Abstract
The nucleocapsid (N) protein is an important antigen for coronavirus, which participate in RNA package and virus particle release. In this study, we expressed the N protein of SARS-CoV-2 and characterized its biochemical properties. Static light scattering, size exclusive chromatography, and small-angle X-ray scattering (SAXS) showed that the purified N protein is largely a dimer in solution. CD spectra showed that it has a high percentage of disordered region at room temperature while it was best structured at 55 °C, suggesting its structural dynamics. Fluorescence polarization assay showed it has non-specific nucleic acid binding capability, which raised a concern in using it as a diagnostic marker. Immunoblot assays confirmed the presence of IgA, IgM and IgG antibodies against N antigen in COVID-19 infection patients’ sera, proving the importance of this antigen in host immunity and diagnostics.
Keywords: COVID-19, SARS-CoV-2, Nucleocapsid protein, Structure and function, SAXS, Antigenicity
Highlights
-
•
SARS-CoV-2 nucleocapsid protein is full of coils and highly disordered.
-
•
SARS-CoV-2 N protein forms a dimer by CTD-CTD interaction.
-
•
SARS-CoV-2 N protein can bind with non-specific nucleic acid with high affinity.
-
•
SARS-CoV-2 N protein can be a good antigen for serological test of COVID-19.
1. Introduction
In December 2019, a new type of coronavirus (SARS-CoV-2 or 2019-nCoV) causing a novel pneumonia now named COVID-19 broke out in Wuhan, China. The virus is rapidly spreading cross the world and caused a great impact on health and economy [1,2]. So far, as of April 16, 2020, there were 83,797 confirmed cases of COVID-19 coronavirus infection in China and over 1,954,724 cases globally in over 200 countries [3,4]. Studies on the virus are urgently needed for such severe situation.
The SARS-CoV-2 genome is composed of approximately 30,000 nucleotides, which encodes four structural proteins include spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein [5]. Among them, N protein is a highly immunogenic and abundantly expressed protein during infection [2,6]. Furthermore, N protein is frequently used in vaccine development and serological assays [7]. At present, there is few reports focus on SARS-CoV-2 N protein, and the updated understanding of SARS-CoV-2 N protein is in urgent need.
After infection, the N protein enters the host cell together with the viral RNA to facilitate its replication and process the virus particle assembly and release [8]. SARS-CoV N protein contains two distinct RNA-binding domains (the N-terminal domain [NTD] and the C-terminal domain [CTD]) linked by a poorly structured linkage region (LKR) containing a serine/arginine-rich (SR-rich) domain (SRD) [9,10]. Due to the positive amino acids, SARS-CoV N-NTD and N-CTD have been reported to bind with viral RNA genome [11,12]. LKR is ability to improve oligomerization [13,14]. However, the molecular properties of SARS-CoV-2 N protein remain to be excavated.
Serological diagnosis detected that the specific antibodies against the N protein in the serum of SARS patients have higher sensitivity and longer persistence than those of other structural proteins of SARS-CoV [15,16]. Moreover, anti-N antibodies have been detected with high specificity in the early stage of infection [17]. Thus, any information generated from the analysis of this protein, whether in vivo or in vitro, will improve our understanding of COVID-19 and help us to design better biological agents for the treatment or diagnostics of diseases.
At present work, we found SARS-CoV-2 N protein a dimer in solution by CTD-CTD interaction. Additionally, N protein can binding with non-specific dsDNA probably by its electrostatic interaction. Furthermore, we analyzed the immunogenicity of antibodies which specific for N protein. Our work reveals new information of the mechanism and characterization of N protein, which may provide a prospection for the vaccine or diagnostic kit development of N protein.
2. Results
2.1. SARS-CoV-2 N protein profile
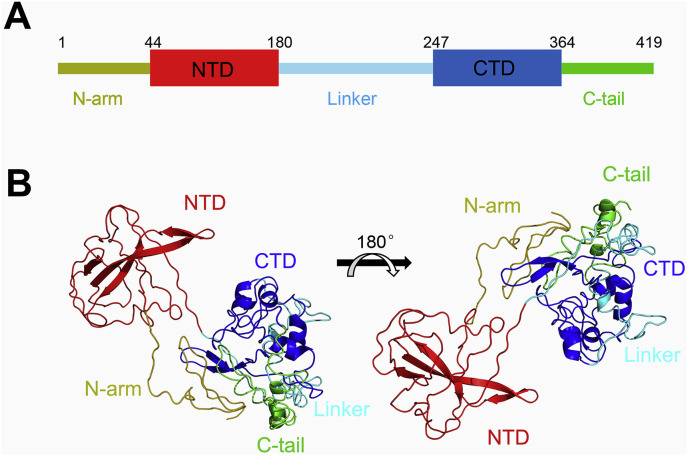
To gain insights into the structural and functional relationships of the SARS-CoV-2 N protein, we purified full-length protein with 419 amino acids (Fig. 1 A). It is predicted to have two well-folded domain, both of the NTD and CTD of SARS-CoV-2 N protein are rich in β-strands while CTD has some short helices (Fig. 1B).
