Abstract
The impact of fungal pathogens on human health is devastating. For fungi and other pathogens, a key determinant of virulence is the capacity to thrive at host temperatures, with elevated temperature in the form of fever as a ubiquitous host response to defend against infection. A prominent feature of cells experiencing heat stress is the increased expression of heat shock proteins (Hsps) that play pivotal roles in the refolding of misfolded proteins in order to restore cellular homeostasis. Transcriptional activation of this heat shock response is orchestrated by the essential heat shock transcription factor, Hsf1. Although the influence of Hsf1 on cellular stress responses has been studied for decades, many aspects of its regulation and function remain largely enigmatic. In this review, we highlight our current understanding of how Hsf1 is regulated and activated in the model yeast Saccharomyces cerevisiae, and highlight exciting recent discoveries related to its diverse functions under both basal and stress conditions. Given that thermal adaption is a fundamental requirement for growth and virulence in fungal pathogens, we also compare and contrast Hsf1 activation and function in other fungal species with an emphasis on its role as a critical regulator of virulence traits.
Keywords: Hsf1, temperature response, Saccharomyces cerevisiae, Candida albicans, virulence, stress response
The authors describe how the heat shock responsive transcription factor Hsf1 is regulated in diverse fungal species, and highlight recent discoveries related to its role in governing fungal virulence traits.
INTRODUCTION
The ability to sense and respond to environmental stress is critical for the survival of all organisms. This includes thermal insults that are experienced by animals and plants inhabiting environments with fluctuating temperatures, as well as thermal stress experienced by microbial pathogens in a mammalian host with febrile episodes. The heat shock response is a highly conserved process across archaeal, bacterial and eukaryotic domains, which enables adaptation to these thermal stresses (Ritossa 1962; Lindquist and Craig 1988). In eukaryotes, this involves a rapid response from heat shock transcription factors (HSFs) that regulate the expression of heat shock proteins (Hsps), molecular chaperones involved in the folding, stabilization, trafficking and degradation of proteins (Wu 1995). The transcriptional response to heat shock was first characterized in Drosophila, where a puffing pattern in polytene chromosomes that formed upon exposure to elevated temperature was correlated with the production of Hsps (Ritossa 1962; Lindquist 1986; Wu et al.1987). We now understand that HSFs are broadly conserved across eukaryotes from invertebrates like the nematode Caenorhabditis elegans, which has one HSF gene (hsf-1), to humans which have six HSF genes (HSF1, HSF2, HSF4, HSF5, HSFX and HSFY) (Pirkkala, Nykänen and Sistonen 2001; Gomez-Pastor, Burchfiel and Thiele 2017), and plants such as Arabidopsis thaliana whose genome encodes 21 HSF genes (Nover et al.2001). Although all HSFs maintain the basic functions of coordinating the transcriptional response to temperature, these regulators exhibit remarkable complexity in their structure, DNA-binding selectivity, post-translational modifications (PTMs), interacting partners and regulation in response to a myriad of environmental stresses (Anckar and Sistonen 2011; Gomez-Pastor, Burchfiel and Thiele 2017).
Given the critical roles of HSFs in governing protein homeostasis in response to cellular stress, it is not surprising that compromised HSF activity has been linked to several human protein folding diseases, including Huntington disease and Parkinson disease (Anckar and Sistonen 2011; Gomez-Pastor, Burchfiel and Thiele 2017). In contrast, greater levels of activated HSFs are associated with various cancers, likely due its capacity to promote survival despite enhanced proliferation and proteotoxic stress (Whitesell and Lindquist 2009; Dai and Sampson 2016). Although studies with the model yeast Saccharomyces cerevisiae have been instrumental in exploring the molecular mechanisms that enable these transcription factors to orchestrate diverse human diseases, there is a growing appreciation that HSFs also play pivotal roles in the biology and virulence of fungal pathogens. In this review, we focus on our understanding of the sole HSF in fungi, the essential transcription factor Hsf1. We describe our current understanding of how Hsf1 is regulated and activated in S. cerevisiae, and highlight its diverse functions under both basal and stress conditions. Given that thermal adaption is not only a fundamental requirement for growth but also for virulence in fungal pathogens (Klein and Tebbets 2007; Leach and Cowen 2014b), we also highlight the distinct facets of Hsf1 activation and function in fungal pathogens with an emphasis on its role as a critical virulence regulator in diverse fungal species.
Saccharomyces cerevisiae Hsf1 Structure
Given the genetic tractability and availability of diverse molecular tools in S. cerevisiae, the foundational characterization of fungal HSFs has been performed in the model yeast (Sorger and Pelham 1987). Unlike mammalian HSF1, which exists as an inactive monomer until it is induced to trimerize and accumulate in the nucleus upon stress (Whitesell and Lindquist 2009; Åkerfelt, Morimoto and Sistonen 2010; Anckar and Sistonen 2011), yeast Hsf1 is constitutively nuclear and bound to DNA (Sorger and Pelham 1987; Jakobsen and Pelham 1988). In response to thermal stress, induction of Hsf1 causes an increase in binding to its basal targets (Erkine et al.1999), as well as in binding to additional targets (Hahn et al.2004), leading to a rapid and dramatic global transcriptional remodeling required for survival and adaptation to stress (Sorger and Pelham 1988) (Fig. 1). As in other organisms, yeast Hsf1 binds to DNA as a homotrimer within the promoter region of its targets, with each individual monomer binding to the major groove of cis-acting heat shock elements (HSEs) with the binding motif nGAAn, where n is any nucleotide, in alternating orientations (Sorger and Pelham 1987; Amin, Ananthan and Voellmy 1988; Trinklein et al.2004; Yamamoto, Mizukami and Sakurai 2005). As a consequence of co-operative binding, Hsf1 tolerates many variations of this HSE motif, including non-alternating orientations of nTTCn or nGAAn or insertions of five base pairs between the repeating units, especially under stress conditions (Xiao, Perisic and Lis 1991; Erkine et al.1999; Hahn et al.2004; Yamamoto, Mizukami and Sakurai 2005). Due to the low complexity and flexibility of this binding motif, ∼30% of promoters in the yeast genome contain this sequence (Hahn et al.2004), highlighting the potential of Hsf1 to govern genome-wide transcriptional changes.
