Abstract
Substrates with high sulfate levels pose problems for biogas production as they allow sulfate reducing bacteria to compete with syntrophic and methanogenic members of the community. In addition, the end product of sulfate reduction, hydrogen sulfide, is toxic and corrosive. Here we show how sulfate addition affects physiological processes in a thermophilic methanogenic system by analyzing the carbon flow and the microbial community with quantitative PCR and amplicon sequencing of the 16s rRNA gene. A sulfate addition of 0.5 to 3 g/L caused a decline in methane production by 73–92%, while higher sulfate concentrations had no additional inhibitory effect. Generally, sulfate addition induced a shift in the composition of the microbial community towards a higher dominance of Firmicutes and decreasing abundances of Bacteroidetes and Euryarchaeota. The abundance of methanogens (e.g., Methanoculleus and Methanosarcina) was reduced, while sulfate reducing bacteria (especially Candidatus Desulforudis and Desulfotomaculum) increased significantly in presence of sulfate. The sulfate addition had a significant impact on the carbon flow within the system, shifting the end product from methane and carbon dioxide to acetate and carbon dioxide. Interestingly, methane production quickly resumed, when sulfate was no longer present in the system. Despite the strong impact of sulfate addition on the carbon flow and the microbial community structure during thermophilic biogas production, short-term process disturbances caused by unexpected introduction of sulfate may be overcome due to the high resilience of the engaged microorganisms.
Keywords: Sulfate reducing bacteria, Methanogens, Inhibition, Community composition, Hydrogen sulfide, Acetate
Introduction
The anaerobic digestion of organic waste of changing composition and quantity for biogas production involves the risk of introducing undesirable substances to the system, which endanger optimal process performance (Illmer and Gstraunthaler 2009; Wagner et al. 2014). One of these substances is sulfate (SO42−), which can be introduced in the reactor when digesting wastes from the food industry, particularly from the production of alcohol, yeast, citric acid and edible oils, and the paper industry (Colleran et al. 1995). The input of high levels of SO42− into biogas fermenters results in lower methane (CH4) production rates and the evolution of hydrogen sulfide (H2S), which causes odors and corrosion. In the presence of SO42− as a terminal electron acceptor, SO42− reducing bacteria (SRB) compete with syntrophic bacteria and methanogens for their common substrates (lactate, acetate, propionate, butyrate, and H2) and thereby produce cytotoxic H2S (Muyzer and Stams 2008). Data on inhibiting SO42− levels vary largely as the effect of SO42− in an anaerobic digestion process depends on various factors, such as the type of reactor, the operation temperature, the used substrates, the pH, and the native degrading community (Colleran and Pender 2002; Chen et al. 2008). Nevertheless, it is generally agreed that the ratio of available carbon to SO42− is more decisive than the SO42− concentration itself and that at a chemical oxygen demand (COD) to SO42− ratio of 10 or larger methanogenesis is not inhibited, while below this value both, successful and unsuccessful anaerobic digestion may occur (Hulshoff Pol et al. 1998).
SRB affect the anaerobic community at multiple degradation levels, although it is assumed that SRB cannot compete with fast-growing acidogenic, fermenting organisms (Chen et al. 2008). From a thermodynamic point of view, SRB should out-compete acetogenic and methanogenic organisms for hydrogen (H2), acetate, propionate, and butyrate (Stams et al. 2005). In reality, however, the dominance of SRB is, depending on the substrate, less distinct, as acetogens or methanogens may be advantageous concerning their pH and temperature optimum, sensitivity to H2S toxicity, initial abundance, or growth rate (Colleran and Pender 2002; Paulo et al. 2015; Chen et al. 2008). Therefore, experimental data on the competition of SRB with acetogenic and methanogenic organisms are often contradictory (Chen et al. 2008). Nevertheless, it was mostly found that SRB outcompete their opponents for H2 and propionate, while the outcome of the competition for acetate and butyrate is rather unclear (Colleran et al. 1995; Paulo et al. 2015; Chen et al. 2008). Under SO42− limiting conditions, SRB can grow as acetogens, meaning that they may be abundant in biogas reactors even after long periods of SO42− absence (Visser et al. 1993). The toxicity of H2S is dependent on the pH, as unionized H2S can diffuse through the cell membrane and cause damage to proteins, coenzymes and can interfere with the assimilatory sulfur metabolism (Chen et al. 2008). The susceptibility to sulfide inhibition differs between the trophic groups and can be sorted as follows: hydrogenotrophic methanogens < hydrogenotrophic SRB < propionate oxidizing SRB < acetotrophic methanogens < syntrophic propionate oxidizing bacteria < acetate oxidizing SRB (Maillacheruvu and Parkin 1996). The inhibitory levels for dissolved sulfides in the literature range from 100 to 800 mg/L (Chen et al. 2008).
Most research on SO42− in anaerobic digestion is descriptively investigating real-life substrates at mesophilic conditions. The present study combines a defined substrate with an undefined mixed methanogenic community at thermophilic conditions to investigate competition-related and inhibitory effects of SO42− addition to a reproducible system. In a first step, we examined the effects of various SO42− levels on the applied methanogenic system regarding CH4 production, SO42− reduction, microbial community composition, and abundance of the relevant physiological groups. In a second step, we focused on SO42− induced shifts in carbon flow over a period of 4 weeks of anaerobic digestion.
Material and methods
Experimental design
The investigation was composed of two main experiments and a viability test. The aim of the first experiment was to determine the inhibition threshold of SO42− on the applied methanogenic system. The tested SO42− concentrations were 0, 0.5, 1.5, 3, and 5 g SO42−/L. The variants were incubated for 4 weeks at 50 °C and evaluated concerning CH4, carbon dioxide (CO2) and H2 production, reduction of SO42−, pH, production of H2S, abundance of methanogens and SRB, as well as shifts in the microbial community composition (see below). In a second experiment, the variants supplemented with 0 and 3 g SO42−/L were repeated and analyzed more comprehensively regarding changes in the carbon flow. To examine if methanogens and SRB in the SO42− containing samples were still alive on day 28, a viability test was performed. For this purpose, 0.1 mL of a parallel from day 28 containing 3 g SO42−/L was used to inoculate fresh media with 0 or 3 g SO42−/L in triplicate and incubated as described above experiment. Increasing CH4 and H2S concentrations were used as viability proof for methanogens and SRB, respectively.
Medium and inoculum
The anaerobic medium contained per liter: 1 g NaCl, 0.4 g MgCl2 × 6 H2O, 0.5 g KCl, 0.2 g KH2PO4, 0.15 g CaCl × 2H2O, 0.5 g l-cysteine, 1 mL resazurin solution (1.15% w/v), 1 mL trace element solution (Wagner et al. 2019), 10 g carboxymethylcellulose, 2 g yeast extract, 3 g peptone from casein, 8.4 g NaHCO3, and the intended amount of Na2SO42− (0, 0.5, 1.5, 3, or 5 g SO42−/L). The pH was adjusted to 7.75 and the medium autoclaved. Subsequently, 1 mL vitamin solution and 2 mL reducing agent (0.24 g NaS2/L final concentration) were added aseptically; the medium was aliquoted at 50 mL in 120-mL serum flasks and flushed with N2/CO2 (70:30) to ensure anaerobic conditions. The flasks were inoculated with 5 mL 1:5 diluted fermenter sludge from a thermophilic plug flow fermenter treating organic waste in Roppen, Tirol (Austria), as described elsewhere (Wagner et al. 2014).
