The microbial decomposition of animal tissues is an important ecological process that impacts nutrient cycling in natural environments. We studied the microbial decomposition of a common North American fish (rainbow darters) over four time points, combining 16S rRNA gene and shotgun metagenomic sequence data to obtain both taxonomic and functional perspectives. Our data revealed a strong community succession that was reproduced across different fish and environments. Decomposition time point was the main driver of community composition and functional potential; fish environmental origin (upstream or downstream of a wastewater treatment plant) had a secondary effect. We also identified strains related to the putative pathogen Aeromonas veronii as dominant members of the decomposition community. These bacteria peaked early in decomposition and coincided with the metagenomic abundance of hemolytic toxin genes. Our work reveals a strong decomposer succession in wild-caught fish, providing functional and taxonomic insights into the vertebrate necrobiome.
KEYWORDS: necrobiome, microbiome, decomposition, wastewater, rainbow darter, Aeromonas, aerolysin, community profiling, fish, metagenomics, thanatomicrobiome, toxins
ABSTRACT
Despite progress understanding microbial communities involved in terrestrial vertebrate decomposition, little is known about the microbial decomposition of aquatic vertebrates from a functional and environmental context. Here, we analyzed temporal changes in the “necrobiome” of rainbow darters, which are common North American fish that are sensitive indicators of water quality. By combining 16S rRNA gene and shotgun metagenomic sequence data from four time points, we studied the progression of decomposers from both taxonomic and functional perspectives. The 16S rRNA gene profiles revealed strong community succession, with early decomposition stages associated with Aeromonas and Clostridium taxa and later stages dominated by members of the Rikenellaceae (i.e., Alistipes/Acetobacteroides genera). These results were reproducible and independent of environmental perturbation, given that exposure to wastewater treatment plant effluent did not substantially influence the necrobiome composition of fish or the associated water sample microbiota. Metagenomic analysis revealed significant changes throughout decomposition in degradation pathways for amino acids, carbohydrates/glycans, and other compounds, in addition to putrefaction pathways for production of putrescine, cadaverine, and indole. Binning of contigs confirmed a predominance of Aeromonas genome assemblies, including those from novel strains related to the pathogen Aeromonas veronii. These bins of Aeromonas genes also encoded known hemolysin toxins (e.g., aerolysin) that were particularly abundant early in the process, potentially contributing to host cell lysis during decomposition. Overall, our results demonstrate that wild-caught fish have a reproducible decomposer succession and that the fish necrobiome serves as a potential source of putative pathogens and toxigenic bacteria.
IMPORTANCE The microbial decomposition of animal tissues is an important ecological process that impacts nutrient cycling in natural environments. We studied the microbial decomposition of a common North American fish (rainbow darters) over four time points, combining 16S rRNA gene and shotgun metagenomic sequence data to obtain both taxonomic and functional perspectives. Our data revealed a strong community succession that was reproduced across different fish and environments. Decomposition time point was the main driver of community composition and functional potential; fish environmental origin (upstream or downstream of a wastewater treatment plant) had a secondary effect. We also identified strains related to the putative pathogen Aeromonas veronii as dominant members of the decomposition community. These bacteria peaked early in decomposition and coincided with the metagenomic abundance of hemolytic toxin genes. Our work reveals a strong decomposer succession in wild-caught fish, providing functional and taxonomic insights into the vertebrate necrobiome.
INTRODUCTION
The decomposition of animal tissues is a fundamental ecological process that impacts nutrient cycling and species composition in terrestrial and aquatic ecosystems. Vertebrate tissue decomposition creates a unique ecological niche supporting a wide variety of specialized decomposer species, including insects, predators, and microorganisms. These species form an interconnected community whose combined activities lead to the decomposition of an organism from its initial death to the complete degradation of its exterior and internal contents.
The microbial communities involved in decomposition, including bacteria derived from the surrounding environment (e.g., water, soil) and the host (e.g., digestive tract and lungs), are collectively referred to as the “necrobiome” (from nekrós, the Greek word for dead body) (1), or alternatively, the “thanatomicrobiome” (from Thanatos, the Greek god of death) (2). Studies of necrobiome structure and function in several model systems (e.g., human, cow, pig, and mouse) have revealed strong microbial succession with distinct taxonomic and functional shifts linked to the phases of tissue decomposition (3–8). After cellular autolysis breaks down tissue following death, anaerobic bacteria such as Clostridium spp. increase in relative abundance and metabolize available carbohydrates and proteins from the body, producing organic acids and gas (9). Functional shifts occur; these shifts include increases in catabolic pathways, carbohydrate and energy metabolism, nitrogen cycling, and processes related to bacterial invasion. Foul-smelling compounds associated with the process of putrefaction are also produced as by-products of fermentation and amino acid decomposition, including putrescine, cadaverine, and indole. Because putatively pathogenic bacteria proliferate within vertebrate necrobiomes, such as Clostridium botulinum (10), it has been proposed that bacterial toxins secreted by these bacteria may play roles in decomposition by interfering with host cellular functions (11).
Although much knowledge of necrobiome community structure and function has come from studies of terrestrial mammals, less is known about the structure, function, and dynamics of decomposition in aquatic ecosystems. Previous studies of fish carcass decomposition demonstrate that as in terrestrial systems, both macroinvertebrates and microorganisms play important roles as aquatic decomposers (12, 13). Microbial analysis of forage fish (i.e., menhaden) carrion decomposing for 48 h in an estuary-like environment revealed a necrobiome dominated by members of the Firmicutes, Bacteroidetes, and Gammaproteobacteria (14). Similarly, a freshwater study of salmon carcasses reported that members of the Proteobacteria, Firmicutes, and Bacteroidetes dominated carcass decomposition (15). Despite these previous studies characterizing the taxonomic shifts associated with fish decomposition, much more work is needed to explore the microbial communities and their functions within fish necrobiomes and their associated aquatic ecosystems. Particularly important research questions include the following. (i) What is the composition of aquatic vertebrate necrobiomes and how does it change over time? (ii) How do changes in environmental parameters affect necrobiome communities? (iii) What metabolic activities/functions are present in necrobiome communities and how do they change over time?
In this study, to gain insights into these questions, we focused on the rainbow darter (Etheostoma caeruleum) as a model organism to profile microbial decomposer community succession from both taxonomic and functional perspectives. Rainbow darters are a common North American fish found in streams and small- to medium-sized river, and they have high site fidelity and sensitivity to changes in water quality (16–18). Thus, they represent ideal targets for exploring the role of host-associated and location-specific microbial communities present before and after death. Rainbow darters are affected physiologically by disturbances of the river ecosystem, such as wastewater treatment plant (WWTP) effluent inputs and urbanization (16–18). These anthropogenic factors affect rivers by introducing pollutants and altering the balance of available nutrients (18–20), thus influencing fish physiology and their associated microbiomes. Using 16S rRNA sequencing combined with metagenomics, we study both the taxonomic and functional succession of the rainbow darter necrobiome community. We also compare necrobiomes between two different locations in the Grand River (southwestern Ontario, Canada), upstream and downstream of a WWTP, allowing us to analyze community members and their functional potential both spatially and temporally. Studying necrobiome-associated microbial communities provides a unique way to better understand links to aquatic health, fish physiology, and ecosystem dynamics.
RESULTS AND DISCUSSION
Time series community profiling of fish necrobiomes.
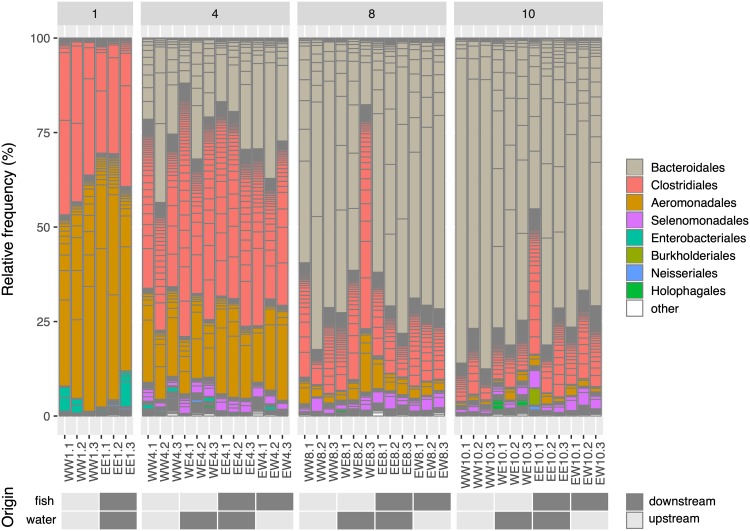
To examine the structure and temporal succession of aquatic vertebrate necrobiomes, we performed a 16S rRNA-based study of decomposing fish at different time points and locations. We collected female rainbow darters (Etheostoma caeruleum) from the Grand River in Waterloo, Ontario, Canada, both upstream and downstream of the Waterloo wastewater treatment plant (WWTP) (Fig. 1). Individual fish were subjected to decomposition with river water and sediment at room temperature for 1, 4, 8, and 10 days in sterile containers that acted as microcosms of a natural decomposition environment. Sample 16S rRNA gene profiles for fish decomposition microbiomes (“necrobiomes”) for these four time points and two water/sediment sources revealed reproducible microbial communities among independent replicates and also between environments (i.e., fish and water source; Fig. 2 and 3). This microbial succession was apparent at the order level of taxonomy (Fig. 2) and at the level of amplicon sequence variants (ASVs) (Fig. 3), although variation in ASV composition was evident among fish samples and environments (Fig. 3).
