Abstract
Autism spectrum disorder (ASD) is a group of neurodevelopmental disorders with multiple biological etiologies and highly variable symptoms. Using a novel analytical framework that integrates cortex-wide MRI markers of vertical (i.e., thickness, tissue contrast) and horizontal (i.e., surface area, geodesic distance) cortical organization, we could show that a large multi-centric cohort of individuals with ASD falls into 3 distinctive anatomical subtypes (ASD-I: cortical thickening, increased surface area, tissue blurring; ASD-II: cortical thinning, decreased distance; ASD-III: increased distance). Bootstrap analysis indicated a high consistency of these biotypes across thousands of simulations, while analysis of behavioral phenotypes and resting-state fMRI showed differential symptom load (i.e., Autism Diagnostic Observation Schedule; ADOS) and instrinsic connectivity anomalies in communication and social-cognition networks. Notably, subtyping improved supervised learning approaches predicting ADOS score in single subjects, with significantly increased performance compared to a subtype-blind approach. The existence of different subtypes may reconcile previous results so far not converging on a consistent pattern of anatomical anomalies in autism, and possibly relate the presence of diverging corticogenic and maturational anomalies. The high accuracy for symptom severity prediction indicates benefits of MRI biotyping for personalized diagnostics and may guide the development of targeted therapeutic strategies.
Keywords: autism, biomarker, biotype, computational neuroanatomy, magnetic resonance imaging, subtype
Introduction
Autism spectrum disorder (ASD) is a pervasive group of neurodevelopmental conditions currently diagnosed in >1% of children (CDC 2014). ASD is characterized by impairments in social cognition, communication, together with narrow and repetitive behaviors and interests (American Psychiatric Association 2013). A key challenge for the development of objective diagnostics and targeted therapy in ASD is high heterogeneity across individuals, not only with respect to behavioral symptoms but also in terms of etiological factors (Betancur 2011).
Structural magnetic resonance imaging (MRI) is suitable to describe brain anatomy with high resolution, versatility, and biological validity (Miller 2004; Toga et al. 2006). Several MRI features have been applied to study atypical anatomy in autism, including morphological markers such as cortical thickness and surface area (Ecker, Ginestet et al. 2013; Hazlett et al. 2017; Yang et al. 2016), measures of tissue intensity and contrast at the cortical interface (Petropoulos et al. 2006; Boddaert et al. 2009; Andrews et al. 2017), as well as markers of distance between different regions to index wiring cost (Ecker, Ronan et al. 2013). Collectively, these indices capture important aspects of both vertical as well as horizontal cortical organization, and may thus reveal manifestations of different corticogenic and neurodevelopmental disruptions (Hong, Bernhardt, Caldairou et al. 2017). However, previous studies that compared cohorts with ASD to typical controls have reported rather diverse morphological anomalies, with only little agreement of results (DeRamus and Kana 2015; Bernhardt et al. 2017; Haar et al. 2016).
One explanation for inconsistent findings might be the existence of different ASD subtypes which may each have a specific neuroanatomical profile (Ecker and Murphy 2014). Conventional analyses that compare ASD to control groups may average across important inter-individual variability, ultimately representing a suboptimal approach. Conversely, unsupervised statistical learning techniques provide an appropriate framework to detect groups of individuals with similar profiles across multiple feature dimensions. Notably, these approaches are not biased towards the involvement of particular regions, and may identify complex inter-feature relations not apparent from standard univariate analyses. In other neurodevelopmental conditions, including attention-deficit hyperactivity disorder, schizophrenia, and epilepsy, these techniques have recently discovered subtypes with divergent histopathological profiles (Bernhardt et al. 2015; Hong, Bernhardt, Caldairou et al. 2017; Hong, Bernhardt, Gill et al. 2017) and behavioral outcomes (Fair et al. 2012; Dias et al. 2015; Sun et al. 2015; Yang et al. 2012; Van Dam et al. 2016), suggesting benefits for early clinical prognosis.
We developed a novel multidimensional imaging framework to address neuroanatomical variability across the autism spectrum. Our approach was developed on a subsample of the Autism Brain Imaging Data Exchange I (ABIDE-I) repository, which aggregates MRI and phenotypic information of individuals with ASD and controls across multiple sites (Di Martino et al. 2014). In brief, we profiled single individuals using a battery of MRI features that tap into complementary aspects of brain anatomy (i.e., cortical thickness, surface area, geodesic distance, tissue contrast). Following the correction of age- and site-specific effects, we applied agglomerative hierarchical clustering on these profiles to partition our ASD group into new classes with distinct neuroanatomical phenotypes. We assessed symptom severity load and intrinsic functional network anomalies in each class to determine the clinical and functional significance of the MRI-driven subtypes. To furthermore evaluate our approach in a diagnostic setting, we assessed whether the subtyping procedure improved individualized prediction of symptom severity using a supervised pattern-learning paradigm.
Materials and Methods
Subjects
The current work was based on a previously described ABIDE-I subsample (Valk et al. 2015). Briefly, we selected those sites that included both children and adults with ASD and typical controls, with ≥10 individuals/group (n = 297). We restricted the analysis to males given the low prevalence of female data. A detailed quality control (Valk et al. 2015) included only cases with acceptable MRI and surface-extraction quality. This resulted in 220 individuals from 3 sites: 1) NYU Langone Medical Center (NYU, 52/40 ASD/controls); 2) University of Utah, School of Medicine (USM, 20/22 ASD/controls); 3) University of Pittsburg, School of Medicine (PITT, 35/51 ASD/controls).
Individuals with ASD had DSM-IV-TR diagnosis of Autistic Disorder, Asperger’s Disorder, or Pervasive Developmental Disorder Not-Otherwise-Specified, established by expert clinical opinion aided by “gold standard” diagnostic instruments: the Autism Diagnostic Observation Schedule, ADOS (Lord et al. 2000), and/or the Autism Diagnostic Interview-Revised, ADI-R (Lord et al. 1994). Both instruments focus on 3 domains including reciprocal social interactions, communication and language, and restricted/repeated behaviors. In PITT and USM datasets, individuals diagnosed with disorders such as Fragile-X or tuberous sclerosis were explicitly excluded. In the NYU dataset, individuals with current chronic systemic conditions were excluded. Full-scale/performance/verbal intelligence quotation (IQ) was measured by WASI, WAIS III, and/or WISC III (Wechsler 1999). Controls had no history of mental disorders and were statistically matched for age to the ASD group at each site.
MRI Acquisition
High-resolution T1-weighted images (T1w) and resting-state functional MRIs (rs-fMRI) were available from all sites; they were collected on 3 T Siemens scanners. NYU data were acquired on an Allegra using 3D-TurboFLASH for T1w (TR = 2530 ms; TE = 3.25 ms; TI = 1100 ms; flip angle = 7°; matrix = 256 × 256; 1.3 × 1.0 × 1.3 mm3 voxels) and GRE-EPI for rs-fMRI (TR = 2000 ms; TE = 15 ms; flip angle = 90°; matrix = 80 × 80; 180 vols, 3.0 × 3.0 × 4.0 mm3 voxels). PITT data were acquired on an Allegra using 3D-MPRAGE for T1w (TR = 2100 ms; TE = 3.93 ms; TI = 1000 ms; flip angle = 7°; matrix = 269 × 269; 1.1 × 1.1 × 1.1 mm3 voxels) and 2D EPI for rs-fMRI (TR = 1500 ms; TE = 35 ms; flip angle = 70°; matrix size = 64 × 64; 200 volumes, 3.1 × 3.1 × 4.0 mm3 voxels). USM data were acquired on a TrioTim using 3D-MPRAGE for T1w (TR = 2300 ms; TE = 2.91 ms; TI = 900 ms; flip angle = 9°; matrix = 240 × 256; 1.0 × 1.0 × 1.2 mm3 voxels) and 2D EPI for rs-fMRI fMRI (TR = 2000 ms; TE = 28 ms; flip angle = 90°; matrix = 64 × 64; 240 volumes; 3.4 × 3.4 × 3.0 mm3 voxels).