Biochemical analysis
To calculate the total biogas production the overpressure was measured with a digital precision manometer (GDH 200-13, GREISINGER electronic, Germany) and normalized to ambient pressure (data from Zentralanstalt für Meterologie und Geodynamik, Austria). The gas composition was analyzed on a Shimadzu GC2010, as described in Wagner et al. (2011). Gas samples were diluted with N2 prior the measurement to protect the column of the gas chromatograph (GC) from high H2S concentrations. CO2 and H2 concentrations were quantified via thermal conductivity detector, while CH4 concentrations were measured with a flame ionization detector. Volatile fatty acids (VFAs) were determined in undiluted liquid samples following the procedure in (Wagner et al. 2017). The pH was measured with a glass electrode. For dissolved organic carbon (DOC) contents, liquid samples were diluted, filtered (filter papers 615 1/4, Macherey-Nagel, Germany), and measured with a TOC/NPOC analyzer (Shimadzu, Japan) according to manufacturer’s protocol. SO42− was measured via ion chromatography (Metrohm, Herisau, Switzerland) by using a Metrosep A-Supp 5-250 column at 35 °C and a 753 Suppressor Module (Metrohm, Herisau, Switzerland). The solvent, containing 3.2 mM Na2CO3, and 1 mM NaHCO3 was used at a flow rate of 0.7 mL/min. The COD was measured with a Nanocolor® COD 1500 test (Macherey-Nagel, Germany) according to the user guide.
Carbon flow diagrams
To create the carbon flow diagrams, all measurable carbon fractions were converted into millimoles, considering temperature and pressure. CO2 and CH4 values represent the cumulative gas amounts including day 0. Dissolved CH4 and CO2 were not included in the diagrams, because they could not be determined analytically or mathematically, and due to their relatively low solubility at 50 °C (Diamond and Akinfiev 2003). Acetate, propionate, and butyrate amounts quantified via high-performance liquid chromatography were subtracted from the DOC values. Diagrams exclude inorganic carbon and microbial biomass.
Gibbs free energy calculations
To assess the thermodynamic properties of the common reactions under the prevailing conditions, biochemical data from day 14 were used together with the standard free Gibbs enthalpy (ΔG0) from (Amend and Shock 2001). H2S concentrations were estimated on basis of SO42− decrease, and the H2 partial pressure was assumed to be 0.002 mbar, which is the substrate threshold of SRB (Lovley et al. 1982).
DNA extraction and quantification
For molecular analyses, 1 mL of culture fluid was centrifuged at 11,000×g for 10 min. Subsequently, 0.8 mL of supernatant was removed; the pellet was resuspended in the remaining fluid and used for DNA extraction. DNA extraction was done with a NucleoSpin® Soil kit (Macherey-Nagel, Germany) according to the manufacturer’s protocol. DNA was quantified using a Quant-iT™ PicoGreen™ dsDNA Assay Kit (Invitrogen, USA) and a multimode fluorometer Zenyth 3100 (Anthos, Austria).
qPCR
The quantitative PCR (qPCR) to quantify methanogens and SRB targeted the functional genes methyl coenzyme M reductase α-subunit (mcrA) and dissimilatory sulfite reductase α-subunit (dsrA), respectively. The primers for the mcrA gene were mlas and mcrA-rev (Steinberg and Regan 2008) and as a standard served Methanosarcina thermophila (DSM 1825, DSMZ-German Collection of Microorganisms and Cell Cultures, Germany). The qPCR mix for mcrA contained per 20 μL 5.28 μL PCR grade water, 10 μL SensiFAST™ Probe No-ROX (Bioline, UK), 0.40 μL MgCl2 (50 mM), 0.38 μL of each primer (10 μM), 0.8 μL Enhancer (5×), and 2 μL template (diluted to 0.9–1.8 ng/μL). The primers used for SRB were dsrA_290F and dsrA_660R, and Desulfovibrio vulgaris (DSM 644, DSMZ, Germany) was used for the standards (Pereyra et al. 2010). The qPCR mix for dsrA contained per 20 μL 6.4 μL PCR grade water, 10 μL SensiFAST™ Probe No-ROX (Bioline, UK), 0.4 μL of each primer (10 μM), 0.8 μL Enhancer (5×), and 2 μL template (diluted to 0.9–1.8 ng/μL). The reactions were executed in a Rotor-Gene Q (Qiagen Bioinformatics, Germany) with an initial denaturation at 95 °C for 10 min (mcrA) or 3 min (dsrA), 45 (mcrA) or 50 (dsrA) cycles of 30 s (mcrA) or 40 s (dsrA) at 95 °C, 30 s at 66 °C (mcrA) or 60 °C (dsrA), and 30 s at 72 °C.
Amplicon sequencing and data processing
Sequencing targeted the V4 region of the 16s rRNA gene using the primer pair 515f/806r (Apprill et al. 2015; Parada et al. 2016). To validate the performance of the entire sequencing process, a mock community was included in all steps beginning from DNA extraction until final data processing. The mock consisted of a ZymoBIOMICS Microbial Community Standard (Zymo Research, USA) mixed with a pure culture of Methanosarcina thermophila (DSM 1825, DSMZ, Germany) to also include archaeal DNA. In the first PCR step, the primers 515f/806r were used to attach an adapter sequence to the PCR fragment, conducting 30 cycles of 45 s at 95 °C, 45 s at 57 °C, and 60 s at 72 °C in a Flexcycler (analytikjena, Germany). The PCR mix contained per 25 μL reaction volume 12.5 μL Red Taq DNA Polymerase 2× Mastermix (VWR, USA), 0.8 μL 50 mM MgCl2, 0.5 μL of each primer, 1 μL enhancer (5×), and 1 μL template (diluted to 2.5 ng dsDNA/μL). In a second PCR step, individual barcodes for each sample were attached to the adapter sequence, running 7 cycles of 45 s at 95 °C, 45 s at 56 °C, and 90 s at 72 °C. Subsequently, the library was prepared with equal DNA amounts per sample to reach a final DNA concentration of 15 ng/μL. The library was cleaned up using a Hi Yield® Gel/PCR DNA Fragment Extraction Kit (Germany) and sent to Microsynth AG, Switzerland for sequencing (Illumina MiSeq 2 × 250 bp paired-end read).