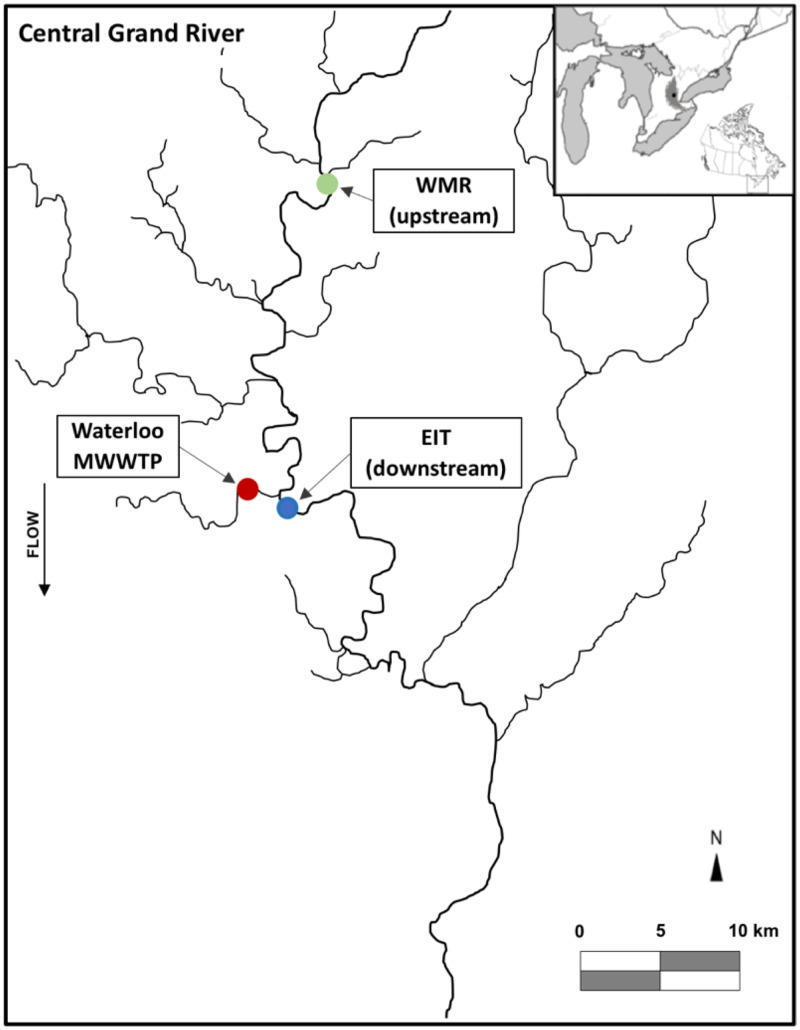
FIG 1.
Map showing sampling locations of Grand River fish for metagenomic analysis. The municipal wastewater treatment plant (WWTP) for the city of Waterloo, Canada, and the two sampling locations, upstream at West Montrose (WMR) and downstream at the Economic Insurance Trail (EIT), are displayed.
FIG 2.
Relative frequency of ASVs within each sample colored by taxonomic order. Samples are sorted by decomposition time (1 day, 4 days, 8 days, and 10 days). The fish and water/sediment origin of the samples are displayed at the bottom of the figure, with upstream referring to the WMR site and downstream referring to the EIT site. Low-relative-abundance taxonomic orders are grouped into “other.”
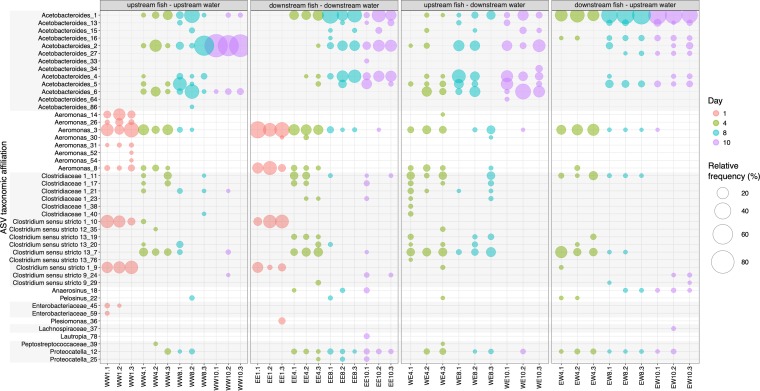
FIG 3.
Bubble-plot depicting the relative abundance (as a percentage) of ASVs in the fish necrobiome at four time points. Light gray boxes indicate shared family level taxonomic affiliation. Bubbles are displayed only if the ASV taxonomic affiliation was ≥2%. For other ASVs, see Data Set S1A in the supplemental material. Bubbles are colored by decomposition time (days).
(A) ASV table; (B) differential relative abundances; (C) Bin_4 VFanalyzer results; (D) Bin_11 VFanalyzer results. Download Data Set S1, XLSX file, 0.4 MB (398.8KB, xlsx) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although infectious lesions began to form on fish sampled on day 1, “bloat-stage” decomposition associated with anaerobic microbial decomposition was not visually apparent until later time points (see Fig. S1b in the supplemental material). The day 1 decomposer communities were composed predominantly of taxa affiliated with Aeromonas and Clostridium, and to a lesser extent by members of the Enterobacteriales. All of these bacterial groups are commonly associated with fish gut microbial communities (21–24), especially Aeromonas within freshwater fish microbiota (25, 26). Species of Clostridium sensu stricto 1 have proteolytic activities and thus may be associated with a more carnivorous diet (24), consistent with the predominantly insectivorous diets of rainbow darters (27). The taxa that were abundant in day 1 fish (e.g., Clostridium sensu stricto 1 sp. ASV 9 and 10, and Aeromonas sp. ASV 3 and 8) decreased in relative abundance over the course of decomposition from 11 to 28% on day 1 to < 2% on day 10.
Decomposition setup and images of fish decay. (a) Example of the mason jars used for decay of female rainbow darter (Etheostoma caeruleum) in a microcosm of the Grand River. (b) Representative images of decay of female rainbow darter for enrichment of the necrobiome at 1, 4, 8, and 10 days. Download FIG S1, TIF file, 2.2 MB (2.2MB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fish sampled on day 4 were associated with anaerobic bloat-stage decomposition and advanced tissue degradation (Fig. S1b). Consistent with previous studies of decomposition using other fish species (14, 15), dominant day 4 bacterial phyla detected were Proteobacteria, Firmicutes, and Bacteroidetes (Fig. 2). Compared to day 1 samples, there were considerable changes in degrader community composition for day 4 profiles, with substantial increases in Acetobacteroides ASVs (family Rikenellaceae, order Bacteroidales) from < 2% on day 1 up to 21% for some ASVs (Fig. 2 and 3). The Acetobacteroides genus was associated with 45 distinct ASVs across all samples, with 13 ASVs at a relative abundance of ≥2%. Characterized Acetobacteroides members are fermentative, mesophilic, strictly anaerobic, and capable of metabolizing carbohydrates and producing acetate, H2, and CO2 as end products (28). These bacteria also classify under the “Blvii28 wastewater-sludge group” according to the SILVA database. Following day 4, taxa affiliated with Acetobacteroides were the dominant decomposer group, increasing to a relative abundance of as much as 87% in the decomposer community by the final day 10 sampling. Distinct Acetobacteroides ASVs dominated the day 10 decomposer community for fish collected from each of the two river locations (Fig. 3). Acetobacteroides ASV 2 increased to a relative abundance 77% in the fish and water pairing collected upstream of the Waterloo WWTP, whereas Acetobacteroides ASV 1 increased to a relative abundance of 51% in fish collected downstream of the WWTP and left to decompose in water/sediment collected from upstream. These results indicate an influence of the sampled environment on species- or strain-level variations in decomposer communities. The day 4 decomposer communities were also associated with an increase in taxa affiliated with the Selenomonadales order, including Pelosinus and Anaerosinus genera (Fig. 3), which persisted throughout the decomposition process but at low relative abundance. The relative abundance of Selenomonadales increased from 0.0037% on day 1 to 3.8% on day 4, 3.5% on day 8, and 3.3% on day 10.
Influence of spatial location on fish necrobiome succession.
