Abstract
Killer immunoglobulin-like receptor (KIR) and KIR-ligand (KIRL) interactions play an important role in natural killer (NK) cell mediated graft versus leukemia effect following hematopoietic cell transplantation (HCT). However, there is considerable heterogeneity in the KIR gene and KIRL content in individuals, making it difficult to estimate the full clinical impact of NK cell reconstitution following HCT. Here, a novel adaptive mathematical model designed to quantify these interactions is presented to better assess the influence of NK cell-mediated alloreactivity on transplant outcomes. Ninety-eight HLA matched unrelated donor (URD) HCT recipients were retrospectively studied. The KIR-KIRL interactions were quantified using a system of matrix equations. Unit values were ascribed to each KIR-KIRL interaction and directionality of interactions was denoted by, either a positive (activating) or negative symbol (inhibition); these interactions were then summed. The absolute values of both the missing KIRL as well as inhibitory KIR-KIRL interactions were significantly associated with overall survival and relapse. These score components were initially used to develop a weighted (w-KIR Score) and subsequently a simplified, non-weighted KIR-KIRL interaction scores (IM-KIR Score). Increased w-KIR Score and IM-KIR Score were both predictive of all-cause mortality and relapse; w-KIR score HR of 0.37 (P=0.001) and 0.44 (P=0.044) respectively; IM-KIR score HR of 0.5 (P=0.049) and 0.44 (P=0.002) respectively. IM-KIR score was also associated with NK cell reconstitution post HCT. KIR-KIRL interactions as reflected by the w-KIR and IM-KIR scores influence both relapse risk and survival in recipients of HLA matched URD HCT with hematological malignancies.
Graphical Abstract
Introduction
Hematopoietic cell transplantation (HCT) provides curative therapy for high-risk hematological malignancies1; however, relapse and transplant related mortality rates remain high2,1. Therapeutic benefit of a stem cell allograft is predominantly mediated through the alloreactivity of donor immune effectors directed at a recipient’s malignant cells and is termed the graft versus leukemia (GVL) effect3. Natural Killer (NK) cells are the first immune effector cells to reconstitute after allogeneic HCT, and are capable of affecting GVL4,5; largely through germline-encoded receptors expressed on the NK cells, and inherited independently of human leukocyte antigens (HLA)6. These properties give NK cells a unique advantage, allowing them to mediate early GVL effects in a HLA matched environment, prior to the emergence of T cell-mediated GVL, without causing GVHD.
Human NK cells possess a multitude of different cell surface receptors classes; of these, the largest and most well studied are the killer immunoglobulin-like receptors (KIR). KIRs transduce either inhibitory or activating signals to the NK cell after interacting with, or in the absence of interactions with HLA and HLA-like ligands on the target cell surface7. The balance of these signals may lead to inhibition, or activation of the NK cell and target cell destruction through multiple mechanisms, including the release of cytotoxic granules containing mediators like perforin and granzyme8,9–11. Previous studies evaluating the role of KIR and HLA interactions in NK cell alloreactivity and HCT outcomes have included specific KIR-KIR ligand interactions or have considered the donor KIR haplotypes. Further, initial studies of KIR alloreactivity in HCT examined the ‘missing self-phenomenon’ in haploidentical transplantation where recipients who lacked an HLA ligand for their donor’s inhibitory KIR (iKIR) genotype exhibited a decreased risk of relapse12,13. Further studies examined the similar case of missing KIR ligand interactions in the HLA matched setting. In these instances, the donor possessed iKIR for which both the donor and recipient lacked the corresponding HLA KIR ligand (KIRL). This missing KIRL (mKIRL) effect was also shown to decrease relapse in HLA matched related (MRD) HCT14 and unrelated donor (URD) HCT in myeloid malignancies15.
The KIR gene locus is highly polymorphic and has been classified into 2 haplotypes based on KIR gene content6, haplotype B containing a larger complement of activating KIR (aKIR) and haplotype A containing only one activating gene. HCT transplantation with KIR haplotype B donors generally yield favorable outcomes with less relapse compared to donors with KIR haplotype A, possibly due to the increased NK activation potential and greater GVL capabilities8. Single activating KIRs have also been studied in relation to the recipient’s HLA status. It has been reported that relapse risk for AML is reduced when recipients with a HLA C1+ phenotype are transplanted using donors with activating KIR2DS116. While these clinical associations are well characterized, as more evidence has been gathered, conflicting data have emerged17–21, in some instances disputing the NK cell mediated alloreactivity in HLA-matched HCT. Further, these studies have not fully accounted for the variability in donor KIR gene complement and recipient HLA types. It is very likely that this variability introduces a high degree of heterogeneity in donor NK cell-recipient target cell interactions. The lack of knowledge regarding these interactions compromises optimal donor selection for allogeneic HCT. Here, we propose a novel analytical approach to quantify the interactions that may occur between variables controlling NK cell function and have developed a system of scores to mathematically quantify cumulative KIR-KIRL interactions in individual transplant recipients. In this system, missing KIR ligand, and the inhibitory and activating KIR-KIR ligand interactions are considered summative in their effect in mediating NK cell influence on clinical outcomes. Such a scoring system may allow prediction of NK cell mediated GVL effect than might be expected from different HLA-matched donors for the same recipient. The resulting scores -- if validated in a large cohort of patients -- may be used to select optimal HCT donors that yield an adequate GVL effect.
Methods
Patients
Virginia Commonwealth University (VCU) Institutional Review Board gave approval to conduct this retrospective study. All 8/8 HLA-matched unrelated donor-recipient pairs (DRP) who had KIR genotyping performed and were transplanted at VCU between 2014 and 2017 were retrospectively studied. Most of the patients underwent in vivo T cell depletion with rabbit anti-thymocyte globulin (Thymoglobulin, Sanofi Aventis) 5 mg/kg in three divided doses, as a part of the pre-transplant conditioning. GVHD prophylaxis was with tacrolimus (n=96) or cyclosporine (n=2) given along with methotrexate (n=51) or mycophenolate mofetil (n=42). Patient characteristics are given in Table 1. Peripheral blood NK cell counts were measured by flow cytometry on days +30, +60 and +100 posttransplant using a BD FACSCanto™ II flow cytometer and BD Multitest™ 6-color TBNK reagent (BD Biosciences, 2350 Qume Drive, San Jose, CA 95131). This is a six-color direct immunofluorescence reagent containing FITC-labeled CD3, PE-labeled CD16 and CD56, PerCP-Cy™5.5–labeled CD45, PE-Cy™7–labeled CD4, APC-labeled CD19, and APC-Cy7–labeled CD8, to identify and determine the percentages and absolute counts of T, B, and natural killer (NK) cells. NK cells, identified as CD3– and CD16+ and/or CD56+, were determined by analysis with BD FACSCanto clinical software v2.4 (BD Biosciences, 2350 Qume Drive, San Jose, CA 95131) and the FCS files imported into FCS Express 4 (De Novo Software, 400 N Brand Blvd, Glendale, CA 91203) for final reports. Acute and chronic GVHD were assigned utilizing the Glucksberg and NIH consensus criteria, respectively.
TABLE 1.
