Abstract
Chronic hepatitis B virus (HBV) infection results in 887,000 deaths annually. The central challenge in curing HBV is eradication of the stable covalently closed circular DNA (cccDNA) form of the viral genome, which is formed by the repair of lesion-bearing HBV relaxed circular DNA (rcDNA) delivered by the virions to hepatocytes. A complete and minimal set of host factors involved in cccDNA formation is unknown, largely due to the lack of a biochemical system that fully reconstitutes cccDNA formation. Here, we have developed experimental systems where various HBV rcDNA substrates are repaired to form cccDNA by both cell extracts and purified human proteins. Using yeast and human extract screenings, we identified five core components of lagging strand synthesis as essential for cccDNA formation: PCNA, the replication factor C (RFC) complex, DNA polymerase δ (POLδ), FEN-1, and DNA ligase 1 (LIG1). We reconstituted cccDNA formation with purified human homologs, establishing these as a minimal set of factors for cccDNA formation. We further demonstrated that treatment with DNA polymerase inhibitor aphidicolin diminishes cccDNA formation both in biochemical assays and in HBV-infected human cells. Altogether, our findings define key components in HBV cccDNA formation.
Hepatitis B virus (HBV) afflicts two billion people worldwide, with the 257 million chronically infected patients at increased risk for developing hepatocellular carcinoma (1). HBV has a very compact 3.2 kb partially double-stranded DNA genome that is organized into five partially overlapping open reading frames (ORFs). These encode the HBV surface polypeptides (HBs), HBV E antigen (HBeAg), HBV core protein (HBc), the X protein, and the viral polymerase (2). Viral entry depends on interaction of the large HBs with the bile acid transporter sodium-taurocholate cotransporting polypeptide (NTCP) expressed on hepatocytes (3). Following entry, the HBV virion delivers its 3.2 kb relaxed circular DNA (rcDNA) genome into the host cell nucleus. rcDNA contains four known lesions, including an HBV polymerase-5’-DNA covalent adduct, a 5’ DNA flap on the minus strand and, on the plus strand, a 5’-capped RNA primer and ssDNA gap (4,5) (Extended Data Fig. 1a). To establish infection, these lesions must be repaired by elusive host factors to form the episomal covalently closed circular DNA (cccDNA), which serves as the transcriptional template for HBV viral transcripts (2). Current HBV therapies rarely achieve a cure due to the refractory nature of the stable cccDNA. Blocking cccDNA formation and purging existing cccDNA pools are widely regarded as crucial for curing patients (6). Thus, the elucidation of factors and mechanisms involved in cccDNA formation are of utmost importance in determining new drug targets (6).
The repair of rcDNA is a multi-step process (Extended Data Fig. 1a). Removal of the covalently attached HBV polymerase could occur by poorly understood processes whereby tyrosyl-DNA Phosphodiesterase 2 (TDP2) (7), (an) unknown endonuclease(s), and (a) unknown protease(s) generate three types of deproteinated rcDNA (dp-rcDNA) intermediates with, respectively, a clean-end 5’ DNA flap (type A), flap-free (type B), or a ‘dirty’-end 5’ DNA flap with a remnant peptide attached (type C). The possibility of very low level of removal by HBV polymerase itself (auto-release) has also been proposed, however, it may be due to contamination during sample preparation (8). These dp-rcDNA substrates are proposed to be key intermediates that undergo additional repair steps, including removal of the DNA flap and RNA primer, gap filling and nick ligation, to form HBV cccDNA (8,9).
Despite decades-long efforts, HBV cccDNA formation has never been reconstituted with purified human proteins, and a complete set of factors required for cccDNA formation has remained unknown. Recent genetics studies have implicated TDP2, translesional DNA polymerase kappa (POLκ), DNA polymerase alpha (POLα), DNA ligases 1 and 3 (LIG1, LIG3), and flap endonuclease 1 (FEN-1) in cccDNA formation (2,7,10–15). However, direct evidence of these proteins’ involvement in rcDNA repair has been hindered by the lack of a biochemical system for reconstituting cccDNA formation. Here, we have established biochemical systems that fully support repair of various HBV rcDNA substrates to form cccDNA. We identified five core factors involved in the DNA lagging strand synthesis to be essential for cccDNA formation in yeast extracts. We then validated the effects of factors that could be depleted in human extracts. Ultimately, we reconstituted rcDNA repair with five purified human proteins/complexes – PCNA, RFC complex, POLδ, FEN-1, and LIG1 – and define them as a minimal set of factors essential for reconstitution of HBV cccDNA formation.
Results
Generation of recombinant HBV rcDNA substrates that mimic authentic rcDNA or its deproteinated intermediates.
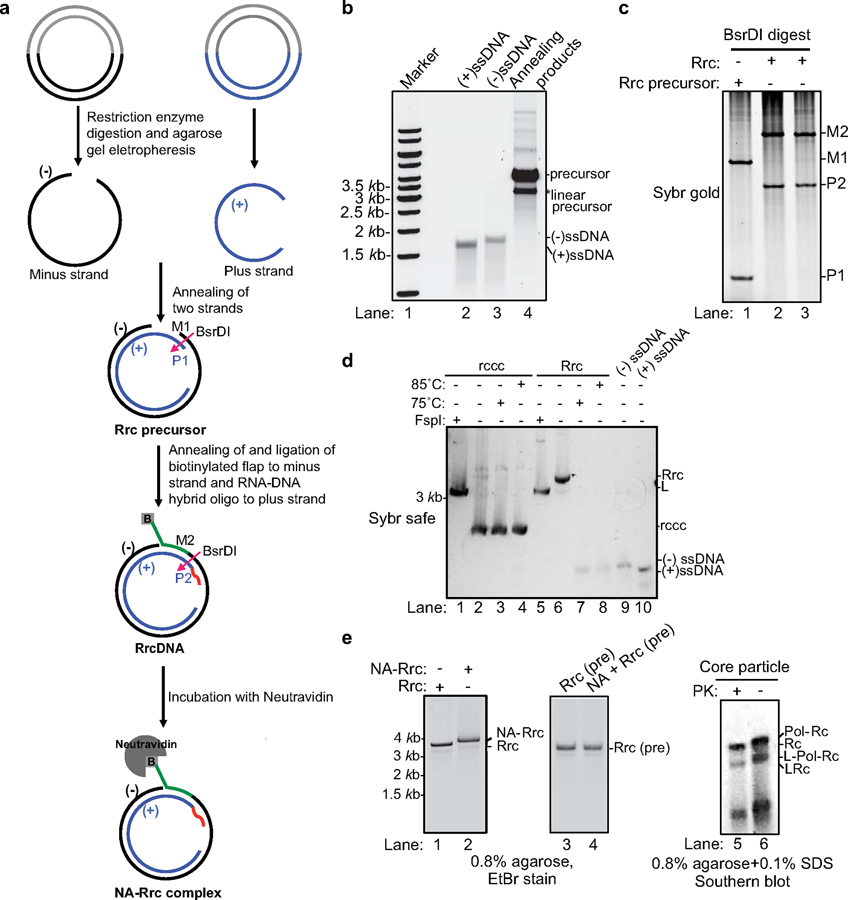
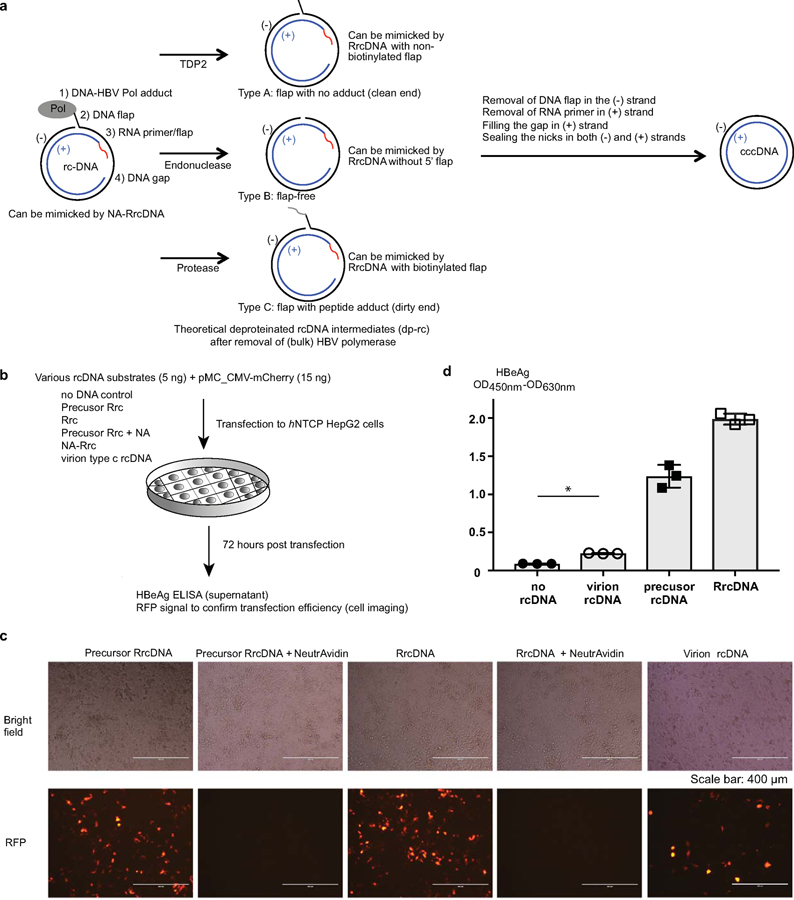
A key to biochemical assays is the generation of substrates. Currently, it is technically impossible to obtain pure, authentic HBV rcDNA for biochemical studies, since purified HBV rcDNA with the covalently attached HBV polymerase is unstable and can only be purified with 0.1% SDS solution, which denatures HBV polymerase and renders it unsuitable for biochemical assays (5). As an alternative, treatment of authentic HBV rcDNA with proteinase K can remove bulk HBV polymerase and generate stable type C deproteinated rcDNA (dp-rcDNA, Extended Data Fig. 1a) intermediate, which contains a short remnant peptide covalently attached to the flap on the minus strand and could be used to study subsequent repair steps. However, purification of dp-rcDNA is notoriously laborious and generates low yields. To overcome this problem, we generated recombinant synthetic substrates to mimic both authentic HBV rcDNA with protein adduct and three types of dp-rcDNA (Extended Data Fig. 1a). We generated recombinant rcDNA (RrcDNA) by annealing purified recombinant minus and plus strand ssDNA and ligating to each strand a DNA flap and RNA-DNA oligo, respectively (Fig. 1a–c). RrcDNA is indistinguishable from dp-rcDNA in its sensitivity to heat and change in electrophoretic mobility upon linearization (16) (Fig. 1d). We also generated a RrcDNA without a 5’ adduct to mimic “type A” dp-rcDNA, without a flap to mimic “type B” dp-rcDNA or with a biotinylated flap to mimic “type C” dp-rcDNA, which contains a ‘dirty’ end requiring further processing (Extended Data Fig. 1a). For simplicity, RrcDNA refers to recombinant rcDNA with a biotinylated 5’ flap while the other RrcDNA substrates will be specified. Unless stated otherwise, dp-rcDNA refers to virion-derived “type C” dp-rcDNA.
Fig. 1 |. Generation of recombinant rcDNA (RrcDNA) and the NeutrAvidin RrcDNA (NA-RrcDNA) complex.
(a) Schematic representation of the generation of recombinant HBV rcDNA. RrcDNA and NA-RrcDNA are mimics of dp-rcDNA and HBV polymerase covalently linked rcDNA, respectively. BsrDI cleavage was used to monitor the efficiency of RrcDNA formation in (c) (BsrDI restriction sites are indicated by magenta arrows); green line, biotinylated flap; red line, RNA primer. Note that M1 and M2 are two oligos released from the minus strands of RrcDNA precursor and RrcDNA after BsrDI digestion, while P1 and P2 are two oligos released from the plus strands after BsrDI digestion. (b) Annealing products of recombinant minus and plus strand ssDNA to form RrcDNA precursor were monitored by SYBR™ safe staining. (c) Efficiency of RrcDNA generation is analyzed by formation of M2 and P2 oligos after BsrDI digestion and urea-PAGE gel electrophoresis followed by SYBR™ gold staining. Repeated three times independently. (d) Electrophoretic mobility of RrcDNA and recombinant cccDNA (rccc) after heat and restriction enzyme FspI treatment are compared on an agarose gel stained by SYBR™ safe. Repeated twice independently. (e) Both NA-RrcDNA (lanes 1–2) and the authentic HBV protein-rcDNA complexes (lanes 5–6) display a characteristic shift in mobility as compared to their deproteinated counterparts (5,17). NeutrAvidin does not cause non-specific mobility shift of precursor RrcDNA (lanes 3–4). PK, proteinase K treatment; L, linear; Pol, HBV polymerase. Lanes 1–4 were repeated independently three times; lanes 5–6 twice.
Incubation of RrcDNA with NeutrAvidin (NA) led to near complete formation of specific NA-RrcDNA complex (Fig. 1a, e), a close approximation of authentic HBV rcDNA. NA-RrcDNA migrated slower than RrcDNA in a manner indistinguishable from that of authentic rcDNA isolated from HBV virions (5,17) (Fig. 1e, lanes 1–2, 5–6).
Furthermore, we confirmed that transfection of RrcDNA, precursor RrcDNA, and dp-rcDNA to a human HBV permissive cell line, led to production of HBeAg, a key indicator of cccDNA formation (Extended Data Fig. 1b–d). NA-RrcDNA could not be evaluated since the presence of NeutrAvidin strongly inhibited DNA transfection (Extended Data Fig. 1c, 1st and 2nd panel). Of note, RrcDNA and precursor RrcDNA led to more robust HBeAg production than dp-rcDNA (Extended Data Fig. 1d). This could be due to lower transfection efficiency and higher toxicity of dp-rcDNA samples (Extended Data Fig. 1c, 3rd and 5th panel), which are not as pure as RrcDNA.
Yeast cell extract repair system fully supports conversion of dp-rcDNA to cccDNA.