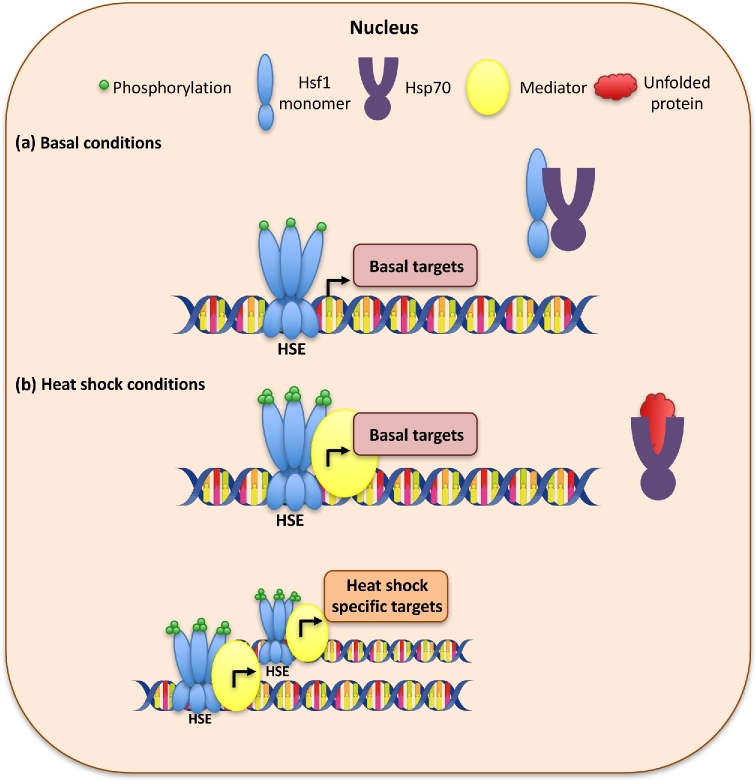
Figure 1.
Model for Hsf1-dependent transcriptional regulation under basal and heat shock conditions in Saccharomyces cerevisiae. (a) Under basal conditions, S. cerevisiae Hsf1 is phosphorylated and binds as a trimer at heat shock elements (HSEs) within the promoter regions of its basal target genes. Hsf1 is bound and repressed by interactions with Hsp70. (b) Under temperature stress, an accumulation of unfolded proteins titrates Hsp70 away from Hsf1. This allows free Hsf1 to bind to additional heat shock targets at their HSEs and increase target gene expression in order to adapt to the thermal insults. Many of these promoter regions can also be bound by the general stress response transcription factors Msn2 and Msn4. Hsf1 is hyperphosphorylated in response to the elevated temperature, which serves to tune the levels of Hsf1 activity by recruiting Mediator to the promoters of Hsf1 target genes. This mediates interactions with RNA polymerase II. After the heat shock, the temperature response is attenuated through the dissociation of Mediator from Hsf1-dependent promoters and the binding of Hsp70 to Hsf1 to restore Hsf1 repression.
The conservation in the binding motif of Hsf1 across eukaryotes is likely due to the high degree of conservation of the central regulatory domains. This includes a conserved winged-helix-turn-helix DNA-binding domain (DBD) (Harrison, Bohm and Nelson 1994), and a hydrophobic repeat coiled-coil domain responsible for Hsf1 oligomerization (Sorger and Nelson 1989; Hashikawa, Yamamoto and Sakurai 2007) (Fig. 2). Yeast Hsf1 also contains N-terminal and C-terminal transcriptional activation domains (AR1 and AR2), which are repressed by the central regulatory domains and are not necessary for growth under basal conditions. However, the N-terminal AR1 domain is important for responses to transient temperature shifts, and the C-terminal AR2 domain enables responses to sustained changes in temperature (Nieto-Sotelo et al.1990; Sorger 1990; Bulman, Hubl and Nelson 2001). It has been proposed that these activation domains become exposed upon increased temperature due to a stress-induced conformational change in Hsf1 (Bonner, Heyward and Fackenthal 1992; Lee et al.2000). However, no full-length Hsf1 crystal structure has been reported to date, and uncertainties remain with this model based on results from in vitro DNA-binding assays and genetic reporter assays (Bonner, Heyward and Fackenthal 1992; Lee et al.2000). Finally, yeast Hsf1 contains a unique regulatory domain called conserved element 2 (CE2) that represses Hsf1 activity (Jakobsen and Pelham 1991), and a C-terminal modulator (CTM), which is necessary for binding to atypical HSEs and for regulation of CE2 (Sakurai and Fukasawa 2001; Hashikawa and Sakurai 2004) (Fig. 2). The CE2 and CTM domains are subject to regulation by hyperphosphorylation (Hoj and Jakobsen 1994; Hashikawa and Sakurai 2004), the role of which will be discussed in subsequent sections.
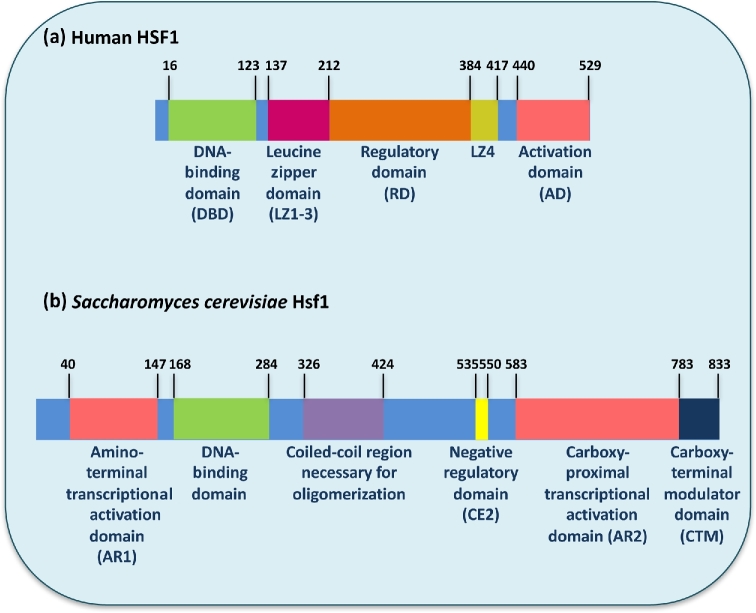
Figure 2.
Domain structure of Hsf1 in humans and Saccharomyces cerevisiae. (a) Schematic of the domain structure of human HSF1, adapted from Gomez-Pastor et al. (2017). HSF1 contains a winged helix-turn-helix DNA-binding domain (DBD). The leucine zipper oligomerization domain (LZ1–3) contains two heptad repeats composed of hydrophobic and charged residues that are predicted to form intermolecular leucine zippers when aligned upon oligomerization. Deletion of this domain produces a constitutively monomeric HSF1. The intrinsically disordered regulatory domain (RD) is post-translationally modified and regulates HSF1 activity and stability. In the LZ4 domain is another heptad repeat that interacts with LZ1–3 to repress oligomerization. HSF1 also contains an activation domain (AD). (b) Schematic of the domain structure of S. cerevisiae Hsf1, adapted from Nicholls et al. (2011) and Hashikawa et al. (2007). Unlike the human ortholog, S. cerevisiae Hsf1 contains two activation domains, AR1 and AR2, located at the N- and C-terminal ends of the protein. S. cerevisiae Hsf1 contains a DBD, as well as a coiled-coil region required for oligomerization. Hsf1 also contains a short conserved element close to the activator (CE2) that is proposed to hold Hsf1 in an inactive configuration. Finally, S. cerevisiae Hsf1 contains a C-terminal modulator (CTM) domain rich in basic amino acids at the extreme C-terminus which is required for the efficient induction of heat shock genes.