Fig. 1.
Structural organization of SARS-CoV-2 N protein and sequence alignment.
(A) Domain structure of SARS-CoV-2 N protein. The domain boundaries were shown on the top and the different domains were labeled in different colors. (B) The predicted structure of SARS-CoV-2 N protein was presented. The NTD and CTD were highlighted in red and blue, respectively.
Sequence analysis showed that it has 90.52% identity to that of SARS-CoV, with the most conserved region in the two core domains and the linker (Fig. S1A). Molecular evolutionary analysis of the N proteins showed that SARS-CoV-2 belongs to lineage B betacoronavirus which lies in the same branch as SARS-CoV and two bat coronaviruses (Fig. S1B). They are well-separated with other coronaviruses, which is generally in agreement with the evolution tree of these coronaviruses [18].
2.2. The solution oligomerization state of SARS-CoV-2 N protein
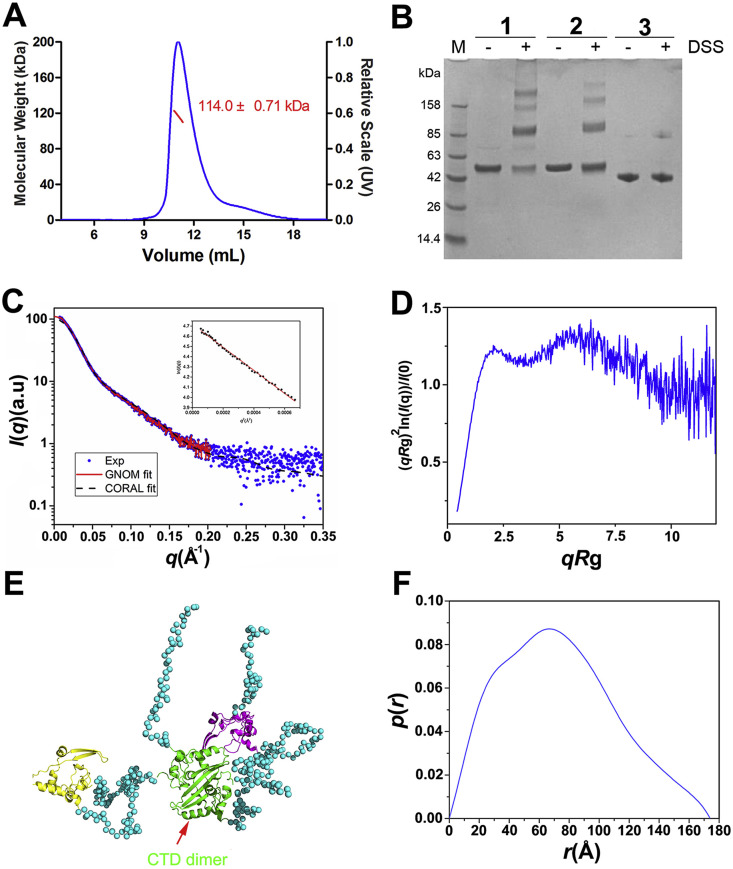
To access the oligomerization state of N-protein in solution, we used static light scattering (SLS) to determine the molecular weight. Our data showed that the SARS-CoV-2 N protein formed dimers according to a calculated molecular weight of the protein was 114 ± 0.7 kDa by SLS (Fig. 2 A). Furthermore, DSS cross-linking verified that the N protein, with a theoretical molecular weight of 49.5 kDa including an extra 20 residues at the N-terminus, could form dimers (Fig. 2B). A small portion of higher-order oligomers was also observed by cross-linking.
Fig. 2.
Oligomerization state and conformation analysis of the N protein.
