Abstract
Tumor cells demonstrate substantial plasticity in their genotypic and phenotypic characteristics. Epithelial-mesenchymal plasticity (EMP) can be characterized into dynamic intermediate states and can be orchestrated by many factors, either intercellularly via epigenetic reprograming, or extracellularly via growth factors, inflammation and/or hypoxia generated by the tumor stromal microenvironment. EMP has the capability to alter phenotype and produce heterogeneity, and thus by changing the whole cancer landscape can attenuate oncogenic signaling networks, invoke anti-apoptotic features, defend against chemotherapeutics and reprogram angiogenic and immune recognition functions. We discuss here the role of phenotypic plasticity in tumor initiation, progression and metastasis and provide an update of the modalities utilized for the molecular characterization of the EMT states and attributes of cellular behavior, including cellular metabolism, in the context of EMP. We also summarize recent findings in dynamic EMP studies that provide new insights into the phenotypic plasticity of EMP flux in cancer and propose therapeutic strategies to impede the metastatic outgrowth of phenotypically heterogeneous tumors.
Keywords: EMT, EMP, stem cell, CTCs, hybrid EMT states, metastasis, metabolism, tumor cell heterogeneity
Introduction (EMT-MET)
Epithelial–mesenchymal transition (EMT), in which epithelial cells undergo dynamic cellular transition from a sessile epithelial state to a motile mesenchymal state allowing the formation of new tissues, is considered one of the pivotal processes during embryogenesis and organogenesis (Chaffer et al., 2007; Yang and Weinberg, 2008). The process of EMT (classified as three different subtypes) has been implicated in a broad range of normal and pathophysiological processes from development, wound healing and tissue regeneration (type I), to organ fibrosis (type 2), and cancer progression (type 3) (Kalluri and Weinberg, 2009). During cancer progression, it is postulated that epithelial-derived carcinoma cells undergo a reversible, trans-differentiation process with changes in cell–cell adhesion and polarity, cytoskeletal remodeling, migratory and invasive enhancement, and dissemination into secondary organs via local invasion, intravasation and transfer through the blood stream and lymphatics (Polyak and Weinberg, 2009). In addition to cellular migration during metastasis, EMT also influences resistance to anoikis and apoptosis, blocks senescence, enhances survival, facilitates genomic instability, causes cancer stem cell (CSC) activity, alters metabolism, and induces drug resistance and immune suppression (Przybylo and Radisky, 2007; Ansieau et al., 2008; Gal et al., 2008; Kumar et al., 2011; Huang et al., 2013; Dongre et al., 2017; Lee et al., 2018; Redfern et al., 2018).
After invasion and spread, cancer recurrence at the metastatic site is thought to require the reverse process, termed mesenchymal to epithelial transition (MET) (Chaffer et al., 2007; Hugo et al., 2007; Brabletz, 2012). The reversal of EMT, referred to as MET, has received less attention than EMT in the establishment of metastasis. Microenvironmental cues are considered a major deterministic factor for the reversion of the migratory mesenchymal neoplastic cells and the subsequent development of macrometastases. However, the re-expression of E-cadherin, inhibition of SNAIL, and β-catenin sequestration have provided evidence of MET in liver metastasis from MDA-MB-231 (Chao et al., 2010; Brabletz, 2012), as has the anti-metastatic effects of sustained pro-mesenchymal signals (Ocana et al., 2012; Tsai et al., 2012). The concept of MET in metastasis is refuted in some of the cancer recurrence studies as no definitive proof of a MET requirement was obtained in the MMTV-PyMT genetically engineered mouse model (GEMMs) of metastatic breast cancer or in the KPC GEMM for metastatic pancreatic cancer (Fischer et al., 2015; Zheng et al., 2015). Nevertheless, recent data on EMP phenomena during metastatic cancer colonization is emerging (Chao et al., 2010; Rhim et al., 2012; Nieto, 2013; Beerling et al., 2016; Pastushenko et al., 2018) and could be of particular interest in breast and pancreatic carcinomas where EMT is considered an early event in tumorigenesis (Hüsemann et al., 2008; Rhim et al., 2012). Moreover, other studies have reported at least partial involvement of EMP in the breast model (Ye et al., 2015) and Zeb1 has been shown to contribute to metastasis in the pancreatic model (Krebs et al., 2017).
Considerably less information is available on the key intrinsic factors that drive MET in vivo and in vitro, while the drivers and transcriptional mediators of EMT are quite comprehensively documented (Stemmler et al., 2019). Bone morphogenetic protein 7 (BMP7) is reported to trigger MET in renal fibroblasts during kidney development (Zeisberg et al., 2005), and also in breast cancer cells, reducing their capability to form bone metastases (Buijs et al., 2007). Protein Kinase A was recently identified as an inducer of MET in human mammary epithelial cells (Pattabiraman et al., 2016). The role of Notch4 in melanoma cells to induce MET and suppress malignancy in mice has also been reported (Bonyadi Rad et al., 2016). The course of epigenetic reprograming is also supporting EMT and MET acquisition (Tamura et al., 2000). Reversible epigenetic changes acquired during EMT underpin the emergence of self-renewal and chemo-refractory stem cell-like features, which can revert to the MET phenotype for establishing metastasis (Voulgari and Pintzas, 2009; Sharma et al., 2010). Here, we discuss the role and the regulatory mechanisms of EMP, with the focus on recent emerging concepts that highlights the bidirectional dynamics of this phenomenon and the hybrid intermediate states. We also provide a brief overview of various techniques/modalities employed to analyze EMP in cancer. Understanding the phenotypic plasticity will provide insights for various therapeutic strategies that can be implemented to prevent/restrict spread of cancer by metastasis.
Significance of EMP and Hybrid EMT States
Epithelial–mesenchymal transition, however, is not a two-step event through which cancer cells lose epithelial markers and acquire mesenchymal traits between two rigid phenotypes. Rather, studies performed within the last decade increasingly show that cancer cells sequentially acquire mesenchymal traits, but don’t automatically dissipate all of their previously expressed epithelial features (Tam and Weinberg, 2013; Aiello and Kang, 2019). The term “epithelial–mesenchymal plasticity” (EMP) is more favored recently as compared to EMT-MET (Bhatia et al., 2017; Williams et al., 2019). The multiple signal transduction cascade for EMT-MET programing results in dynamic and intermediate transitional states wherein, the cancer cells can reside in all three EMP phenotypes (epithelial, mesenchymal and hybrid phenotype). EMP reflects the bidirectional flux often in a continuum across the full spectrum (Lee et al., 2006; Oltean et al., 2006). Thus, a full spectrum of EMP endows the formation of a new carcinomatous tumor at distant organ sites with similar histopathology as observed in primary tumor (Gunasinghe et al., 2012).
Hybrid epithelial-mesenchymal features of carcinoma cells have indeed been observed in various invasive carcinoma model systems (Lee et al., 2006; Klymkowsky and Savagner, 2009), in which individual cells co-express markers of both epithelial and mesenchymal lineages, and circulating tumor cells (CTCs) in particular have been shown to exhibit a spectrum of EMP states (Armstrong et al., 2011; Yu et al., 2014; Khoo et al., 2015; Bourcy et al., 2016); reviewed in McInnes et al. (2015); Hassan et al. (2020). The hybrid EMP state seen in carcinomas and CTCs, in which individual cells co-express markers of both epithelial and mesenchymal lineages, is predicted to have the highest tumourigenicity and metastatic potential (Lee et al., 2006; Klymkowsky and Savagner, 2009; Jolly et al., 2016; Kroger et al., 2019; Pastushenko and Blanpain, 2019). An emerging challenge is also to decipher correctly the contribution that intermediate states of the EMT spectrum make to tumor evolution for therapeutic interventions.
Extrinsic and Intrinsic Mechanisms and Regulators Involved in Plasticity
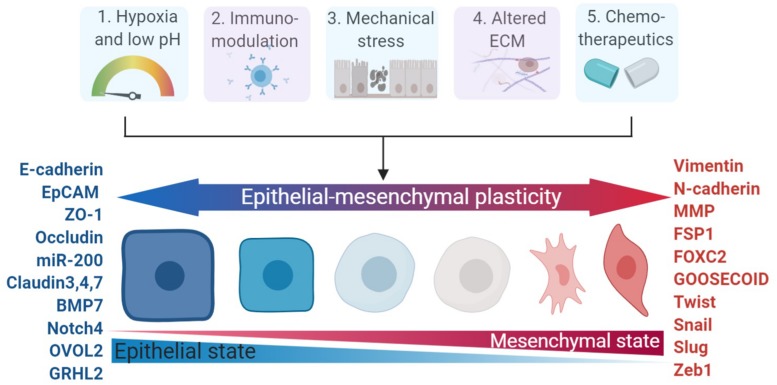
The crosstalk mediated by autocrine and/or paracrine factors secreted by cancer cells and tumor stroma has been widely proven to occur via extracellular mediators of EMT (Scheel et al., 2011). A host of extracellular mediators secreted by tumor stromal cells are already proven to elicit EMT induction. Examples of validated extracellular mediators as EMT inducers include TGF-β (Buonato et al., 2015), EGF (Hugo et al., 2009), FGF (Kurimoto et al., 2016), PDGF (Devarajan et al., 2012), HGF (Suarez-Causado et al., 2015), IGF (Wang et al., 2016), Interleukin-6 (IL-6) (Miao et al., 2014), WNT (Ochoa-Hernandez et al., 2012), Hedgehog (Yoo et al., 2011), and Notch (Yuan et al., 2014). Other inducers of EMT include collagen types I and III, matrix metalloproteinases-2 (MMP-2), MMP-3, MMP-9, and MMP14/MT1-MMP (Thiery et al., 2009). YAP and TAZ are also emerging as key modulators in inducing plasticity and skin cancer initiation (Moroishi et al., 2015; Debaugnies et al., 2018). EMT of tumor cells can also be induced by various stimuli from the tumor microenvironment (Marcucci et al., 2014); Fabrizio Marcucci and his colleagues proposed five major classes of these stimuli in 2016 (Marcucci et al., 2016): hypoxia and low pH, innate and adaptive immune responses, mechanical stress, altered ECM and treatment with chemotherapeutics (Figure 1).
FIGURE 1.
Major categories of EMP stimuli and markers involved in EMP. The dynamics of the epithelial – mesenchymal spectrum can be induced by five major stimulii (hypoxia, immuno-modulators, mechanical stress, altered ECM, and chemotherapeutics), which involve changes in various functional and morphological states and enlisted markers across the spectrum of epithelial–mesenchymal plasticity. ECM, extracellular matrix.
Interestingly, hypoxic features in the tumor microenvironment can stimulate EMT as a downstream consequence of upregulated hypoxia-inducible factor 1α (HIF1α) (Wong et al., 2012). Apart from tumor microenvironment stimuli for EMT induction, stimulus-independent activation of signaling pathways, caused by mutations or epigenetic modifications leading to overexpression of certain pathway components, can also trigger EMT (Wallin et al., 2012; Serrano-Gomez et al., 2016). Gain-of-function mutations in P53 has been reported to induce EMT via modulation of miR-130b-Zeb1 axis (Dong P. et al., 2013).
Epigenetic modifications can also cause a shift of epithelial to mesenchymal state; for example, aberrant DNA CpG island methylation correlated with the repression of the miR-200 cluster, which promotes EMT and contributes to tumor progression (Vrba et al., 2010). LSD1-dependent genome-scale epigenetic reprograming was also observed during EMT (McDonald et al., 2011; Tang et al., 2013; Boulding et al., 2019). Various other chromatin regulators (e.g., DNMT1, KDM6B, PHF8, EZH2, and HDAC) are also reported to regulate EMT, genomic stability and metastasis (Suvà et al., 2013; Lu and Kang, 2019). Apart from epigenetics and mutations, EMT can also be modulated at transcriptional, post-transcriptional, translational and post-translational levels. The intrinsic gene network regulators, via alternate splice isoforms of ESRP1/2, microRNAs and long non-coding RNAs, also acts as other distinctive mechanisms to induce EMT (Aiello et al., 2018; Aiello and Kang, 2019). It has been postulated that during chemotherapy regimens, undifferentiated cancer cells also commence EMT, causing therapy resistance, CSC-like behavior, and a high propensity for metastasize. Tumor relapse after drug treatment cessation is due to persistence of disseminated CSC with mesenchymal features (Witta et al., 2006). Redfern et al. (2018) have also recently shown shorter overall survival times in patients treated with EMT-inducing agents compared to agents known to inhibit EMT.
The expression changes of various key molecular markers during EMT, are represented in Figure 1 (Christiansen and Rajasekaran, 2006; Pastushenko and Blanpain, 2019). The transition of epithelial cells to a more mesenchymal state is also characterized by reduced intracellular adhesion through the downregulation of E-cadherin (CDH1) and EpCAM, and gain of mesenchymal markers such as N-cadherin (CDH2), vimentin and FSP1/S100A4 (Francart et al., 2018). Repressors of E-cadherin can be divided into groups that modulate either directly or indirectly effects on gene transcription by binding to promoter sites. ZEBs, SNAIL1 and KLF8 repress expression by binding the E-cadherin promotor, thereby inactivating transcription, while E2.2, FOXC2, GOOSECOID, and TWIST repress E-cadherin transcription as indirect repressors (Peinado et al., 2004, 2007; Xu et al., 2019). These factors also share an elaborate interactome, in that SNAIL1 upregulates SNAIL2 and TWIST (Thuault et al., 2008; Smit et al., 2009), SNAIL1 and TWIST then induce ZEB1 and SNAIL2 (Casas et al., 2011; Dave et al., 2011), and SNAIL2 induces ZEB2 (Thuault et al., 2008). Although commonly serving as repressors of E-cadherin, these broader mechanisms also selectively modulate other programs involved in cell division, cell survival, and cell attachment, thereby resulting in a motile, invasive and resistant cell phenotype (Barrallo-Gimeno and Nieto, 2005).
