USA, RPN2132) and Super RX film (Fujifilm, Tokyo, Japan, 03G01) (Immobilon-P) or using the Odyssey CLx and Image Studio software (Li-COR; Immobilon-FL). Quantitation of Western blots was performed using ImageJ software (version 1.46r).
Total RNA sequencing and 3′RNA-seq
Cells (1 × 106 per treatment condition) were resuspended in 350 μl of TRIzol (Thermo Fisher Scientific, 15596026), and RNA was isolated using a Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA, R2052). For SLAM-seq analysis (39), cells were incubated with 500 μM 4-thiouridine (4SU; Sigma-Aldrich, T4509) for 30 min before RNA isolation. Chloroform (one-fifth volume) was added to TRIzol lysates, and samples were mixed by shaking followed by centrifugation at 4°C (16,000g, 15 min). The aqueous phase was isolated, and dithiothreitol (DTT) was added (final concentration of 0.1 mM). RNA was precipitated with one volume of isopropanol at room temperature for 10 min followed by centrifugation at 4°C (16,000g, 20 min). The RNA pellet was washed with 500 μl of 75% ethanol supplemented with 0.1 mM DTT and centrifuged at room temperature (7500g, 5 min). The RNA pellet was dried at room temperature for 10 min before resuspension in 1 mM DTT and incubation for 10 min at 55°C. RNA (8 μg) was treated with iodoacetamide (IAA) in a 50-μl reaction volume at 50°C for 15 min [final concentration of 10 mM IAA, 50 mM NaPO4 (pH 9.9), and 50% (v/v) DMSO] followed by addition of 1 μl of 1 M DTT to stop the reaction. RNA was precipitated at −80°C for 30 min with 1 μl of GlycoBlue (Ambion, Thermo Fisher Scientific, AM9515), 5 μl of 3 M NaOAc (pH 5.2), and 125 μl of 100% ethanol. RNA was pelleted at 4°C (16,000g, 30 min), washed with 1 ml of 75% ethanol, and centrifuged at 4°C (16,000g). The RNA pellet was dried at room temperature for 10 min and resuspended in 12 μl of nuclease-free H2O. RNA yield and quality were assessed using the TapeStation 2200 (Agilent, Santa Clara, CA, USA). For total RNA-seq, samples underwent ribodepletion using the NEBNext rRNA Depletion Kit (New England BioLabs, Ipswich, MA, USA, E6310) or poly-dT enrichment (New England BioLabs, Ipswich, MA, USA, E7490), and sequencing libraries were prepared from 500 ng of RNA using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina. Paired-end base pair reads (75 or 150) were sequenced using the Illumina NextSeq 500 (Illumina, San Diego, CA, USA). For 3′RNA-seq, sequencing libraries were prepared from 500 ng of RNA using the QuantSeq 3′mRNA-seq Library Prep Kit FWD for Illumina (Lexogen, Vienna, Austria).Single-end base pair reads (75 or 150) were sequenced using the Illumina NextSeq 500 (Illumina, San Diego, CA, USA).
Analysis of total RNA-seq for validation of AS cell lines
HISAT2 (Galaxy version 2.1.0) was used to map paired FASTQ file to human genome (hg19; stranded information was not considered). Reads mapped to exons were counted using featureCounts (Galaxy version 1.6.2), and Limma-Voom (Galaxy version 3.36.5) was used to generate normalized counts.
Analysis of SLAM-seq and APA
The first 12 bases of each read were clipped using Cutadapt (v1.18), and clipped reads were processed by the Tail Tools pipeline (v1.5) (39) using the ENSEMBL Homo sapiens reference (release 93) to generate differential gene expression matrices and perform alternative polyadenylation analysis. For differential gene expression analysis, reads were counted at the level of whole genes, considering reads anywhere within the annotated gene region and up to 2 kb down-strand of the annotated TES. Reads that covered a down-strand annotated gene on the same strand were excluded, and genes for which no relevant sample had at least 10 reads were removed. Differential gene expression was tested between experimental groups using the Fitnoise library (https://github.com/pfh/fitnoise) as part of the Tail Tools pipeline, following log transformation and weighting of normalized counts using voom (40) from the limma R package (v3.40.6). For alternative polyadenylation analysis, peaks were called if at least 10 reads ending in a poly(A) sequence of at least six bases were present. Peaks where genomic sequences consisting of 60% or more “A”s could account for the poly(A) sequence were regarded as mispriming events and excluded. To assign peaks to genes, peaks called anywhere within an annotated gene region and up to 2 kb down-strand of the annotated TES were included. Peaks that covered a down-strand annotated gene on the same strand were excluded. For genes where more than two peaks were detected, the two peaks with the highest number of reads across all samples were selected. To compare between two experimental groups (e.g., DMSO treated and 1-NM-PP1 treated), a test was performed for each gene with two peaks to detect an interaction between peak usage and experimental group. This test was performed by the Fitnoise library, following log transformation and weighting of peak counts using voom (40) from the limma R package (v3.40.6). Fitnoise is an implementation of Empirical Bayes moderated t tests on weighted linear models as described in (41). Specifically, this test sought to reject that the log ratio of ratios of peak usage was zero. Volcano