MRI Processing
Structural MRI. We used FreeSurfer (5.3; http://surfer.nmr.mgh.harvard.edu/) to process T1w MRI. Details on the processing are described elsewhere (Fischl et al. 1999). Individual surfaces were aligned to an average spherical representation, fsaverage5 (a triangular mesh with 20 484 surface points or “vertices”), improving point-correspondence with regards to sulcation. Extractions were visually inspected and segmentation inaccuracies manually corrected by 2 raters blind to diagnosis (SLV, BCB). Subjects with faulty segmentations, movement, or other artifacts were excluded (n = 77/297).
rs-fMRI. We examined the ABIDE I rs-fMRI data, preprocessed by the ABIDE Preprocessed Connectome Project (http://preprocessed-connectomes-project.org/abide/) initiative. Processing was based on C-PAC (https://fcp-indi.github.io/) and included slice-time and head motion correction, skull stripping, and intensity normalization. Statistical corrections furthermore removed effects of head motion (Friston et al. 1996) and the top 5 principal components from white matter and cerebro-spinal fluid (using “CompCor” (Behzadi et al. 2007)), and linear/quadratic trends. After band-pass filtering (0.01–0.1 Hz), we registered the functional data to the MNI152 template through a combination of linear and non-linear transformations. We non-linearly registered the T1w to the same template, and projected cortical surfaces onto rs-fMRI. The position of surfaces in rs-fMRI was visually evaluated and confirmed for each case. Finally, we mapped voxel-wise fMRI time-series to surface models running at 50% thickness using nearest neighbor interpolation and applied a surface-based 5 mm full-width-at-half-maximum Gaussian diffusion kernel.
MRI quality control was complemented by assessment of signal-to-noise ratio (SNR) and visual scoring of surface extraction for structural MRI, and evaluation of temporal derivatives of time courses and mean frame-wise displacement (FD) for rs-fMRI (Jenkinson et al. 2002; Power et al. 2012) (Supplementary Fig. 1).
Surface-based Feature Generation
For subject-wise neuroanatomical profiling (Fig. 1), we computed 4 features previously reported as abnormal in neuroimaging and postmortem studies of ASD (Avino and Hutsler 2010; Wegiel et al. 2010; Ecker, Ginestet et al. 2013; Ecker, Ronan et al. 2013; Bernhardt et al. 2014; Wallace et al. 2013; Yang et al. 2016). Notably, while cortical thickness and intensity contrasts are common features to describe vertical cortical organization, surface area and geodesic distance index properties of the cortical ribbon in horizontal direction (Ecker, Ronan et al. 2013; Hong et al. 2016):
“Cortical thickness” was measured as the distance of corresponding vertices between the gray-white and pial bounary. It has previously been related to neuronal/synaptic density (Huttenlocher 1990; la Fougere et al. 2011).
“Intensity contrast” was quantified as the intensity gradient between voxels 0.5 mm above/below the gray-white matter boundary in surface-normal direction. Reduced contrast may signify “cortical blurring”, a remnant of atypical neuronal migration and cortical organization (Wegiel et al. 2010; Casanova et al. 2013; Andrews et al. 2017; Hong, Bernhardt, Caldairou et al. 2017).
“Cortical surface area” was measured as the area of triangles surrounding a vertex along the white matter interface (Winkler et al. 2012; Ecker, Ginestet et al. 2013; Wallace et al. 2013). Areal changes possibly reflect alterations in cortical columnar organization (Casanova et al. 2002) and atypical proliferation of radial unit progenitor cells (Ecker and Murphy 2014).
“Geodesic distance” was measured as length of the shortest path between 2 points (i.e., 2 surface vertices) running through the cortical mantle using an approach invariant to mesh configuration (Griffin 1994). This metric has been related to “intrinsic cortico-cortical connectivity”, with closer vertices being more closely integrated, and structural wiring cost (Ecker, Ronan et al. 2013).
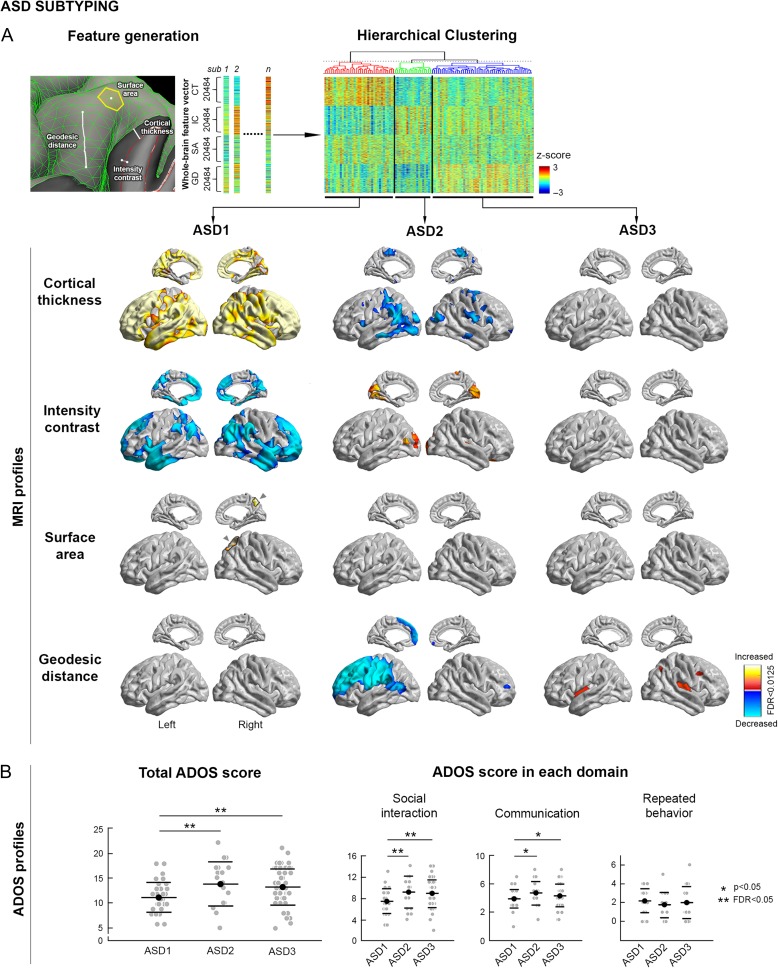
Figure 1.
ASD subtyping. (A) Top: The 4 MRI-based features (cortical thickness, surface area, intensity contrast, geodesic distance) used for subject-wise profiling. Bottom: Three classes (ASD-I, II, III) optimally described our multi-centric cohort. Features are corrected for age and site effects. Shown are differences in MRI features compared to controls (increases/decreases in red/blue). Cortex-wide significances were corrected using the false discovery rate procedure, furthermore adjusted by the number of features (qFDR < 0.05/4 = 0.0125). (B) Symptom severity profiles of ASD classes based on total and domain-specific ADOS scores (i.e., social interaction, communication, repeated behavior). **Difference between subgroups at qFDR < 0.05; *Difference at P < 0.05 uncorrected.