(A) Static light scattering analysis of the oligomerization of the N protein. The molecular weight was calculated by Astra software and is shown in red.
(B) DSS cross-linking analysis of the oligomerization forms of the N-protein (1). The protein used for positive control was mCARD9-CARD with an MBP tag (52 kDa) which was reported to form dimers in solution (2) [33]. The MBP was used as a negative control (42 kDa) (3). (C) SAXS results for the protein. Scattering profile (points) and fitting with GNOM (solid lines). I, scattering intensity; q, scattering angle vector. Insert: the guinier region with fitting line of the scattering profile.
(D) Dimensionless Kratky plot showed that the protein was partially extended in solution. (E) A representative CORAL model in which the NTDs are shown in yellow and purple, respectively, and the CTD dimer is shown in green. The coiled coil regions are represented as dots. (F) Results from GNOM showing the pairwise distance distribution [P(r)] and the maximum distance. The radius of gyration is fitted to 59 Å, and r represents the pairwise distances.
2.3. The flexible linker is partially extended in solution
The confirmation of the full-length protein, we further studied by the SAXS technique to provide information on its shape. As shown in Fig. 2C, the radius of gyration of the molecule was 59 Å, much larger than that expected for a 99 kDa globular protein (Fig. 2F), and Kratky plot showed that the protein was partially extended in solution (Fig. 2D). This is in consistent with the model that the NTD and CTD do not interact, and the two NTDs in the dimer are likely to move freely in solution. A representative structure of NP45-365 based on CORAL simulations is shown in Fig. 2E. Due to the flexible nature of the linker region, this structure represents only a model of the conformational ensemble and does not represent a structure per se. However, the model captures features of the conformational ensemble and allows for the qualitative analysis of gross structural features. The most prominent feature of the model is that the flexible linker does not adopt a fully extended conformation, suggesting the existence of residual structures within the linker.
2.4. Circular dichroism (CD) spectroscopic analysis
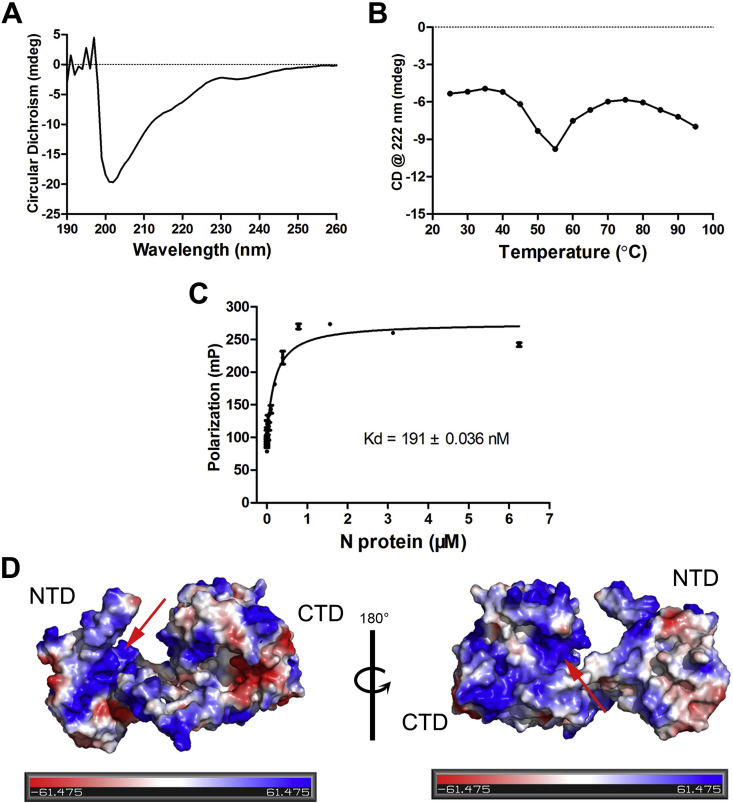
In order to characterize the conformational properties of the N protein, CD spectroscopy was used to analyze the secondary structures. The spectra shown in Fig. 3 A demonstrated that the N protein is mainly composed of coils, which consistent with the structural model in Fig. 1B and the SAXS results (Fig. 2E). Interestingly, the content of secondary structures increase with temperature and then started decreasing when it above 55 °C.
Fig. 3.
Conformational and functional analysis of the N protein
(A) CD spectrum analysis of the N protein (right) and thermal denaturation of the N protein monitored at Θ222 nm (left).
