ABSTRACT
Acetylene (IUPAC name: ethyne) is a colorless, gaseous hydrocarbon, composed of two triple bonded carbon atoms attached to hydrogens (C2H2). When microbiologists and biogeochemists think of acetylene, they immediately think of its use as an inhibitory compound of certain microbial processes and a tracer for nitrogen fixation. However, what is less widely known is that anaerobic and aerobic microorganisms can degrade acetylene, using it as a sole carbon and energy source and providing the basis of a microbial food web. Here, we review what is known about acetylene degrading organisms and introduce the term 'acetylenotrophs' to refer to the microorganisms that carry out this metabolic pathway. In addition, we review the known environmental sources of acetylene and postulate the presence of an hidden acetylene cycle. The abundance of bacteria capable of using acetylene and other alkynes as an energy and carbon source suggests that there are energy cycles present in the environment that are driven by acetylene and alkyne production and consumption that are isolated from atmospheric exchange. Acetylenotrophs may have developed to leverage the relatively high concentrations of acetylene in the pre-Cambrian atmosphere, evolving later to survive in specialized niches where acetylene and other alkynes were produced.
Keywords: acetylene degraders, acetylenotrophs, acetylene, ethyne
The current knowledge of acetylene degrading organisms is reviewed and the authors introduce the term 'acetylenotrophs' to refer to the organisms that carry out this metabolic pathway.
INTRODUCTION
Acetylene (C2H2, IUPAC name: ethyne, CAS 74-86-2) is the simplest alkyne, composed of two triple bonded carbon atoms attached to hydrogen. It is best known in the microbiology community as an inhibitor of microbial processes and for its use in nitrogen fixation and denitrification bioassays, where nitrogenase converts acetylene to ethylene or blocks the reduction of N2O to N2, respectively (Stewart, Fitzgerald and Burris 1967; Knowles 1982). However, this colorless, quite soluble (KH = 4.15 × 10−2 moles L−1 atm−1 at 298 K; e.g., comparable to CO2 (Sander 2015)), gaseous hydrocarbon with its characteristic garlic-like odor can be degraded by anaerobic and aerobic microorganisms that use acetylene as a sole carbon and energy source. Acetylene degrading microorganisms were first observed in 1932 (Birch-Hirschfeld 1932) and since then, numerous environmental samples have demonstrated acetylene consumption (Table 1) and yielded strains that use acetylene for growth (Table 2). Here, we introduce the term 'acetylenotrophs' to refer to the organisms that carry out this metabolic growth pathway. In addition, we review what is known about the genetics, ecology and biogeochemistry of acetylenotrophs and discuss how these organisms could be supporting a microbial food web, subsisting on hidden sources of acetylene in the environment.
Table 1.
Summary of environments where acetylene degradation has been observed.
| Sample location | Habitat Type | Character/ Salinity (g L−1) | Incubation condition | C2H2 uptake | AH PCR result | Reference |
|---|---|---|---|---|---|---|
| Los Baños, Philippines | Rice paddy soil | Fresh | Anaerobic | + | N/Aa | Watanabe and De Guzman (1980) |
| San Francisco Bay, California USA | Intertidal mud flat sediment | Estuarine | Anaerobic | + | N/Aa | Culbertson, Zehnder and Oremland (1981) |
| San Francisquito Creek, California USA | Freshwater sediment | Fresh | Anaerobic | − | N/Aa | Culbertson, Zehnder and Oremland (1981) |
| Unknown | Fox sandy loam soil | Fresh | Anaerobic | + | N/Aa | Yeomans and Beauchamp (1982) |
| Brookston silty clay | Fresh | Anaerobic | + | N/Aa | Yeomans and Beauchamp (1982) | |
| Floradale, Ontario, Canada | Stream sediment and water | Aerobic | + | N/Aa | Tam, Mayfield and Inniss (1983) | |
| Fresh | Anaerobic | − | N/Aa | Tam, Mayfield and Inniss (1983) | ||
| Ithaca, New York USA | Collamer silty clay loam soil | Aerobic | + | N/Aa | Terry and Duxbury (1986) | |
| Fresh | Anaerobic | + | N/Aa | Terry and Duxbury (1986) | ||
| Spanish Fork, Utah USA | Timpanogos clay loam soil | Aerobic | + | N/Aa | Terry and Duxbury (1986) | |
| Fresh | Anaerobic | + | N/Aa | Terry and Duxbury (1986) | ||
| Belle Glade, Florida USA | Pahokee muck soil | Fresh | Aerobic | + | N/Aa | Terry and Duxbury (1986) |
| Dijon, France | Agricultural soil | Aerobic | + | N/Aa | Topp and Germon (1986) | |
| Fresh | Anaerobic | − | N/Aa | Topp and Germon (1986) | ||
| Unknown | Estuarine sediment | Estuarine | Anaerobic | + | N/Aa | Culbertson, Strohmaier and Oremland (1988) |
| Hajdów Research Station (Lublin, Poland) | Peat muck soil | Fresh | Aerobic | + | N/Aa | Brzeziska et al.(2011) |
| Cambisol soil | Fresh | Aerobic | + | N/Aa | Brzeziska et al.2011) | |
| San Francisco Bay, California USA | Intertidal sediment | 25 | Anaerobic | + | + | Miller et al.(2013) |
| Saltmarsh sediment | 25 | Anaerobic | + | − | Miller et al.(2013) | |
| Searsville Lake, California USA | Freshwater sediment | <1 | Anaerobic | + | − | Miller et al.(2013) |
| Mono Lake, California USA | Littoral sediment | 80 | Anaerobic | − | − | Miller et al.(2013) |
| Hot spring biofilm | 25 | Anaerobic | − | − | Miller et al.(2013) | |
| Gulf of Mexico | Cold hydrocarbon seep brine | 60 | Anaerobic | − | − | Miller et al.(2013) |
| Long Valley, California USA | Hot spring biofilm | 4 | Anaerobic | − | − | Miller et al.(2013) |
| Western Pennsylvania USA | Peat bog sediment | <1 | Anaerobic | − | − | Miller et al.(2013) |
| Piedmont, Virginia USA | Constructed wetland sediment | <1 | Anaerobic | + | − | Miller et al. (2013) |
| Pillar Point, California USA | Saltmarsh sediment | 20 | Anaerobic | + | − | Miller et al.(2013) |
| Great Bay, New Hampshire USA | Estuarine sediment (Portsmouth) | 28 | Anaerobic | + | + | Miller et al. (2013) |
| Estuarine sediment (Squamscott) | 28 | Anaerobic | − | + | Miller et al. (2013) | |
| Trenton, New Jersey USA (Naval Air Warfare Centerb) | TCE-contaminated groundwater (well 36BR-A, 2009c) | <1 | Anaerobic | + | + | Miller et al. (2013) |
| TCE-contaminated groundwater (well 36BR-A, 2015) | <1 | Anaerobic | + | Not measured | This study | |
| TCE-contaminated groundwater (well 73BR-D2, 2015) | <1 | Anaerobic | + | Not measured | This study | |
| TCE-contaminated groundwater (well 36BR-A, 2017) | <1 | Anaerobic | + | + | This study | |
| Mountain View, California (NASA Ames Research Center) | Soil above TCE-contaminated groundwater (2017) | <1 | Aerobic | + | − | This study |
| Soil above TCE-contaminated groundwater (2017) | <1 | Anaerobic | + | + | This study | |
| Trace TCE-contaminated groundwater (well 14D26A1) | <1 | Anaerobic | − | Not measured | This study | |
| Trace TCE-contaminated groundwater (well 11N22A1) | <1 | Anaerobic | − | Not measured | This study | |
| Trace TCE-contaminated groundwater (well 11MO3A) | <1 | Anaerobic | − | Not measured | This study |
N/A=not applicable because the reference pre-dated the development of Pelobacter acetylenicus-specific acetylene hydratase (AH) PCR primers.
For additional information about the NAWC site see (Révész et al.2014).
Indicates year groundwater samples were collected for acetylene uptake experiments.
Table 2.
Summary of known acetylenotrophic bacterial strains.
| Type | Phylogenetic group | Organism | Habitat/Isolation source | Requirements for acetylenotrophy | Products of acetylenotrophy | Reference |
|---|---|---|---|---|---|---|
| Aerobe | Actinobacteria | Mycobacterium lacticola a,b | Soil (Germany) | C2H2 alone | Acetaldehyde | Birch-Hirschfeld (1932) |
| Rhodococcus rhodochrous strain A1a,c | Dutch soil (The Netherlands) | C2H2 alone | Acetaldehyde | de Bont and Peck (1980) and de Bont et al.(1980) | ||
| Rhodococcus rhodochrous (ATCC 33258) (isolated and deposited as: Nocardia rhodochrousa) | Agricultural field soil and lake sediment (New Jersey, USA) | C2H2 alone | Acetaldehyde | (Kanner and Bartha (1979) and Kanner and Bartha (1982) | ||
| Rhodococcus rhodochrous strain E5d | Agricultural soil | C2H2 alone# | Acetaldehyde, ethanol and acetate | Topp and Germon (1986) and Germon and Knowles (1988) | ||
| Rhodococcus opacus strain MoAcy1 (DSM 44186) | Dry garden soil sample near a parking lot (Tübingen, Germany) | C2H2 alone | Not measured | Rosner et al.(1997) | ||
| Rhodococcus opacus strain TueAcy1 (DSM 44188) | Dry soil sample (Tübingen, Germany) | C2H2 alone | Not measured | Rosner et al.(1997) | ||
| Rhodococcus zopfii strain TueAcy3 (DSM 44189) | Dry soil sample (Tübingen, Germany) | C2H2 and yeast extract | Not measured | Rosner et al.(1997) | ||
| Gordonia alkanivorans strain MoAcy2 (DSM 44187) | Dry garden soil sample near a parking lot (Tübingen, Germany) | C2H2 and yeast extract | Not measured | Rosner et al.(1997) | ||
| Rhodococcus rhodochrous strain PNKb1e | Freshwater | C2H2 alone | Trace acetaldehyde detected | Woods (1988) | ||
| Firmicutes | Bacillus spp.a | Stream sediment and water (Ontario, Canada) | C2H2 alone | Not measured | Tam, Mayfield and Inniss (1983) | |
| Anaerobe | Deltaproteobacteria | Pelobacter acetylenicus strain WoAcy1 (DSM 3246T) | Freshwater creek sediment (Germany) | C2H2 alone | Ethanol, acetate and acetaldehyde | Schink (1985) |
| Pelobacter acetylenicus strain KoAcy23 | Sewage sludge (Germany) | C2H2 alone | Ethanol, acetate and acetaldehyde | Schink (1985) | ||
| Pelobacter acetylenicus strain GhAcy1 (DSM3247) | Marine sediment (Venice, Italy) | C2H2 alone | Ethanol, acetate and acetaldehyde | Schink (1985) | ||
| Pelobacter acetylenicus strain GhAcy3 | Marine sediment (Venice, Italy) | C2H2 alone | Ethanol, acetate and acetaldehyde | Schink (1985) | ||
| Pelobacter sp. strain SFB93 | Estuarine sediment (San Francisco Bay, California, USA) | C2H2 alone | Acetaldehyde, ethanol, acetate and hydrogen | Culbertson, Strohmaier and Oremland (1988) and Miller et al. (2013) |
Strains were identified using phenotypic characteristics and the listed name corresponds to that used in the original publication. It is important to note that the correct name of the organism may have changed since the original date of publication.