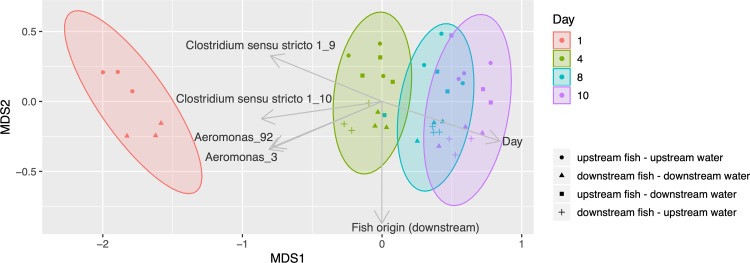
Based on sample ordination, necrobiome 16S rRNA gene profiles separated primarily by time point, with distinct microbial communities associated with different stages of tissue decomposition (Fig. 4). Necrobiomes also separated according to sample type, demonstrating distinct profiles for fish that originated upstream versus downstream of the WWTP, but this separation was less apparent than separation based on time. There was no strong effect of water/sediment origin on sample separation. This pattern was found for samples in which fish decomposed in water from the same location and in “swap” experiments in which fish were transferred into sediment/water samples derived from different original locations (e.g., upstream WWTP fish decomposing in downstream WWTP water). Even when isolating the effect of decomposition time, no effect of water/sediment sample origin was detected (Fig. S2). Thus, necrobiomes appear to be influenced primarily by factors occurring prior to decomposition, such as the living fish microbiomes, physiological states, or other fish-environment interactions.
FIG 4.
A nonmetric multidimensional scaling (NMDS) ordination of necrobiomes based on microbial community composition, using Bray-Curtis distances generated from ASV frequency profiles. Stress is 0.098. Together, 99% of the variance is represented based on the R2 value between distance in ordination space and distance in the original matrix. Vectors with R2 values greater than 0.7 were shown on the plot. Ellipses are colored by decomposition time (days).
NMDS ordination of necrobiomes after 4, 8, and 10 days of decomposition based on microbial community composition with Bray-Curtis distances. The ordinations in the top and bottom row for each time point are the same. The ellipses for the top row were calculated for sample fish origin, while the ellipses for the bottom row were calculated for sample water/sediment origin. A strong agreement between the ordination space and the distance matrix was observed for time points 4 days, 8 days, and 10 days (R2 = 0.984, 0.989, and 0.991, respectively) and the stress values are 0.127, 0.103, and 0.097, respectively. Download FIG S2, TIF file, 1.0 MB (1,017.7KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
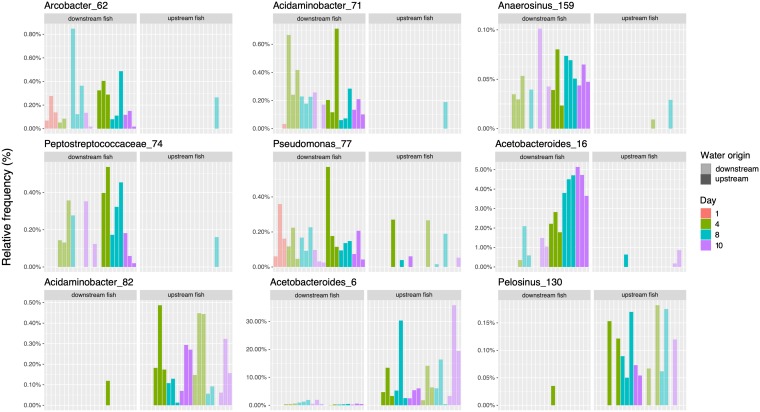
In order to further investigate the influence of fish origin location on decomposer microbial community differences, we calculated differentially abundant taxa (P < 0.05, Mann-Whitney U test) among fish necrobiomes associated with upstream and downstream WWTP fish sources (Fig. 5; for additional ASVs, see Table S2 in the supplemental material; for additional statistical methods, see Data Set S1B). Necrobiome-associated taxa with significantly increased relative abundances (P < 0.05 and fold changes of >3) in fish collected upstream of the WWTP include species of Acetobacteroides (ASV 6) and Pelosinus (ASV 130) (Fig. 5). Taxa with significantly increased relative abundances in downstream fish samples include Anaerosinus (ASV 159), Peptostreptococcaceae (ASV 74), Arcobacter (ASV 62), and Pseudomonas (ASV 77). However, these ASVs were low-abundance organisms with a relative abundance of less than 1%. Characterized Peptostreptococcaceae species are anaerobic bacteria that include pathogens associated with tissue infections and antibiotic resistance (29). Known Arcobacter species are aerotolerant and include human and animal pathogens that have been found in groundwater and water reservoirs (30–32). An additional taxonomic group that increased in relative abundance in necrobiomes originating from fish collected from downstream of the WWTP was Tolumonas (e.g., ASV 176), a genus with member species associated with the production of toluene from phenol precursors, which are known wastewater pollutants (33, 34). Further investigation into the contribution of wastewater effluent on fish decomposition would be helpful to confirm that these differences were indeed linked to WWTP effluent, especially because these putative pathogenic bacteria may contribute to a portion of the energetic costs of fish living downstream of WWTPs (18).
FIG 5.
Differentially abundant ASVs between necrobiomes from upstream versus downstream WWTP fish samples. Differential relative abundance was calculated based on pairwise rank sum tests of relative frequency (community proportion). The top ASVs with adjusted P values of less than 0.05 are shown. Bars are colored by decomposition time (days), with opacity determined by water origin.
In samples with both fish and water/sediment originating downstream of the WWTP, we also observed higher relative abundance of ASVs associated with Plesiomonas and Lautropia genera (Fig. 3). Again, these ASVs had low (<1%) relative abundance. Known Plesiomonas species associate with aquatic habitats, cause human infections associated with uncooked shellfish, and have been implicated in infectious outbreaks in regions, including Canada (35, 36). Lautropia species have been isolated from the oral cavities of immunocompromised individuals suffering from HIV and cystic fibrosis (37, 38).
Metagenomic binning and analysis of decomposition pathways.
To explore the genomes and genome-encoded metabolic/functional potential of the necrobiomes, we performed metagenomic sequencing on one replicate for each condition (14 total). Subsequent assembly and binning resulted in four MAGs (metagenome-assembled genomes) with >85% completion and <5% redundancy. We examined the taxonomic composition of the MAGs using MetAnnotate (39). These MAGs included two genomes affiliated with Alistipes (Rikenellaceae), a genome annotated as Aeromonas veronii, and a Selenomonadaceae-associated genome (Table 1). The bins are consistent with ASVs identified by 16S rRNA gene sequencing, corresponding to Acetobacteroides (Rikenellaceae), Aeromonas, and various members of the Selenomonadales (Fig. 2 and 3). Other ASVs identified by 16S rRNA gene sequencing were also recovered in the lower-quality MAGs (Table 1). One bin was affiliated with the genus Pseudomonas, and another bin was affiliated with the family Rikenellaceae.
TABLE 1.
Bins obtained from metagenomic sequencing of fish necrobiomes
| Bin name |
Completion (%) |
Redundancy (%) |
GC content (%) |
Total length (Mb) |
Gene count |
Contig count |
Taxonomic affiliation (predicted) |
|---|---|---|---|---|---|---|---|
| Bin_4 | 98.6 | 0.7 | 60.7 | 3.85 | 3,855 | 784 |
Bacteria, Proteobacteria, Gammaproteobacteria, Aeromonadales, Aeromonadaceae, Aeromonas, Aeromonas veronii |
| Bin_9 | 97.1 | 1.4 | 47.5 | 2.25 | 2,216 | 402 |
Bacteria, Firmicutes, Negativicutes, Selenomonadales, Selenomonadaceae, Propionispira |
| Bin_3 | 87.1 | 2.2 | 47.0 | 2.64 | 2,467 | 801 |
Bacteria, Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae, Alistipes |
| Bin_10 | 92.8 | 2.2 | 44.0 | 3.26 | 2,882 | 368 |
Bacteria, Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae, Alistipes |
| Bin_7 | 38.8 | 7.9 | 61.4 | 0.78 | 1,187 | 628 |
Bacteria, Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae, Pseudomonas |
| Bin_2 | 25.2 | 1.4 | 48.2 | 1.71 | 1,872 | 960 |
Bacteria, Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae |
The relative abundance of Bin_4 (Aeromonas veronii) decreased throughout decomposition from an average relative abundance of 3.7 (day 1) to an average relative abundance of 0.14 (day 10), consistent with our 16S rRNA data (Fig. S3a). Because Aeromonas has been associated with fish gut microbiomes (40–44), it is possible that Bin_4 and other Aeromonas taxa were initially derived from the fish guts and were important only for early stage decomposition. In contrast, Bin_3 (Rikenellaceae family) may represent a late-stage decomposer because its relative abundance increased in metagenomes from days 8 to 10 of decomposition (average relative abundance of 3.9 on day 8 to an average relative abundance 5.1 on day 10; Fig. S3a). In the downstream fish-upstream sediment/water set, both Rikenellaceae-affiliated bins (Bin_3 and Bin_10) were similar in relative abundance, implying site-specific influences on the relative abundance of different Rikenellaceae-affiliated taxa, consistent with 16S rRNA gene data for Acetobacteroides ASVs (Fig. 3). Phylogenetic analysis of the two Rikenellaceae-associated bins revealed that Bin_3 was more closely related to Acetobacteroides hydrogenigenes RL-C and Bin_10 was more closely related to Alistipes sp. strain ZOR0009 (Fig. S3b). Bin_9 (Propionispira) was present at low (0.0 to 0.54 average on days 1 to 10; Fig. S3a) relative abundance, close to the sample’s mean coverage across the entire course of decomposition, consistent with the abundance patterns seen for Selenomonadales based on 16S rRNA gene data (Fig. 2).