plots, log-ratio versus mean-average (MA) plots, and heat maps were generated using ggplot2 (v3.2.1) and pheatmap (v1.0.12) in R (v3.5.1). For differential gene expression Venn diagrams, differentially expressed genes with logFC < −1 and adjusted P < 0.05 (1-NM-PP1 relative to DMSO), excluding genes with adjusted P < 0.05 in 1-NM-PP1–treated WT clones, were presented. A hypergeometric test on differentially expressed genes indicated in the Venn diagram were performed using the Graeber Lab hypergeometric P value calculator (https://systems.crump.ucla.edu/hypergeometric/), where N (population size) is the total number of expressed transcripts [15515], M (number of successes in the population) is the number of genes differentially expressed in CDK13AS/AS [809], s (sample size) is the number of genes differentially expressed in CDK12AS/NULL [305], and k (number of successes) is the number of differentially expressed genes overlapping between CDK12AS/NULL and CDK13AS/AS [105]. Differentially expressed genes with absolute logFC > 1 and adjusted P < 0.01 in any of the groups were included in the heat map. For GO analysis, protein coding genes that were exclusively down-regulated (logFC < −1 and adjusted P < 0.05) in 1-NM-PP-1–treated CDK12AS/NULL and CDK13AS/AS, and excluding genes with adjusted P < 0.05 in 1-NM-PP1–treated WT clones, were analyzed by Metascape (release 3.5) (42). Overlapping down-regulated genes in 1-NM-PP1 CDK12AS/NULL and CDK13AS/AS groups were not considered in the analysis. Gene set enrichment analysis (GSEA; 4.0.1) was performed using GSEAPreranked function testing against hallmark gene sets (v7.0). Gene lists were ranked by –log10(P value) with the sign of logFC from the differential gene expression analysis. For APA Venn diagrams, the number of significant (adjusted P = 0.05 relative to DMSO) proximal or distal 3′UTR peak shifts in 1-NM-PP1–treated MV4;11 clones, THZ531-treated HEK293T cells, or R749H- and H1108Y-Rpb1 mutant compared to WT-Rpb1–expressing Flp-In HEK293 cells was presented. Venn diagrams were generated using BioVenn (43). Strand-specific bigWig files were also generated by Tail Tools pipeline (v1.5) and were visualized using IGV (v2.3.92/2.4.19).
Total RNA-seq and MISO analysis
Total RNA-seq paired-end reads were trimmed to 100 base pairs (bp) with Cutadapt (v1.18) to remove adapter sequences. Reads were aligned to GRCh37/hg19 with HISAT2 (v2.1.0) using settings for reverse-stranded library. Reads with MAPping Quality (MAPQ) values of <10 and secondary alignments were filtered. MISO (v0.5.4) alternative events analysis was performed using the annotation supplied by MISO. As MISO does not handle replicates, each replicate experiment was independently analyzed, and events were required to pass the following thresholds in all three replicate experiments: --num-inc 1 --num-exc 1 --num-sum-inc-exc 10 --delta-psi 0.20 --bayes-factor 10 --votes 3. As tandem 3′UTR events do not have exclusion reads (the shorter isoform is contained within the longer isoform), the following filter was used for those events: --num-inc 2 --num-sum-inc-exc 10 --delta-psi 0.20 --bayes-factor 10 --votes 3. To filter for expressed genes in MV4;11 parental cells (see below for ChIP-seq, TT-seq, and PRO-seq analysis), total RNA-seq analysis Illumina TruSeq Universal adapters were trimmed from the FASTQ files using Cutadapt (Galaxy version 1.16.1). Trimmed FASTQ files were mapped to human genome (hg19) using HISAT2 (Galaxy version 2.1.0). Reads mapping to exons were counted by featureCounts (Galaxy version 1.6.2), and genes that displayed greater than 5 counts per million (CPM) in at least three of the nine samples were considered expressed genes in MV4;11 cells. The gene lists were subtracted from the normalized count matrix generated by limma-voom (Galaxy version 3.36.5). BigWig files were generated using deepTools bamCoverage (Galaxy version 3.0.2.0).
To visualize polyadenylation events, 3′RNA-seq was performed from the same RNA as used for total RNA-seq. Reads were trimmed with Cutadapt (v 1.14) to remove priming bias (5 bp at 5′ end) of random hexamers and to remove polyA stretches (20 bp at 3′ end). Subsequently, the trimmed reads were mapped to GRCh37/hg19 with HISAT2 (v 2.1.0) using settings for forward-stranded library. SAMtools (v 1.4.1) was used to convert the SAM files to BAM files using view command, and the sorted BAM files were further converted to TDF files using IGVtools (v 2.3.95).
Quantitative PCR
Total RNA was isolated from 1 × 106 to 2 × 106 cells per treatment condition as described previously, and complementary DNA (cDNA) was synthesized from 1 μg of RNA using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit as per the manufacturer’s instructions (Thermo Fisher Scientific, 4368814). qPCRs were performed in 384-well plates using the SensiFAST SYBR Hi-ROX Kit (Bioline, London, UK, BIO-92005) using 0.75 μM oligo primers per reaction (table S2) and the LifeScience LightCycler 480 Instrument II (Roche, Basel, Switzerland, 05015243001). For cDNA, qPCR data were analyzed using the ΔΔCt method normalizing to the ribosomal RNA 5′ external transcribed spacer (ETS) as the housekeeping gene.