Kanner and Bartha (1979) suggested based on phenotypic characteristics that this organism was mostly likely a Rhodococcus rhodochrous.
de Bont et al. (1980) identified strain A1 as Rhodococcus rhodochrous using DNA–DNA hybridization.
Although originally isolated and cultivated under aerobic conditions Germon and Knowles (1988) showed that strain E5 can degrade acetylene under anaerobic conditions.
Isolation of and metabolism of propane by this organism was reported in Woods and Murrell (1989).
ACETYLENE ON EARTH AND EXTRATERRESTRIAL BODIES
Acetylene’s ability to support microbial growth and a microbial food web has not been a major interest in environmental microbiology due to its apparently inconsequential trace environmental concentrations (e.g., ∼ 20–40 parts per trillion in the Earth’s atmosphere, Cronn and Robinson 1979; Goldman et al.1981; Rudolph, Ehhalt and Khedim 1984). The spatial distribution, sources, chemical sinks and variability of acetylene in the Earth’s atmosphere have been extensively studied usually in association with a broader survey of atmospheric volatiles (Whitby and Altwicker 1978; Cronn and Robinson 1979; Bonsang, Kanakidou and Lambert 1987; Plass, Koppmann and Rudolph 1992; Jobson et al.1994; Blake et al.1997; Gupta et al.1998; Heard et al.2006; Hudson and Ariya 2007; XiaoJacob and Turquety 2007; Baker et al.2008). The lower tropospheric mixing ratio of acetylene ranges from its typical higher abundance over urban areas (e.g., 1–2 parts per billion by volume (ppbv)) presumably from anthropogenic sources to 10-fold lower abundances (e.g. tens of pptv) over remote areas of the ocean. Acetylene decreases markedly (∼10-fold) with the upward vertical transition from the lower troposphere to the tropopause (Blake et al.1997; Gupta et al.1998; Xiao, Jacob and Turquety 2007; Baker et al.2008). In the atmosphere, acetylene is a short-lived tracer of combustion and air mass photochemical age, with oxidation by the OH radical being the major loss pathway, leading to a tropospheric residence time for acetylene of approximately 12 days (Atkinson et al.1997; Talbot et al.1997; Shim et al.2007). This is very short indeed when compared with the 8–12 year residence time for the less reactive but far more abundant (∼ 1.8 parts per million by volume (ppmv)) and climatically relevant methane for which OH radical oxidation is also its major sink (e.g., Cicerone and Oremland 1988). The dominant source of acetylene to the atmosphere is anthropogenic combustion of fossil fuel (1.7 Tg yr−1) and biofuel (3.3 Tg yr−1), accompanied by a biomass burning (1.6 Tg yr−1) component (Xiao, Jacob and Turquety 2007).
Several early studies suggested that the global ocean represents a minor source of acetylene (Kanakidou et al.1988; Plass, Koppmann and Rudolph 1992; Plass-Dülmer et al.1995). However, subsequent studies have shown that enhancements in acetylene mixing ratios observed over remote oceanic locations are most likely attributable to long range atmospheric transport rather than any significant in situ marine production (Blake et al.1997; Talbot et al.1997; Gupta et al.1998; Lewis, Carpenter and Pilling 2001; Hopkins et al.2002; Hudson and Ariya 2007; Shim et al.2007). Global atmospheric chemistry modeling shows that any emissions from an oceanic source to the atmosphere would have to be less than 0.5 Tg yr−1 ( < 7.5% of the global budget), in order to reconcile observed global distributions (Xiao, Jacob and Turquety 2007). However, a natural biogenic source of acetylene cannot be definitively ruled out as microorganisms have been shown to generate acetylene (Fukuda, Fujii and Ogawa 1984; Belay and Daniels 1987). Belay and Daniels (1987) showed that pure cultures of methanogenic bacteria can generate acetylene from 1,2-dibromoethene (Belay and Daniels 1987). While this was demonstrated only in the laboratory, the fact that the precursor, 1,2-dibromoethene, is produced by marine algae (Gribble 1996) suggests the possibility of a natural marine source of acetylene. Furthermore, work by Fukuda, Fujii and Ogawa (1984) screened C2-hydrocarbon production by 166 microbial strains and found that 2% of the organisms tested produced acetylene. The acetylene producing strains included Fungi (Phycomyces nitens IFO 9421) and Bacteria (Proteus mirabilis IFO 3849 and Streptomyces fradiae IFO 3360) and they had maximal rates of acetylene production of 0.2 nl ml−1 hr−1.
Little to date is known about naturally occurring terrestrial (i.e., soil and subsurface aquifer) sources of acetylene. However, numerous reports have shown that acetylene can be formed by abiotic reactions of minerals with chlorinated solvents (as reviewed in He et al.2015). Laboratory studies showed that zero-valent (Fe0) and metallic iron can react with dichloroethylene, trichloroethylene (TCE) and tetrachloroethylene to form acetylene as an end-product (Roberts et al.1996; Campbell et al.1997; Arnold and Roberts 2000). Further studies revealed that a diverse array of reduced iron minerals can degrade chlorinated compounds, including magnetite (Fe3O4), mackinawite (FeS), pyrite (FeS2) and green rusts (mixed Fe(II)/(III) minerals) (as reviewed in He et al.2015). These reactive minerals catalyze chlorinated solvent degradation via reductive elimination, hydrogenolysis, dehydrohalogenation and hydrolysis reactions. These reactions were shown to occur in field studies at chlorinated solvent contaminated sites (Puls, Blowes and Gillham 1999; Han et al.2012; Schaefer et al.2015) and accumulation of acetylene can thus be used as a proxy for the success of remediation strategies employing zero valent iron or reduced iron minerals (He et al.2015).
With respect to early Earth and planet(oid)s of the outer Solar System, acetylene is hypothesized to play an important role in the evolution of life. Acetylene may have reached an abundance of 5 ppm (Zahnle 1986; Trainer et al.2004) in the anoxic atmosphere of early Earth (Kasting 1993; Kasting 2004) due to atmospheric photochemical reactions that originate with methane (Zahnle 1986; Trainer et al.2004). Such reactive atmospheric chemistry derived from simple constituents formed the basis of the complex pre-biotic chemistry thought to lead to the origin of life (Miller 1953; Miller and Urey 1959; Scheidler et al.2016).
Analogous processes of complex organic formation occur in the methane-rich planet(oid)s of the outer Solar System, including Saturn’s moon Titan (Abbas and Schulze-Makuch 2002; Shemansky et al.2005; Clark et al.2010). It has been proposed that life on Titan could be sustained energetically by hydrogen’s reductive splitting of acetylene to form methane at ∼ 90 K (McKay and Smith 2005). Indeed, this possibility was supported by Titan’s atmospheric profile of diminishing vertical H2 and C2H2 abundances with depth as measured by the descent of the Huygens probe facet of the Cassini–Huygens Mission (Strobel 2010) and the absence of detectable acetylene just above its surface (Clark et al.2010). These observations suggested the exciting possibility of a biological mechanism for removal of these gases from the atmosphere that existed on Titan’s frigid surface, perhaps something akin to what had been postulated by McKay and Smith (2005). Molecular hydrogen and C1–C5 light hydrocarbons, possibly including acetylene, have been detected emanating from the 'Tiger Stripe' fissures prominent on the south pole of Enceladus (Waite et al.2006; Kopparla et al.2016; Waite et al.2017). It is hypothesized that a global anoxic ocean lies beneath Enceladus’ ice sheet and represents a potential habitable environment for microbial life that could be sustained on light hydrocarbons or the accompanying H2 also evident in the plumes (McKay et al.2008; Waite et al.2017). Thus, sampling its plume for live microbes and/or their biomarker molecules is an exciting prospect (McKay et al.2014; Porco, Dones and Mitchell 2017).
