Abstract
Eimeria spp. infection was investigated in 10 free-roaming European bison aged three months to 26 years by anatomopathological, histopathological, coproscopic and PCR-RFLP examination. The coproscopic study identified Eimeria oocysts in the faeces of five bison. The most prevalent morphotypes were E. bovis, present in all positive samples, and E. zuernii, in all but one. Additionally, mixed infections consisting of E. bovis, E. zuernii, E. alabamensis, E. auburnensis, E. canadensis, E. cylindrica, E. ellipsoidalis and E. subspherica were diagnosed in two bison calves. Besides being the most prevalent form, E. bovis also demonstrated the highest OPG (2,750). The presence of oocysts in the faeces was associated with those of macrogamonts, microgamonts and oocysts in the epithelium of the large intestine. Intestinal coccidiosis associated with lymphoplasmacytic enteritis was observed in many bison, not only those with positive OPG. Four animals with negative coproscopy results demonstrated early-stage gametogony in the large intestine; one case presented no endogenous stages of coccidians in the histopathological sections of the intestine, nor oocysts in the faecal samples. A 530 bp product of E. bovis 18S rDNA (GenBank: MK951685) was obtained from both the colon wall and oocysts; this was subjected to PCR-RFLP analysis based on AluI and Hin1II (NlaIII) restriction enzymes. Both samples yielded a consistent seven-band pattern, four of which (270 bp, 40 bp, 180 bp and 84 bp) were expected, and the other three represented undigested fragments. The obtained digestion pattern is indicative of Eimeria spp. infection, and can serve as a first-step diagnostic approach in detection of infection. The result of computer-based virtual digestion of the PCR product suggests that double digestion with Mval (BstNI) and KpnI restriction enzymes may be used as a second-step tool to distinguish between E. bovis, E. zuernii and E. alabamensis, all of which are highly-pathogenic species.
Keywords: Protozoa, Eimeriidae, Wildlife, Histopathology, PCR-RFLP
Graphical abstract
Highlights
-
•
Picture of intestinal coccidiosis in 9 of 10 examined European bison.
-
•
Coccidian oocysts with predominance of Eimeria bovis in the faeces of 5 individuals.
-
•
Exclusively immature gamonts in the intestine of bison with negative coproscopy.
-
•
PCR-RFLP detects Eimeria spp. developmental stages in the wall of colon of wisent.
-
•
PCR-RFLP enable to distinguish between pathogenic species of Eimeria.
1. Introduction
Protozoans of the genus Eimeria are well known to be causative agents of coccidiosis in domestic ruminants, mainly in calves younger than one year of age (Pellérdy, 1974). In severe cases, the infected animals demonstrate hemorrhagic, devastating diarrhea and wasting, with a possible fatal outcome (Jolley and Bardsley, 2006). Hence, epidemic-like outbreaks of acute coccidiosis can lead to serious economic losses to livestock production worldwide (Fitzgerald, 1980).
Although coccidiosis is a well-recognized threat to livestock, relatively little is known about its pathogenicity and course, or of the species involved in the coccidiosis of wild ruminants. While Eimeria parasites are considered strictly host specific (McDougald, 1979), common Eimeria species have been identified in cattle and wild bovids such as water buffalo (Dubey, 2018; Sayin, 1969), American bison (Penzhorn et al., 1994) and European bison (Pyziel et al., 2014). Besides E. bareillyi, the most pathogenic species in buffalo, which is not transmittable to cattle (Dubey et al., 2008), the most commonly-observed infective agents among wild bovids are E. bovis and E. zurenii, both of which infect domestic cattle, and are considered to be the most pathogenic species of Eimeria in this regard (Pyziel et al., 2011; De Noronha et al., 2009; Penzhorn et al., 1994). Eimeria bovis also appears to be the most prevalent coccidian of European bison (prevalence = 29.7%), and E. zuernii the second most prevalent one (prevalence = 9.9%) (Pyziel and Demiaszkiewicz, 2015; Pyziel et al., 2014).
Eimeria bovis and E. zuernii exert their pathogenic potential by damaging the crypt cells of the large intestinal mucosa during their sexual developmental stages (Stockdale, 1977a, 1977b; Hammond et al., 1964). However, as it is difficult to distinguish E. bovis or E. zuernii based on the morphology of these histopathological lesions (Stockdale, 1977b), accurate identification requires the use of molecular biology techniques. One potential candidate is PCR - restriction fragment length polymorphism (PCR-RFLP), which has been successfully used to identify other pathogenic protozoans, including Toxoplasma gondii and Cryptosporidium spp. (Su et al., 2006; Spano et al., 1997). The aim of the study was to compare the histopathological picture of intestinal coccidiosis in European bison with the outcome of coprological examination, and to evaluate the potential of PCR – RFLP for the diagnosis of potentially pathogenic E. bovis infection in European bison, a mammal recognized as being vulnerable to extinction (IUCN, 2019).