Phosphoproteomic MS
Five replicate samples of 10 × 106 cells per treatment condition were prepared for phosphopeptide MS. Cells were washed twice with cold PBS, and cell pellets were snap-frozen in Eppendorf protein LoBind tubes (Sigma-Aldrich, Z666505). Cell pellets were incubated for 10 min at 95°C in 5% (w/v) SDS, 10 mM tris(2-carboxyethyl)phosphine (TCEP), 40 mM 2-chloroacetamide (CAA), and 10 mM tris-HCl (pH 7.5). DNA was hydrolyzed using 1% trifluoroacetic acid (TFA) before sample neutralization through the addition of 3 M tris-HCl (pH 7.5; final concentration of 160 mM). Neutralized samples were incubated at room temperature with PureCUBE carboxy agarose magnetic beads (Cube Biotech, Germany, 50201) and 70% (v/v) acetonitrile for 20 min, and beads were washed twice with 70% ethanol and once with 100% acetonitrile. Beads were lyophilized to remove residual acetonitrile before addition of 50 μl of lysis buffer (10% trifluoroethanol and 100 mM ammonium bicarbonate) containing 1:25 enzyme:substrate ratio of trypsin (Promega, V5280) and lysyl endopeptidase (Wako, 125-05061). Beads were placed in an ultrasonic water bath (Unisonics, Australia) at room temperature for 2 min and then incubated for 4 hours in a Thermomixer (37°C, 1200 rpm). The supernatant was isolated, and beads were washed once with an equivalent volume of ultrapure water. The wash was pooled with the supernatant as the peptide-containing fraction. TFA was added to peptide solution to a final concentration of 1% (v/v). Peptide solution was centrifuged at 20,000g, and the peptide-containing supernatant was isolated. TFA-supplemented acetonitrile was added to the peptide-containing supernatant to a final concentration of 80% (v/v) acetonitrile and 0.1% (v/v) TFA. Peptide-containing samples were incubated at room temperature with 50 μl of PureCUBE Fe(III)-NTA beads (Cube Biotech, 31501-Fe) for 30 min with gentle shaking. Beads were washed three times (80% acetonitrile and 0.1% TFA) and transferred in wash buffer onto acetonitrile prewetted C8 stage tips. C8 tips were centrifuged (500g, 1 min) to remove wash buffer. Phosphopeptides were eluted into tubes containing 30 μl of 10% TFA using 2 × 20 μl of elution buffer [50% acetonitrile and 2.5% ammonium hydroxide (pH 10)]. Eluted samples were lyophilized until near to dryness, and peptides were resuspended in 50 μl of 5% formic acid. Phosphopeptides were transferred to prewetted and equilibrated (100% isopropyl alchohol (IPA), then 60% acetonitrile, 5% formic acid, then 5% formic acid) C18 stage tips and centrifuged (500g, 1 min). C18 stage tips were washed twice with 5% formic acid, and phosphopeptides were eluted in 50 μl of 60% acetonitrile and 5% formic acid. Phosphopeptides were lyophilized to dryness and stored at −80°C. Phosphopeptides were resuspended in 2% acetonitrile and 1% formic acid. Samples were injected and separated by reversed-phase liquid chromatography using an Easy-nLC 1200 ultrahigh-performance liquid chromatography (UHPLC) system (Thermo Fisher Scientific) using a 1.6-μm C18 packed emitter tip column (250 mm × 75 μm, IonOpticks, Australia) with linear 120-min gradient at a flow rate of 400 nl/min from 98% solvent A (0.1% formic acid in ultrapure water) to 37% solvent B (0.1% formic acid in acetonitrile). The UHPLC was coupled online to a Q-Exactive HF Orbitrap mass spectrometer equipped with a nanoelectrospray ionization source (Thermo Fisher Scientific) and a column oven at 50°C (Sonation, Germany). The Q-Exactive HF was operated in a data-dependent mode, switching automatically between one full scan and subsequent MS/MS scans of the 10 most abundant peaks. The instrument was controlled using Exactive series version 2.9 and Xcalibur 4.1.31.9. Full scans [350 to 1850 mass/charge ratio (m/z)] were acquired with a resolution of 60,000 at 200 m/z. The 10 most intense ions were sequentially isolated with a target value of 10,000 ions and an isolation width of 1.4 m/z and fragmented using higher-energy collisional dissociation (HCD) with normalized collisional energy (NCE) of 27. Maximum ion accumulation times were set to 100 ms for full MS scan and 110 ms at 17,500 resolution for MS/MS. Raw files were analyzed using MaxQuant (version 1.5.8.3). The database search was performed using the UniProt Homo sapiens database and common contaminants with strict trypsin specificity allowing up to three missed cleavages. The minimum peptide length was seven amino acids. Carbamidomethylation of cysteine was a fixed modification, while N-acetylation of proteins N termini, oxidation of methionine, and phosphorylation of serine/threonine/tyrosine were set as variable modifications. During the MaxQuant main search, precursor ion mass error tolerance was set to 4.5 parts per million (ppm), and fragment ions were allowed a mass deviation of 20 ppm. Peptide spectrum match (PSM) and protein identifications were filtered using a target-decoy approach at a false discovery rate (FDR) of 1%, with the match between runs option enabled.
Further analysis was performed using a custom pipeline developed in R, which uses the MaxQuant output file evidence.txt. A feature was defined as the combination of peptide sequence, charge, and modification. Features not found in at least 80% of the replicates in one group were removed. To correct for injection volume variability, feature intensities were normalized by converting to base 2 logarithms and then multiplying each value by the ratio of maximum median intensity of all replicates over median replicate intensity. Missing values were imputed using a random normal distribution of values with the mean set at mean of the real distribution of values minus 2.2 SD, and an SD of 0.15 times the SD of the distribution of the measured intensities. The probability of differential peptide expression between groups was calculated using the Limma R package (version 3.34.9). Probability values were corrected for multiple testing using Benjamini-Hochberg method. The heat map for differentially phosphorylated peptides (adjusted P < 0.3, 1-NM-PP1 treatment relative to DMSO treatment) was generated in R (v 3.5.1) using the gplots heatmap.2 function (v3.0.1.1). Differentially phosphorylated peptides with adjusted P ≤ 0.3 and logFC < 0 relative to DMSO treatment in 1-NM-PP1–treated CDK13AS/AS and #2 CDK12AS/NULL;CDK13AS/AS (excluding peptides with an adjusted P ≤ 0.3 in 1-NM-PP1–treated WT clone) were included in the Venn diagram. GO analysis was performed for proteins with differentially phosphorylated peptides in #2 CDK12AS/NULL;CDK13AS/AS cells treated with 1-NM-PP1 (adjusted P ≤ 0.3 and logFC < 0 relative to DMSO treatment and excluding proteins with peptides with and adjusted P ≤ 0.3 in WT) with Metascape using default expressed analysis (42). Interaction networks for proteins included in the GO analysis for #2 CDK12AS/NULL;CDK13AS/AS cells were analyzed in Cytoscape (v3.7.1) using the STRING network analysis function.