ACETYLENE TRANSFORMATIONS
Acetylene is only known to be biologically transformed via nitrogenase (N2ase) in a non-metabolic reaction and acetylene hydratase (AH) that catalyzes the first step in acetylene metabolism (Fig. 1). Nitrogenase (N2ase) is well characterized compared to AH: a simple SCOPUS search yields 8,592 and 43 citations, respectively (www.scopus.com, accessed 13 February 2018). Nitrogenase catalyzes the reduction of di-nitrogen (N2) to ammonia (2 NH3) and is found in an array of diazotrophic (nitrogen-fixing) bacteria. N2ase will also use acetylene as substrate, resulting in the production of ethylene (C2H4) and it is also known to bind and reductively react with other triple-bonded molecules such as cyanide. Since acetylene is inhibitory to several cellular pathways (e.g., nitrogen fixation, methanogenesis, methane oxidation, denitrification, reductive dechlorination, nitrification, certain hydrogenases and anaerobic ammonium oxidation, known as 'anammox' (Stewart, Fitzgerald and Burris 1967; Oremland and Taylor 1975; Balderston, Sherr and Payne 1976; Dalton and Whittenbury 1976; Yoshinari, Hynes and Knowles 1977; Payne and Grant 1982; Payne 1984; Oremland and Capone 1988; Pon, Hyman and Semprini 2003; Jensen, Thamdrup and Dalsgaard 2007)), it is possible that other enzymes are also degrading acetylene as a side-reaction rather than part of a metabolic pathway; particularly those that already use triple bonded substrates. A third enzyme, 4-hydroxy-3-methyl-butenyl diphosphate reductase (IspH), responsible for reductive dehydration to form isopentenyl diphosphate and dimethylallyl diphosphate on the methylerythritol phosphate pathway, has been shown experimentally to hydrate terminal acetylene groups on larger hydrocarbons (Span et al.2012). It is unclear whether this enzyme has the ability to hydrate C2H2 (Span et al.2012), but it does serve as another example of an enzyme interacting in a non-metabolic fashion with acetylene groups.
Figure 1.
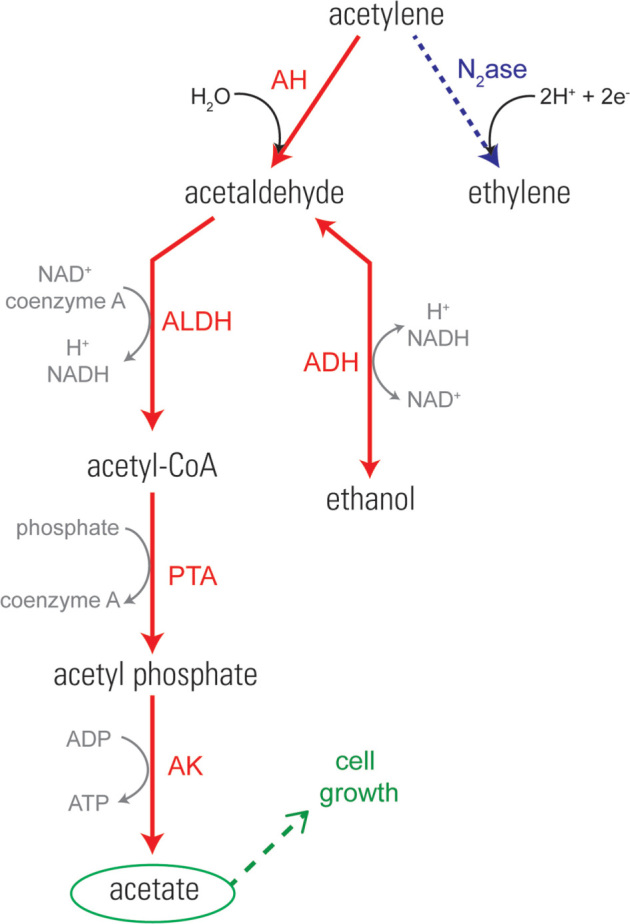
Pathways of acetylene transformation via acetylene fermentation (left process, red) and N2ase (right process, blue). Genes in the acetylene fermentation pathway include acetylene hydratase (AH), ALDH, ADH, phosphate acetyltransferase (PTA) and acetate kinase (AK). Modified from (Caspi et al.2010; Akob et al.2017).
Growth-linked metabolism of acetylene by a bacterial strain was first reported in 1932 for a mycobacterium (Birch-Hirschfeld 1932) and was followed 50 years later by several reports documenting the consumption of acetylene over the course of incubations of soils and sediments employing acetylene-based assays for denitrification or nitrogen-fixation (Watanabe and De Guzman 1980; Culbertson, Zehnder and Oremland 1981; Yeomans and Beauchamp 1982; Tam, Mayfield and Inniss 1983) (Table 1). Building on these investigations, Pelobacter acetylenicus was the first isolated and most extensively studied acetylenotrophic anaerobe that coupled growth to the fermentation of acetylene (Schink 1985; Boll et al.2016; Kroneck 2016). From work with P. acetylenicus, the enzyme responsible for the first step in acetylene fermentation, acetylene hydratase (AH; EC 4.2.1.112), was identified and extensively characterized. AH catalyzes the hydration of acetylene in an exergonic, non-redox reaction to form acetaldehyde (Fig. 1, Schink 1985; ten Brink 2014):
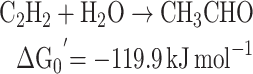 |
(1) |
AH, a member of the dimethyl sulfoxide reductase protein family, contains a tungsten pyranopterin cofactor (Seiffert et al.2007; ten Brink, Schink and Kroneck 2010) and its activity is dependent on the presence of a strong reducing agent, such as titanium(III) citrate or dithionite (Rosner and Schink 1995; ten Brink 2014). The AH enzyme, encoded by the ahy gene, is a monomer of 730 amino acids with four domains. Domain I contains a 4Fe-4S cluster coordinated by four cysteine residues. Domains I, II and IV coordinate the bis-molybdopterin-gunanine-dinucletide cofactors that coordinates a W ion (Seiffert et al.2007). AH is specific for acetylene and does not react with other triple bonded molecular analogues; for a detailed description of the AH enzyme from P. acetylenicus see the review by ten Brink (2014).
Acetylene fermentation continues via disproportionation of acetaldehyde to acetate and ethanol via aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH), respectively (Fig. 1, Schink 1985; ten Brink 2014). Together, these enzymes catalyze this reaction:
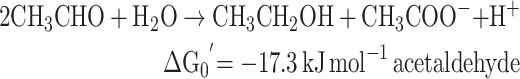 |
(2) |
The acetate can be used directly by acetylenotrophs as a carbon source to support cell growth, whereas the ethanol is used by cells via a reverse reaction of alcohol dehydrogenase converting ethanol back to acetaldehyde where it can then be converted to acetate (Fig. 1). The resulting acetaldehyde, acetate, ethanol and H2 from acetylene metabolism are secreted into the external milieu by pure cultures, and in mixed associations can then serve as carbon and energy substrates for other opportunist microorganisms present (Table 1 and Fig. 2).
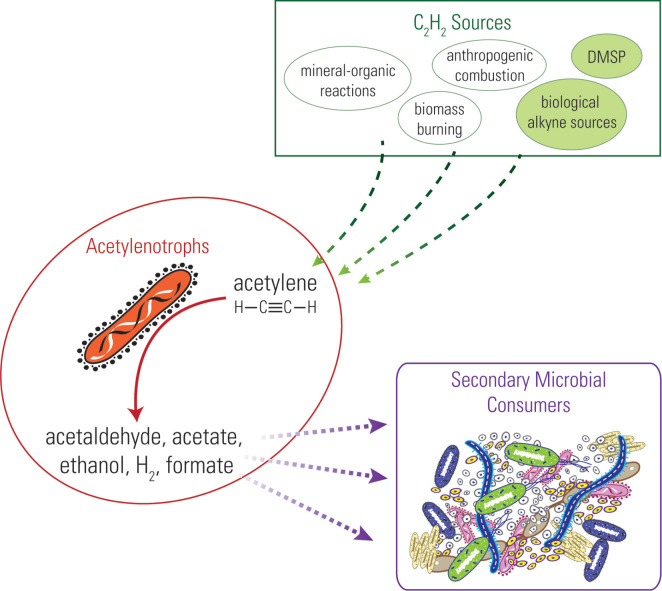
Figure 2.
Acetylenotrophy-driven microbial food web. Sources of acetylene include known and hypothetical (green circles) sources.
Studies to date have shown that the AH of P. acetylenicus and Pelobacter sp. strain SFB93 favors 12C-acetylene over 13C-acetylene (Miller, Baesman and Oremland 2015). This results in kinetic isotope effects (KIEs) ranging from ∼ 2.0 to 9.0‰ enrichments of 12C, thereby leaving the residual, as yet un-metabolized acetylene enriched in 13C. These KIE were also seen when acetylenotrophy was carried out by the anaerobic enrichment culture SV7 and sediments from San Francisco Bay, California (Miller, Baesman and Oremland 2015). The ecological and astrobiological implication of this work was that the 'δ13C' composition of acetylene and its metabolic products (e.g., acetaldehyde, acetate) could be used to evaluate environmental acetylenotrophy or be employed as possible biomarkers in the search for extant microbial life in the icy worlds of the outer Solar System.
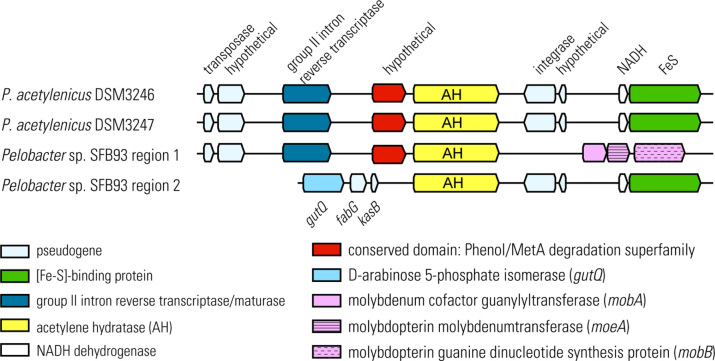
Interestingly, we recently reported that AH and N2ase are present in the annotated genomes of three Pelobacter strains (Akob et al.2017) including the two deposited type reference strains of P. acetylenicus (DSM3246 and DSM3247) and Pelobacter sp. strain SFB93, isolated from San Francisco Bay (Culbertson, Strohmaier and Oremland 1988; Miller et al.2013; Sutton et al.2017a,b). Pelobacter sp. SFB93 contained two copies of the ahy gene, which were flanked by horizontal gene transfer (HGT) elements (Fig. 3). In addition, we experimentally showed that Pelobacter sp. strain SFB93 was capable of diazotrophic growth, during which time both enzymes were active (Akob et al.2017).