PATIENT CHARACTERISTICS N=98
| AGE | 52 (SD +/− 24) (Range 8–73) |
| MALE/FEMALE | 41/57 (41.8%/58.2%) |
| ETHNICITY N (MEAN) | |
| CAUCASIAN | 87 (89%) |
| AFRICAN AMERICAN | 8 (8%) |
| OTHER | 3 (3%) |
| DISEASE | |
| AML | 29 (30%) |
| MDS | 14 (14%) |
| CML/MF | 15 (15%) |
| ALL | 17 (17%) |
| CLL/MM/NHL/T CELL DISEASE | 22 (23%) |
| SAA | 1 (1%) |
| SCORING SYSTEMS MEDIAN (RANGE) | |
| KIR-KIRL SCORE | 0 (range −5 – 3) |
| IKIR COMPONANT SCORE | 2 (1 – 5) |
| AKIR COMPONANT SCORE | 0 (0 – 4) |
| MISSING LIGAND SCORE | 2 (0 – 4) |
| COMPONANT | |
| WKIR-KIRL SCORE | 3.8 (2.07–4.85) |
| INHIBITORY MISSING LIGAND | 4 (2–5) |
| SCORE | |
| CD 56 CELL RECONSTITUTION MEDIAN | |
| DAY 30 N= 80 | 288 (3 – 950) |
| DAY 60 N=79 | 198 (4 – 864) |
| DAY 100 N=72 | 167 (16 – 600) |
| DISEASE STATE AT TRANSPLANT | |
| CR1 | 44 (45%) |
| CR2 | 16 (16%) |
| OTHER | 33 (33%) |
| N/A | 5 (5%) |
| CONDITIONING | |
| FLU/MEL FLUDERABINE BASED+ | 28 (29%) |
| BU/CY BUSULFANE BASED+ | 27 (28%) |
| BUSULFAN AND FLUDERABINE | 19 (19%) |
| CYTOXAN BASED | 15 (15%) |
| TBI IN CONJUNCTION WITH OTHER | 33 (34%) |
| PREREATORY REGAMINS | |
| ATG IN CONJUCTION WITH OTHER | 86 (88%) |
| PREREATORY REGAMINS | |
| OTHER TRANSPLANT CHARACTERISTICS | |
| PBSC/BM | 83/15 (85/15%) |
| HLA MATCH 8/8 | 98 (100%) |
| CONDITIONING INTENSITY M/RIT | 50/48 (51/49%) |
| CD 34+ CELL DOSE | 4.78 mm3/L (+/− 1.7) |
| DONOR HAPLOTYPE AA/BX | 30/68 (30/70%) |
KIR and KIR Ligand Assignment
Patients and their donors were matched at 8/8 loci, including HLA-A, HLA-B, HLA-C, HLA-DRB1. High throughput HLA sequence-based typing was performed after DNA was isolated from blood or buccal samples (Protrans, Ketschau, Germany). KIR genotyping was determined by intermediate resolution, quantitative polymerase chain reaction (Immucor, Norcross, GA) using LinkSeq KIR 384 (One Lambda Canoga Park, CA). KIR genotyping was not considered during donor selection. HLA epitopes for HLA-B and HLA-C recognized as KIRL by KIR were determined using the European bioinformatics KIR Ligand calculator (https://www.ebi.ac.uk/ipd/kir/ligand.html). Every HLA-C allotype was designated as either C1 or C2. Similarly, HLA-B allotypes were also divided into 2 epitopes Bw4 and Bw6. Bw4 is a KIR epitope, as are HLA-A3 and -A11. Frequencies of KIR and KIRL in our patient cohort are given in supplementary table 1. KIR and KIRL interactions were determined as described in supplementary table 2 (adapted from S. Cooley et al 20188).
KIR-KIRL interaction scores
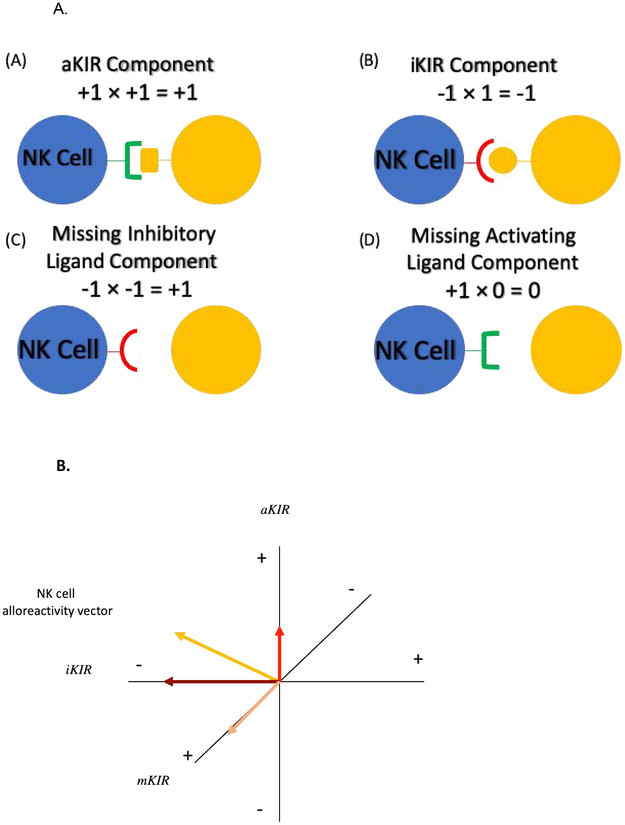
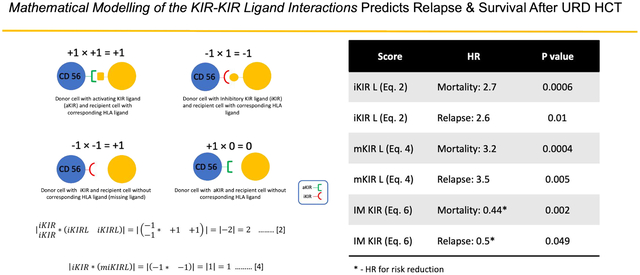
To mathematically derive the KIR-KIRL interaction score, the interactions were viewed from the frame of reference of the donor NK cells (Figure 1A). Based on known KIR-KIRL interactions, hypothetical values were assigned in the following manner: if an inhibitory KIR (iKIR) had a ligand, this resulted in an interaction (−1) × (1) = −1, which gave the NK cell an inhibitory signal. In this equation, the first term is the iKIR, given a value of −1 to denote its inhibition of NK cell activity, the second term is the KIRL which in this case is given a value of +1 which means that the KIR is engaged. The product of these variables has a negative symbol, consistent with inhibition of the NK cell upon engagement of the relevant ligand. If the inhibitory KIR does not have its ligand (mKIRL), its inhibitory effect is abrogated, and since it is assumed that under basal conditions NK cell are constitutively active, the missing KIRL situation was described mathematically by the interaction (−1) × (−1) = +1. In this instance the second term is now negative denoting missing KIRL, and the product is a positive number, consistent with NK cell activation. Interactions between activating KIR (aKIR) and their ligands analogously were given, (1) × (1) = +1 when the ligand was present and (1) × (0) = 0, when the ligand was absent. The first term is now positive because of the activation of the NK cells under baseline conditions. In the latter instance however, the absence of a ligand is characterized by a 0 rather than −1, because rather than an inhibitory signal being abrogated by the absence of its ligand, in this instance the activating signal is simply not given, since the aKIRL is not present (Figure 1). With these values assigned to each KIR-KIRL interaction, the aggregate KIR effect on the NK cells may be quantified by a system of matrix, vector-operator equations (adapted from Abdul Razzaq et al, 201622 and Koparde et al, 201723) to yield a hypothetical NK cell alloreactivity vector. In this system the multicomponent, NK cell-KIR-vector is composed of the KIRs (given in a column matrix) which recognize specific KIRL present on the target cell-operator (given in a row matrix), which transforms the afore-mentioned vector. This vector transformation describes the interaction of KIR with their specific KIRL, and the total magnitude of these interactions may be derived by a matrix multiplication operation (absence of an aKIR cognate ligand is designated noKIRL in these equations),
| [1] |
Figure 1A.