ChIP sequencing
Cells (40 × 106 to 60 × 106 per treatment condition) were resuspended in PBS and crossed-linked with fresh formaldehyde solution [50 mM Hepes-KOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 11% formaldehyde] for 30 min at room temperature. Excess formaldehyde was quenched by the addition of glycine to 125 mM for 5 min. Cross-linked cells were washed once with ice-cold PBS and were then incubated three times (ice, 5 min) with nuclear extraction buffer [20 mM tris-HCl (pH 8), 10 mM NaCl, 2 mM EDTA, and 0.5% (v/v) IGEPAL CA-630] containing Roche cOmplete protease inhibitors (Merck, 04693159001). Nuclear extracts were resuspended in sonication buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% (v/v) IGEPAL CA-630, and 0.3% (w/v) SDS] containing protease and Pierce phosphatase inhibitors (Thermo Fisher Scientific, A32957) and were sonicated at maximum power for 16 min using the Covaris S220 Focused-ultrasonicator. Sonicated lysates were diluted with one volume of ChIP dilution buffer [20 mM tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, and 1% (v/v) Triton X-100] containing protease and phosphatase inhibitors. D. melanogaster chromatin was isolated from S2 cells using the same protocol, and S2 and human chromatin lysates were mixed at 1:10 (1-NM-PP1 experiments) and 1:5 (THZ531 experiments) ratios by DNA content. Protein A and protein G Dynabeads (Thermo Fisher Scientific, 10002D and 10004D) were mixed 1:1 (50 μl total per IP) and washed in blocking buffer [20 mM tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, 1% (v/v) Triton X-100, 0.15% (w/v) SDS, and 0.1% (w/v) bovine serum albumin (BSA)] containing protease inhibitors at 4°C. Protein A/G beads were resuspended in ChIP IP buffer [20 mM tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, 1% (v/v) Triton X-100, 0.15% (w/v) SDS; ~0.5 ml per IP] containing protease and phosphatase inhibitors and added to diluted human/S2 chromatin lysates with 5 μg of antibody (table S3) and 0.45% (w/v) BSA. IP samples were incubated overnight at 4°C while tumbling. Protein A/G beads were washed twice with ChIP IP buffer on ice before washing once with ChIP wash buffer 1 [20 mM tris-HCl (pH 8), 500 mM NaCl, 2 mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) SDS] and wash buffer 2 [20 mM tris-HCl (pH 8), 250 mM LiCl, 2 mM EDTA, 0.5% (v/v) IGEPAL CA-630, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate], each containing protease and phosphatase inhibitors, and washing twice with tris-EDTA buffer [10 mM tris-HCl (pH 7.5) and 1 mM EDTA]. Washed beads were incubated with reverse crosslinking buffer [1% (w/v) SDS, 100 mM NaHCO3, and 200 mM NaCl] containing 300 μg of proteinase K (Sigma-Aldrich, P2308) at 55°C for 1 hour before incubation of the supernatant at 65°C overnight. DNA was isolated using the ChIP DNA Clean & Concentrator Kit (Zymo Research, D5205). DNA was measured with the Qubit dsDNA HS (Thermo Fisher Scientific), and sequencing libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs, E7645). Fragments between 200 and 500 bp were selected using a Pippin Prep 1.5% agarose cassette (Sage Science, Beverly, MA, USA) and pooled, and 25 × 106 75 single-end base pair reads were sequenced using the Illumina NextSeq 500.