2. Materials and methods
2.1. Study area and materials collected
Tissue samples were taken from the ileum, colon and cecum of 10 free-roaming European bison, aged three months to 26 years, from the Białowieża Primeval Forest (52° 41′ N, 23° 43’ E). All animals had previously been killed for sanitary or breeding reasons. The tissue samples intended for DNA extraction were preserved in 70% ethanol, and those for histopathological investigation in 10% formalin.
At the same time, fresh faecal samples were collected directly from the rectum of each individual. These were placed separately in a labeled tube, stored in a portable refrigerator and transported to the laboratory.
2.2. Histopathological study
The intestinal specimens preserved in formalin were embedded in paraffin, cut into 4 μm-thick sections and stained with hematoxylin and eosin (H-E). Histopathological lesions were investigated with the use of an Olympus BX43 light microscope (Olympus, Tokyo, Japan) coupled to a computer equipped with CellSens software (Olympus).
2.3. Coprological study
Briefly, the oocyst count per gram (OPG) value was calculated for 3 g of each faecal sample using the McMaster technique (Taylor et al., 2007a, Taylor et al., 2007b) in sucrose solution (SG = 1.27) (Pyziel and Demiaszkiewicz, 2013). Following this, the oocysts were sporulated and identified on the basis of their morphological characters according to Duszynski and Wilber (1997) and Pyziel et al. (2014). The samples with the highest E. bovis OPG values were selected for oocyst collection; these were taken from the McMaster chamber using a pipette with a capillary tip as described previously (Pyziel et al., 2019). Finally, 30 oocysts of E. bovis were collected prior to DNA extraction.
2.4. Extraction of genomic DNA
Genomic DNA was extracted from the colon tissue of the European bison and E. bovis oocysts with the use of a Nucleospin Tissue DNA Extraction Kit (Macherey-Nagel, Düren, Germany). Initial breakage of the oocyst wall was performed according to Hnida and Duszynski (1999) and Zhao et al. (2001), with minor modifications (Pyziel et al., 2019).
2.5. Amplification of partial 18S rRNA gene, restriction fragment length polymorphism and sequencing of the PCR products
In order to amplify the partial region of the 18S rRNA of Eimeria spp., the following set of primers was designed: forward F8 (5’ – TTT CGA CGG TAG GGT ATT GGC CT – 3′) and reverse ER24 (5’ – ACG AAT GCC CCC AAC TGT CC – 3’). The primers were designed based on sequences of the Eimeria spp. 18S rDNA available in GenBank (GenBank: AF291427; MK691697; U67115; U67116; U67117; U67117; U67118; U67119; U67119; U67120; U67121; U76748) using FastPCR, version 5.4 (PrimerDigital, Helsinki, Finland).
The PCR reaction itself was performed in a Techne TC-512 thermal cycler (Bibby Scientific Limited, UK) in a total reaction volume of 50 μl comprising 70 mM Tris-HCl at pH 8.3, 16.6 mM (NH4)2SO4, 2.5 mM MgCl2, 100 μM of each dNTP, 1 μl of each primer at 20 pM/μl, 1 unit HiFiTaq polymerase (5 units/μl) (Novazym, Poznań, Poland) and 1 μl of template DNA. The thermal profile comprised an initial denaturation at 94 °C for 2 min, followed by 34 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 30 s, and extension at 72 °C for 30 s, this was followed by prolongation of final extension for 3 min.
Following DNA amplification, double digestion of PCR products was performed according to the manufacturer's instructions using the AluI enzyme recognising AG∧CT sites (Thermo Fisher Scientific, Waltham, Massachusetts, USA, catalogue no. ER0011) and the Hin1II (NlaIII) enzyme recognising CATG∧ sites (Thermo Fisher Scientific, catalogue no. ER1831). Briefly, 10 μl of the PCR reaction mixture was mixed with 18 μl of Molecular Biology Reagent Water (Sigma-Aldrich, Saint Louis, Missouri, USA), 2 μl of 10X Buffer Tango, 2 μl of AluI and 4 μl of Hin1II. The mixture was spun down for 30 s and incubated at 37 °C overnight.
The effects of the DNA amplification, and the subsequent digestion, were verified by electrophoresis in 1.5% agarose gel with ethidium bromide (0.5 μg/ml). The resulting bands were visualised under UV light using a Gel Logic 200 Imaging System (Kodak, Rochester, New York, USA) equipped with Kodak 1D ver. 3.6 software.
Additionally, the PCR products obtained from the intestinal tissue wall and the oocysts were purified with a Nucleospin Gel and PCR Clean-up Kit (Macherey-Nagel) and eluted with 30 μl of Molecular Biology Reagent Water (Sigma-Aldrich). They were then sequenced in both directions by Genomed S.A. (Warsaw, Poland) with the use of primers F8 and EF24, as described above. The obtained sequences were assembled into contigs using Contig-Express software (Thermo Fisher Scientific), aligned using GeneDoc – Multiple Sequence Alignment Editor (Nicholas et al., 1997) and submitted into the GenBank.