Data analysis and statistics
Sequencing data were processed with the software platform mothur v.1.39.5, 64 Bit executable (Schloss et al. 2009) using the SILVA database release 132 ribosomal SSU (release date 12.12.17) as a reference database (Quast et al. 2013; Yilmaz et al. 2014). Quality filtering of trimmed reads included screening for and removal of contigs longer than 300 bp, containing any ambiguities or more than 7 homopolymers. Chimera were identified and removed with VSEARCH 2.3.4 (Rognes et al. 2016). Unique sequences with identical taxonomic classification were assigned to operational taxonomic units (OTUs). To ensure equal sample sizes, all samples were subsampled to the smallest read number (41326). Prokaryotic 16s rRNA read numbers are treated as relative abundance data throughout the present study. Furthermore, OTUs representing in average less than 0.1% were summarized as “rare OTUs” and all abundance data were Box-Cox transformed before statistical analysis. Statistics and graphs were done in PAST 3 (Hammer et al. 2001) and STATISTICA 12 (StatSoft®). The analyses included non-metric multidimensional scaling using Euclidean distances as well as multivariate and univariate Permanova (9999 permutations, Euclidean distances); significances of multiple tests were corrected according to Bonferroni. All taxa of the mock community were recovered at genus level and sequencing depth was sufficient according to rarefraction. Quality filtered sequencing data has been deposited in the NCBI database as BioProject (ID: PRJNA540929).
Data analysis of physiochemical data was performed using STATISTICA 12 (StatSoft®). All experiments were deducted in three replicates. Normality of the data was tested with Shapiro-Wilk test. Significant differences were defined by one-way ANOVA, the alpha level used throughout was 0.05, and homogenous groups were calculated using Bonferroni correction.
Results
Addition of various SO42− concentrations
The COD in the inoculated medium at day 0 was 15.05 g/L, and the pH value at day 28 was 7.5 ± 0.1 over all samples. As expected, increasing initial SO42− concentrations of up to 3 g/L resulted in increasing inhibition of CH4 production, while concentrations above 3 g/L had no additional inhibitory effect during 4 weeks of incubation (Fig. 1). Cumulative CO2 production was slightly, but significantly, decreased in the two high-SO42− samples compared with the control at day 28 (0 g SO42−/L: 6.17 ± 0.12 mmol; 3 g SO42−/L: 4.86 ± 0.21 mmol; 5 g SO42−/L: 4.59 ± 0.05 mmol), whereas H2 could not be detected at any sampling time (detection limit 0.05%). SO42− levels decreased throughout the incubation (Fig. 2), leading to final H2S concentrations in the gas phase of 0.10% ± 0.06 (0 g/L), 1.83% ± 0.17 (0.5 g/L), 3.00% ± 0.29 (1.5 g/L), 4.67% ± 0.60 (3 g/L), and 5.83% ± 0.44 (5 g/L). The viability test of methanogens and SRB, in which microorganisms from a 3 g SO42−/L sample at day 28 were re-cultivated, was positive for both groups as 10.4% ± 0.6 CH4 and 4.2% ± 0.6 H2S could be detected after 28 days of re-incubation in fresh SO42−-free medium and fresh SO42−-containing medium, respectively.
Fig. 1.
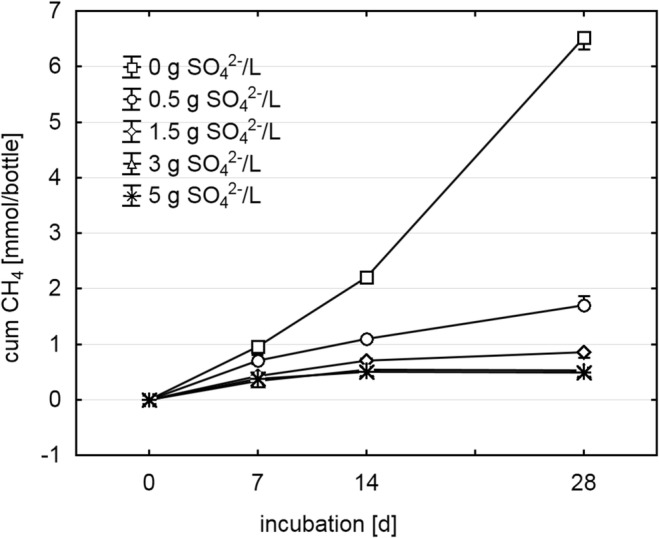
Cumulative CH4 production during 28 days of anaerobic digestion at various initial SO42− concentrations (n = 3, mean ± standard deviation)
Fig. 2.
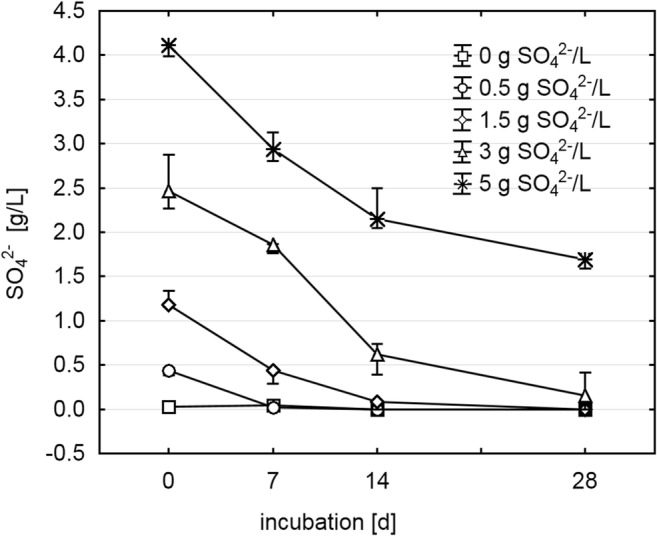
Measured SO42−concentration during the anaerobic digestion at various initial SO42− concentrations (n = 3, mean ± standard deviation)
Microbial community
Illumina sequencing yielded more than 40,000 reads per sample after quality filtering and subsampling to a common read size. The microbial community of the inoculum was mainly composed by Firmicutes (74%), Bacteroidetes (8%), Thermotogae (6%), Haloanaerobiaeota (5%), and Synergistes (4%). At genus level, the composition of the inoculum clearly differed from all samples at the end of the experiment, with the two clostridial genera DTU014 (20%) and MBA03 (12%) being most abundant, followed by Lentimicrobiaceae (8%), Defluviitoga (6%), Syntrophaceticus (6%), Halocella (5%), and Tepidimicrobium (5%). Methanogens represented only 0.3%, while SO42− reducing genera were rare (< 0.1%).
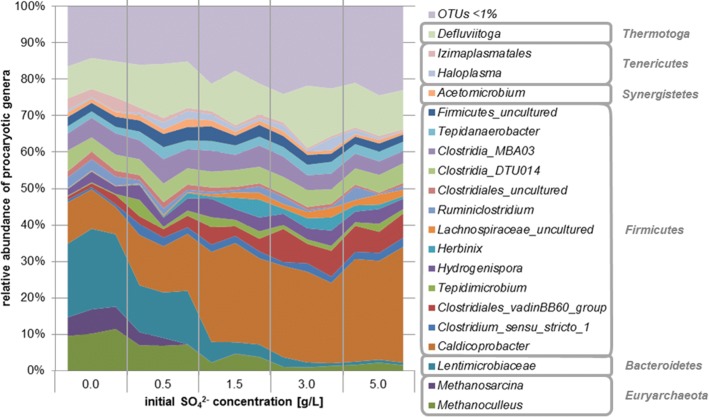
After 28 days of anaerobic digestion, the microbial community was dominated by members of the phylum Firmicutes (45–84%), followed by Thermotogae (7–17%), Bacteroidetes (1–23%), and Euryarchaeota (1–18%) (Fig. 3). Less abundant phyla included Atribacteria, Chloroflexi, Synergistes, Haloanaerobiaeota, and Tenericutes. According to multivariate permanova analysis, the structure of the microbial consortia was significantly influenced by the initial SO42− concentration at all taxonomic levels. nmMDS analysis revealed that on genus level, the communities of each SO42− variant formed distinctive clusters mainly separated along coordinate 1, which showed a high correlation with the decrease in SO42− concentration in the medium during 4 weeks of anaerobic incubation (data not shown).