Metagenomic bin relative abundance and phylogenetic analysis of Bin_3 and Bin_10. (a) Relative abundance of four high-quality binned genomes across each necrobiome sample. Relative abundance was computed as mean bin coverage/mean sample coverage. Mean coverage was calculated per base pair using Anvi’o (66). (b) RAxML tree using the LG likelihood model made from concatenated single-copy core protein sequences detected with Anvi’o (Campbell et al. set [70]). The tree outgrouped with Lentimicrobium saccharophilum. Acetobacteroides hydrogenigenes, representatives of Alistipes strains, and all uncharacterized Alistipes isolates were used for this tree and sourced from NCBI Genome. This tree was visualized with iTOL (69). Download FIG S3, TIF file, 0.6 MB (650.7KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
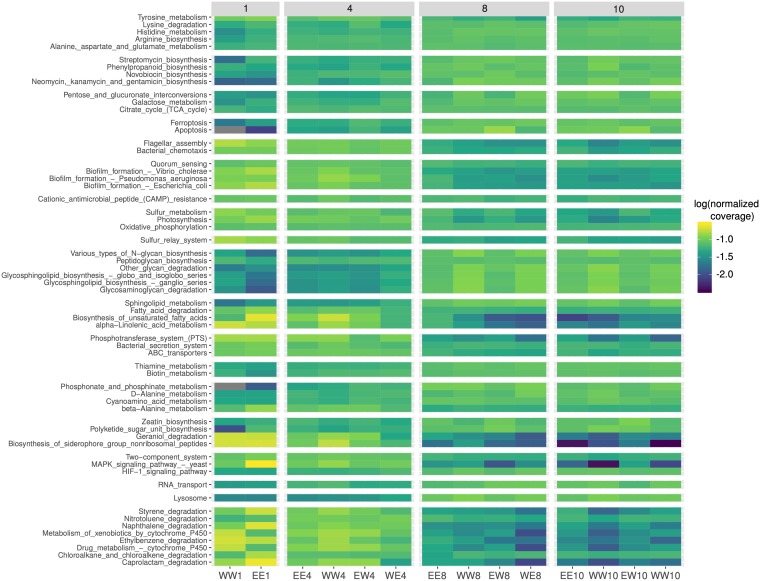
Using a KEGG analysis of assembled contigs and binned metagenomes, we examined metabolic pathway potentials associated with decomposition samples. The resulting functional profiles had a highly similar grouping in ordination space compared to the 16S rRNA gene community profiles, whereby samples grouped primarily based on decomposition time point (Fig. S4). Analysis of specific KEGG pathways revealed patterns consistent with a functional succession, mirroring the taxonomic succession described earlier (Fig. 6). Pollutant degradation pathways for polyaromatic hydrocarbons such as naphthalene, styrene, and nitrotoluene showed increased relative abundances on day 1 (13% on average) compared to subsequent time points (6.2% on average). The initial fish bacterial community may have been enriched for microorganisms that could degrade river water contaminants, which can originate from both anthropogenic and natural sources and bioaccumulate in fish (45–47). Naphthalene degradation in polluted sediment-water systems can be accomplished through several bacterial pathways, and bioremediation of this toxic molecule by native organisms is currently being studied (48–50). Various biofilm formation pathways were also proportionally abundant (13%) within day 1 metagenomes (Fig. 6), possibly reflecting skin and gut community functions originating prior to decomposition. Degrading river water contaminants and skin and gut biofilm formation may be functions that are more important for the bacterial communities living with their fish host and dealing with possibly contaminated river water than for the necrobiome that formed in our closed system after the fish’s death.
FIG 6.
Selected KEGG pathways displaying significant differential relative abundance across the course of decomposition. Pathways were selected that had an unadjusted P value of <0.03 after a Kruskall-Wallis test comparing decomposition time (1, 4, 8 , and 10 days). Shown is the log10 value of the fractional coverage of the pathway with respect to the total coverage across all the pathways in the sample. Total pathway coverage is also proportionally normalized across every sample. Note that some pathways are based on a few representative genes. For example, coverage of the photosynthesis pathway is mainly derived from genes encoding sodium ion pumps.
NMDS ordination of metagenomic functional profiles with Bray-Curtis distances calculated based on KEGG pathway frequencies. A strong agreement between the ordination space and the distance matrix was observed (R2 = 0.996), and the stress value is 0.063. Download FIG S4, TIF file, 0.5 MB (483.5KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Glycan metabolism generally increased in coverage from early stages (2.4% on day 1) to later stages of decomposition (10%). Glycan degradation pathways (e.g., glycosaminoglycans) increased in coverage by days 8 and 10, which may be involved in decomposition of fish skin and intestinal mucins. Late-stage increases in streptomycin, phenylpropanoid, novobiocin, neomycin, kanamycin, and gentamicin biosynthesis pathways (2.4-fold change from day 1 to 10) were also detected, implying that the remaining microorganisms by day 10 possess increased potential for antibiotic synthesis.
These metagenome-wide functional patterns closely matched the functional potentials of individual Aeromonas (early stage) and Rikenellaceae (late stage) bins, when taking into consideration their shifts in relative abundance through the time course (Fig. S5). Genes belonging to pollutant degradation pathways were present in the Aeromonas bin yet mostly absent from other MAGs with lower relative abundance from days 1 and 4 metagenomes. Likewise, biofilm formation pathway genes had a 6.2-fold-higher frequency in the Aeromonas bin compared to the Acetobacteroides/Alistipes bins. In contrast, antibiotic biosynthesis pathway genes had a 2.5-fold-higher frequency in the Rikenellaceae-associated bins, in addition to multiple key glycan degradation genes. Thus, the detected shifts in functional profiles were in part due to the hand-off microbial community dominance from Aeromonas to Rikenellaceae. It is important to note that these apparent late-stage functional shifts could also be important for earlier phases when Rikenellaceae initially began to increase in relative abundance.
Count of KEGG annotations mapping to the corresponding KEGG pathways in Fig. 6 across each MAG. Shown is the log10 value of the fractional frequency of the pathway with respect to the total across all the pathways in the sample. The total pathway coverage is also proportionally normalized across every sample. Download FIG S5, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our data suggest strong Acetobacteroides dominance in late-stage rainbow darter necrobiomes (Fig. 2 and 3). Because related species have been implicated in anaerobic sugar fermentation (28), we investigated the two MAGs affiliated with these bacteria for glycolytic enzymes. Both Bin_3 and Bin_10 possess a complete glycolysis pathway as well as l-lactate dehydrogenase for anaerobic fermentation (Fig. S6). Bin 3 genes also encode pyruvate dehydrogenase, aldehyde dehydrogenase, and enzymes for conversion of d-fructose, d-fructose-1-phosphate (d-fructose-1P), and d-mannose-6P to glycolysis precursors. Based on a previous analysis of decomposition pathways (51), Bin_3 and Bin_10 genes also encode components of potential pathways for production of indole (EC 4.1.99.1), putrescine (EC 3.5.3.11), and spermidine (EC 2.5.1.6 and 2.5.1.16), in addition to histidine degradation (EC 4.3.1.3, 4.2.1.49, and 3.5.3.8, Bin_10 only).
End of the KEGG glycolysis/gluconeogenesis pathway for Rikenellaceae Bin_10 and Bin_3. Green indicates the presence of a match to that enzyme. Images were generated using KEGG (67). Download FIG S6, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Previous research showed that Acetobacteroides hydrogenigenes RL-C can produce acetate and carbon dioxide from glucose fermentation (28). Because these metabolites could potentially be converted to methane by methanogenic archaea (52), we analyzed the assembled metagenomic data for archaea-associated contigs. Contigs taxonomically affiliated with methanogenic archaea were identified from almost all samples, including species of the classes Methanobacteria, Methanococci, and Methanomicrobia (Fig. S7a). A 1.7-fold-higher relative abundance of these contigs was observed after day 1 (Fig. S7a), indicating that methanogenic archaea may have increased in relative abundance early in decomposition, coinciding with anoxic conditions and the generation of acetate or carbon dioxide by Rikenellaceae bacteria. However, no archaeal taxa were identified in the 16S rRNA. This potentially reflects an increased ability to detect low-abundance archaeal organisms in our metagenomic data set, perhaps due to the availability of more taxonomic markers and/or lower archaeal 16S copy numbers (53).
Relative abundance of methanogen contigs and metagenomic bins. (a) Relative abundance (calculated by mean contig coverage/mean sample coverage) of archaeal contigs belonging to the methanogen classes Methanobacteria, Methanococci, and Methanomicrobia. (b) Relative abundance (calculated by mean bin coverage/mean sample coverage) of the four high-quality bins assembled from the fish necrobiome community as well as a small incomplete bin, Bin_11. Download FIG S7, TIF file, 0.8 MB (791.4KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A toxigenic strain of Aeromonas veronii is a dominant member of the necrobiome.