Role of EMT in Tumor Initiation, Progression and Metastasis
Although much less studied than later tumor stages, a number of studies have made a connection between the linkage of EMT to stemness and tumor-initiating capacity (Mani et al., 2008; Morel et al., 2008). In some carcinoma cells, overexpression of EMT transcription factors (EMT-TFs) has been observed to drive and enhance tumorigenicity (Wellner et al., 2009), and in particular, EMT has been shown to cause avoidance of oncogene-induced senescence (Ansieau et al., 2008). In a mouse skin SCC model, low levels of TWIST was explicitly responsible for the tumor initiation process, whereas higher levels of TWIST induced EMT and tumor progression (Beck et al., 2015). In recent lineage tracing studies along with transcriptional and epigenomic profiling, Latil et al found disparities in the tumors generated from interfollicular epidermis (IFE) and hair follicle (HF) stem cells (Lgr5CreER). While IFE tumors showed a well-differentiated phenotype, tumors generated from HF stem cells displayed an EMT spectrum and increased metastatic potential (Latil et al., 2017).
The profound role of EMP in tumor progression and metastasis in vivo has remained a topic with various controversies (Brabletz et al., 2018; Williams et al., 2019). The number of mesenchymal cells observed in primary cancers in many xenograft studies had been observed to be less than 10%. Although the specific dissemination process of these cells is not yet well documented (Bhatia et al., 2019; Lourenco et al., 2020), enrichment of EMT in circulating tumor cells has supported a role for EMT in the initial steps of metastasis. Various studies have highlighted the role of key EMT TFs, such as Slug and Zeb1, in promoting metastasis of breast and colorectal cancer to liver and lung, respectively (Spaderna et al., 2008; Guo et al., 2012). Downregulation of TWIST expression in highly metastatic mammary carcinoma cells was found to inhibit their metastatic seeding ability in the lung (Yang et al., 2004). However, these studies are nuanced by observations that enforced overexpression or downregulation of EMT-TFs doesn’t recapitulate the dynamic spectrum of transitional and/or partial EMT states discovered in vivo (Pastushenko et al., 2018). Similarly, the studies from the genetic abrogation of Twist or Snail in mouse models of pancreatic adenocarcinoma and from EMT lineage tracing using Fsp1 and β-actin promoter in breast cancer mouse model have questioned the indispensability of full mesenchymal transition in the metastasis process (Fischer et al., 2015; Zheng et al., 2015). The conclusions of these studies have been subsequently refuted by other studies where genetic depletion of Zeb1 in the same pancreatic model resulted in strong suppression of metastasis. Therefore, caution is required while interpreting such results as the context of EMT and other compensatory mechanisms may significantly influence their role in promoting metastasis (Aiello et al., 2017; Ye et al., 2017). With the advent of cell fate mapping studies using intra-vital imaging, plasticity was revealed in mouse breast tumor cells from primary site to its re-epithelisation upon metastasis (Beerling et al., 2016). Several other studies have also reported the direct evidence of EMP under physiological conditions (Rhim et al., 2012; Chaffer et al., 2013; Ye et al., 2015). Multiple tumor subpopulations screened from mammary and skin tumors suggested that tumor cells with hybrid phenotypes were more efficient in dissemination and metastasis (Pastushenko et al., 2018; Thompson and Nagaraj, 2018; Pastushenko and Blanpain, 2019; Rios et al., 2019). Similar, other relevant studies are also emerging to suggest that cancer cells mostly transition between epithelial/mesenchymal and hybrid intermediate states, but rarely undergo complete EMT during metastasis (Kroger et al., 2019).
EMP Analysis of Circulating Tumor Cells (CTCs)
Generation of CTCs is regarded as a consequential effect of the multi-step processes that constitute the metastasic cascade (Lambert et al., 2017), and have become a particularly rich source of evidence and information regarding the role of EMP in cancer progression. Understanding the biology and characteristics of CTCs can provide important insights into the molecular and cellular requirements of cancer cells during metastatic spread. Observations of enriched levels of mesenchymal genes (e.g., N-cadherin, vimentin and Twist) and reduced expression of epithelial genes (e.g., E-cadherin, EpCAM and CK8/18/19) has been reported in the CTCs relative to cells in the tumors of origin in the breast cancer patients (Yu et al., 2013; Wang et al., 2018). Although many CTCs exhibit a mesenchymally enriched phenotype, some researchers have revealed that a small population of CTCs co-expressed both epithelial and mesenchymal (E/M) hybrid phenotype traits, which likely promoted cell migration, cell invasion and cell survival capabilities (Lecharpentier et al., 2011; Milano et al., 2018). Hence hybrid CTCs may be more metastatic than mesenchymal CTCs.
High numbers of CTCs in blood is significantly associated with poor prognosis in several carcinoma types, such as prostate cancer (Wang et al., 2011), breast cancer (Bulfoni et al., 2016), pancreatic cancer (Han et al., 2014), lung cancer (Naito et al., 2012), and increasingly these have taken account of CTC phenotypes (Tachtsidis et al., 2016). Pan et al. (2019) conducted a correlation study between CTC phenotypes and clinicopathological features of early cervical cancer, finding lower CTC counts in stage I patients than stage II patients with pelvic lymph node metastasis, but also that mesenchymal CTCs expressing vimentin and TWIST were more commonly found in the latter. Consistently, Markiewicz et al. (2014) selectively found of VIM, SNAI1, and UPAR expression in mesenchymal CTCs derived from breast cancer patient with lymph nodes metastases. Due to the low number of CTCs in blood, the greatest challenge in studying CTCs is the detection and isolation of these cells from patients’ blood (Kowalik et al., 2017). Molecular profiling of EMT markers in CTCs has been used to establish tools to isolate and classify CTCs. RNA in situ hybridization (RNA-ISH) is a detection method that employs specific probes targeting different epithelial and mesenchymal genes to detect multiple transcripts simultaneously (Lopez-Munoz and Mendez-Montes, 2013). An enhanced RNA-ISH-based detection system, CTCscope, was innovated to detect eight epithelial markers and three EMT markers (Payne et al., 2012), and has been employed successfully in the landmark breast cancer CTC study (Yu et al., 2013; Wang et al., 2018). The FDA-approved CELLSEARCH® system (Menarini-Silicon Biosystems, Inc.), which immunocaptures EpCAM-expressing CTCs for patient prognosis (Riethdorf et al., 2007), is intrinsically biased toward predominantly epithelial CTCs. However, recent CTC studies have employed microfluidic devices to capture and isolate CTCs according to their size and deformability, which allows for better coverage of different phenotypic states (Lemaire et al., 2018; Ribeiro-Samy et al., 2019).
Although the devices used to isolate CTCs have improved the quality and quantity assessment of CTCs, there are still limitations when studying CTCs. Over the past few years, use of the revolutionary single- cell RNA sequencing (scRNA-seq) has emerged to assess genome-wide expression profiles of isolated CTC populations and CTC clusters. Aceto et al. (2014) conducted scRNA-seq on endogenous CTCs generated using tumor xenografts of LM2 variant of MDA-MB-231 human breast cancer cells, showing that CTC clusters are oligoclonal and highly metastatic compared to single CTCs. It was found that the cell junction protein plakoglobin (JUP) mediates cell cluster formation, enhancing the metastatic potential of CTCs. Ting et al. (2014) performed scRNA-seq analysis on CTCs in a mouse pancreatic cancer model, and revealed a universal loss of the epithelial markers E-cadherin (Cdh1) and Mucin-1 (Muc1) across all CTCs compared with the primary xenograft tumors. Hugo et al. (2017) showed that both in vitro and in vivo knockdown of Cdh1 in MDA- MB-468 breast cancer cells reduced proliferation, and this was also reported by Padmanaban et al. (2019), who further indicated that the loss of Cdh1 increased invasion capacity while reducing cell survival, CTC number and metastasis spread in the breast cancer.
The interconnection between CTC, EMT and CSC has been actively studied and reported to harbor important mechanisms underlying tumourigenicity (Agnoletto et al., 2019). EMT generates stem-like cells (Mani et al., 2008) and tumor cells that features both EMT and stem-like characters are better equipped to induce metastasis (May et al., 2011; Barriere et al., 2014), while some CTCs have dynamic cellular plasticity expressing EMT traits and stemnicity (Alonso-Alconada et al., 2014). A minor fraction of EMT hybrid phenotype CTCs have been shown to exhibit stem-like features, and these cells have been shown to promote collective migration Kaigorodova et al. (2017); Quan et al. (2020), as well as enhanced survivability and chemoresistant (Papadaki et al., 2019). Papadaki et al. (2019) modeled four CTC subpopulations based on the co-expression of three different markers; cytokeratin (epithelial marker), ALDH1 (stemness marker) and TWIST1 (partial EMT marker), and revealed that CTCs co-expressing cytokeratin, high levels of ALDH1, and nuclear TWIST1 (CSC+/partial-EMT+) were enriched after the first-line chemotherapy, implying that they were the most chemoresistant subpopulation, and had a favored prognostic value in patients with metastatic breast cancer. Another study has showed that EpCAMhigh CTCs were significantly associated with poor prognosis compared to EpCAMlow CTCs in patients with breast and prostate cancer (de Wit et al., 2018), however the level of mesenchymal co-expression was not measured. Ting et al. (2014) showed that the stem cell markers Aldh1a1 and Aldh1a2 were enriched in pancreatic CTCs, and they also demonstrated that Igfbp5 (a transport protein of epithelial stroma) and SPARC (a collagen-binding glycoprotein related to ECM reorganization) were highly expressed in the CTCs. Although they stated that there was no intrinsic correlation between EMP state and stemness in their CTCs, other reports have shown expression of these genes were associated with Cdh1 reduction (Bradshaw, 2009; Sureshbabu et al., 2012). There still remains a lack of evidence to fully elucidate the mechanistic relationship between CTCs, EMT and CSCs through the association of their existing markers with functional features, although it seems clear that they represent only a small fraction of CTCs.
Understanding Dynamics of EMT
In the last two decades, many new concepts and findings have flourished around the dynamics of EMP. The dynamics of the stochastic state transitions, which allows cancer cells to switch between phenotypic states, is not yet explicitly described. However, novel concepts of dynamic equilibrium, asymmetrical dynamics of EMT-MET conversions, bet hedging, and hysteresis/cellular memory of cancer cells have heralded a deeper understanding of the phenotypic heterogeneity that cancer cells endow/possess (Jolly and Celià-Terrassa, 2019). This intrinsic mechanism of bi-directional transitions between epithelial (differentiated) and mesenchymal (stem-like) states is reported in different kinds of cancer (Polyak and Weinberg, 2009; Chaffer et al., 2011; Gupta et al., 2011; Yang et al., 2012; Ruscetti et al., 2016; Bhatia et al., 2019). Sequencing of breast cancer stem cell populations also indicates a dynamic conversion between differentiation states in vivo (Klevebring et al., 2014). A phenotypically stable equilibrium was observed in breast cancer cell lines, differentially segregated across cell state proportions (Gupta et al., 2011; Bhatia et al., 2019). DNA barcoding and subsequently high-throughput sequencing of breast cancer cell clones had also been employed to quantify the extent of intrinsic phenotypic plasticity exhibiting epithelial or mesenchymal phenotypes (Mathis et al., 2017; Rios et al., 2019). Various mechanism-based mathematical modeling and data-based statistical modeling approaches have been developed in an attempt to uncover the presence of these metastable states (Lu et al., 2013; Jolly et al., 2016; Jolly and Celià-Terrassa, 2019).
The presence of “multiple attractor states” based on Waddington landscape and intrinsic cellular variability also contributes to phenotypic plasticity (Huang et al., 2009; Ferrell, 2012; Li et al., 2016). The studies pertaining to EMT and MET reversion have also explained explicitly that the dynamics achieved for its reversion back may not follow the same path. For example, studies with a Snail-inducible expression system in prostate cancer cells has identified metabolic plasticity and asymmetrical dynamics during their EMT-MET cycle (Stylianou et al., 2019). Other studies, where re-expression of significant epithelial markers such as E-cadherin, OVOL2 and GRHL2 after their knockout may not obtain the same spectrum of reversion also suggests asymmetrical dynamics (Qi et al., 2018; Chung et al., 2019; Jolly et al., 2019). The concept of bet hedging had been observed in bacterial persistence under different environmental stimulations by generating mutation-independent phenotypic heterogeneity (Veening et al., 2008). This pre-existing phenotypic heterogeneity is thought to be exploited by cancer cells in generating drug-persistence cells via non-genetic mechanism, which might lead to anti-drug resilience in clinical scenarios (Jolly et al., 2018). The property of hysteresis and “cellular memory” allows cells from the same clonal population to respond differently to the same strength and duration of a signal. The differential response again can be attributed to the cellular placement across different “attractor states” or the possibility of history of input stimuli (Chang et al., 2006; Jolly and Celià-Terrassa, 2019). The possibility of EMT occurring via non-linear hysteretic mode had been recently observed to result in different dynamics and increased metastasis in a breast cancer model (Celià-Terrassa et al., 2018). Thus, these dynamics impart a further layer of intricacies in understanding the causes and reasons of non-genetic heterogeneity in cancer in regard to phenotypic plasticity. An integrative understanding of the approaches to block this phenotypic plasticity and EMP dynamics could further aid in combating cancer resistance.