Fig. 3.
Composition of the microbial community at genus level at varying initial SO42− concentrations after 28 days of anaerobic incubation. Individual samples shown; OTUs comprising < 1% were summarized
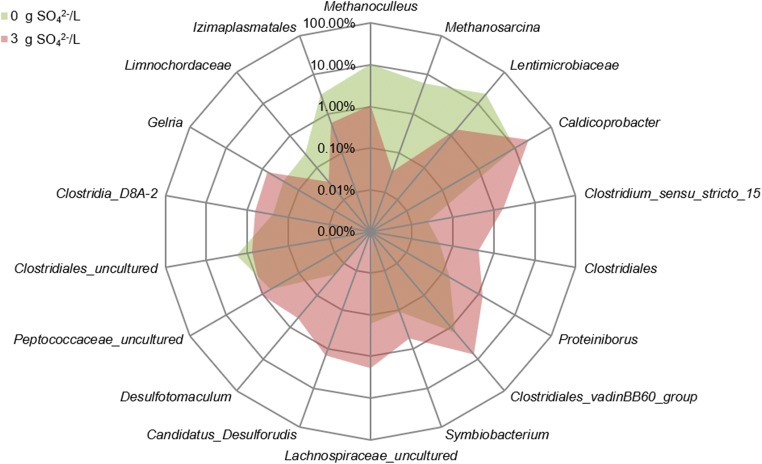
SO42− addition clearly promoted Firmicutes at the expense of Bacteroidetes and Euryarchaeota, whereas Thermotogae seemed to be unaffected by varying SO42− levels. At genus level, 18 of 60 OTUs with relative abundances > 0.1% showed significant differences between the SO42− variants (univariate Permanova, Bonferroni corrected; Fig. 4). The genera Caldicoprobacter, unclassified Clostridiales_vadinBB60_group, Lachnospiraceae_uncultured, Candidatus_Desulforudis, Proteiniborus, Clostridium_sensu_stricto_15, Peptococcaceae_uncultured, Gelria, unclassified Clostridia_D8A-2, Symbiobacterium, Desulfotomaculum, and unclassified Clostridiales (all Firmicutes) increased significantly with the addition of SO42−, while unclassified Lentimicrobiaceae (Bacteroidetes), Methanoculleus (Euryarchaeota), Methanosarcina (Euryarchaeota), unclassified Izimaplasmatales (Tenericutes), unclassified Clostridiales_uncultured (Firmicutes), and unclassified Limnochordaceae (Firmicutes) were negatively influenced. The most abundant archaeal genera were Methanoculleus (1–11%) and Methanosarcina (0–7%), with the latter being completely inhibited at concentrations higher than 0.5 g SO42−/L (Fig. 3). The most abundant bacteria were Caldicoprobacter (8–32%), Defuviitoga (7–17%), unclassified Lentimicrobiaceae (1–22%), unclassified Clostridia_MBA03 (3–6%), and unclassified Clostridia_DTU014 (4–6%), while genera known to reduce SO42− were found to a considerably lesser extent, comprising Candidatus Desulforudis (0–2%) and Desulfotomaculum (0–1%) (Fig. 3).
Fig. 4.
Radar chart of relative abundances [%] of genera significantly (univariate permanova, Bonferroni corrected) affected by the initial SO42− concentration. Means of triplicates
According to qPCR analysis, the mcrA copy number, a functional gene for methanogenesis, was up to one magnitude lower in samples containing 3 or 5 g SO42−/L than in the controls at day 28 (Fig. 5). By contrast, the dsrA copy numbers, used to quantify SRB, increased more than 3 magnitudes with rising initial SO42− concentration (Fig. 5). As also seen in CH4 production, there was no significant difference in both functional groups between samples amended with 3 or 5 g SO42−/L. The 1:5 diluted inoculum contained 106.3 mcrA copies/mL (corresponds to 105.3 copies/mL batch), while dsrA copies were below detection limit of 103 copies/mL).
Fig. 5.
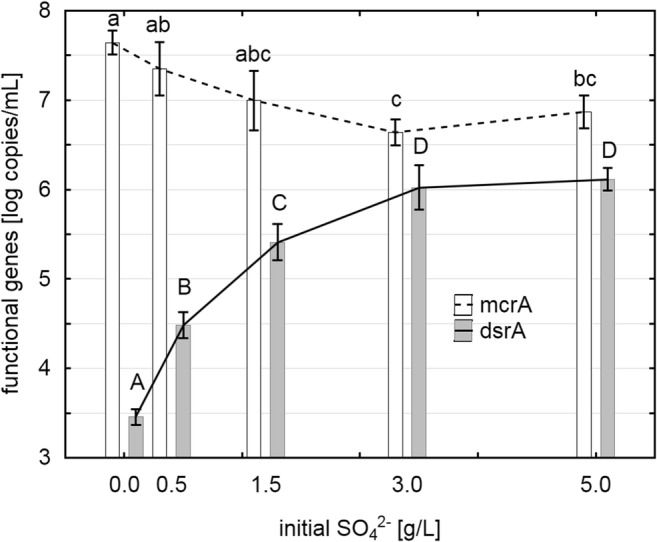
Abundance of functional genes for methanogenesis (mcrA: methyl coenzyme M reductase α-subunit) and SO42− reduction (dsrA: dissimilatory sulfite reductase α-subunit) at varying initial SO42− concentration after 28 days of incubation (mean ± standard deviation; letters show homogenous groups of ANOVA)
Carbon flow
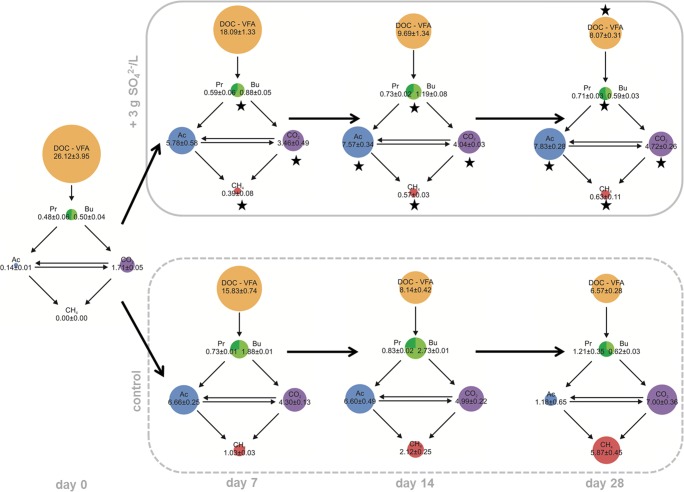
The carbon flow through the mixed microbial community system was calculated for controls and samples containing 3 g SO42−/L (Fig. 6). During the first 2 weeks, concentrations of DOC decreased in both samples with and without SO42−, leading to an accumulation of mainly acetate and CO2 (Fig. 6). CH4 production lagged behind the DOC mineralization and was significantly higher in control samples. Contrary, propionate and butyrate levels were significantly lower in SO42− containing samples at day 14. In the second half of the incubation, the accumulated acetate in control samples was converted to CH4 and CO2, whereas it prevailed in the in SO42− containing samples. At day 28, dissolved non-VFA carbon concentrations (DOC-VFA) and acetate levels were significantly higher, whereas propionate, butyrate, CH4, and CO2 concentrations were significantly lower in the SO42−-containing samples compared with the control.