Feature maps were smoothed on tessellated surfaces using a 20 mm full-width-at-half-maximum kernel. Surface-based smoothing reduces measurement noise but preserves sensitivity for localization, as it respects cortical topology (Lerch and Evans 2005). Prior to subtyping, we statistically corrected features for age and site effects. Following previous recommendations, we additionally corrected intensity contrast for mean gray/white matter intensity (Davatzikos and Resnick 2002; Salat et al. 2011), surface area for total white matter volume (Ecker, Ginestet et al. 2013), and geodesic distance for total surface area (Ecker, Ronan et al. 2013). Site effects were indeed removed from all features (Supplementary Fig. 2), and there were no significant site-by-group interactions following the correction.
We furthermore assessed test–retest correlations for our features, that is, cortical thickness, surface area, contrast, and geodesic distance. We used data from 25 healthy individuals that were scanned twice in the same session and openly shared through the Consortium for Reliability and Reproducibility repository, CoRR [site: NYU-2; (Zuo et al. 2014)]. Generating MRI features using identical processing as in our original study, we computed surface-wide correlations between the first and second scan for all individuals. Correlation coefficients were above 0.9 across all features (mean±SD r: cortical thickness = 0.91±0.07, surface area = 0.97±0.09, intensity = 0.91 ± 0.08, distance = 0.99±0.03), indicating high stability.
Neuroanatomical ASD Subtyping
At each surface point, we normalized feature data in each individual with ASD against the corresponding distribution in controls using vertex-wise z-scoring (Bernhardt et al. 2015). Concatenating normalized feature vectors across subjects yielded a multi-variate profile matrix with 113 rows (one per subject) and 81 936 columns (4 features with 20 484 entries each). Prior to clustering, we evaluated feature cross-correlations for each subject. Correlation coefficients were small-to-moderate (“thickness-vs-contrast” = −0.16 ± 0.14, “thickness-vs-area” = −0.07 ± 0.09, “thickness-vs-distance” = −0.10 ± 0.13, “contrast-vs-area” = 0.01 ± 0.11, “contrast-vs-distance” = 0.05 ± 0.2, “area-vs-distance” = 0.34 ± 0.09), supporting feature complementarity.
Agglomerative hierarchical clustering partitioned our ASD group into subclasses, with cases in the same class having similar profiles, while those in different classes had different profiles. Between-cluster distances were estimated using “Ward’s” minimum variance approach (Ward 1963). The clustering solution depends on the number of a priori selected classes k. After evaluating k = 1–20, we determined the optimal k as the one minimizing the ratio between intra-cluster to inter-cluster distance (Davies and Bouldin 1979).
For each newly detected ASD subtype, we compared structural profiles across all 4 features relative to controls using surface-based Student’s t-tests (Worsley et al. 2009). To evaluate reproducibility of these profiles, we carried bootstrap-based stability tests with 1000 iterations. In each iteration, we re-clustered randomly subsampled subjects with replacement from our ASD cohort and comparing them to healthy controls. Results were corrected for multiple comparisons at a false discovery rate of qFDR = 0.0125 = 0.05/4, additionally accounting for the number of features assessed (Benjamini and Hochberg 1995).
To assess specificity, we repeated the clustering of multidimensional MRI features in controls, where feature z-scores were computed using a leave-one-out strategy, in which a single individual was normalized based on the distribution of all remaining controls.
Relation to Behavioral Symptoms and Functional Networks
We complemented class-wise structural mapping by comparing demographic and clinical variables between ASD subtypes, including age, full-scale IQ, and ADOS symptom severity using Student’s t-tests. We verified that classes were composed out of similar proportions of individuals per site, based on Fisher’s exact tests.
We related our classes to intrinsic functional networks thought to contribute to social cognition/interactions and communication-key processes known to be impaired in ASD. We extracted regions-of-interest (ROIs) from Neurosynth.org, a platform for large-scale synthesis of previous fMRI results (Yarkoni et al. 2011). To approximate the network involved in social cognition/interaction, we applied reverse inference on the terms “mentalizing”, “social interactions”, and “social cognition” and formed the geometric union of findings. To approximate the network involved in verbal/non-verbal communication, we applied reverse inference on the terms “language”, “gesture”, and “facial expression” and formed their geometric union. After projecting ROIs on the cortical surface of individual subjects, we systematically extracted mean functional time-series for each ROI and computed inter-ROI time-series correlations to construct mentalizing and communication networks for each subject. Functional connectivity estimates in each class were finally compared to controls, with age, site, and mean FD as nuisance variables (Power et al. 2012). Similar to the structural analyses, we corrected findings at qFDR = 0.05/2, accounting for the 2 networks tested.
Automated Prediction of Symptom Severity
We developed a supervised learning framework predicting ADOS symptom severity scores in individual patients based on MRI profiles. To evaluate benefits of our subtyping, we compared prediction performance between learners operating on each newly discovered class independently to that of a learner operating on the full dataset without clustering. To reduce data dimensionality, we parcellated the brain into 1000 regions-of-interest (ROIs) with comparable surface area (Cammoun et al. 2012). This approach first identifies macroscopic parcels using automated anatomical labeling (AAL) (Tzourio-Mazoyer et al. 2002). We estimated the number of ROIs per AAL parcel according to its surface area relative to the whole-cortex. Through repeated k-means clustering with different random seeds within each AAL parcel, we systematically subdivided each parcel 100 times and determined the consensus parcellation across all initializations. Calculating mean feature values in these parcels resulted in 4000 features per subject (i.e., 4 features × 1000 parcels).
Prediction encompassed 2 stages: “model building” (feature selection, training, validation) and “testing” (performance evaluation). Both stages relied on “ensemble” methods that combine several weaker base classifiers to generate a final, strong classifier (Dietterich 2000). For feature selection, we used a “random forest”, which constructs multiple decision trees and reduces overfitting by randomly choosing subsets of features (Breiman 2001). For actual prediction, we applied “gradient boosting”, which optimizes performance by iteratively giving higher weights to previously misclassified cases (Friedman 2002). Optimal parameters were determined through grid search (“random forest”: 100 trees, 100 randomly sampled features, minimum leaf size = 5; “gradient boosting”: 100 boosts, 5 features at each split, minimum leaf size = 5, least square regression learner).
Model building and testing followed nested 10-fold cross-validation with 100 iterations. At each iteration, 9-folds of dataset were used for model training and validation in an out-of-bag mode (i.e., 75% of data in the 9-folds were subsampled for feature selection and training, the remaining 25% for validation and parameter optimization). The remaining fold was used for testing. Notably, feature selection iteratively collected metrics giving a positive contribution to prediction during out-of-bag validations, and stopped the procedure if >20 features were found.
We carried out 1000 permutation tests with randomly shuffled ADOS scores to determine whether classifier performance exceeded chance. Moreover, observed and predicted ADOS scores were compared using the Pearson correlation coefficient, r, and mean absolute error, MAE. Two-sample t-tests compared indices between a framework with/without clustering.