2.6. Simulation of restriction digestion of the 18S rDNA sequence of highly-pathogenic species of Eimeria
To distinguish between three highly-pathogenic species of Eimeria [E. bovis (GenBank: MK951685), E. zuernii (GenBank: KU351734), E, alabamensis (GeneBank: KT184335)] their 18S rRNA gene (sequence data including the region between the primers F8 + ER24) was subjected to virtual double digestion by two endonucleases: Mval (BstNI) recognising CC∧WGG sites and KpnI recognising GGTAC∧C sites. Both restriction enzymes are most effective at a temperature of 37 °C. The digestion was simulated with SnapGene version 5.0.6 (GSL Biotech LLC, Chicago, USA). The simulated enzymatic digest pattern of the hypothetical products was visualised on a virtual 1% agarose gel.
3. Results
3.1. Macroscopic, histopathological and coproscopic examination
Gross lesions were observed in nine dissected individuals, and they were mainly present in the mucosa of the large intestine. The mucosa of the colon and cecum was severely congested and petechial or ecchymotic haemorrhages were observed. Additionally, the mucosa was thickened, edematous and covered with thick, viscous and, in some cases, murky mucus. Nevertheless, the content of the intestinal tracts showed proper color and consistency.
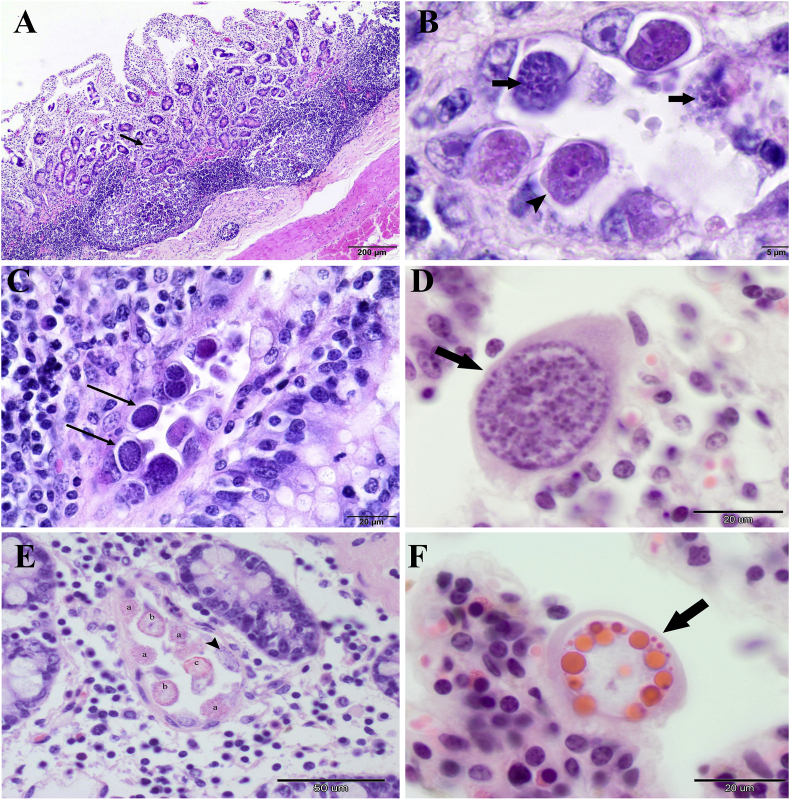
Histopathological investigation of these nine animals revealed intestinal coccidiosis associated with severe lymphoplasmacytic enteritis in the intestines (Fig. 1A). Numerous cross-sections of Eimeria spp. were observed. Various developmental stages could be identified and although most individuals were observed in the dilated lumen of the crypts in the deep mucosa of the intestine, various stages were also seen throughout the villi. Other lesions consisted of moderate blunting of the intestinal villi (villous atrophy), with visible proliferative changes, edema and congestion. Additionally, lymphocytic depletion was observed in the lymphoid follicles of the submucosal tissue and numerous tingible body macrophages in the germinal centers (Fig. 1A). The cytoplasm of the epithelial cells of the large intestine glands was massively parasitized with various sexual stages of Eimeria; however, schizonts and merozoites were rarely observed in the same location (Fig. 1B and C).
Fig. 1.