Fig. 6.
Carbon flow in samples with and without (control) 3 g SO42−/L during 28 days of anaerobic digestion (DOC dissolved organic carbon, VFA volatile fatty acids, Pr propionate, Bu butyrate, Ac acetate). The areas of the colored circles correspond to the amount of carbon in the specific fraction. Values are given in millimoles of carbon (mean ± standard deviation). Stars depict significant differences in the specific carbon fraction between the SO42−-samples and the controls at the same day
Discussion
The addition of SO42− to the system affected the anaerobic digestion process on multiple levels, including gas production, VFA concentrations, the microbial community structure, and the absolute abundance of SRB and methanogens. The threshold for maximal inhibition of methanogenesis was 3 g SO42−/L, which corresponds to a COD/SO42− ratio of 5. Data on inhibitory SO42− levels at thermophilic conditions are less available than at mesophilic conditions, but existing values are in a similar range as in our study. Siles et al. (2010) observed a total inhibition of CH4 production at 1.8 g SO42−/L when fermenting glucose (6 COD g/L), McFarland and Jewell (McFarland and Jewell 1990) found that CH4 production decreased to 30% of the maximum at 0.8 g SO42−/L when fermenting sucrose (10 g COD/L), and the CH4 content in biogas from the digestion of sugar beet molasses (12 g COD/L) was reduced by half at a SO42− concentration of 3 g/L (Colleran and Pender 2002).
In general, the inhibitory effect of SO42− addition could be caused by both toxic and competitive effects. The competition for carbon sources and electron donors between microorganisms is determined by Gibbs free energy, substrate affinities, cell numbers and growth rates, and the availability of electron acceptors (Lovley et al. 1982; Stams et al. 2005; Elferink et al. 1998; Visser et al. 1993). In the present investigation, SO42− was available in excess, at least in the samples containing 3 and 5 g SO42−/L, in which it was not depleted until the end of the incubation. Initial abundances favored methanogens over SRB, as the inoculum derived from a well-performing biogas reactor with low SO42− levels, which was reflected by dsrA copy numbers below detection limit and a low relative abundance of SRB found during NGS analysis.
In the early phase of incubation, the main inhibitory mechanism of SO42− addition was probably the successful competition of SRB for H2, explaining the H2 levels below detection limit. H2 consumption combined with SO42− reduction yields more free energy than hydrogenotrophic methanogenesis, and SRB have a lower threshold for H2 uptake than methanogens, being 0.002 and 0.011 mbar, respectively (Lovley et al. 1982). Therefore, SRB outcompeted methanogens by decreasing the H2 partial pressure below the level necessary for successful methanogenesis, making thermodynamics irrelevant for the outcome of the competition as long as SO42− was not limited (Lovley et al. 1982). Subsequently, during SO42− reduction, toxic levels of H2S were reached (up to 6%), which enhanced the inhibition of CH4 production until the end of the incubation. In this context, the pH in the present study was 7.5 ± 0.1 at day 28, meaning that approximately 20% of the total dissolved sulfide should have been present as free H2S (Koster et al. 1986). This two-step inhibition mechanism was also suggested before (Karhadkar et al. 1987; McFarland and Jewell 1990).
The composition of the microbial community on phylum level was similar to results of other investigations of thermophilic methanogenic microbiomes. Considering all samples from day 28 regardless of the SO42− concentration, Firmicutes was the dominant prokaryotic taxon (45–84%), with comparable relative abundances also found by (De Vrieze et al. 2018; Liang et al. 2018; Lin et al. 2017; Maus et al. 2016; Tuan et al. 2014), ranging from 37 to 96%. Also, Thermotogae, Bacteroidetes, Tenericutes, Synergistes, and Chloroflexi were found, which are common members of a thermophilic CH4 producing community (De Vrieze et al. 2018; Liang et al. 2018; Lin et al. 2017; Maus et al. 2016; Tuan et al. 2014), with Bacteroidetes being exceptional abundant in our control samples. Generally, microorganisms can be affected by increased SO42− concentrations on three levels: (i) they are able to directly use SO42− as terminal electron acceptor, (ii) they are positively or negatively influenced by the change in abiotic conditions (including pH and substrate availability) caused by ongoing SO42− reduction, and (iii) they are inhibited by H2S or they benefit from the inhibition of competing microorganisms.
To our knowledge, no NGS data on SO42−-influenced communities under thermophilic conditions has been available until now. Under mesophilic conditions, the dominant SRB were diverse, with Desulfomicrobium, Desulfobulbus, and Desulfovibrio (all Proteobacteria) when digesting maize silage and pig manure (Kushkevych et al. 2017); Desulfosporosinus (Firmicutes) when digesting manure (St-Pierre and Wright 2017); and Desulfonauticus (Proteobacteria) when digesting synthetic wastewater containing sucrose and ethanol under a high phenol load. In the present investigation, the candidate genus Desulforudis (Firmicutes) was most abundant, with up to 2% in samples with 3 or 5 g SO42−/L. The description of this genus is based on the genome analysis of a single uncultivated bacterium found in a South African gold mine, as no cultivated representative is available at present (Chivian et al. 2008). Furthermore, with increasing SO42− concentrations, species belonging to the OTU Desulfotomaculum (Firmicutes) occurred in increasing abundances up to 1% of total prokaryotes. Until 2018, the genus Desulfotomaculum comprised a heterogeneous and polyphyletic group of thermophilic spore-forming SRB, which were reclassified into five genera by Watanabe et al. (2018). Since the SILVA database used in this investigation was from December 2017, the OTU “Desulfotomaculum” contains the present-day genera Desulfotomaculum, Desulfallas, Desulfofundulus, Desulfofarcimen, and Desulfohalotomaculum (Watanabe et al. 2018). Both OTUs, Candidatus Desulforudis and Desulfotomaculum, are thermophilic and are able to oxidize H2 among other substrates, including various sugars, VFAs, and alcohols (Muyzer and Stams 2008; Chivian et al. 2008). Besides, it should be considered that further members of the microbial community could be able to reduce SO42− but were not isolated or described as such this far. Especially, the family Peptococcaceae contains versatile physiological groups including SRB, as the two genera mentioned above (Stackebrandt 2014).