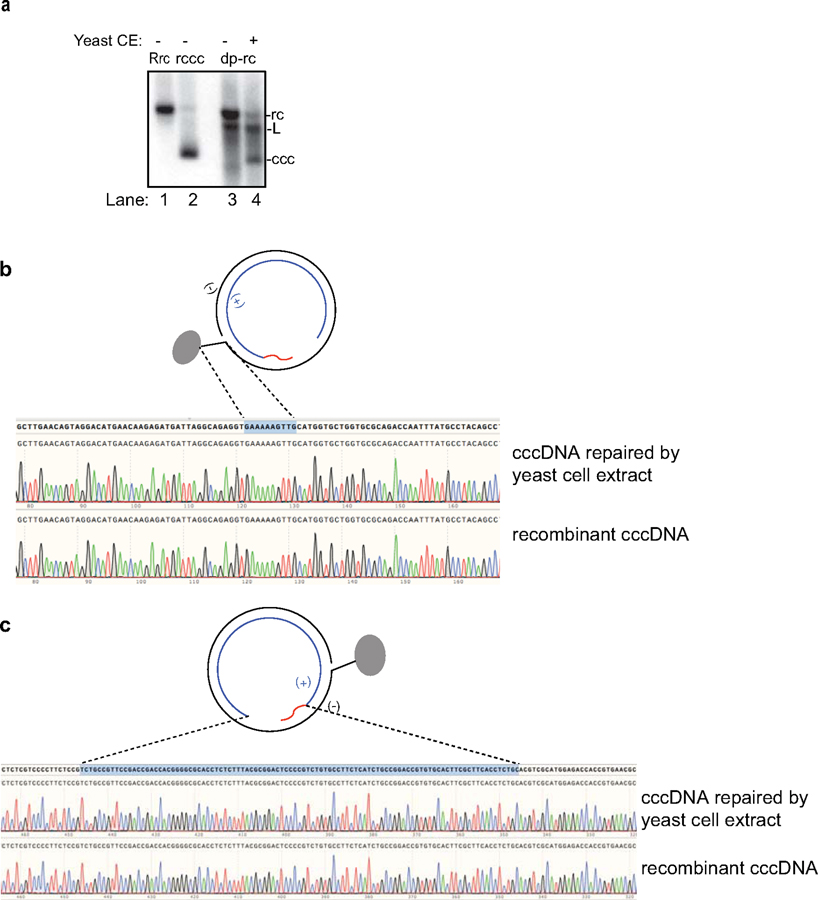
Yeast genetics are a powerful tool to deplete key factors involved in various biological pathways, including DNA repair and replication. Since most factors involved in DNA repair and replication are conserved from yeast to human, the factors identified in yeast most likely correspond to those in humans. Therefore, we aimed to first establish a yeast cell extract repair system that supports HBV rcDNA to cccDNA conversion to identify factors critical for this process followed by validation in human cell extract and purified human protein systems. As shown in Extended Data Fig. 2a, yeast cell extract fully supports repair of dp-rcDNA.
Yeast cell extract repair system supports repair of RrcDNA.
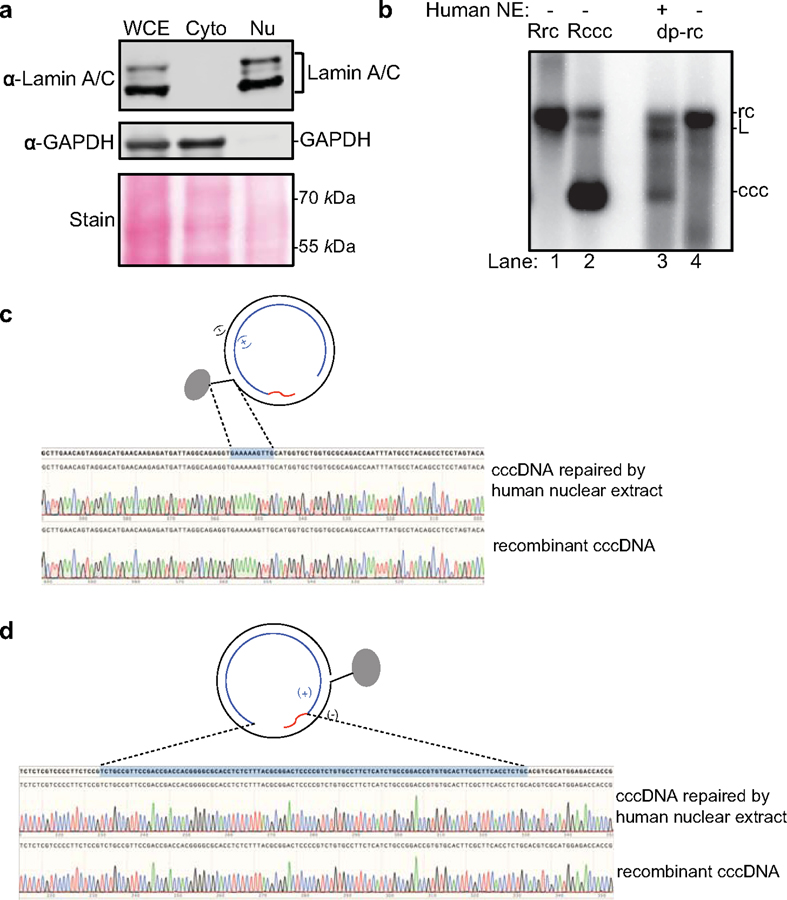
Next, we tested the repair of RrcDNA by yeast extracts. Since most repair proteins are not exclusively localized in the nucleus, it is unclear why HBV cccDNA only exists in the nucleus. This is likely because cytoplasmic components fail to repair rcDNA or rcDNA is not exposed to cytoplasmic repair factors due to protection by HBV capsids. Thus, we used both cytoplasmic and nuclear extracts to study the repair process of RrcDNA. Incubation of RrcDNA with either yeast cytoplasmic or nuclear extracts led to cccDNA formation that was readily monitored by ethidium bromide (EtBr) staining, making detection much less laborious than Southern blotting (Fig. 2a). We employed three methods to confirm that the repair product cccDNA was identical to recombinant cccDNA (referred to as Rccc), which has been shown to behave like its authentic counterpart (18). Indeed, our repaired cccDNA i) was resistant to heat treatment at 85°C (16) (Fig. 2a, lanes 3–4, 7–8); ii) had reduced electrophoretic mobility upon linearization (16) (Fig. 2a, lanes 3, 5; 7, 9); and iii) sequences of the repair cccDNA and RcccDNA were identical (Extended Data Fig. 2b,c). Our data also indicate that both cytoplasmic and nuclear extracts are capable of catalyzing cccDNA formation, and that rcDNA is most likely shielded from cytoplasmic repair factors during natural infection. We next employed the yeast extract repair system to screen for factors essential for repairing RrcDNA to form cccDNA.
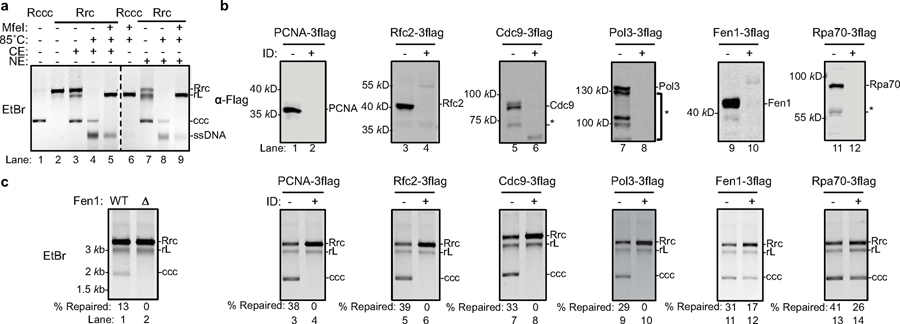
Fig. 2 |. PCNA, RFC complex, Cdc9, Polδ, and Fen1 are required to repair recombinant rc-DNA to form cccDNA in yeast cell extracts.
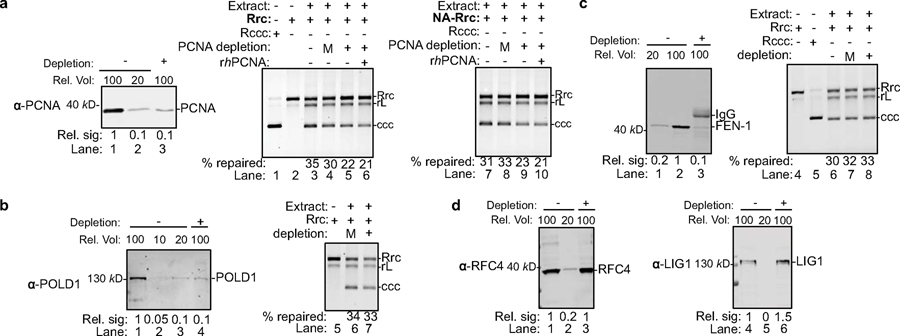
(a) cccDNA formation assay (see methods for details) in yeast cytoplasmic (CE) and nuclear extracts (NE) were carried out to examine the repair of recombinant rcDNA (Rrc) to form cccDNA. Repair products were resolved on an agarose gel and stained with ethidium bromide (EtBr). Both yeast CE and NE fully support cccDNA formation (lanes 3 and 7). Untreated recombinant cccDNA (Rccc) and Rrc serve as controls (lanes 1–2). Repair products of CE (lanes 4–5) or NE (lanes 8–9) were treated with 85°C for 5 min alone or 85°C for 5 min and followed by MfeI digestion. Repaired cccDNA is heat resistant and shows mobility shift upon linearization. rL, recombinant linear DNA. Dashed line indicates removal of superfluous lanes. Repeated twice independently. (b) Near complete immuno-depletion of Flag-tagged-DNA repair proteins in yeast cell extracts examined by western blotting. ID, Immuno-depletion; *, indicates degradation bands. Repeated twice independently. (c) Depletion of factors involved in lagging strand synthesis abrogates repair of RrcDNA to form cccDNA. Lanes 1–2, cccDNA formation assay using extracts from WT or Fen1 null yeast strains. Lanes 3–14, same as lanes 1–2, except immunodepleted extracts from (b) are analyzed. % repaired, the percentage of total RrcDNA that is repaired to form cccDNA was calculated by the intensity of the ccc band divided by the sum intensities of Rrc, rL and ccc bands. Absolute values are shown above each lane number. Repeated twice independently.
Five core factors of the DNA lagging strand synthesis machinery are essential for HBV cccDNA formation in yeast extract.
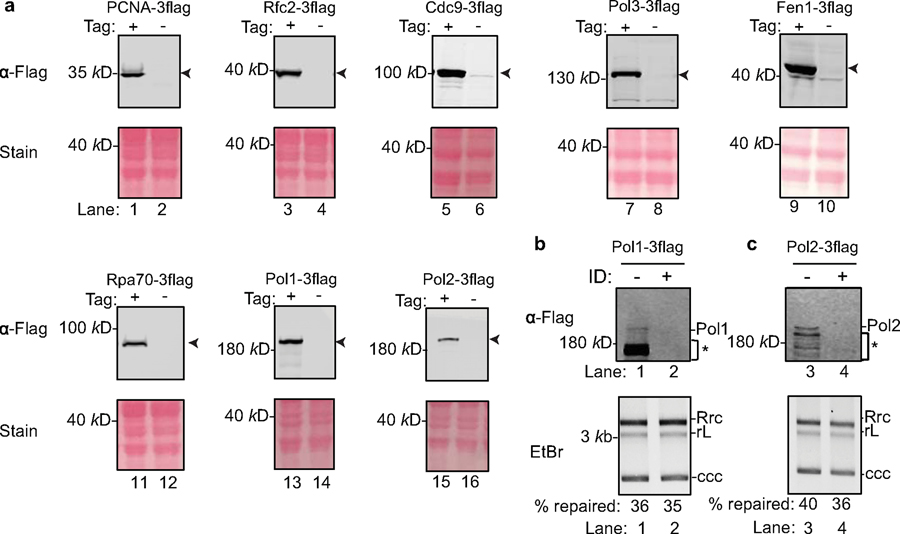
We reasoned that DNA lesions/structures on rcDNA resemble those of Okazaki fragments in DNA replication; therefore, we set out to determine whether components of the Okazaki fragment synthesis or other factors with similar functions could repair rcDNA. These eight factors that were tested include the PCNA clamp, the PCNA clamp loader RFC complex, DNA polymerases Polδ, Polα and Polε, the structure-specific endonuclease Fen-1, DNA ligase Cdc9, and the ssDNA binding protein RPA. Since antibodies capable of achieving near complete protein depletion are not always available, we took advantage of the fact that yeast genes could be readily tagged chromosomally and that some DNA repair factors are not essential in yeast, permitting the use of null mutants. We knocked Flag-tags into candidate chromosomal loci in yeast (Extended Data Figs. 3, 4a) and assessed the effects of these host factors on cccDNA formation by incubating cell extracts immuno-depleted of individual factors with RrcDNA (Fig. 2b, c and Extended Data Fig. 4b, c). Of the eight factors we tested, five demonstrated essential roles in cccDNA formation: PCNA, RFC, Polδ, Fen-1, and Cdc9 – five core components of DNA lagging strand synthesis (Fig. 2c). The effects of these factors were specific, Polδ could not be replaced by the other two B family polymerases, as depletion of Pol1 and Pol2, the catalytic subunits of Polα and Polε, respectively, had no effect on cccDNA formation (Extended Data Fig. 4b, c). Depletion of the Rpa70 subunit of the ssDNA binding protein RPA only marginally reduced the repair efficiency by 30% (Fig. 2c, lanes 13–14). Notably, Fen-1 is not essential in yeast, and although cell extract from a null mutant did not support cccDNA formation (Fig. 2c, lanes 1–2), cccDNA formation was reduced by only 50% in extracts with Flag-tagged Fen-1 immuno-depleted to undetectable levels, lending credence to the high activity of this enzyme (Fig. 2c, lanes 11–12). These results also indicate that genetic analysis of cccDNA formation is prone to false negatives and underscore the need for caution when conducting genetic analysis of HBV cccDNA formation.