Figure 3.
Syntenic map of acetylene hydratase (ahy) and flanking genes in Pelobacter sp. strain SFB93 and P. acetylenicus DSM3246 and DSM3247. Strain SFB93 contains two ahy genes in two regions, separated by ∼18,000 bp. Reprinted from (Akob et al.2017).
Aerobic, acetylene-degrading bacterial strains have been obtained that use acetylene either as their sole source of carbon and energy, or consume it in conjunction with low concentrations of supplemental yeast extract (Birch-Hirschfeld 1932; de Bont and Peck 1980; Kanner and Bartha 1982; Germon and Knowles 1988; Rosner et al.1997) (Table 1). The acetylene-metabolizing enzymes from aerobic acetylenotrophs have not been fully characterized. However, experiments by Rosner et al. (1997) showed that aerobic AHs from strains Rhodococcus opacus strain MoAcy1 and TueAcy1, Rhodococcus zopfii strain TueAcy3, and Gordonia alkanivorans strain MoAcy2 differ from those of P. acetylenicus. The AH of these strains had a lack of cross-reactivity with antibodies raised to the AH of P. acetylenicus (Rosner et al.1997). In addition, expression of AH in these aerobes was dependent on molybdenum, unlike the AH of P. acetylenicus which is tungsten-dependent. Together, these observations indicate that the AH of aerobic acetylenotrophs are unique and that this is a heterogeneous group of enzymes.
ECOLOGY OF ACETYLENOTROPHS
As stated above, the first report of acetylene-fueled growth dates to 1932 for an aerobic mycobacterium isolated from soils in Germany (Birch-Hirschfeld 1932). Since that finding, 9 papers have documented the consumption of acetylene from soil, sediment, and groundwater samples (Table 1) and resulted in the isolation of 15 pure cultures of acetylenotrophs (Table 2). Many of the early incubations of environmental samples where acetylene uptake was observed were originally designed for acetylene-based denitrification or nitrogen-fixation assays, with acetylene consumption being a spurious observation.
The observations of acetylenotrophy in environmental samples, summarized in Table 1, show that this metabolism is distributed widely amongst geochemical conditions, from freshwater to hypersaline, surface to subsurface, soils to aquifers, and anoxic to oxic ecosystems. The first observations of anaerobic acetylenotrophy was in incubations of rice paddy soils from the Philippines (Watanabe and De Guzman 1980) and further studies expanded on this work demonstrating anaerobic acetylene uptake in soils, sediments and groundwaters from around the globe (Culbertson, Zehnder and Oremland 1981; Yeomans and Beauchamp 1982; Terry and Duxbury 1986; Topp and Germon 1986; Culbertson, Strohmaier and Oremland 1988; Miller et al.2013). In addition, further studies investigated the potential for aerobic or anaerobic acetylene uptake in soils with varying results. Terry and Duxbury (1986) found that soils could consume acetylene under both anaerobic and aerobic conditions. Brzezinska et al. (2011) showed that soils from Poland consumed acetylene under aerobic conditions. Tam, Mayfield and Inniss (1983) and Topp and Germon (1986) compared the ability of stream sediments and soils to consume acetylene and found that only aerobic incubations could do so, whereas under anaerobic conditions no uptake was observed. Interestingly, Tam, Mayfield and Inniss (1983) reported that 33% of their sediment incubations were positive for acetylene consumption. This observation has held in other surveys of the ubiquity of acetylene consumption.
Over 3 decades after the first report of anaerobic acetylenotrophy, a survey was undertaken to determine if this metabolism was a common or rare phenomenon amongst a diverse array of anoxic sediment samples and one groundwater sample (Miller et al.2013). Acetylenotrophy, e.g., consumption, was clearly evident in 9 of 42 (21.4%) of incubated samples tested (Table 1, Miller et al.2013). Primers were also developed in the Miller et al. (2013) study that were specific for the ahy gene in P. acetylenicus. Application of this primer set to DNA extracts from numerous field samples revealed that ahy was even less common, with only 63 of 645 samples testing (9.8%) positive for the gene (Table 1, Miller et al.2013). Samples that were positive for anaerobic acetylenotrophy in the Miller et al. (2013) study included TCE-contaminated groundwaters taken from the Naval Air Warfare Center (NAWC) in Trenton, NJ an in situ bioremediation study site, and the amplicons were confirmed to be AH genes. A likely source of acetylene at the NAWC site could be attributed to abiotic dehalogenation reactions with TCE and reduced iron minerals. Consumption of acetylene by NAWC groundwater microorganisms was observed in samples collected in 2015 and 2017 (Table 1). Newer work by our group, has shown that soils and groundwater from a TCE-impacted site, namely the NASA Ames Research Center (ARC) can consume acetylene (Table 1).
To date, 15 acetylenotrophic bacterial strains have been obtained that either use acetylene as either their sole carbon and energy source or with low concentrations of supplemental yeast extract during anaerobic or aerobic metabolism (Table 2). However, this is at odds with the frequency of acetylene degradation observed in environmental samples. Acetylenotrophs span the Actinobacteria, Proteobacteria, and Firmicutes phyla (Table 2). However, many of the early strains were identified by phenotypic characteristics and not available in culture collections, therefore, their phylogeny cannot be confirmed use DNA molecular markers. Of particular interest, is the isolation of an acetylenotrophic strain from the Firmicutes phyla (Tam, Mayfield and Inniss 1983). This strain is not available in culture collections, so little is known about its ability to transform acetylene or its metabolic versatility. Acetylenotrophic members of the Actinobacteria have been repeatedly isolated under aerobic conditions, although not always reported in the literature, for example, Rhodococcus rhodochrous strain PNKb1 was isolated by Woods in 1988 but no peer-reviewed paper documented the finding. We hypothesize that more acetylenotrophs may be out there but the lack of testing for this metabolism has hindered their discovery.
The acetylenotrophy study by Miller et al. (2013) resulted in the re-isolation of Pelobacter sp. strain SFB93 from San Francisco Bay, which allowed for further characterization of this organism via genome sequencing (Akob et al.2017; Sutton et al.2017a,b). In addition, Miller et al. (2013) obtained a stable anaerobic enrichment culture (SV7) from freshwater Searsville Lake which to date has defied purification into a single acetylene-fermenting species but contained members of the Actinobacteria (Oremland, Baesman, and Akob, personal observation). Presence of members of the Actinobacteria was initially surprising as these taxa are typically characterized as aerobic and includes the majority of known aerobic acetylenotrophs (Table 2). However, the aerobic acetylenotroph Rhodococcus rhodochrous strain E5 was originally isolated and cultivated under aerobic conditions but was later shown to degrade acetylene under anaerobic conditions (Topp and Germon 1986). Together this suggests that members of the Actinobacteria may be more versatile in their oxygen requirements and that acetylenotrophy may confer metabolic versatility on these organisms if oxygen becomes limiting. In addition, although SV7 grew at the expense of acetylene, its DNA did not amplify with Pelobacter-specific ahy primers, and the culture formed different intermediates and end products (acetate, formate, carbon dioxide, methane) than P. acetylenicus or Pelobacter sp. strain SFB93 (Miller et al.2013). The lack of amplification with Pelobacter-specific ahy primers is not surprising based on the findings of Rosner et al. (1997) that aerobic AHs are different. Further, we recently sequenced the genomes of Rosner’s strains and found that Pelobacter-specific ahy primers do not bind in silico to the genomes (Sutton and Fierst, unpublished data). Lastly, SV7 manifests higher KIEs than other Pelobacter strains or San Francisco Bay sediments (Miller, Baesman and Oremland 2015).
The presence of so many acetylene-consuming environmental samples and strains suggests that this metabolism might be more wide spread than previously known (discussed more below). However, to date we lack quantitative information on the abundance of acetylenotrophic organisms as estimates for population size using most-probable-number (MPN) assays or quantitative PCR for AH genes have not been performed. To fully understand the contribution of acetylenotrophs estimates of population size are needed. In addition, from these observations we hypothesize that acetylenotrophs could play an important ecological and biogeochemical role by fueling a microbial food web on modern Earth (Fig. 2). Acetylenotrophic strains produce ethanol, acetate, acetaldehyde, and H2 (Table 1B) as byproducts of their metabolism. These substrates can, in mixed systems (e.g., co-cultures or sediments), further serve to fuel secondary microbial consumers and thereby support a microbial food web (Fig. 2). Indeed, acetylenotrophs were shown to support an anaerobic food-chain including sulfate-reducers (Culbertson, Zehnder and Oremland 1981), methanogens (Seitz et al.1990; Miller et al.2013), and organo-halide reducers (Mao et al.2017). The presence of other organisms might be required for acetylenotrophy to be favorable through a syntrophic relationship.