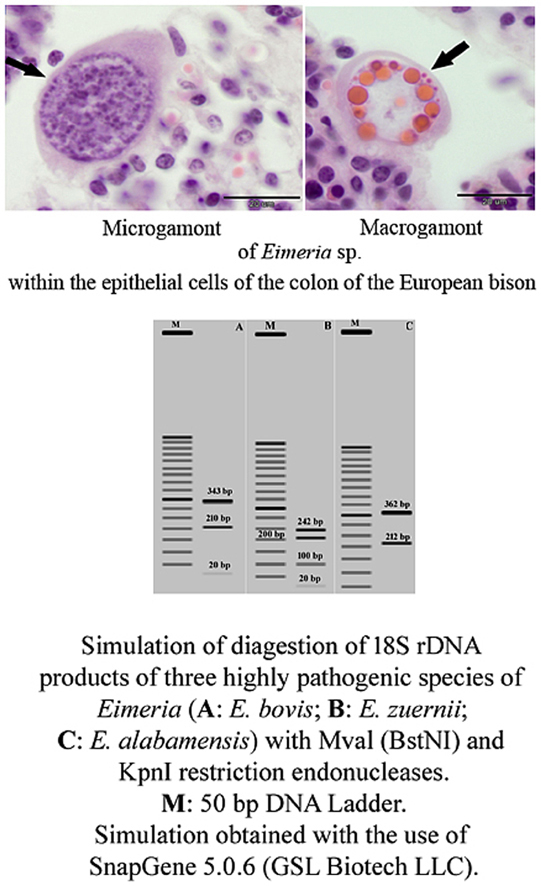
Histopathological lesions associated with endogenous stages of Eimeria spp. in sections of the ileum and colon of European bison (H-E staining). (A) Shortening and blunting of the intestinal villi of the ileum with diffuse infiltration of mononuclear inflammatory cells within the lamina propria, edematous stroma, dilated crypt containing necrotic debris (arrow), and atrophy of submucosal lymphoid follicles (× 20 magnification). (B) Schizonts and degenerating merozoites in the crypt lumen of the colon (arrows); immature macrogamont with a central nucleus (arrowhead) (× 1000 magnification). (C) Immature microgamonts in the epithelial cells of the colon crypt (arrows) (× 400 magnification). (D) Mature microgamont in the epithelial cells of the colon crypt (arrow) (× 1000 magnification). (E) Gametogonic stages of Eimeria development in the epithelial cells of the colon. Microgamont with peripheral microgames (arrowhead), (a) nearly mature microgamonts, (b) macrogamont with eosinophilic wall-forming bodies, (c) early oocyst (× 400 magnification). (F) Mature macrogamont in the epithelial cells of the cecum (arrow) (× 1000 magnification).
Infection was accompanied with necrosis and sloughing of the intestinal epithelial cells, and the crypts were distended and focally filled with cellular debris (Fig. 1B and C). Sexually differentiated male gamonts (microgamonts) were identified based on the presence of several nuclei, whereas female gamonts (macrogamonts) possessed central nuclei and granules (Fig. 1C, D, E and F). Both microgamonts and macrogamonts were observed together in the same regions of infected crypts (Fig. 1C). In addition, the intestinal mucosa was expanded by a cellular infiltrate composed of various mononuclear cells (lymphocytes, plasma cells, macrophages), accompanied by focally scattered Mott cells and eosinophils.
Although the histopathological examination revealed intestinal coccidiosis in nine of the ten investigated animals, only five of these demonstrated positive OPG results. Interestingly, no mature stages of gametogony were noted in the large intestine of individuals with negative coproscopic results (Fig. 1B and C). In one case, a 25-year-old male, no endogenous stages of coccidians were found in histopathological sections of the intestine, nor were any oocysts observed in the faecal sample.
In all cases, shedding oocysts in the faeces was associated with the presence of macrogamonts, microgamonts and oocysts among the host enterocytes (Fig. 1D, E and F). The mature macrogamonts and microgamonts measured up to 25 μm and 32 μm, respectively, whereas the oocysts were around 17 μm in diameter. Among five of the faecal samples positive for Eimeria spp., all were positive for the occurrence of E. bovis oocysts, four for E. zuernii oocysts, and two derived from calves for multi-mixed infection of four species in three-month-old individual (E. alabamensis, E. bovis, E. subspherica, E. zuernii), and of five species in eight-month-old bison (E. alabamensis, E. bovis, E. cylindrica, E. ellipsoidalis, E. zuernii). Animals over one year of age shedded oocysts of two or three species of Eimeria (Table 1). The highest OPG score was obtained for E. bovis (2,750).
Table 1.