Implications of Metabolic Plasticity and EMP
During the processes of EMP, there are numerous adaptations, not only in cell morphology and epigenetic changes, but also in metabolism (Cha et al., 2015). Among them, glucose and lipid metabolism alterations are crucial for the EMT induction (Kondaveeti et al., 2015; Sánchez-Martínez et al., 2015; Morandi et al., 2017; Kang et al., 2019). In terms of carbohydrate metabolism, it is well known that cancer cells prefer to reply on the glycolysis to generate ATP instead of oxidative phosphoruylation (OXPHOS), even under the well-oxygenated conditions, according to the Warburg effect (Warburg, 1956). However, apart from the Warburg effect, other glucose metabolic pathway adaptations have been observed during the last decade. When cancer cells undergo an EMP process, their metabolism will reprogram from aerobic glycolysis for proliferation to EMT-like metabolism to meet the increased energy needs. Both enhanced glucose and lipid uptake and increased glycolytic mediated biosynthesis and lipid synthesis are the characteristics of EMT-like metabolism. The correlation between metabolism and EMP is dynamic. EMP-associated genetic changes can stimulate metabolic adaptations, while the higher metabolic rate can support and facilitate the EMP process.
A number of studies illustrate the EMT-associated metabolic changes and their implications. According to the research of Dong et al., up-regulation of the EMT-driving transcription factor Snail-1 in basal-like breast cancer cells leads to the formation of a Snail-G9a-Dnmt1 complex to silence the expression of fructose-1,6-bisphosphatase (FBP1), which is an important enzyme of gluconeogenesis (Dong C. et al., 2013). The loss of the FBP1 caused an increase in glucose uptake for ATP production and glycolytic mediated biosynthesis, like the pentose phosphate pathway (PPP), serine and glycerol-3-phosphate. The reprogramed metabolism offers enough energy to fuel the invasion and metastasis processes.
For lipid metabolism, higher expression levels of lipid synthesis enzymes such as ATP-citrate lyase (ACLY), stearoyl-CoA desaturase (SCD), fatty acid synthase (FASN) and HMG-CoA reductase, have been detected in more aggressive tumor cells (Sánchez-Martínez et al., 2015). Jing et al. reported that overexpression of these proteins in association with mutated p53 in mostly mesenchymal cancer cells, along with aberrant expression of sterol regulatory element-binding proteins (SREBPs) (Hu et al., 2013). In normal tissue, wild type p53 can inhibit the expression of SREBP-1c, a transcription factor of FASN and ACLY (Horton et al., 2002), while the mutated p53 loses this capacity. Moreover, the mutated p53 can bind with SREBP-2 to enhance the cholesterol biosynthesis (Freed-Pastor et al., 2012). Thus, mutated p53 significantly upregulates both fatty acid (FA) and cholesterol levels in cancer cells, which generate more membrane lipid rafts to support cell motility during the EMT process. High levels of SREBP1 can also induce EMT, via recruiting a SNAIL1/HDAC1/2 complex to stop E-cadherin mRNA expression (Zhang et al., 2019). Chen et al., has proposed that drugs targeting SREBPs could suppress cancer cell metastasis (Chen et al., 2018).
Growth factors from the tumor microenvironment can also reprogram cancer cells from the Warburg-like metabolism to EMT-like metabolism. Activated PI3K/AKT/mTOR signaling due to growth factor stimulation can enhance the uptake of glucose and lipid, as well as the synthesis of FA and protein (Chen et al., 2018). The study of EMP relative metabolism changes can offer a promising target for cancer therapy.
Current Modalities to Investigate Plasticity
Many techniques recently employed in the field of cancer cellular plasticity have corroborated not only the epithelial and mesenchymal phenotypic states, but also the spectrum of intermediate and hybrid E/M states (Pastushenko et al., 2018; Karacosta et al., 2019). The molecular approaches widely used in the cancer EMT field are broadly divided into two categories: in vitro based molecular and functional assays and in vivo based cancer models. The in vitro assays routinely performed in EMP studies involve various molecular and functional assays. Molecular assays, using FACS and immunocytochemistry staining with microscopy analysis, relies on various validated EMP markers that are used to delineate the phenotypic state of cells (Celià-Terrassa et al., 2018; Pastushenko et al., 2018; Risom et al., 2018; Bhatia et al., 2019). Microscopy based snap-shot and real time analysis in conjunction with quantitative assessment is an imperative technique. These optic techniques are widely employed to study the cellular localization of various molecular markers, such as E-cadherin presence at the cell junctions, and also the subtle dynamic changes of various markers in the absence or presence of various stimuli or inducers can be studied (Hirata et al., 2014; Labernadie et al., 2017; Liu et al., 2018). Microscopy approaches are also well integrated in various functional assays, such as in vitro wound closure, Transwell migration studies performed in the presence or absence of ECM, quantification of single cell migration and invasion studies in culture medium, spheroid assessment and co-culture assays with cancer associated fibroblasts or endothelial cells (Kramer et al., 2013; Tanner and Gottesman, 2015; Mitchell and O’Neill, 2016; Klymenko et al., 2017; Reynolds et al., 2017; Mason et al., 2019). Other in vitro assessment also include “soft agar assay” for anchorage independent growth studies, “ECM degradation assays” to measure MMP and other protease activity, and “trans-epithelial resistance” assays to study monolayer integrity and permeability (Narai et al., 1997; Anderl et al., 2012; Borowicz et al., 2014). In studies relevant to single cell colonization, plasticity generated from single cell clonal culture is also examined for differences in migration, invasion and chemoresistance assays, which can be extrapolated to the metastatic cascade (Kramer et al., 2013; Harner-Foreman et al., 2017; Bhatia et al., 2019). While in vitro studies are important to study cellular behavior in context of phenotypic plasticity and tumoural non-genetic heterogeneity, these routinely performed assays have the drawback of not presenting the whole landscape of cancer and the real EMP spectrum, where cancer cells are infiltrated with stromal and immune microenvironment.
Researchers in the field of EMP have employed various animal models, including as C. elegans, Drosophila Melanogaster, chick embryos, zebrafish and mice to study the in vivo dynamics of phenotypic plasticity in developmental EMT and cancer EMP (Jimenez et al., 2016; Gómez-Cuadrado et al., 2017; Nieto, 2018; Stuelten et al., 2018; Campbell et al., 2019). Genetically engineered mouse models and patient-derived xenografts (PDXs) have been observed to recapitulate metastatic and organ homing properties similar to clinical specimens (Sikandar et al., 2017). Orthotopic implantation strategies, such as inoculation into the mammary fat pad, has also improved the recapitulation of the breast cancer in mice (Proia et al., 2011). In conjunction with intravital imaging and fluorophore chemistry, various Cre-Lox lineage tracing approaches have been employed in cell lines, and in injected mouse and zebrafish models, to delineate EMP status of the cells at primary and metastatic sites, and also of encaptured CTCs (Lourenco et al., 2020). These reporter tags are valuable in identification of CTCs and in scenarios of low numbers of cells seeding at secondary niches during metastasis (Zheng et al., 2015; Sikandar et al., 2017). The inducible system utilized for Twist1 induction or deletion at different stages of skin carcinogenesis allowed flexibility in spatio-temporal tuning (Tsai et al., 2012; Beck et al., 2015). The use of confetti mouse models and lineage tracing can also aid in the determination of intratumoural heterogeneity owing to clonal variations, and in fate mapping of the cancer evolution studies (Janiszewska and Polyak, 2018; Marx, 2018; Rios et al., 2019). Technological advances in the fields of single cell transcriptomic analysis (Patel et al., 2014; Ting et al., 2014; Horning et al., 2018; Kim et al., 2018; Puram et al., 2018; Cook and Vanderhyden, 2019), single-cell methylome profiling or ChIP sequencing (Rotem et al., 2015; Angermueller et al., 2016; Grosselin et al., 2019) and multiplex in situ imaging (Tsujikawa et al., 2017; Schulz et al., 2018) has allowed researchers to gain insightful information of cellular phenotypic status from clinical specimens. Microfluidic modalities are also gaining attention recently and are of great help not only in detection and capturing of label-free CTCs from patients, but also to gauge the effects of fluid pressures, cancer cell motility assessment associated with single cell or collective migration, and for co-culture studies (Sarioglu et al., 2015; Ma et al., 2018; Shang et al., 2019; Truong et al., 2019). Similarly, various mathematical approaches and modeling have been helpful in deciphering the significant genes and molecular networks associated with the spectrum of epithelial and mesenchymal states, as well as phenotypic plasticity (Jolly et al., 2017; Bocci et al., 2018; Jia et al., 2019; Yang et al., 2019). However, it is crucial to acknowledge that the modalities and analytical approaches utilized in the field of EMP present context-specific studies, such that inferences derived will not provide an overarching conclusion (Henkel et al., 2019). Inherent limitations of the employed assays should always be taken into consideration while extrapolating from the data.
Therapeutic Strategies for Targeting EMP
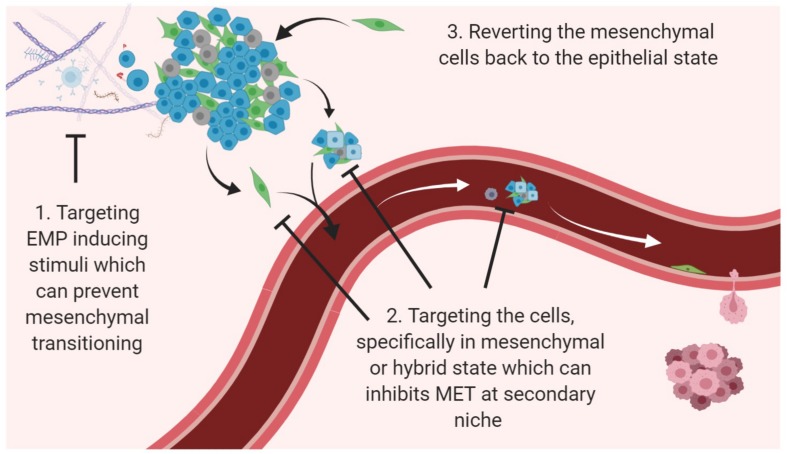
The presence of plasticity in tumor cells and resultant heterogeneity is one of the utmost challenges in targeting cancer on a whole (Bhatia et al., 2017; Redfern et al., 2018). EMT and/or CSC have been reported to confer drug resistance characteristics against a number of conventional therapeutics like taxol, vincristine, oxaliplatin, gemcitabine, cisplatin and 5-fluorouracil in human pancreatic cell lines, and against EGFR-targeted therapies erlotinib, cetuximab and gefitinib in lung cancer (Fuchs et al., 2008; Sabbah et al., 2008; Arumugam et al., 2009). Similarly, studies have also reported that an active EMT program in breast cancer cell lines makes them unresponsive to tamoxifen, paclitaxel, and adriamycin treatment (Kajita et al., 2004; Hiscox et al., 2006; Cheng et al., 2007; Li Q. Q. et al., 2009). Breast cancer cells with EMT-associated CSC features (CD44high, CD24low) have been reported to remain after neoadjuvant chemotherapy and HER2 pharmacological inhibition, suggesting that they encode resistance (Li et al., 2008; Blick et al., 2010). Many reports have also shown basal, mesenchymal-like neoplasms to be more resistant to neoadjuvant chemotherapy than epithelial, luminal-like tumors (Yauch et al., 2005; Carey et al., 2007; Liedtke et al., 2008), and reversal of the EMT phenotype in resistant cell lines has re-established drug sensitivity (Arumugam et al., 2009; Li Y. et al., 2009). Therefore, three main strategies as combinatorial therapies that are being widely acknowledged and/or proposed in the field of combating plasticity are (i) Targeting EMP inducing stimuli which can prevent mesenchymal transitioning, (ii) Targeting the cells, specifically in mesenchymal or hybrid state which can inhibit MET at secondary niche, and (iii) Reverting the mesenchymal cells back to the epithelial state (Bhatia et al., 2017; Williams et al., 2019; Figure 2).
FIGURE 2.
Potential avenues to target EMP. Three main strategies for targeting cancer progression and recurrence with relevance to EMP dynamics are to use agents/compounds (i) that can target the inducers to prevent EMT; (ii) that can selectively kill mesenchymal phenotype and cells present within multiple transition states; (iii) that can revert the cells via MET.
In the first scenario to target EMP inducing stimuli, many different approaches have been utilized to inhibit different signaling pathways that contribute to the induction and maintenance of EMT, such as TGFβ/TGFβR, EGF/EGFR, FGF/FGFR, IGF/IGFR, IL-6/IL-6R, HGF/MET, PDGF/PDGFR, TNFα, Wnt and Notch signaling (Marcucci et al., 2016; Bhatia et al., 2017). Of all, TGFβ and EGF pathway inhibitors have been most extensively studied and investigated, as these have been found to be common inducers of EMT in different cancer types (Li et al., 2015). Table 1 details the current active clinical trials inhibiting these two EMT-inducing pathways in combination with chemotherapeutics.
TABLE 1.