Saccharomyces cerevisiae Hsf1 function under basal and stress conditions
Unlike other stress-responsive transcription factors in yeast that tend to be dispensable for viability under basal conditions (Estruch and Carlson 1993), Hsf1 is essential for growth even in the absence of stress, highlighting its critical role in regulating protein homeostasis independent of thermal stress (Jakobsen and Pelham 1988; Sorger and Pelham 1988; Wiederrecht, Seto and Parker 1988). This is in contrast to mammalian HSF1, which is dispensable for growth under basal conditions (Sarge, Murphy and Morimoto 1993). Despite these differences, the biological function of Hsf1 remains conserved between yeast and mammals, so far as a constitutively trimerized and active form of human HSF1 is sufficient to rescue the essential functions of S. cerevisiae Hsf1 (Liu et al.1997), supporting Hsf1 as an evolutionarily conserved regulator of proteostasis. Hsf1 direct targets were initially identified via chromatin immunoprecipitation coupled to microarray (ChIP-chip) in combination with expression profiling, with binding identified upstream of nearly 3% of yeast genes (165 genes), many of which are bound in a heat-inducible manner 5 to 15 minutes post heat shock (Hahn et al.2004). These targets are involved in diverse processes integral for stress responses, including functions in chaperoning proteins, ubiquitination and proteolysis, vesicular transport, carbohydrate metabolism and maintenance of the cell wall (Hahn et al.2004).
Recently, elegant work using a yeast strain where export of Hsf1 from the nucleus was induced using an ‘anchor-away’ approach (Haruki, Nishikawa and Laemmli 2008) revealed that Hsf1 actually drives a highly compact transcriptional program in basal conditions (Solis et al.2016). Using a combination of ChIP coupled with sequencing (ChIP-seq), native elongating transcript sequencing (NET-seq) and RNA sequencing, only 18 genes were identified as exquisitely Hsf1-dependent under basal conditions (Solis et al.2016). These included previously known targets of Hsf1, such as both the constitutive and inducible forms of Hsp90 (HSC82 and HSP82), the Hsp70 chaperone genes (SSA1 and SSA2), HSP104, as well as many other genes with GO terms associated with protein folding and refolding, response to heat and response to stress (Solis et al.2016). Specifically, the essentiality of Hsf1 under basal conditions was attributed to its role in regulating the basal expression of just two genes, the molecular chaperones Hsp70 (SSA2) and Hsp90 (HSC82), which are essential for proteostasis (Solis et al.2016). Depletion of HSF1 reduced Hsp70 and Hsp90 protein levels, leading to proteotoxic stress that ultimately resulted in cell death (Solis et al.2016). Thus, Hsf1 and its associated Hsps not only have pivotal roles as stress response factors, but they are also core housekeeping genes required to maintain eukaryotic proteostasis under basal conditions.
Although Hsf1 has been implicated in regulating transcriptional changes in response to many conditions, including osmotic stress, oxidative stress, glucose starvation and proteotoxic stress (Tamai et al.1994; Amoros and Estruch 2001; Hahn and Thiele 2004; Brandman et al.2012), the transcriptional response of Hsf1 to heat shock has been the most deeply characterized (Hahn et al.2004; Yamamoto, Mizukami and Sakurai 2005; Eastmond and Nelson 2006; Solis et al.2016). Although heat shock induces a dramatic and sustained induction of genes necessary for protein folding and transport that is specific and characteristic of the Hsf1-mediated heat shock response (Causton et al.2001), the overall transcriptional changes that S. cerevisiae undergoes in response to elevated temperature largely mirrors the response to other environmental stresses, such as changes in pH, osmolarity, nutrient depletion or the presence of oxidizing agents (Gasch et al.2000; Causton et al.2001). Approximately 10% of the yeast genome is misregulated in this common environmental stress response, which includes many heat shock genes, factors necessary for protein degradation and transcripts responsible for carbohydrate metabolism (Causton et al.2001). Many of these common stress response genes contain stress response elements in their promoters, which are known to be bound and regulated by the non-essential general response regulators Msn2 and Msn4 (Gasch et al.2000; Causton et al.2001). To separate the functions of Hsf1 and Msn2/Msn4 in regulating the heat shock response, numerous groups have characterized the transcriptional changes in strains harboring temperature-sensitive Hsf1 alleles or upon deletion of Msn2 and Msn4. These studies identified that while Hsf1 and Msn2/Msn4 share some overlapping targets, each complex has distinct roles in the heat shock response, with Hsf1 specifically promoting the expression of chaperone and heat shock genes (Treger et al.1998; Boy-Marcotte et al.1999; Eastmond and Nelson 2006). In fact, it has been estimated as many as 3%–7% of genes in the S. cerevisiae genome are induced upon heat shock in an Hsf1-dependent manner (Hahn et al.2004; Eastmond and Nelson 2006), reinforcing the paradigm that Hsf1 is a master regulator of the heat shock response.
With technological advancements in genome-wide sequencing methodologies coupled with novel and innovative ways to inactivate Hsf1 in yeast, recent work has suggested that the role of Hsf1 as a master regulator of the heat shock response may be greatly overestimated. Using a chemical genetics approach that allowed for rapid Hsf1 inactivation, anchoring away of Hsf1 from the nucleus did not significantly alter the heat shock response (Solis et al.2016). Although a small suite of Hsf1-dependent genes were repressed upon compromise of Hsf1 activity, the remaining genes, many of which were targets of Msn2 and Msn4, were still induced upon exposure to elevated temperature (Solis et al.2016). This finding is consistent with recent work in mammalian cells showing that mammalian HSF1 plays a limited but specialized role in coordinating the heat shock response (Mahat et al.2016; Solis et al.2016). Notably, these conclusions do not preclude the possibility that Hsf1 may engage with many other targets under both basal conditions and heat shock than those described by Solis et al., akin to what has traditionally been described (Hahn et al.2004). It merely points to the fact that other transcription factors or compensatory mechanisms could be involved in the co-regulation of targets important for survival upon exposure to elevated temperature. Overall, these latest findings suggest a model in which Hsf1’s role in response to heat shock is not to dramatically expand its target gene repertoire to control global transcriptional changes, but rather to upregulate a critical but small subset of genes dedicated to protein folding as an essential adaptive mechanism. Future studies will be necessary to determine whether this paradigm holds true for the other stresses under which Hsf1 transcriptional activity is stimulated.
Consistent with work in mammalian cells, there is an emerging body of evidence to suggest there are novel and distinct roles of Hsf1 that are independent of regulating protein homeostasis. Studies have suggested that Hsf1 may regulate the cell cycle (Zarzov, Boucherie and Mann 1997), as strains with defective Hsf1 undergo reversible cell cycle arrest at the G2/M phase at elevated temperatures, which is suppressed by the heat-inducible Hsp90, HSP82 (Morano et al.1999). Studies have also shown that Hsf1 has a role in translational control, where Hsf1 is activated by the ribosome quality control complex when polypeptides are stalled during translation (Brandman et al.2012). Hsf1 may also be involved in regulating virulence of the normally benign S. cerevisiae, as the ability to grow at supraoptimal temperatures is associated with, but not entirely responsible for, virulence of clinical isolates in mice (Clemons et al.1994; McCusker et al.1994). As a key regulator of growth at elevated temperatures, Hsf1 likely contributes to the proliferation of clinical isolates in the host.