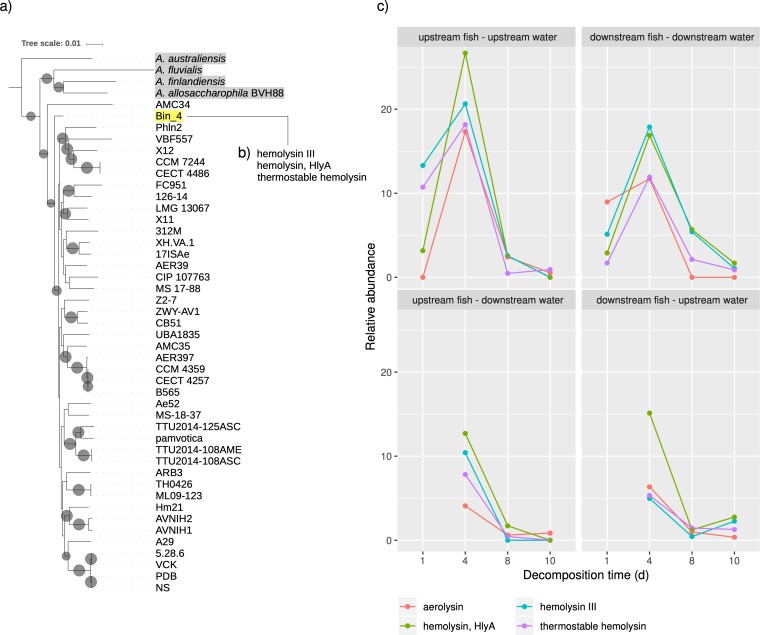
Because Bin_4 affiliated with A. veronii, a well-established pathogen of fish and humans (36, 54–60), and a common inhabitant of the fish gut microbiome (40–43), we explored its phylogenetic position, functional profile, and virulence repertoire. A maximum likelihood phylogeny of A. veronii and other related Aeromonas genomes from the NCBI was constructed based on a concatenated alignment of conserved ribosomal marker genes (Fig. 7a). Within this phylogeny, Bin_4 grouped with a clade of A. veronii genomes but as a basal lineage outgrouping all A. veronii species except AMC34.
FIG 7.
A toxigenic Aeromonas veronii-like strain is a dominant species in early decomposition. (a) RAxML tree using the GTR+GAMMA model made from concatenated single-copy core gene nucleotide sequences detected with Anvi’o (Campbell et al. set [70] ). The tree was outgrouped on Aeromonas hydrophila. Gray circles are scaled to bootstrap support of ≥85, with the largest size representing 100. Aeromonas species outside Aeromonas veronii are highlighted in gray. Representative Aeromonas veronii strains from the NCBI Genome Tree report were chosen to display here (not highlighted), and only their strain name is shown. This tree was visualized with iTOL (69). (b) Bin_4’s predicted toxin repertoire from VFDB. (c) Relative abundance (mean gene coverage/mean sample coverage) of Aeromonas hemolysin toxin genes. Decomposition time is shown in days.
We used the VFanalyzer from the Virulence Factor Database (VFDB) to detect virulence factors within Bin_4 and compared it to a reference Aeromonas strain, A. veronii B565. This bin contained virulence-related genes for adherence, iron uptake, and secretion systems (Data Set S1C). Indeed, a total of 54 genes that were associated with secretion systems were identified, compared to only 15 in A. veronii B565. In addition, we identified 13 genes associated with endotoxin production. Like A. veronii B565, Bin_4 genes encoded hemolysin III, hemolysin HlyA, and a thermostable hemolysin gene (Fig. 7b). We also recovered a relatively small incomplete bin (Bin_11, 0.64 Mb, 717 coding sequences [CDSs], 321 contigs) that correlated with Bin_4 in relative abundance (Fig. S7b). This small bin affiliated with Aeromonas veronii and also included a gene encoding aerolysin toxin production (Data Set S1D). Based on metagenomic mapped read coverage, the relative abundance of genes encoding Aeromonas toxins increased on day 4 of decomposition (Fig. 7c), indicating an enrichment in Aeromonas strains carrying hemolytic proteins. A possible explanation for this is that lytic toxins, including those from Aeromonas, may function in host cell lysis during decomposition and therefore peak in relative abundance during earlier stages of decomposition. Bin_4 also possessed genomic potential for decomposition-related pathways, including histidine degradation (contains EC 4.3.1.3, 4.2.1.49, 3.5.2.7, and 3.5.3.8) and the production of putrescine (EC 4.1.1.19, 3.5.3.12, and 3.5.1.53), indole (EC 4.1.99.1), and cadaverine (EC 4.1.1.18).
Conclusion.
Overall, our microcosm study of rainbow darter fish decomposition revealed a highly reproducible microbial succession throughout the time course, even across different fish and water/sediment sources. The location of the fish when sampled (upstream or downstream of the WWTP) also affected its decomposition profile, suggesting that necrobiomes may be influenced by prior fish-environment interactions. Together, our data suggest that environmental interactions may shape the initial gut community and/or the physiological state of the fish, which then seeds or impacts the later necrobiome community and its succession.
Both 16S and metagenomic analysis revealed a strong succession in which initial time points were dominated by Clostridiaceae and Aeromonas, with Rikenellaceae species appearing by day 4 and becoming major community members by day 10. Analysis of functional profiles inferred from the metagenomic data revealed common decomposition pathways, as well as temporal shifts in function that mirrored taxonomic succession. Notably, pollutant degradation pathways and biofilm formation pathways were enriched in the early stages of decomposition and associated with Clostridiaceae and Aeromonas, and glycan metabolism and antibiotic synthesis increased in later stages and associated with Rikenellaceae. Last, we identified a toxigenic Aeromonas strain that was a dominant member of the necrobiome community. The presence of numerous hemolytic toxin genes in this organism suggests a potential role for toxins in the decomposition of host tissues as proposed previously (11). Further work investigating the prevalence and function of toxigenic bacterial species in decomposer communities will be important to explore their broader ecological roles and niches within natural ecosystems.
MATERIALS AND METHODS
Fish collection.
On 24 October 2016, female rainbow darters (Etheostoma caeruleum) were collected from the Grand River (Fig. 1), both upstream (Westmontrose [WMR]; 43°35′08′′N; 80°28′53′′W) and downstream (Economic Insurance Trail [EIT]; 43°28′24′′N; 80°28′22′′W) of the Waterloo wastewater treatment plant (WWTP) (43°29′16′′N; 80°30′25′′W). Forty-two fish (21 from each site) were collected using a backpack electrofisher (Smith Root, LR-20) and euthanized quickly with a sharp blow to the head. Then each fish was placed in an autoclaved 250-ml mason jar microcosm that contained a mixture of water and river substrate (see Table S1 for river water quality metadata and Fig. S1a for an example mason jar setup). The lids were closed, but not sealed, in order to ensure oxic conditions that would accompany natural in-river decay events. The jars were then left to decay in a fume hood at room temperature. Three samples containing both fish and water/sediment from the same site were left to decompose for 1 day (24 h), 4 days, 8 days, and 10 days for both the WMR and EIT sites, totaling 24 fish. For additional treatments to assess differences in water quality and aquatic microorganisms, three samples containing fish and water/sediment from different sites (i.e., WMR fish in EIT conditions and EIT fish in WMR conditions) were allowed to decay for 4, 8, and 10 days, totaling 18 fish. At each time point, decay was documented (Fig. S1b), and fish were removed from the replicate jars, then rinsed with sterile water, and ground with liquid nitrogen using a clean mortar and pestle. The powdered tissue was stored at –80°C prior to genomic DNA extraction.
Water quality parameters from collection sites taken on 24 October 2016 upstream (43°35′08′′N; 80°28′53′′W) and downstream (43°28′24′′N; 80°28′22′′W) from the Waterloo wastewater treatment plant (WWTP; 43°29′16′′N; 80°30′25′′W). Data are presented as the means plus 1 standard deviation (STD) from 24 samples (hourly) from remote monitoring stations located along the Grand River. Data were obtained through the Grand River Conservation Authority (www.grandriver.ca). Download Table S1, PDF file, 0.05 MB (51.9KB, pdf) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially abundant ASVs between downstream WWTP and upstream WWTP necrobiome samples. Download Table S2, PDF file, 0.08 MB (78.1KB, pdf) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Experimental procedures and the use of animals in this study were approved by the University of Waterloo Animal Care Committee and within Canadian Council on Animal Care (CCAC) guidelines (AUPP 40318).
DNA extraction.