Individual Eimeria species composition and their OPGs in investigated European bison.
| Investigated individuals |
Eimeria species + OPG |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Age (in months) | E. alabamensis | E. auburnensis | E. bovis | E. canadensis | E. cylindrica | E. ellipsoidalis | E. subspherica | E. zuernii |
| 1. | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2. | 300 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3. | 3 | 2600 | 0 | 2750 | 0 | 0 | 0 | 550 | 650 |
| 4. | 30 | 0 | 0 | 150 | 50 | 0 | 0 | 0 | 50 |
| 5. | 312 | 0 | 0 | 100 | 0 | 0 | 50 | 0 | 0 |
| 6. | 18 | 0 | 100 | 250 | 350 | 0 | 0 | 0 | 0 |
| 7. | 72 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8. | 8 | 50 | 0 | 150 | 0 | 50 | 200 | 0 | 50 |
| 9. | 240 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10. | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
3.2. Nucleotide sequence of the isolated partial 18S rDNA and its digestion by restriction enzymes: AluI and Hin1II (NlaIII)
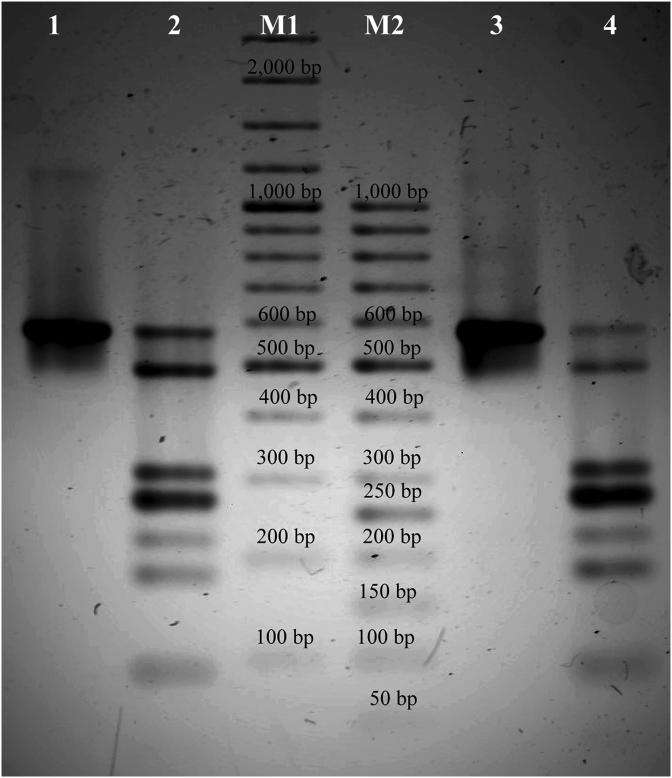
Amplification of the partial 18S rRNA gene of the European bison colon wall and E. bovis oocysts from the same host yielded a 530 bp sequence of E. bovis (excluding primers) (GenBank: MK951685) (Fig. 2). PCR-RFLP analysis of the sequence yielded a consistent seven-band restriction pattern for both the colon wall and the oocysts (Fig. 2). Four of the bands (270 bp, 40 bp, 180 bp and 84 bp) were predicted based on the nucleotide sequence of the 18S rDNA of Eimeria spp., whereas the other three (310 bp, 490 bp and 573 bp) represented undigested rDNA fragments. Virtual digestion of partial sequences of 18S rDNA of E. bovis and other bovine eimerians available in the GenBank database revealed that the obtained pattern of restriction endonuclease products is common for various Eimeria spp., including three highly pathogenic species known to undergo sexual development in the large intestine of a bovine host (Stockdale, 1977a; Hammond et al., 1964; Davis et al., 1957).
Fig. 2.
The PCR products identified within the 18S rRNA of Eimeria bovis following digestion with two restriction endonucleases: AluI recognising AG∧CT and Hin1II recognising CATG∧. M1: GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher Scientific); M2: GeneRuler 50bp DNA Ladder (Thermo Fisher Scientific); lane 1: European bison colon wall tissue; lane 2: European bison colon wall tissue after digestion; lane 3: E. bovis oocysts of European bison; lane 4: E. bovis oocysts of European bison after digestion.
3.3. Simulation of differential digestion of 18S rDNA sequence of highly-pathogenic species of bovine coccidia with the use of Mval (BstNI) and KpnI restriction enzymes
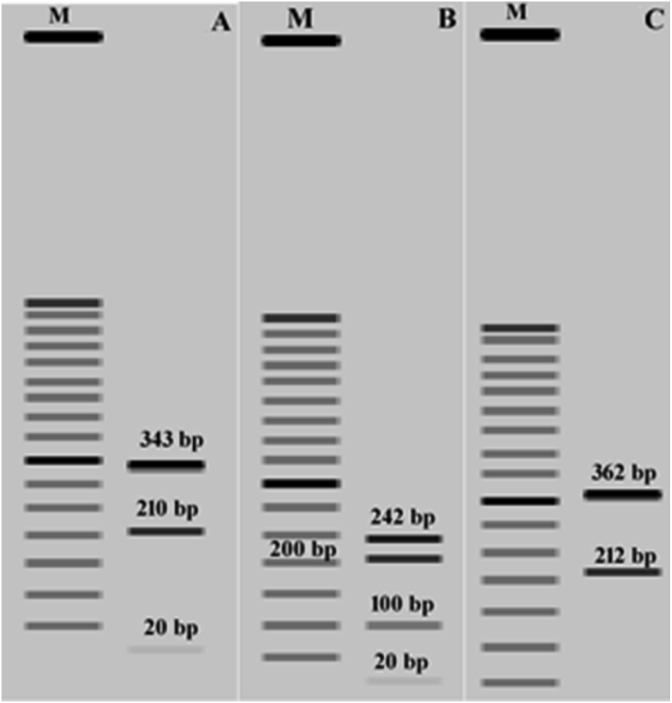
The virtual double digestion of the 18S rRNA gene of eimerians infecting the large intestine with the restriction enzymes Mval (BstNI) and KpnI yielded a unique band pattern for each species (Fig. 3). The simulated separation of digestion products on 1% agarose gel resulted in a three-band pattern for E. bovis (20 bp, 210 bp, 343 bp) (Fig. 3A), a four-band pattern for E, zuernii (20 bp, 100 bp, 210 bp, 242 bp) (Fig. 3B), and finally two-band pattern for E. alabamensis (212 bp, 362 bp) (Fig. 3C).
