Abstract
Sec-dependent protein translocation is an essential process in bacteria. SecA is a key component of the translocation machinery and has multiple domains that interact with various ligands. SecA acts as an ATPase motor to drive the precursor protein/peptide through the SecYEG protein translocation channels. As SecA is unique to bacteria and there is no mammalian counterpart, it is an ideal target for the development of new antimicrobials. Several reviews detail the assays for ATPase and protein translocation, as well as the search for SecA inhibitors. Recent studies have shown that, in addition to the SecA-SecYEG translocation channels, there are SecA-only channels in the lipid bilayers, which function independently from the SecYEG machinery. This mini-review focuses on recent advances on the newly developed SecA inhibitors that allow the evaluation of their potential as antimicrobial agents, as well as a fundamental understanding of mechanisms of SecA function(s). These SecA inhibitors abrogate the effects of efflux pumps in both Gram-positive and Gram-negative bacteria. We also discuss recent findings that SecA binds to ribosomes and nascent peptides, which suggest other roles of SecA. A model for the multiple roles of SecA is presented.
Keywords: SecA inhibitors, SecA-only channels, SecA-SecYEG channels, efflux pumps, Rose Bengal-thiouracil-triazole-pyrimidine analogs, SecA-ribosomes
SecA inhibitors as potential antimicrobial agents
INTRODUCTION
The role of SecYEG as a protein-conducting channel in Sec-dependent protein translocation in bacteria is well established by both genetics and biochemical studies (Davis and Tai 1980; Danese and Silhavy 1998; Mori and Ito 2001; Papanikolau et al.2007; Driessen and Nouwen 2008). Details of the mechanism of function of the SecA-SecYEG-SecDF•YajC complex and the essential roles of SecA have also been reviewed extensively (Kusters and Driessen 2011; Lycklama and Driessen 2012; Bauer et al.2014; Chatzi et al.2014). Although the SecYEG translocation pathway is homologous to the Sec61 pathway in mammals (Rapoport 2007; Park and Rapoport 2012), SecA is unique to bacteria (and chloroplasts in plants, Hu et al.2007) and is essential for their cell growth; it is present in E. coli in both soluble and membrane-bound forms (Oliver and Beckwith 1981). X-ray structures of soluble SecA and SecA-SecYEG have been resolved (Hunt et al.2002; Papanikou, Karamanou and Economou 2007; Zimmer, Nam and Rapoport 2008). Soluble SecA is an ATPase (intrinsic ATPase) that is regulated by its C-terminal domain, an activity that is increased by interaction with anionic lipids (lipid/membrane-ATPase), and further increased by precursors and membrane embedded with SecYEG (translocation ATPase) (Lill, Dowhan and Wickner 1990). The findings that (1) precursors of periplasmic alkaline phosphatase (proPhoA) and outer membrane OmpA (proOmA) can be translocated post-translationally into membrane vesicles (Chen, Rhoads and Tai 1985), (2) ATP hydrolysis is essential for protein translocation (Chen and Tai 1985), and (3) SecA is required for protein translocation into membrane vesicles (Cabelli et al.1988), paved the way for the complete reconstitution of anionic liposomes with purified SecA and SecYEG defining functional protein translocation complexes (Kusters and Driessen 2011; Lycklama and Driessen 2012; Bauer et al.2014; Chatzi et al.2014). Proton motive force, SecD-SecF-YajC, SecB and other chaperones, while not essential, contribute to the efficiency of the protein translocation process in vitro and in the cells It should be noted that the reconstituted proteo-liposome system has not addressed another important component, signal peptidase which cleaves the signal peptide from precursors.