Human hepatoma cell extract efficiently supports conversion of various HBV rcDNA substrates to cccDNA.
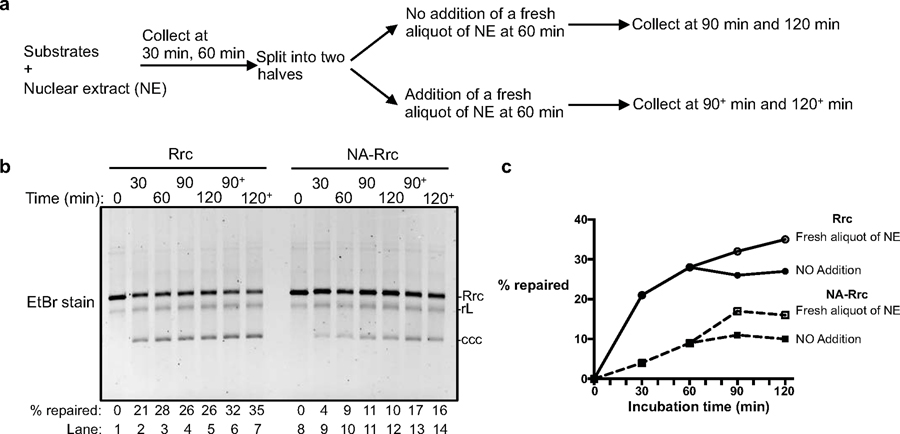
Since the factors and molecular mechanisms of DNA lagging strand synthesis are conserved from yeast to mammals (19), we next aimed to ascertain the relevance of these host repair factors on cccDNA formation using human cell extracts. Successful separation of cytoplasmic and nuclear fractions was confirmed by western blotting using the cytoplasmic and nuclear markers GAPDH and Lamin A/C, respectively (Extended Data Fig. 5a). Indeed, human cytoplasmic and nuclear extracts from HBV-permissive human NTCP-expressing HepG2 cells (3,20) also robustly repaired dp-rcDNA purified from HBV virions, RrcDNA, and NA-RrcDNA (Extended Data Fig. 5b and Fig. 3a, b). We verified that the repaired cccDNA was identical to RcccDNA by the aforementioned three criteria (Fig. 3a and Extended Data Fig. 5c, d). Assessing the kinetics of cccDNA formation using RrcDNA or NA-RrcDNA with human nuclear extract revealed that the repair of both RrcDNA and NA-RrcDNA peaked at 60 min, but the repair efficiency of NA-RrcDNA was only about 40% of RrcDNA (Fig. 3b, c), suggesting that removal of the attached protein is time limiting. Addition of a fresh aliquot of nuclear extract 60 min after the start of the reaction led to a marginal increase in repair efficiency, indicating that the exhaustion of certain repair factors most likely only play a minor role in the plateau observed at 60 min (Extended Data Fig. 6).
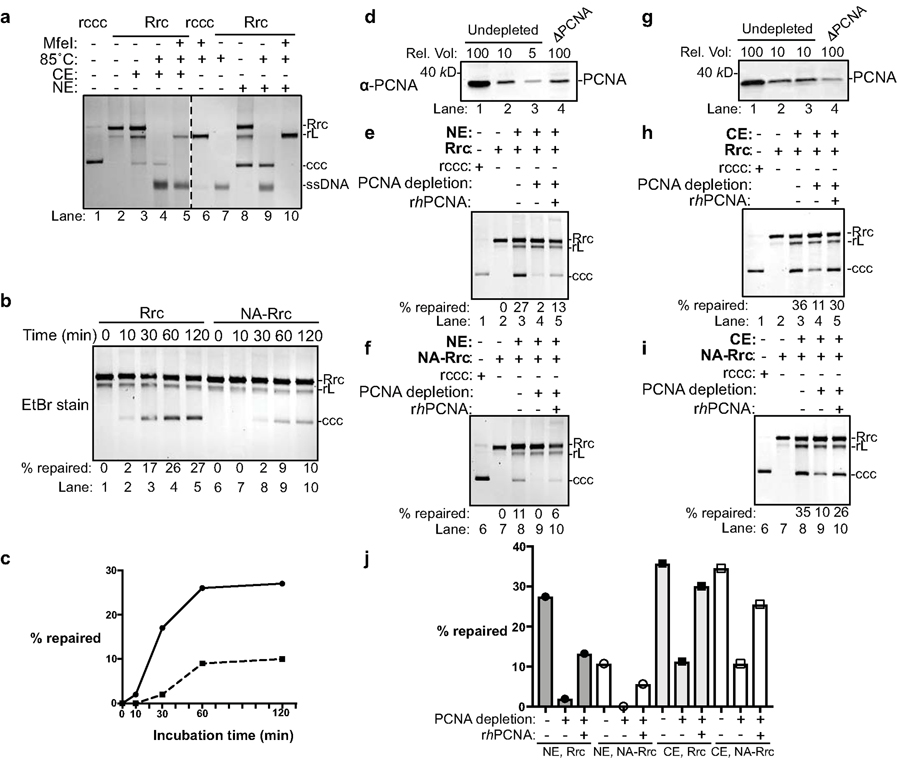
Fig. 3 |. PCNA is required for cccDNA formation in human cell extracts.
(a) Cytoplasmic and nuclear extracts from hNTCP-expressing HepG2 cells fully support cccDNA formation. Repair of RrcDNA was carried out and analyzed as in Fig. 2a, except the incubation temperature was 37°C instead of 30°C. Dashed line indicates removal of superfluous lanes. (b) Time course assay showing the kinetics of cccDNA formation from both RrcDNA (lanes 1–5) and NA-RrcDNA complex (lanes 6–10) in nuclear extracts. Experiments were performed as in (a), except that reactions were terminated at indicated time points. Efficiency of cccDNA formation was calculated as in Fig. 2c and indicated in row ‘% repaired’. (c) The efficiency of cccDNA formation from (b) is plotted against incubation time. (d) Immuno-depletion of more than 90% of PCNA from human nuclear extracts. Un-depleted and PCNA-depleted extracts were analyzed by western blotting using an anti-PCNA antibody. A relative volume (rel. vol) of 100 corresponds to 0.5 μl extract. Depletion of PCNA drastically reduced repair efficiency of RrcDNA (e) and NA-RrcDNA (f) in human nuclear extracts. Untreated recombinant cccDNA (Rccc) and Rrc were used as control (lanes 1, 2). Lanes 3 and 8, mock-depleted extract with mouse IgG; lanes 4 and 9, PCNA-depleted extract from (d) was used; lanes 5 and 10, addition of recombinant human PCNA (rhPCNA, shown in Fig. 4a) to PCNA-depleted extract restored cccDNA repair. (g–i) same to (d–f), except that human cytoplasmic extracts were used. (j) The efficiency of cccDNA formation from (e–f, g–i) is plotted. All data shown were repeated twice independently.
PCNA is required for cccDNA formation in human cell extracts.
We next investigated the effect on cccDNA formation of depleting these five factors from human extracts. Unlike in yeast extracts, many human factors could not be depleted to more than 90% due to lack of antibodies and difficulty in chromosomal tagging. Among the five factors, we successfully immuno-depleted PCNA by 90% from human nuclear extracts and found that cccDNA formation was nearly abrogated, which could be partially restored by addition of recombinant human PCNA (Figs. 3d–f, j, and 4a). Similarly, immuno-depletion of PCNA by more than 90% from human cytoplasmic extracts diminished the repair efficiency by 70%, which could be restored by addition of recombinant PCNA (Figs. 3g–j and 4a). However, less efficient depletion of PCNA – even by 80% – did not impair cccDNA formation (Extended Data Fig. 7a). These observations are reminiscent of the discrepancy between Fen-1 depletion versus complete gene knock-out of Fen-1 in yeast extracts (Fig. 2c, compare lanes 1–2 with lanes 11–12) and suggest that even trace amounts of certain factors are sufficient to catalyze the formation of cccDNA. Indeed, less efficient depletion of human FEN-1 and POLD1, the catalytic subunit of POLδ, by 80% also did not affect cccDNA formation (Extended Data Fig. 7b, c), while complete depletion in yeast extracts abolished cccDNA formation. Unfortunately, we could not achieve more efficient depletion of these factors in human cell extracts by commercially available antibodies, because too many rounds of depletion non-specifically compromise the extract activity. The effects of the human RFC and LIG1, the human homolog of yeast Cdc9, on cccDNA formation could not be analyzed, as we did not achieve depletion of these factors despite several trials using commercially available antibodies (Extended Data Fig. 7d). Collectively, our data recapitulate those found in yeast extracts and fully confirm that PCNA is required for cccDNA formation in human cell extracts. To complement the depletion study and further validate the functions of these factors in cccDNA formation, we next aimed to biochemically reconstitute cccDNA formation with these purified components.
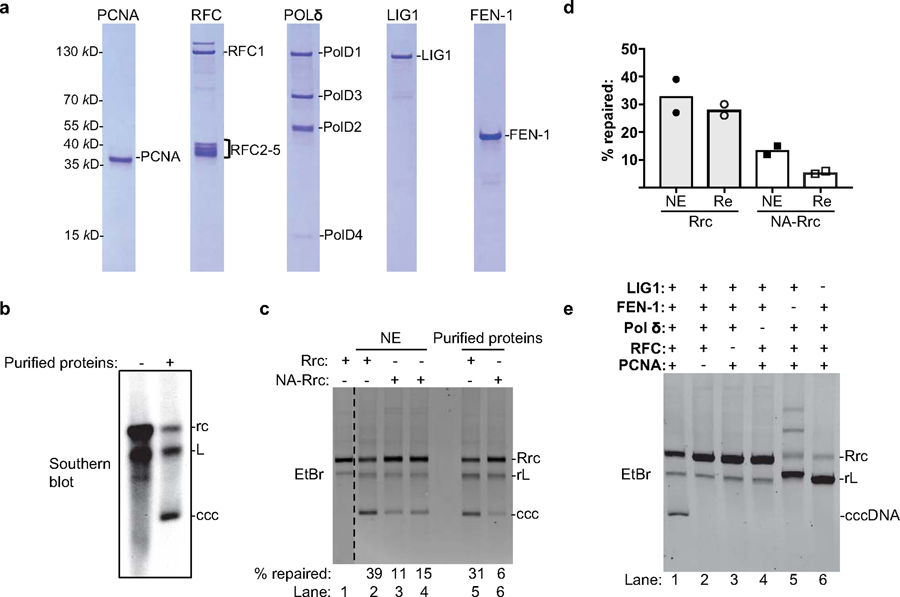
Fig. 4 |. Purified human PCNA, RFC, LIG1, POLδ, and FEN-1 define the minimal set of factors for cccDNA formation.
(a) Purified human proteins involved in DNA lagging strand synthesis analyzed by SDS-PAGE with Coomassie staining. Repeated three times independently. (b) cccDNA formation assay using virion derived dp-rcDNA and purified human proteins from (a). Repeated twice independently. (c) The repair efficiency of RrcDNA and NA-RrcDNA by human nuclear extract (lanes 2–4) versus by purified human proteins (lanes 5–6) were compared. Untreated RrcDNA was used as a control (lane 1). Dashed line indicates removal of superfluous lanes. Repeated three times independently. (d) The repair efficiency from (c) is plotted (n=2). NE, nuclear extract. PP, purified proteins. Bar value indicates mean of two measurements. (e) All five factors are required for cccDNA formation. Omission of factors is indicated by ‘–’. All data presented are representatives of two to three independent experiments. Repeated twice independently.
PCNA, RFC, POLδ, FEN-1 and LIG1 constitute a minimal set of factors for cccDNA formation in vitro.
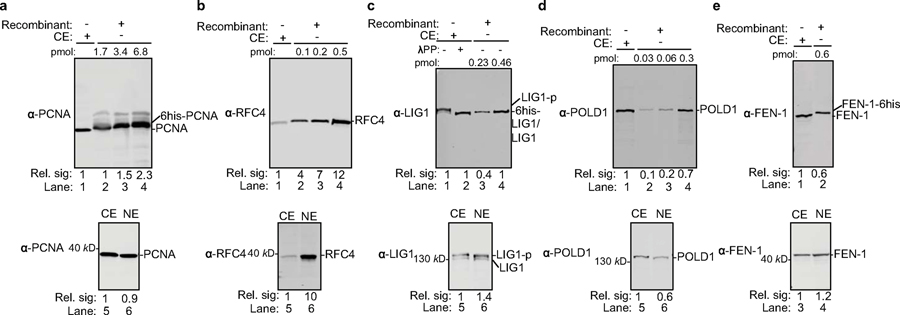
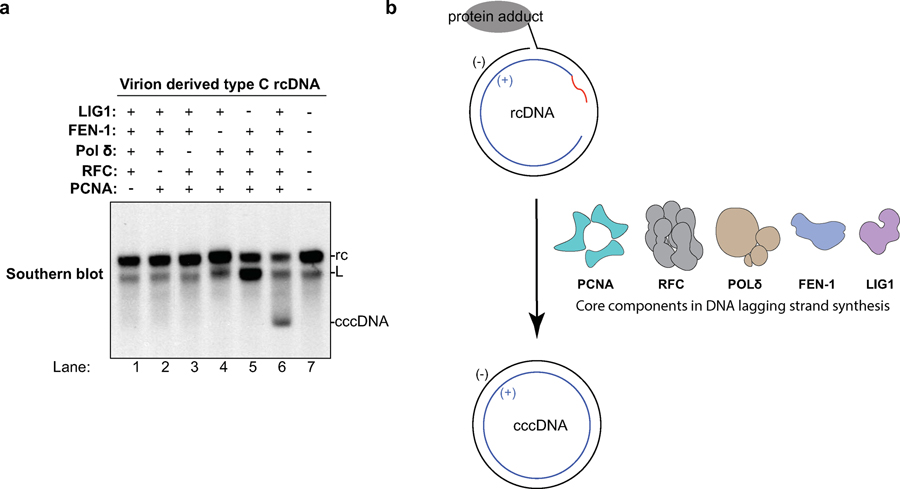
We purified human recombinant PCNA, RFC, POLδ, FEN-1 and LIG1 to near homogeneity and incubated them with different rcDNA substrates at concentrations similar to those found in human cell extracts (Extended Data Fig. 8). Remarkably, these five factors fully reconstituted the repair of dp-rcDNA, RrcDNA and NA-RrcDNA to cccDNA with efficiency comparable to that of human extracts (Fig. 4b–d), demonstrating that these five factors define a minimal set of activities required to repair all lesions on rcDNA. Omitting any of these five factors abrogated repair of RrcDNA and dp-rcDNA (Fig. 4e, Extended Data Fig. 9a), indicating that all five factors are required for cccDNA formation. Taken together, these data demonstrate that PCNA, RFC, POLδ, FEN-1, and LIG1 function as a minimal set of host repair factors for cccDNA formation (Extended Data Fig. 9b).