Apart from known SRB, seven bacterial OTUs were positively affected by the SO42− addition and showed more than 1% relative abundance at high SO42− levels. The most abundant of those was Caldicoprobacter (up to 29%), a genus, comprised of only four cultivated species, known to ferment a range of sugars including glucose and cellobiose (both intermediates of CMC mineralization) to lactate, acetate, ethanol, CO2, and H2 under thermophilic conditions (Yokoyama et al. 2010; Bouanane-Darenfed et al. 2013, 2011, 2015). Up to date, no SO42− reducing Caldicoprobacter species was found and the genus was rarely detected during NGS investigations of biogas sludge, e.g., De Vrieze et al. (2018). The OTU unclassified Clostridiales_vadinBB60_group was less abundant (up to 7%) and is based on a 16s rRNA gene fragment sequenced, when investigating the microbial diversity in an anaerobic digester fermenting vinasses (Godon et al. 1997). Further, positively affected OTUs include the following: the genus Proteiniborus, mainly known for fermenting proteins to acetate, ethanol, and H2/CO2 (Hahnke et al. 2018; Niu et al. 2008); members of the Clostridium sensu stricto 15 group; and uncultured members of the family Peptococcaceae. The latter is, as mentioned above, an ecologically and physiologically heterogeneous group of obligate anaerobes, including besides SRB also syntrophic, autotrophic as well as heterotrophic bacteria (Stackebrandt 2014).
Negatively influenced bacterial OTUs (> 1% abundance in control samples) were Lentimicrobiaceae, Izimaplasmatales, and uncultured Clostridiales. At the moment, the family Lentimicrobiaceae, representing the third abundant OTU (up to 21%) in the present study, includes only one described species, Lentimicrobium saccharophilum, which was isolated from a mesophilic UASB reactor, fermenting sugars to acetate, malate, propionate, formate and H2 (Sun et al. 2016). The order Izimaplasmatales belongs to the phylum Tenericutes, whose members are cell wall free bacteria typically found as parasites or commensals of eukaryotic hosts (Skennerton et al. 2016). The order is based on the genomes of two free-living bacteria extracted from a deep-sea CH4 seep sediment (Skennerton et al. 2016).
In contrast, some relatively abundant OTUs were not affected by the SO42− concentrations. The second most abundant OTU Defluviitoga comprises only one cultivated species, Defluviitoga tunisiensis, isolated from a mesophilic anaerobic reactor, fermenting among others cellobiose and glucose to acetate, H2 and CO2 (Hania et al. 2012). Members of the genus Defluviitoga have been detected in great abundances in thermophilic reactors (Liang et al. 2018; Maus et al. 2016). Further on, both OTUs Clostridia_DTU014 and Clostridia MBA03 showed average relative abundance of 5% and are based only on sequences isolated from thermophilic biogas reactors (Campanaro et al. 2016).
The archaeal community in the control samples consisted mainly of two methanogenic genera, Methanoculleus and Methanosarcina, comprising 10% and 6% of prokaryotic 16 s DNA, respectively. Both were inhibited by SO42− addition, with Methanosarcina being more sensitive and totally inhibited at more than 0.5 g SO42−/L, while Methanoculleus preserved a relative abundance of 4% at 1.5 g SO42−/L. The higher susceptibility towards sulfide inhibition of acetoclastic compared with hydrogenotrophic methanogens was also observed previously by Maillacheruvu and Parkin (1996).
As expected, the absolute abundance of SRB was pushed by the SO42− addition to 106 copies/mL but surprisingly did not exceed the abundance of methanogens, even in samples with excessive SO42−. Moestedt et al. (2013), who investigated the abundance of SRB in 25 industrial biogas fermenters with various SO42− loads, found 105 to 107 copies/mL. In our investigation, mcrA copy numbers, which were used to quantify methanogen, of more than 106 could be found even in samples in which methanogenesis had been totally inhibited for at least 2 weeks, probably deriving from inactive or dead cells (Wagner et al. 2008). This result highlights the discrepancy between DNA-based abundance measurements and physiologically determined activity parameters and underlines the importance of biochemical data. To determine if the measured methanogen-specific DNA derived from free DNA, inactive cells or dead cells, a viability test was performed. It could be shown that methanogenesis restarted immediately when a small aliquot of organisms of a 3 g SO42−/L variant was transferred into new SO42− free medium, meaning that inhibition by H2S was not lethal but reversible. The same is valid for the SRB, which also seemed inhibited by H2S, referring to the low CO2 production in week three and four of cultivation; transferred into fresh SO42−-containing medium, SRB instantly resumed their metabolic activities.
In the present study, the addition of SO42− led to a decrease in propionate and butyrate concentrations and to an accumulation of acetate, although both, methanogens and SRB, should be able to utilize acetate (Muyzer and Stams 2008). This phenomenon was also observed by other authors under mesophilic, but also thermophilic conditions (O’Flaherty et al. 1998; Colleran and Pender 2002). As SRB have a lower threshold for H2, these organisms create a lower H2 partial pressure than methanogens, which improves the thermodynamics for VFA-oxidizing H2-producing acetogens (Stams et al. 2005). Furthermore, SRB themselves incompletely oxidize propionate and butyrate to acetate, with especially propionate oxidation being thermodynamically extremely favorable (O’Flaherty et al. 1998). Under the physiochemical conditions at day 14, the oxidation of propionate and butyrate with SO42− yields a ΔG of − 265 and − 94 kJ/mol, respectively. The preference of SRB for propionate over other organic acids was experimentally shown previously by Visser et al. (1993). This means, SRB directly and indirectly cause low levels of propionate and high levels of acetate. The higher the SO42− level, the more decisive is the direct effect. In this context, Li et al. (2015) successfully tested the application of low additions of SO42− (COD/SO42− 200 to 350) to fight propionate accumulation during a thermophilic co-digestion of coffee grounds, milk, and waste activated sludge.
The prevalence of acetate in the SO42− samples in the present study might be explained by the inhibition of the acetoclastic methanogenic genus Methanosarcina by H2S, as seen in the NGS results. Moreover, the SO42− samples were metabolically not very active in the last 2 weeks, indicating that all organisms including the SRB were at least partially inhibited by the high H2S concentrations. Moreover, it is possible that acetate-utilizing SRB were not present in the applied inoculum, although thermophilic acetate-oxidizing SRB have previously been isolated from methanogenic reactors (Min and Zinder 1990; Hattori et al. 2000; Balk et al. 2007). The OTU Desulfotomaculum comprises species able as well as species unable to oxidize acetate, while corresponding information is not available for Candidatus Desulforudis. These possible explanations were also mentioned by Colleran and Pender (2002), who also observed high effluent levels of acetate during thermophilic digestion of SO42−-rich wastewater. Contrary to that, Visser et al. (1992) found that SO42− reducers could outcompete acetoclastic methanogens under thermophilic conditions fed on an acetate/propionate/butyrate substrate mixture.
In conclusion, our results show that SRB are able to strongly influence their abiotic environment and thus the entire microbial community even in low abundances. Finally, the altered community and chemical properties lead to a distinct shift in the carbon flow of the entire system.
Acknowledgments
We would like to thank Dr. Thomas Pümpel for his valuable technical support on the sulfate measurements.