Regulation of Hsf1 activity
Despite the conservation of Hsf1 across eukaryotic organisms, many questions remain regarding how Hsf1 is regulated during periods of cellular stress. Some aspects of Hsf1 regulation are species-specific, such as trimerization, which is a regulated event in mammalian cells but constitutive in yeast (Sorger and Pelham 1987; Sorger and Nelson 1989). However, two common features are thought to contribute to Hsf1 regulation in all organisms: interactions with other cellular proteins and PTM via phosphorylation (Anckar and Sistonen 2011). The transcriptional activity of Hsf1 in response to temperature is dependent on recruitment of the Mediator complex to Hsf1 target genes (Fan, Chou and Struhl 2006). Upon heat shock, Mediator is rapidly and selectively recruited to the promoters of Hsf1 target genes, mediating interactions with RNA polymerase II (Kim and Gross 2013). This binding is co-operative and dependent on physical interactions with Hsf1, as truncation of either its N- or C-terminal activation domain significantly reduces Mediator occupancy and removal of both activation domains completely abolishes it (Kim and Gross 2013). After heat shock, Mediator dissociates and the RNA polymerase II recruitment diminishes, turning off the transcription of Hsf1-dependent genes (Kim and Gross 2013).
Further, Hsf1 activity is regulated through interactions with molecular chaperones in a feedback loop commonly referred to as the chaperone titration model (Voellmy and Boellmann 2007). Specifically, under stress conditions an accumulation of misfolded proteins titrate chaperones away from Hsf1, enabling its transcriptional activity. However, once proteostasis is restored, client-free chaperones again bind to Hsf1 in order to deactivate it. There is biochemical, pharmacological and genetic evidence to support roles for the Hsp70 and Hsp90 chaperones, as well as their co-chaperones, in regulating HSF1 in mammalian systems and Hsf1 in yeast (Duina, Kalton and Gaber 1998; Zou et al.1998; Guo et al.2001; Gomez-Pastor, Burchfiel and Thiele 2017). A recent study in S. cerevisiae provided additional support for this model in which Hsp70 (SSA1) was shown to act as a dynamic switch that binds and inhibits Hsf1 in the absence of stress, dissociates immediately following heat shock to promote Hsf1 activity, and then re-associates and inhibits Hsf1 at a later point to turn off the temperature response (Zheng et al.2016). The interaction between Hsp70 and Hsf1 was abrogated by the presence of an aggregation prone peptide, demonstrating that unfolded proteins can titrate Hsp70 away from Hsf1 (Zheng et al.2016). Finally, although hyperactivation of the Hsf1 transcriptional program by overexpression of Hsf1 leads to impaired growth, this was alleviated by increasing expression of Hsp70 or Hsp40 (YDJ1), which assists Hsp70 in repressing Hsf1 activity (Zheng et al.2016). A subsequent study identified that Hsp70 binds directly to the CE2 domain of Hsf1 with a weak association that allows for the disruption of the complex upon stress (Krakowiak et al.2018). Binding of Hsp70 to the CE2 domain of Hsf1 represses Hsf1 transactivation by causing the C-terminal activation domain to be closed, preventing the recruitment of the transcriptional machinery (Krakowiak et al.2018). Surprisingly, these studies did not find any evidence of Hsp90 inhibiting Hsf1 function directly (Zheng et al.2016). This is in contrast to previous genetic studies that have suggested that Hsp90 represses Hsf1 activity such that Hsp90 inhibition activates Hsf1 (Duina, Kalton and Gaber 1998; Brandman et al.2012). Thus, it remains unclear whether the effects of compromising Hsp90 function on Hsf1 activation are direct, or potentially through an increase in unfolded proteins that require Hsp70 function for their stabilization.
In addition to Hsf1 regulation by protein interactions, Hsf1 activity is also controlled by PTMs. Although mammalian HSF1 is regulated by multiple modifications including phosphorylation, sumolyation and acetylation (Vihervaara and Sistonen 2014), most of the literature describing Hsf1 regulation in yeast has focused on the role of phosphorylation, which has recently become a topic of much debate. Hsf1 is phosphorylated under basal conditions and hyperphosphorylated upon exposure to stresses such as heat shock (Sorger and Pelham 1988). Traditionally, this increase in phosphorylation was thought to be required for the activation of the transcription factor. However, a recent report provided evidence suggesting that Hsf1 phosphorylation does not act as a switch to turn on its transcriptional response, but rather that it tunes or adjusts the level of Hsf1 activity (Zheng et al.2016). This was demonstrated by generating a phosphorylation-deficient strain in which all but one of the serine and threonine residues of Hsf1 were mutated to alanine, with the remaining residue being required for DNA binding. With this allele as the sole source of Hsf1 in the cell, Hsf1 maintained its activity under basal conditions, but displayed a dampened transcriptional response upon exposure to elevated temperature (Zheng et al.2016). This is consistent with observations in mammalian cells that mutating 15 HSF1 phosphorylation sites to alanine residues did not abolish the transcriptional activity of HSF1 upon heat shock (Budzyński et al.2015). In addition, mutations that mimicked hyperphosphorylation caused increased Hsf1 transcriptional activity (Zheng et al.2016). While hyper-phosphorylation did not affect the interactions between Hsf1 and Hsp70, it did increase the recruitment of the Mediator complex to Hsf1-dependent genes (Zheng et al.2016). Together, this suggests that Hsp70 acts as switch to turn Hsf1 activity off and on, while the phosphorylation status of Hsf1 acts as a mechanism to tune the level of activation. While it is known that Hsf1 is phosphorylated by Yak1 (Lee et al.2008), Rim15 (Lee et al.2013) and Snf1 in response to glucose depletion (Hahn and Thiele 2004), the kinase that regulates Hsf1 under basal conditions or upon heat shock remains elusive. Further work will be necessary to identify the specific kinases and phosphatases that regulate Hsf1 phosphorylation in response to the different conditions, in order to better understand the dynamic and environmentally contingent regulation of Hsf1 activity.
Hsf1 in the fungal pathogen Candida albicans
So far, we have outlined our understanding of how Hsf1 is regulated and functions in the model yeast S. cerevisiae. Yet Hsf1 also plays an integral role in temperature-dependent responses of one of the leading human fungal pathogens, Candida albicans, which has been separated from S. cerevisiae by ∼200–800 million years of evolution (Heckman et al.2001). C. albicans is a leading causal agent of mycotic death worldwide (Pfaller and Diekema 2007, 2010). Further, the CDC has classified Candida species as a serious threat to human health due to the dramatic rise in antimicrobial resistance (CDC 2013). The pathogenic prowess of C. albicans is due to numerous factors, including the ability to survive at physiological temperatures within a mammalian host, and the ability to transition between distinct morphological states that play a key role in the virulence of the organism (Whiteway and Oberholzer 2004; Kumamoto and Vinces 2005). There is significant conservation of Hsf1 between the two organisms with the DBDs of C. albicans and S. cerevisiae Hsf1 sharing 71.9% identity (Nicholls et al.2009). Like in S. cerevisiae, C. albicans Hsf1 binds to its basal and heat shock specific targets at HSEs containing inverted repeats of nGAAn with variable spacing between (Leach et al.2016). Intriguingly, over two thirds of the genes in the C. albicans genome contain HSEs within their promoter region but no appreciable Hsf1 binding is observed at many of these genes under basal or heat shock conditions, indicating that the presence of a HSE alone cannot predict Hsf1 binding (Leach et al.2016). Hsf1 binding is strongly influenced by nucleosome positioning at Hsf1 motifs, where Hsf1-bound regions have significantly lower nucleosome occupancy than unbound regions in the genome (Leach et al.2016). This demonstrates a large capacity for Hsf1 binding under different conditions, which is limited by nucleosome positioning.