Unless noted, all chemicals and reagents were purchased from Sigma-Aldrich (Mississauga, Ontario, Canada). For DNA extraction, 100 mg of ground tissue was added to 1.2 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), 100 μl of 10% sodium dodecyl sulfate (SDS), 20 μl of proteinase K, 8 μl of RNase A, and 200 μl of 5 M NaCl. This mixture was vortexed quickly and incubated at 55°C for 30 min. Then 160 μl of CTAB extraction solution (2% cetrimonium bromide, 100 mM Tris, 20 mM EDTA, 1.4 M NaCl [pH 8.0]) was added, and the samples were further incubated at 65°C for 1.5 h. Following this lysis incubation, 700 μl of the lysate was extracted with an equal volume of phenol and centrifuged at 10,000 × g for 5 min. The aqueous phase was retained and twice extracted with equal volumes of phenol-chloroform-isoamyl alcohol (25:24:1), followed each time with centrifugation at 10,000 × g for 5 min. One volume of isopropanol was used to precipitate aqueous phase DNA in a new ultracentrifuge tube, followed by centrifugation at 13,000 × g for 10 min at room temperature. The resulting pellet was washed twice with 70% ethanol, dried, and then dissolved in 50 μl of DNase- and RNase-free H2O (Sigma) at 50°C for 15 min. The quantity and quality of DNA were determined with a SpectraDrop (Molecular Devices) and stored at –20°C prior to sequencing.
16S rRNA gene and metagenomic sequencing.
Extracted DNA was amplified in triplicate using Pro341F and Pro805R universal prokaryotic primers (61). Triplicate amplicons were pooled, gel quantified, and sequenced to a depth of at least 30,000 paired-end reads per sample using the MiSeq reagent kit v3 (2 × 300 cycles; Illumina).
For metagenomic sequencing, genomic DNA (1 ng) was fragmented and individually barcoded using the Nextera XT DNA Library Prep kit (Illumina) following the supplier’s guidelines. Small fragments of library DNA were removed by adding 0.6 volumes of AMPure XP beads (Beckman Coulter). After washing twice with 80% ethanol and air drying for 10 min, DNA was eluted from the beads with 10 mM Tris-HCl (pH 8.5). Purified library DNA was quantified with the Qubit dsDNA (double-stranded DNA) HS (high-sensitivity) assay kit, diluted to 4 nM with the Tris-HCl buffer and then pooled in an equal volume. Library DNA was denatured with equal volumes of 0.2 N NaOH, diluted to 7 pM with hybridization buffer HT1, and sequenced with MiSeq reagent kit v2 (2 × 250 cycles; Illumina).
16S rRNA gene analysis.
Demultiplexed sequences were processed using DADA2 v1.4 (62), managed through QIIME2 v.2017.10 (63). Briefly, forward and reverse reads were truncated with decreasing quality metrics while maintaining sequence overlap (∼250 bases). Primers were removed, and paired reads were assembled after error modeling and correction, creating amplicon sequence variants (ASVs). Chimeric ASVs were removed by reconstruction against more abundant parent ASVs. The resulting ASV table was constructed for downstream analysis (see Data Set S1A in the supplemental material).
Taxonomy was assigned to representative sequence variants using a naive Bayesian classifier implemented in QIIME2 with scikit-learn (v.0.19.0), trained against SILVA release 128 (64), clustered at 99% identity, and trimmed to the amplified region. Assignments were accepted above a 0.7 confidence threshold.
For ordination, we used a proportion matrix of ASVs across each sample with a sparsity cutoff (i.e., ASV detected in at least 3 of 42 samples). The metaMDS() and envfit() scripts from vegan package v2.4-2 in R were used to calculate ordination coordinates and data vectors. A stress or Shepard diagram was generated with stressplot() from the vegan package to determine the nonmetric fit. The ASVs with significant rank sum differences in sample proportion were calculated with the Mann-Whitney test in R. Multiple hypothesis correction of P values was performed using the p.adjust() function in R with the Benjamini-Hochberg model. We also calculated differential taxon relative abundance using a variety of methods (metagenomeSeq, edgeR, DESeq2, and LEfSe) as implemented in the Marker Data Profiling pipeline from MicrobiomeAnalyst (65) with default settings on 20 March 2020.
Metagenomic data analysis.
Raw reads were processed with TrimGalore v0.5.0, coassembled with metaSPAdes (SPAdes v3.12.0), and eukaryotic contigs were identified with Centrifuge v1.0.4 using their NCBI nr preindexed database (last updated 3 March 2018) and subsequently removed. Reads were mapped with Bowtie 2 v2.3.4.3 using default settings and binned using CONCOCT with Anvi’o v5.2 (minimum 1-kb contig cutoff). Mean coverage data for the metagenomic functional analyses and for the methanogen analysis were extracted from Anvi’o (66) using all contigs (no contig length cutoff).
For metagenomic and bin functional analysis, KEGG (Kyoto Encyclopedia of Genes and Genomes) annotations were identified with GhostKOALA (67). The average coverage for each gene (per base pair), normalized by dividing by the average sample coverage (per base pair), was summed to give a total coverage value for each KEGG pathway. The decostand() function from the vegan package v2.4-2 in R was used to determine the fractional value of each pathway with respect to the total summed coverage across all KEGG pathways detected in the sample. A Kruskall-Wallis test was done in R to identify KEGG pathways with significantly different distributions by day of decomposition. The decostand() function was also used to proportionally normalize each pathway value across every sample for plotting. For the bin functional analysis, the frequency of each KEGG orthology (KO) annotation in each MAG bin was counted. These counts were summed for each KEGG pathway, and fractional values were calculated across all KEGG pathways detected in the bins as before.
The VFanalyzer software from the Virulence Factor Database (VFDB) (68) identified virulence factors in the predicted coding sequences of Bin_4 using Aeromonas veronii B565 as a representative genome. We also used the domain architecture from the Aeromonas toxin gene set from the VFDB to identify Aeromonas toxin genes in the coassembly. Putative toxins longer than 150 amino acids were assessed with BLASTp for Aeromonas taxonomy and gene annotation.
Data availability.
All 16S rRNA gene and metagenomic sequencing data for this project were deposited into the NCBI Short Read Archive (SRA) under BioProject accession no. PRJNA604775.
ACKNOWLEDGMENTS
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) through Discovery Grants awarded to A.C.D. (RGPIN-2019-04266) and P.M.C. (RGPIN-2015-05643) as well as the Canada Foundation for Innovation (CFI 34317).
REFERENCES
- 1.Cobaugh KL, Schaeffer SM, DeBruyn JM. 2015. Functional and structural succession of soil microbial communities below decomposing human cadavers. PLoS One 10:e0130201. doi: 10.1371/journal.pone.0130201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Javan GT, Finley SJ, Can I, Wilkinson JE, Hanson JD, Tarone AM. 2016. Human thanatomicrobiome succession and time since death. Sci Rep 6:29598. doi: 10.1038/srep29598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metcalf JL, Wegener Parfrey L, Gonzalez A, Lauber CL, Knights D, Ackermann G, Humphrey GC, Gebert MJ, Van Treuren W, Berg-Lyons D, Keepers K, Guo Y, Bullard J, Fierer N, Carter DO, Knight R. 2013. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. Elife 2:e01104. doi: 10.7554/eLife.01104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pechal JL, Crippen TL, Benbow ME, Tarone AM, Dowd S, Tomberlin JK. 2014. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int J Legal Med 128:193–205. doi: 10.1007/s00414-013-0872-1. [DOI] [PubMed] [Google Scholar]
- 5.Hyde ER, Haarmann DP, Petrosino JF, Lynne AM, Bucheli SR. 2015. Initial insights into bacterial succession during human decomposition. Int J Legal Med 129:661–671. doi: 10.1007/s00414-014-1128-4. [DOI] [PubMed] [Google Scholar]
- 6.Guo J, Fu X, Liao H, Hu Z, Long L, Yan W, Ding Y, Zha L, Guo Y, Yan J, Chang Y, Cai J. 2016. Potential use of bacterial community succession for estimating post-mortem interval as revealed by high-throughput sequencing. Sci Rep 6:24197. doi: 10.1038/srep24197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metcalf JL, Xu ZZ, Weiss S, Lax S, Van Treuren W, Hyde ER, Song SJ, Amir A, Larsen P, Sangwan N, Haarmann D, Humphrey GC, Ackermann G, Thompson LR, Lauber C, Bibat A, Nicholas C, Gebert MJ, Petrosino JF, Reed SC, Gilbert JA, Lynne AM, Bucheli SR, Carter DO, Knight R. 2016. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 351:158–162. doi: 10.1126/science.aad2646. [DOI] [PubMed] [Google Scholar]