NK cell - Target Cell interactions were assigned values (A) Donor cell with activating KIR ligand (aKIR) and recipient cell with corresponding HLA ligand (B) Donor cell with Inhibitory KIR ligand (iKIR) and recipient cell with corresponding HLA ligand (C) Donor cell with iKIR and recipient cell without corresponding HLA ligand (mKIR) (D) Donor cell with aKIR and recipient cell without corresponding HLA ligand. 1B. Hypothetical NK cell alloreactivity vector and its components. Yellow arrow depicts the hypothetical total NK cell alloreactivity vector. The various vector components are depicted along the x, y & z axes.
Solving this equation yields
The total KIR-KIRL interaction score, or the magnitude of the hypothetical NK cell alloreactivity vector in this model is the sum of all these interactions, and in this example equals 1, but may range from −5 to 9 based on KIR KIRL interactions evaluated in this iteration of the adaptive model presented. It is also important to note that KIR-KIRL interactions for this formula are entered in a KIR-specific manner, for this paper we have used the interactions given in supplementary table 2. These scores were calculated for all donor-recipient pairs. Since each of these interactions may behave in an additive manner i.e., aKIR-KIRL24,25, missing KIR ligand26 and iKIR-KIRL interactions9–11, these different classes of KIR-KIRL interactions may also be considered in isolation; therefore, the combined scores for individual patients were resolved into their components, as in vector addition interactions (Figure 1B). In other words, while the total magnitude and direction of the KIR-KIRL interaction may in all likelihood represent a nonlinear product of the activating, inhibitory and missing KIR-KIRL interactions, these components may also be considered individually. These components of the total KIR-KIRL interaction scores will then give an estimate of the NK cell mediated alloreactivity resulting from a specific class of interactions. To accomplish this, the total inhibitory KIR score was calculated, and the absolute value (designated as |…|) of the interaction between iKIR (vector) and the corresponding KIRL in the (operator) was determined to estimate the total magnitude of each class of interactions, regardless of the direction of effect (NK cell inhibition or activation)
| [2] |
Total aKIR score component was similarly calculated by taking the absolute value of the interaction between aKIR (vector) and corresponding KIRL in the (operator)
| [3] |
The missing KIRL component was computed by taking the absolute value of the product of the iKIR (vector) present without the corresponding KIRL in the (operator)
| [4] |
Statistical Methods:
Time to relapse and death were determined from the day of transplant. Associations between KIR-KIRL interaction scores or the score components outlined above, and time-to-event outcomes (relapse and mortality) were estimated using parametric survival analysis. Given the exploratory nature of the work with this novel scoring system, the choice of distribution which minimized the Bayesian Information Criterion (BIC) was chosen from either exponential, Weibull, or gamma distributions; models were fit separately for each outcome and for the KIR score or its components. KIR-KIRL component score models were also fit in both unadjusted and adjusted manners, where in the latter case the models included CD34+ cell dose infused, recipient age at transplantation, recipient sex, conditioning intensity, whether ATG was administered or not, donor KIR haplotype, myeloid disease, and disease status at transplant. The UNIVARIATE, FREQ, CORR and GLIMMIX procedures from the SAS statistical software (version 9.4, Cary, NC, USA) are used for all KIR-KIRL component summaries and analyses.
Next, unique weights for inhibitory and activating KIR, and missing KIR ligand score components were generated separately to determine relapse-free survival using Cox proportional hazards models. These weights were used to generate weighted donor-recipient KIR-KIRL interaction scores for each individual to predict all-cause mortality and relapse. Subsequently, the two clinical endpoints (mortality and relapse) were studied by generating a single score by the unweighted addition of total inhibitory KIR and total missing ligand scores. Association of weighted and unweighted scores with both mortality and relapse, were examined using Cox proportional hazards models. To further examine the significance of these weighted and unweighted scores, we examined their association with NK cell count using mixed linear models with unstructured covariance. These analyses were performed using Stata 14.1 for MS Windows (StataCorp. 2015: Release 14. College Station, TX: StataCorp LP).
Results
Demographics
The study cohort comprised 98 patients who underwent 8/8 HLA matched unrelated donor HCT for hematologic malignancy (Table 1). KIR gene and KIRL frequencies within our population are given in supplementary table 127.
KIR-KIRL interaction score
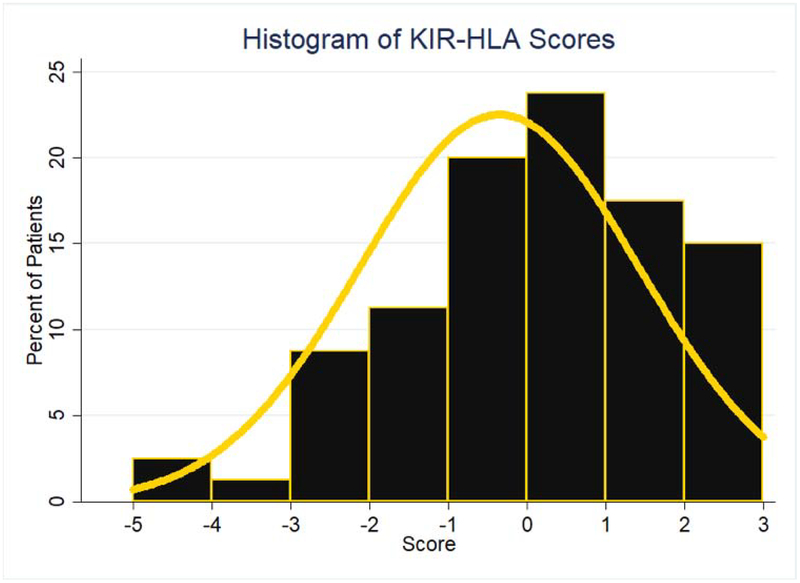
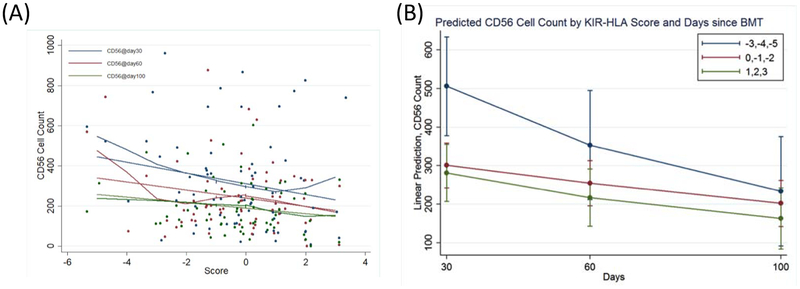
Total KIR-KIRL interaction scores (Equation 1) ranged between −5 to +3, with a median of 0 (Figure 2) and a distribution approximating a normal distribution. There was no significant difference in the total KIR-KIRL interaction scores between donors with KIR haplotype A/A or B/x. This variability in the derived KIR-KIRL interaction scores implies that within HLA identical donors there is considerable heterogeneity in the potential for NK cell mediated alloreactivity. Relationships between NK cell reconstitution and this score were explored in a subset of DRPs who had NK cell counts measured at days +30, +60, and +100 post-transplant. A linear relationship between the total KIR-KIRL score and NK cell count recovery was demonstrated in the patients examined (Figure 3A). When divided into 3 groups based on the magnitude of the score -- i.e., with negative (scores ranging from −5 to −3), a neutral (−2 to 0) or positive scoring group (1 to 3) -- the NK cell counts were significantly different between the three score groups. When compared to the lowest scoring group which had the highest NK cell counts, the neutral scoring group (p=0.049) and high scoring groups (p=0.013) had significantly lower counts (Figure 3B). There was no association of NK cell count recovery with CMV or EBV reactivation. These data suggest a relationship between the magnitude of KIR-KIRL interaction post-transplant and NK cell reconstitution.
Figure 2.
Frequency histogram utilized to represent the distribution scores for the 98 patient cohort when scored using the KIR-KIRL interaction scoring system. Yellow line used to represent an overlay of normal distribution
Figure 3.