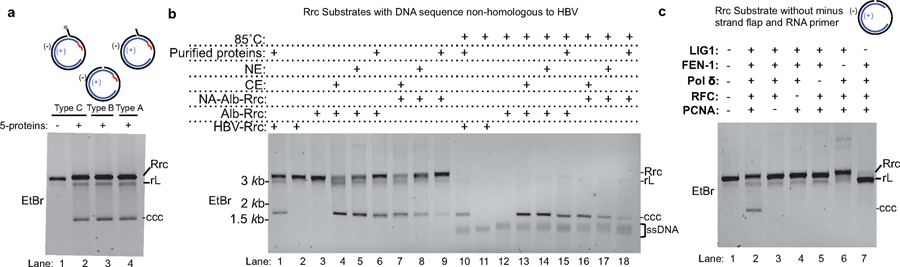
Repair of recombinant rcDNA substrates composed of various lesions or non-HBV DNA sequence.
The recombinant rcDNA substrates can be manipulated to harbor various DNA lesions and sequences and thus hold great potential to be utilized by the HBV field to delineate the detailed molecular mechanisms of rcDNA repair. These could not be achieved by using authentic rcDNA due to the fact that the strict sequence requirement of the HBV genome precludes their isolation from cell culture-derived HBV virions. We have shown that our five-factor reconstitution system can repair NA-RrcDNA, RrcDNA, and virion-derived “type C” dp-rcDNA. We also confirmed that RrcDNA substrates that mimic “type A” and “type B” dp-rcDNA could also be fully repaired by the five-protein system (Fig. 5a), indicating that these proteins are sufficient to repair all three types of dp-rcDNA, which are potential repair intermediates after HBV polymerase removal.
Fig. 5 |. Repair of RrcDNA with various lesion structure and sequences with purified proteins and human cell extracts.
(a) Analogous to RrcDNA that mimics type C dp-rcDNA (lane 2), RrcDNA substrates that mimic type A (no flap) and type B dp-rcDNA (clean-ended flap) can also be repaired by 5 purified human factors (lanes 3–4). Repeated twice independently. (b) A 3.2 kb RrcDNA or NA-RrcDNA containing a flap and RNA primer, and with all HBV sequence replaced by albumin promoter and mCherry sequences, was subject to cccDNA formation reaction in human cytoplasmic extract (lanes 4, 7), nuclear extract (lanes 5, 8), and purified protein system (lanes 6, 9). HBV-RrcDNA was used as a control (lane 1). The cccDNA reaction products from lanes 1–9 were subjected to heat treatment at 85°C for 5 min and loaded to lanes 10–18 to show cccDNA bands. Alb: human Albumin and mCherry sequences; NA: NeutrAvidin. Repeated twice independently. (c) Rrc Substrate without DNA flap and RNA primer/flap was incubated with 5 purified human factors (lane 2) or with factors omitted (lanes 3–7), and cccDNA formation was detected by agarose gel electrophoresis. All data presented are representatives of two to three independent experiments. Repeated twice independently.
It remains unclear whether the requirement of lagging strand synthesis components for the repair of rcDNA is solely determined by Okazaki-like structures in rcDNA or whether HBV-specific DNA sequences also play a role. Our recombinant rcDNA system allows us to generate such non-HBV rcDNA containing Okazaki-like structures and bearing sequences not related to HBV. As shown in Fig. 5b, a 3.2 kb, non-HBV rcDNA was generated, with all HBV-specific sequences, including flap and RNA primer, replaced by the human albumin promoter and mCherry sequences. This non-HBV rcDNA was efficiently repaired by human cytoplasmic and nuclear extracts as well as the minimal set of five factors. These results demonstrate that HBV-specific DNA sequences do not have an overt effect in cccDNA formation, which is in concordance with the fact that repair factors usually recognize the type of lesion instead of the specific sequence.
One feature of HBV rcDNA is the DNA flap and RNA primer/flap structures on the minus and plus strands, respectively. The removal of these flaps is most likely mediated by the flap endonuclease FEN-1 in our five minimal protein repair system. Thus, we tested if a RrcDNA substrate without the DNA and RNA flaps could obviate the need for FEN-1. Surprisingly, the flap-free RrcDNA substrate still required all five factors for cccDNA formation (Fig. 5c). This is most likely because DNA polymerase displaces the DNA strand, generating a flap that needs to be removed.
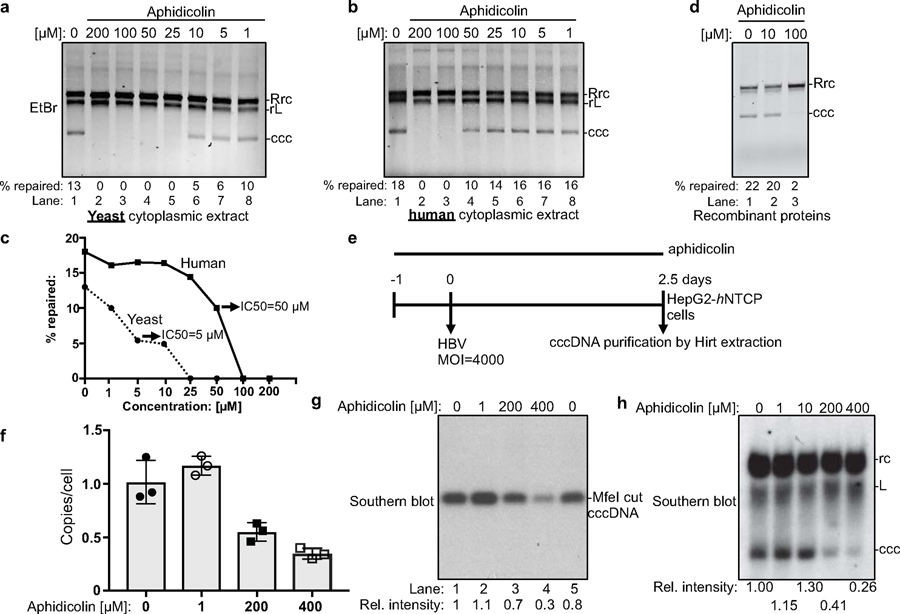
Aphidicolin, an inhibitor of DNA POLδ, ε, and α, inhibits cccDNA formation in cell extracts, purified-protein system, and HBV-challenged cell culture system.
Since POLδ is a key component in our repair system, we tested whether aphidicolin, an inhibitor of DNA POLδ, ε, and α (21), also inhibited cccDNA formation. Indeed, cccDNA formation is abrogated in yeast cell extracts treated with 25–200 μM of aphidicolin (Fig. 6a, c). Treatments of 100–200 μM aphidicolin in human extracts as well as in our purified system, where POLδ is the only DNA polymerase, are required to abrogate cccDNA formation (Fig. 6b–d), supporting our finding that (an) aphidicolin-sensitive polymerase(s) is essential for cccDNA formation. Most importantly, aphidicolin also diminished cccDNA formation in human HepG2-hNTCP cells challenged with HBV at doses comparable to our biochemical studies (Fig. 6e–h). Continuous treatment with aphidicolin at 400 μM reduced cccDNA formation by 70%, as measured by quantitative PCR and Southern blotting (22–24) (Fig. 6e–h). These effects are not simply due to the diminished viability of host cells, as we did not observe any overt cytotoxicity at the time of harvest. Collectively, these data are consistent with our finding that (an) aphidicolin-sensitive polymerase(s) play(s) a critical role in cccDNA formation in both cell extracts and HBV-infected cells.
Fig. 6 |. DNA polymerase inhibitor aphidicolin inhibits cccDNA formation in cell extracts, in the purified protein system, and HBV-infected human hepatoma cells.
Robust inhibition of cccDNA formation by aphidicolin in yeast ((a), at 30°C, protein concentration ~15 mg/ml) and human HepG2-hNTCP cytoplasmic extract ((b), at 37°C, protein concentration ~30 mg/ml). (c) Efficiencies of cccDNA formation from (a) and (b) are plotted against aphidicolin concentration. (d) Aphidicolin inhibits cccDNA formation by the purified protein components (at 37°C). (e) Schematic for testing the effect of aphidicolin on cccDNA formation in hNTCP-expressing HepG2 cells infected with HBV. ‘–1’ indicates aphidicolin treatment started one day before HBV challenge. MOI, multiplicity of infection. (f) ExoI/ExoIII/T5 nuclease treated cccDNA copy numbers from HBV-infected cells detected by quantitative PCR (n=3). Bar value indicates mean of three measurements, and error bars are s.d. (g) cccDNA samples from (f) were linearized by MfeI and analyzed by Southern blotting. The relative intensities of cccDNA bands were normalized to those of mock-treated cells (lane 1). (h) Southern blotting of cccDNA extracted by Hirt method without ExoI/ExoIII/T5 nuclease treatment. Southern blotting was performed as in (g). All data presented are representatives of two to three independent experiments. All data shown [except for (f, n=3)] are repeated twice independently.
Discussion
There are two main ways our RrcDNA differs from native HBV rcDNA. First, the RNA primers in RrcDNA are not capped. Although a cap is unlikely to significantly affect the repair process, this possibility has not been completely ruled out. Second, authentic HBV rcDNA contains the HBV polymerase covalently linked to a 5’ flap via a tyrosyl-phosphodiester bond, which is potentially processed by TDP2 (7). NA-RrcDNA contains NA attached to the 10-nt flap of the minus strand through a non-covalent NA-biotin interaction, which cannot be processed by TDP2. Therefore, the effect of TDP2 on cccDNA formation could not be directly assessed by our recombinant substrates. However, NA-RrcDNA is suitable to study the removal of protein adduct by endonucleases and proteases (Extended Data Fig. 1a), which is rarely dependent on the identity of the adduct per se. Indeed, NA-RrcDNA could be repaired by these five factors to form cccDNA (Fig. 4c, lane 6), indicating that the nuclease FEN-1 can remove the protein adduct by cleaving the 5’ flap. It has been shown that TDP2 knockout does not abolish cccDNA formation (7,13), most likely because FEN-1 can fulfill the role of HBV Polymerase removal.
Our findings indicate that the aphidicolin-sensitive polymerase POLδ is critical for cccDNA formation. However, previous work demonstrated that treatment with up to 10 μM aphidicolin did not affect cccDNA formation in HBV-infected hepatoma cells, and suggested that an aphidicolin-insensitive translesional DNA polymerase, POLκ, instead played a key role in cccDNA formation (11). Our data showed that 10 μM aphidicolin was not sufficient to inhibit POLδ in purified protein or in human cell extract (Fig. 6b, d). As a matter of fact, a much higher dose of aphidicolin (150–300 μM) is routinely used to inhibit aphidicolin-sensitive polymerases (25,26). Therefore, we used 200–400 μM aphidicolin in HBV-infected cells and found that cccDNA formation is dramatically reduced, indicating that (an) aphidicolin-sensitive polymerase(s) is critical for cccDNA formation. However, it is conceivable that cccDNA formation in hepatocytes infected with HBV may be more complex and be regulated by more factors, and our findings do not exclude the involvement of some previously implicated factors in cccDNA formation, such as POLκ, LIG3, and POLα, (11,12,15).
The biochemical approaches and reagents from this study are valuable tools to further unravel the detailed molecular mechanisms of cccDNA formation. Meanwhile, future efforts are needed to fully validate and elucidate the function of these replication/repair factors in cccDNA formation both in cell culture and animal models. However, three main hurdles exist when using knock-out/down approaches to evaluate their effects: i) many DNA repair factors are essential and cannot readily be knocked out in mammalian cells; ii) many DNA repair enzymes are present in excess in cells, so the residual levels of enzymes after transient knockdown are sufficient to execute their functions, potentially leading to false negatives; iii) knockdown of repair factors can lead to corollary effects, including DNA damage checkpoint activation, which could indirectly affect cccDNA formation and generate false positives. Therefore, highly specific inhibitors for these repair factors and/or stringent genetic approaches that allow acute depletion of these repair factors would be desirable in future studies.
Methods
Yeast strains and techniques.
Saccharomyces cerevisiae strains (kindly provided by Xiaolan Zhao, Memorial Sloan–Kettering Cancer Center, New York, NY) were isogenic to W1588–4C, a RAD5 derivative of W303 (MATa ade2–1 can1–100 ura3–1 his3–11,15 leu2–3,112 trp1–1 rad5–535) (27). Strains were grown in standard Yeast extract-Peptone-Dextrose (YPD) media. Strains used in this study are listed in Extended Data Fig. 10. Proteins were fused with 3x Flag tag at their endogenous chromosomal loci by standard methods, and correct tagging was verified by Sanger sequencing and western blotting. All genetic and biochemical experiments were performed with two different spore clones for each genotype.
Cell lines.
HepG2 cells (American Tissue Culture Collection, ATCC® Number: HB-8065™, Manassas, VA) and the hNTCP-expressing HepG2 cells clone 3B10 (20) were maintained in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Hyclone), 100 units/ml of penicillin, and 100 μg/ml streptomycin (Thermo Fisher, Waltham, MA). HepG2.2.15 cells (28) (kindly provided by Christoph Seeger, Fox Chase Cancer Center, PA) were maintained in DMEM/F-12 media (Thermo Fisher Scientific, Waltham, MA) supplemented with 5% (vol/vol) FBS, 100 units/ml of penicillin, and 100 μg/ml streptomycin (Thermo Fisher, Waltham, MA). All cell lines are tested to be free of mycoplasma contamination. hNTCP-expressing HepG2 3B10 cell line was authenticated by its susceptibility to HBV infection. HepG2.2.15 cell line has been authenticated by its ability to produce HBV.