EVOLUTION AND GENETICS OF ACETYLENOTROPHS
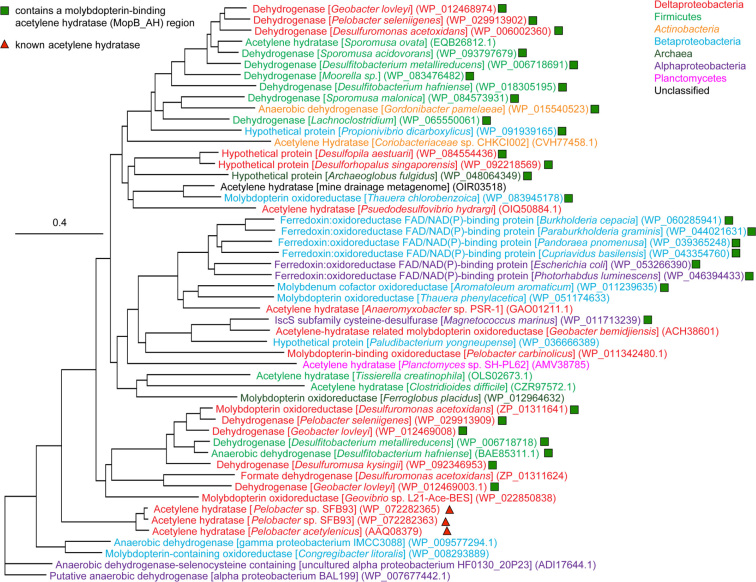
The fact that both anaerobic and aerobic acetylenotrophs have been found around the world in diverse environments and that their capacity to grow on acetylene is dependent on different AHs implies that this trait has been around for quite some time. It is tempting to pose a scenario of primordial anoxic food chains developing around acetylene, growing more complex over time, and with the advent of oxygenic photosynthesis (∼ 2.4 Gya), came the AH found in aerobic acetylenotrophs and the draw-down of atmospheric acetylene to present day 'background' levels. Admittedly, until very recently these ideas seem a bit tenuous considering that there are so few acetylenotrophic strains (Table 2), and that much of the work on this group of organisms ceased well before the genomics revolution came into its own. Yet it is from our genetic studies of P. acetylenicus and Pelobacter sp. strain SFB93 (Akob et al.2017) that certain key facets supporting these notions can be discerned: 1) their ahy genes are flanked by group II intron reverse transcriptases/maturases and integrases (Fig. 3) suggesting a fluid influx or efflux via HGT of AH amongst microbes and 2) that AH shares with other molybdopterin oxido-reductases an ancestral evolution from dehydrogenases (Fig. 4). It is further supported by the diverse phylogeny of acetylenotrophs based on SSU rRNA genes and phenotypic characterization (Table 2).
Figure 4.
Neighbor-joining tree showing the relationship between amino acid sequences for known and select putative AH and dehydrogenase proteins. All proteins shown contain both a molybdopterin-binding domain (conserved protein domain family cd00368) and a molybdopterin-binding C-terminal (cd02775). Putative AH containing a molybdopterin-binding acetylene hydratase (cd02759) and a molybdopterin-binding acetylene hydratase C-terminal (cd02781) are indicated with green squares and known AH are indicated by red triangles. The phylogenetic affiliation of the organisms is indicated by color. The tree was constructed by downloading amino acid (aa) sequences that matched a key-word search of 'acetylene hydratase' in the NCBI GenBank database. Additional dehydrogenase sequences were included for reference. Sequences were aligned in Geneious v. 9.1.8 (Kearse et al.2012) using the ClustalW aligner then trimmed to a final length of 980 aa to compare overlapping regions of sequence. The tree was constructed using the Geneious Tree Builder with neighbor-joining methods and Jukes-Cantor distance model.
Many complete bacterial genomes and environmental metagenomes now reside in genetic databases where no such information existed until quite recently, providing a wealth of data that can be interrogated for the presence of new acetylenotrophs. Indeed, merely entering the keywords 'acetylene hydratase' into the National Center for Biotechnology Information (NCBI) database keyword search results in over 2,400 word-annotations for putative AH proteins whereas there were less than five as recently as two years ago (NCBI Resource Coordinators 2016). These annotations span the Bacteria and Archaea kingdoms, including uncultured organisms from metagenomes (Fig. 4). Interestingly, these annotated sequences share fairly low nucleotide and amino acid identities with the sequences of Pelobacter ahy. For example, the ahy of P. acetylenicus and Pelobacter sp. strain SFB93 are 86%–99% identical but the next best alignment in the NCBI protein database is from a Burkholderia species and shares just 46% amino acid similarity. These putative AH genes were identified through computational predictions and it is not clear if these sequences actually encode a functional AH or if this diverse array of microorganisms can be sustained by an acetylenotrophic metabolism. Nonetheless, the fact that there are now so many plausible 'hits' highlights the need for closer investigation as to the gene sequence and structural nature of these ahy genes and AH proteins. If they were found to be reasonable candidates for viable ahy genes and functional proteins, further experimental effort demonstrating a capacity for acetylenotrophy in some of the annotated species having annotated ahy genes would be warranted. Preliminary efforts have begun to investigate whether this growing list of annotated ahy genes in the NCBI database are viable or a just a mis-annotation by the computational pipelines in place to annotate bacterial and archaeal genomes (e.g. RAST and PGAP). The challenge here lies in the low nucleotide identities. If related, these genes may be highly diverged from one another, making their computational identification more difficult than a simple BLAST inquiry (Madden 2002; Johnson et al.2008).
A CRYPTIC ACETYLENE CYCLE?
An intriguing question is why acetylenotrophy has been maintained in multiple microorganisms when acetylene is found at very low concentrations in the environment. It is particularly interesting that Pelobacter strains are maintaining genes for acetylene metabolism that are flanked by HGT elements. We hypothesize that in addition to the known anthropogenic and combustion sources that contribute to low concentrations of acetylene on modern Earth (as described above), there may be additional unidentified sources. This transient acetylene could be so rapidly degraded by proximately located acetylenotrophs in nature that it forms a 'hidden' acetylene cycle. Such a hidden acetylene cycle would then favor the maintenance of heterogeneous enzymes for acetylene metabolisms that are found in the phylogenetically diverse aerobic and anaerobic acetylenotrophs. In addition, this would also provide an explanation for the fact that Pelobacter sp. strain SFB93 has active copies of both AH and N2ase (Akob et al.2017). The presence of both AH and N2ase could provide a competitive advantage for these organisms with each providing the organism carbon and energy and nitrogen source, respectively. In addition, the presence of a hidden acetylene cycle could provide the base of a food web for other organisms, such as methanogens, that utilize end products or intermediates from acetylenotrophy (Fig. 2).
Potential natural sources of acetylene
Absent a strong environmental source of free acetylene to the environment, there are several possible routes for acetylene to be created from intermediate precursor materials that after being metabolized is readily consumed by acetylenotrophs, hereby precluding its release into the open environment. Marine algae produce trace amounts 1,2-dibromoethene (Gribble 1996; Gribble 2003), which can be degraded to acetylene by pure cultures of methanogenic bacteria (Belay and Daniels 1987). While acetylene formation from 1,2-dibromoethene has only been shown in the laboratory and has not been observed in the ocean, we cannot rule out the possibility that this transformation is occurring naturally in marine systems (Singh and Fabian 1999; Simpson et al.2015). An additional potential oceanic source of acetylene-precursors is dimethylsulfoniopropionate (DMSP; Fig. 5). In the ocean, DMSP is released as a product of phytoplankton cell lysis, which is then broken down into more volatile compounds by other bacteria into dimethyl sulfide (DMS), acetate, acetaldehyde, acrylate and ethene (Plettner, Steinke and Malin 2005; Reisch, Moran and Whitman 2011). One possible cryptic source of acetylene is conversion of acrylate to acetylene and CO2 (decomposition) (Fig. 5).
Figure 5.
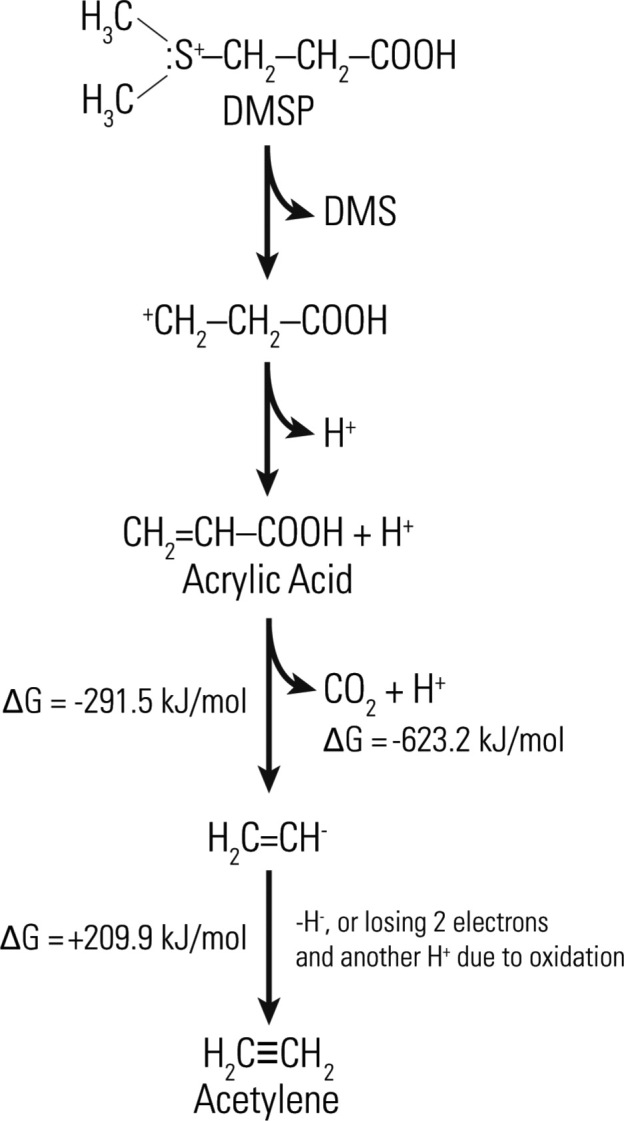
Acrylic acid and DMS are decomposition products of DMSP. Acrylic acid is hypothesized to be microbially oxidized to acetylene thereby potentially fueling a hidden acetylene cycle. The reaction from acrylic acid to acetylene yields a free energy of ΔGreaction = −623.2 + 209.9 - (−291.5) = −121.8 kJ mol−1.