SecA-DEPENDENT PROTEIN-CONDUCTING CHANNELS WITH OR WITHOUT SecYEG
E. coli SecA in solution is a dimer of 901 residues that binds to diverse partner ligands (Schmidt et al.1988; Woodbury, Hardy and Randall 2002; Cooper et al.2008). Most considerations of SecA-SecYEG translocation machinery view SecA as a peripheral motor that uses ATP hydrolysis to drive precursors through SecYEG translocons (Karamanou et al.1999). SecA undergoes ATP-driven cycles of membrane insertion and de-insertion (Economou and Wickner 1994). However, SecA can integrate into membranes by itself. Thus, many SecA domains are exposed to the periplasmic side of membranes (Ramamurthy and Oliver 1997; Eichler and Wickner 1998; Jilaveanu and Oliver 2007). A significant fraction of SecA is permanently associated with the cytoplasmic membrane; it does not cycle on and off (Chen, Xu and Tai 1996). There are two asymmetric forms of SecA in the membrane (Tang et al.2010) (Fig. 1b): SecAS and SecAM as revealed by the observations upon trypsin treatments, yielding 68 kD and 34 kD fragments (similar to treated soluble SecA) of the former, and 39 kD and 48 kD that are specific for membrane-bound SecA for the latter (Chen, Brown and Tai 1998). All four fragments are integrated into membranes as shown by their resistance to chemical treatments. SecA is present as a symmetric dimer in solution (Chen et al.2008; Fig. 1a), and forms asymmetric SecAS and SecAM upon interaction with anionic phospholipids (You et al.2013; Fig. 1b). Computing analyses predict that SecA can embed into membranes (Hu et al.2007) and can bind to phospholipids at multiple sites (Keller 2011). The structural role of SecA has been shown by drastic conformational changes and formation of “ring-pore” structures by soluble SecA when it interacts with anionic phospholipids, as revealed by both Transmission-Electronic-Microscopic and Atomic-Force-Microscopic observations (Wang et al.2003). These ring-pore structures have diameters approximating 80–90Å and pore diameters about 20–26 Å; they are active upon interaction with chaperone SecB (Chen, Tai and Sui 2007). Such ring-pore structures provide the morphological support for the observations that in the presence of ATP, SecA-liposomes alone form active ion channels and promote translocation of proOmpA (Hsieh et al.2011; Fig. 1c). It has been suggested that, in addition to the SecA-SecYEG protein-conducting channel (Fig. 1d), there is a second SecA-only protein-conducting channel (You et al.2013) (Fig. 1c). It should be noted that Sec-YEG is not active without SecA, but SecA-only channels can function without SecYEG (Hsieh et al.2011). The two channels have different activities: the one (the SecA-SecYEG channel) is a high-affinity protein-conducting channel with considerable specificity and high efficiency, while the other (the SecA-only channel) is a primitive low-affinity channel with less specificity. In addition to SecA-dependent pathways (Chatzi et al.2014), there are other high- and low-affinity transport systems in bacteria (Anderson and Oxender 1978).
Figure 1.