Generation of recombinant cccDNA by minicircle technology.
Recombinant cccDNA (Rccc) was generated as previously described (18,29). The HBV genotype D genome (GenBank accession number: U95551.1) was generated by gene synthesis (Integrated DNA Technologies, Coralville, IA). The parental Rccc production plasmid (pLW25) was constructed by inserting the whole 3182 bp HBV genome into pMC.CMV-GFP-SV40-polyA (System Biosciences, Palo Alto, CA) between the bacteriophage ΦC31 integrase-dependent attB and attP recombination sites. DNA adenosine (Dam) and cytosine (Dcm) methylases from the producer E.coli strain ZYCY10P3S2T (System Biosciences, Palo Alto, CA) were deleted by P1 phage-mediated gene transfer using dam- and dcm- strains from the Keio collection (30) by standard methods, resulting in a new producer E.coli strain ZYCY10P3S2T dam-/dcm-. The parental plasmid pLW25 was transformed into ZYCY10P3S2T dam-/dcm- and RcccDNA was produced according to the manufacturer’s instructions. Briefly, overnight E. coli cultures were grown in Terrific Broth (TB) supplemented with 50 μg/ml kanamycin and RcccDNA was induced by addition of L-arabinose (Sigma Aldrich, St. Louis, MO) to a final concentration of 0.01% (w/vol). RcccDNA was further purified by NucleoBond Maxi prep kit (Macherey Nagel, Bethlehem, PA) according to the manufacturer’s instructions. The final RcccDNA contains a 39 bp insertion (CCCCAACTGGGGTAACCTTTGGGCTCCCCGGGCGCGACC) in the polymerase domain between nt 2849 and nt 2850 (nucleotide numbers correspond to those in U95551.1), which does not cause a frame shift nor affect its function, and the 3221 bp recombinant cccDNA behaves like authentic HBV cccDNA (18).
Generation of recombinant rcDNA and NeutrAvidin-RrcDNA complex.
The schematic to generate recombinant rcDNA (RrcDNA) and NeutrAvidin-RrcDNA complex is described in Fig. 1a. Plasmids pLW213 and pLW227 were generated to produce minus and plus strand ssDNA, respectively. All agarose gel electrophoresis described below were carried out in RNase-free buffer unless otherwise indicated.
For pLW213, the 3208 bp HBV genome starting from nt 1819 to nt 1805 (nucleotide numbers correspond to those in U95551.1) of RcccDNA containing the same 39bp insertion as described above was amplified by PCR and inserted into the pMiniT 2.0 plasmid (New England Biolabs, NEB, Ipswich, MA) between the newly introduced restriction enzyme sites Nt.BspQI and BssHII. To generate minus strand ssDNA, pLW213 was digested with BssHII and Nt.BspQI, and the digestion products were subsequently denatured and subjected to electrophoresis to separate the minus strand ssDNA from the other digest products. The gel fragment containing the minus strand ssDNA was dissolved and the ssDNA extracted, dissolved in RNase-free water (Thermo Fisher Scientific, Waltham, MA) and stored at −80°C.
pLW227 was constructed as pLW213 except that the HBV genome starting from nt 1624 to nt 1501 (nucleotide numbers correspond to those in U95551.1) of RcccDNA was inserted into the pMiniT 2.0 plasmid. The plus strand ssDNA was generated as the minus strand ssDNA as described above, except for using pLW227 instead of pLW213.
Non-HBV RrcDNA was generated with plasmid pLW343 and pLW363. For pLW343, the 1969 bp Albumin promoter sequence from nt 16023–17991 (Genbank: EF649953.1), 63bp self-cleaving 2A sequence (GGCAGTGGAGAGGGCAGAGGAAGTCTGCTAACATGCGGTGACGTCGAGGAGAATCCTGGCCCA), 705 bp mCherry sequence from nt 1808–2512 (Genbank: MK024392.1), and 462 bp albumin promoter sequence from nt 17992–18453 (Genbank: EF649953.1) was amplified and inserted to the pMiniT 2.0 plasmid between the BssHII and nt.BspQI sites. For pLW363, the 196 bp and 1768 bp Albumin promoter sequence from nt 18280–18475 and nt 16023–17991 (Genbank: EF649953.1), 63bp self-cleaving 2A sequence (GGCAGTGGAGAGGGCAGAGGAAGTCTGCTAACATGCGGTGACGTCGAGGAGAATCCTGGCCCA), 705 bp mCherry sequence from nt 1808–2512 (Genbank: MK024392.1), and 150 bp albumin promoter sequence from nt 17992–18141 (Genbank: EF649953.1) was amplified and inserted to the pMiniT 2.0 plasmid between the nt.BspQI and BssHII sites. ssDNA from pLW343 and pLW363 were generated as described above for pLW213 and 227.
The minus and plus strand ssDNAs were mixed at a 1:1 molar ratio and subsequently annealed. The annealing product containing recombinant rcDNA (RrcDNA) precursor (Fig. 1b) was resolved on a 0.7% (w/vol) agarose gel, which was visualized by SYBR™ Safe staining (Thermo Fisher Scientific, Waltham, MA). The gel fragment containing the RrcDNA precursor was recovered, purified, and dissolved in RNase-free water (Thermo Fisher Scientific, Waltham, MA) and stored at −80°C.
To generate RrcDNA, RrcDNA precursor was incubated with oligo PU-O-5573 (5’ Biotin-GAAAAAGTTGCATGGTGCTGGTG, Integrated DNA Technologies, Coralville, IA) and oligo PU-O-5670 (5’ rGrCrArArCrUrUrUrUrUrCrArCrCrUrCrUrGrCACGTCGCATGGAGACCACCGT, Integrated DNA Technologies, Coralville, IA) in buffer containing 50 mM KOAc, 20 mM Tris-OAc (pH 8.0 at 25°C), 10 mM Mg(OAc)2 to restore the flap structure and RNA primer on the minus and plus strands, respectively. For non-HBV rcDNA, oligo PU-O-6861 (5’ biotin-ATGCAGTAAGATGAGTTGAACTATTAGGTTGAAC, Integrated DNA Technologies, Coralville, IA) and oligo PU-O-6862 (5’rGrCrUrUrArCrUrGrCrArUrGrUrGrUrUrUrGrUTTCCAAAGGGTCAAAATTCAAAATTG, Integrated DNA Technologies, Coralville, IA) were used.
The mixture was subsequently supplemented with T4 DNA ligase (Thermo Fisher Scientific, Waltham, MA) and 0.5 mM ATP (Sigma Aldrich, St. Louis, MO) to seal the nicks. The ligase was removed by phenol-chloroform (1:1, vol/vol) extraction and the ligation product was purified and dissolved in RNAse-free water containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. The ligation efficiency was determined as described in Fig. 1a–c. Briefly, purified RrcDNA precursor and RrcDNA were digested with restriction enzyme BsrDI (NEB, Ipswich, MA) in buffer NEB2.1 (NEB, Ipswich, MA) at 65°C for 1 hour. The digestion product was purified by phenol-chloroform (1:1, vol/vol) extraction followed by ethanol precipitation and dissolved in RNase-free water. The digest was mixed with equal volume denaturing loading dye containing 95% (vol/vol) formamide, 0.025% (w/vol) bromophenol blue, 0.025% (w/vol) xylene cyanol, and 5 mM EDTA. The solution was heated at 95°C for 5 min and resolved on a 10% (w/vol) urea-PAGE gel, which was subsequently stained with SYBR™ Gold (Thermo Fisher Scientific, Waltham, MA) to visualize the M1/P1 single stranded oligos generated by RrcDNA precursor and M2/P2 single stranded oligos generated by RrcDNA after BsrDI digestion as shown in Fig. 1a,c. The ligation efficiency of RrcDNA from RrcDNA precursor consistently approaches 100% as shown in Fig. 1c.
To generate the NeutrAvidin RrcDNA complex, RrcDNA was incubated with NeutrAvidin (Thermo Fisher, Waltham, MA) at a final concentration of 4 mg/ml in buffer containing 20 mM HEPES-KOH (pH 8.0) and 50 mM KCl at 37°C for 30 min. The efficiency of NeutrAvidin-RrcDNA complex formation was determined by subjecting RrcDNA and NeutrAvidin-RrcDNA complex to 0.7% (w/vol) agarose gel electrophoresis at 3.5 V/cm for 2 hours, and the gel is stained with SYBR™ safe and visualized on Typhoon™ TLA 9500 (GE Healthcare Lifesciences, Chicago, IL) according to manufacturer’s instructions. The efficiency of NeutrAvidin-RrcDNA complex formation consistently approaches 100% as shown in Fig. 1e.
Preparation of yeast cell extracts.
Yeast cells were grown in yeast extract peptone dextrose (YPD) media until cell density reached 1.6–2×107 cells/ml. Cells were pelleted, washed once with media, and then re-suspended in spheroplasting buffer (1.2 M sorbitol, 10 mM EDTA, 10 mM HEPES-KOH (pH 8.0) and 0.1% (vol/vol) 2-mercaptoethanol). Zymolyase (US Biological, Salem, MA) was then added to the mixture, which was incubated at 30°C for 30 min. The spheroplasts were spun at 2,500 x g for 2 min at 4°C and washed once with buffer containing 25 mM HEPES-KOH (pH 8.0), 5 mM MgCl2, and 0.5 M sucrose. The mixture was spun at 2,500 x g for 2 min at 4°C, and lysis buffer containing 10 mM HEPES-KOH (pH 8.0), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, and supplemented with 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail (Sigma Aldrich, St. Louis, MO) was added to the pellet, which was subsequently lysed using a Dounce homogenizer (DWK, Millville, NJ). The lysed cells were then spun at 2,500 x g for 5 min; the supernatant was collected and supplemented with glycerol and KCl to a final concentration of 5% (vol/vol) and 50 mM, respectively. The solution was spun at 100,000 x g for 1 hour, and the supernatant was dialyzed against dialysis buffer containing 20 mM HEPES (pH 8.0), 1.5 mM MgCl2, 50 mM KCl, 5% (vol/vol) glycerol, 1 mM DTT, and 0.5 mM proteinase inhibitor PMSF. After dialysis, the solution was spun at 20,000 x g for 10 min. The supernatant contained yeast cytoplasmic extract, which was subsequently concentrated to reach a protein concentration between 15–30 mg/ml. Note that the protein concentration was measured on a Nanodrop spectrophotometer (Thermo Fisher, Waltham, MA) by A280 absorbance, using the default setting that 1 Abs=1 mg/ml. The pellet from the 2,500 x g spin, which contained nuclei, was resuspended in high salt solution containing 20 mM HEPES (pH 8.0), 1.5 mM MgCl2, 700 mM KCl, 5% (vol/vol) glycerol, and 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail. The mixture was incubated at 4°C for 30 min before being subjected to centrifugation at 100,000 x g for 1 hour. The nuclear extract was subsequently concentrated and dialyzed as described for cytoplasmic extract. The final protein concentrations of nuclear extracts were 7.5–14 mg/ml. It is worth noting that the repair activity of the extracts may have batch-to-batch variation. Some extracts may be more active than others.
Preparation of human cell extracts.
Human hepatoma cell line HepG2 expressing human NTCP (hNTCP, clone 3B10) (20) was grown in DMEM/10% (vol/vol) FBS medium and extracts were prepared as previously described with modifications (31). Cells were harvested, resuspended in hypotonic buffer (10 mM HEPES-KOH (pH 8.0), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, supplemented with 0.5 mM PMSF and 1x Protease Inhibitor Cocktail (Sigma Aldrich, St. Louis, MO)) and lysed using a Dounce homogenizer (DWK, Millville, NJ). The lysed cells were then spun at 1,500 x g for 5 min. The supernatant (cytoplasmic fraction) was collected and supplemented with glycerol and KCl to a final concentration of 5% (vol/vol) and 50 mM, respectively. The supernatant was then spun at 100,000 x g for 1 hour and subsequently dialyzed. The cytoplasmic extract was then concentrated to reach a protein concentration between 20–40 mg/ml. The cell pellet (the nuclear fraction) was resuspended in high salt solution containing 20 mM HEPES (pH 8.0), 1.5 mM MgCl2, 700 mM KCl, 5% (vol/vol) glycerol, 0.5 mM PMSF, and 1x Protease Inhibitor Cocktail (Sigma Aldrich, St. Louis, MO). The mixture was incubated at 4°C for 30 min before being subjected to centrifugation at 20,000 x g for 30 min. The supernatant was recovered, concentrated, and dialyzed as described above. The final protein concentrations of nuclear extracts were between 15–30 mg/ml. It is worth noting that the repair activity of the extracts may have batch-to-batch variation, so some extracts may be more active than others.
Preparation of HBV derived from cell culture.
The preparation of cell culture-derived HBV was carried out as previously described (32). Briefly, HepG2.2.15 cells were cultured in tissue culture treated 150 mm plates containing DMEM/F-12, supplemented by 5% FBS (vol/vol), 100 U/ml of penicillin, and 100 μg/ml streptomycin. Supernatant was collected every 3–4 days, pooled, and filtered through 0.45 μm filter units (VWR, Radnor, PA). 40% PEG 8,000 (w/vol) (Sigma Aldrich, St. Louis, MO) was added to the clarified supernatant to a final concentration of 8% (w/vol), and the mixture was incubated overnight at 4°C. The mixture was centrifuged at 3,000 x g for 20 min using a swing-bucket rotor at 4°C, and the supernatant was discarded. The pellet was resuspended in 1x PBS supplemented with 10% (vol/vol) FBS, and the mixture was incubated overnight at 4°C. The solution was spun at 3,000 x g for 10 min at 4°C, and the supernatant was transferred to a clean tube and the pellet discarded. The supernatant was overlaid onto a solution containing 20% sucrose in 1x PBS for further purification and was spun at 100,000 x g in a SW28 rotor (Beckman Coulter, Brea, CA) for 4 hours at 4°C. After centrifugation, the supernatant was discarded and the pellet resuspended in 1x PBS supplemented with 10% (vol/vol) FBS and stored at −80°C.