List of the current active clinical trials targeting EGF and TGF-β signaling pathways in combination with chemotherapeutics.
| Target class | Functional class | Drug name | chemotherapeutics combination | Cancer type | Clinical status (first posted, recruitment status) | Intervention/treatment |
| TGF-β/TGF-β Receptor inhibitors | Tyrosine kinase inhibitor | LY-2152799 (Galunisertib) | Fluorouracil/Capecitabine + Tumor specific mesorectal excision | Locally Advanced Rectal Adenocarcinoma | NCT02688712 (2016, Recruiting) | Drug: LY2157299Drug: Capecitabine Drug: Fluorouracil Procedure: Tumor specific mesorectal excision |
| Paclitaxel/Carboplatin | Carcinosarcoma of the Uterus or Ovary | NCT03206177 (2017, Recruiting) | Drug: Galunisertib Drug: Paclitaxel Drug: Carboplatin | |||
| Sorafenib | Advanced Hepatocellular Carcinoma | NCT02178358 (2014, Active, not recruiting) | Drug: LY2157299 Drug: Sorafenib Drug: Placebo | |||
| TGF-β receptor inhibitor | TEW-7197 | Pomalidomide | Relapsed or Relapsed and Refractory Multiple Myeloma | NCT03143985 (2017, Recruiting) | Drug: TEW-7197 Drug: Pomalidomide | |
| EGF/EGFR inhibitors | EGFR tyrosine kinase inhibitor | Gefitinib | Pemetrexed | Advanced Non-Small Cell Lung Cancer | NCT01982955 (2013, Active, not recruiting) | Drug: Tepotinib Drug: Gefitinib Drug: Pemetrexed Drug: Cisplatin Drug: Carboplatin |
| Icotinib | Pemetrexed, Carboplatin | III B/IV Non-Small Cell Lung Cancer | NCT03151161 (2017, Not yet recruiting) | Drug: Icotinib, Pemetrexed, Carboplatin Drug: Icotinib | ||
| Cisplatin or Carboplatin | metastatic non-squamous non-small cell lung cancer who did not progress after pemetrexed combined with platinum chemotherapy | NCT03992885 (2019, Recruiting) | Drug: Icotinib Drug: Cisplatin Drug: Carboplatin | |||
| Apatinib | Pemetrexed, Gemcitabine, Docetaxel | NSCLC Patients Without T790M Mutation | NCT03758677 (2018, Not yet recruiting) | Drug: Apatinib Drug: Chemotherapy with platinum-based double drugs (Pemetrexed, Gemcitabine, Docetaxel) | ||
| Pemetrexed Plus Carboplatin | Advanced Non-small Cell Lung Cancer | NCT03164694 (2018, Recruiting) | Drug: Apatinib + Pemetrexed + Carboplatin Drug: Pemetrexed + Carboplatin | |||
| Osimertinib | Cisplatin or Carboplatin | Metastatic EGFR Mutant Lung Cancers | NCT03567642 (2018, Recruiting) | Drug: Osimertinib Drug: Platinum Drug: Etoposide | ||
| Platinum-based Doublet-Chemotherapy | Locally Advanced or Metastatic Non-Small Cell Lung Cancer | NCT02151981 (2018, Active, not recruiting) | Drug: Chemotherapy Drug: Cross-over to Osimertinib | |||
| Pemetrexed + Cisplatin or Pemetrexed + Carboplatin | Locally Advanced Non-Small Cell Lung Cancer | NCT04035486 (2019, Recruiting) | Drug: Osimertinib Drug: Osimertinib + Pemetrexed + Cisplatin Drug: Osimertinib + Pemetrexed + Carboplatin | |||
| Monoclonal antibody | Panitumumab | Carboplatin and Paclitaxel | Invasive Triple Negative Breast Cancer | NCT02876107 (2016, recruiting) | Drug: Carboplatin Other: Laboratory Biomarker Analysis Drug: Paclitaxel Biological: Panitumumab | |
| HLX07 | Gemcitabine + Cisplatin/Paclitaxel + Carboplatin/mFOLFOX6 | Advanced Solid Tumors | NCT03577704 (2018, Recruiting) | Drug: HLX07 + Gemcitabine + Cisplatin Drug: HLX07 + Paclitaxel + Carboplatin Drug: HLX07 + mFOLFOX6 |
Secondly, for therapies specifically targeting mesenchymal cells, different novel strategies such as EMP-targeting vaccines against transcription factors such as TWIST1 and Brachyury; nutraceuticals; and the repurposing of drugs such as metformin, salinomycin and resveratrol, have been extensively discussed in our previous review (Bhatia et al., 2017). Table 2 details current clinical trials (2015 onward) with the focus on targeting EMP in cancer patients, as an update from our previous review (Bhatia et al., 2017). New combinatorial approaches combining EMT inhibitors alongside targeting immunotherapy blockade are also being developed, as EMT is reported to induce PDL1 expression in carcinoma cells (Chen et al., 2014; Noman et al., 2017), and an EMT signature was seen in tumors that responded to anti PD1/PD-L1- and CTLA4-associated treatments (Lou et al., 2016).
TABLE 2.
Different categories of inhibitors that target stimuli and signaling pathways associated with EMT and are targeted in current clinical trials.
| Target class | Functional class | Drug Name | Cancer type | Clinical status (first posted) |
| Inhibitors of extracellular mediators and their corresponding receptors | ||||
| TGF-β–TGF-β receptor inhibitors | TGF-β receptor inhibitor | TEW-7197 | Urothelial Carcinoma Recurrent, Advanced Urothelial Carcinoma, Myelodysplastic Syndromes | NCT04064190(2019); NCT03074006(2017) |
| TGFβ receptor ectodomain-IgG Fc fusion protein | AVID200 | refractory advanced and metastatic malignancies, Myelofibrosis (Myeloproliferative Neoplasms Research Consortium [MPN-RC] 118) | NCT03834662(2019); NCT03895112(2019) | |
| a bifunctional fusion protein targeting PD-L1 and TGF-β | MSB0011359C (M7824) | Stage II-III HER2 Positive Breast Cancer, Locally Advanced or Metastatic Second Line (2L) Biliary Tract Cancer (Cholangiocarcinoma and Gallbladder Cancer), Solid Tumors, Recurrent Respiratory Papillomatosis, HPV Associated Malignancies | NCT03620201(2018); NCT03833661(2019); NCT02699515(2016); NCT02517398(2015); NCT03707587(2018); NCT03427411(2018) | |
| CAR-T cells that target GPC3 (GPC3-CART cell) and/or soluble TGFβ (GPC3/TGFβ-CART) | GPC3-T2-CAR-T | Hepatocellular Carcinoma, Squamous Cell Lung Cancer | NCT03198546(2017) | |
| IL-6/IL-6R inhibitors | Monoclonal antibody | Siltuximab (CNTO-328, Tocilizumab) | Metastatic Pancreatic Cancer; multiple myeloma (MM) and systemic AL amyloidosis (AL) | NCT04191421(2019); NCT03315026(2017) |
| EGF/EGFR inhibitors | Tyrosine kinase inhibitor | Afatinib (BIBW2992) | Chordoma, | NCT03083678(2018) |
| Dacomitinib (PF00299804) | EGFR Mutant Lung Cancer | NCT03755102(2018) | ||
| Osimertinib | Stage I-IIIA EGFR-mutant Non-small Cell Lung Cancer, stage IIIB-IV or Recurrent Non-small Cell Lung Cancer | NCT03586453(2018); NCT03434418(2018); NCT03433469(2018); NCT03191149(2018) | ||
| Brigatinib (AP26113) | Advanced Non-small Cell Lung Cancer (NSCLC), Anaplastic Large Cell Lymphoma, Advanced Malignant Neoplasm | NCT02737501(2016); NCT02706626(2017); NCT03719898(2018); NCT03868423(2019); NCT03707938(2018); NCT03596866(2019); | ||
| inhibitor for (EGFR, HER2, and ErbB4) | Poziotinib (HM781-36B) | EGFR Exon 20 Mutant Advanced NSCLC, Breast Cancer, Stage IV Lung Adenocarcinoma with HER2 Mutation | NCT03066206(2017); NCT03066206(2017); NCT03744715(2018); NCT03318939(2017); NCT02979821(2016) | |
| Monoclonal antibody | Panitumumab | Anaplastic Lymphoma Kinase-Positive (ALK +), Advanced Non-Small-Cell Lung Cancer (NSCLC) | NCT03535740(2019) | |
| HLX07 | Advanced Solid Cancers | NCT02648490(2016) | ||
| PDGF/PDGFR inhibitors | Tyrosine kinase inhibitor | Axitinib | Pheochromocytoma, Paraganglioma, Renal Cell Carcinoma, Hepatobiliary Neoplasm, Liver Neoplasm, Biliary Tract Neoplasms, Cervical Cancer, Non-Small Cell Lung Cancer, Urothelial Cancer | NCT03839498(2019); NCT03494816(2018); NCT04010071(2019); NCT03826589(2019); NCT03472560(2018); NCT03341845(2017) |
| FGF/FGFR inhibitors | Tyrosine kinase inhibitor | Lenvatinib | Advanced Biliary Tract Cancer, Thyroid Neoplasms, Advanced Gastric Cancer, Non-small Cell Lung Cancer, Solid Tumor, Thyroid Cancer | NCT04211168(2019); NCT03573960(2018); NCT03609359(2018); NCT03829332(2019); NCT03009292(2017); NCT03139747(2017) |
| Nintedanib (BIBF1120) | Appendix Cancer, Lymphangioleiomyomatosis, Adenocarcinoma of the Lung | NCT03287947(2017); NCT03062943(2017); NCT04046614(2019) | ||
| Pazopanib | Refractory Solid Tumors, Metastatic Sarcoma, Recurrent Sarcoma, Resectable Sarcoma, Advanced Renal Cell Carcinoma Metastatic Renal Cell Carcinoma | NCT02691767(2016); NCT04199026(2019); NCT03200717(2017) | ||
| Ponatinib | Medullary Thyroid Cancer, Acute Myeloid Leukemia, Accelerated Phase Chronic Myeloid Leukemia, Blast Phase Chronic Myeloid Leukemia, GIST, Malignant, Chronic Myeloid Leukemia, Acute Lymphoblastic Leukemia, Philadelphia Chromosome-positive Acute Lymphoblastic Leukemia | NCT03838692(2019); NCT03934372(2019); NCT03171389(2017); NCT04233346(2020); NCT03709017(2018) | ||
| TNFα inhbitors | Monoclonal antibody | Infliximab | Advanced Melanoma | NCT03293784(2017) |
| Hedgehog/Smoothened inhibitors | Smoothened antagonists (small-molecule inhibitor) | Vismodegib | Stomach Neoplasms, Basal Cell Carcinoma, Metastatic Basal Cell Carcinoma, Locally Advanced Basal Cell Carcinoma, Advanced Solid Tumors | NCT03052478(2017); NCT03035188(2017); NCT03610022(2018); NCT03297606(2017) |
| Sonidegib | Clinical Stage III Cutaneous Melanoma AJCC v8, Clinical Stage III Gastric Cancer AJCC v8, Basal Cell Carcinoma | NCT04007744(2019); NCT04066504(2019) | ||
| Notch/Notch ligand (Delta-like and Jagged) inhibitors | Small-molecule inhibitor | γ-secretase inhibitor: LY3039478 | Advanced Solid Tumor | NCT02836600(2016) |
| γ-secretase inhibitor: PF-03084014 | Desmoid Tumor, Aggressive Fibromatosis, Desmoid-Type Fibromatosis, Recurrent Desmoid-Type Fibromatosis, Unresectable Desmoid-Type Fibromatosis | NCT03785964(2018); NCT04195399(2019) | ||
| PAN-Notch inhibitor BMS-906024 | recurrent or metastatic Adenoid Cystic Carcinoma | NCT03691207(2018) | ||
| WNT/Frizzled inhibitors | Wnt5a mimetic | Foxy-5 | Colon Cancer | NCT03883802(2019) |
| Peptidomimetics | CWP232291 | Acute Myeloid Leukemia | NCT03055286(2017) | |
| Inhibits the recruiting of β-catenin with its co-activator CBP | PRI-724 | Liver Cirrhosis | NCT03620474(2018) | |
| Inhibitors of intracellular signaling pathways | ||||
| SRC inhibitors | Tyrosine Kinase inhibitor | Dasatinib (BMS-354825) | Relapsed AML, Waldenstrom Macroglobulinemia, Relapsed CML | NCT03560908(2018); NCT04115059(2019); NCT03573596(2018) |
| Bosutinib (SKI-606) | Metastatic Breast Cancer, Chronic Myeloid Leukemia, Advanced Solid Tumors | NCT03854903(2019); NCT02810990(2016); NCT03297606(2017) | ||
| FAK inhibitors | Tyrosine Kinase inhibitor | Defactinib (VS-6063) | Malignant Pleural Mesothelioma, Advanced Solid Tumors | NCT04201145(2019); NCT02546531(2015) |
| PI3K/AKT/mTOR inhibitors | PI3K inhibitor | Idelalisib | Follicular Non-Hodgkin’s Lymphoma Refractory, Relapsed Diffuse Large B-cell Lymphoma, B-cell Lymphoma Recurrent, B-cell Chronic Lymphocytic Leukemia | NCT03568929(2018); NCT03576443(2018); NCT03757000(2018) |
| AKT inhibitor | AZD5363 | Advanced Solid Tumors, Advanced Breast Cancer | NCT03310541(2017); NCT03182634(2017) | |
| Temsirolimus | Non-muscle Invasive Bladder Cancer, | NCT02753309(2016) | ||
| Tyrosine kinase inhibitor | CX-4945 | Recurrent Medulloblastoma | NCT03904862(2019) | |
| AURKA/SYK | Tyrosine kinase inhibitor | Midostaurin | Acute Myeloid Leukemia, AML/MDS | NCT03951961(2019); NCT04097470(2019) |
| AXL inhibitors | Tyrosine Kinase inhibitor | BGB324 | Recurrent Glioblastoma Undergoing Surgery, Advanced NSCLC | NCT03965494(2019); NCT03184571(2017) |
| RAS/RAF/MAPK inhibitors | RAF inhibitor | Sorafenib | Recurrent or Metastatic Triple Negative Breast Cancer, Advanced Liver Cancer, Advanced Hepatic Carcinoma | NCT02624700(2015); NCT04163237(2019); NCT03164382(2017); NCT03211416(2017) |
| MEK inhibitor | Trametinib | Advanced ALK-Positive NSCLC, Advanced/Metastatic Colorectal Cancer | NCT03087448(2017); NCT03714958(2018) | |
| Inhibitors of transcription factors that indirectly induce EMP | ||||
| JAK and STAT3 inhibitors | Small molecule inhibitor | STAT3: BB1608 (Napabucasin) | Metastatic Colorectal Cancer, Metastatic Pancreatic Cancer | NCT03522649(2018); NCT03647839(2018); NCT03721744(2018) |
| Compounds acting on epigenetic modulators | ||||
| Histone deacetylase inhibitor | Vorinostat | Mutated Advanced Melanoma, Breast Cancer Metastatic | NCT02836548(2016); NCT03742245(2018) | |
| Romidepsin | Survey- Relapsed or Refractory Peripheral T-Cell Lymphoma | NCT03742921(2018); NCT03547700(2018) | ||
| Mocetinostat | Advanced Lung Cancer, Unresectable Stage III or Stage IV Melanoma | NCT03220477(2017); NCT03565406(2018) | ||
| Panobinostat | Multiple Myeloma | NCT02722941(2016); NCT04150289(2019); | ||
| Histone methyl transferases inhibitor | EZH2 inhibitor | E7438 (Tazemetostat, EPZ-6438) | Relapsed or Refractory B-cell Non-Hodgkin’s Lymphoma, Relapsed/Refractory Follicular Lymphoma | NCT03009344(2017); NCT03456726(2018); NCT04224493(2020) |
| EZH1/2 inhibitor | DS-3201b | Relapsed or Refractory Adult T-cell Leukemia/Lymphoma, Acute Leukemia Myeloid Leukemia, Acute Lymphocytic, Recurrent Small Cell Lung Cancer | NCT04102150(2019); NCT03110354(2017); NCT03879798(2019) | |
| Inhibitors of stimuli from the tumor microenvironment | ||||
| HIF-1α inhibitors | Small molecule inhibitor | PT2385 | Von Hippel-Lindau Disease-Associated Clear Cell Renal Cell Carcinoma, Recurrent Glioblastoma | NCT03108066(2017); NCT03216499(2017) |
| Digoxin | Breast Cancer, Circulating Tumor Cells (CTCs), Advanced Pancreatic Cancer, Advanced Solid Tumor | NCT03928210(2019); NCT03889795(2019) | ||
For the third strategy, the detailed molecular knowledge of MET regulation will provide opportunities to curtail this event and prevent the development of metastasis, which is of high clinical relevance. Depending on the clinical scenario, MET-inducing/stabilizing factors may inhibit metastasis if they block the initial EMT stages that allow the dissemination, or promote the later stages of metastasis, which can cause some conflicting considerations (van Denderen and Thompson, 2013; Pattabiraman and Weinberg, 2016). An emerging challenge is then to determine the correct timings for therapeutic interventions, and also to decipher correctly the contribution that intermediate states of the EMT spectrum make to tumor evolution for therapeutic interventions (McGranahan and Swanton, 2015). A high-throughput screening approach is required to identify suitable drugs or “repurposable” small molecular agents in context of specifically targeting hybrid and/or partial EMP cells. The concept of intermittent dosing (drug holidays) is also resurfacing to prevent the plasticity and transitioning of cells in carcinoma. For example, resistance to the BRAF inhibitor vemurafenib in melanoma is remodeled to forestall drug resistance (Das Thakur et al., 2013). Thus, the development of combinatorial therapeutic interventions that can target dynamics and plasticity alongside proliferative tendency of cancer cells may pave the way to more promising treatment strategies.