- 8.Burcham ZM, Pechal JL, Schmidt CJ, Bose JL, Rosch JW, Benbow ME, Jordan HR. 2019. Bacterial community succession, transmigration, and differential gene transcription in a controlled vertebrate decomposition model. Front Microbiol 10:745. doi: 10.3389/fmicb.2019.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter DO, Yellowlees D, Tibbett M. 2007. Cadaver decomposition in terrestrial ecosystems. Naturwissenschaften 94:12–24. doi: 10.1007/s00114-006-0159-1. [DOI] [PubMed] [Google Scholar]
- 10.Can I, Javan GT, Pozhitkov AE, Noble PA. 2014. Distinctive thanatomicrobiome signatures found in the blood and internal organs of humans. J Microbiol Methods 106:1–7. doi: 10.1016/j.mimet.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 11.Mansfield MJ, Doxey AC. 2018. Genomic insights into the evolution and ecology of botulinum neurotoxins. Pathog Dis 76:fty040. doi: 10.1093/femspd/fty040. [DOI] [PubMed] [Google Scholar]
- 12.Minshall GW, Hitchcock E, Barnes JR. 1991. Decomposition of rainbow trout (Oncorhynchus mykiss) carcasses in a forest stream ecosystem inhabited only by nonanadromous fish populations. Can J Fish Aquat Sci 48:191–915. doi: 10.1139/f91-026. [DOI] [Google Scholar]
- 13.Premke K, Fischer P, Hempel M, Rothhaupt KO. 2010. Ecological studies on the decomposition rate of fish carcasses by benthic organisms in the littoral zone of Lake Constance, Germany. Ann Limnol 46:157–168. doi: 10.1051/limn/2010017. [DOI] [Google Scholar]
- 14.Burkepile DE, Parker JD, Woodson CB, Mills HJ, Kubanek J, Sobecky PA, Hay ME. 2006. Chemically mediated competition between microbes and animals: microbes as consumers in food webs. Ecology 87:2821–2831.2.0.co;2] doi: 10.1890/0012-9658(2006)87[2821:CMCBMA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 15.Pechal JL, Benbow ME. 2016. Microbial ecology of the salmon necrobiome: evidence salmon carrion decomposition influences aquatic and terrestrial insect microbiomes. Environ Microbiol 18:1511–1522. doi: 10.1111/1462-2920.13187. [DOI] [PubMed] [Google Scholar]
- 16.Fuzzen MLM, Bennett CJ, Tetreault GR, McMaster ME, Servos MR. 2015. Severe intersex is predictive of poor fertilization success in populations of rainbow darter (Etheostoma caeruleum). Aquat Toxicol 160:106–116. doi: 10.1016/j.aquatox.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Hicks KA, Servos MR. 2017. Site fidelity and movement of a small-bodied fish species, the rainbow darter (Etheostoma caeruleum): implications for environmental effects assessment. River Res Applic 33:1016–1025. doi: 10.1002/rra.3161. [DOI] [Google Scholar]
- 18.Mehdi H, Dickson FH, Bragg LM, Servos MR, Craig PM. 2018. Impacts of wastewater treatment plant effluent on energetics and stress response of rainbow darter (Etheostoma caeruleum) in the Grand River watershed. Comp Biochem Physiol B Biochem Mol Biol 224:270–279. doi: 10.1016/j.cbpb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Guillette LJ Jr, Gunderson M. 2001. Alterations in development of reproductive and endocrine systems of wildlife populations exposed to endocrine-disrupting contaminants. Reproduction 122:857–864. doi: 10.1530/rep.0.1220857. [DOI] [PubMed] [Google Scholar]
- 20.Carey RO, Migliaccio KW. 2009. Contribution of wastewater treatment plant effluents to nutrient dynamics in aquatic systems: a review. Environ Manage 44:205–217. doi: 10.1007/s00267-009-9309-5. [DOI] [PubMed] [Google Scholar]
- 21.Trust TJ, Bull LM, Currie BR, Buckley JT. 1979. Obligate anaerobic bacteria in the gastrointestinal microflora of the grass carp (Ctenopharyngodon idella), goldfish (Carassius auratus), and rainbow trout (Salmo gairdneri). J Fish Res Board Canada 36:1174–1179. doi: 10.1139/f79-169. [DOI] [Google Scholar]
- 22.Sugita H, Tokuyama K, Deguchi Y. 1985. The intestinal microflora of carp Cyprinus carpio, grass carp Ctenopharyngodon idella and tilapia Sarotherodon niloticus. Nippon Suisan Gakkaishi 51:1325–1329. doi: 10.2331/suisan.51.1325. [DOI] [Google Scholar]
- 23.Nayak SK. 2010. Role of gastrointestinal microbiota in fish. Aquaculture Res 41:1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x. [DOI] [Google Scholar]
- 24.Li T, Long M, Gatesoupe F-J, Zhang Q, Li A, Gong X. 2015. Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb Ecol 69:25–36. doi: 10.1007/s00248-014-0480-8. [DOI] [PubMed] [Google Scholar]
- 25.Ringø E, Strøm E. 1994. Microflora of Arctic charr, Salvelinus alpinus (L.): gastrointestinal microflora of free-living fish and effect of diet and salinity on intestinal microflora. Aquaculture Res 25:623–629. doi: 10.1111/j.1365-2109.1994.tb00726.x. [DOI] [Google Scholar]
- 26.Tarnecki AM, Burgos FA, Ray CL, Arias CR. 2017. Fish intestinal microbiome: diversity and symbiosis unravelled by metagenomics. J Appl Microbiol 123:2–17. doi: 10.1111/jam.13415. [DOI] [PubMed] [Google Scholar]
- 27.French JRP, Jude DJ. 2001. Diets and diet overlap of nonindigenous gobies and small benthic native fishes co-inhabiting the St. Clair River, Michigan. J Great Lakes Res 27:300–311. doi: 10.1016/S0380-1330(01)70645-4. [DOI] [Google Scholar]
- 28.Su X-L, Tian Q, Zhang J, Yuan X-Z, Shi X-S, Guo R-B, Qiu Y-L. 2014. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol 64:2986–2991. doi: 10.1099/ijs.0.063917-0. [DOI] [PubMed] [Google Scholar]
- 29.Murphy EC, Frick I-M. 2013. Gram-positive anaerobic cocci–commensals and opportunistic pathogens. FEMS Microbiol Rev 37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 30.Jacob J, Woodward D, Feuerpfeil I, Johnson WM. 1998. Isolation of Arcobacter butzleri in raw water and drinking water treatment plants in Germany. Zentralbl Hyg Umweltmed 201:189–198. [PubMed] [Google Scholar]
- 31.Rice EW, Rodgers MR, Wesley IV, Johnson CH, Tanner SA. 1999. Isolation of Arcobacter butzleri from ground water. Lett Appl Microbiol 28:31–35. doi: 10.1046/j.1365-2672.1999.00483.x. [DOI] [PubMed] [Google Scholar]
- 32.Diergaardt SM, Venter SN, Spreeth A, Theron J, Brözel VS. 2004. The occurrence of campylobacters in water sources in South Africa. Water Res 38:2589–2595. doi: 10.1016/j.watres.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Chertkov O, Copeland A, Lucas S, Lapidus A, Berry KW, Detter JC, Del Rio TG, Hammon N, Dalin E, Tice H, Pitluck S, Richardson P, Bruce D, Goodwin L, Han C, Tapia R, Saunders E, Schmutz J, Brettin T, Larimer F, Land M, Hauser L, Spring S, Rohde M, Kyrpides NC, Ivanova N, Göker M, Beller HR, Klenk H-P, Woyke T. 2011. Complete genome sequence of Tolumonas auensis type strain (TA 4). Stand Genomic Sci 5:112–120. doi: 10.4056/sigs.2184986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mrowiec B. 2014. Toluene in sewage and sludge in wastewater treatment plants. Water Sci Technol 69:128–134. doi: 10.2166/wst.2013.563. [DOI] [PubMed] [Google Scholar]
- 35.Potasman I, Paz A, Odeh M. 2002. Infectious outbreaks associated with bivalve shellfish consumption: a worldwide perspective. Clin Infect Dis 35:921–928. doi: 10.1086/342330. [DOI] [PubMed] [Google Scholar]
- 36.Janda JM, Abbott SL, McIver CJ. 2016. Plesiomonas shigelloides revisited. Clin Microbiol Rev 29:349–374. doi: 10.1128/CMR.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Dekhil SM, Peel MM, Lennox VA, Stackebrandt E, Sly LI. 1997. Isolation of Lautropia mirabilis from sputa of a cystic fibrosis patient. J Clin Microbiol 35:1024–1026. doi: 10.1128/JCM.35.4.1024-1026.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossmann SN, Wilson PH, Hicks J, Carter B, Cron SG, Simon C, Flaitz CM, Demmler GJ, Shearer WT, Kline MW. 1998. Isolation of Lautropia mirabilis from oral cavities of human immunodeficiency virus-infected children. J Clin Microbiol 36:1756–1760. doi: 10.1128/JCM.36.6.1756-1760.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrenko P, Lobb B, Kurtz DA, Neufeld JD, Doxey AC. 2015. MetAnnotate: function-specific taxonomic profiling and comparison of metagenomes. BMC Biol 13:92. doi: 10.1186/s12915-015-0195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugita H, Tanaka K, Yoshinami M, Deguchi Y. 1995. Distribution of Aeromonas species in the intestinal tracts of river fish. Appl Environ Microbiol 61:4128–4130. doi: 10.1128/AEM.61.11.4128-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molinari LM, De Oliveira Scoaris D, Pedroso RB, De Lucas Rodrigues Bittencourt N, Nakamura CV, Ueda-Nakamura T, De Abreu Filho BA, Dias Filho BP. 2003. Bacterial microflora in the gastrointestinal tract of Nile tilapia, Oreochromis niloticus, cultured in a semi-intensive system. Acta Sci Biol Sci 25:267–271. [Google Scholar]
- 42.Goldschmidt-Clermont E, Wahli T, Frey J, Burr SE. 2008. Identification of bacteria from the normal flora of perch, Perca fluviatilis L., and evaluation of their inhibitory potential towards Aeromonas species. J Fish Dis 31:353–359. doi: 10.1111/j.1365-2761.2008.00912.x. [DOI] [PubMed] [Google Scholar]
- 43.Jiang Y, Xie C, Yang G, Gong X, Chen X, Xu L, Bao B. 2011. Cellulase-producing bacteria of Aeromonas are dominant and indigenous in the gut of Ctenopharyngodon idellus (Valenciennes). Aquaculture Res 42:499–505. doi: 10.1111/j.1365-2109.2010.02645.x. [DOI] [Google Scholar]