Results
Demographic and Clinical Data
Across sites, ASD and controls had similar age (mean ± SD in years: ASD: 20.9 ± 8.0; Controls: 19.3 ± 7.3; P = 0.14). Conversely, individuals with ASD had lower full-scale IQ (ASD: 104 ± 16; Controls: 114 ± 12; P < 0.001), performance IQ (ASD: 106 ± 15; Controls: 112 ± 13; P < 0.002), and verbal IQ (ASD: 102 ± 17; Controls: 113 ± 12; P < 0.001) compared to controls. ADOS was available for raw score across all patients (total [social and communication]: 12.7 ± 3.7; social: 8.4 ± 2.7; communication: 4.3 ± 1.5; repeated behavior: 2.0 ± 1.5). ADOS calibrated severity scores (CSS), which account for subject characteristics that may contribute to ADOS score variability, were provided for some but not all cases in the ABIDE dataset. We therefore followed a previously proposed heuristic (Moradi et al. 2017) to approximate CSS consistently across all cases using established conversion criteria (Gotham et al. 2009; Hus and Lord 2014). For each individual, we used the information of a raw ADOS score (i.e., a total of the social and communication items), age, and ADOS modules used and retrieved an approximate severity score using a lookup table provided by Gotham et al. (module 1–3) and Hus and Lord (module 4) (calibrated total score: 7.8 ± 1.7; modules: module 1 [none], module 2 [n = 1], module 3 [n = 40], module 4 [n = 66]). Demographic patterns were similar across the 3 sites, except for no difference of IQ compared to controls (P > 0.3) in PITT, while IQ was lower in ASD than in controls in the other sites. For breakdown by site, please see Table 1.
Table 1.
Demographic and clinical data
| Controls (113) | ASD (107) | USM | PITT | NYU | ||||
|---|---|---|---|---|---|---|---|---|
| Controls (40) | ASD (52) | Controls (22) | ASD (20) | Controls (51) | ASD (35) | |||
| Age | 19.3 ± 7.3 | 20.9 ± 8.0 | 21.5 ± 7.8 | 23.6 ± 7.6 | 19.7 ± 7.0 | 20.8 ± 7.3 | 17.5 ± 6.7 | 16.8 ± 7.5 |
| IQ | 114 ± 12.3 | 104 ± 15.7 | 115 ± 13.9 | 101 ± 16.5 | 110 ± 8.95 | 113 ± 13.5 | 115 ± 12.1 | 105 ± 13.8 |
| Type (A/AS/P) | — | 94/10/3 | — | 51/0/1 | — | 20/0/0 | — | 23/10/2 |
| ADOS | ||||||||
| Totala | — | 12.7 ± 3.7 | — | 13.6 ± 3.3 | — | 12.7 ± 3.1 | — | 11.3 ± 4.2 |
| Social | — | 8.4 ± 2.7 | — | 8.9 ± 2.5 | — | 8.5 ± 2.3 | — | 7.7 ± 3.0 |
| Comm | — | 4.3 ± 1.5 | — | 4.7 ± 1.4 | — | 4.3 ± 1.1 | — | 3.6 ± 1.7 |
| Repeat | — | 2.0 ± 1.5 | — | 1.8 ± 1.8 | — | 2.7 ± 1.2 | — | 2.1 ± 1.1 |
Age, full-scale IQ, and severity symptom scores are presented in mean ± SD. Abbreviation: “ASD”, autism spectrum disorder; “A”, “AS”, “P” in Type represents Autism, Asperger Syndrome and Pervasive developmental disorder—not otherwise specified, respectively; “ADOS”, Autism Diagnostic Observation Schedule; “IQ”, intelligence quotient; “Social”, social interaction; “Comm”, communication; “Repeat”, stereotyped behaviors and restricted interests.
aTotal score = social interaction + communication.
ASD Subtyping: MRI Findings
Results of the hierarchical clustering are shown in Fig. 1. Our ASD cohort was optimally partitioned by 3 classes, based on the ratio of intra-/inter-cluster distances (Supplementary Fig. 3), which showed distinctive MRI profiles compared to controls. ASD-I (n = 36) displayed diffusely increased thickness (qFDR < 0.0125; mean ± SD Cohen’s d in regions of findings = 0.67±0.17), together with cortical interface blurring in frontal, parietal, and temporal lobes (qFDR < 0.0125; d = 0.65±0.07). Separate analysis additionally correcting thickness measures for blurring showed virtually identical findings (Supplementary Fig. 4). Moreover, while geodesic distance was not altered in this group, we observed increased medial parietal surface area (qFDR < 0.0125; d = 0.74±0.06). Conversely, ASD-II (n = 19) presented with widespread thinning in central and posterior cortices, together with increased contrast in lateral occipital cortices and decreased distance of a large left frontal territory spanning from frontopolar to central regions (qFDR < 0.0125; d > 0.60 across features). Last, ASD-III (n = 52) presented with a selective increase in distance of insular-opercular regions (qFDR < 0.0125; d = 0.58±0.03), but otherwise normal profiles.
Bootstrap stability analyses supported high reproducibility of our biotyping, confirming the signature anomalies in the discovered biotypes (i.e., increased cortical thickening/blurring in ASD-I; decreased distance in ASD-II; increased distance in ASD-III) in 75–100% of the simulations (Supplementary Fig. 5).
Given findings suggesting effects of subtle head motion on structural MRI markers (Reuter et al. 2015), we adopted a recent approach to approximate the tendency of a subject to move in the scanner based on head motion parameters derived from resting-state fMRI (Savalia et al. 2017). Here, we corrected structural MRI profiles for mean FD and the number of TRs with excessive FD (>0.2 mm) (Jenkinson et al. 2002; Power et al. 2012). Repeating the clustering for corrected features yielded 3 classes with similar overall imaging phenotypes, suggesting only little influence of motion on the subtyping solution (Supplementary Fig. 6).
Clustering in controls revealed 2 classes. Notably, alterations were not significantly different from zero following correction for multiple comparisons, supporting that control classes likely fell into the spectrum of normal brain variability (Supplementary Fig. 7).
ASD Subtyping: Clinical Findings and Functional Network Anomalies
While newly discovered ASD classes showed similar age, IQ, and site composition (P > 0.2) (Table 2), they differed with regards to total ADOS symptom severity (ANOVA F = 4.02, P = 0.021), particularly when comparing ASD-II and ASD-III to ASD-I (qFDR < 0.04). Findings were primarily related to selective deficits in ADOS social interaction subscores (qFDR < 0.05), and marginally to ADOS communication subscores (uncorrected P < 0.03). Conversely, there was no between-class difference with respect to repetitive behavioral ADOS subscores.
Table 2.
Demographic and clinical profiles across subgroups
| ASD1 (36) | ASD2 (19) | ASD3(52) | |
|---|---|---|---|
| Age | 21.1 ± 10.5 | 21.3 ± 7.7 | 20.5 ± 6.1 |
| IQ | 105 ± 17.8 | 107 ± 14.5 | 103 ± 14.6 |
| Site (USM/PITT/NYU) | 17/5/14 | 9/6/4 | 26/9/17 |
| Type (A/AS/P) | 31/4/1 | 19/0/0 | 44/6/2 |
| ADOS | |||
| Total | 11.3 ± 3.0 | 13.7 ± 4.4 | 13.2 ± 3.6 |
| Social | 7.4 ± 2.3 | 9.1 ± 3.0 | 8.9 ± 2.6 |
| Comm | 3.9 ± 1.3 | 4.7 ± 1.6 | 4.4 ± 1.6 |
| Repeat | 2.2 ± 1.3 | 1.7 ± 1.4 | 2.0 ± 1.7 |
See Table 1 for further details.
Repeating group comparisons based on approximated calibrated symptom severity scores resulted in between-class differences that were similar to findings based on raw ADOS scores (ASD-I vs. ASD–II: 7.2±1.7 vs. 8.3±1.9, qFDR < 0.05, t = 2.16; ASD-I vs. ASD-III: 7.2±1.7 vs. 8.2±1.6, qFDR < 0.02, t = 2.61).