(B) Fluorescence polarization analysis of the N protein. The concentration of 5′-FAM double stranded 14mer DNA was 20 nM, and the apparent Kd value was 191 ± 0.036 nM. (C) The electrostatic surface of the N protein generated by PyMOL, where the negatively charged region are represented in red, neutral regions in white, and positively charged regions in blue.
2.5. The N protein is potent to bind non-specific nucleic acid with high affinity
In order to characterize the nuclei acid binding ability of SARS-CoV-2 N protein, we used fluorescence polarization to assess the binding affinity of the protein to a non-specific nucleic acid (a double stranded 14mer DNA probe with a fluorescence label). As shown in Fig. 3C, the N protein is potent to bind the dsDNA, the apparent Kd value is 191 ± 0.036 nM. Additionally, the electrostatic surface potential map generated with PyMOL (Fig. 3D) confirms SARS CoV-2 N protein is a highly basic protein. The surface of both NTD and CTD displayed highly positively charged regions which may facilitate binding to nucleic acids.
2.6. The N protein is an important viral antigen for SARS-CoV-2
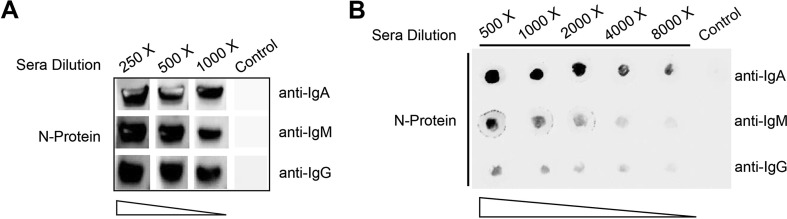
To pinpoint the possibility of the N protein as a diagnosis marker in COVID-19, we used Western Blotting and Dot Blotting to identify the antibodies which specifically bind with the N antigen. WB analysis (Fig. 4 A) and Dot blot analysis (Fig. 4B) showed the presence of IgG, IgA and IgM antibodies against the N protein were detected in the confirmed COVID-19 patients’ sera pool with different dilution. This result further confirmed that the N protein is a potent antigen for host immunity and for disease diagnosis.
Fig. 4.
Antigenicity of the N antigen
Western Blot (A) and Dot Blot (B) analysis of specific IgA, IgM, IgG antibodies against the N
-protein after incubated with different dilution of COVID-19 recovering patients’ serum pool using anti-human IgA-Fc/IgM-μ chain/IgG-Fc secondary antibodies, respectively.
3. Discussion
The nucleocapsid protein is an important structural protein for the coronaviruses. It is highly abundant in the viruses. Its function involves entering the host cell, binding to the viral RNA genome, and forms the ribonucleoprotein core. The SARS-CoV-2 N protein shares high homology with the SARS-CoV N protein, with a sequence identity of 90.52%.
Our structural characterization of recombinant full length N protein showed that it has high content of disordered region without bound nucleic acid (Fig. 1B/3A). Noticeably, the linker of SARS-CoV N protein is also highly disordered, as reported before [19,20]. This disordered region may facilitate the protein to transiently bind to different partners and maintain a correct conformation of the N protein [13,21,22].
According to SAXS modeling, the NTD seems to move freely in solution, and the flexible linker is partially extended in solution (Fig. 2E), while the CTD forms a dimer similar to other N proteins [23]. Static light scattering and DSS cross-linking were strongly corroborating the results (Fig. 2 A/B). Furthermore, N protein of SARS-CoV-2 is highly positively charged (Fig. 3C), which may facilitate the binding ability of non-specific nucleic acid (Fig. 3B). These results further confirmed the functional conformation of N protein gain the ability to bind nuclei acids.
Guo et al. confirmed that the IgG in SARS-CoV and SARS-CoV-2 infected patients’ sera can bind with the N antigen by WB and ELISA [24]. Another report showed that antibodies against SARS-CoV-2 N protein and RBD protein began to rise at the 10th day after COVID-19 symptoms onset [25]. Importantly, by using COVID-19 patients’ sera, we found the existence of IgG, IgA, and IgM antibodies against N antigen in recovering patients (Fig. 4).
Overall, our study increased the understanding of the SARS-CoV-2 nucleocapside protein and provided the basis for future vaccine and diagnostic kits development.