TT sequencing
Cells (30 × 106 per treatment condition) were incubated with 1 mM 4SU (Sigma-Aldrich, T4509) for 5 min at 37°C, followed by lysis in TRIzol. Chloroform (one-fifth volume) was added, and samples were vortexed and centrifuged at 4°C (13,000g, 15 min). The aqueous phase was isolated, and total RNA was extracted by isopropanol precipitation at room temperature (10 min) followed by centrifugation at 4°C (13,000g, 10 min). The isolated RNA pellet was washed with 70% ethanol followed by centrifugation at 4°C (13,000g, 15 min). The RNA pellet was dissolved in 100 μl of nuclease-free H2O and denatured at 65°C for 10 min. Total RNA (150 μg per treatment condition) (plus 15 μg of D. melanogaster S2 RNA spike-in) in 100-μl final volume was sonicated in microTUBE AFA Fiber screw cap tubes (6 mm × 16 mm, Covaris, 520096) using the Covaris S220 Focused-ultrasonicator at a maximum power for 15 s. Thiol-specific biotinylation of fragmented RNA was performed by adding 750-μl volume nuclease-free water followed by incubation with 10 mM tris (pH 7.4), 1 mM EDTA, dimethylformamide (200 μg/ml; Sigma-Aldrich, 227056), and 300 μg of EZ-Link HPDP-Biotin (Thermo Fisher Scientific, 21341) at room temperature for 1.5 hours with constant rotation (1500-μl final volume; “biotin labeling reaction”). One volume of chloroform was added to the biotin labeling reaction, and the reaction was vortexed and centrifuged at 4°C (20,000g, 5 min). The aqueous phase was isolated, and the chloroform isolation was repeated twice. NaCl (to 0.5 M) and one volume of isopropanol were added to the aqueous phase, and fragmented RNA was isolated by centrifugation at 4°C (20,000g, 20 min). The RNA pellet was washed with 75% ethanol, centrifuged at 4°C (20,000g, 20 min), and dissolved in nuclease-free water (1 μl/1 μg of input RNA). Fragmented RNA was denatured at 65°C for 10 min before incubation with streptavidin beads (μMACs Streptavidin Kit, Miltenyi Biotec, Bergisch Gladbach, Germany, 130-074-101) at room temperature for 15 min with constant rotation. Biotinylated RNA was isolated using μMACs columns that had been pre-equilibrated with room temperature wash buffer [100 mM tris-HCl (pH 7.4), 10 mM EDTA, 1 M NaCl, and 0.1% (v/v) Tween 20]. Following incubation with biotinylated RNA/streptavidin beads, the columns were washed with five times, each with 0.9 ml of prewarmed (65°C) and room temperature wash buffer. Biotinylated RNA was eluted through two additions of 100 μl of 100 mM DTT 3 min apart and collected into 700 μl of RLT buffer from the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany, 74204). Biotinylated RNA was isolated using the Qiagen RNeasy MinElute Cleanup Kit using 12 μl of elution buffer to elute RNA. RNA was quantified using the Agilent TapeStation High Sensitivity RNA Kit (Agilent, 5067-5576). Sequencing libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, and 75 single-end base pair reads were sequenced using the Illumina NextSeq 500.
PRO sequencing
Cells (1 × 107) were used per treatment condition. For DRB-release PRO-seq assays, cells were pretreated for 4 hours with 100 μM DRB, washed three times with ice-cold PBS, and resuspended in prewarmed culture medium. For 1-NM-PP1/DMSO treatment, 1-NM-PP1/DMSO was added at the first PBS wash step and the release medium. For THZ531/DMSO treatment, THZ531 was added at the same time as DRB. Cells were harvested at 25 min (1-NM-PP1) or at 12.5, 25, and 45 min (THZ531) after addition of fresh culture medium. For 0-min (T0) samples (THZ531), cells were harvested before DRB washout. Cells were washed in ice-cold PBS before incubation in permeabilization buffer [1 ml/106 cells; 100 μl/107 cells; 10 mM tris-HCl (pH 7.4), 200 mM sucrose, 10 mM KCl, 5 mM MgCl2, 1 mM EDTA, 0.05% (v/v) Tween 20, 0.15% (v/v) NP-40, 0.5 mM DTT, and SUPERase•In (2 U/ml) (Thermo Fisher Scientific, AM2694)] containing Roche cOmplete protease inhibitors for 5 min on ice. The cell pellet was washed twice with permeabilization buffer (0.5 ml per 106 cells per wash) and then resuspended in a storage buffer [10 mM tris-HCl (pH 8), 25% glycerol, 5 mM MgCl2, 0.1 mM EDTA, and 5 mM DTT]. For the nuclear run-on reaction, permeabilized cells were incubated with one volume of 2× nuclear run-on reaction mastermix containing Biotin 11-CTP [1.4 mM tris-HCl (pH 8), 0.7 mM MgCl2, 0.14 mM DTT, 42 mM KCl, 0.05 mM Biotin 11-CTP (PerkinElmer, NEL544001EA), 2.5 μM CTP, 0.25 mM ATP, 0.25 mM guanosine triphosphate (GTP), 0.25 mM uridine triphosphate (UTP), 40 U SUPERase•In, and 1% Sarkosyl (Sigma-Aldrich, L5125)] at 37°C for 3 min with gentle agitation before the addition of TRIzol-LS (Thermo Fisher Scientific, 10296010). TRIzol samples were mixed with chloroform, and the aqueous phase was isolated after centrifugation. One microliter of GlycoBlue and 2.5 volume of ethanol were added to the aqueous phase for RNA precipitation (incubation at room temperature for 10 min, 14,000g centrifugation at 4°C for 20 min). Precipitated RNA was washed with 70% ethanol and resuspended in nuclease-free water. RNA was denatured at 65°C for 40 s and then was incubated on ice with 0.2 M NaOH for 12.5 min before the addition of 1 volume of 1 M tris-HCl (pH 6.8). Base hydrolyzed RNA was exchanged to 10 mM tris solution using a Micro Bio-Spin 30 Column (Bio-Rad, 7326250) as per the manufacturer’s instruction. For enrichment of biotin-incorporated RNA, hydrolyzed RNA was incubated with Dynabeads M-280 streptavidin (Thermo Fisher Scientific, 11206D) in PRO-seq binding buffer [10 mM tris-HCl (pH 7.4), 300 mM NaCl, and 0.1% (v/v) Triton X-100] at room temperature for 20 min with gentle rotation. Streptavidin beads were washed (1 min with rotation per wash) three times each in high-salt wash buffer [50 mM tris-HCl (pH 7.4), 2 M NaCl, and 0.5% (v/v) Triton X-100], binding buffer, and low-salt wash buffer [5 mM tris-HCl (pH 7.4), 0.1% Triton X-100] and then resuspended in TRIzol. Following incubation at room temperature for 3 min, TRIzol samples were mixed with chloroform and the aqueous phase was isolated after centrifugation. TRIzol/chloroform treatment of washed streptavidin beads was repeated once, and beads were incubated with chloroform for 3 min at room temperature before centrifugation. One microliter of GlycoBlue and 900 μl of ethanol were added to the pooled aqueous phase solution, and samples were incubated at room temperature for 10 min before centrifugation. Precipitated biotin-enriched RNA was washed with 70% ethanol and resuspended in nuclease-free water. Samples (AS cell lines) underwent ribodepletion using the NEBNext rRNA Depletion Kit, sequencing libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, and 75 single-end base pair reads were sequenced using the Illumina NextSeq 500.