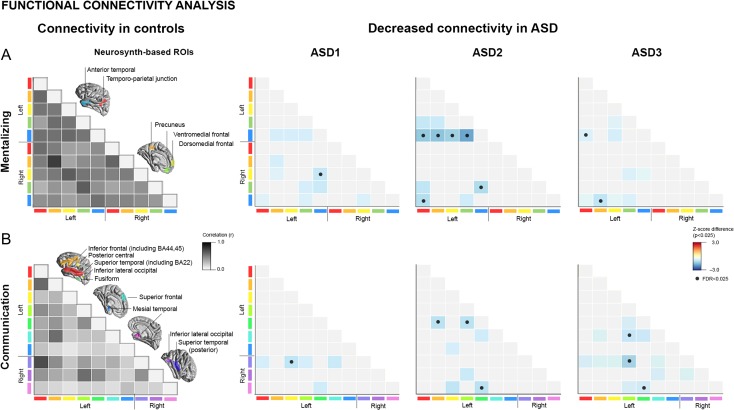
Mirroring symptom load, classes presented with differential intrinsic functional network anomalies (Fig. 2). Considering functional connectivity related to mentalizing NeuroSynth ROIs, we observed marked decreases in ASD-II and ASD-III compared to controls, particularly between temporal and medial frontal ROIs (qFDR < 0.025; d = 0.61±0.12), whereas ASD-I presented with only sporadic connectivity decreases (qFDR < 0.025; d = 0.50). Similar findings were observed in language and communication networks, with marked decreases in ASD-II and III(qFDR < 0.025; d > 0.46) and only marginal differences in ASD-I.
Figure 2.
Functional connectivity analyses. (A) We generated functional networks between regions-of-interest (ROIs) using an ad hoc meta-analysis of previous studies on mentalizing (upper panel) and communication (lower panel), based on Neurosynth.org. Networks in healthy controls are presented for reference on the left, with intensity values reflecting inter-regional connectivity strength. (B) Functional connectivity differences between the neuroanatomically defined ASD subtypes (see Fig. 1) and controls, with increases/decreases shown in red/blue. •Findings significant after network-wide FDR correction (qFDR < 0.05/2 = 0.025), furthermore adjusted by the number of networks.
Prediction of ADOS Scores
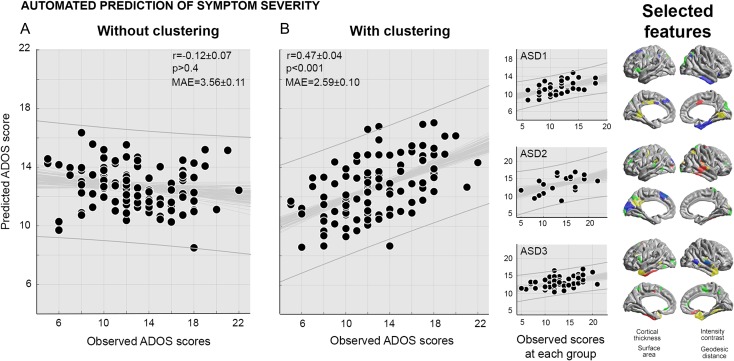
Class-informed learners predicted ADOS severity scores highly correlating with the actual scores (mean ± SD across 100×10-folds: r = 0.47±0.04; MAE = 2.59±0.10; P < 0.001; Fig. 3 shows features contributing to optimal performance). Permutation tests with randomly shuffled ADOS labels showed that our approach exceeded chance levels (P = 0.001). Repeating the prediction experiment using approximated calibrated symptom severity scores demonstrated a slightly lower, but still significant accuracy (r = 0.25±0.05, MAE = 1.46 ± 0.04, P < 0.05).
Figure 3.
Automated prediction of symptom severity. A supervised learning paradigm was used to predict ADOS scores in individual subjects. Shown are observed and predicted ADOS scores for a classifier that was either naïve of the subtyping (A) or informed by it (B). The scatter plots show the mean predicted scores for each ASD individual, averaged across the 100 iterations of the 10-fold cross-validation. The observation-prediction correlation at each iteration is displayed in thin gray, together with a 95% confidence interval. Prediction performance was benchmarked using 3 indices: Pearson correlation [r], statistical significance [p], and mean absolute error [MAE]. Observation-prediction correlations were also presented across subgroups. Finally, selected features (those that were chosen in more than 50% of the 100 cross-validations) were mapped back to cortical surface models (i.e., red: thickness, yellow: intensity contrast, green: surface area, purple: geodesic distance).
Performance of class-based learners was consistently high across all ASD subgroups (ASD-I: r = 0.48±0.07, MAE = 2.08±0.14; ASD-II: r = 0.44±0.11, MAE = 3.26±0.31; ASD-III: r = 0.38±0.08, MAE = 2.71±0.17). Notably, simply splitting the ASD cohort into equally sized random subgroups did not yield comparable accuracy across 100 iterations (Group-I: r = −0.10±0.22, MAE = 3.41±0.45; Group-II: r = −0.11±0.20, MAE = 4.36±0.58; Group-III: r = 0.0±0.10, MAE = 3.44±0.32). This suggests that high performance of the class-wise learner is not simply related to potential overfitting when operating on smaller partitions of the data (Schnack and Kahn 2016; Arbabshirani et al. 2017), but rather due to meaningful data sub-categorization.
We did not observe strong ADOS prediction when the learners were built across entire dataset (i.e., when no class information was used; r = −0.12±0.07; MAE = 3.56±0.11; P > 0.4). Indeed, directly comparing performance between-class-informed and class-free learners revealed markedly increased performance in the former (P < 0.001), suggesting that subtyping in ASD represents an important data preparation prior to development of predictive models.
Discussion
The striking inter-subject heterogeneity characteristic of ASD has been suggested to represent one of the most important obstacles for objective diagnosis and targeted therapy in this condition (Ghosh et al. 2013; Ecker and Murphy 2014). In fact, the current diagnostic system assigns a single, behaviorally defined, label to a population composed of different subgroups with likely different etiologies (Jeste and Geschwind 2014). Our work presented a novel approach for non-invasive ASD biotyping, based on hierarchical clustering of multidimensional MRI profiles. Our large ASD cohort fell into 3 anatomically diverse classes with otherwise similar demographics. Bootstrap stability analysis indicated a high consistency of the discovered subtypes, which also showed a gradual symptom severity and anomalies in intrinsic functional connectivity networks, supporting the validity of our classification on a behavioral and functional level. Moreover, our approach improved data-driven routines to predict symptom severity scores in individual subjects, suggesting potential clinical utility of “divide-and-conquer” approaches to solve difficulties inherent in biomarker discovery in ASD.
Most previous studies evaluated single imaging markers or, if combined, assessed them in a separate fashion, ignoring multi-variate feature relationships. Moreover, prevailing group-level analyses have largely ignored inter-individual variability across the ASD population (Bernhardt et al. 2017). Considering cortical thickness, one of the most widely used MRI markers (Han et al. 2006), our findings indeed suggest that previously reported group-level effects [in our case overall thickening, as shown in an earlier analysis of the same sample (Valk et al. 2015)] might be driven by disproportionally strong effects in a specific subtype, ASD-I, despite other ASD individuals presenting with normal or even inverted patterns. Similarly, when the entire cohort was analyzed as a single group, we did not observe any abnormalities in surface area (previously suggested to reflect columnar/horizontal cortical organization (Buxhoeveden and Casanova 2002)), intensity gradient (designed to detect laminar alterations and ectopic neurons (Avino and Hutsler 2010)), and geodesic distance (thought to capture intrinsic gray-matter connectivity (Ecker, Ronan et al. 2013)). Yet, subtyping could identify a spectrum of alterations, characterized by co-existence of normal and atypical features in ASD. Our results, thus, provide support for different neuroanatomical phenotypes in the larger ASD population, which may partially explain inconsistencies across previous research findings (Haar et al. 2016; Bernhardt et al. 2017).