Another possible source of acetylene could be decomposition of naturally occurring higher alkynes. Alkynes are naturally produced by plants, fungi and microbes throughout diverse ecosystems (Shi Shun and Tykwinski 2006; Yosef Friedjung et al.2013; Peñuelas et al.2014). There are over 600 naturally occurring complex alkynes known (Bohlmann, Burkhardt and Zdero 1973). The ubiquity, low volatility and allelopathic utility of these higher alkynes as a source of acetylene would explain the broad occurrence and persistence of acetylenotrophs in nature’s ecosystems. Indeed, it is known that certain species of bacteria can even produce short-chain acetylenic compounds such as propynoic acid (Bohlmann, Burkhardt and Zdero 1973), and numerous enzymes have been identified that form terminal alkyne bonds during alkyne biosynthesis (Edwards et al.2004; Zhu et al.2014; Zhu, Su and Manickam 2015). Support for this hypothesis is seen by the work of de Bont and Peck (1980) where their aerobic acetylenotroph, Rhodococcus sp. A1, was capable of growing on propyne (e.g., methylacetylene, CH3C≡CH). In addition, Pseudomonas spp. strains were isolated that degraded but-2-ynedioic acid (e.g., acetylene dicarboxylic acid, HO2C-C≡C-CO2H) (Yamada and Jakoby 1958) and prop-2-ynoic acid (e.g., acetylene monocarboxylic (propynoic) acid, HC≡C-CO2H) (Yamada and Jakoby 1959), in what is thought to be a hydration reaction. Bacteria and Fungi were also isolated that could degrade prop-2-ynoic acid (de Bont, Scholten A, van and Tweel 1985) and propynol (CH3C≡C-OH) (de Bont, Scholten and van den Tweel 1985; Van Den Tweel and de Bont 1985). It was postulated from these studies that there are at least three hydratases capable of breaking the CC triple bond that are unique from the AHs described above and specific for acetylene (Hartmans, de Bont and Harder 1989). These findings further support our hypothesis that acetylenotrophy is a ubiquitous microbial process on modern Earth that is a cryptic part of the microbial food web.
Anthropogenic sources of acetylene
Conditions of the Anthropocene could be further selecting for acetylene metabolism whereby the ancient AH enzyme was kept at minimal levels but is now responding to anthropogenic inputs of acetylene, e.g., from pollutant degradation (Roberts et al.1996; Arnold and Roberts 2000; Han et al.2012; Schaefer et al.2015), industrial uses as a synthesis feedstock, in oxy-acetylene torches or from the exhaust of motor vehicles (Seinfeld and Pandis 1998). AH genes were detected in metagenomes of fluidized bed reactors bioremediating a tropical diesel oil spill (Rodríguez-Martínez et al.2006) and as discussed above in a TCE-contaminated aquifer (Miller et al.2013). Although acetylene is not known to be present in diesel fuel, it is formed by abiotic dehalogenation reactions of TCE with reduced iron minerals (Roberts et al.1996; Arnold and Roberts 2000; Han et al.2012; Schaefer et al.2015), and as such would provide a substrate for acetylenotrophic organisms. Moreover, the overall goal of TCE-bioremediation efforts is to stimulate complete dechlorination of this and other halogenated alkenes, resulting in the accumulation of ethylene (C2H4) as the desired innocuous end product (Révész et al.2014; Mao et al.2015). Recent laboratory studies have shown that acetylene-fermenting organisms can be used to overcome acetylene-linked inhibition of TCE bioremediation (Mao et al.2017).
CONCLUSIONS
Here, we reviewed what is known about acetylenotrophic microorganisms and proposed the presence of a hidden acetylene cycle that could be supported by unknown sources of acetylene and fueling a microbial food web. Acetylenotrophy has been observed in a variety of habitats sampled from all over the world and there are likely unknown natural sources of acetylene in the environment. With the paradigm in environmental microbiology that ‘we only know what we’ve looked for’, this review highlights the need to look in more depth at acetylene metabolism and to search for yet unknown sources of naturally-derived acetylene. Research may reveal that acetylenotrophs represent an important control of global biogeochemical cycles by rapidly removing inhibitory acetylene and allowing for critical microbial functions, e.g., methane oxidation and nitrogen cycling, to proceed at the rates observed currently.
We further hypothesize that acetylenotrophy represents an ancient mode of energy metabolism that has persisted in the broad microbial genome through geologic time up to the present day. Indeed, acetylenotrophy may have been sustained by evolution from an anaerobic to aerobic metabolism in response to oxidation of Earth and could help explain the variability in the structure of anaerobic and aerobic AHs. With the genomics revolution, we have an abundance of data that can be interrogated to learn more about existing AHs and the diversity of acetylenotrophs. These data can then be used to drive laboratory studies targeting acetylenotrophs and field studies to fully explore the ubiquity and distribution of these organisms. Field research is likely to reveal the importance of acetylene cycling and acetylenotrophs in bioremediation, perhaps improving our ability to cleanse the environment of toxic chlorinated solvents (e.g., TCE) and other halogenated alkenes. Moreover, the possible detection of acetylene in extraterrestrial bodies, e.g., the moons of Saturn (Titan and Enceladus; (Matson et al.2007), suggests the possibility of a suitable exobiological habitat for extraterrestrial microbial acetylenotrophs.
FUNDING
This work was supported by a NASA Research Opportunities in Space and Earth Science (ROSES-2013), Astrobiology: Exobiology and Evolutionary Biology Program Element grant (grant 13-EXO13-0001) to D.M.A., R.S.O., and L.G.M. Funding was also provided by the U.S. Geological Survey (USGS) Toxic Substances Hydrology Program and Water Mission Area. R.S.O. was partially supported by the NASA/USGS 'From Orbit to Core' program. J.L.F. and J.M.S. were supported by University of Alabama start-up funds and NIH NIGMS grant R01 GM102511.
ACKNOWLEDGEMENTS
We thank J. Colin Murrell, Joseph Suflita, Thomas Hanson, Robert Andrews, Yesha Shrestha, Marisa Mihori, Amy Springfield, Sarah Netteman, and Thomas DiChristina for helpful discussions.
Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the US Government.
Conflict of interest. None declared.
REFERENCES
- Abbas O, Schulze-Makuch D. Acetylene-based pathways for prebiotic evolution on Titan. In: Lacoste H. (ed). Proceedings of the First European Workshop on Exo-Astrobiology, ESA SP-518 Graz, Austria: ESA Publications Division, 2002, 345–8. [Google Scholar]
- Akob DM, Baesman SM, Sutton JM et al. Detection of diazotrophy in the acetylene-fermenting anaerobe Pelobacter sp. strain SFB93. Appl Environ Microbiol. 2017;83:e01198–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold WA, Roberts AL. Pathways and kinetics of chlorinated ethylene and chlorinated acetylene reaction with Fe(0) particles. Environ Sci Technol. 2000;34:1794–805. [Google Scholar]
- Atkinson R, Baulch DL, Cox RA et al. Evaluated kinetic and photochemical data for atmospheric chemistry: supplement VI. IUPAC subcommittee on gas kinetic data evaluation for atmospheric chemistry. J Phys Chem Ref Data. 1997;26:1329–499. [Google Scholar]
- Baker AK, Beyersdorf AJ, Doezema LA et al. Measurements of nonmethane hydrocarbons in 28 United States cities. Atmos Environ. 2008;42:170–82. [Google Scholar]
- Balderston WL, Sherr B, Payne WJ. Blockage by acetylene of nitrous oxide reduction in Pseudomonas perfectomarinus. Appl Environ Microbiol. 1976;31:504–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay N, Daniels L. Production of ethane, ethylene, and acetylene from halogenated hydrocarbons by methanogenic bacteria. Appl Environ Microbiol. 1987;53:1604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Hirschfeld L. Die Umsetzung von Acetylen durch Mycobacterium lacticola. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 2. 1932;86:113–30. [Google Scholar]
- Blake NJ, Blake DR, Chen T-Y et al. Distribution and seasonality of selected hydrocarbons and halocarbons over the western Pacific basin during PEM-West A and PEM-West B. J Geophys Res. 1997;102:28315–31. [Google Scholar]
- Bohlmann F, Burkhardt T, Zdero C. Naturally Occurring Acetylenes. Academic Press, 1973,1–547. [Google Scholar]
- Boll M, Einsle O, Ermler U et al. Structure and function of the unusual tungsten enzymes acetylene hydratase and class II benzoyl-coenzyme A reductase. J Mol Microbiol Biotechnol. 2016;26:119–37. [DOI] [PubMed] [Google Scholar]
- Bonsang B, Kanakidou M, Lambert G. Non methane hydrocarbons chemistry in the atmosphere of an equatorial forest: a case of indirect photochemical production of OH radicals. Geophys Res Lett. 1987;14:1250–3. [Google Scholar]
- Brzeziska M, Rafalski P, Wodarczyk T et al. How much oxygen is needed for acetylene to be consumed in soil?. J Soils Sed. 2011;11:1142. [Google Scholar]