Purification of deproteinated rcDNA from HBV virions.
The purification of deproteinated rcDNA from HBV virions was carried out as previously described (33) with minor modifications. 1010 HBV virions purified by sucrose ultracentrifugation were first treated with DNase I in the presence of 6 mM MgCl2 at 37°C for 2 hours. The reaction was subsequently incubated with 75 mM Tris-HCl, pH 7.5, 75 mM NH4Cl, 30 mM MgCl2, 12 mM 2-mercaptoethanol, 0.5% (vol/vol) NP-40, and 0.4 mM dNTP at 37°C for 4 hours to complete the endogenous polymerase reaction (EPR), since HBV nucleocapsids most likely are subjected to EPR after entry into a cellular environment containing sufficient amount of dNTP. EDTA and SDS were added to a final concentration of 50 mM and 1% (w/vol), respectively, before addition of proteinase K (Sigma Aldrich, St. Louis, MO) at 2 mg/ml. This solution was then incubated at 37°C for 4 hours and extracted twice with phenol and once with phenol-chloroform (1:1, vol/vol). The aqueous phase was recovered and 2 volumes of 100% (vol/vol) ethanol were added and incubated for 30 min on ice to precipitate deproteinated rcDNA. After centrifugation at 20,000 x g for 20 min, the precipitate was washed once with 500 μl of 4°C 70% (vol/vol) ethanol and subsequently dissolved in TE buffer consisting 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA.
Immuno-depletion of yeast cell extracts.
30 μl of anti-FLAG M2 affinity gel (Sigma Aldrich, St. Louis, MO) were washed three times with 100 μl of washing buffer (20 mM HEPES-KOH (pH 8.0), 50 mM KCl, 5% (vol/vol) glycerol, 1 mM DTT) and added to 40 μl of yeast cytoplasmic extract. The mixture was incubated at 4°C for 1 hour on a rotator. The mixture was spun briefly at 1,000 x g for 1 min and the supernatant transferred to a clean Eppendorf tube. This is one round of immuno-depletion. Generally, two rounds of immuno-depletion were required to achieve near complete depletion, as determined by western blotting using anti-FLAG M2 antibody (Sigma Aldrich, St. Louis, MO).
Immuno-depletion of human cell extracts.
50 μl of protein A or G conjugated magnetic beads (Thermo Fisher, Waltham, MA) were washed three times with 100 μl of washing buffer (20 mM HEPES-KOH (pH 8.0), 50 mM KCl, 5% (vol/vol) glycerol, 1 mM DTT) and added to 20–40 μg of commercial antibody solutions. The mixture was incubated at 4°C overnight on a rotator to capture the antibodies on magnetic beads. This was then washed three times with washing buffer and added to 40 μl of human cytoplasmic or nuclear extracts and incubated for 2 hours at 4°C on a rotator. The supernatant was transferred to a clean Eppendorf tube. This is one round of immuno-depletion. Generally, three rounds of immuno-depletion were performed without significant non-specific loss of activity.
cccDNA formation assay in yeast cell extracts.
60 ng of RrcDNA or NeutrAvidin-RrcDNA complex, or 1–5 ng (estimated by quantitative PCR) of deproteinated virion derived rcDNA were incubated with yeast cell extract in reaction buffer consisting of 20 mM Tris-HCl (pH 7.6), 50 mM KCl, 0.1 mM dNTP, 5 mM MgCl2, 1 mM reduced glutathione, 2.6 mM ATP, 26 mM phosphocreatine disodium (Sigma Aldrich, St. Louis, MO), and 6 μg/ml creatine phosphokinase (Sigma Aldrich, St. Louis, MO). The mixture was incubated at 30°C for 60 min before the reaction was stopped by addition of SDS and EDTA to final concentrations of 0.5% (w/vol) and 25 mM, respectively. The solution was subsequently treated with RNase A and proteinase K for 1 hour each. The solution was then extracted by phenol-chloroform (1:1, vol/vol) and repair product precipitated by ethanol and dissolved in TE buffer consisting of 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. To monitor cccDNA formation, the repair products were loaded onto a 0.7% (w/vol) agarose gel containing 0.05 μg/ml ethidium bromide and run at 4 V/cm for 2 hours before the gel was visualized on Typhoon™ FLA 9500 (GE Healthcare Lifesciences, Chicago, IL). The intensity of DNA bands was quantified by ImageJ.
For the repair reaction using deproteinated rcDNA (dp-rcDNA), cccDNA was detected by Southern blotting as describe in the Southern blotting section.
cccDNA formation assay in human cell extracts.
The cccDNA formation assay using human cytoplasmic and nuclear extracts was performed analogously to the one using yeast extract, with one modification – the incubation was carried out at 37°C instead of 30°C.
Preparation of repaired cccDNA from extracts for Sanger sequencing.
The repaired cccDNA bands were cut from agarose gels (Fig. 2a, lane 3; Fig. 3a, lane 8). DNA was purified using the Nucleospin Gel and PCR clean up kit (Macherey Nagel, Bethlehem, PA) and dissolved in TE buffer consisting 10 mM Tris-HCl, pH 8.0 and 1 mM EDTA. 100pg of DNA was used in a 50 μl PCR reaction containing primers PU-O-4195 (Forward: GAACCTTTTCGGCTCCTC) and PU-O-5320 (Reverse: GGAAAGAAGTCAGAAGGCAA). PCR program was run on a thermocycler (Eppendorf, Hamburg, Germany) with the following program: 95 °C for 10 min, followed by 25 cycles of 95 °C for 30 s, 52°C for 30 s, and 72°C for 60 s. The PCR product was resolved on a 1% agarose gel, a single prominent band was observed and DNA was extracted with Nucleospin Gel and PCR clean up kit (Macherey Nagel, Bethlehem, PA) and dissolved in TE buffer consisting 10 mM Tris-HCl, pH 8.0 and 1 mM EDTA to a final concentration of 80 ng/μl.
Purification of human PCNA.
The bacterial protein expression plasmid encoding human PCNA (pLW263) was generated by cloning the PCNA coding sequence from cDNA (ORFeome Collaboration, NCBI reference sequence: NM_002592.2) (34) into the pET15 expression vector. PCNA was expressed as a N-terminal 6x histidine-tagged protein (6xhis-PCNA) in E. coli BL21(DE3). Cells were grown at 37°C until OD600 reached 0.5–0.7. PCNA expression was induced by addition of isopropyl-1-thio-β-D-galactopranoside (IPTG) to a final concentration of 0.5 mM and incubated at 37°C for 3 hours. Cells were harvested by centrifugation and the cell pellet washed once with 1x PBS. The cell pellet was resuspended in lysis buffer containing 20 mM HEPES-KOH (pH 8.0), 400 mM NaCl, 10 mM Imidazole, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail (Sigma Aldrich, St. Louis, MO), 1 mM PMSF, and 1 mM benzamidine. Cells were disrupted by sonication and cell lysate was clarified by centrifugation at 20,000 x g for 30 min. Supernatant was passed through a column containing 1 ml of Ni2+-NTA resin (Qiagen, Hilden, Germany) twice to enrich for 6xhis-PCNA on the resin. After washing the column with 20 ml of lysis buffer, 6xhis-PCNA was eluted with buffer containing 20 mM HEPES-KOH (pH 8.0), 100 mM NaCl, 250 mM Imidazole, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, 1 mM PMSF, and 1 mM benzamidine. The eluent was fractioned on a Mono Q column (GE Healthcare Lifesciences, Chicago, IL) using a gradient of 0.1–1 M NaCl in a buffer of 20 mM HEPES-KOH (pH 8.0), 1 mM DTT, 1 mM PMSF, and 1 mM benzamidine. Peak fractions were analyzed by SDS-PAGE stained with Coomassie blue, and the fractions containing 6xhis-PCNA were pooled, concentrated to 500 μl and loaded onto a Superdex-200 size exclusion column (GE Healthcare Lifesciences, Chicago, IL) equilibrated in 20 mM HEPES-KOH (pH 8.0), 50 mM KCl, 5% (vol/vol) glycerol, and 1 mM DTT. Peak fractions were analyzed by SDS-PAGE stained with Coomassie blue and the fractions containing near homogeneous 6xhis-PCNA were pooled and stored in aliquots of ~1.4 mg/ml at −80°C. Note that the concentrations of all proteins were measured on a Nanodrop spectrophotometer (Thermo Fisher, Waltham, MA) by A280 absorbance, using the default setting that 1 Abs=1 mg/ml, unless otherwise indicated.
Purification of human RFC complex.
The bacterial protein expression plasmids encoding the components of the human RFC complex (pCDF-hRFC140 and pCDF-hRFC2–5) were kindly provided by Paul Modrich (Duke University). Human RFC complex was purified as previously described (35) with minor modifications. Briefly, non-tagged human RFC complex was expressed in E. coli BLR (BL21 DE3) cells (Novagen, Madison, WI). Cells were grown to OD600=0.4 at 37°C, and IPTG was added at a final concentration of 0.5 mM for 15 hours at 15°C to induce expression of RFC subunits. Cells were harvested and washed once with 1x PBS. The pellet was resuspended in lysis solution containing 20 mM HEPES-KOH (pH 7.5), 200 mM NaCl, 2 mM EDTA, 5% glycerol, 0.01% NP-40, 1 mM EDTA, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, 1 mM PMSF, and 1 mM benzamidine. Cells were lysed by sonication and the supernatant was clarified by centrifugation at 20,000 x g for 30 min. Clarified cell lysate was loaded onto SP-Sepharose column (GE Healthcare Lifesciences, Chicago, IL) equilibrated with lysis buffer. Bound proteins were eluted with a gradient of NaCl (200–700 mM) in lysis buffer. Peak fractions were analyzed by SDS-PAGE stained with Coomassie blue, and the fractions containing RFC complex were pooled, the salt concentration adjusted to 150 mM and loaded onto a Mono Q column. A gradient of NaCl (150–500 mM) in buffer containing 20 mM HEPES-KOH (pH 7.5), 5% (vol/vol) glycerol, 0.01% NP-40, and 1 mM DTT was applied to the column and the peak fractions containing RFC complex were pooled and loaded onto a Heparin column (GE Healthcare Lifesciences, Chicago, IL). A linear gradient of NaCl (300–1000 mM) in buffer containing 20 mM HEPES-KOH (pH 7.5), 5% (vol/vol) glycerol, 0.01% NP-40, 1 mM DTT was applied to the column and RFC-containing factions were concentrated to 500 μl and subsequently loaded onto a Superdex-200 column equilibrated with 20 mM HEPES-KOH (pH 8.0), 100 mM KCl, 5% (vol/vol) glycerol, 0.01% (vol/vol) NP-40, and 1 mM DTT. Near-homogenous RFC fractions were pooled, concentrated to 1 mg/ml, and stored at −80°C.
Purification of human POLδ complex.
The protein expression plasmids encoding the POLδ complex (pFBd-p50, 6xhis-p66 and pFBd-p125, p12) were kindly provided by Paul Modrich (Duke University). Human POLδ complex was purified as previously described (35) with minor modifications. Briefly, SF9 cells were grown to a density of 1×106 cells/ml. PolD1-POLD4 and POLD2-(6xhis-POLD3) baculoviruses were each used to infect SF9 cells at a multiplicity of infection of 20. 48 hours post infection at 27°C, cells were harvested and washed once with 1x PBS. The cell pellet was lysed in lysis buffer containing 20 mM HEPES-KOH (pH 7.5), 200 mM NaCl, 5% (vol/vol) glycerol, 0.2% (vol/vol) NP-40, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, and 1 mM PMSF. The cell lysate was clarified by centrifugation at 20,000 x g for 30 min and the supernatant loaded onto a column containing 2 ml of Ni2+-NTA resin (Qiagen, Hilden, Germany) twice to enrich POLδ complex on the resin. POLδ complex was eluted with 250 mM imidazole in 20 mM HEPES-KOH (pH 7.0), 100 mM NaCl, 5% glycerol, 0.01% NP-40, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, and 1 mM PMSF. The eluent was loaded onto a MonoQ column and fractionated with a linear NaCl gradient of 150–500 mM. Peak fractions containing POLδ complex were pooled and loaded onto a Heparin column and fractionated with a linear NaCl gradient of 400–1000 mM in buffer containing 20 mM HEPES-KOH (pH 7.0), 100 mM NaCl, 5% glycerol, 1 mM DTT, 0.01% NP-40, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, and 1 mM PMSF. The peak fractions containing POLδ was concentrated to 500 μl and subsequently loaded onto a Superdex-200 column equilibrated with 20 mM HEPES-KOH (pH 8.0), 100 mM KCl, 5% glycerol, 0.01% NP-40, 1 mM DTT. Near-homogenous POLδ fractions were pooled, concentrated to 1 mg/ml, and stored at −80°C.
Purification of human LIG1.