Funding information
Open access funding provided by University of Innsbruck and Medical University of Innsbruck. The authors received doctoral grant from the Universität Innsbruck.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Amend J, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev. 2001;25:175–243. doi: 10.1111/j.1574-6976.2001.tb00576.x. [DOI] [PubMed] [Google Scholar]
- Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–137. [Google Scholar]
- Balk M, Weijma J, Goorissen HP, Ronteltap M, Hansen TA, Stams AJM. Methanol utilizing Desulfotomaculum species utilizes hydrogen in a methanol-fed sulfate-reducing bioreactor. Appl Microbiol Biotechnol. 2007;73:1203–1211. doi: 10.1007/s00253-006-0590-4. [DOI] [PubMed] [Google Scholar]
- Bouanane-Darenfed A, Fardeau M-L, Grégoire P, Joseph M, Kebbouche-Gana S, Benayad T, Hacene H, Cayol J-L, Ollivier B. Caldicoprobacter algeriensis sp. nov. a new thermophilic anaerobic, xylanolytic bacterium isolated from an Algerian hot spring. Curr Microbiol. 2011;62:826–832. doi: 10.1007/s00284-010-9789-9. [DOI] [PubMed] [Google Scholar]
- Bouanane-Darenfed A, Ben Hania W, Hacene H, Cayol J-L, Ollivier B, Fardeau M-L. Caldicoprobacter guelmensis sp. nov., a thermophilic, anaerobic, xylanolytic bacterium isolated from a hot spring. Int J Syst Evol Microbiol. 2013;63:2049–2053. doi: 10.1099/ijs.0.043497-0. [DOI] [PubMed] [Google Scholar]
- Bouanane-Darenfed A, Ben Hania W, Cayol J-L, Ollivier B, Fardeau M-L. Reclassification of Acetomicrobium faecale as Caldicoprobacter faecalis comb. nov. Int J Syst Evol Microbiol. 2015;65:3286–3288. doi: 10.1099/ijsem.0.000409. [DOI] [PubMed] [Google Scholar]
- Campanaro S, Treu L, Kougias PG, de Francisci D, Valle G, Angelidaki I. Metagenomic analysis and functional characterization of the biogas microbiome using high throughput shotgun sequencing and a novel binning strategy. Biotechnol. Biofuels. 2016;9:26. doi: 10.1186/s13068-016-0441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cheng JJ, Creamer KS. Inhibition of anaerobic digestion process. A review. Bioresour Technol. 2008;99:4044–4064. doi: 10.1016/j.biortech.2007.01.057. [DOI] [PubMed] [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, Gihring TM, Lapidus A, Lin L-H, Lowry SR, Moser DP, Richardson PM, Southam G, Wanger G, Pratt LM, Andersen GL, Hazen TC, Brockman FJ, Arkin AP, Onstott TC. Environmental genomics reveals a single-species ecosystem deep within Earth. Science (New York, NY) 2008;322:275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- Colleran E, Pender S. Mesophilic and thermophilic anaerobic digestion of sulphate-containing wastewaters. Water Sci Technol. 2002;45:231–235. [PubMed] [Google Scholar]
- Colleran E, Finnegan S, Lens P. Anaerobic treatment of sulphate-containing waste streams. Antonie Van Leeuwenhoek. 1995;67:29–46. doi: 10.1007/BF00872194. [DOI] [PubMed] [Google Scholar]
- De Vrieze J, Pinto AJ, Sloan WT, Ijaz UZ. The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome. 2018;6:63. doi: 10.1186/s40168-018-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LW, Akinfiev NN. Solubility of CO2 in water from −1.5 to 100 °C and from 0.1 to 100 MPa: evaluation of literature data and thermodynamic modelling. Fluid Phase Equilib. 2003;208:265–290. [Google Scholar]
- Elferink SJWH, Luppens SBI, Marcelis CLM, Stams AJM (1998) Kinetics of acetate oxidation by two sulfate reducers isolated from anaerobic granular sludge. Appl Environ Microbiol 64 [DOI] [PMC free article] [PubMed]
- Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R (1997) Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol 63 [DOI] [PMC free article] [PubMed]
- Hahnke S, Langer T, Klocke M (2018) Proteiniborus indolifex sp. nov., isolated from a thermophilic industrial-scale biogas plant. Int J Syst Evol Microbiol [DOI] [PubMed]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9pp. [Google Scholar]
- Hania WB, Godbane R, Postec A, Hamdi M, Ollivier B, Fardeau M-L. Defluviitoga tunisiensis gen. nov., sp. nov., a thermophilic bacterium isolated from a mesothermic and anaerobic whey digester. Int J Syst Evol Microbiol. 2012;62:1377–1382. doi: 10.1099/ijs.0.033720-0. [DOI] [PubMed] [Google Scholar]
- Hattori S, Kamagata Y, Hanada S, Shoun H. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int J Syst Evol Microbiol. 2000;50(Pt 4):1601–1609. doi: 10.1099/00207713-50-4-1601. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol LW, Lens PNL, Stams AJM, Lettinga G. Anaerobic treatment of sulphate-rich wastewaters. Biodegradation. 1998;9:213–224. doi: 10.1023/a:1008307929134. [DOI] [PubMed] [Google Scholar]
- Illmer P, Gstraunthaler G. Effect of seasonal changes in quantities of biowaste on full scale anaerobic digester performance. Waste Manag. 2009;29:162–167. doi: 10.1016/j.wasman.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Karhadkar PP, Audic J-M, Faup GM, Khanna P. Sulfide and sulfate inhibition of methanogenesis. Water Res. 1987;21:1061–1066. [Google Scholar]
- Koster IW, Rinzema A, de Vegt AL, Lettinga G. Sulfide inhibition of the methanogenic activity of granular sludge at various pH-levels. Water Res. 1986;20:1561–1567. [Google Scholar]
- Kushkevych I, Vítězová M, Vítěz T, Bartoš M. Production of biogas: relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017;12:1145. [Google Scholar]
- Li Q, Li Y-Y, Qiao W, Wang X, Takayanagi K. Sulfate addition as an effective method to improve methane fermentation performance and propionate degradation in thermophilic anaerobic co-digestion of coffee grounds, milk and waste activated sludge with AnMBR. Bioresour Technol. 2015;185:308–315. doi: 10.1016/j.biortech.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Liang X, Whitham JM, Holwerda EK, Shao X, Tian L, Wu Y-W, Lombard V, Henrissat B, Klingeman DM, Yang ZK, Podar M, Richard TL, Elkins JG, Brown SD, Lynd LR. Development and characterization of stable anaerobic thermophilic methanogenic microbiomes fermenting switchgrass at decreasing residence times. Biotechnol Biofuels. 2018;11:243. doi: 10.1186/s13068-018-1238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Yu Z, Li Y (2017) Sequential batch thermophilic solid-state anaerobic digestion of lignocellulosic biomass via recirculating digestate as inoculum - Part II. Microbial diversity and succession. Bioresour Technol 241:1027–1035 [DOI] [PubMed]
- Lovley DR, Dwyer DF, Klug MJ. Kinetic analysis of competition between sulfate reducers and methanogens for hydrogen in sediments. Appl Environ Microbiol. 1982;43:1373. doi: 10.1128/aem.43.6.1373-1379.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillacheruvu KY, Parkin GF. Kinetics of growth, substrate utilization and sulfide toxicity for propionate, acetate, and hydrogen utilizers in anaerobic systems. Water Environ Res. 1996;68:1099–1106. [Google Scholar]