CD56 + cell count kinetics and KIR-KIRL interaction score (A) represents a scatter plot of CD56 + cells at day 30 (blue) day 60 (red) and day 100 (green) with a local regression, LOWESS line representing CD56 + cell counts with KIR-KIRL interaction score. (B) Predicted CD56 cell counts by KIR-KIRL interaction score and days since HSCT. the lowest KIR-KIRL interaction scorers (−5 to −3) had the highest CD56 reconstitution at day 30.
KIR-KIRL interaction score components and survival
Given the relationship between KIR-KIRL scores and NK cell reconstitution any association between these scores and clinical outcomes was explored. While the total KIR-KIRL score did not have a significant impact on overall survival, there was a trend suggesting that components of the score might influence outcomes. KIR-KIRL score components were then examined for their impact on all-cause mortality. Both the inhibitory KIR-KIRL interaction score (Equation 2) and the missing KIRL score (Equation 4) were significantly associated with protection from all-cause mortality, with a hazard ratio of 2.7 (95% CI: 1.5, 4.6; p-value = 0.0006) and 3.2 (95% CI: 1.7, 6.0; p-value = 0.0004) respectively.
However, the activating KIR component score (Equation 3) did not have this protective effect (p-value = 0.4126). These results demonstrated a positive association between the score component magnitudes and time to death, i.e., the larger the magnitude, the longer patients survived. When adjusting for other measures, (Table 2) both the inhibitory KIR-KIRL interaction score (HR = 2.1, 95% CI: 1.2, 3.6; p-value = 0.006) and the missing KIRL score (HR = 2.7, 95% CI: 1.5, 5.1; p-value = 0.001) remained associated with mortality.
Table 2:
Adjusted Hazard Ratios for Time to Mortality
| Unadjusted | Adjusted | ||||||
|---|---|---|---|---|---|---|---|
| Measure | HR | 95% Cl | p-value | HR | 95% Cl | p-value | |
| Inhibitory KIR | 2.7 | 1.5, 4.6 | 0.0006 | 2.1 | 1.2, 3.6 | 0.0059 | |
| Activating KIR | 1.2 | 0.7, 2.0 | 0.4126 | 1.3 | 0.7, 2.4 | 0.3437 | |
| Missing KIR | 3.2 | 1.7, 6.0 | 0.0004 | 2.7 | 1.5, 5.1 | 0.0015 | |
| CD34 Dose | 1.3 | 1.0, 1.6 | 0.0319 | ||||
| Age | 0.9 | 0.9,1.0 | 0.0003 | ||||
| Sex | F vs M | 1.1 | 0.5, 2.3 | 0.7909 | |||
| Treatment Intensity | M vs RIT | 1.4 | 0.7, 3.0 | 0.3214 | |||
| ATG | Y vs N | 0.8 | 0.2, 3.0 | 0.7373 | |||
| Donor Haplotype | A vs B | 1.6 | 0.7, 3.8 | 0.2928 | |||
| Myeloid Disease | Y vs N | 1.5 | 0.7, 3.2 | 0.2938 | |||
| Disease State | CR1 vs Other | 1.1 | 0.5, 2.4 | 0.8853 | |||
KIR-KIRL interaction score components and risk of relapse and GVHD
KIR-KIRL score components were next studied for association with relapse. Both the inhibitory KIR-KIRL interaction score (Equation 2) and missing KIRL scores (Equation 4) were significantly associated with relapse prevention, demonstrating HR of 2.6 (95% CI: 1.3, 5.4; p-value = 0.01) and 3.5 (95% CI: 1.5, 8.3; p-value = 0.005) respectively. Activating score component (Equation 3) did not have a similar impact (p-value = 0.84). As noted above, these findings imply that the larger the KIR-KIRL interaction component score magnitudes, the less likely relapse is to occur and the longer it takes for it to occur. When adjusting for other measures (Table 3) the association for the inhibitory score was no longer significant (p-value = 0.37), though the missing KIR ligand score component remained significantly associated with relapse with a HR of 2.4 (95% CI: 1.0, 5.8; p-value = 0.04), again with a positive association with time to relapse. It is to be noted that the KIR-KIRL and KIR-KIRL component scores were not significantly associated with acute or chronic GVHD, or CMV reactivation. These data suggest that a higher inhibitory KIR and missing KIRL components conferred greater protection from relapse in HLA matched URD HCT.
Table 3:
Adjusted Hazard Ratios for Time to Relapse
| Unadjusted | Adjusted | ||||||
|---|---|---|---|---|---|---|---|
| Measure | HR | 95% Cl | p-value | HR | 95% Cl | p-value | |
| Inhibitory KIR | 2.6 | 1.3, 534 | 0.0106 | 1.4 | 0.7, 3.1 | 0.3754 | |
| Activating KIR | 0.9 | 0.5, 1.8 | 0.8513 | 1.1 | 0.5, 2.8 | 0.7806 | |
| Missing KIR | 3.5 | 1.5, 8.3 | 0.0047 | 2.4 | 1.0, 5.8 | 0.0443 | |
| CD34 Dose | 1.6 | 1.1, 2.3 | 0.0106 | ||||
| Age | 1.0 | 0.9, 1.0 | 0.3148 | ||||
| Sex | F vs M | 1.0 | 0.3, 3.5 | 0.9458 | |||
| Treatment Intensity | M vs RIT | 6.3 | 1.8, 22.0 | 0.0043 | |||
| ATG | Y vs N | 1.6 | 0.3, 8.2 | 0.5715 | |||
| Donor Haplotype | A vs B | 3.2 | 0.6, 16.7 | 0.1589 | |||
| Myeloid Disease | Y vs N | 4.6 | 1.1, 20.0 | 0.0422 | |||
| Disease State | CR1 vs Other | 2.9 | 0.7, 12.8 | 0.1503 | |||
Weighted KIR-KIRL interaction scores
Given the observed impact of the KIR-KIRL components on survival and relapse the relative weights of these interactions on mortality and relapse were calculated and a weighted total KIR-KIRL interaction score determined. The Cox model for mortality associations (Table 4) was utilized to derive an equation for a weighted KIR score (wKIR score) for each donor recipient pair
| [5] |
Table 4.
Cox Model Mortality Associations
| Coefficient | Std. Err. | 95% Conf. Int | P Value | |
|---|---|---|---|---|
| iKIR-KIRL Component Score | −0.80 | 0.27 | −1.32, −0.27 | 0.003 |
| aKIR-KIRL Component Score | −0.14 | 0.25 | −0.62. 0.35 | 0.58 |
| mlKIR-KIRL Component Score | −0.99 | 0.31 | −1.6, −0.37 | 0.002 |
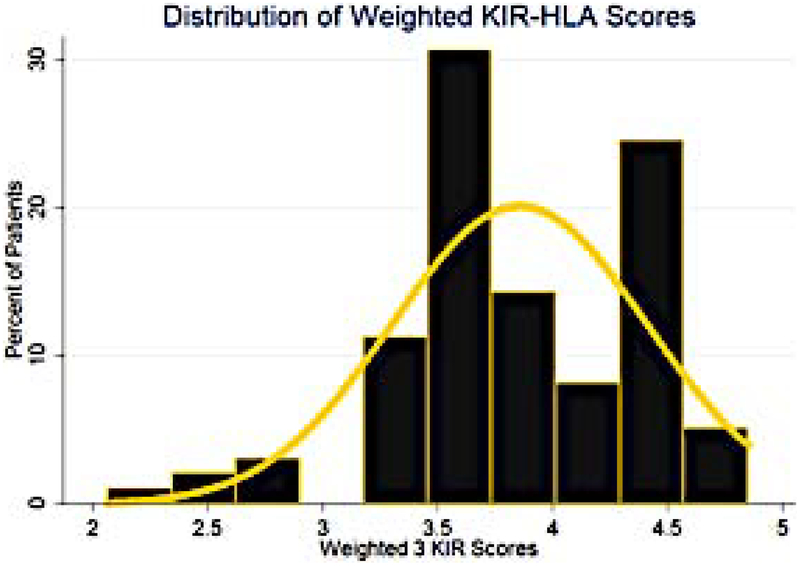
This equation demonstrates the differential impact of the various score components on the final score, the distribution of which was relatively broad in this cohort of 98 HLA matched unrelated DRP (Figure 4). The weighed KIR-KIRL interaction score again demonstrates considerable variability across HLA matched donor-recipient pairs, approximating a normal distribution. Reflecting the earlier findings with KIR component scores, this combined, weighted score (Equation 5) was predictive of all-cause mortality with a HR of 0.37 (95% CI: 0.2, 0.7, P=0.001). This implies that each unit increase in the weighted score results in a 63% decrease in the risk of all-cause mortality following HCT. This weighted score was also predictive of relapse risk with a HR of 0.44 (95% CI: 0.2, 1.0, P=0.044), indicating that for each unit increase in weighted total KIR-KIRL interaction score there was a 56% decrease in risk of relapse. These associations of weighted scores demonstrate the relative influence of the various components of the KIR-KIRL interaction scores with mortality and relapse risk following HCT. As in the previous component analysis, the inhibitory KIR and missing KIRL score components have the largest impact on relapse prevention.