Conclusion and Perspectives
The crucial roles of EMT-MET during embryogenesis and organogenesis is hijacked during tumor progression and metastasis. The roles of various signaling cascades, intrinsic and extrinsic mechanisms, and regulators that contributes to EMP dynamics are reasonably well determined, but more refined studies and techniques need to be employed to recapitulate the MET behavior of cells while extravasating, seeding and colonizing at secondary niches. The intricacies associated with phenotypic plasticity, stemness and intratumoral heterogeneity further sheds light on several unresolved queries. The reliable features of cellular behavior relating to drug persistent states through the spectrum of EMT need to be verified. Is there a ubiquitous molecular feature of partial/hybrid EMT cells that can be identified and targeted across all different cancer context types? What actual mechanisms do cancer cells employ to intravasate from the primary sites, and how do EMT - MET programs cooperate to assist cancer cells through several stages of cancer progression? We are still lagging in obtaining a wider and more complete understanding of the contributions of EMP in cancer. The sophisticated developments in lineage tracing using confetti animal models and implementation of other novel technologies such as high-resolution intravital imaging, live cell imaging, inducible reporter systems and single-cell sequencing techniques will provide great avenues in the fields of plasticity and dynamics around EMP. Finally, it is imperative to determine how phenotypic plasticity can be exploited, as therapeutic interventions that push the conversion of cancer cells to fat cells or apoptosis, for example, (David et al., 2016; Ishay-Ronen et al., 2019) might be promising approaches in clinical settings.
Author Contributions
SB wrote the manuscript, developed the first draft, and contributed significantly toward development of the whole manuscript. PW contributed significantly in content material, figures and tables preparation and drafted metabolomics section. AT contributed in providing content material and drafted CTC section. ET refined, edited the manuscript, and approved the final version to be published.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. During the course of study, SB, PW and AT were supported by a QUTPRA scholarship.
References
- Aceto N., Bardia A., Miyamoto D. T., Donaldson M. C., Wittner B. S., Spencer J. A., et al. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158 1110–1122. 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnoletto C., Corrà F., Minotti L., Baldassari F., Crudele F., Cook W. J. J., et al. (2019). Heterogeneity in circulating tumor cells: the relevance of the stem-cell subset. Cancer 11:483. 10.3390/cancers11040483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N. M., Brabletz T., Kang Y., Nieto M. A., Weinberg R. A., Stanger B. Z. (2017). Upholding a role for EMT in pancreatic cancer metastasis. Nature 547 E7–E8. 10.1038/nature22963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N. M., Kang Y. (2019). Context-dependent EMT programs in cancer metastasis. J. Exp. Med. 216 1016–1026. 10.1084/jem.20181827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N. M., Maddipati R., Norgard R. J., Balli D., Li J., Yuan S., et al. (2018). EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45 681–695. 10.1016/j.devcel.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Alconada L., Muinelo-Romay L., Madissoo K., Diaz-Lopez A., Krakstad C., Trovik J., et al. (2014). Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol. Cancer 13:223. 10.1186/1476-4598-13-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl J., Ma J., Armstrong L., Millipore E. J. N. M. (2012). Fluorescent gelatin degradation assays for investigating invadopodia formation. Biol. Protoc. 121007 1–5. [Google Scholar]
- Angermueller C., Clark S. J., Lee H. J., Macaulay I. C., Teng M. J., Hu T. X., et al. (2016). Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Brief Commun. 13:229. 10.1038/nmeth.3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansieau S., Bastid J., Doreau A., Morel A. P., Bouchet B. P., Thomas C., et al. (2008). Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14 79–89. 10.1016/j.ccr.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Armstrong A. J., Marengo M. S., Oltean S., Kemeny G., Bitting R. L., Turnbull J. D., et al. (2011). Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol. Cancer Res. 9 997–1007. 10.1158/1541-7786.mcr-10-0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam T., Ramachandran V., Fournier K. F., Wang H., Marquis L., Abbruzzese J. L., et al. (2009). Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 69 5820–5828. 10.1158/0008-5472.can-08-2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Nieto M. A. (2005). The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132 3151–3161. 10.1242/dev.01907 [DOI] [PubMed] [Google Scholar]
- Barriere G., Fici P., Gallerani G., Fabbri F., Zoli W., Rigaud M. (2014). Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Ann. Transl. Med. 2:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B., Lapouge G., Rorive S., Drogat B., Desaedelaere K., Delafaille S., et al. (2015). Different levels of twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell 16 67–79. 10.1016/j.stem.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Beerling E., Seinstra D., de Wit E., Kester L., van der Velden D., Maynard C., et al. (2016). Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 14 2281–2288. 10.1016/j.celrep.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S., Monkman J., Blick T., Pinto C., Waltham M., Nagaraj S. H., et al. (2019). Interrogation of phenotypic plasticity between epithelial and mesenchymal states in breast cancer. J. Clin. Med. 8:893. 10.3390/jcm8060893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S., Monkman J., Toh A. K. L., Nagaraj S. H., Thompson E. W. (2017). Targeting epithelial-mesenchymal plasticity in cancer: clinical and preclinical advances in therapy and monitoring. Biochem. J. 474 3269–3306. 10.1042/bcj20160782 [DOI] [PubMed] [Google Scholar]
- Blick T., Hugo H., Widodo E., Waltham M., Pinto C., Mani S. A., et al. (2010). Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J. Mammary Gland Biol. Neoplasia 15 235–252. 10.1007/s10911-010-9175-z [DOI] [PubMed] [Google Scholar]
- Bocci F., Jolly M. K., George J. T., Levine H., Onuchic J. N. (2018). A mechanism-based computational model to capture the interconnections among epithelial-mesenchymal transition, cancer stem cells and Notch-Jagged signaling. Oncotarget 9 29906–29920. 10.18632/oncotarget.25692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonyadi Rad E., Hammerlindl H., Wels C., Popper U., Ravindran Menon D., Breiteneder H., et al. (2016). Notch4 signaling induces a mesenchymal-epithelial-like transition in melanoma cells to suppress malignant behaviors. Cancer Res. 76 1690–1697. 10.1158/0008-5472.can-15-1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowicz S., Van Scoyk M., Avasarala S., Karuppusamy Rathinam M. K., Tauler J., Bikkavilli R. K., et al. (2014). The soft agar colony formation assay. J. Vis. Exp. 9:e51998. 10.3791/51998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulding T., McCuaig R. D., Tan A., Hardy K., Wu F., Dunn J., et al. (2019). Author Correction: LSD1 activation promotes inducible EMT programs and modulates the tumour microenvironment in breast cancer. Sci. Rep. 9:18771 10.1038/s41598-019-55020-55021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourcy M., Suarez-Carmona M., Lambert J., Francart M. E., Schroeder H., Delierneux C., et al. (2016). Tissue factor induced by epithelial-mesenchymal transition triggers a procoagulant state that drives metastasis of circulating tumor cells. Cancer Res. 76 4270–4282. 10.1158/0008-5472.CAN-15-2263 [DOI] [PubMed] [Google Scholar]
- Brabletz T. (2012). To differentiate or not — routes towards metastasis. Nat. Rev. Cancer 12 425–436. 10.1038/nrc3265 [DOI] [PubMed] [Google Scholar]
- Brabletz T., Kalluri R., Nieto M. A., Weinberg R. A. (2018). EMT in cancer. Nat. Rev. Cancer 18 128–134. 10.1038/nrc.2017.118 [DOI] [PubMed] [Google Scholar]
- Bradshaw A. D. (2009). The role of SPARC in extracellular matrix assembly. J. Cell Commun. Signal. 3:239. 10.1007/s12079-009-0062-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs J. T., Henriquez N. V., van Overveld P. G. M., van der Horst G., Que I., Schwaninger R., et al. (2007). Bone morphogenetic protein 7 in the development and treatment of bone metastases from breast cancer. Cancer Res. 67 8742–8751. [DOI] [PubMed] [Google Scholar]
- Bulfoni M., Gerratana L., Del Ben F., Marzinotto S., Sorrentino M., Turetta M., et al. (2016). In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res 18:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonato J. M., Lan I. S., Lazzara M. J. (2015). EGF augments TGFβ-induced epithelial-mesenchymal transition by promoting SHP2 binding to GAB1. J. Cell Sci. 128 3898–3909. 10.1242/jcs.169599 [DOI] [PubMed] [Google Scholar]
- Campbell K., Rossi F., Adams J., Pitsidianaki I., Barriga F. M., Garcia-Gerique L., et al. (2019). Collective cell migration and metastases induced by an epithelial-to-mesenchymal transition in Drosophila intestinal tumors. Nat. Commun. 10:2311. 10.1038/s41467-019-10269-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L. A., Dees E. C., Sawyer L., Gatti L., Moore D. T., Collichio F., et al. (2007). The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 13 2329–2334. 10.1158/1078-0432.ccr-06-1109 [DOI] [PubMed] [Google Scholar]
- Casas E., Kim J., Bendesky A., Ohno-Machado L., Wolfe C. J., Yang J. (2011). Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 71 245–254. 10.1158/0008-5472.can-10-2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celià-Terrassa T., Bastian C., Liu D. D., Ell B., Aiello N. M., Wei Y., et al. (2018). Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat. Commun. 9:5005 10.1038/s41467-018-07538-7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha Y. H., Yook J. I., Kim H. S., Kim N. H. (2015). Catabolic metabolism during cancer EMT. Arch. Pharm. Res. 38 313–320. 10.1007/s12272-015-0567-x [DOI] [PubMed] [Google Scholar]
- Chaffer C. L., Brueckmann I., Scheel C., Kaestli A. J., Wiggins P. A., Rodrigues L. O., et al. (2011). Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. U.S.A. 108 7950–7955. 10.1073/pnas.1102454108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer C. L., Marjanovic N. D., Lee T., Bell G., Kleer C. G., Reinhardt F., et al. (2013). Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154 61–74. 10.1016/j.cell.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer C. L., Thompson E. W., Williams E. D. (2007). Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs 185 7–19. 10.1159/000101298 [DOI] [PubMed] [Google Scholar]
- Chang H. H., Oh P. Y., Ingber D. E., Huang S. (2006). Multistable and multistep dynamics in neutrophil differentiation. BMC Cell Biol. 7:11. 10.1186/1471-2121-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y. L., Shepard C. R., Wells A. (2010). Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Mol. Cancer 9 1–18. 10.1186/1476-4598-9-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gibbons D. L., Goswami S., Cortez M. A., Ahn Y.-H., Byers L. A., et al. (2014). Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5 5241–5241. 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang J., Sampieri K., Clohessy J. G., Mendez L., Gonzalez-Billalabeitia E., et al. (2018). An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat. Genet. 50 206–218. 10.1038/s41588-017-0027-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G. Z., Chan J., Wang Q., Zhang W., Sun C. D., Wang L. H. (2007). Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 67 1979–1987. 10.1158/0008-5472.can-06-1479 [DOI] [PubMed] [Google Scholar]
- Christiansen J. J., Rajasekaran A. K. (2006). Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 66 8319–8326. 10.1158/0008-5472.can-06-0410 [DOI] [PubMed] [Google Scholar]
- Chung V. Y., Tan T. Z., Ye J., Huang R. L., Lai H. C., Kappei D., et al. (2019). The role of GRHL2 and epigenetic remodeling in epithelial-mesenchymal plasticity in ovarian cancer cells. Commun. Biol. 2:272 10.1038/s42003-019-0506-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. P., Vanderhyden B. C. (2019). Comparing transcriptional dynamics of the epithelial-mesenchymal transition. bioRxiv [Preprint], 10.1101/732412 [DOI] [Google Scholar]
- Das Thakur M., Salangsang F., Landman A. S., Sellers W. R., Pryer N. K., Levesque M. P., et al. (2013). Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494 251–255. 10.1038/nature11814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave N., Guaita-Esteruelas S., Gutarra S., Frias A., Beltran M., Peiro S., et al. (2011). Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 286 12024–12032. 10.1074/jbc.M110.168625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. J., Huang Y.-H., Chen M., Su J., Zou Y., Bardeesy N., et al. (2016). TGF-β Tumor Suppression through a Lethal EMT. Cell 164 1015–1030. 10.1016/j.cell.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit S., Manicone M., Rossi E., Lampignano R., Yang L., Zill B., et al. (2018). EpCAMhigh and EpCAMlow circulating tumor cells in metastatic prostate and breast cancer patients. Oncotarget 9:35705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaugnies M., Sánchez-Danés A., Rorive S., Raphaël M., Liagre M., Parent M.-A., et al. (2018). YAP and TAZ are essential for basal and squamous cell carcinoma initiation. EMBO Rep. 19 e45809. 10.15252/embr.201845809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan E., Song Y. H., Krishnappa S., Alt E. (2012). Epithelial-mesenchymal transition in breast cancer lines is mediated through PDGF-D released by tissue-resident stem cells. Int. J. Cancer 131 1023–1031. 10.1002/ijc.26493 [DOI] [PubMed] [Google Scholar]