- Campbell TJ, Burris DR, Roberts AL et al. Trichloroethylene and tetrachloroethylene reduction in a metallic iron–water-vapor batch system. Environ Toxicol Chem. 1997;16:625–30. [Google Scholar]
- Caspi R, Altman T, Dale JM et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicerone RJ, Oremland RS. Biogeochemical aspects of atmospheric methane. Global Biogeochem Cycles. 1988;2:299–327. [Google Scholar]
- Clark RN, Curchin JM, Barnes JW et al. Detection and mapping of hydrocarbon deposits on Titan. J Geophys Res. 2010;115:1–28. [Google Scholar]
- Cronn D, Robinson E. Tropospheric and lower stratospheric vertical profiles of ethane and acetylene. Geophys Res Lett. 1979;6:641–4. [Google Scholar]
- Culbertson CW, Strohmaier FE, Oremland RS. Acetylene as a substrate in the development of primordial bacterial communities. Orig Life Evol Biosphere. 1988;18:397–407. [DOI] [PubMed] [Google Scholar]
- Culbertson CW, Zehnder AJB, Oremland RS. Anaerobic oxidation of acetylene by estuarine sediments and enrichment cultures. Appl Environ Microbiol. 1981;41:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton H, Whittenbury R. The acetylene reduction technique as an assay for nitrogenase activity in the methane oxidizing bacterium Methylococcus capsulatus strain bath. Arch Microbiol. 1976;109:147–51. [Google Scholar]
- de Bont JAM, Peck MW. Metabolism of acetylene by Rhodococcus A1. Arch Microbiol. 1980;127:99–104. [Google Scholar]
- de Bont JAM, Primrose SB, Collins MD et al. Chemical studies on some bacteria which utilize gaseous unsaturated hydrocarbons. J Gen Microbiol. 1980;117:97–102. [Google Scholar]
- de Bont JAM, Scholten A, van den Tweel WJJ. Isolation of microorganisms on 3-butyn-1-ol and other acetylenic compounds. Curr Microbiol. 1985;12:267–71. [Google Scholar]
- Edwards DJ, Marquez BL, Nogle LM et al. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem Biol. 2004;11:817–33. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Fujii T, Ogawa T. Microbial production of C2-hydrocarbons, ethane, ethylene and acetylene. Agric Biol Chem. 1984;48:1363–5. [Google Scholar]
- Germon JC, Knowles R. Metabolism of acetylene and acetaldehyde by Rhodococcus rhodochrous. Can J Microbiol. 1988;34:242–8. [DOI] [PubMed] [Google Scholar]
- Goldman A, Murcray FJ, Blatherwick RD et al. Identification of acetylene (C2H2) in infrared atmospheric absorbtion spectra. J Geophys Res. 1981;86:12143–6. [Google Scholar]
- Gribble GW. Naturally occuring organohalogen compounds—a comprehensive survery. In: Gribble GW, Herz W, Kirby GW et al. (eds). Progress in the Chemistry of Organic Natural Products, Vienna: Springer, 1996, 1–423. [PubMed] [Google Scholar]
- Gribble GW. The diversity of naturally produced organohalogens. Chemosphere. 2003;52:289–97. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Cicerone RJ, Blake DR et al. Global atmospheric distributions and source strengths of light hydrocarbons and tetrachloroethene. J Geophys Res. 1998;103:28219–35. [Google Scholar]
- Han YS, Hyun SP, Jeong HY et al. Kinetic study of cis-dichloroethylene (cis-DCE) and vinyl chloride (VC) dechlorination using green rusts formed under varying conditions. Water Res. 2012;46:6339–50. [DOI] [PubMed] [Google Scholar]
- Hartmans S, de Bont JAM, Harder W. Microbial metabolism of short-chain unsaturated hydrocarbons. FEMS Microbiol Lett. 1989;63:235–64. [DOI] [PubMed] [Google Scholar]
- He YT, Wilson JT, Su C et al. Review of abiotic degradation of chlorinated solvents by reactive iron minerals in aquifers. Groundwater Monitor R. 2015;35:57–75. [Google Scholar]
- Heard DE, Read KA, Methven J et al. The North Atlantic Marine Boundary Layer Experiment (NAMBLEX). Overview of the campaign held at Mace Head, Ireland, in summer 2002. Atmos Chem Phys. 2006;6:2241–72. [Google Scholar]
- Hopkins JR, Jones ID, Lewis AC et al. Non-methane hydrocarbons in the Arctic boundary layer. Atmos Environ. 2002;36:3217–29. [Google Scholar]
- Hudson ED, Ariya PA. Measurements of non-methane hydrocarbons, DOC in surface ocean waters and aerosols over the Nordic seas during polarstern cruise ARK-XX/1 (2004). Chemosphere. 2007;69:1474–84. [DOI] [PubMed] [Google Scholar]
- Jensen MM, Thamdrup B, Dalsgaard T. Effects of specific inhibitors on anammox and denitrification in marine sediments. Appl Environ Microbiol. 2007;73:3151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson BT, Wu Z, Niki H et al. Seasonal trends of isoprene, C2–C5alkanes, and acetylene at a remote boreal site in Canada. J Geophys Res. 1994;99:1589. [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakidou M, Bonsang B, Le Roulley JC et al. Marine source of atmospheric acetylene. Nature. 1988;333:51–2. [Google Scholar]
- Kanner D, Bartha R. Growth of Nocardia rhodochrous on acetylene gas. J Bacteriol. 1979;139:225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanner D, Bartha R. Metabolism of acetylene by Nocardia rhodochrous. J Bacteriol. 1982;150:989–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasting J. Earth’s early atmosphere. Science. 1993;259:920–6. [DOI] [PubMed] [Google Scholar]
- Kasting JF. When methane made climate. Sci Am. 2004;291:78–85. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R. Denitrification. Microbiol Rev. 1982;46:43–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopparla P, Gao P, Shermansky D et al. Organic gas abundances in the plumes of Enceladus as seen by Cassini UVIS. American Geophysical Union, Fall General Assembly 2016. abstract #P33A-2123, 2016. [Google Scholar]
- Kroneck PM. Acetylene hydratase: a non-redox enzyme with tungsten and iron-sulfur centers at the active site. J Biol Inorg Chem. 2016;21:29–38. [DOI] [PubMed] [Google Scholar]
- Lewis AC, Carpenter LJ, Pilling MJ. Nonmethane hydrocarbons in Southern Ocean boundary layer air. J Geophys Res. 2001;106:4987–94. [Google Scholar]
- Madden T. The BLAST sequence analysis tool. In: McEntyre J, Ostell J (eds). The NCBI Handbook. Bethesda, Maryland, USA: National Center for Biotechnology Information, 2002. [Google Scholar]
- Mao X, Oremland RS, Liu T et al. Acetylene fuels TCE reductive dechlorination by defined Dehalococcoides/Pelobacter consortia. Environ Sci Technol. 2017;51:2366–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Stenuit B, Polasko A et al. Efficient metabolic exchange and electron transfer within a syntrophic trichloroethene-degrading coculture of Dehalococcoides mccartyi 195 and Syntrophomonas wolfei. Appl Environ Microbiol. 2015;81:2015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson DL, Castillo JC, Lunine J et al. Enceladus’ plume: compositional evidence for a hot interior. Icarus. 2007;187:569–73. [Google Scholar]
- McKay CP, Anbar AD, Porco C et al. Follow the plume: the habitability of Enceladus. Astrobiology. 2014;14:352–5. [DOI] [PubMed] [Google Scholar]
- McKay CP, Porco CC, Altheide T et al. The possible origin and persistence of life on Enceladus and detection of biomarkers in the plume. Astrobiology. 2008;8:909–19. [DOI] [PubMed] [Google Scholar]
- McKay CP, Smith HD. Possibilities for methanogenic life in liquid methane on the surface of Titan. Icarus. 2005;178:274–6. [Google Scholar]
- Miller LG, Baesman SM, Kirshtein J et al. A biogeochemical and genetic survey of acetylene fermentation by environmental samples and bacterial isolates. Geomicrobiol J. 2013;30:501–16. [Google Scholar]
- Miller LG, Baesman SM, Oremland RS. Stable carbon isotope fractionation during bacterial acetylene fermentation: potential for life detection in hydrocarbon-rich volatiles of icy planet(oid)s. Astrobiology. 2015;15:977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL. A production of amino acids under possible primitive Earth conditions. Science. 1953;117:528–9. [DOI] [PubMed] [Google Scholar]
- Miller SL, Urey HC. Organic compound synthesis on the primitive earth. Science. 1959;130:245–51. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Capone DG. Use of specific inhibitors in biogeochemistry and microbial ecology. Adv Microb Ecol. 1988;10:285–383. [Google Scholar]
- Oremland RS, Taylor BF. Inhibition of methanogenesis in marine sediments by acetylene and ethylene: validity of the acetylene reduction assay for anaerobic microcosms. Appl Environ Microbiol. 1975;30:707–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne WJ. Influence of acetylene on microbial and enzymatic assays. J Microbiol Methods. 1984;2:117–33. [Google Scholar]
- Payne WJ, Grant MA. Influence of acetylene on growth of sulfate-respiring bacteria. Appl Environ Microbiol. 1982;43:727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñuelas J, Asensio D, Tholl D et al. Biogenic volatile emissions from the soil. Plant, Cell Environ. 2014;37:1866–91. [DOI] [PubMed] [Google Scholar]
- Plass C, Koppmann R, Rudolph J. Light hydrocarbons in the surface water of the mid-Atlantic. J Atmos Chem. 1992;15:235–51. [Google Scholar]
- Plass-Dülmer C, Koppmann R, Ratte M et al. Light nonmethane hydrocarbons in seawater. Global Biogeochem Cycles. 1995;9:79–100. [Google Scholar]
- Plettner INA, Steinke M, Malin G. Ethene (ethylene) production in the marine macroalga Ulva (Enteromorpha) intestinalis L. (Chlorophyta, Ulvophyceae): effect of light-stress and co-production with dimethyl sulphide. Plant, Cell Environ. 2005;28:1136–45. [Google Scholar]