Fig. 3.
The virtual double digestion of the 18S rRNA gene of eimerians infecting the large intestine of the European bison with the restriction enzymes Mval (BstNI) recognising CC∧WGG, and KpnI recognising GGTAC∧C, simulated with SnapGene version 5.0.6 (GSL Biotech LLC); M: GeneRuler 50 bp DNA Ladder (Thermo Fisher Scientific). (A) A three-band pattern for E. bovis (20 bp, 210 bp, 343 bp). (B) A four-band pattern for E, zuernii (20 bp, 100 bp, 210 bp, 242 bp). (C) A two-band pattern for E. alabamensis (212 bp, 362 bp).
4. Discussion
Oocysts were identified for eight morphotypes of bovine eimerians, all of which had been previously reported in European bison (Pyziel et al., 2014). Three of these species are considered highly pathogenic as they undergo sexual development in the cells of the large intestine (E. bovis, E. zuernii, E. alabamensis) (Stockdale, 1977a, 1977b; Hammond et al., 1964; Davis et al., 1957). In contrast, the other identified species develop exclusively in the cells of the small intestine, and are hence believed to possess less pathogenic potential (Hammond et al., 1961, 1963): the bovine small intestine has a considerably larger reserve of host cells available for protozoans than the large intestine (Taylor et al., 2007a, Taylor et al., 2007b).
The study presents a two-step approach for rapid detection of Eimeria spp. in the intestinal tissue of European bison based on the PCR-RFLP method. In the first-step, the PCR product was subjected to double digestion with AluI and Hin1II (NlaIII) restriction enzymes to detect Eimeria spp. infection. In the second step, a double digestion with the Mval (BstNI) and KpnI restriction enzymes was suggested to distinguish between highly-pathogenic E. bovis, E. zuernii and E. alabamensis; the results of the second step were interpreted using simulation software. Those potentially useful diagnostic tool should be used empirically; a field of research clearly requiring further examination. Consequently, additional investigation will be conducted by us as soon as the additional biological material is available.
Presented technique has the potential for rapid, clear differentiation of the highly-pathogenic species of bovine eimerians in both, faecal samples and the intestine tissue of the strictly-protected European bison. Traditional approach requires determination of Eimeria species after time-consuming sporulation of the faecal oocysts (Duszynski and Wilber, 1997). The advantage of the method includes the possibility of rapid monitoring of the health status of free-roaming and captive herds of the European bison both intravitally (faeces) and post-mortem (the large intestine tissue). The obtained results revealed that the most of the individuals was infected with Eimeria spp., although their OPGs are negative. Thus, the presented technique can be significantly useful to control the levels of infections in dewormed, captive herds on an example of any dead individual. The PCR-RFLP was successfully adapted as a rapid, accurate technique to detect infections of various protozoans of cattle, including Trypanosoma spp. (Geysen et al., 2003) and Cryptosporidium spp. (Patel et al., 1999). The developed PCR-RFLP method allows readily identification of three the most pathogenic species of bovine Eimeria on the basis of a presence of the corresponding band on the agarose gel after electrophoretic separation of the DNA fragments without a need of their sequencing. Another useful method for pathogenic determination of E. bovis is a multiplex PCR technique with the use of primers developed by us previously (Pyziel et al., 2019). In contrast to multiplex PCR, suggested PCR-RFLP can be used more routinely with the use of commercial restriction enzymes. To conclude, the PCR-RFLP is simple, rapid and inexpensive method, and can be routinely used is the regional laboratories, as observed previously to determine various pathogens (Demkin and Zimin, 2005).
In this study animals older than one year of age were shedding oocysts of two or three species of Eimeria, whereas more species were noted in the faeces of younger individuals. Moreover, E. bovis infection was predominant regardless the age of examined animal. These observations are consistence with the previous study (Pyziel et al., 2011, 2014). According to Pyziel et al. (2014), grater eimerian species richness was presented among calves in comparison with older bison. Additionally, the same authors noted that multiple-species infections of examined individuals consisted of two to seven species of Eimeria, usually including E. bovis.