- Dong C., Yuan T., Wu Y., Wang Y., Fan T. W., Miriyala S., et al. (2013). Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23 316–331. 10.1016/j.ccr.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., Karaayvaz M., Jia N., Kaneuchi M., Hamada J., Watari H., et al. (2013). Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene 32 3286–3295. 10.1038/onc.2012.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre A., Rashidian M., Reinhardt F., Bagnato A., Keckesova Z., Ploegh H. L., et al. (2017). Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 77 3982–3989. 10.1158/0008-5472.can-16-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E., Jr. (2012). Bistability, bifurcations, and Waddington’s epigenetic landscape. Curr. Biol. 22 R458–R466. 10.1016/j.cub.2012.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K. R., Durrans A., Lee S., Sheng J., Li F., Wong S. T., et al. (2015). Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527 472–476. 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francart M.-E., Lambert J., Vanwynsberghe A. M., Thompson E. W., Bourcy M., Polette M., et al. (2018). Epithelial-mesenchymal plasticity and circulating tumor cells: travel companions to metastases. Dev. Dyn. 247 432–450. 10.1002/dvdy.24506 [DOI] [PubMed] [Google Scholar]
- Freed-Pastor W. A., Mizuno H., Zhao X., Langerod A., Moon S. H., Rodriguez-Barrueco R., et al. (2012). Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148 244–258. 10.1016/j.cell.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. C., Fujii T., Dorfman J. D., Goodwin J. M., Zhu A. X., Lanuti M., et al. (2008). Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 68 2391–2399. 10.1158/0008-5472.can-07-2460 [DOI] [PubMed] [Google Scholar]
- Gal A., Sjoblom T., Fedorova L., Imreh S., Beug H., Moustakas A. (2008). Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene 27 1218–1230. 10.1038/sj.onc.1210741 [DOI] [PubMed] [Google Scholar]
- Gómez-Cuadrado L., Tracey N., Ma R., Qian B., Brunton V. G. (2017). Mouse models of metastasis: progress and prospects. Dis. Mod. Mech. 10 1061–1074. 10.1242/dmm.030403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosselin K., Durand A., Marsolier J., Poitou A., Marangoni E., Nemati F., et al. (2019). High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51 1060–1066. 10.1038/s41588-019-0424-429 [DOI] [PubMed] [Google Scholar]
- Gunasinghe N. P., Wells A., Thompson E. W., Hugo H. J. (2012). Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metast. Rev. 31 469–478. 10.1007/s10555-012-9377-9375 [DOI] [PubMed] [Google Scholar]
- Guo W., Keckesova Z., Donaher J. L., Shibue T., Tischler V., Reinhardt F., et al. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148 1015–1028. 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P. B., Fillmore C. M., Jiang G., Shapira S. D., Tao K., Kuperwasser C., et al. (2011). Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146 633–644. 10.1016/j.cell.2011.07.026 [DOI] [PubMed] [Google Scholar]
- Han L., Chen W., Zhao Q. J. T. B. (2014). Prognostic value of circulating tumor cells in patients with pancreatic cancer: a meta-analysis. Tumour Biol. 35 2473–2480. 10.1007/s13277-013-1327-5 [DOI] [PubMed] [Google Scholar]
- Harner-Foreman N., Vadakekolathu J., Laversin S. A., Mathieu M. G., Reeder S., Pockley A. G., et al. (2017). A novel spontaneous model of epithelial-mesenchymal transition (EMT) using a primary prostate cancer derived cell line demonstrating distinct stem-like characteristics. Sci. Rep. 7:40633. 10.1038/srep40633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S., Blick T., Williams E. D., Thompson E. W. (2020). Applications of RNA from circulating tumor cells. Front. Biosci. 25 874–892. 10.2741/4838 [DOI] [PubMed] [Google Scholar]
- Henkel L., Rauscher B., Boutros M. (2019). Context-dependent genetic interactions in cancer. Curr. Opin. Genet. Dev. 54 73–82. 10.1016/j.gde.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Hirata E., Park D., Sahai E. (2014). Retrograde flow of cadherins in collective cell migration. Nat. Cell Biol. 16:621. 10.1038/ncb2995 [DOI] [PubMed] [Google Scholar]
- Hiscox S., Jiang W. G., Obermeier K., Taylor K., Morgan L., Burmi R., et al. (2006). Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of β-catenin phosphorylation. Int. J. Cancer 118 290–301. 10.1002/ijc.21355 [DOI] [PubMed] [Google Scholar]
- Horning A. M., Wang Y., Lin C.-K., Louie A. D., Jadhav R. R., Hung C.-N., et al. (2018). Single-Cell RNA-seq reveals a subpopulation of prostate cancer cells with enhanced cell-Cycle-Related transcription and attenuated androgen response. Cancer Res. 78 853–864. 10.1158/0008-5472.can-17-1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., Goldstein J. L., Brown M. S. (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109 1125–1131. 10.1172/jci15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Liu Z., Wang X. (2013). Does TP53 mutation promote ovarian cancer metastasis to omentum by regulating lipid metabolism? Med. Hypotheses 81 515–520. 10.1016/j.mehy.2013.06.009 [DOI] [PubMed] [Google Scholar]
- Huang R. Y. J., Wong M. K., Tan T. Z., Kuay K. T., Ng A. H. C., Chung V. Y., et al. (2013). An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis. 4:e915. 10.1038/cddis.2013.442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Ernberg I., Kauffman S. (2009). Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Semin. Cell Dev. Biol. 20 869–876. 10.1016/j.semcdb.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo H., Ackland M. L., Blick T., Lawrence M. G., Clements J. A., Williams E. D., et al. (2007). Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J. Cell. Physiol. 213 374–383. 10.1002/jcp.21223 [DOI] [PubMed] [Google Scholar]
- Hugo H., Gunasinghe N., Hollier B., Tanaka T., Blick T., Toh A., et al. (2017). Epithelial requirement for in vitro proliferation and xenograft growth and metastasis of MDA-MB-468 human breast cancer cells: oncogenic rather than tumor-suppressive role of E-cadherin. Breast Cancer Res. 19 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo H. J., Wafai R., Blick T., Thompson E. W., Newgreen D. F. (2009). Staurosporine augments EGF-mediated EMT in PMC42-LA cells through actin depolymerisation, focal contact size reduction and Snail1 induction - a model for cross-modulation. BMC Cancer 9:235. 10.1186/1471-2407-9-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüsemann Y., Geigl J. B., Schubert F., Musiani P., Meyer M., Burghart E., et al. (2008). Systemic spread is an early step in breast cancer. Cancer Cell 13 58–68. 10.1016/j.ccr.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Ishay-Ronen D., Diepenbruck M., Kalathur R. K. R., Sugiyama N., Tiede S., Ivanek R., et al. (2019). Gain fat-lose metastasis: converting invasive breast cancer cells into adipocytes inhibits cancer metastasis. Cancer Cell 35 17–32. 10.1016/j.ccell.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Janiszewska M., Polyak K. (2018). A confetti trail of tumour evolution. Nat. Cell Biol. 20 639–641. 10.1038/s41556-018-0110-117 [DOI] [PubMed] [Google Scholar]
- Jia D., Li X., Bocci F., Tripathi S., Deng Y., Jolly M. K., et al. (2019). Quantifying cancer epithelial-mesenchymal plasticity and its association with stemness and immune response. J. Clin. Med. 8:725. 10.3390/jcm8050725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez L., Wang J., Morrison M. A., Whatcott C., Soh K. K., Warner S., et al. (2016). Phenotypic chemical screening using a zebrafish neural crest EMT reporter identifies retinoic acid as an inhibitor of epithelial morphogenesis. Dis. Model. Mech. 9 389–400. 10.1242/dmm.021790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M. K., Celià-Terrassa T. (2019). Dynamics of phenotypic heterogeneity associated with EMT and stemness during cancer progression. J. Clin. med. 8:1542. 10.3390/jcm8101542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M. K., Kulkarni P., Weninger K., Orban J., Levine H. (2018). Phenotypic plasticity, bet-hedging, and androgen independence in prostate cancer: role of non-genetic heterogeneity. Front. Oncol. 8:50. 10.3389/fonc.2018.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M. K., Tripathi S. C., Jia D., Mooney S. M., Celiktas M., Hanash S. M., et al. (2016). Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 7 27067–27084. 10.18632/oncotarget.8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M. K., Tripathi S. C., Somarelli J. A., Hanash S. M., Levine H. (2017). Epithelial/mesenchymal plasticity: how have quantitative mathematical models helped improve our understanding? Mol. Oncol. 11 739–754. 10.1002/1878-0261.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M. K., Ware K. E., Xu S., Gilja S., Shetler S., Yang Y., et al. (2019). E-cadherin represses anchorage-independent growth in sarcomas through both signaling and mechanical mechanisms. Mol. Cancer Res. 17 1391–1402. 10.1158/1541-7786.Mcr-18-0763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaigorodova E., Tarabanovskaya N., Staheeva M., Savelieva O., Tashireva L., Denisov E., et al. (2017). Effect of small and radical surgical injury on the level of different populations of circulating tumor cells in the blood of breast cancer patients. Neoplasma 64 437–443. 10.4149/neo_2017_315 [DOI] [PubMed] [Google Scholar]
- Kajita M., McClinic K. N., Wade P. A. (2004). Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol. Cell. Biol. 24 7559–7566. 10.1128/MCB.24.17.7559-7566.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., Weinberg R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119 1420–1428. 10.1172/jci39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Kim H., Lee S., Youn H., Youn B. (2019). Role of metabolic reprogramming in epithelial?mesenchymal transition (EMT). Intern. J. Mol. Sci. 20:2042. 10.3390/ijms20082042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karacosta L. G., Anchang B., Ignatiadis N., Kimmey S. C., Benson J. A., Shrager J. B., et al. (2019). Mapping lung cancer epithelial-mesenchymal transition states and trajectories with single-cell resolution. Nat. Commun. 10:5587 10.1038/s41467-019-13441-13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo B. L., Lee S. C., Kumar P., Tan T. Z., Warkiani M. E., Ow S. G., et al. (2015). Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget 6 15578–15593. 10.18632/oncotarget.3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Gao R., Sei E., Brandt R., Hartman J., Hatschek T., et al. (2018). Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevebring D., Rosin G., Ma R., Lindberg J., Czene K., Kere J., et al. (2014). Sequencing of breast cancer stem cell populations indicates a dynamic conversion between differentiation states in vivo. Breast Cancer Res. 16:R72. 10.1186/bcr3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko Y., Johnson J., Bos B., Lombard R., Campbell L., Loughran E., et al. (2017). Heterogeneous cadherin expression and multicellular aggregate dynamics in ovarian cancer dissemination. Neoplasia 19 549–563. 10.1016/j.neo.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymkowsky M. W., Savagner P. (2009). Epithelial-mesenchymal transition: a cancer researcher’s conceptual friend and foe. Am. J. Pathol. 174 1588–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondaveeti Y., Guttilla Reed I. K., White B. A. (2015). Epithelial-mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett. 364 44–58. 10.1016/j.canlet.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Kowalik A., Kowalewska M., Góźdź S. J. T. R. (2017). Current approaches for avoiding the limitations of circulating tumor cells detection methods—implications for diagnosis and treatment of patients with solid tumors. Transl. Res. 185 58.–84. [DOI] [PubMed] [Google Scholar]
- Kramer N., Walzl A., Unger C., Rosner M., Krupitza G., Hengstschlager M., et al. (2013). In vitro cell migration and invasion assays. Mutat. Res. 752 10–24. 10.1016/j.mrrev.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Krebs A. M., Mitschke J., Lasierra Losada M., Schmalhofer O., Boerries M., Busch H., et al. (2017). The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19 518–529. 10.1038/ncb3513 [DOI] [PubMed] [Google Scholar]
- Kroger C., Afeyan A., Mraz J., Eaton E. N., Reinhardt F., Khodor Y. L., et al. (2019). Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116 7353–7362. 10.1073/pnas.1812876116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Park S. H., Cieply B., Schupp J., Killiam E., Zhang F., et al. (2011). A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol. Cell. Biol. 31 4036–4051. 10.1128/MCB.01342-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto R., Iwasawa S., Ebata T., Ishiwata T., Sekine I., Tada Y., et al. (2016). Drug resistance originating from a TGF-beta/FGF-2-driven epithelial-to-mesenchymal transition and its reversion in human lung adenocarcinoma cell lines harboring an EGFR mutation. Int. J. Oncol. 48 1825–1836. 10.3892/ijo.2016.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A., Kato T., Brugues A., Serra-Picamal X., Derzsi S., Arwert E., et al. (2017). A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 19 224–237. 10.1038/ncb3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A. W., Pattabiraman D. R., Weinberg R. A. J. C. (2017). Emerging biological principles of metastasis. Cell 168 670–691. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil M., Nassar D., Beck B., Boumahdi S., Wang L., Brisebarre A., et al. (2017). Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell 20 191–204. 10.1016/j.stem.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharpentier A., Vielh P., Perez-Moreno P., Planchard D., Soria J., Farace F. J. B. (2011). Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br. J. Cancer 105 1338–1341. 10.1038/bjc.2011.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Dedhar S., Kalluri R., Thompson E. W. (2006). The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 172 973–981. 10.1083/jcb.200601018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Ju M. K., Jeon H. M., Lee Y. J., Kim C. H., Park H. G., et al. (2018). Oncogenic metabolism acts as a prerequisite step for induction of cancer metastasis and cancer stem cell phenotype. Oxid. Med. Cell Longev. 2018:1027453. 10.1155/2018/1027453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire C. A., Liu S. Z., Wilkerson C. L., Ramani V. C., Barzanian N. A., Huang K.-W., et al. (2018). Fast and label-free isolation of circulating tumor cells from blood: from a research microfluidic platform to an automated fluidic instrument, VTX-1 liquid biopsy system. SLAS Technol. 23 16–29. 10.1177/2472630317738698 [DOI] [PubMed] [Google Scholar]