Consistent with S. cerevisiae Hsf1, C. albicans Hsf1 is essential for growth in basal conditions and is necessary for the transcriptional response to heat shock (Nicholls et al.2009; Leach et al.2016). Under basal conditions, Hsf1 binds within the promoter region of a core set of target genes that are necessary for protein homeostasis, including the chaperones HSP104, HSP70, and HSP90, as well as Hsp90 co-chaperones such as CPR6 and CDC37 (Leach et al.2016). C. albicans Hsf1 also participates in an autoregulatory circuit by binding to its own promoter and regulating its own expression to enable thermal adaptation (Leach et al.2016). Although C. albicans is an opportunistic pathogen which primarily grows within a thermally buffered human host, C. albicans Hsf1 maintains its role in regulating the heat shock response and enables responses to slow thermal transitions involved in febrile episodes associated with infection (Leach et al.2012b). Hsf1 senses temperature changes by responding to alterations in membrane fluidity, which are regulated by the unsaturated fatty acid desaturase Ole1, the fatty acid synthase Fas2 and the E3 ubiquitin ligase Rps5 (Leach and Cowen 2014a) (Fig. 3). In response to temperature fluctuations, Hsf1 is phosphorylated (Nicholls et al.2009), its basal targets are transcriptionally induced (Nicholls et al.2009), and it binds and induces additional targets involved in diverse processes, including protein folding, protein transport and adhesion (Leach et al.2016) (Fig. 3). This activation of Hsf1 is transient, as once the cells have adapted to the heat shock, Hsf1 phosphorylation is lost (Leach et al.2012a,b). Whether Hsf1 acts as a global regulator of the heat shock response or whether it has a more specialized role in regulation of a small set of core genes required for proteostasis, as has recently been revealed in S. cerevisiae (Solis et al.2016), will be fascinating to uncover in other fungal pathogens.
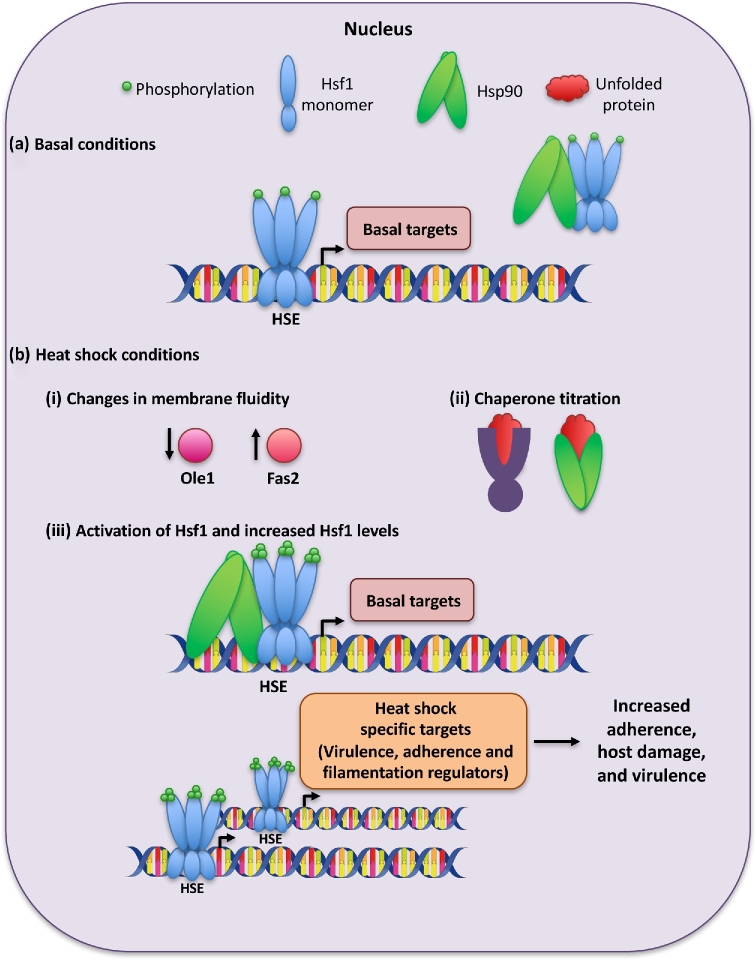
Figure 3.
Model for Hsf1-dependent transcriptional regulation under basal and heat shock conditions in Candida albicans. (a) Under basal conditions, C. albicans Hsf1 binds at the HSEs within the promoter regions of its basal targets and regulates their expression. Hsf1 is bound by Hsp90, which represses Hsf1 function. (b) (i) Upon temperature upshifts, the expression of the unsaturated fatty acid desaturase Ole1 decreases, causing an increased expression of the fatty acid synthase, Fas2. This leads to an alteration in the ratio of saturated to unsaturated fats, which alters membrane fluidity. (ii) Upon elevated temperature there is also an increase in unfolded and misfolded proteins that require chaperoning, leading to a titration of Hsp90 away from repressing Hsf1. While it is possible that other chaperones like Hsp70 are also involved in this regulation, this has yet to be explored. (iii) Hsf1 is activated by the de-repression of Hsp90 and changes in membrane fluidity, and is phosphorylated. Hsf1 induces the expression of its basal targets, including upregulating its own expression. Upon heat shock, Hsf1 also binds and induces the expression of additional heat shock-specific targets involved in adherence, host cell damage and filamentation; this ultimately leads to increased cellular adherence and host cell damage.
Unlike S. cerevisiae, C. albicans Hsf1 specifically responds to changes in temperature and not to other environmental stresses such as heavy metal, weak acid or pH stress, although there is a slight induction of activity in response to SDS treatment (Nicholls et al.2009). This is consistent with findings that the roles of the general stress response regulators Msn2 and Msn4 have diverged in C. albicans (Nicholls et al.2004; Ramsdale et al.2008), and that C. albicans lacks a general stress response (Enjalbert, Nantel and Whiteway 2003). Together, this demonstrates that although Hsf1 maintains its role as an integral regulator of thermal stress responses in C. albicans, substantial transcriptional rewiring has led to distinct contributions of Hsf1 to C. albicans stress adaptation.