ChIP-seq, TT-seq, and PRO-seq analysis
The sequenced reads were demultiplexed using bcl2fastq (v2.17.1.14), and basic quality control (QC) was performed on the resulting FASTQ files using FASTQC (v0.11.5). FASTQ reads were mapped to the GRCh37/hg19 genome using Bowtie2 (v2.3.3) using the standard settings. The resulting SAM files were converted to BAM files using SAMtools (v 1.4.1) using the view command, after which the BAM files were sorted and indexed, and potential PCR duplicates were removed using the rmdup function. DeepTools (v3.0.0) was used to generate occupancy heat maps, and the resulting CPM normalized occupancy matrix was used as input for custom R scripts to generate average profile plots and to calculate processivity indices. Briefly, the BAM files were converted into BigWig files using the BamCoverage function (--normalizeUsing CPM --smoothLength 150 --binSize 50 -e 200 scaleFactor 1). Where indicated, normalization to the Drosophila S2 spike-in was performed at this stage in a manner similar as described previously (44). In this case, the reads were mapped to a combined hg19/dm3 genome, after which the reads mapping to fly and human genomes were calculated using FeatureCounts from the Subread package (v1.5.0). The scaleFactor was subsequently determined by calculating the reads mapping to the fly genome relative to total mapped reads (combined hg19/dm3 reference) and applied in the BamCoverage command. The bigwigCompare function was used to calculate log2 ratio plots, and the computeMatrix function was used to quantify occupancy of either TT-seq, ChIP-seq, or PRO-seq reads across the specified intervals. Rstudio (v1.1.46) and R (v3.5.1) were used for subsequent analysis.
Calculation of POLII elongation rates from DRB-release PRO-seq analysis
Long genes, identified as protein-coding genes with a length of >100 kb (from gene start to end) in the Ensembl GRCh37 v87 GTF, were identified. These genes were filtered to include genes identified as expressed in MV4;11 cells in the RNA-seq analysis, as described above (ChIP-seq). Genes were further filtered to identify genes that demonstrated an increase in transcription over time in the DMSO control sample for the DRB-release PRO-seq time course experiment (fig. S5A) and to remove genes showing interference (e.g., from downstream promotors). The following criteria were used to filter genes: The 0-min time point DMSO control sample was required to show expression of the gene (mean expression of >1 CPM in 250 bp after TSS (beyond the pausing peak region)] and was required to have a wavefront called within 10 kb of pausing peak region to remove artefacts. The wavefront of transcription for each gene was identified by considering the region from the TSS to 80-kb downstream (TSS to 90 kb was considered for the #2 CDK12AS/NULL;CDK13AS/AS clone, where the DRB-release assay was performed at a different time to other clones), generating z scores of the normalized counts and using 5-kb bins in sliding windows from the end of the region to identify the bin where the mean first rises above zero. Genes showing an increase in transcription in the DMSO control sample for the time course were identified by requiring the wavefront in the 0-min sample to be less than the wavefront in the 12.5-min time point wavefront, and the wavefront in the 12.5-min sample to be less than the wavefront in the 25-min time point. This resulted in the identification of 368 genes, for which elongation rates were calculated for the DMSO control, THZ531-treated, or 1-NM-PP1–treated samples by dividing the wavefront position by the time point.
Mathematical modeling and simulations of POLII activity
A simulation package was written in Julia and visualized in Javascript/D3. To model polymerase dynamics numerically, a process-oriented simulator, continuous in time and location along gene but discrete in events, was implemented. A predefined set of biological events (table S5) transcription initiation, promoter proximal pausing, and transcription completion was each associated with a time. The dynamics and the impact each event has are tunable by the set of parameters given in table S5. The overall state of the system is a small number of representative genes, and a number of polymerases, each either positioned along one of the genes or available for initiation on any of the genes. There is an initial period to achieve steady state, where polymerases are allowed to initiate on the genes (weighted, to allow different proportions of the gene classes) and progress along the gene body. When equilibrium is achieved in terms of initiation and completion, the statistics are then recorded to calculate polymerase density at subsequent time intervals. Between events, transcription proceeds in a continuous manner, but when an event occurs (e.g., if POLII’s speed is reduced), an abrupt update is made, and a fresh set of future events is calculated to reflect this. Moving polymerases have a probability of being removed from the gene body at any moment in time through processivity errors. For each event in temporal turn, the state of the system is updated using natural mathematical rules (such as a polymerase position being offset by the product of its speed). Then, the event is handled (e.g., polymerase’s speed is reduced if the event is an elongation defect), and the list of events is updated in light of the previous two steps. The events are then resorted to find the event that is going to happen next in time, and the process is repeated until the required amount of time has elapsed.