Histopathology studies indicated the presence of E. bovis infection in the large intestine, based on the size and location of the majority of identified oocysts. E.bovis was also found to demonstrate the highest OPG scores in faeces. The dominance of the potentially pathogenic E. bovis in the intestine was confirmed by the results of the sequencing of the PCR products. In addition, four individuals found to be negative for coprological investigation were confirmed positive for E. bovis by molecular analysis. Further confirmation was provided by the presence of pathological lesions such as congestion, hemorrhage, destruction of the intestinal glands and mucosa in the colon and cecum (Hammond et al., 1964). Despite this, it is possible that the dissected animals did not manifest any disease symptoms intravitally. According to De Noronha et al. (2009) and Arslan and Tuzer (1998), cases of clinical coccidiosis among cattle and water buffalo are associated with high OPG values of over 5000. Our findings confirm that the developmental stages of the protozoan E. bovis were present in the intestinal mucosa of the examined European bison.
Conflicts of interest
X. This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue.
Acknowledgments
The authors would like to express their gratitude to Mr. Ed Lowczowski for English correction of the text.
References
- Arslan O.M., Tuzer E. Prevalence of bovine eimeriosis in Thracia, Turkey. Turk. J. Vet. Anim. Sci. 1998;22:161–164. [Google Scholar]
- Davis L.R., Bowman G.W., Boughton D.C. The endogenous development of Eimeria alabamensis Christensen, 1941, an intranuclear coccidium of cattle. J. Eukaryot. Microbiol. 1957;4:219–225. [Google Scholar]
- De Noronha A.C.F., Starke-Buzetti W.A., Duszynski D.W. Eimeria spp. in Brazilian water buffalo. J. Parasitol. 2009;95:231–234. doi: 10.1645/GE-1605.1. [DOI] [PubMed] [Google Scholar]
- Demkin V., Zimin A.L. A new amplification target for PCR-RFLP detection and identification of Chlamydiaceae species. Arch. Microbiol. 2005;183:169–175. doi: 10.1007/s00203-004-0757-2. [DOI] [PubMed] [Google Scholar]
- Dubey J.P. A review of coccidiosis in water buffaloes (Bubalus bubalis) Vet. Parasitol. 2018;256:50–57. doi: 10.1016/j.vetpar.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Dubey J.P., Wouda W., Muskens J. Fatal intestinal coccidiosis in a three-week-old buffalo calf (Bubalus bubalus) J. Parasitol. 2008;94:1289–1294. doi: 10.1645/GE-1660.1. [DOI] [PubMed] [Google Scholar]
- Duszynski D.W., Wilber P.G. A guideline for the preparation of species description in the Eimeriidae. J. Parasitol. 1997;83:333–336. [PubMed] [Google Scholar]
- Fitzgerald P.R. The economic impact of coccidiosis in domestic animals. Adv. Vet. Sci. Comp. Med. 1980;24:121–143. [PubMed] [Google Scholar]
- Geysen D., Delespaux V., Geerts S. PCR-RFLP approach using the Ssu-rDNA sequence amplification as an easy method for species-specific diagnosis of Trypanosoma species in cattle. Vet. Parasitol. 2003;110:171–181. doi: 10.1016/s0304-4017(02)00313-8. [DOI] [PubMed] [Google Scholar]
- Hammond D.M., Clark W.N., Miner M.L. Endogenous phase of the life cycle of Eimeria auburnensis in calves. J. Parasitol. 1961;47:591–596. [PubMed] [Google Scholar]
- Hammond D.M., Sayin F., Miner M.L. Kalbern. Berl. Munch. Tierarztl. Vol. 76. 1963. Über den Entwicklungszyklus und die Pathogenität von Eimeria ellipsoidalis Becker und Frye, 1929; pp. 331–332. (In German) [Google Scholar]
- Hammond D.M., Bowman G.W., Davis L.R., Simms B.T. The endogenous phase of the life cycle of Eimeria bovis. J. Parasitol. 1964;32:409–427. [PubMed] [Google Scholar]
- Hnida J.A., Duszynski D.W. Taxonomy and systematics of some Eimeria species of Murid rodents as determined by the ITS1 region of the ribosomal gene complex. Parasitology. 1999;1119 doi: 10.1017/s0031182099004849. 349-257. [DOI] [PubMed] [Google Scholar]
- IUCN 2019. https://www.iucnredlist.org
- Jolley W.R., Bardsley K.D. Ruminant coccidiosis. Vet. Clin. North Am. Food Prod. Anim. 2006;22:613–621. doi: 10.1016/j.cvfa.2006.07.004. [DOI] [PubMed] [Google Scholar]