Figure 4.
Frequency histogram utilized to represent the distribution scores for the 98 patient cohort when scored using the w-KIR scoring system. Yellow line used to represent an overlay of normal distribution
A simplified 2 component score for predicting HCT outcomes
Given the influence of inhibitory KIR and missing KIRL score components (Equations 2 & 4; Table 4), a simple, non-weighted new score was developed using these heavily weighted score components. For these calculations both the components were given equal weights (Equation 6) to calculate the inhibitory-missing KIR ligand score (IM KIR Score)
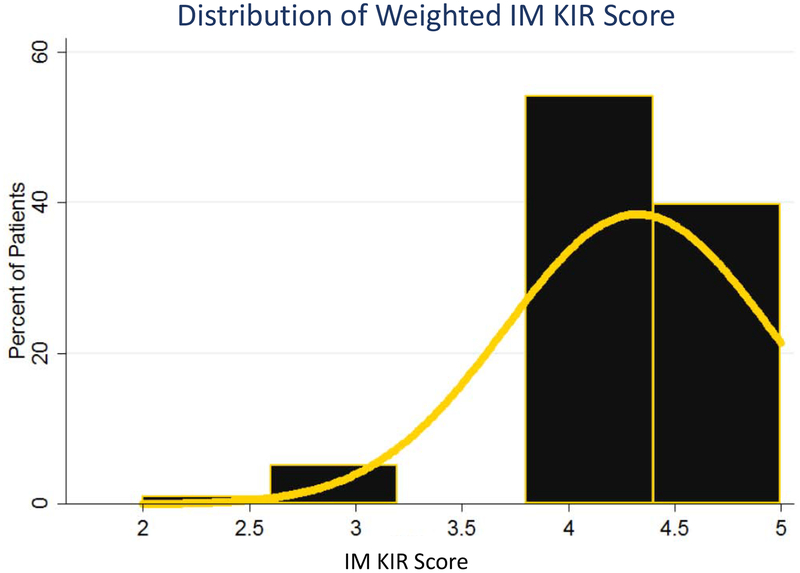
| [6] |
This IM KIR score took on the values 4.3±0.6 (Figure 5). Using Cox proportional hazards model, it was found that for each unit increase in the IM-KIR score, there was a 56% decrease in risk of all-cause mortality (HR = 0.44; 95%CI: 0.26 to 0.73; P=0.002), and a 50% decrease in risk of relapse (HR = 0.5; 95%CI: 0.25 to 1.0; P=0.049), confirming the importance of inhibitory KIR-KIRL and missing KIRL interactions in determining clinical outcomes following HCT, in the cohort reported here.
Figure 5.
frequency histogram utilized to represent the distribution scores for the 98 patient cohort when scored using the IM KIR scoring system. Yellow line used to represent a normal distribution
The association of IM KIR Score with NK cell count recovery
NK cell reconstitution was examined as a function of the IM KIR Score and demonstrated that an increase in the score of 1 unit was associated with an increase of fifty NK cells/μL (SE 25.2, P= 0.046). This suggests that inhibitory and missing KIR-KIRL interactions strongly influence NK cell reconstitution after HCT.
Discussion
This study was designed to develop a logic-based quantitative method to understand the variation in, and the cumulative effect of KIR and KIRL interactions on clinical outcomes following HLA-matched unrelated HCT. In doing so, this method departs from the conventional analytic methodology employed in examining the impact of pretransplant variables on clinical outcomes, where such characteristics are used to statistically derive probabilities of specific clinical outcomes, without consideration of direct interactions between these variables. The findings reported in this paper demonstrate that within a cohort of HLA identical patients with both myeloid and lymphoid malignancies, the magnitude of the KIR-KIRL interactions vary considerably, approximating a normal distribution. Further, both the inhibitory KIR-KIRL interactions, as well as the missing KIR ligands when mathematically determined are associated with mortality and relapse risk, albeit in a heterogeneous cohort of patients. When examined cumulatively in a patient cohort which primarily received ATG for GVHD prophylaxis, both a weighted total KIR-KIRL interaction score, as well as a non-weighted IM-KIR score (combining inhibitory KIR and missing KIRL interaction magnitudes) were similarly associated with improved survival and decreased relapse. These KIR-KIRL interactions are also associated with the magnitude of NK cell reconstitution. This novel formalized, and adaptive mathematical framework for quantifying KIR-KIRL interactions presented here may therefore be predictive of clinical outcomes in recipients of HLA matched unrelated donor allografts and may help identify optimal donors from amongst equally well HLA matched donors and merits further study in a larger cohort of patients.
The KIR gene locus shows a very high degree of variability, similar to the major histocompatibility locus encoding HLA molecules28. In order to understand and quantify the effect KIR-KIRL interactions have on clinical outcomes following HCT, one must consider a model that accounts for variability in and interactions between the genes on these loci in the donors as well as the recipients. The variation between KIR and KIRL population frequencies means that within HLA matched donor-recipient pairs there may be significant heterogeneity in NK cell mediated alloreactivity, and thus disease relapse and mortality risk. This was borne out in the analysis reported here where HLA matched donor recipient pairs demonstrated an approximately normal frequency distribution curve for the various KIR-KIRL interaction scores. It is to be noted that for this cohort KIR typing information was not utilized in donor identification, eliminating a potential source of bias.
The initial studies of NK cell alloreactivity using missing ligand analysis did not consider KIR gene variability. There are more than 30 KIR genotypes known6, which can be functionally split into 2 haplotypes, A and B as previously described8. This simple distinction was first used to look at the effect of additional aKIR gene content on clinical outcomes in HCT. The presence of KIR haplotype B alone was associated with reduced risk of relapse and increasing survival (RR relapse or death, 0.70 95% CI 0.55–0.88) in AML, however it did not have the same effect in ALL, and was not always reproducible29. This clinical benefit was then found to increase when donors homozygous for centromeric, B cen-specific gene content were utilized (RR relapse or death 0.85 95% CI 0.73–0.99). One theory for this phenomenon is the presence of the stronger binding affinity, KIR2DL2 and the absence of the low binding affinity, KIR2DL3. However others have reported reduced overall survival with higher B cen content17. And while it has been shown that allografts from donors positive for aKIR, KIR2DS1 decrease the risk of relapse with a hazard ratio of 0.76, further studies on in vivo T cell depleted patients have shown an increased risk of relapse with increasing aKIR genes with a HR of 1.37. The mathematical model reported here, demonstrating variability in KIR-KIRL interactions between donor recipient pairs, regardless of their haplotypes may help understand some of these inconsistencies reported in the literature. In addition, the magnitude of effect reported here in a combined myeloid and lymphoid disease population far exceeds the effect previously reported in the KIR haplotype studies. One reason for this may be related to the loss of information that occurs when the haplotype is considered, and the expected HLA interactions are not accounted for, diminishing the signal strength one may get from KIR haplotype analyses. A mathematical framework accounting for all the individual KIR-KIRL interactions is not susceptible to such loss of information.