The different ASD subgroups likely relate to differential neurodevelopment disruptions driven by both genetic anomalies and external influences. A recent study estimated that approximately 20% of individuals with ASD present with de-novo genetic mutations (Iossifov et al. 2014). Atypical genetic expressions may selectively affect developmental processes (Krishnan et al. 2016) and result in heterogeneous cortical morphology. Recent imaging-genetic studies in mouse models suggested that ASD-related genes may be differentially expressed across several brain networks and contribute to anatomical variations (Steadman et al. 2014; Ellegood et al. 2015). In addition, environmental impacts during development may lead epigenetic modifications (Grafodatskaya et al. 2010) and interact with genetic mutations (Berko et al. 2014; Nardone et al. 2014). Interestingly, co-occurring cortical thickness increases and cortical interface blurring that were observed in ASD-I have been also observed in the study of malformations of cortical development (MCDs), a spectrum of congenital anomalies associated with drug-resistant epilepsy. Despite emerging genetic studies showing common risk factors for both autism and MCDs (Blackmon 2015; Lee et al. 2015), MRI investigations in ASD that specifically target neuroimaging signs of MCD are virtually inexistent. Given their resemblance to focal cortical dysplasia type-II (i.e., an early stage malformation associated dyslamination and the presence of dysmorphic neurons, resulting in a blurred and thick cortex on MRI), it is tempting to speculate that our findings in ASD-I may possibly reflect the remnants of anomalies in proliferative and migratory stages of corticogenesis (Wegiel et al. 2010). Notably, both ASD and focal forms of MCD-related epilepsy may relate to genetic alteration in the mTOR pathway, which may regulate cell growth and apoptosis during development (Crino 2016). Collectively, a more targeted analysis through MRI subtyping may guide future investigations in perinatal and early postnatal stages, in both humans and animal models, potentially providing a helpful subgroup identification to better elucidate genetic mechanisms that could foster the development of novel treatments (Belinson et al. 2016).
Parallel analysis of ADOS symptom severity and resting-state connectivity indicated a spectrum of behavioral and functional anomalies across ASD subclasses. Indeed, ASD-I presented with lowest ADOS scores and only marginal disruptions in mentalizing and language networks, while ASD-II and III presented with higher severity and marked functional network reorganization. These findings, derived from modalities not contributing to the MRI-based clustering, provide independent support for our subtyping. Our results, therefore, support the existence of different neuroanatomical subtypes in ASD, which may explain, in part, variable effects reported across previous studies (Haar et al. 2016).
Despite bootstrap analysis suggesting high stability of the discovered subtypes in the current dataset, our findings will require further generalizability tests on large-scale prospective samples. Irrespective of the possibility that other subtypes may exist in more broadly sampled populations, the current work demonstrated that MRI biotyping may generally improve individualized diagnostic/stratification procedures, which suggests clinical utility beyond the mere description of anatomical variability. Supervised learning platforms similar to the employed ADOS prediction routines have recently made a significant impact on cognitive and clinical neurosciences, due to their ability to identify salient data relationships and to perform predictive tasks for previously unseen cases (Gabrieli et al. 2015). Notably, our framework minimized bias, by operating at a cortex-wide level without the use of spatial priors and by bench testing the algorithm only on cases not used for classifier training. Given the markedly improved accuracy of subtype-informed learners compared to “class-naive” procedures, our findings suggest that MRI-based stratification is a useful and biologically meaningful avenue to identify ASD subgroups where tailored diagnostic procedures can be employed and that may benefit from targeted therapy.
Despite the potential implications of our findings in the understanding and management of ASD, several strategies may further optimize the proposed platform. Firstly, by developing our approach on an aggregate and heterogeneous data sample, we may have given priority to features that are robust to site-related data variations (Abraham et al. 2016). Heterogeneity in such datasets may lead to underperformance, as well as between-site effects contaminating the subtypes solution. Increased availability of data collected from a single site or harmonized multisite collections may offer additional scenarios for subtype discovery. Notably, we statistically controlled for potential between-site effects and sites were proportionally represented across subtypes, alleviating concerns about potential propagation of site effects into our findings. Secondly, while the current work is among the first studies to describe neuroanatomical subtypes that showed group differences in terms of symptom severity, we noted overlap in ADOS scores across classes at the individual level. Incorporation of functional imaging or cognitive batteries may improve future subtyping efforts and calibrate individualized syndrome stratification. Several studies have already begun to identify autism subtypes based on symptom scores (Marquand et al. 2016) and measures of mentalizing performance (Lombardo et al. 2016). In a similar context, given that a body of evidence suggests subcortical anomalies in ASD (Hollander et al. 2005), inclusion of both cortical and subcortical structures in the subtyping model may enable an enriched whole-brain phenotyping. Thirdly, in brain-symptom studies as the current work, criterion (i.e., ADOS scores) and predictor measures (i.e., MRI data) should be collected close in time. Unfortunately, the ABIDE dataset does not provide this information systematically, making it impossible to address the time interval between MRI acquisition and ADOS collection. In the NYU cohort, where additional information was available to the authors, the ADOS interview was generally carried out within one month from MRI scan in >90% of cases. In the remaining cases, the interval was at most 6 months. Future data sharing initiatives may benefit from minimizing and consistently reporting these intervals, to achieve robust concurrent validity. Lastly, while cross-sectional profiles have clear subtyping value, they only capture a single point in time. Enriched profiles can be derived from longitudinal data, which allow the consideration of dynamic trajectories in subtyping and prediction of outcomes.
Supplementary Material
Footnotes
We would like to thank the numerous contributors at each site (http://fcon_1000.projects.nitrc.org/indi/abide/) and the NITRC (http://www.nitrc.org) for providing the data sharing platform for the ABIDE initiative as well as the other informatics databases for providing additional platforms. The NYU site was supported by NIH (K23MH087770; R21MH084126; R01MH081218; R01HD065282), Autism Speaks, The Stavros Niarchos Foundation, The Leon Levy Foundation, an endowment provided by Phyllis Green and Randolph Cowen. The PITT site was supported by Autism Speaks Grant 04 593, KO1 NIMH MH081191, NIMH MH67924, NIH HD55748. National Institutes of Health (grant numbers: K08 MH092697, RO1MH080826, P50MH60450, T32DC008553, R01NS34783). The USM site was supported by NIMH (R01MH080826; K08MH092697) and Autism Speaks Mentor-based Predoctoral Fellowship (grant number: 1677), University of Utah Multidisciplinary Research Seed Grant, NRSA Predoctoral Fellowship (grant number: F31 DC010143), Ben B. and Iris M. Margolis Foundation. Conflict of Interest: None declared.
Funding
B.C.B. was supported by startup funds of the Montreal Neurological Institute (MNI), SickKids Foundation, the Canadian Institutes of Health Research (CIHR-Foundation), the National Sciences and Engineering Research Council of Canada (NSERC-Discovery), and the Fonts de la Reserche du Quebec—Sante (FRQS-chercheur boursier). S.L.V. was supported by the International Max Planck Research School (IMPRS). M.P.M. was supported by the NIH (U01MH099059 and R01 AG047596).
References
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, Varoquaux G. 2016. Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. NeuroImage. 147:736–745. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (2013) Diagnostic and statistical manual of mental disorders: DSM-5.