- Li L., Qi L., Liang Z., Song W., Liu Y., Wang Y., et al. (2015). Transforming growth factor-beta1 induces EMT by the transactivation of epidermal growth factor signaling through HA/CD44 in lung and breast cancer cells. Int. J. Mol. Med. 36 113–122. 10.3892/ijmm.2015.2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wennborg A., Aurell E., Dekel E., Zou J.-Z., Xu Y., et al. (2016). Dynamics inside the cancer cell attractor reveal cell heterogeneity, limits of stability, and escape. Proc. Natl. Acad. Sci. U.S.A. 113:2672. 10.1073/pnas.1519210113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Q., Xu J. D., Wang W. J., Cao X. X., Chen Q., Tang F., et al. (2009). Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin. Cancer Res. 15 2657–2665. 10.1158/1078-0432.ccr-08-2372 [DOI] [PubMed] [Google Scholar]
- Li Y., VandenBoom T. G., II, Kong D., Wang Z., Ali S., Philip P. A., et al. (2009). Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 69 6704–6712. 10.1158/0008-5472.can-09-1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lewis M. T., Huang J., Gutierrez C., Osborne C. K., Wu M. F., et al. (2008). Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 100 672–679. 10.1093/jnci/djn123 [DOI] [PubMed] [Google Scholar]
- Liedtke C., Mazouni C., Hess K. R., Andre F., Tordai A., Mejia J. A., et al. (2008). Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 26 1275–1281. 10.1200/jco.2007.14.4147 [DOI] [PubMed] [Google Scholar]
- Liu T. L., Upadhyayula S., Milkie D. E., Singh V., Wang K., Swinburne I. A., et al. (2018). Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360:6386. 10.1126/science.aaq1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Munoz E., Mendez-Montes M. (2013). Markers of circulating breast cancer cells. Adv. Clin. Chem. 61 175–224. 10.1016/b978-0-12-407680-8.00007-5 [DOI] [PubMed] [Google Scholar]
- Lou Y., Diao L., Cuentas E. R., Denning W. L., Chen L., Fan Y. H., et al. (2016). Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. 22 3630–3642. 10.1158/1078-0432.Ccr-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco A. R., Ban Y., Crowley M. J., Lee S. B., Ramchandani D., Du W., et al. (2020). Differential contributions of pre- and post-EMT tumor cells in breast cancer metastasis. Cancer Res. 80 163–169. 10.1158/0008-5472.Can-19-1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Jolly M. K., Levine H., Onuchic J. N., Ben-Jacob E. J. P. (2013). MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. PNAS 110 18144–18149. 10.1073/pnas.1318192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Kang Y. (2019). Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49 361–374. 10.1016/j.devcel.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.-H. V., Middleton K., You L., Sun Y. (2018). A review of microfluidic approaches for investigating cancer extravasation during metastasis. Microsyst. Nanoeng. 4:17104. [Google Scholar]
- Mani S. A., Guo W., Liao M.-J., Eaton E. N., Ayyanan A., Zhou A. Y., et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133 704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci F., Bellone M., Caserta C. A., Corti A. (2014). Pushing tumor cells towards a malignant phenotype: stimuli from the microenvironment, intercellular communications and alternative roads. Int. J. Cancer 135 1265–1276. 10.1002/ijc.28572 [DOI] [PubMed] [Google Scholar]
- Marcucci F., Stassi G., De Maria R. (2016). Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat. Rev. Drug Discov. 15 311–325. 10.1038/nrd.2015.13 [DOI] [PubMed] [Google Scholar]
- Markiewicz A., Ksiazkiewicz M., Welnicka-Jaskiewicz M., Seroczynska B., Skokowski J., Szade J., et al. (2014). Mesenchymal phenotype of CTC-enriched blood fraction and lymph node metastasis formation potential. PLoS One 9:e93901. 10.1371/journal.pone.0093901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. (2018). Stem cells: lineage tracing lets single cells talk about their past. Nat. Methods 15 411–414. 10.1038/s41592-018-0016-10 [DOI] [PubMed] [Google Scholar]
- Mason D. E., Collins J. M., Dawahare J. H., Nguyen T. D., Lin Y., Voytik-Harbin S. L., et al. (2019). YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J. Cell Biol. 218 1369–1389. 10.1083/jcb.201806065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis R. A., Sokol E. S., Gupta P. B. (2017). Cancer cells exhibit clonal diversity in phenotypic plasticity. Open Biol. 7:160283. 10.1098/rsob.160283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C. D., Sphyris N., Evans K. W., Werden S. J., Guo W., Mani S. A. (2011). Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 13:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald O. G., Wu H., Timp W., Doi A., Feinberg A. P. (2011). Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat. Struct. Mol. Biol. 18 867–874. 10.1038/nsmb.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N., Swanton C. (2015). Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27 15–26. 10.1016/j.ccell.2014.12.001 [DOI] [PubMed] [Google Scholar]
- McInnes L. M., Jacobson N., Redfern A., Dowling A., Thompson E. W., Saunders C. M. (2015). Clinical implications of circulating tumor cells of breast cancer patients: role of epithelial-mesenchymal plasticity. Front. Oncol. 5:42. 10.3389/fonc.2015.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J. W., Liu L. J., Huang J. (2014). Interleukin-6-induced epithelial-mesenchymal transition through signal transducer and activator of transcription 3 in human cervical carcinoma. Int. J. Oncol. 45 165–176. 10.3892/ijo.2014.2422 [DOI] [PubMed] [Google Scholar]
- Milano A., Mazzetta F., Valente S., Ranieri D., Leone L., Botticelli A., et al. (2018). Molecular detection of EMT markers in circulating tumor cells from metastatic non-small cell lung cancer patients: potential role in clinical practice. Anal. Cell Pathol. 2018:3506874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C. B., O’Neill G. M. (2016). Cooperative cell invasion: matrix metalloproteinase-mediated incorporation between cells. Mol. Biol. Cell 27 3284–3292. 10.1091/mbc.E16-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi A., Taddei M. L., Chiarugi P., Giannoni E. (2017). Targeting the metabolic reprogramming that controls epithelial-to-mesenchymal transition in aggressive tumors. Front. Oncol. 7:40. 10.3389/fonc.2017.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A.-P., Lièvre M., Thomas C., Hinkal G., Ansieau S., Puisieux A. (2008). Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 3:e2888. 10.1371/journal.pone.0002888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Hansen C. G., Guan K.-L. (2015). The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15 73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T., Tanaka F., Ono A., Yoneda K., Takahashi T., Murakami H., et al. (2012). Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J. Thor. Oncol. 7 512–519. [DOI] [PubMed] [Google Scholar]
- Narai A., Arai S., Shimizu M. (1997). Rapid decrease in transepithelial electrical resistance of human intestinal Caco-2 cell monolayers by cytotoxic membrane perturbents. Toxicol. Vitro 11 347–354. 10.1016/s0887-2333(97)00026-x [DOI] [PubMed] [Google Scholar]
- Nieto M. A. (2013). Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342:6159. [DOI] [PubMed] [Google Scholar]
- Nieto M. A. (2018). A snail tale and the chicken embryo. Int. J. Dev. Biol. 62 121–126. 10.1387/ijdb.170301mn [DOI] [PubMed] [Google Scholar]
- Noman M. Z., Janji B., Abdou A., Hasmim M., Terry S., Tan T. Z., et al. (2017). The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 6:e1263412. 10.1080/2162402x.2016.1263412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocana O. H., Corcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S., et al. (2012). Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22 709–724. 10.1016/j.ccr.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Ochoa-Hernandez A. B., Juarez-Vazquez C. I., Rosales-Reynoso M. A., Barros-Nunez P. (2012). WNT-beta-catenin signaling pathway and its relationship with cancer. Cir. Cir. 80 389–398. [PubMed] [Google Scholar]
- Oltean S., Sorg B. S., Albrecht T., Bonano V. I., Brazas R. M., Dewhirst M. W., et al. (2006). Alternative inclusion of fibroblast growth factor receptor 2 exon IIIc in Dunning prostate tumors reveals unexpected epithelial mesenchymal plasticity. Proc. Natl. Acad. Sci. U.S.A. 103 14116–14121. 10.1073/pnas.0603090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban V., Krol I., Suhail Y., Szczerba B. M., Aceto N., Bader J. S., et al. (2019). E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573 439–444. 10.1038/s41586-019-1526-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Yan G., Chen W., Sun L., Wang J., Yang J. (2019). Distribution of circulating tumor cell phenotype in early cervical cancer. Cancer Manag. Res. 11:5531. 10.2147/cmar.s198391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki M. A., Stoupis G., Theodoropoulos P. A., Mavroudis D., Georgoulias V., Agelaki S. J. (2019). Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol. Cancer Ther. 18 437–447. 10.1158/1535-7163.mct-18-0584 [DOI] [PubMed] [Google Scholar]
- Pastushenko I., Blanpain C. (2019). EMT Transition states during tumor progression and metastasis. Trends Cell Biol. 29 212–226. 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., et al. (2018). Identification of the tumour transition states occurring during EMT. Nature 556 463–468. 10.1038/s41586-018-0040-43 [DOI] [PubMed] [Google Scholar]
- Patel A. P., Tirosh I., Trombetta J. J., Shalek A. K., Gillespie S. M., Wakimoto H., et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344 1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D. R., Bierie B., Kober K. I., Thiru P., Krall J. A., Zill C., et al. (2016). Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351:3680. 10.1126/science.aad3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D. R., Weinberg R. A. (2016). Targeting the Epithelial-to-Mesenchymal Transition: the Case for differentiation-based therapy. Cold. Spring Harb. Symp. Quant. Biol. 81 11–19. 10.1101/sqb.2016.81.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne R., Wang F., Su N., Krell J., Zebrowski A., Yagüe E., et al. (2012). Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br. J. Cancer 106 1790–1797. 10.1038/bjc.2012.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Marin F., Cubillo E., Stark H. J., Fusenig N., Nieto M. A., et al. (2004). Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. J. Cell Sci. 117(Pt 13), 2827–2839. 10.1242/jcs.01145 [DOI] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., Cano A. (2007). Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7 415–428. 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Polyak K., Weinberg R. A. (2009). Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9 265–273. 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Proia T. A., Keller P. J., Gupta P. B., Klebba I., Jones A. D., Sedic M., et al. (2011). Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 8 149–163. 10.1016/j.stem.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybylo J. A., Radisky D. C. (2007). Matrix metalloproteinase-induced epithelial-mesenchymal transition: tumor progression at Snail’s pace. Int. J. Biochem. Cell Biol. 39 1082–1088. 10.1016/j.biocel.2007.03.002 [DOI] [PubMed] [Google Scholar]
- Puram S. V., Parikh A. S., Tirosh I. (2018). Single cell RNA-seq highlights a role for a partial EMT in head and neck cancer. Mol. Cell Oncol. 5:e1448244. 10.1080/23723556.2018.1448244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X. K., Han H. Q., Zhang H. J., Xu M., Li L., Chen L., et al. (2018). OVOL2 links stemness and metastasis via fine-tuning epithelial-mesenchymal transition in nasopharyngeal carcinoma. Theranostics 8 2202–2216. 10.7150/thno.24003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Q., Wang X., Lu C., Ma W., Wang Y., Xia G., et al. (2020). Cancer stem-like cells with hybrid epithelial/mesenchymal phenotype leading the collective invasion. Cancer Sci. 111 467. 10.1111/cas.14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern A. D., Spalding L. J., Thompson E. W. (2018). The Kraken Wakes: induced EMT as a driver of tumour aggression and poor outcome. Clin. Exp. Metastasis 35 285–308. 10.1007/s10585-018-9906-x [DOI] [PubMed] [Google Scholar]
- Reynolds D. S., Tevis K. M., Blessing W. A., Colson Y. L., Zaman M. H., Grinstaff M. W. (2017). Breast cancer spheroids reveal a differential cancer stem cell response to chemotherapeutic treatment. Sci. Rep. 7:10382 10.1038/s41598-017-10863-10864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim A. D., Mirek E. T., Aiello N. M., Maitra A., Bailey J. M., McAllister F., et al. (2012). EMT and dissemination precede pancreatic tumor formation. Cell 148 349–361. 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Samy S., Oliveira M. I., Pereira-Veiga T., Muinelo-Romay L., Carvalho S., Gaspar J., et al. (2019). Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci. Rep. 9 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S., Fritsche H., Müller V., Rau T., Schindlbeck C., Rack B., et al. (2007). Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin. Cancer Res. 13 920–928. 10.1158/1078-0432.ccr-06-1695 [DOI] [PubMed] [Google Scholar]