- McDougald L.R. Attempted cross-transmission of coccidia between sheep and goats and description of Eimeria ovinoidalis sp. J. Protozool. 1979;26:109–113. doi: 10.1111/j.1550-7408.1979.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Nicholas K.B., Nicholas H.B., JR., Deerfield D.W. GeneDoc: analysis and visualisation of genetic variation. Embnet News. 1997;4:1–4. [Google Scholar]
- Patel S., Pedraza-Díaz S., McLauchlin J. The identification of Cryptosporidium species and Cryptosporidium parvum directly from the whole faeces by analysis of a multiplex PCR of the 18S rRNA gene and by PCR/RFLP of the Cryptosporidium outer wall protein (COWP) gene. Int. J. Parasitol. 1999;29:1241–1247. doi: 10.1016/s0020-7519(99)00079-x. [DOI] [PubMed] [Google Scholar]
- Pellérdy L., editor. Coccidia and Coccidiosis. Akademia Kiado; Budapest: 1974. pp. 723–760. [Google Scholar]
- Penzhorn B.L., Knapp S.E., Speer C.A. Enteric coccidia in free-ranging American bison (Bison bison) in Montana. J. Wildl. Dis. 1994;30:267–269. doi: 10.7589/0090-3558-30.2.267. [DOI] [PubMed] [Google Scholar]
- Pyziel A.M., Demiaszkiewicz A.W. Coccidia (Apicomplexa: Eimeriidae) of elk (Alces alces) in Poland. Parasitol. Res. 2013;112:2083–2085. doi: 10.1007/s00436-012-3262-6. [DOI] [PubMed] [Google Scholar]
- Pyziel A.M., Demiaszkiewicz A.W. Observations on sporulation of Eimeria bovis (Apicomplexa: Eimeriidae) from the European bison Bison bonasus:effect of themperature and potassium dichromate solution. Folia Parasitol. 2015;62 doi: 10.14411/fp.2015.020. 010. [DOI] [PubMed] [Google Scholar]
- Pyziel A.M., Kowalczyk R., Demiaszkiewicz A.W. The annual cycle of shedding Eimeria oocysts by European bison (Bison bonasus) in the Białowieża Primeval Forest, Poland. J. Parasitol. 2011;97:737–739. doi: 10.1645/GE-2567.1. [DOI] [PubMed] [Google Scholar]
- Pyziel A.M., Jóźwikowski M., Demiaszkiewicz A.W. Coccidia (Apicomplexa: Eimeriidae) of the lowland European bison Bison bonasus bonasus (L.) Vet. Parasitol. 2014;202:138–144. doi: 10.1016/j.vetpar.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Pyziel A.M., Demiaszkiewicz A.W., Klich D., Laskowski Z. A morphological and molecular comparison of Eimeria bovis-like oocysts (Apicomplexa: Eimeriidae) from European bison, Bison bonasus L., and cattle, Bos taurus L., and the development of two multiplex PCR assays for their identification. Vet. Parasitol. 2019 doi: 10.1016/j.vetpar.2019.08.011. (in press) [DOI] [PubMed] [Google Scholar]
- Sayin F. The sporulated oocysts of Eimeria ankarensis n. sp. and of other species of Eimeria of buffalo in Turkey and transmission of four species of Eimeria from buffalo to cow calves. Ankara Univ. Vet. Fak. 1969;15:282–300. [Google Scholar]
- Spano F., Putignani L., McLauchlin J., Casemore D.P., Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- Stockdale P.H.G. Proposed life cycle of Eimeria zuernii. Br. Vet. J. 1977;133:471–473. doi: 10.1016/s0007-1935(17)33987-8. [DOI] [PubMed] [Google Scholar]
- Stockdale P.H.G. The pathogenesis of the lesions produced by Eimeria zuernii in calves. Can. J. Comp. Med. 1977;41:338–344. [PMC free article] [PubMed] [Google Scholar]
- Su C., Zhang X., Dubey J.P. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int. J. Parasitol. 2006;36:841–848. doi: 10.1016/j.ijpara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Taylor M.A., Coop R.L., Wall R.L. Parasites of cattle. In: Taylor M.A., Coop R.L., Wall R.L., editors. Veterinary Parasitology. Blackwell Publishing; Oxford: 2007. pp. 69–73. [Google Scholar]
- Taylor M.A., Coop R.L., Wall R.L. The laboratory diagnosis of parasitism. In: Taylor M.A., Coop R.L., Wall R.L., editors. Veterinary Parasitology. Blackwell Publishing; Oxford: 2007. pp. 798–799. [Google Scholar]
- Zhao X., Duszynski D.W., Loker E.S. A simple method of DNA extraction for Eimeria species. J. Microbiol. Methods. 2001;44:131–137. doi: 10.1016/s0167-7012(00)00249-9. [DOI] [PubMed] [Google Scholar]