- 44.Turner LA, Bucking C. 2019. The role of intestinal bacteria in the ammonia detoxification ability of teleost fish. J Exp Biol 222:jeb209882. doi: 10.1242/jeb.209882. [DOI] [PubMed] [Google Scholar]
- 45.Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- 46.D’Adamo R, Pelosi S, Trotta P, Sansone G. 1997. Bioaccumulation and biomagnification of polycyclic aromatic hydrocarbons in aquatic organisms. Mar Chem 56:45–49. doi: 10.1016/S0304-4203(96)00042-4. [DOI] [Google Scholar]
- 47.Meador JP, Stein JE, Reichert WL, Varanasi U. 1995. Bioaccumulation of polycyclic aromatic hydrocarbons by marine organisms. Rev Environ Contam Toxicol 143:79–165. [DOI] [PubMed] [Google Scholar]
- 48.Tomás-Gallardo L, Gómez-Álvarez H, Santero E, Floriano B. 2014. Combination of degradation pathways for naphthalene utilization in Rhodococcus sp. strain TFB. Microb Biotechnol 7:100–113. doi: 10.1111/1751-7915.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J, Hroudova M, Vlcek C, Cardenas E, Mackova M, Macek T. 2012. Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS One 7:e40653. doi: 10.1371/journal.pone.0040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, Lee Y, Jeon CO. 2019. Biodegradation of naphthalene, BTEX, and aliphatic hydrocarbons by Paraburkholderia aromaticivorans BN5 isolated from petroleum-contaminated soil. Sci Rep 9:860. doi: 10.1038/s41598-018-36165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur H, Das C, Mande SS. 2017. In silico analysis of putrefaction pathways in bacteria and its implication in colorectal cancer. Front Microbiol 8:2166. doi: 10.3389/fmicb.2017.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enzmann F, Mayer F, Rother M, Holtmann D. 2018. Methanogens: biochemical background and biotechnological applications. AMB Express 8:1. doi: 10.1186/s13568-017-0531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun D-L, Jiang X, Wu QL, Zhou N-Y. 2013. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol 79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joseph SW, Carnahan AM, Brayton PR, Fanning GR, Almazan R, Drabick C, Trudo EW, Colwell RR. 1991. Aeromonas jandaei and Aeromonas veronii dual infection of a human wound following aquatic exposure. J Clin Microbiol 29:565–569. doi: 10.1128/JCM.29.3.565-569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.González-Serrano CJ, Santos JA, García-López ML, Otero A. 2002. Virulence markers in Aeromonas hydrophila and Aeromonas veronii biovar sobria isolates from freshwater fish and from a diarrhoea case. J Appl Microbiol 93:414–419. doi: 10.1046/j.1365-2672.2002.01705.x. [DOI] [PubMed] [Google Scholar]
- 56.Martins LM, Marquez RF, Yano T. 2002. Incidence of toxic Aeromonas isolated from food and human infection. FEMS Immunol Med Microbiol 32:237–242. doi: 10.1111/j.1574-695X.2002.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 57.Rahman M, Colque-Navarro P, Kühn I, Huys G, Swings J, Möllby R. 2002. Identification and characterization of pathogenic Aeromonas veronii biovar sobria associated with epizootic ulcerative syndrome in fish in Bangladesh. Appl Environ Microbiol 68:650–655. doi: 10.1128/aem.68.2.650-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozińska A. 2007. Dominant pathogenic species of mesophilic aeromonads isolated from diseased and healthy fish cultured in Poland. J Fish Dis 30:293–301. doi: 10.1111/j.1365-2761.2007.00813.x. [DOI] [PubMed] [Google Scholar]
- 59.Dong HT, Techatanakitarnan C, Jindakittikul P, Thaiprayoon A, Taengphu S, Charoensapsri W, Khunrae P, Rattanarojpong T, Senapin S. 2017. Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.). J Fish Dis 40:1395–1403. doi: 10.1111/jfd.12617. [DOI] [PubMed] [Google Scholar]
- 60.Ran C, Qin C, Xie M, Zhang J, Li J, Xie Y, Wang Y, Li S, Liu L, Fu X, Lin Q, Li N, Liles MR, Zhou Z. 2018. Aeromonas veronii and aerolysin are important for the pathogenesis of motile aeromonad septicemia in cyprinid fish. Environ Microbiol 20:3442–3456. doi: 10.1111/1462-2920.14390. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. 2014. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. 2017. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 68.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, Söll D, Podar M. 2013. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci U S A 110:5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) ASV table; (B) differential relative abundances; (C) Bin_4 VFanalyzer results; (D) Bin_11 VFanalyzer results. Download Data Set S1, XLSX file, 0.4 MB (398.8KB, xlsx) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Decomposition setup and images of fish decay. (a) Example of the mason jars used for decay of female rainbow darter (Etheostoma caeruleum) in a microcosm of the Grand River. (b) Representative images of decay of female rainbow darter for enrichment of the necrobiome at 1, 4, 8, and 10 days. Download FIG S1, TIF file, 2.2 MB (2.2MB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NMDS ordination of necrobiomes after 4, 8, and 10 days of decomposition based on microbial community composition with Bray-Curtis distances. The ordinations in the top and bottom row for each time point are the same. The ellipses for the top row were calculated for sample fish origin, while the ellipses for the bottom row were calculated for sample water/sediment origin. A strong agreement between the ordination space and the distance matrix was observed for time points 4 days, 8 days, and 10 days (R2 = 0.984, 0.989, and 0.991, respectively) and the stress values are 0.127, 0.103, and 0.097, respectively. Download FIG S2, TIF file, 1.0 MB (1,017.7KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metagenomic bin relative abundance and phylogenetic analysis of Bin_3 and Bin_10. (a) Relative abundance of four high-quality binned genomes across each necrobiome sample. Relative abundance was computed as mean bin coverage/mean sample coverage. Mean coverage was calculated per base pair using Anvi’o (66). (b) RAxML tree using the LG likelihood model made from concatenated single-copy core protein sequences detected with Anvi’o (Campbell et al. set [70]). The tree outgrouped with Lentimicrobium saccharophilum. Acetobacteroides hydrogenigenes, representatives of Alistipes strains, and all uncharacterized Alistipes isolates were used for this tree and sourced from NCBI Genome. This tree was visualized with iTOL (69). Download FIG S3, TIF file, 0.6 MB (650.7KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NMDS ordination of metagenomic functional profiles with Bray-Curtis distances calculated based on KEGG pathway frequencies. A strong agreement between the ordination space and the distance matrix was observed (R2 = 0.996), and the stress value is 0.063. Download FIG S4, TIF file, 0.5 MB (483.5KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Count of KEGG annotations mapping to the corresponding KEGG pathways in Fig. 6 across each MAG. Shown is the log10 value of the fractional frequency of the pathway with respect to the total across all the pathways in the sample. The total pathway coverage is also proportionally normalized across every sample. Download FIG S5, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
End of the KEGG glycolysis/gluconeogenesis pathway for Rikenellaceae Bin_10 and Bin_3. Green indicates the presence of a match to that enzyme. Images were generated using KEGG (67). Download FIG S6, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundance of methanogen contigs and metagenomic bins. (a) Relative abundance (calculated by mean contig coverage/mean sample coverage) of archaeal contigs belonging to the methanogen classes Methanobacteria, Methanococci, and Methanomicrobia. (b) Relative abundance (calculated by mean bin coverage/mean sample coverage) of the four high-quality bins assembled from the fish necrobiome community as well as a small incomplete bin, Bin_11. Download FIG S7, TIF file, 0.8 MB (791.4KB, tif) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Water quality parameters from collection sites taken on 24 October 2016 upstream (43°35′08′′N; 80°28′53′′W) and downstream (43°28′24′′N; 80°28′22′′W) from the Waterloo wastewater treatment plant (WWTP; 43°29′16′′N; 80°30′25′′W). Data are presented as the means plus 1 standard deviation (STD) from 24 samples (hourly) from remote monitoring stations located along the Grand River. Data were obtained through the Grand River Conservation Authority (www.grandriver.ca). Download Table S1, PDF file, 0.05 MB (51.9KB, pdf) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially abundant ASVs between downstream WWTP and upstream WWTP necrobiome samples. Download Table S2, PDF file, 0.08 MB (78.1KB, pdf) .
Copyright © 2020 Lobb et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All 16S rRNA gene and metagenomic sequencing data for this project were deposited into the NCBI Short Read Archive (SRA) under BioProject accession no. PRJNA604775.