3.1. Experimental procedures
3.1.1. Patient serum samples
Serum samples were collected from recovering COVID-19 patients admitted to the First Affiliated Hospital of USTC between Jan 30 and Feb 23, 2020. All patients were confirmed to be infected with SARS-CoV-2 by use of real-time RT- PCR (rRT-PCR) on throat swab samples from the respiratory tract. Serum preparation as [26].
3.1.2. Molecular cloning, protein expression and purification
The coding sequence of the core N protein factor homology region (A1-A419) (NCBI accession code: ADI66791.1) was ligated into pET28a with a His∗6 on the N-terminus. The recombinant plasmid was transformed into BL21 (DE3) bacteria for protein overexpression. The lysis supernatant was purified by a 5 ml Hisprep™ IMAC column (GE Healthcare), and eluted protein was added with Ammonium sulfate to a final concentration of 0.5 M. The final protein was further purified with a 24 ml Superdex-200 gel filtration column. The UV–vis spectrum was acquired on the purified protein using a spectrophotometer (Jena).
3.1.3. Circular dichroism spectroscopic study
Circular dichroism (CD) spectra were acquired on a Chirascan Spectrometer (Applied Photophysics, Leatherhead, UK). The proteins were changed buffer into PBS. The procedure is as reported before [27].
3.1.4. Small-angle X-ray scattering (SAXS) and low-resolution model building
Purified full-length N protein was concentrated to 5 mg/ml using Amicon centrifugal concentrators (Millipore). To exclude concentration dependence, two different concentrations, 1 mg/ml and 5 mg/ml of purified N protein (corresponding to 21.7 μM, 101 μM, respectively) were prepared and measured. The procedure followed previous report [28]. The parameter is shown in Table S1.
3.1.5. Rigid body modeling using SAXS data
Modeling of the N protein was performed using three rigid bodies. The model of residues 46–171 was built by the 1.7 Å resolution crystal structure of the RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 (PDB: 6M3M.1.A) with SWISS-MODEL [29]. The model of the CTD dimer was built using the structure of SARS Coronavirus Nucleocapsid Protein (PDB: 2CJR.1.A) as a template.
SAXS-based rigid body modeling of complexes was performed by CORAL (COmplexes with RAndom Loops) [30]. CORAL fixed the CTD dimer, translated and rotated the atomic models of NTD domains. The NTER and CTER loops were randomly generated by a library of self-avoiding random loops. A simulated annealing protocol was employed to find the optimal positions and orientations of available high-resolution models of domains and the approximate conformations of the missing portions of the polypeptide chain(s).
3.1.6. Cross-linking
Disuccinimidyl suberate (DSS, Pierce) was used to cross-link closely spaced surface-exposed active amino groups of interacting proteins. The experimental protocol is according to previous report [31].
3.1.7. Immuno-blotting
For Western Blot, 0.5 μg per well of the N proteins were analyzed with SDS-PAGE and transferred to a PVDF membrane (Millipore); for Dot Blot, 0.1 μg per drop of the N protein were spotted on a nitrocellulose membrane (Pall). The protein-coupled membranes were blocked with defatted milk at room temperature for 1 h and then incubated with different dilutions of virus-free sera of COVID-19 patients overnight at 4 °C. On the next day, the membranes were washed with PBST (0.1%v/v Tween 20). After that, the membranes were incubated with a secondary antibody, anti-IgA (Boster biological technology), anti-IgM-μ (Boster biological technology), or anti-IgG-Fc (Sino biological), for 1 h. Last, the membranes were washed with PBST and detected with an ECL kit (abpbiotech) using a chemiluminescence apparatus (Bio-Rad).
3.1.8. Fluorescence polarization (FP)
The purified N protein and 5′-FAM fluorescently labeled dsDNA (5′-FAM-TCG TCG TTT TGT CG) were mixed together with the final concentration of 6.25 μM and 20 nM, respectively, in PBS with 15 mM MgCl2. Then, the mixture was serially diluted with PBS containing 15 mM MgCl2 and 20 nM 5′-FAM dsDNA to different protein concentrations. The procedure was reported before [32].
Author contributions
T.J. and H.H. provided the funding, designed the study, participated in data analysis and extensively reviewed the manuscript. W.Z. and H.M. designed the study, performed the experiments, analyzed the data and drafted the manuscript. Other authors participated in the experiments and reviewed the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
T.J. is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No.: XDB29030104), National Natural Science Foundation of China (Grant No.: 31870731 and U1732109), the Fundamental Research Funds for the Central Universities (Grant No.: WK2070000108) and Chinese Academy of Science Clinical Research Hospital (Hefei) (Grant No.: YD2070002017). H.H. is supported by the Hefei Comprehensive National Science Center. H.M. is supported by the New Medical Science Fund of USTC (Grant No.: WK2070000130).