- Pon G, Hyman MR, Semprini L. Acetylene inhibition of trichloroethene and vinyl chloride reductive dechlorination. Environ Sci Technol. 2003;37:3181–8. [DOI] [PubMed] [Google Scholar]
- Porco CC, Dones L, Mitchell C. Could it be snowing microbes on Enceladus? Assessing conditions in its plume and implications for future missions. Astrobiology. 2017;17:876–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls RW, Blowes DW, Gillham RW. Long-term performance monitoring for a permeable reactive barrier at the U.S. Coast Guard Support Center, Elizabeth City, North Carolina. J Hazard Mater. 1999;68:109–24. [DOI] [PubMed] [Google Scholar]
- Reisch CR, Moran MA, Whitman WB. Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front Microbiol. 2011;2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Révész KM, Lollar BS, Kirshtein JD et al. Integration of stable carbon isotope, microbial community, dissolved hydrogen gas, and 2HH2O tracer data to assess bioaugmentation for chlorinated ethene degradation in fractured rocks. J Contam Hydrol. 2014;156:62–77. [DOI] [PubMed] [Google Scholar]
- Roberts AL, Totten LA, Arnold WA et al. Reductive elimination of chlorinated ethylenes by zero-valent metals. Environ Sci Technol. 1996;30:2654–9. [Google Scholar]
- Rodríguez-Martínez EM, Pérez EX, Schadt CW et al. Microbial diversity and bioremediation of a hydrocarbon-contaminated aquifer (Vega Baja, Puerto Rico). Int J Env Res Public Health. 2006;3:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner BM, Rainey FA, Kroppenstedt RM et al. Acetylene degradation by new isolates of aerobic bacteria and comparison of acetylene hydratase enzymes. FEMS Microbiol Lett. 1997;148:175–80. [DOI] [PubMed] [Google Scholar]
- Rosner BM, Schink B. Purification and characterization of acetylene hydratase of Pelobacter acetylenicus, a tungsten iron-sulfur protein. J Bacteriol. 1995;177:5767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Ehhalt DH, Khedim A. Vertical profiles of acetylene in the troposphere and stratosphere. J Atmos Chem. 1984;2:117–24. [Google Scholar]
- Sander R. Compilation of Henry’s law constants (version 4.0) for water as solvent. Atmos Chem Phys. 2015;15:4399–981. [Google Scholar]
- Schaefer CE, Towne RM, Lippincott DR et al. Abiotic dechlorination in rock matrices impacted by long-term exposure to TCE. Chemosphere. 2015;119:744–9. [DOI] [PubMed] [Google Scholar]
- Scheidler C, Sobotta J, Eisenreich W et al. Unsaturated C3,5,7,9-monocarboxylic acids by aqueous, one-pot carbon fixation: possible relevance for the origin of life. Sci Rep. 2016;6:27595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B. Fermentation of acetylene by an obligate anaerobe, Pelobacter acetylenicus sp. nov. Arch Microbiol. 1985;142:295–301. [Google Scholar]
- Seiffert GB, Ullmann GM, Messerschmidt A et al. Structure of the non-redox-active tungsten/[4Fe:4S] enzyme acetylene hydratase. Proc Natl Acad Sci USA. 2007;104:3073–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinfeld JH, Pandis SN. Atmospheric composition, global cycles, and lifetimes. Atmos Chem Phys, 1st edn New York: John Wiley and Sons, Inc, 1998,49–124. [Google Scholar]
- Seitz HJ, Siñeriz F, Schink B et al. Hydrogen production during fermentation of acetoin and acetylene by Pelobacter acetylenicus. FEMS Microbiol Lett. 1990;71:83–7. [Google Scholar]
- Shemansky DE, Stewart AIF, West RA et al. The Cassini UVIS stellar probe of the Titan atmosphere. Science. 2005;308:978–82. [DOI] [PubMed] [Google Scholar]
- Shi Shun ALK, Tykwinski RR. Synthesis of naturally occurring polyynes. Angewandte Chemie International Edition. 2006;45:1034–57. [DOI] [PubMed] [Google Scholar]
- Shim C, Wang Y, Singh HB et al. Source characteristics of oxygenated volatile organic compounds and hydrogen cyanide. J Geophys Res. 2007;112:D10305. [Google Scholar]
- Simpson WR, Brown SS, Saiz-Lopez A et al. Tropospheric halogen chemistry: sources, cycling, and impacts. Chem Rev. 2015;115:4035–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ON, Fabian P. Reactive bromine compounds. In: The Handbook of Environmental Chemistry, Springer-Verlag, 1999;1–43. [Google Scholar]
- Span I, Wang K, Wang W et al. Discovery of acetylene hydratase activity of the iron-sulphur protein IspH. Nat Commun. 2012;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart WD, Fitzgerald GP, Burris RH. In situ studies on N2 fixation using the acetylene reduction technique. Proc Natl Acad Sci USA. 1967;58:2071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel DF. Molecular hydrogen in Titan’s atmosphere: implications of the measured tropospheric and thermospheric mole fractions. Icarus. 2010;208:878–86. [Google Scholar]
- Sutton JM, Baesman SM, Fierst JL et al. Complete genome sequence of the acetylene fermenting Pelobacter strain SFB93. Genome Announc. 2017a;5:e01573–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton JM, Baesman SM, Fierst JL et al. Complete genome sequences of two acetylene fermenting Pelobacter acetylenicus strains. Genome Announc. 2017b;5:e01572–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot RW, Dibb JE, Lefer BL et al. Chemical characteristics of continental outflow from Asia to the troposphere over the western Pacific Ocean during February-March 1994: results from PEM-West B. J Geophys Res. 1997;102:28255–74. [Google Scholar]
- Tam T-Y, Mayfield CI, Inniss WE. Aerobic acetylene utilization by stream sediment and isolated bacteria. Curr Microbiol. 1983;8:165–8. [Google Scholar]
- ten Brink F. Living on acetylene. A primordial energy source. In: The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. vol .14 Dordrecht, Netherlands: Springer, 2014;15–35. [DOI] [PubMed] [Google Scholar]
- ten Brink F, Schink B, Kroneck PMH. Exploring the active site of the tungsten, iron-sulfur enzyme acetylene hydratase. J Bacteriol. 2010;193:1229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry RE, Duxbury JM. Acetylene decomposition in soils. Soil Sci Soc Am J. 1986;49:90–4. [Google Scholar]
- Topp E, Germon J-C. Acetylene metabolism and stimulation of denitrification in an agricultural soil. Appl Environ Microbiol. 1986;52:802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainer MG, Pavlov AA, Curtis DB et al. Haze aerosols in the atmosphere of early Earth: manna from heaven. Astrobiology. 2004;4:409–19. [DOI] [PubMed] [Google Scholar]
- Van Den Tweel WJJ, de Bont JAM. Metabolism of 3-Butyn-1-ol by Pseudomonas BB1. J Gen Microbiol. 1985;131:3155–62. [Google Scholar]
- Waite JH, Glein CR, Perryman RS et al. Cassini finds molecular hydrogen in the Enceladus plume: evidence for hydrothermal processes. Science. 2017;356:155–9. [DOI] [PubMed] [Google Scholar]
- Waite JH Jr., Combi MR, Ip WH et al. Cassini ion and neutral mass spectrometer: enceladus plume composition and structure. Science. 2006;311:1419–22. [DOI] [PubMed] [Google Scholar]
- Watanabe I, De Guzman MR. Effect of nitrate on acetylene disappearance from anaerobic soil. Soil Biol Biochem. 1980;12:193–4. [Google Scholar]
- Whitby RA, Altwicker ER. Acetylene in the atmosphere: sources, representative ambient concentrations and ratios to other hydrocarbons. Atmos Environ. 1978;12:1289–96. [Google Scholar]
- Woods NR. The bacterial metabolism of propane. Ph.D. Thesis United Kingdom: University of Warwick, 1988. [Google Scholar]
- Woods NR, Murrell JC. The metabolism of propane in Rhodococcus rhodochrous PNKb1. Microbiology. 1989;135:2335–44. [Google Scholar]
- Xiao Y, Jacob DJ, Turquety S. Atmospheric acetylene and its relationship with CO as an indicator of air mass age. J Geophys Res. 2007;112::D12305 [Google Scholar]
- Yamada E, Jakoby W. Enzymatic utilization of acetylenic compounds II. Acetylenemonocarboxylic acid hydrase. J Biol Chem. 1959;234:941–5. [PubMed] [Google Scholar]
- Yamada EW, Jakoby WB. Enzymatic utilization of acetylenic compounds I. An enzyme converting acetylenedicarboxylic acid to pyruvate. J Biol Chem. 1958;233:706–11. [PubMed] [Google Scholar]
- Yeomans JC, Beauchamp EG. Acetylene as a possible substrate in the denitrification process. Can J Soil Sci. 1982;62:139–44. [Google Scholar]
- Yosef Friedjung A, Choudhary SP, Dudai N et al. Physiological conjunction of allelochemicals and desert plants. PLoS One. 2013;8:e81580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari T, Hynes R, Knowles R. Acetylene inhibition of nitrous oxide reduction and measurement of denitrification and nitrogen fixation in soil. Soil Biol Biochem. 1977;9:177–83. [Google Scholar]
- Zahnle KJ. Photochemistry of methane and the formation of hydrocyanic acid (HCN) in the Earth’s early atmosphere. J Geophys Res. 1986;91:2819–34. [Google Scholar]
- Zhu X, Liu J, Zhang W. De novo biosynthesis of terminal alkyne-labeled natural products. Nat Chem Biol. 2014;11:115–20. [DOI] [PubMed] [Google Scholar]
- Zhu X, Su M, Manickam K et al. Bacterial genome mining of enzymatic tools for alkyne biosynthesis. ACS Chem Biol. 2015;10:2785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]