The model reported here is robust in that its simple adaptive nature will allow incorporation of other NK cell receptors, alleles and their interactions, as additional data become available in the future. At the time of publication the most current list of KIR with known HLA ligands has been utlized8. There are variations in the expression and/or affinity of certain KIR which may be accounted for by ascribing appropriate weights to specific interactions as more data become available and appropriate coding of specific interactions can be undertaken. As an example, in vitro models have shown that certain alleles of KIR2DL2 and KIR2DL3 interact with C2 alleles30. However, these interactions are not of the same strength or affinity, as the dominant clinical impact for these KIR reported in contemporary literature is for C1+ targets31–33. This provides rationale for performing high resolution KIR typing in the future. Further, different alleles of KIR2DL1, 2DL2 and 2DL3 have also been shown to alter KIR expression on the NK cell surface34,35. Such information from high resolution KIR typing may allow better characterization of KIR-KIRL interactions as the model incorporates these given its ability to account for multiple interactions.
An important question raised by these findings is how inhibitory KIR have such a profound effect on clinical outcomes and activating KIRs less so? First off, the activating KIR-KIRL interactions in our cohort were not as frequent as the inhibitory effect (Supplementary Table 1), which diminishes the statistical power to identify an effect associated with these interactions. Secondly, the strong inhibitory KIR effect may be understood in terms of NK cell education. NK cells undergo education to ensure that if an individual is missing the iKIRL or has a corresponding aKIRL for their own HLA, their NK cells will not be continually activated and cause autologous tissue injury, or alternatively, if they have a high inhibitory KIR complement, they do adequately proliferate when faced with an appropriate stimulus. Education of NK cells causes them to dampen their proliferation in the former setting, and amplify it in the latter. It is logical that this education (or signal modulation in physical terms) will be proportional to the magnitude of the activating or inhibitory signals. It has also been shown that NK cells which express a higher numbers of iKIR, when exposed to their requisite HLA ligand display increased cytokine production and cell degranulation9–11. Consistent with this, a more robust NK cell reconstitution at day +30 was seen with increased negative KIR-KIRL scores and a larger IM KIR score. While this model does not account for education, because it is based on genotyping information, it does demonstrate the potential impact of NK cell education on clinical outcomes i.e., superior outcomes in patients with a larger complement of iKIR. It should also be noted that in this cohort every donor-recipient pair had the potential for education with at least 1 iKIR KIRL interaction. The data presented here, support the notion that high iKIR content promotes a robust NK cell response in the face of an appropriate stimulus40,11 and that donor iKIR gene content may have the most significant effect on clinical outcomes after HCT (see Appendix for further discussion of education). It is also evident from the data reported here that missing KIR ligands have a more pronounced effect on clinical outcomes than having a large complement of iKIR with the cognate ligands.
In conclusion, KIR-KIRL interactions in an HLA-matched URD HCT setting are variable and influence both the risk of all-cause mortality and relapse risk in patients with hematological malignancies. If verified in a larger cohort of patients these findings have the potential to alter the current practice of donor selection in the HLA matched setting, and potentially in the haploidentical related donor setting as well.
Supplementary Material
Highlights.
An adaptive mathematical model is developed to quantify KIR-KIRL interactions.
The magnitude of inhibitory and missing KIR-KIRL interactions predicts overall survival and relapse risk following HLA matched unrelated donor (URD) hematopoietic cell transplantation (HCT).
Cumulative known KIR-KIRL interactions predict clinical outcomes after URD HCT.
High inhibitory KIR-KIRL interaction scores correlates with robust NK cell recovery after URD HCT.
Acknowledgments:
The authors gratefully acknowledge Ms. Dana Broadway for her role in managing the HLA & KIR typing of the unrelated donor transplant recipients at VCU.
Appendix.
There are several interesting questions raised by our model, one of which is, while weighted scores are predictive of clinical outcomes, total score in and of itself is not. It is known that NK cells express their KIR in a stochastic fashion28, where within an individual’s NK cell repertoire some NK cells express no KIR, while others express every KIR within their genotype, as well as every combination of KIR expression in-between. This implies that there may be as many as x NK cell ‘clones’, each expressing a different KIR complement.
This may be simply stated, that there are
| [7] |
possible NK cell clones found within every person’s NK cell repertoire. In this equation, n represents the number of KIR genes in the patient’s genome, and k represents the number of KIR genes expressed in a given NK cell clone. The sum of all possible expressed KIR gene combinations then constitutes the number of possible NK cell clones, x. The NK cell clone number can be calculated by taking the n possible values of k, and calculating the number of combinations for each k. The model reported herein scores the KIR-KIRL interaction score on the extreme end of this spectrum, assuming that all of the known KIR are expressed on all the NK cells which may account for variable predictability within scores and score components. At this time, it is unknown what effect allogeneic transplantation will have on the distribution of KIR expression on the NK cells39. This is an area which needs to be prospectively explored, to determine the effect KIR expression has on KIR-KIRL interactions in the post HCT setting. This may allow more accurate weights to be ascribed to each category of the interactions in the model reported here and improve its predictive value.
The other question raised by our findings is why do inhibitory KIR have such a profound effect on clinical outcomes? To develop a quantitative understanding of KIR-KIRL interactions one may take a dynamical systems view of NK cell responses. In this system the future states of the system are dependent on the preceding states of the systems, and differential equations describe the evolution of such systems. The different score components constitute variables in these equations describing NK cell function and proliferation. One method to accomplish this includes using the logistic equation of growth which may be used to model NK cell proliferation
In this equation: N0, is the NK cell count at the outset (N0=1 at t=1); Nt, is the NK cell count at time t following transplant (t modeled as iterations); Nt−1 represents the T cell count at the previous iteration; K is the NK cell count at the asymptote (steady state conditions after infinite iterations and may be considered a proliferation constant), and represents the maximum NK cell count the system would support (carrying capacity); r is the growth rate. The NK cell population, at time t, Nt, depends on the preceding T cell populations, such that
In performing these calculations, K is dependent on several factors. For NK cells an important variable will be the effect of KIR-KIRL, since the KIR dependent signals drives NK activity (Skir calculated in equation 1). The signal from the KIR (e^Skir) will either amplify or diminish the proliferation constant depending on the input from KIR, and given the growth kinetics generally observed in immune cell proliferation, would take an exponential form. For simplicity it may be assumed that proliferation is a surrogate for activation and function.
| [8] |
The value Skir represents the cumulative effect of activating KIR (akir) or inhibitory kir (ikir) and missing KIR ligands. Substituting the negative values of ikir and positive values of akir and missing KIRL from the earlier calculations will result in shrinking or growth of the NK cell population as time passes following HCT. This becomes clear if we model the effect of the different KIR-KIRL interactions separately, in other words use the score components (activating, a; inhibitory, I; missing ligand, m) to determine signal strength, Sakir, Sikir, and Smkir. These equations however raise the question as to why inhibitory KIR with a negative exponent should have such a profound effect on clinical outcomes, and activating KIR with a positive exponent, such a limited impact. The answer to this lies in understanding the NK cell education and thinking of post-transplant events as a function of time following HCT.