Supplementary Material
Acknowledgments
We thank the Peter MacCallum Cancer Centre Molecular Genomics Core (T. Semple and G. Mir Arnau), I. Kong and E. Hawkins (Walter and Eliza Hall Institute), and members of the Johnstone laboratory for technical help and support. We thank D. Bentley (UC Denver) for providing reagents. Funding: This work was supported by a project grant (to R.W.J.) and fellowship (to S.J.H.) from the Cancer Council Victoria, project grant support from the NHMRC (to R.W.J.), NHMRC Program (grant 454569 to R.W.J.), NHMRC Senior Principal Research Fellowship (to R.W.J.), Victorian Cancer Agency Early Career Seed Grant (to J.R.D.), Melbourne Research Scholarship (to Z.F.), and The Kids’ Cancer Project (to R.W.J. and S.J.V.). S.J.V. was supported by a Rubicon Fellowship from the Netherlands Organization for Scientific Research (NWO, 019.161LW.017). We acknowledge support from the Peter MacCallum Cancer Centre Foundation, the Australian Cancer Research Foundation, and the Victorian Government’s Operational Infrastructure Support Program. Work in the Svejstrup laboratory was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001166), the UK Medical Research Council (FC001166), and the Wellcome Trust (FC001166). Author contributions: R.W.J. and S.J.V. conceptualized and designed the study. J.R.D., S.J.V., and R.W.J. wrote the manuscript. Z.F., J.R.D., I.T., L.A.C., D.A.K., J.J.S., and S.J.V. performed the experiments. Z.F., J.R.D., S.J.H., M.A.D., P.F.H., J.J.S., A.F., T.H.B., A.T.V., G.P.K., and S.J.V. analyzed the data. G.G., N.K., N.E.S., J.Q.S., M.G., and N.S.G. edited the manuscript and provided intellectual advice. Competing interests: The Johnstone laboratory has received project support from AstraZeneca, Roche, MecRx, and BMS. R.W.J. is a paid consultant of MecRx. N.S.G. and N.K. are inventors on patent applications covering THZ531 (WO 2015/058126, WO/2014/063068, and WO 2015/058140), which are licensed to a company founded by N.S.G. Data and materials availability: Phosphoproteomic data are available on the PRIDE database (identification number PXD015453). Next-generation sequencing data have been deposited at the Gene Expression Omnibus (accession no. GSE141377). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. THZ531 can be provided by the Gray laboratory pending scientific review and a completed material transfer agreement. Requests for THZ531 should be submitted to N.S.G.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/18/eaaz5041/DC1
REFERENCES AND NOTES
- 1.Smith E., Shilatifard A., Transcriptional elongation checkpoint control in development and disease. Genes Dev. 27, 1079–1088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malumbres M., Cyclin-dependent kinases. Genome Biol. 15, 122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradner J. E., Hnisz D., Young R. A., Transcriptional addiction in cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Fisher R. P., Cdk7: A kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription 10, 47–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory G. P., Hogg S. J., Kats L. M., Vidacs E., Baker A. J., Gilan O., Lefebure M., Martin B. P., Dawson M. A., Johnstone R. W., Shortt J., CDK9 inhibition by dinaciclib potently suppresses Mcl-1 to induce durable apoptotic responses in aggressive MYC-driven B-cell lymphoma in vivo. Leukemia 29, 1437–1441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker A., Gregory G. P., Verbrugge I., Kats L., Hilton J. J., Vidacs E., Lee E. M., Lock R. B., Zuber J., Shortt J., Johnstone R. W., The CDK9 inhibitor dinaciclib exerts potent apoptotic and antitumor effects in preclinical models of MLL-rearranged acute myeloid leukemia. Cancer Res. 76, 1158–1169 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Galbraith M. D., Bender H., Espinosa J. M., Therapeutic targeting of transcriptional cyclin-dependent kinases. Transcription 10, 118–136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenleaf A. L., Human CDK12 and CDK13, multi-tasking CTD kinases for the new millennium. Transcription 10, 91–110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B. M., The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25, 2158–2172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajrami I., Frankum J. R., Konde A., Miller R. E., Rehman F. L., Brough R., Campbell J., Sims D., Rafiq R., Hooper S., Chen L., Kozarewa I., Assiotis I., Fenwick K., Natrajan R., Lord C. J., Ashworth A., Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 74, 287–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi P. M., Sutor S. L., Huntoon C. J., Karnitz L. M., Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J. Biol. Chem. 289, 9247–9253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubbury S. J., Boutz P. L., Sharp P. A., CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564, 141–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krajewska M., Dries R., Grassetti A. V., Dust S., Gao Y., Huang H., Sharma B., Day D. S., Kwiatkowski N., Pomaville M., Dodd O., Chipumuro E., Zhang T., Greenleaf A. L., Yuan G. C., Gray N. S., Young R. A., Geyer M., Gerber S. A., George R. E., CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 10, 1757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang K., Gao X., Gilmore J. M., Florens L., Washburn M. P., Smith E., Shilatifard A., Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol. Cell. Biol. 35, 928–938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eifler T. T., Shao W., Bartholomeeusen K., Fujinaga K., Jäger S., Johnson J. R., Luo Z., Krogan N. J., Peterlin B. M., Cyclin-dependent kinase 12 increases 3′ end processing of growth factor-induced c-FOS transcripts. Mol. Cell. Biol. 35, 468–478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y. M., Cieślik M., Lonigro R. J., Vats P., Reimers M. A., Cao X., Ning Y., Wang L., Kunju L. P., de Sarkar N., Heath E. I., Chou J., Feng F. Y., Nelson P. S., de Bono J. S., Zou W., Montgomery B., Alva A.; PCF/SU2C International Prostate Cancer Dream Team, Robinson D. R., Chinnaiyan A. M., Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greifenberg A. K., Hönig D., Pilarova K., Düster R., Bartholomeeusen K., Bösken C. A., Anand K., Blazek D., Geyer M., Structural and functional analysis of the Cdk13/Cyclin K complex. Cell Rep. 14, 320–331 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kim H. E., Kim D. G., Lee K. J., Son J. G., Song M. Y., Park Y. M., Kim J. J., Cho S. W., Chi S. G., Cheong H. S., Shin H. D., Lee S. W., Lee J. K., Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas. PLOS ONE 7, e43223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton M. J., Caswell R. C., Canham N., Cole T., Firth H. V., Foulds N., Heimdal K., Hobson E., Houge G., Joss S., Kumar D., Lampe A. K., Maystadt I., Kay V. M., Metcalfe K., Newbury-Ecob R., Park S.