Many questions remain regarding how C. albicans Hsf1 is regulated upon exposure to different environmental insults, including what cellular proteins orchestrate Hsf1 phosphorylation and how PTMs modulate Hsf1 activity. Akin to what is described in many eukaryotes, C. albicans Hsf1 activity is regulated via titration of chaperones by misfolded proteins, although the key chaperone that modulates Hsf1 function in this context may be distinct from what is described in S. cerevisiae. A feedback loop exists between Hsf1 and Hsp90, in which Hsf1 positively regulates the expression of HSP90 (Nicholls et al.2009; Leach et al.2016), and Hsp90 physically interacts with Hsf1 to repress its function (Leach et al.2012a) (Fig. 3). In S. cerevisiae, Hsp70 has been implicated as the central chaperone that represses Hsf1 activation under basal conditions (Zheng et al.2016); however, this remains to be explored in C. albicans. Genetic depletion or pharmacological inhibition of Hsp90 alleviates repression of Hsf1, leading to phosphorylation of Hsf1 and an induction of its targets (Leach et al.2012a). Upon exposure to thermal stress, global protein misfolding compromises the functional capacity of Hsp90, which is thought to release Hsf1 from Hsp90’s repressive effects and allow for increased expression of HSP90 and other genes necessary for protein folding and refolding (Leach et al.2012b). Upon heat shock, the interaction between Hsf1 and Hsp90 is increased, likely because of increased protein levels of Hsf1 and Hsp90 (Fig. 3). This leads to an accumulation of Hsp90 in the nucleus and after prolonged periods of heat shock, Hsp70 is recruited to the Hsf1/Hsp90 complex (Leach et al.2012a). This is consistent with a model in which Hsp90 dampens the heat shock response once adaptation to the increased temperature has been achieved. Hsp90 also influences the activity of Hsf1 upon heat shock by modifying nucleosome architecture, affecting nucleosome occupancy and binding at Hsf1 targets (Leach et al.2016). Thus, complex interactions between Hsf1 and cellular chaperones in C. albicans are required to enable thermotolerance.
Temperature sensing is critical for C. albicans to survive in the host and cause infection, as temperature affects many key C. albicans virulence traits, including the morphogenetic switch between the white and mating-competent opaque growth state (Noble, Gianetti and Witchley 2017), and the transition from yeast to filamentous growth (Shapiro and Cowen 2010; Sudbery 2011). As a critical temperature sensor, Hsf1 is essential for virulence in C. albicans, as Hsf1 activity is induced in response to murine systemic infection and loss of this induction results in attenuated virulence (Nicholls et al.2011). The induction of Hsf1 target genes during infection could provide a mechanism for C. albicans to exploit thermal insults in the host to upregulate its virulence program and cause host damage. This is consistent with findings that Hsf1 regulates many virulence genes specifically under heat shock conditions, including genes involved in filamentation, biofilm formation and adhesion (Leach et al.2016). In further support of this model, C. albicans cells that have experienced a heat shock show greater adherence to human cell lines, cause more host cell damage, and are associated with a higher mortality of zebrafish and the greater wax moth Galleria mellonella in infection models (Leach et al.2016). Recent work has also shown that homeostasis of Hsf1 levels is required to maintain yeast form growth in C. albicans, as both overexpression and depletion of Hsf1 induce filamentation, albeit through distinct mechanisms (Veri et al.2018). Hsf1 depletion leads to compromised function of Hsp90, which is a key regulator of morphogenesis (Shapiro et al.2009; O’Meara and Cowen 2014), while Hsf1 overexpression induces filamentous growth by influencing the filamentation transcriptional program directly through an expanded set of targets (Veri et al.2018). HSF1 levels in a wild-type strain were observed to change dramatically in response to different environmental conditions, including being induced in response to heat shock and during growth in biofilm conditions (Veri et al.2018). Together, this work provides the premiere example of a protein that acts both as a positive and negative regulator of filamentation, and highlights that homeostasis in the levels of an environmentally responsive cellular regulator is required to maintain yeast form growth in C. albicans. Hsf1 has also been shown to be necessary for filamentation in response to solid inducing cues (Nair et al.2017). Finally, reports have suggested that Hsf1 modulates the susceptibility of C. albicans to diverse antifungal drugs (Dhamgaye et al.2014), and influences iron homeostasis (Nair et al.2017). The next frontier is to define the scope of impact of Hsf1 on diverse virulence traits, and explore the potential of targeting Hsf1 in the development of novel strategies to treat fungal infections.
Hsf1 in other fungal pathogens
For environmental fungi that infect humans including Cryptococcus neoformans, Aspergillus fumigatus and Histoplasma capsulatum, the capacity to respond to temperature fluctuations is required for their pathogenic lifecycle, as they adapt to the thermal shift during the transition from ambient temperature in the environment to physiological temperatures in their mammalian host. C. neoformans is an environmentally ubiquitous pathogen that infects humans when infectious cells or spores are inhaled, allowing for colonization of the lungs, dissemination through the bloodstream and, in severe cases, infection of the brain manifesting as meningoencephalitis (May et al.2016). Studies of the signaling pathways important for thermal adaptation in C. neoformans revealed that mutations that cause thermosensitivity also attenuate virulence (Odom et al.1997; Alspaugh et al.2000; Kraus et al.2003), demonstrating the potential for thermotolerance regulators to serve as novel antifungal targets. Transcriptional analyses established that at 37°C, C. neoformans induces the expression of heat shock genes, genes for translation machinery components, mitochondrial genes and stress genes including superoxide dismutase (Steen et al.2002). Although the specific role of C. neoformans Hsf1 in thermotolerance is largely elusive, Hsf1 is phosphorylated in response to elevated temperature, although surprisingly it becomes downregulated, while the levels of HSP90 and heat shock genes are induced (Yang et al.2017). C. neoformans Hsf1 is essential under basal conditions, while its overexpression promotes growth at higher temperatures by regulating temperature-responsive genes such as HSP104 and SSA1 (Yang et al.2017). Finally, C. neoformans Hsf1 does not regulate its own expression (Yang et al.2017), as is observed in C. albicans (Leach et al.2016). Thus, although Hsf1 is a conserved and essential determinant of thermal adaptation in C. neoformans, there is considerable rewiring in the regulatory circuitry governing its activation. Exploring the transcriptional targets of C. neoformans Hsf1 and the impact of this transcriptional regulator on C. neoformans virulence is poised to reveal fascinating insights.
Even less is known about Hsf1 in other pathogenic fungi, such as A. fumigatus or H. capsulatum. A. fumigatus is found ubiquitously in soil or compost, and causes devastating disease when spores are inhaled by humans. As with all fungal pathogens of mammals, thermotolerance is essential for A. fumigatus pathogenicity; given that this mold grows in composts with temperatures up to 70°C, making this species well adapted for growth at high temperatures (Bhabhra et al.2004; Bhabhra and Askew 2005; Nierman et al.2005). Transcriptional analyses have established that heat shock genes are upregulated upon elevated temperatures (Nierman et al.2005; Dinamarco et al.2012), including HSF1 (Dinamarco et al.2012). In addition, proteomic analyses have highlighted that increased temperature induces a heat shock response that upregulates factors involved in protein folding, organization of the cytoskeletion and transcription, many of which have HSEs in their promoters and could be targets of Hsf1 (Albrecht et al.2010). H. capsulatum is a thermally dimorphic fungus that grows in the soil as filamentous mold, and converts to the virulent yeast morphology when spores are inhaled by a mammalian host. The yeast cells proliferate within macrophages and cause respiratory and systemic histoplasmosis, which affect both healthy and immunocompromised individuals (Klein and Tebbets 2007). This morphological transition is a thermally regulated virulence trait (Shearer et al.1987), with correlation between the level of thermosensitivity and pathogenesis in mice (Medoff et al.1986). The temperature change associated with morphogenesis induces the expression of heat shock proteins (Lambowitz et al.1983; Caruso et al.1987; Shearer et al.1987), and deletion of the Hsp90 family member HSP82 impairs thermotolerance and virulence (Edwards, Zemska and Rappleye 2011). Although some of the transcriptional network that regulates H. capsulatum responses to thermal shifts has been defined (Beyhan et al.2013; Gilmore et al.2015), Hsf1 has yet to be implicated. As a core hub of regulatory circuitry, Hsf1 is poised to play a central role in governing thermal adaptation across the fungal kingdom.