- Maus I, Koeck DE, Cibis KG, Hahnke S, Kim YS, Langer T, Kreubel J, Erhard M, Bremges A, Off S, Stolze Y, Jaenicke S, Goesmann A, Sczyrba A, Scherer P, König H, Schwarz WH, Zverlov VV, Liebl W, Pühler A, Schlüter A, Klocke M. Unraveling the microbiome of a thermophilic biogas plant by metagenome and metatranscriptome analysis complemented by characterization of bacterial and archaeal isolates. Biotechnol Biofuels. 2016;9:171. doi: 10.1186/s13068-016-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland MJ, Jewell WJ. The effect of sulfate reduction on the thermophilic (55°C) methane fermentation process. J Ind Microbiol. 1990;5:247–257. [Google Scholar]
- Min H, Zinder SH. Isolation and characterization of a thermophilic sulfate-reducing bacterium Desulfotomaculum thermoacetoxidans sp. nov. Arch Microbiol. 1990;153:399–404. [Google Scholar]
- Moestedt J, Nilsson Påledal S, Schnürer A. The effect of substrate and operational parameters on the abundance of sulphate-reducing bacteria in industrial anaerobic biogas digesters. Bioresour Technol. 2013;132:327–332. doi: 10.1016/j.biortech.2013.01.043. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6 [DOI] [PubMed]
- Niu L, Song L, Dong X. Proteiniborus ethanoligenes gen. nov., sp. nov., an anaerobic protein-utilizing bacterium. Int J Syst Evol Microbiol. 2008;58:12–16. doi: 10.1099/ijs.0.65108-0. [DOI] [PubMed] [Google Scholar]
- O’Flaherty V, Lens P, Leahy B, Colleran E. Long-term competition between sulphate-reducing and methane-producing bacteria during full-scale anaerobic treatment of citric acid production wastewater. Water Res. 1998;32:815–825. [Google Scholar]
- Parada AE, Needham DM, Fuhrman JA. Every base matters. Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- Paulo LM, Stams AJM, Sousa DZ. Methanogens, sulphate and heavy metals. A complex system. Rev Environ Sci Biotechnol. 2015;14:537–553. [Google Scholar]
- Pereyra LP, Hiibel SR, Prieto Riquelme MV, Reardon KF, Pruden A. Detection and quantification of functional genes of cellulose- degrading, fermentative, and sulfate-reducing bacteria and methanogenic archaea. Appl Environ Microbiol. 2010;76:2192–2202. doi: 10.1128/AEM.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siles JA, Brekelmans J, Martín MA, Chica AF, Martín A. Impact of ammonia and sulphate concentration on thermophilic anaerobic digestion. Bioresour Technol. 2010;101:9040–9048. doi: 10.1016/j.biortech.2010.06.163. [DOI] [PubMed] [Google Scholar]
- Skennerton CT, Haroon MF, Briegel A, Shi J, Jensen GJ, Tyson GW, Orphan VJ. Phylogenomic analysis of Candidatus ‘Izimaplasma’ species: free-living representatives from a Tenericutes clade found in methane seeps. ISME J. 2016;10:2679–2692. doi: 10.1038/ismej.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E. The emended family Peptococcaceae and description of the families Desulfitobacteriaceae, Desulfotomaculaceae, and Thermincolaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes. Berlin Heidelberg: Springer; 2014. pp. 285–290. [Google Scholar]
- Stams AJM, Plugge CM, de Bok FAM, van Houten BHGW, Lens P, Dijkman H, Weijma J. Metabolic interactions in methanogenic and sulfate-reducing bioreactors. Water Sci Technol. 2005;52:13–20. [PubMed] [Google Scholar]
- Steinberg LM, Regan JM. Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl Environ Microbiol. 2008;74:6663–6671. doi: 10.1128/AEM.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, Wright A-DG. Implications from distinct sulfate-reducing bacteria populations between cattle manure and digestate in the elucidation of H2S production during anaerobic digestion of animal slurry. Appl Microbiol Biotechnol. 2017;101:5543–5556. doi: 10.1007/s00253-017-8261-1. [DOI] [PubMed] [Google Scholar]
- Sun L, Toyonaga M, Ohashi A, Tourlousse DM, Matsuura N, Meng X-Y, Tamaki H, Hanada S, Cruz R, Yamaguchi T, Sekiguchi Y. Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:2635–2642. doi: 10.1099/ijsem.0.001103. [DOI] [PubMed] [Google Scholar]
- Tuan NN, Chang Y-C, Yu C-P, Huang SL (2014) Multiple approaches to characterize the microbial community in a thermophilic anaerobic digester running on swine manure. A case study. Microbiol Res 169:717–724 [DOI] [PubMed]
- Visser A, Gao Y, Lettinga G. Anaerobic treatment of synthetic sulfate-containing wastewater under thermophilic conditions. Water Sci Technol. 1992;25:193–202. [Google Scholar]
- Visser A, Beeksma I, Zee F, Stams AJM, Lettinga G (1993) Anaerobic degradation of volatile fatty acids at different sulphate concentrations. Appl Microbiol Biotechnol 40
- Wagner AO, Malin C, Knapp BA, Illmer P. Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Appl Environ Microbiol. 2008;74:2537–2539. doi: 10.1128/AEM.02288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AO, Malin C, Lins P, Illmer P. Effects of various fatty acid amendments on a microbial digester community in batch culture. Waste Manag. 2011;31:431–437. doi: 10.1016/j.wasman.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Wagner AO, Malin C, Lins P, Gstraunthaler G, Illmer P. Reactor performance of a 750 m3 anaerobic digestion plant: varied substrate input conditions impacting methanogenic community. Anaerobe. 2014;29:29–33. doi: 10.1016/j.anaerobe.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Wagner AO, Markt R, Puempel T, Illmer P, Insam H, Ebner C. Sample preparation, preservation, and storage for volatile fatty acid quantification in biogas plants. Eng Life Sci. 2017;17:132–139. doi: 10.1002/elsc.201600095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AO, Prem EM, Markt R, Kaufmann R, Illmer P. Formation of phenylacetic acid and phenylpropionic acid under different overload conditions during mesophilic and thermophilic anaerobic digestion. Biotechnol Biofuels. 2019;12:359–383. doi: 10.1186/s13068-019-1370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kojima H, Fukui M. Review of Desulfotomaculum species and proposal of the genera Desulfallas gen. nov., Desulfofundulus gen. nov., Desulfofarcimen gen. nov. and Desulfohalotomaculum gen. nov. Int J Syst Evol Microbiol. 2018;68:2891–2899. doi: 10.1099/ijsem.0.002915. [DOI] [PubMed] [Google Scholar]
- Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Wagner ID, Wiegel J. Caldicoprobacter oshimai gen. nov., sp. nov., an anaerobic, xylanolytic, extremely thermophilic bacterium isolated from sheep faeces, and proposal of Caldicoprobacteraceae fam. nov. Int J Syst Evol Microbiol. 2010;60:67–71. doi: 10.1099/ijs.0.011379-0. [DOI] [PubMed] [Google Scholar]