The bacterial protein expression plasmid encoding human LIG1 (pLW264) was generated by cloning the LIG1 coding sequence from cDNA (ORFeome Collaboration, GenBank ACC: KJ897129) (34) into the pET15 expression vector. LIG1 was expressed as a N-terminal 6x histidine tagged protein (6xhis-LIG1) in E. coli BL21 (DE3)RP cells (VWR, Radnor, PA). Cells were grown at 24°C until OD600 reached 0.6–0.8. Expression of 6xhis-LIG1 was induced by addition of IPTG to a final concentration of 0.1 mM and incubation at 16°C for 18 hours. Cells were harvested by centrifugation and the cell pellet washed once with 1x PBS. The cell pellet was resuspended in lysis buffer containing 20 mM HEPES-KOH (pH 8.0), 50 mM NaCl, 1% (vol/vol) NP-40, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, 1 mM PMSF, and 1 mM benzamidine. Cells were disrupted by sonication and cell lysate was clarified by centrifugation at 20,000 x g for 30 min. Supernatant was loaded onto a column containing 1 ml of Ni2+-NTA resin (Qiagen, Hilden, Germany). After washing the column with 20 ml of lysis buffer, 6xhis-LIG1 was eluted with elution buffer containing 20 mM HEPES-KOH (pH 8.0), 50 mM NaCl, 150 mM Imidazole, 1x cOmplete™ EDTA-free Protease Inhibitor Cocktail, 1 mM PMSF, and 1 mM benzamidine. The eluent was loaded on a Mono Q column (GE Healthcare Lifesciences, Chicago, IL), and a gradient of 0.1–1 M NaCl in buffer containing 20 mM HEPES-KOH (pH 8.0), 1 mM DTT, 1 mM PMSF, and 1 mM benzamidine was used to elute LIG1. Peak fractions were analyzed by SDS-PAGE stained with Coomassie blue, and the fractions containing 6xhis-LIG1 were pooled, concentrated to 500 μl and loaded onto a Superdex-200 size exclusion column (GE Healthcare Lifesciences, Chicago, IL) equilibrated in 20 mM HEPES-KOH, (pH 8.0), 50 mM KCl, 5% (vol/vol) glycerol, and 1 mM DTT. Peak fractions were analyzed by SDS-PAGE stained with Coomassie blue and the fractions containing near-homogeneous 6xhis-LIG1 were pooled and stored in aliquots of ~5 mg/ml at −80°C.
Purification of human FEN-1.
The bacterial protein expression plasmid encoding human FEN-1 (pLW290) was generated by cloning the FEN-1 coding sequence from cDNA (ORFeome Collaboration GenBank ACC: L37374.1) (34) into the pET15 expression vector. C-terminal 6x histidine tagged human FEN-1 (FEN-1–6xhis) protein was expressed in E. coli BL21(DE3) cells (Novagen, Madison, WI). The purification of FEN-1–6xhis was carried out as described for PCNA purification. Near-homogeneous FEN-1–6xhis fractions were pooled and stored in aliquots of ~5 mg/ml at −80°C.
Reconstitution of cccDNA formation by purified human proteins.
60 ng (29 fmol, ~2×1010 molecules) of RrcDNA or NeutrAvidin-RrcDNA complex were incubated with 1.5 μM PCNA, 35 nM RFC, 20 nM POLδ, 100 nM LIG1, and 20 nM FEN-1 in repair buffer containing 20 mM Tris-HCl (pH 7.6), 50 mM KCl, 0.1 mM each dNTP, 5 mM MgCl2, 1 mM reduced glutathione, 2.6 mM ATP, 26 mM phosphocreatine disodium (Sigma Aldrich, St. Louis, MO), and 6 μg/ml creatine phosphokinase (Sigma Aldrich, St. Louis, MO). This mixture was incubated at 37°C for 60 min, and the reaction was terminated by addition of SDS and EDTA to a final concentration of 0.5% (w/vol) and 25 mM, respectively. Purification of repair product was carried out as described above for cccDNA formation in yeast and human extracts.
HBV infection of HepG2-hNTCP cells in the presence aphidicolin.
HBV infection of HepG2 cells overexpressing hNTCP (3B10) was performed as previously described (20) with minor modifications. Tissue culture-derived HBV from HepG2.2.15 cells (sucrose ultracentrifugation purified, see preparation of HBV derived from cell culture section for details) was used for all experiments. 1.2×106 3B10 cells/well were plated in a 6-well plate, and after an overnight incubation, cells were treated for 12 hours with various concentrations of aphidicolin (Catalog number 102513, VWR, Radnor, PA) (0 μM, 1 μM, 10 μM, 200 μM, or 400 μM) in the presence of DMEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) DMSO (Sigma Aldrich, St. Louis, MO). Note that the aphidicolin used in Figure 6a–b were from MP Biomedicals (Catalog number 215988301). The cells were then challenged with HBV virus at a multiplicity of infection (MOI) of 4000 in the presence of aphidicolin and DMEM supplemented with 10% (vol/vol) FBS, 4% (vol/vol) PEG 8,000 (Sigma Aldrich, St. Louis, MO), and 1% (vol/vol) DMSO (Sigma Aldrich, St. Louis, MO). After 18 hours, the inoculum was aspirated and the cells washed three times with medium. The cells were subsequently maintained in aphidicolin and DMEM supplemented with 10% (vol/vol) FBS and 1% (vol/vol) DMSO. To ensure the efficacy of aphidicolin, the media was replaced every 12 hours with fresh aphidicolin-containing media until cells were harvested at day 2.5 post-infection.
HBV cccDNA isolation from HBV-infected HepG2-hNTCP cells.
HBV cccDNA was purified from HBV-infected HepG2-hNTCP cells by the Hirt extraction method as previously described (23,24). For the cccDNA in Fig. 6g, HBV-infected 3B10 cells in a 6-well plate were washed twice with 1x PBS and subsequently lysed in 1 ml of lysis buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 150 mM NaCl, and 1% (w/vol) SDS. The cell lysate was incubated for 10 min at 20°C and transferred to a clean Eppendorf tube. KCl was added to a final concentration of 500 mM and the lysate was incubated at 4°C overnight with gentle agitation. The lysate was then clarified by centrifugation at 20,000 x g at 4°C, and the supernatant was transferred to a clean Eppendorf tube and extracted three times with phenol and once with phenol-chloroform (1:1, vol/vol). The aqueous phase was collected and DNA precipitated and washed with ethanol. DNA was then dissolved in 20 μl TE solution containing 10 mM Tris-HCl and 1 mM EDTA. To further purify cccDNA, the DNA eluent was treated with ExoI, ExoIII, and T5 exonuclease, as previously described (22,23). Briefly, 10 μl DNA eluent was treated with 5U of ExoI and 25U of ExoIII in 1x Cutsmart buffer (NEB, Ipswich, MA) for 2 hours at 37°C. The reaction mix was extracted with phenol-chloroform and DNA precipitated by ethanol and dissolved in 20 μl TE solution. The eluent was subsequently treated with 5U of T5 exonuclease for 60 min and cccDNA was purified by phenol-chloroform extraction as described above. The purified cccDNA was dissolved in 20 μl TE solution. cccDNA in Fig. 6h was purified as previously described (24), and was not digested by exonucleases.
Determination of HBV viral copy number by quantitative PCR.
HBV DNA sample preparation and quantitative PCR procedures were performed as previously described (20) with minor modifications. Briefly, HBV DNA from samples purified by sucrose ultracentrifugation was extracted by the QIAamp MinElute Virus spin kit (Qiagen, Hilden, Germany) per the manufacturer’s instructions. Briefly, 50 μl HBV samples were digested with 25 μl of proteinase K in AL lysis buffer (Qiagen, Hilden, Germany) at 56 °C for 20 min. Following this step, 250 μl of 100% (vol/vol) ethanol were added and after a 5 min incubation, the suspension was applied to a Qiamp DNA mini kit column and centrifuged for 1 min at 6,000 x g. After washes with buffers AW1 and AW2 (contained in the kit), HBV DNA was eluted in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA). Serial dilutions of standards and HBV DNA isolated from purified HBV virions were used to determine the copy number by quantitative PCR. To amplify HBV DNA, the following primers and probes were used: CCGTCTGTGCCTTCTCATCTG (forward primer), AGTCCAAGAGTCCTCTTATGTAAGACCTT (reverse primer). Primers were used at a concentration of 400 nM in a 12 μl reaction containing 5 μl of HBV DNA or standard and 1x SYBR Green PCR master mix (Thermo Fisher, Waltham, MA). The following PCR program was run on a Step One Plus qPCR machine (Life Technologies, Carlsbad, CA): 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s.
HBV cccDNA quantification by real-time quantitative PCR.
HBV cccDNA quantification by quantitative PCR was performed as previously described (22,23) with minor modifications. Serial dilution of HBV cccDNA samples and HBV standards were performed in a dilution solution containing 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 10 ng/ml yeast tRNA, and 0.01% (vol/vol) Tween-20. 5 μl of DNA samples from HBV cccDNA or standard serial dilutions were mixed with cccDNA-specific primers [forward: GTCTGTGCCTTCTCATCTGC (nt 1553–1572, GenBank accession number U95551.1), Reverse: ACAAGAGATGATTAGGCAGAGG (nt 1830–1851, GenBank accession number U95551.1)] at a concentration of 400 nM and 1x SYBR green master mix (Thermo Fisher, Waltham, MA) in a 12 μl reaction. The following PCR program was run on a Step One Plus qPCR machine (Life Technologies, Carlsbad, CA): 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 60 s. Amplification specificity was confirmed by melting curve analysis or agarose gel electrophoresis.
HBV cccDNA detection and quantification by Southern blotting.
HBV cccDNA detection by Southern blotting was carried out as previously described (14,24) with minor modifications. For Fig. 6h, purified cccDNA were not treated with exonucleases. For Fig. 6g, purified HBV cccDNA treated with ExoI, ExoIII, and T5 exonuclease was digested with MfeI to linearize cccDNA to dsDNA, which could be more robustly detected by Southern blotting (14). cccDNA samples were resolved on a 1% (w/vol) agarose gel as described in cccDNA formation assay section. In-gel DNA was depurinated with 0.1 M HCl for 10 min and subsequently denatured with 0.5 M NaOH, 1.5 M NaCl, and then neutralized by 1 M Tris (pH 7.4), 1.5 M NaCl. DNA was then transferred onto a positively charged nylon membrane (Sigma Aldrich, St. Louis, MO) by 20x SSC buffer. DNA was cross-linked to the membrane by UV irradiation at 120,000 μJ/cm2 and incubated with pre-hybridization solution supplied in the digoxigenin (DIG) High Prime DNA Labeling and detection Starter Kit II (Sigma Aldrich, St. Louis, MO) for 1 hour at 42°C. The membrane was subsequently hybridized with alkaline-labile DIG-labeled HBV probes (558 bp, nt 2625–3182, GenBank accession number U95551.1) at 42°C for 14 hours. HBV DNA bands were visualized following manufacturer’s instructions. Briefly, the hybridized membrane was washed with washing buffer (0.1 M maleic acid (pH 7.5), 150 mM NaCl, 0.3% (vol/vol) Tween-20). The membrane was then blocked and subsequently incubated with alkaline phosphatase-conjugated DIG antibody at a 1:10,000 dilution. After a 1-hour incubation at room temperature, the membrane was washed with washing buffer for 15 min. The membrane was subsequently subjected to chemiluminescent detection by incubation with the substrate CSPD and the signal captured on autoradiography film (Thermo Fisher, Waltham, MA). The film was then scanned with an Epson scanner at 400 dpi resolution and the cccDNA band quantified with ImageJ.
Immunoblotting analysis and antibodies.
Protein samples were resolved by SDS-PAGE and transferred to a 0.2-μm nitrocellulose membrane (Biorad, Hercules, CA). Primary antibodies used were anti-flag (1:1000 dilution, clone M2, Sigma Aldrich, St. Louis, MO), anti-GAPDH (1:1000 dilution, sc-47724, lot#K2817, Santa Cruz Biotechnology, Dallas, TX), anti-Lamin A/C (1:1000 dilution, sc-7292, lot#C1918, Santa Cruz Biotechnology, Dallas, TX), anti-PCNA (for western blotting, 1:500 dilution, clone PC10, sc-56, lot# E2418, Santa Cruz Biotechnology, Dallas, TX), anti-PCNA (for immune-depletion, clone F2, sc-25280, lot #H1417, Santa Cruz Biotechnology, Dallas, TX) anti-POLD1 (1:1000 dilution, rabbit polyclonal, 15646–1-AP, Proteintech, Rosemont, IL), anti-FEN-1 (1:1500 dilution, rabbit polyclonal, A300–256A, Bethyl Laboratories, Montgomery, TX), anti-LIG1 (1:1000 dilution, clone 10H5, GTX70141, lot#809703999, GeneTex, Irvine, CA), and anti-RFC4 (1:1000 dilution, clone 1320, GTX70285, lot#14130, GeneTex, Irvine, CA). Secondary antibodies used were DyLight 800-conjugated goat anti-Mouse IgG (1:3000 dilution, Thermo Fisher Scientific, Waltham, MA) and DyLight 680-conjugated goat anti-Rabbit IgG (1:3000 dilution, Thermo Fisher Scientific, Waltham, MA). For quantification, membranes were scanned with an Odyssey CLx Imager (Li-COR Biosciences, Lincoln, NE). Quantification of blots and generation of figures was performed with ImageJ and Adobe Illustrator. Only un-saturated signals were used in quantification.
Lambda phosphatase treatment.
10 μl of yeast cytoplasmic extract were supplemented with 1 mM MnCl2 and treated with 200 U of lambda phosphatase (NEB, Ipswich, MA) at 30°C for 30 min. 1 μl of reaction was withdrawn and 10 μl 2x Laemmli buffer was added to stop the reaction and the solution was boiled at 95°C for 5 min before SDS-PAGE and western blotting analysis.
Data availability
All data that support the findings of this study are available in the manuscript and the Extended Data File. All unprocessed images of western blots, Southern blots, and stained agarose gels, raw numerical data are available in the source data file.
Extended Data
Extended Data Fig. 1. Characteristics of HBV rcDNA and effects of transfection of various rcDNA substrates to human hepatoma cells.