- Andrews DS, Avino TA, Gudbrandsen M, Daly E, Marquand A, Murphy CM, Lai MC, Lombardo MV, Ruigrok AN, Williams SC, et al. 2017. In vivo evidence of reduced integrity of the gray-white matter boundary in autism spectrum disorder. Cereb Cortex. 27:877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbabshirani MR, Plis S, Sui J, Calhoun VD. 2017. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. NeuroImage. 145:137–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avino TA, Hutsler JJ. 2010. Abnormal cell patterning at the cortical gray-white matter boundary in autism spectrum disorders. Brain Res. 1360:138–146. [DOI] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. 2007. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 37:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinson H, Nakatani J, Babineau BA, Birnbaum RY, Ellegood J, Bershteyn M, McEvilly RJ, Long JM, Willert K, Klein OD, et al. 2016. Prenatal beta-catenin/Brn2/Tbr2 transcriptional cascade regulates adult social and stereotypic behaviors. Mol Psychiatry. 21:1417–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 57:289–300. [Google Scholar]
- Berko ER, Suzuki M, Beren F, Lemetre C, Alaimo CM, Calder RB, Ballaban-Gil K, Gounder B, Kampf K, Kirschen J, et al. 2014. Mosaic epigenetic dysregulation of ectodermal cells in autism spectrum disorder. PLoS Genet. 10:e1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt BC, Di Martino A, Valk SL, Wallace GL. 2017. Neuroimaging-based phenotyping of the autism spectrum. Curr Topic Behav Neurosci. 30:341–355. [DOI] [PubMed] [Google Scholar]
- Bernhardt BC, Hong SJ, Bernasconi A, Bernasconi N. 2015. Magnetic resonance imaging pattern learning in temporal lobe epilepsy: classification and prognostics. Ann Neurol. 77:436–446. [DOI] [PubMed] [Google Scholar]
- Bernhardt BC, Valk SL, Silani G, Bird G, Frith U, Singer T. 2014. Selective disruption of sociocognitive structural brain networks in autism and alexithymia. Cereb Cortex. 24:3258–3267. [DOI] [PubMed] [Google Scholar]
- Betancur C. 2011. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 1380:42–77. [DOI] [PubMed] [Google Scholar]
- Blackmon K. 2015. Structural MRI biomarkers of shared pathogenesis in autism spectrum disorder and epilepsy. Epilepsy Behav. 47:172–182. [DOI] [PubMed] [Google Scholar]
- Boddaert N, Zilbovicius M, Philipe A, Robel L, Bourgeois M, Barthelemy C, Seidenwurm D, Meresse I, Laurier L, Desguerre I, et al. 2009). MRI findings in 77 children with non-syndromic autistic disorder. PloS one. 4:e4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn. 45:5–32. [Google Scholar]
- Buxhoeveden DP, Casanova MF. 2002. The minicolumn hypothesis in neuroscience. Brain J Neurol. 125:935–951. [DOI] [PubMed] [Google Scholar]
- Cammoun L, Gigandet X, Meskaldji D, Thiran JP, Sporns O, Do KQ, Maeder P, Meuli R, Hagmann P. 2012. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J Neurosci Methods. 203:386–397. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Brown C. 2002. Clinical and macroscopic correlates of minicolumnar pathology in autism. J Child Neurol. 17:692–695. [DOI] [PubMed] [Google Scholar]
- Casanova MF, El-Baz AS, Kamat SS, Dombroski BA, Khalifa F, Elnakib A, Soliman A, Allison-McNutt A, Switala AE. 2013. Focal cortical dysplasias in autism spectrum disorders. Acta Neuropathol Comm. 1:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC 2014. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 63:1–21. [PubMed] [Google Scholar]
- Crino PB. 2016. The mTOR signalling cascade: paving new roads to cure neurological disease. Nature reviews. Neurology. 12:379–392. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Resnick SM. 2002. Degenerative age changes in white matter connectivity visualized in vivo using magnetic resonance imaging. Cereb Cortex. 12:767–771. [DOI] [PubMed] [Google Scholar]
- Davies DL, Bouldin DW. 1979. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell. 1:224–227. [PubMed] [Google Scholar]
- DeRamus TP, Kana RK. 2015. Anatomical likelihood estimation meta-analysis of grey and white matter anomalies in autism spectrum disorders. Neuroimage Clin. 7:525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, et al. 2014. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 19:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias TGC, Iyer SP, Carpenter SD, Cary RP, Wilson VB, Mitchell SH, Nigg JT, Fair DA. 2015. Characterizing heterogeneity in children with and without ADHD based on reward system connectivity. Dev Cogn Neuros-Neth. 11:155–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich TG. 2000. Ensemble methods in machine learning. Mult Class Sys. 1857:1–15. [Google Scholar]
- Ecker C, Ginestet C, Feng Y, Johnston P, Lombardo MV, Lai MC, Suckling J, Palaniyappan L, Daly E, Murphy CM, et al. 2013. Brain surface anatomy in adults with autism: the relationship between surface area, cortical thickness, and autistic symptoms. JAMA Psychiatry. 70:59–70. [DOI] [PubMed] [Google Scholar]
- Ecker C, Murphy D. 2014. Neuroimaging in autism-from basic science to translational research. Nat Rev Neurol. 10:82–91. [DOI] [PubMed] [Google Scholar]
- Ecker C, Ronan L, Feng Y, Daly E, Murphy C, Ginestet CE, Brammer M, Fletcher PC, Bullmore ET, Suckling J, et al. 2013. Intrinsic gray-matter connectivity of the brain in adults with autism spectrum disorder. Proc Natl Acad Sci USA. 110:13222–13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood J, Anagnostou E, Babineau BA, Crawley JN, Lin L, Genestine M, DiCicco-Bloom E, Lai JKY, Foster JA, Penagarikano O, et al. 2015. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol Psychiatry. 20:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Bathula D, Nikolas MA, Nigg JT. 2012. Distinct neuropsychological subgroups in typically developing youth inform heterogeneity in children with ADHD. Pro Natl Acad Sci USA. 109:6769–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 9:195–207. [DOI] [PubMed] [Google Scholar]
- Friedman JH. 2002. Stochastic gradient boosting. Comput Stat Data An. 38:367–378. [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. 1996. Movement-related effects in fMRI time-series. Magn Reson Med. 35:346–355. [DOI] [PubMed] [Google Scholar]
- Gabrieli JDE, Ghosh SS, Whitfield-Gabrieli S. 2015. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 85:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Michalon A, Lindemann L, Fontoura P, Santarelli L. 2013. Drug discovery for autism spectrum disorder: challenges and opportunities. Nat Rev Drug Discov. 12:777–790. [DOI] [PubMed] [Google Scholar]
- Gotham K, Pickles A, Lord C. 2009. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord. 39:693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D, Chung B, Szatmari P, Weksberg R. 2010. Autism spectrum disorders and epigenetics. J Am Acad Child Adolesc Psychiatry. 49:794–809. [DOI] [PubMed] [Google Scholar]
- Griffin LD. 1994. The intrinsic geometry of the cerebral-cortex. J Theor Biol. 166:261–273. [DOI] [PubMed] [Google Scholar]
- Haar S, Berman S, Behrmann M, Dinstein I. 2016. Anatomical abnormalities in autism? Cereb Cortex. 26:1440–1452. [DOI] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Busa E, Pacheco J, Albert M, Killiany R, et al. 2006. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NeuroImage. 32:180–194. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Gu HB, Munsell BC, Kim SH, Styner M, Wolff JJ, Elison JT, Swanson MR, Zhu HT, Otteron KNB, et al. 2017. Early brain development in infants at high risk for autism spectrum disorder. Nature. 542:348. -+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander E, Anagnostou E, Chaplin W, Esposito K, Haznedar MM, Licalzi E, Wasserman S, Soorya L, Buchsbaum M. 2005. Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol Psychiatry. 58:226–232. [DOI] [PubMed] [Google Scholar]
- Hong SJ, Bernhardt BC, Caldairou B, Hall JA, Guiot MC, Schrader D, Bernasconi N, Bernasconi A. 2017. Multimodal MRI profiling of focal cortical dysplasia type II. Neurology. 88:734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Bernhardt BC, Gill RS, Bernasconi N, Bernasconi A (2017) Connectome-based pattern learning predicts histology and surgical outcome of epileptogenic malformations of cortical development. In: Medical Image Computing and Computer-Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec city, Canada, September 10–14, 2017, Processings Part II (Duchesne S, Descoteaux M, Collins DL, Jannin P, eds.). Cham: Springer International Publishing.