NK cells undergo education to ensure that if an individual is missing the iKIRL or has a corresponding aKIRL for their own HLA, their NK cells will not be continually activated and cause autologous tissue injury, or alternatively, if they have a high inhibitory KIR complement, they do adequately proliferate when faced with an appropriate stimulus. Education of NK cells causes them to dampen their proliferation in the former setting, and amplify it in the latter. It is logical that this education (or signal modulation in physical terms) will be proportional to the magnitude of the activating or inhibitory signals. So while, homeostatic NK cell proliferation in response to cytokines can be modelled as a function of K*e^Skir, a second term describing the process of NK cell education is necessary. This term makes NK cell proliferation in response to a target inversely proportional to the magnitude of the activating or inhibitory signals (1/e^Skir). With inhibitory KIR this term will counteract the negative growth effect upon encountering the target and with activating KIR, dampen the signal. This relationship serves to balance the signal input with the genetically hardwired information that NK cells have to work with.
In other words, the education can render these cells inert, and drive down the whole term K*e^Skir, if one is missing KIR Ligands for the inhibitory KIR expressed, or if the NK cells have activating KIR for KIR ligands they possess. An increase in the number of iKIR interacting with HLA molecules results in a stronger target cell response9. And when there are a number of inhibitory KIRs, with the NK cells getting a large inhibitory input, it may set them to a higher threshold level of activation at base line, such that when the inhibitory signal is removed the NK cell responses are augmented. This is analogous to a rubber band being imparted with potential energy as it is stretched, such that the more it is stretched, the greater the force of its rebound when it is released. Similarly, when iKIR with corresponding iKIRL are abundant, homeostatic proliferation in response to cytokines is very high, and when the inhibitory signals are turned off proliferation is rapid. So for the iKIR component the NK cell proliferation equation will be modified as follows
| [9] |
For a missing KIR ligand component the equation will be modified
| [10] |
Notice that for the mKIR situation, K has a positive exponent for e and negative exponent for the coefficient 1/e, making the proliferation more robust. The activating KIR will have a similar component determining equation,
| [11] |
These equations together define the components of the hypothetical NK cell proliferation vector, which may be used to understand NK cell responses to stimuli (Figure 1B). When determining final NK cell responses, the variation in Skir components and the effect of education, as well as impact of cytokine mediated proliferation will have to be taken into consideration simultaneously. It is important to recognize that the hypothetical NK cell proliferation vector in this instance is a mathematical surrogate for NK cell activation and effector functions.
Another way to understand NK cell education is if one makes an assumption that over time, NK cells have an equal basal proliferation in a cytokine-free environment. This implies that to compensate the inhibitory KIR input the proliferation coefficient will vary as a function of the KIR input (through the term 1/e^ikir), in this instance the larger the inhibitory input with lower values of e^Skir, the larger the coefficient becomes with time to maintain steady state NK cell levels. Once the inhibitory influence is removed this results in exponential growth, analogous to releasing the brake on a car which has its engine revved up as it starts motion on a slope. For the activating KIR the opposite effect will hold, that education dampens the NK cell proliferation through a coefficient which lowers the basal activity. So for a high inhibitory input, e−x, the proliferation coefficient will have to be high (1/e−x), and alternatively for a high activating input, ex, the coefficient will be low (1/ex). This also helps understand why in the model reported here, the total KIR-KIRL interaction score as calculated in Equation 1 did not predict outcomes as well as the absolute magnitude of KIR score components calculated in Equations 2–4.
This concept of KIR signal inputs applied to the NK cells to maintain steady state NK cell populations at an arbitrary basal level works well for both, when the signals are removed, so there is a commensurate rebound activation, and for the inhibitory KIR input education and for activating education. Cytokine effects are on top of this KIR mediated growth. This implies that that the proliferation coefficient is a variable quantity which starts at a certain basal value and then varies because of education, such that despite the KIR input the NK cell function may be adjusted to prevent either autologous killing or inertness in the face of a threat. In other words, to begin with proliferation coefficient is determined by the cytokine milieu and the KIR input decides NK cell function early after transplant. However, when education takes effect over time, the response to KIR input is adjusted as this process is completed. The magnitude of this effect may vary amongst the NK cell ‘clones’ depending on the expressed KIR complement of these cells.
Footnotes
Conflict-of-Interest Disclosure: The authors have no relevant conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Rautenberg C, Germing U, Haas R, Kobbe G, Schroeder T. Relapse of Acute Myeloid Leukemia after Allogeneic Stem Cell Transplantation : Prevention, Detection, and Treatment. Int J Mol Sci. 2019;20(1). doi: 10.3390/ijms20010228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wingard JR, Hsu J, Hiemenz JW. Hematopoietic Stem Cell Transplantation : An Overview of Infection Risks and Epidemiology. Infect Dis Clin North Am. 2010;24:257–272. doi: 10.1016/j.idc.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 3.Bleakley M, Riddell SR, Cancer FH. Molecules and Mechanisms of the Graft-Versus-Leukemia. Nat Rev. 2004;4(May):1–10. doi: 10.1038/nrc1365 [DOI] [PubMed] [Google Scholar]
- 4.Mehta RS, Rezvani K. Immune reconstitution post allogeneic transplant and the impact of immune recovery on the risk of infection. Virulence. 2016;7(8):901–916. doi: 10.1080/21505594.2016.1208866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill S, Olson JA, Negrin RS. Natural Killer Cells in Allogeneic Transplantation: Effect on Engraftment, Graft-versus-Tumor, and Graft-versus-Host Responses. Biol Blood Marrow Transplant. 2009;15(7):765–776. doi: 10.1016/j.bbmt.2009.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bashirova AA, Martin MP, McVicar DW, Carrington M. KIRn gene cluster. Annu Rev Genomics Hum Genet. 2006;7(1):277–300. http://www.ncbi.nlm.nih.gov/books/NBK10134/. [DOI] [PubMed] [Google Scholar]
- 7.Farag SS, Fehniger TA, Ruggeri L, Velardi A, Caligiuri MA. Natural killer cell receptors: New biology and insights into the graft-versus-leukemia effect. Blood. 2002;100(6):1935–1947. doi: 10.1182/blood-2002-02-0350 [DOI] [PubMed] [Google Scholar]
- 8.Cooley S, Parham P, Miller JS. Strategies to activate NK cells to prevent relapse and induce remission following hematopoietic stem cell transplantation. Blood. 2018;131(10):1053–1062. doi: 10.1182/blood-2017-08-752170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodin P, Ka K. NK cell education : not an on-off switch but a tunable rheostat. 2009;(March):143–149. doi: 10.1016/j.it.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 10.Sleiman M, Brons NHC, Kaoma T, et al. NK Cell Killer Ig-like Receptor Repertoire Acquisition and Maturation Are Strongly Modulated by HLA Class I Molecules. J Immunol. 2014;192(6):2602–2610. doi: 10.4049/jimmunol.1302843 [DOI] [PubMed] [Google Scholar]