We would like to thank the staff at BL19U2 beamline at Shanghai Synchrotron Radiation Facility (SSRF) for the assistance during SAXS data collection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2020.04.136.
Contributor Information
Hongliang He, Email: hhl725@ustc.edu.cn.
Tengchuan Jin, Email: jint@ustc.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data to this article:
figs1.
References
- 1.WHO Novel coronavirus – China. http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ accessed Jan 19, 2020.
- 2.Shang B., Wang X.Y., Yuan J.W., Vabret A., Wu X.D., Yang R.F., Tian L., Ji Y.Y., Deubel V., Sun B. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 2005;336:110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Coronavirus (COVID-19) https://covid19.who.int/
- 5.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S.J., Leng C.H., Lien S.P., Chi H.Y., Huang C.Y., Lin C.L., Lian W.C., Chen C.J., Hsieh S.L., Chong P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020:12. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayanan K., Chen C.-J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83:7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 11.Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang C.-k., Chen C.-M.M., Chiang M.-h., Hsu Y.-l., Huang T.-h. Transient oligomerization of the SARS-CoV N protein–implication for virus ribonucleoprotein packaging. PloS One. 2013;8 doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Y.-J., Goh P.-Y., Fielding B.C., Shen S., Chou C.-F., Fu J.-L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41:5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung D.T.M., Chi Hang T.F., Chun Hung M., Sheung Chan P.K., Cheung J.L.K., Niu H., Tam J.S.L., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. JID (J. Infect. Dis.) 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lee S.J., Hsiao H.H., Wu W.J., Chang W.L., Lin C.H., Huang T.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu I.M., Oldham M.L., Zhang J.Q., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between Corona- and Arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T., Cao J., Booth T.F., Plummer F.A., Tyler S., Li X. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H.B., Chen Q., Chen J., Chen K.X., Shen X., Jiang H.L. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–2628. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q., Xu S., Zhu H., Xu Y., Jin Q., Sharma L., Wang L., Wang J. Clinical Infectious Diseases : an Official Publication of the Infectious Diseases Society of America. 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horowitz B., Minor P., Morgenthaler J.J., Burnouf T., McIntosh R., Padilla A., Thorpe R., van Aken W.G. vol. 924. World Health Organization technical report series; 2004. pp. 1–232. (WHO Expert Committee on Biological Standardization). (backcover) [PubMed] [Google Scholar]
- 27.Zeng W., Ma H., Fan W., Yang Y., Zhang C., Arnaud Kombe Kombe J., Fan X., Zhang Y., Dong Z., Shen Z., Zhou Y., Yang M., Jin T. Structure determination of CAMP factor of Mobiluncus curtisii and insights into structural dynamics. Int. J. Biol. Macromol. 2020;150:1027–1036. doi: 10.1016/j.ijbiomac.2019.10.107. [DOI] [PubMed] [Google Scholar]
- 28.Jia X., Yao J., Gao Z., Liu G., Dong Y.H., Wang X., Zhang H. Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system. J. Biol. Chem. 2018;293:6812–6823. doi: 10.1074/jbc.RA118.002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:w296–w303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin X., Li W., Ma H., Zeng W., Peng C., Li Y., Liu M., Chen Q., Zhou R., Jin T. Crystal structure and activation mechanism of DR3 death domain. FEBS J. 2019;286:2593–2610. doi: 10.1111/febs.14834. [DOI] [PubMed] [Google Scholar]
- 32.Samad A., Li Y., Zhang C., Chen F., Zeng W., Fan X., Jin T. X-ray crystal structure of putative transcription regulator lmo2088 from Listeria monocytogenes. Biochem. Biophys. Res. Commun. 2019;520:434–440. doi: 10.1016/j.bbrc.2019.10.033. [DOI] [PubMed] [Google Scholar]
- 33.Holliday M.J., Witt A., Gama A.R., Walters B.T., Arthur C.P., Halfmann R., Rohou A., Dueber E.C., Fairbrother W.J. Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation. Nat. Commun. 2019;10:1–17. doi: 10.1038/s41467-019-10953-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.