- Hong SJ, Bernhardt BC, Schrader DS, Bernasconi N, Bernasconi A. 2016. Whole-brain MRI phenotyping in dysplasia-related frontal lobe epilepsy. Neurology. 86:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hus V, Lord C. 2014. The autism diagnostic observation schedule, module 4: revised algorithm and standardized severity scores. J Autism Dev Disord. 44:1996–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR. 1990. Morphometric study of human cerebral-cortex development. Neuropsychologia. 28:517–527. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 515:216–U136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. 2002. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 17:825–841. [DOI] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH. 2014. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 10:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Zhang R, Yao V, Theesfeld CL, Wong AK, Tadych A, Volfovsky N, Packer A, Lash A, Troyanskaya OG. 2016. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 19:1454–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Fougere C, Grant S, Kostikov A, Schirrmacher R, Gravel P, Schipper HM, Reader A, Evans A, Thiel A. 2011. Where in-vivo imaging meets cytoarchitectonics: the relationship between cortical thickness and neuronal density measured with high-resolution [F-18]flumazenil-PET. NeuroImage. 56:951–960. [DOI] [PubMed] [Google Scholar]
- Lee BH, Smith T, Paciorkowski AR. 2015. Autism spectrum disorder and epilepsy: disorders with a shared biology. Epile Behav. 47:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Evans AC. 2005. Cortical thickness analysis examined through power analysis and a population simulation. NeuroImage. 24:163–173. [DOI] [PubMed] [Google Scholar]
- Lombardo MV, Lai MC, Auyeung B, Holt RJ, Allison C, Smith P, Chakrabarti B, Ruigrok AN, Suckling J, Bullmore ET, et al. 2016. Unsupervised data-driven stratification of mentalizing heterogeneity in autism. Sci Rep. 6:35333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH Jr., Leventhal BL, DiLavore PC, Pickles A, Rutter M. 2000. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 30:205–223. [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. 1994. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 24:659–685. [DOI] [PubMed] [Google Scholar]
- Marquand AF, Wolfers T, Mennes M, Buitelaar J, Beckmann CF. 2016. Beyond lumping and splitting: a review of computational approaches for stratifying psychiatric disorders. Biol Psychiatry Cogn Neurosci Neuroimaging. 1:433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI. 2004. Computational anatomy: shape, growth, and atrophy comparison via diffeomorphisms. Neuroimage. 23(Suppl 1):S19–S33. [DOI] [PubMed] [Google Scholar]
- Moradi E, Khundrakpam B, Lewis JD, Evans AC, Tohka J. 2017. Predicting symptom severity in autism spectrum disorder based on cortical thickness measures in agglomerative data. NeuroImage. 144:128–141. [DOI] [PubMed] [Google Scholar]
- Nardone S, Sams DS, Reuveni E, Getselter D, Oron O, Karpuj M, Elliott E. 2014). DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry. 4:e433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos H, Friedman SD, Shaw DW, Artru AA, Dawson G, Dager SR. 2006. Gray matter abnormalities in autism spectrum disorder revealed by T2 relaxation. Neurology. 67:632–636. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Tisdall MD, Qureshi A, Buckner RL, van der Kouwe AJW, Fischl B. 2015. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. NeuroImage. 107:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat DH, Chen JJ, van der Kouwe AJ, Greve DN, Fischl B, Rosas HD. 2011. Hippocampal degeneration is associated with temporal and limbic gray matter/white matter tissue contrast in Alzheimer’s disease. NeuroImage. 54:1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savalia NK, Agres PF, Chan MY, Feczko EJ, Kennedy KM, Wig GS. 2017. Motion-related artifacts in structural brain images revealed with independent estimates of in-scanner head motion. Hum Brain Mapp. 38:472–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, Kahn RS. 2016. Detecting neuroimaging biomarkers for psychiatric disorders: sample size matters. Front Psychiatry. 7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadman PE, Ellegood J, Szulc KU, Turnbull DH, Joyner AL, Henkelman RM, Lerch JP. 2014. Genetic effects on cerebellar structure across mouse models of autism using a magnetic resonance imaging atlas. Autism Research. 7:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lui S, Yao L, Deng W, Xiao Y, Zhang W, Huang X, Hu J, Bi F, Li T, et al. 2015. Two patterns of white matter abnormalities in medication-naive patients with first-episode schizophrenia revealed by diffusion tensor imaging and cluster analysis. JAMA Psychiatry. 72:678–686. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Mori S, Amunts K, Zilles K. 2006. Towards multimodal atlases of the human brain. Nat Rev Neurosci. 7:952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 15:273–289. [DOI] [PubMed] [Google Scholar]
- Valk SL, Di Martino A, Milham MP, Bernhardt BC. 2015. Multicenter mapping of structural network alterations in autism. Hum Brain Mapp. 36:2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam NT, O’Connor D, Marcelle ET, Ho EJ, Cameron Craddock R, Tobe RH, Gabbay V, Hudziak JJ, Xavier Castellanos F, Leventhal BL, et al. 2016. Data-driven phenotypic categorization for neurobiological analyses: beyond DSM-5 labels. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GL, Robustelli B, Dankner N, Kenworthy L, Giedd JN, Martin A. 2013. Increased gyrification, but comparable surface area in adolescents with autism spectrum disorders. Brain J Neurol. 136:1956–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH. 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 58:236–244. [Google Scholar]
- Wechsler D. (1999) Wechsler Adult Intelligence Scale—3rd UK Edition (WAIS- III UK). In: (Corporation TP, ed.). London.
- Wegiel J, Kuchna I, Nowicki K, Imaki H, Wegiel J, Marchi E, Ma SY, Chauhan A, Chauhan V, Wierzba Bobrowicz T, et al. 2010. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 119:755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Sabuncu MR, Yeo BT, Fischl B, Greve DN, Kochunov P, Nichols TE, Blangero J, Glahn DC. 2012. Measuring and comparing brain cortical surface area and other areal quantities. NeuroImage. 61:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley K, Taylor J, Carbonell F, Chung MK, Duerden E, Bernhardt B, Lyttelton O, Boucher M, Evans A (2009) SurfStat: a Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. NeuroImage, OHBM poster.
- Yang DY, Beam D, Pelphrey KA, Abdullahi S, Jou RJ. 2016. Cortical morphological markers in children with autism: a structural magnetic resonance imaging study of thickness, area, volume, and gyrification. Mol Autism. 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zuo XN, Wang PP, Li ZH, LaConte SM, Bandettini PA, Hu XPP. 2012. Generalized RAICAR: discover homogeneous subject (sub)groups by reproducibility of their intrinsic connectivity networks. NeuroImage. 63:403–414. [DOI] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. 2011. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 8:665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, Breitner JC, Buckner RL, Calhoun VD, Castellanos FX, et al. 2014. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data. 1:140049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.