(a) Four types of structures on HBV rcDNA need to be repaired by host repair factors to form cccDNA. Red line, a RNA primer/flap; Pol, HBV polymerase. A model describing the steps of rcDNA repair is as following: Removal of HBV Polymerase adduct from rcDNA can be achieved by TDP2, unknown endonuclease and protease digestion, generating 3 types (type A–type C) of deproteinated rcDNA (dp-rcDNA). Dp-rc DNA needs to be further processed to remove DNA flap and RNA primer/flap, fill the gap, and ligate the nicks. HBV rcDNA, and type A–type C dp-rcDNA can be mimicked by indicated recombinant rcDNA substrates (see Fig.1 for details) to study cccDNA formation in our biochemical repair system. (b–d) Examination of HBeAg production after transfection with X-tremeGENE™ reagent of various HBV rcDNA substrates (described in (a) and Fig. 1a) into hNTCP HepG2 cells. (b) Schematics of transfection experiments. 5 ng of individual control and rcDNA substrates together with 15 ng of mCherry expressing plasmids were transfected to hNTCP HepG2 cells seeded in a 96-well plate. 72 hours post transfection, transfection efficiency of each rcDNA substrates was estimated by mCherry fluorescence (c, scale bars indicate 400 μm) and HBeAg production in the supernatant was detected by ELISA (n=3, independent experiments) (d). Note that addition of NeutrAvidin strongly inhibited transfection efficiency shown in (c, compare 1st and 2nd panel). Statistic comparisons between ‘no rcDNA’ and ‘virion rcDNA’ samples were made using student-t test (two-tailed), *p value is 8×10-5. Bar value indicates mean of three measurements, and error bars are s.d. All data shown are representatives of three independent experiments.
Extended Data Fig. 2. Yeast cell extract fully supports repair of rcDNA to form cccDNA.
(a) Repair of HBV virion-derived deproteinated rcDNA to cccDNA in yeast cell extract. Repair products are analyzed by Southern blotting (see methods for details). Un-treated recombinant rcDNA (RrcDNA) (lane 1), recombinant cccDNA (RcccDNA) (lane 2), and virion-derived rcDNA (lane 3) were used as controls. Lane 4, repair product following incubation of rcDNA with yeast cell extract (CE). L, linearized rcDNA. (b–c) Both minus and plus strands in RrcDNA are faithfully repaired by yeast cytoplasmic extract to form cccDNA. (b) DNA sequences of the minus strand of repaired cccDNA (Fig. 2a, lane 3) and recombinant cccDNA. The 10 nt flap region on the minus strand of rcDNA is indicated by a blue shaded box and dashed lines. (c) DNA sequences of the plus strand of repaired cccDNA (Fig. 2a, lane 3) and recombinant cccDNA. The ssDNA gap and RNA primer-containing region on the plus strand of rcDNA is indicated by a blue shaded box and dashed lines. All data shown are representatives of two independent experiments.
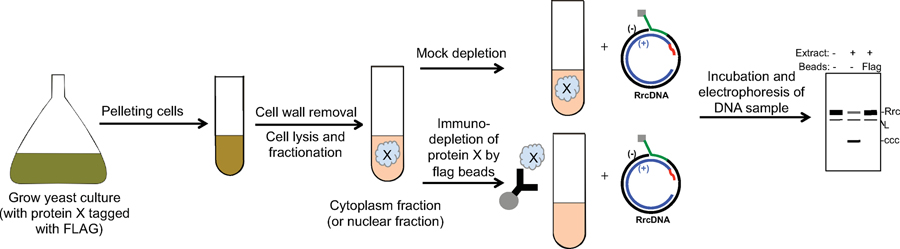
Extended Data Fig. 3. Schematic of a screening system using yeast cell extract to identify host factors essential for HBV rc- to cccDNA conversion.
Culture of yeast strains with a given protein ‘X’ tagged with Flag epitope are grown and pelleted. Cell pellets are treated with zymolyase to remove the cell wall and subsequently lysed. The cell lysate is separated into either cytoplasmic or nuclear fractions. Flag-tagged protein X is either mock depleted or depleted with anti-Flag antibody-conjugated beads. Both cell extracts are then incubated with recombinant rcDNA (RrcDNA), and the DNA from the repair reactions is subsequently extracted and separated on an agarose gel and visualized by EtBr staining or Southern blotting. A hypothetical gel pattern is shown – repair of RrcDNA will lead to the appearance of a cccDNA band that is expected to migrate faster. L stands for linearized RrcDNA due to nicking and shearing.
Extended Data Fig. 4. Tagging of yeast replication and repair factors with a C-terminal Flag epitope and effects of immuno-depletion of Pol1 and Pol2 on cccDNA formation.
(a) Successful fusion of a 3x Flag tag at the endogenous chromosomal loci of various repair genes was confirmed by immunoblots using anti-Flag M2 antibody. Arrowheads indicate 3xFlag fusion gene products. Ponceau stains indicate equal loading. Repeated four times independently. (b) Depletion of Pol1 (top) does not affect cccDNA formation (bottom). (c) Immuno-depletion of Pol2 to undetectable levels (top) does not affect cccDNA formation (bottom). * indicates degraded proteins; ID stands for immunodepletion. Repeated twice independently.
Extended Data Fig. 5. Repair of rcDNA to form cccDNA using human nuclear extract.
(a) Total cell, cytoplasmic, and nuclear extract from human HepG2 hepatoma cells were analyzed by western blotting to confirm separation of cytoplasmic and nuclear contents. Nuclear factor Lamin A/C, and cytoplasmic factor GAPDH were detected by specific antibodies. Cyto, cytoplasmic fraction; Nu, nuclear fraction. (b) HBV virion-derived deproteinated rcDNA is repaired with human nuclear extract (NE) and cccDNA formation is analyzed by Southern blotting. Un-treated recombinant rcDNA (RrcDNA) (lane 1), recombinant cccDNA (RcccDNA) (lane 2), and virion-derived rcDNA (lane 4) were used as controls. Lane 3, repair product following incubation of rcDNA with human nuclear extract (NE). (c) DNA sequences of the minus strand of repaired cccDNA (extracted from Fig. 3a, lane 8) and recombinant cccDNA. The 10 nt flap region on the minus strand of rcDNA is indicated by a blue shaded box and dashed lines. (d) DNA sequences of the plus strand of repaired cccDNA (extracted from Fig. 3a, lane 8) and recombinant cccDNA. The ssDNA gap and RNA primer-containing region on the plus strand of rcDNA is indicated by a blue shaded box and dashed lines. All data shown are representatives of two independent experiments.
Extended Data Fig. 6. Replenishment of fresh nuclear extract at cccDNA formation plateau only marginally improves cccDNA formation.
Related to Fig. 3b–c. (a) Schematics showing a time course for cccDNA formation reaction in human nuclear extracts. At 60 min, the reaction solution was split into two parts; one part was supplemented with a fresh aliquot of nuclear extract, while the other was not. (b) Time course assay showing the kinetics of cccDNA formation from both RrcDNA (lanes 1–7) and NeutrAvidin (NA)-RrcDNA complex (lanes 8–14) as described in (a). Efficiency of cccDNA formation was calculated as in Fig. 2c and indicated in row ‘% repaired’. (c) The efficiency of cccDNA formation from (b) is plotted against incubation time. All data shown are representatives of two independent experiments.
Extended Data Fig. 7. 80% depletion of PCNA and POLD1 and 90% depletion of FEN-1 do not affect cccDNA formation in human cytoplasmic extracts.
(a) Depletion of 80% of PCNA in human cytoplasmic extract (left) does not affect repair of RrcDNA (middle) and NA-RrcDNA (21). Experiments were carried out as in Fig. 3g–i. M, Mock depletion using mouse IgG. (b–c) same as (a), except that POLD1 and FEN-1 are examined, respectively. M, Mock depletion using rabbit IgG. (d) Antibodies against the RFC4 subunit of the RFC complex and LIG1 fail to achieve protein depletion in human cytoplasmic extracts. (a–d) a relative volume (rel. vol) of 100 corresponds to 0.5 μl extract. All data shown are representatives of two independent experiments.
Extended Data Fig. 8. Estimation of the concentration of repair factors in human cell extract.
(a) The concentrations of PCNA in cytoplasmic extract (CE, top) and nuclear extract (NE, bottom) are compared by immunoblot with purified 6xhistidine-tagged PCNA of the indicated amount. 1 μl of extracts were contained in CE and NE. Relative signal of PCNA bands was calculated by setting that of cytoplasmic extract to 1. (b–e) Same as (a), except that RFC4, LIG1, POLD1, and FEN-1 are examined. LIG1-p, phosphorylated form of LIG1. λPP, lambda phosphatase. Note that λPP treatment (lane 2) in (c) reduces the mobility of phosphorylated LIG1 (lane 1) to that of the non-phosphorylated form (lanes 2–4). All data shown are representatives of two independent experiments.
Extended Data Fig. 9. All five factors are necessary and sufficient for repair of virion derived rcDNA and a summary of human factors involved in cccDNA formation in this study.
(a) The repair of virion derived type C rcDNA requires all five factors as recombinant rcDNA substrates used in Fig. 4e. (b) The core components involved in DNA lagging strand synthesis – PCNA, RFC, POLδ, FEN-1 and LIG1 – constitute a minimal set of factors for cccDNA formation in this study. All data shown are representatives of two independent experiments.
Extended Data Fig. 10. Strains and plasmids used in this study.
Saccharomyces cerevisiae strains were isogenic to W1588–4C, a RAD5 derivative of W303 (MATa ade2–1 can1–100 ura3–1 his3–11,15 leu2–3,112 trp1–1 rad5–535) (27). Only one strain is listed for each genotype, but at least two independent isolates of each genotype were used in the experiments.
Supplementary Material
Acknowledgments
General: We are grateful to Xiaolan Zhao (MSKCC) for sharing WT yeast stains and yeast tagging plasmids; Paul Modrich (Duke University) for sharing plasmids for expression of the RFC and POLδ complexes; Bingbing Wan (MSKCC) for discussions on yeast tagging strategies; the Zakian and Shenk lab (Princeton University) for sharing equipment and reagents for the yeast work; Sarah Port and Sophie Travis (Hughson lab, Princeton University), Xin Gong (Yan lab, Princeton University), Michael Estrella (Korennykh lab, Princeton University), Raymundo Alfaro-Aco (Petry lab, Princeton University), and the Remus lab (MSKCC) for advice on protein purification and extract preparation; Benjamin Bratton and Joseph Sheehan (Gitai lab, Princeton University) for advice on P1 phage transduction. We thank Jenna Gaska and other members of the Ploss lab for providing critical feedback on this manuscript.
Funding: This work was supported in part by grants from the National Institutes of Health (R01 AI138797 to A.P), a Research Scholar Award from the American Cancer Society (RSG-15–048-01-MPC to A.P.), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (to A.P.), and funding from Princeton University. L.W. is a recipient of a postdoctoral fellowship award from the New Jersey Commission on Cancer Research (NJCCR, DFHS17PPC011).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Winer BY & Ploss A. Determinants of hepatitis B and delta virus host tropism. Curr Opin Virol 13, 109–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Yan H. et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1, e00049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GH & Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71, 663–670 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Gerlich WH & Robinson WS. Hepatitis B virus contains protein attached to the 5’ terminus of its complete DNA strand. Cell 21, 801–809 (1980). [DOI] [PubMed] [Google Scholar]
- 6.Lucifora J & Protzer U. Attacking hepatitis B virus cccDNA--The holy grail to hepatitis B cure. J Hepatol 64, S41–S48 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Koniger C. et al. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci U S A 111, E4244–4253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao W & Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol 81, 6164–6174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H. et al. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 81, 12472–12484 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J & Seeger C. Hepadnavirus Genome Replication and Persistence. Cold Spring Harb Perspect Med 5, a021386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Y. et al. DNA Polymerase kappa Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog 12, e1005893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long Q. et al. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog 13, e1006784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui X. et al. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS One 10, e0128401 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura K. et al. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog 14, e1007124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang L, Sheraz M, McGrane M, Chang J & Guo JT. DNA Polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog 15, e1007742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Zhao J, Yuan Q & Xia N. Detection of HBV Covalently Closed Circular DNA. Viruses 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molnar-Kimber KL, Summers J, Taylor JM & Mason WS. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J Virol 45, 165–172 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Z. et al. HBVcircle: A novel tool to investigate hepatitis B virus covalently closed circular DNA. J Hepatol 66, 1149–1157 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Waga S & Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem 67, 721–751 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Winer BY. et al. Long-term hepatitis B infection in a scalable hepatic co-culture system. Nat Commun 8, 125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright GE, Hubscher U, Khan NN, Focher F & Verri A. Inhibitor analysis of calf thymus DNA polymerases alpha, delta and epsilon. FEBS Lett 341, 128–130 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Qu B, Ni Y, Lempp FA, Vondran FWR & Urban S. T5 Exonuclease Hydrolysis of Hepatitis B Virus Replicative Intermediates Allows Reliable Quantification and Fast Drug Efficacy Testing of Covalently Closed Circular DNA by PCR. J Virol 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo J, Cui X, Gao L & Hu J. Identification of Intermediate in Hepatitis B Virus CCC DNA Formation and Sensitive and Selective CCC DNA Detection. J Virol (2017). [DOI] [PMC free article] [PubMed]
- 24.Cai D. et al. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol 1030, 151–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parlanti E, Fortini P, Macpherson P, Laval J & Dogliotti E. Base excision repair of adenine/8-oxoguanine mispairs by an aphidicolin-sensitive DNA polymerase in human cell extracts. Oncogene 21, 5204–5212 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Thomas DC, Roberts JD & Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem 266, 3744–3751 (1991). [PubMed] [Google Scholar]
- 27.Zhao X, Muller EG & Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2, 329–340 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Sells MA, Chen ML & Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84, 1005–1009 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kay MA, He CY & Chen ZY. A robust system for production of minicircle DNA vectors. Nat Biotechnol 28, 1287–1289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006 0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folco EG, Lei H, Hsu JL & Reed R. Small-scale nuclear extracts for functional assays of gene-expression machineries. J Vis Exp (2012). [DOI] [PMC free article] [PubMed]
- 32.Ni Y & Urban S. Hepatitis B Virus Infection of HepaRG Cells, HepaRG-hNTCP Cells, and Primary Human Hepatocytes. Methods Mol Biol 1540, 15–25 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Kaplan PM, Greenman RL, Gerin JL, Purcell RH & Robinson WS. DNA polymerase associated with human hepatitis B antigen. J Virol 12, 995–1005 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collaboration OR. The ORFeome Collaboration: a genome-scale human ORF-clone resource. Nat Methods 13, 191–192 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Kadyrov FA. et al. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci U S A 106, 8495–8500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available in the manuscript and the Extended Data File. All unprocessed images of western blots, Southern blots, and stained agarose gels, raw numerical data are available in the source data file.