-M., Robert L., Rustad C. F., Wakeling E., Wilkie A. O. M.; The Deciphering Developmental Disorders Study, Twigg S. R. F., Suri M., Heterozygous mutations affecting the protein kinase domain of CDK13 cause a syndromic form of developmental delay and intellectual disability. J. Med. Genet. 55, 28–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T., Kwiatkowski N., Olson C. M., Dixon-Clarke S. E., Abraham B. J., Greifenberg A. K., Ficarro S. B., Elkins J. M., Liang Y., Hannett N. M., Manz T., Hao M., Bartkowiak B., Greenleaf A. L., Marto J. A., Geyer M., Bullock A. N., Young R. A., Gray N. S., Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 12, 876–884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iniguez A. B., Stolte B., Wang E. J., Conway A. S., Alexe G., Dharia N. V., Kwiatkowski N., Zhang T., Abraham B. J., Mora J., Kalev P., Leggett A., Chowdhury D., Benes C. H., Young R. A., Gray N. S., Stegmaier K., EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma. Cancer Cell 33, 202–216.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williamson L., Saponaro M., Boeing S., East P., Mitter R., Kantidakis T., Kelly G. P., Lobley A., Walker J., Spencer-Dene B., Howell M., Stewart A., Svejstrup J. Q., UV irradiation induces a non-coding rna that functionally opposes the protein encoded by the same gene. Cell 168, 843–855.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tien J. F., Mazloomian A., Cheng S. W. G., Hughes C. S., Chow C. C. T., Canapi L. T., Oloumi A., Trigo-Gonzalez G., Bashashati A., Xu J., Chang V. C. D., Shah S. P., Aparicio S., Morin G. B., CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 45, 6698–6716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz Y., Wang E. T., Airoldi E. M., Burge C. B., Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamieniarz-Gdula K., Proudfoot N. J., Transcriptional control by premature termination: A forgotten mechanism. Trends Genet. 35, 553–564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X. D., Bentley D. L., Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 28, 2663–2676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashiguchi T., Bruss N., Best S., Lam V., Danilova O., Paiva C. J., Wolf J., Gilbert E. W., Okada C. Y., Kaur P., Drew L., Cidado J., Hurlin P., Danilov A. V., Cyclin-dependent kinase-9 is a therapeutic target in MYC-expressing diffuse large B-cell lymphoma. Mol. Cancer Ther. 18, 1520–1532 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Chirackal Manavalan A. P., Pilarova K., Kluge M., Bartholomeeusen K., Rajecky M., Oppelt J., Khirsariya P., Paruch K., Krejci L., Friedel C. C., Blazek D., CDK12 controls G1/S progression by regulating RNAPII processivity at core DNA replication genes. EMBO Rep. 20, e47592 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y. E., Manteiga J. C., Henninger J. E., Sabari B. R., Dall’Agnese A., Hannett N. M., Spille J. H., Afeyan L. K., Zamudio A. V., Shrinivas K., Abraham B. J., Boija A., Decker T. M., Rimel J. K., Fant C. B., Lee T. I., Cisse I. I., Sharp P. A., Taatjes D. J., Young R. A., Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eick D., Geyer M., The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456–8490 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Bowman E. A., Kelly W. G., RNA polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: A tail of two kinases. Nucleus 5, 224–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S. H., Singh I., Tisdale S., Abdel-Wahab O., Leslie C. S., Mayr C., Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 561, 127–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bösken C. A., Farnung L., Hintermair C., Schachter M. M., Vogel-Bachmayr K., Blazek D., Anand K., Fisher R. P., Eick D., Geyer M., The structure and substrate specificity of human Cdk12/Cyclin K. Nat. Commun 5, 3505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devany E., Park J. Y., Murphy M. R., Zakusilo G., Baquero J., Zhang X., Hoque M., Tian B., Kleiman F. E., Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. Cell Discov. 2, 16013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paculova H., Kohoutek J., The emerging roles of CDK12 in tumorigenesis. Cell Div. 12, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartkowiak B., Greenleaf A. L., Expression, purification, and identification of associated proteins of the full-length hCDK12/CyclinK complex. J. Biol. Chem. 290, 1786–1795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartkowiak B., Yan C., Greenleaf A. L., Engineering an analog-sensitive CDK12 cell line using CRISPR/Cas. Biochim. Biophys. Acta 1849, 1179–1187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herzog V. A., Reichholf B., Neumann T., Rescheneder P., Bhat P., Burkard T. R., Wlotzka W., von Haeseler A., Zuber J., Ameres S. L., Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison P. F., Powell D. R., Clancy J. L., Preiss T., Boag P. R., Traven A., Seemann T., Beilharz T. H., PAT-seq: A method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA 21, 1502–1510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., Benner C., Chanda S. K., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hulsen T., de Vlieg J., Alkema W., BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orlando D. A., Chen M. W., Brown V. E., Solanki S., Choi Y. J., Olson E. R., Fritz C. C., Bradner J. E., Guenther M. G., Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 9, 1163–1170 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/18/eaaz5041/DC1