CONCLUSION/OUTLOOK
Over the last 30 years, we have developed a deep but ever expanding appreciation of Hsf1 function, which has highlighted that Hsf1 is a complex transcriptional regulator essential for diverse protective responses to stress. The HSF-dependent transcriptional response to thermal stress is one of the most conserved and ancient stress responses in nature, central to survival in yeast, plants, fruit flies, worms and humans. Although much work remains to be done in order to understand the complexities of Hsf1 function, it is clear that Hsf1 plays a critical role in governing temperature-dependent virulence traits across the fungal kingdom, and that targeting Hsf1 and the associated regulatory circuitry could provide an exciting avenue for antifungal drug development. Given that HSFs regulate such core cellular functions, it is not surprising that they are emerging as targets for many human diseases, including cancer and neurodegenerative disorders (Anckar and Sistonen 2011; Vihervaara and Sistonen 2014; Dai and Sampson 2016; Li, Labbadia and Morimoto 2017). Although a crystal structure for the full-length Hsf1 remains elusive, there is a crystal structure for the DBD of Hsf1 (Harrison, Bohm and Nelson 1994), as well as a full-length structure for one of the mammalian orthologs, HSF2 (Jaeger et al.2016). Advances with crystallography of fungal Hsf1 will be instrumental for structure-guided drug design. Intervention could be achieved at the level of impairing trimerization, nuclear localization or DNA binding. Exploiting the differences between HSFs in mammals and in fungi will be critical for the development of fungal-selective Hsf1 inhibitors to eradicate many devastating fungal diseases. It is exquisitely clear that exploration of the broader context of the protein homeostasis network governing cellular stress responses provides rich opportunities to gain fundamental insights into biology, development and disease.
Acknowledgements
We thank all the members of the Cowen lab for helpful discussions.
FUNDING
This work was supported by the Canadian Institutes of Health Research Frederick Banting and Charles Best Canada Graduate Scholarship [to AOV]; by Ontario Graduate Scholarships [to AOV]; Canadian Institutes of Health Research Operating Grants [MOP-86452 and MOP-119520 to LEC] and Foundation Grant [FDN-154288 to LEC]; the Natural Sciences and Engineering Council (NSERC) of Canada Discovery Grants [06261 and 462167 to LEC]; an NSERC E.W.R. Steacie Memorial Fellowship [477598 to LEC], and National Institutes of Health NIAID R01 Grants [1R01AI127375–01 and 1R01AI120958-01 to LEC].
Conflict of interest. None declared.
REFERENCES
- Åkerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 2010;11:545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht D, Guthke R, Brakhage AA et al. Integrative analysis of the heat shock response in Aspergillus fumigatus. BMC Genomics 2010;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Cavallo LM, Perfect JR et al. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol 2000;36:352–65. [DOI] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol 1988;8:3761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoros M, Estruch F. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol Microbiol 2001;39:1523–32. [DOI] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 2011;80:1089–115. [DOI] [PubMed] [Google Scholar]
- Beyhan S, Gutierrez M, Voorhies M et al. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol 2013;11:e1001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabhra R, Askew DS. Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med Mycol 2005;43Suppl 1:87–93. [DOI] [PubMed] [Google Scholar]
- Bhabhra R, Miley MD, Mylonakis E et al. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun 2004;72:4731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JJ, Heyward S, Fackenthal DL. Temperature-dependent regulation of a heterologous transcriptional activation domain fused to yeast heat shock transcription factor. Mol Cell Biol 1992;12:1021–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy-Marcotte E, Lagniel G, Perrot M et al. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol Microbiol 1999;33:274–83. [DOI] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 2012;151:1042–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzyński MA, Puustinen MC, Joutsen J et al. Uncoupling stress-inducible phosphorylation of Heat Shock Factor 1 from its activation. Mol Cell Biol 2015;35:2530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulman AL, Hubl ST, Nelson HC. The DNA-binding domain of yeast heat shock transcription factor independently regulates both the N- and C-terminal activation domains. J Biol Chem 2001;276:40254–62. [DOI] [PubMed] [Google Scholar]
- Caruso M, Sacco M, Medoff G et al. Heat shock 70 gene is differentially expressed in Histoplasma capsulatum strains with different levels of thermotolerance and pathogenicity. Mol Microbiol 1987;1:151–8. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS et al. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 2001;12:323–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: CDC, 2013, 114. [Google Scholar]
- Clemons KV, McCusker JH, Davis RW et al. Comparative pathogenesis of clinical and nonclinical isolates of Saccharomyces cerevisiae. J Infect Dis 1994;169:859–67. [DOI] [PubMed] [Google Scholar]
- Dai C, Sampson SB. HSF1: Guardian of proteostasis in cancer. Trends Cell Biol 2016;26:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamgaye S, Devaux F, Vandeputte P et al. Molecular mechanisms of action of herbal antifungal alkaloid berberine, in Candida albicans. PLoS One 2014;9:e104554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinamarco TM, Almeida RS, de Castro PA et al. Molecular characterization of the putative transcription factor SebA involved in virulence in Aspergillus fumigatus. Eukaryot Cell 2012;11:518–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem 1998;273:18974–8. [DOI] [PubMed] [Google Scholar]
- Eastmond DL, Nelson HC. Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J Biol Chem 2006;281:32909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, Zemska O, Rappleye CA. Discovery of a role for Hsp82 in Histoplasma virulence through a quantitative screen for macrophage lethality. Infect Immun 2011;79:3348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Nantel A, Whiteway M. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell 2003;14:1460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine AM, Magrogan SF, Sekinger EA et al. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol Cell Biol 1999;19:1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol 1993;13:3872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol 2006;13:117–20. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000;11:4241–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore SA, Voorhies M, Gebhart D et al. Genome-wide reprogramming of transcript architecture by temperature specifies the developmental states of the human pathogen Histoplasma. PLoS Genet 2015;11:e1005395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 2017;19:4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Guettouche T, Fenna M et al. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem 2001;276:45791–9. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem 2004;279:5169–76. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ et al. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 2004;24:5249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HC. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 1994;263:224–7. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 2008;31:925–32. [DOI] [PubMed] [Google Scholar]
- Hashikawa N, Sakurai H. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol Cell Biol 2004;24:3648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa N, Yamamoto N, Sakurai H. Different mechanisms are involved in the transcriptional activation by yeast heat shock transcription factor through two different types of heat shock elements. J Biol Chem 2007;282:10333–40. [DOI] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR et al. Molecular evidence for the early colonization of land by fungi and plants. Science 2001;293:1129–33. [DOI] [PubMed] [Google Scholar]