- Rios A. C., Capaldo B. D., Vaillant F., Pal B., van Ineveld R., Dawson C. A., et al. (2019). Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35 618–632. 10.1016/j.ccell.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Risom T., Langer E. M., Chapman M. P., Rantala J., Fields A. J., Boniface C., et al. (2018). Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat. Commun. 9:3815. 10.1038/s41467-018-05729-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A., Ram O., Shoresh N., Sperling R. A., Goren A., Weitz D. A., et al. (2015). Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. J. Cell Sci. 33:1165. 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti M., Dadashian E. L., Guo W., Quach B., Mulholland D. J., Park J. W., et al. (2016). HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene 35 3781–3795. 10.1038/onc.2015.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M., Emami S., Redeuilh G., Julien S., Prevost G., Zimber A., et al. (2008). Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist. Updat. 11 123–151. 10.1016/j.drup.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Sánchez-Martínez R., Cruz-Gil S., Gómez de Cedrón M., Álvarez-Fernández M., Vargas T., Molina S., et al. (2015). A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget 6 38719–38736. 10.18632/oncotarget.5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarioglu A. F., Aceto N., Kojic N., Donaldson M. C., Zeinali M., Hamza B., et al. (2015). A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 12 685–691. 10.1038/nmeth.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C., Eaton E. N., Li S. H., Chaffer C. L., Reinhardt F., Kah K. J., et al. (2011). Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145 926–940. 10.1016/j.cell.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D., Zanotelli V. R. T., Fischer J. R., Schapiro D., Engler S., Lun X.-K., et al. (2018). Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 6 25–36. 10.1016/j.cels.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Gomez S. J., Maziveyi M., Alahari S. K. (2016). Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 15:8. 10.1186/s12943-016-0502-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang M., Soon R. H., Lim C. T., Khoo B. L., Han J. (2019). Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab. Chip. 19 369–386. 10.1039/c8lc00970h [DOI] [PubMed] [Google Scholar]
- Sharma S. V., Lee D. Y., Li B., Quinlan M. P., Takahashi F., Maheswaran S., et al. (2010). A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141 69–80. 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikandar S. S., Kuo A. H., Kalisky T., Cai S., Zabala M., Hsieh R. W., et al. (2017). Role of epithelial to mesenchymal transition associated genes in mammary gland regeneration and breast tumorigenesis. Nat. Commun. 8:1669 10.1038/s41467-017-01666-1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit M. A., Geiger T. R., Song J. Y., Gitelman I., Peeper D. S. (2009). A Twist-Snail axis critical for TrkB-induced epithelial-mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol. Cell. Biol. 29 3722–3737. 10.1128/mcb.01164-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S., Schmalhofer O., Wahlbuhl M., Dimmler A., Bauer K., Sultan A., et al. (2008). The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 68 537–544. 10.1158/0008-5472.Can-07-5682 [DOI] [PubMed] [Google Scholar]
- Stemmler M. P., Eccles R. L., Brabletz S., Brabletz T. (2019). Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21 102–112. 10.1038/s41556-018-0196-y [DOI] [PubMed] [Google Scholar]
- Stuelten C. H., Parent C. A., Montell D. J. (2018). Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat. Rev. Cancer 18 296–312. 10.1038/nrc.2018.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou N., Lehman M. L., Wang C., Fard A. T., Rockstroh A., Fazli L., et al. (2019). A molecular portrait of epithelial-mesenchymal plasticity in prostate cancer associated with clinical outcome. Oncogene 38 913–934. 10.1038/s41388-018-0488-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Causado A., Caballero-Diaz D., Bertran E., Roncero C., Addante A., Garcia-Alvaro M., et al. (2015). HGF/c-Met signaling promotes liver progenitor cell migration and invasion by an epithelial-mesenchymal transition-independent, phosphatidyl inositol-3 kinase-dependent pathway in an in vitro model. Biochim. Biophys. Acta 1853(10 Pt A), 2453–2463. 10.1016/j.bbamcr.2015.05.017 [DOI] [PubMed] [Google Scholar]
- Sureshbabu A., Okajima H., Yamanaka D., Tonner E., Shastri S., Maycock J., et al. (2012). IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J. Cell Sci. 125 1693–1705. 10.1242/jcs.092882 [DOI] [PubMed] [Google Scholar]
- Suvà M. L., Riggi N., Bernstein B. E. (2013). Epigenetic reprogramming in cancer. Science 339:1567. 10.1126/science.1230184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachtsidis A., McInnes L. M., Jacobsen N., Thompson E., Saunders C. M. J. (2016). Minimal residual disease in breast cancer: an overview of circulating and disseminated tumour cells. Clin. Exp. Metast. 33 521–550. 10.1007/s10585-016-9796-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. L., Weinberg R. A. (2013). The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19 1438–1449. 10.1038/nm.3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura G., Yin J., Wang S., Fleisher A. S., Zou T., Abraham J. M., et al. (2000). E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J. Natl. Cancer Inst. 92 569–573. 10.1093/jnci/92.7.569 [DOI] [PubMed] [Google Scholar]
- Tang M., Shen H., Jin Y., Lin T., Cai Q., Pinard M. A., et al. (2013). The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition. J. Biol. Chem. 288 27680–27691. 10.1074/jbc.M113.482349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K., Gottesman M. M. (2015). Beyond 3D culture models of cancer. Sci. Transl. Med. 7:283s289. 10.1126/scitranslmed.3009367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009). Epithelial-mesenchymal transitions in development and disease. Cell 139 871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Thompson E. W., Nagaraj S. H. (2018). Transition states that allow cancer to spread. Nature 556 442–444. 10.1038/d41586-018-04403-x [DOI] [PubMed] [Google Scholar]
- Thuault S., Tan E. J., Peinado H., Cano A., Heldin C. H., Moustakas A. (2008). HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 283 33437–33446. 10.1074/jbc.M802016200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting D. T., Wittner B. S., Ligorio M., Jordan N. V., Shah A. M., Miyamoto D. T., et al. (2014). Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8 1905–1918. 10.1016/j.celrep.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D. D., Kratz A., Park J. G., Barrientos E. S., Saini H., Nguyen T., et al. (2019). A Human organotypic microfluidic tumor model permits investigation of the interplay between patient-derived fibroblasts and breast cancer cells. Cancer Res. 79 3139–3151. 10.1158/0008-5472.Can-18-2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. H., Donaher J. L., Murphy D. A., Chau S., Yang J. (2012). Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22 725–736. 10.1016/j.ccr.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa T., Kumar S., Borkar R. N., Azimi V., Thibault G., Chang Y. H., et al. (2017). Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 19 203–217. 10.1016/j.celrep.2017.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Denderen B. J., Thompson E. W. (2013). Cancer: the to and fro of tumour spread. Nature 493 487–488. 10.1038/493487a [DOI] [PubMed] [Google Scholar]
- Veening J.-W., Smits W. K., Kuipers O. P. (2008). Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62 193–210. 10.1146/annurev.micro.62.081307.163002 [DOI] [PubMed] [Google Scholar]
- Voulgari A., Pintzas A. (2009). Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim. Biophys. Acta 1796 75–90. 10.1016/j.bbcan.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Vrba L., Jensen T. J., Garbe J. C., Heimark R. L., Cress A. E., Dickinson S., et al. (2010). Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One 5:e8697. 10.1371/journal.pone.0008697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin J. J., Guan J., Edgar K. A., Zhou W., Francis R., Torres A. C., et al. (2012). Active PI3K pathway causes an invasive phenotype which can be reversed or promoted by blocking the pathway at divergent nodes. PLoS One 7:e36402. 10.1371/journal.pone.0036402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.-B., Yang X.-Q., Yang S., Wang B.-C., Feng M.-H., Tu J.-C. (2011). A higher number of circulating tumor cells (CTC) in peripheral blood indicates poor prognosis in prostate cancer patients–a meta-analysis. Asian Pac. J. Cancer Prev. 12 2629–2635. [PubMed] [Google Scholar]
- Wang R., Li H., Guo X., Wang Z., Liang S., Dang C. (2016). IGF-I induces epithelial-to-mesenchymal transition via the IGF-IR-Src-MicroRNA-30a-E-cadherin pathway in nasopharyngeal carcinoma cells. Oncol. Res. 24 225–231. 10.3727/096504016x14648701447931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-C., Zhang X.-F., Peng J., Li X.-F., Wang A.-L., Bie Y.-Q., et al. (2018). Survival mechanisms and influence factors of circulating tumor cells. Biomed. Res. Interna. 2018:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science 123 309–314. 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., et al. (2009). The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11 1487–1495. 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- Williams E. D., Gao D., Redfern A., Thompson E. W. (2019). Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer 19 716–732. 10.1038/s41568-019-0213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witta S. E., Gemmill R. M., Hirsch F. R., Coldren C. D., Hedman K., Ravdel L., et al. (2006). Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 66 944–950. 10.1158/0008-5472.can-05-1988 [DOI] [PubMed] [Google Scholar]
- Wong C. C., Zhang H., Gilkes D. M., Chen J., Wei H., Chaturvedi P., et al. (2012). Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J. Mol. Med. 90 803–815. 10.1007/s00109-011-0855-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Won J.-Y., Kim C.-H., Kim D.-E., Yim H. (2019). Roles of the phosphorylation of transcriptional factors in epithelial-mesenchymal transition. J. Oncol. 2019:5810465. 10.1155/2019/5810465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Quan Y., Wang W., Fu Q., Wu J., Mei T., et al. (2012). Dynamic equilibrium between cancer stem cells and non-stem cancer cells in human SW620 and MCF-7 cancer cell populations. Br. J. Cancer 106 1512–1519. 10.1038/bjc.2012.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., et al. (2004). Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117 927–939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Yang J., Weinberg R. A. (2008). Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14 818–829. 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yang Y., Jolly M. K., Levine H. (2019). Computational modeling of collective cell migration: mechanical and biochemical aspects. Adv. Exp. Med. Biol. 1146 1–11. 10.1007/978-3-030-17593-1_1 [DOI] [PubMed] [Google Scholar]
- Yauch R. L., Januario T., Eberhard D. A., Cavet G., Zhu W., Fu L., et al. (2005). Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin. Cancer Res. 11(24 Pt 1), 8686–8698. 10.1158/1078-0432.ccr-05-1492 [DOI] [PubMed] [Google Scholar]
- Ye X., Brabletz T., Kang Y., Longmore G. D., Nieto M. A., Stanger B. Z., et al. (2017). Upholding a role for EMT in breast cancer metastasis. Nature 547 E1–E3. 10.1038/nature22816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Tam W. L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., et al. (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525 256–260. 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y. A., Kang M. H., Lee H. J., Kim B. H., Park J. K., Kim H. K., et al. (2011). Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 71 7061–7070. 10.1158/0008-5472.can-11-1338 [DOI] [PubMed] [Google Scholar]
- Yu M., Bardia A., Aceto N., Bersani F., Madden M. W., Donaldson M. C., et al. (2014). Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345 216–220. 10.1126/science.1253533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Bardia A., Wittner B. S., Stott S. L., Smas M. E., Ting D. T., et al. (2013). Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339 580–584. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu H., Han N., Xu H., Chu Q., Yu S., et al. (2014). Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J. Hematol. Oncol. 7:87. 10.1186/s13045-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M., Shah A. A., Kalluri R. (2005). Bone morphogenic protein-7 induces mesenchymal to epithelial transition in adult renal fibroblasts and facilitates regeneration of injured kidney. J. Biol. Chem. 280 8094–8100. 10.1074/jbc.M413102200 [DOI] [PubMed] [Google Scholar]
- Zhang N., Zhang H., Liu Y., Su P., Zhang J., Wang X., et al. (2019). SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death Differ. 26 843–859. 10.1038/s41418-018-0158-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Carstens J. L., Kim J., Scheible M., Kaye J., Sugimoto H., et al. (2015). Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527 525–530. 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]