- 11.Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK Cell Responsiveness Is Tuned Commensurate with the Number of Inhibitory Receptors for Self-MHC Class I: The Rheostat Model. J Immunol. 2009;182(8):4572–4580. doi: 10.4049/jimmunol.0803900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljunggren HG, Karre K. Host Resistance Directed Selectively Against H-2-Deficient Lymphoma Varients Analysis of the Mechanism. J Exp Med. 1985;162(December):1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of Donor Natural Killer Cell Alloreactivity in Mismatched Hematopoietic. Vol 295; 2002. http://www.jstor.org/stable/3076290%5Cnhttp://www.jstor.org/stable/. [DOI] [PubMed] [Google Scholar]
- 14.Hsu KC, Keever-Taylor CA, Wilton A, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005. doi: 10.1182/blood-2004-12-4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JS, Cooley S, Parham P, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood. 2007;109(11):5058–5061. doi: 10.1182/blood-2007-01-065383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venstrom JM, Pittari G, Gooley TA, et al. HLA-C-Dependent Prevention of Leukemia Relapse by Donor Activating KIR2DS1 Abstract. N Engl J Med. 2012;367:805–821. doi: 10.1056/NEJMoa1200503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escudero A, Martínez-Romera I, Fernández L, et al. Donor KIR Genotype Impacts on Clinical Outcome after T Cell–Depleted HLA Matched Related Allogeneic Transplantation for High-Risk Pediatric Leukemia Patients. Biol Blood Marrow Transplant. 2018;000:1–8. doi: 10.1016/j.bbmt.2018.08.009 [DOI] [PubMed] [Google Scholar]
- 18.Blazar BR, Noreen H, Weisdorf DJ, et al. Negative effect of KIR alloreactivity in recipients of umbilical cord blood transplant depends on transplantation conditioning intensity. Blood. 2009;113(22):5628–5634. doi: 10.1182/blood-2008-12-197467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies SM, Ruggieri L, DeFor T, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Blood. 2002;100(10):3825–3827. doi: 10.1182/blood-2002-04-1197 [DOI] [PubMed] [Google Scholar]
- 20.Arima N, Kanda J, Tanaka J, et al. Homozygous HLA-C1 is Associated with Reduced Risk of Relapse after HLA-Matched Transplantation in Patients with Myeloid Leukemia. Biol Blood Marrow Transplant. 2018;24(4):717–725. doi: 10.1016/j.bbmt.2017.11.029 [DOI] [PubMed] [Google Scholar]
- 21.Giebel S, Nowak I, Dziaczkowska J, et al. Activating killer immunoglobulin-like receptor incompatibilities enhance graft-versus-host disease and affect survival after allogeneic hematopoietic stem cell transplantation. Eur J Haematol. 2009;83(4):343–356. doi: 10.1111/j.1600-0609.2009.01280.x [DOI] [PubMed] [Google Scholar]
- 22.Abdul Razzaq B, Scalora A, Koparde VN, et al. Dynamical System Modeling to Simulate Donor T Cell Response to Whole Exome Sequencing-Derived Recipient Peptides Demonstrates Different Alloreactivity Potential in HLA-Matched and -Mismatched Donor-Recipient Pairs. Biol Blood Marrow Transplant. 2016;22(5):850–861. doi: 10.1016/j.bbmt.2015.11.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koparde V, Razzaq BA, Suntum T, et al. Dynamical system modeling to simulate donor T cell response to whole exome sequencing-derived recipient peptides: Understanding randomness in alloreactivity incidence following stem cell transplantation. PLoS One. 2017;12(12):1–24. doi: 10.1371/journal.pone.0187771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beksac K, Beksac M, Dalva K, Karaagaoglu E, Tirnaksiz MB. Impact of “killer immunoglobulin-like receptor/ligand” genotypes on outcome following surgery among patients with colorectal cancer: Activating KIRs are associated with long-term disease free survival. PLoS One. 2015;10(7):1–11. doi: 10.1371/journal.pone.0132526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verheyden S, Schots R, Duquet W, Demanet C. A defined donor activating natural killer cell receptor genotype protects against leukemic relapse after related HLA-identical hematopoietic stem cell transplantation. Leukemia. 2005;19(8):1446–1451. doi: 10.1038/sj.leu.2403839 [DOI] [PubMed] [Google Scholar]
- 26.Clausen J, Wolf D, Petzer AL, et al. Impact of natural killer cell dose and donor killer-cell immunoglobulin-like receptor (KIR) genotype on outcome following human leucocyte antigen-identical haematopoietic stem cell transplantation. Clin Exp Immunol. 2007;148(3):520–528. doi: 10.1111/j.1365-2249.2007.03360.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39(9):1114–1119. doi: 10.1038/ng2077 [DOI] [PubMed] [Google Scholar]
- 28.Gardiner CM. Killer cell immunoglobulin-like receptors on NK cells: The how, where and why. Int J Immunogenet. 2008;35(1):1–8. doi: 10.1111/j.1744-313X.2007.00739.x [DOI] [PubMed] [Google Scholar]
- 29.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia Plenary paper Donor selection for natural killer cell receptor genes leads to superior survival after unrelate. Blood. 2012;116(14):2411–2419. doi: 10.1182/blood-2010-05-283051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moesta AK, Norman PJ, Yawata M, et al. Synergistic Polymorphism at Two Positions Distal to the Ligand-Binding Site Makes KIR2DL2 a Stronger Receptor for HLA-C Than KIR2DL3. 2019. doi: 10.4049/jimmunol.180.6.3969 [DOI] [PubMed] [Google Scholar]
- 31.Sekine T, Marin D, Cao K, et al. Specific combinations of donor and recipient KIR-HLA genotypes predict for large differences in outcome after cord blood transplantation. Blood. 2016;128(2):297–312. doi: 10.1182/blood-2016-03-706317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marra J, Greene J, Hwang J, et al. KIR and HLA Genotypes Predictive of Low-Affinity Interactions Are Associated with Lower Relapse in Autologous Hematopoietic Cell Transplantation for Acute Myeloid Leukemia. J Immunol. 2015;194(9):4222–4230. doi: 10.4049/jimmunol.1402124 [DOI] [PubMed] [Google Scholar]
- 33.Körner C, Granoff ME, Amero MA, et al. Increased frequency and function of KIR2DL1–3+ NK cells in primary HIV-1 infection are determined by HLA-C group haplotypes. Eur J Immunol. 2014;44(10):2938–2948. doi: 10.1002/eji.201444751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunphy SE, Guinan KJ, Chorcora CN, et al. 2DL1, 2DL2 and 2DL3 all contribute to KIR phenotype variability on human NK cells. Genes Immun. 2015;16(5):301–310. doi: 10.1038/gene.2015.15 [DOI] [PubMed] [Google Scholar]
- 35.Leung W, Iyengar R, Triplett B, et al. Comparison of Killer Ig-Like Receptor Genotyping and Phenotyping for Selection of Allogeneic Blood Stem Cell Donors. J Immunol. 2005;174(10):6540–6545. doi: 10.4049/jimmunol.174.10.6540 [DOI] [PubMed] [Google Scholar]
- 36.Bosch M, Dhadda M, Hoegh-Petersen M, et al. Immune reconstitution after anti-thymocyte globulin-conditioned hematopoietic cell transplantation. Cytotherapy. 2012;14(10):1258–1275. doi: 10.3109/14653249.2012.715243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eyrich M, Leiler C, Lang P, et al. A prospective comparison of immune reconstitution in pediatric recipients of positively selected CD34+ peripheral blood stem cells from unrelated donors vs recipients of unmanipulated bone marrow from related donors. Bone Marrow Transplant. 2003;32(4):379–390. doi: 10.1038/sj.bmt.1704158 [DOI] [PubMed] [Google Scholar]
- 38.Andersson S, Fauriat C, Malmberg JA, Ljunggren HG, Malmberg KJ. KIR acquisition probabilities are independent of self-HLA class I ligands and increase with cellular KIR expression. Blood. 2009;114(1):95–104. doi: 10.1182/blood-2008-10-184549 [DOI] [PubMed] [Google Scholar]
- 39.Sternberg-Simon M, Brodin P, Pickman Y, et al. Natural killer cell inhibitory receptor expression in humans and mice: A closer look. Front Immunol. 2013;4(MAR):1–12. doi: 10.3389/fimmu.2013.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006;24(3):249–257. doi: 10.1016/j.immuni.2006.03.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.