- Hoj A, Jakobsen BK. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO 1994;13:2617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger AM, Pemble CWt, Sistonen L et al. Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nat Struct Mol Biol 2016;23:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen BK, Pelham HR. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol 1988;8:5040–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen BK, Pelham HR. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J 1991;10:369–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Gross DS. Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and Mediator tail subunits Med15 and Med16. J Biol Chem 2013;288:12197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BS, Tebbets B. Dimorphism and virulence in fungi. Curr Opin Microbiol 2007;10:314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak J, Zheng X, Patel N et al. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. Elife 2018;7:e31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus PR, Fox DS, Cox GM et al. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol Microbiol 2003;48:1377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol 2005;7:1546–54. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Kobayashi GS, Painter A et al. Possible relationship of morphogenesis in pathogenic fungus, Histoplasma capsulatum, to heat shock response. Nature 1983;303:806–8. [DOI] [PubMed] [Google Scholar]
- Leach MD, Cowen LE. Membrane fluidity and temperature sensing are coupled via circuitry comprised of Ole1, Rsp5, and Hsf1 in Candida albicans. Eukaryot Cell 2014a;13:1077–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Cowen LE. To sense or die: mechanisms of temperature sensing in fungal pathogens. Curr Fungal Infect Rep 2014b;8:185–91. [Google Scholar]
- Leach MD, Budge S, Walker L et al. Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog 2012a;8:e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Farrer RA, Tan K et al. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Comms 2016;7:11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Tyc KM, Brown AJP et al. Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS One 2012b;7:e32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Carlson T, Christian N et al. The yeast heat shock transcription factor changes conformation in response to superoxide and temperature. Mol Biol Cell 2000;11:1753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Cho BR, Joo HS et al. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol Microbiol 2008;70:882–95. [DOI] [PubMed] [Google Scholar]
- Lee P, Kim MS, Paik SM et al. Rim15-dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae. FEBS Lett 2013;587:3648–55. [DOI] [PubMed] [Google Scholar]
- Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol 2017;27:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem 1986;55:1151–91. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet 1988;22:631–77. [DOI] [PubMed] [Google Scholar]
- Liu XD, Liu PC, Santoro N et al. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J 1997;16:6466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker JH, Clemons KV, Stevens DA et al. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42 degrees C and form pseudohyphae. Infect Immun 1994;62:5447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM et al. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell 2016;62:63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RC, Stone NR, Wiesner DL et al. Cryptococcus: from environmental saprophyte to global pathogen. Nat Rev Microbiol 2016;14:106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medoff G, Maresca B, Lambowitz AM et al. Correlation between pathogenicity and temperature sensitivity in different strains of Histoplasma capsulatum. J Clin Invest 1986;78:1638–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Santoro N, Koch KA et al. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol Cell Biol 1999;19:402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R, Shariq M, Dhamgaye S et al. Non-heat shock responsive roles of HSF1 in Candida albicans are essential under iron deprivation and drug defense. BBA-Mol Cell Res 2017;1864:345–54. [DOI] [PubMed] [Google Scholar]
- Nicholls S, Leach MD, Priest CL et al. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol Microbiol 2009;74:844–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, MacCallum DM, Kaffarnik FA et al. Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans. Fungal Genet Biol 2011;48:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, Straffon M, Enjalbert B et al. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot Cell 2004;3:1111–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005;438:1151–6. [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Wiederrecht G, Okuda A et al. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 1990;62:807–17. [DOI] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 2017;15:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P et al. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 2001;6:177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Cowen LE. Hsp90-dependent regulatory circuitry controlling temperature-dependent fungal development and virulence. Cell Microbiol 2014;16:473–81. [DOI] [PubMed] [Google Scholar]
- Odom A, Muir S, Lim E et al. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 1997;16:2576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 2007;20:133–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 2010;36:1–53. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 2001;15:1118–31. [DOI] [PubMed] [Google Scholar]
- Ramsdale M, Selway L, Stead D et al. MNL1 regulates weak acid-induced stress responses of the fungal pathogen Candida albicans. Mol Biol Cell 2008;19:4393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 1962;18:571–3. [Google Scholar]
- Sakurai H, Fukasawa T. A novel domain of the yeast heat shock factor that regulates its activation function. Biochem Bioph Res Co 2001;285:696–701. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol 1993;13:1392–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Cowen LE. Coupling temperature sensing and development. Virulence 2010;1:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Uppuluri P, Zaas AK et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol 2009;19:621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer G Jr, Birge CH, Yuckenberg PD et al. Heat-shock proteins induced during the mycelial-to-yeast transitions of strains of Histoplasma capsulatum. J Gen Microbiol 1987;133:3375–82. [DOI] [PubMed] [Google Scholar]
- Solis EJ, Pandey JP, Zheng X et al. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell 2016;63:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 1990;62:793–805. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Nelson HC. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell 1989;59:807–13. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J 1987;6:3035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 1988;54:855–64. [DOI] [PubMed] [Google Scholar]
- Steen BR, Lian T, Zuyderduyn S et al. Temperature-regulated transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res 2002;12:1386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol 2011;9:737–48. [DOI] [PubMed] [Google Scholar]
- Tamai KT, Liu X, Silar P et al. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol 1994;14:8155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger JM, Schmitt AP, Simon JR et al. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J Biol Chem 1998;273:26875–9. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Murray JI, Hartman SJ et al. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell 2004;15:1254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veri AO, Miao Z, Shapiro RS et al. Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet 2018;14:e1007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Sistonen L. HSF1 at a glance. J Cell Sci 2014;127:261–6. [DOI] [PubMed] [Google Scholar]
- Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol 2007;594:89–99. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets 2009;13:469–78. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Oberholzer U. Candida morphogenesis and host-pathogen interactions. Curr Opin Microbiol 2004;7:350–7. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 1988;54:841–53. [DOI] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 1995;11:441–69. [DOI] [PubMed] [Google Scholar]
- Wu C, Wilson S, Walker B et al. Purification and properties of Drosophila heat shock activator protein. Science 1987;238:1247–53. [DOI] [PubMed] [Google Scholar]
- Xiao H, Perisic O, Lis JT. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell 1991;64:585–93. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Mizukami Y, Sakurai H. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J Biol Chem 2005;280:11911–9. [DOI] [PubMed] [Google Scholar]
- Yang DH, Jung KW, Bang S et al. Rewiring of signaling networks modulating thermotolerance in the human pathogen Cryptococcus neoformans. Genetics 2017;205:201–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzov P, Boucherie H, Mann C. A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J Cell Sci 1997;110:1879–91. [DOI] [PubMed] [Google Scholar]
- Zheng X, Krakowiak J, Patel N et al. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 2016;5:e18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T et al. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 1998;94:471–80. [DOI] [PubMed] [Google Scholar]