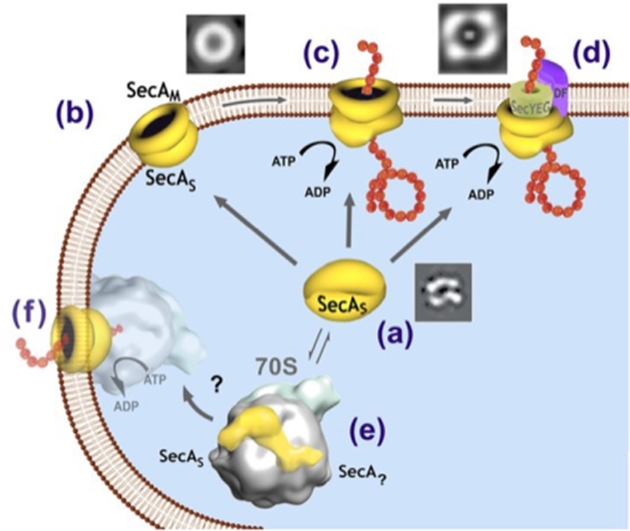
Proposed multiple roles of SecA; (a) Soluble, symmetric dimer SecA (cryo-EM image; Chen et al.2008); (b) Membrane-associated, asymmetric ring-pore structures (TEM image of SecA on the lipids, Wang et al.2003); (c) Membrane-associated, SecA-only protein conducting channels, which are active for proteins with or without signal peptides (albeit inefficiently) in ion channel activity and protein translocation (Hsieh et al.2011; You et al.2013), (d) More efficient SecA-SecYEG-DFC channels require precursors with signal peptides, and can be converted from SecA-only channels (TEM image of SecA-SecYEG, Tang et al.2011); (e) SecA binds to ribosomes (cryo-EM-based SecA-ribosome structures from Singh et al.2014), presumably leading to (f) SecA-mediated co-translational translocation with the exiting nascent peptide providing the sole source for stable association of ribosome and membrane? (Smith et al., 1977, 1978; Herskovits and Bibi 2000; Halbedel et al.2014; Rawat et al.2015; Wang, Yang and Shan 2017)
The possible existence of a protein-conducting channel lacking SecYEG was from the original observations that certain precursors were efficiently translocated into reconstituted membrane vesicles deprived of SecY/PrlA, but still requiring SecA (Watanabe, Nicchitta and Blobel 1990; Watanabe and Blobel 1993). The membrane vesicles from cells depleted in SecE and SecY were also able to translocate certain precursor proteins, including proOmpA and proLpp (Yang, Lian and Tai 1997a; Yang, Yu and Tai 1997b), but not proPhoA. Moreover, Baars et al. (2008) showed that bacterial cells depleted in SecE and SecY could translocate and assemble OmpA, but not all membrane proteins. These observations on the differential translocation of substrates through protein-conducting channels with or without SecYEG reveal fundamental differences between the two SecA-dependent translocation pathways. In most post-translational translocation studies the yardstick has been translocation of precursors inside membrane vesicles or proteo-liposomes as the translocated proteins become resistant to protease digestion. Blobel's group showed that E. coli membrane vesicles possess large, aqueous ion channels, whose activity is opened by signal peptides (Simon, Blobel and Zimmerberg 1989; Simon and Blobel 1992). To provide additional and alternative assays for protein-conducting channel activity, a semi-physiological assay of ion-channel activity was developed by injecting E. coli membranes into frog oocytes, in which SecA and functional precursors (or signal peptides) are also required (Lin et al.2006). Reconstituted membrane vesicles deprived of SecYEG actively elicit both anion and cation channel activity, but have no signal peptide specificity (Lin et al.2012). Both resistance to protease in liposomes and ion channel activity in oocytes are important assays that have established SecA-only channels as being functional protein translocators (Hsieh et al.2011). As expected, chaperone SecB also enhances translocation activity (Hsieh et al.2011). Even though ion channel activity relates only to channel opening, not protein translocation, measuring channel activity in semi-physiological oocytes has great advantages, as discussed (Lin et al.2006; Hsieh et al.2015).
In comparison with SecA-SecYEG (Fig. 1c,d), SecA-only channels (1) are smaller (Tang et al.2011), (2) less efficient (about 35%), (3) require more ATP-Mg2+, (4) require SecA at an optimal concentration of 1–2 μM (Hsieh et al.2011), 10 times higher than SecYEG-containing membranes, though still in the physiological range (Mizushima, Tokuda and Matsuyama 1992; Moran, Phillips and Milo 2010); (5) lose signal-peptide specificity (like Prl mutants, Derman et al.1993) and can translocate unfolded signal-peptide-less proteins (Hsieh et al.2011); (6) lose ion-size selectivity (Lin et al.2012 and unpublished data). The addition of SecYEG-SecDFC to lower-efficiency, SecA-only-liposome changes the shape and increases size of the pore structure (Tang et al.2011) (Fig. 1d), and restores all activities (Hsieh et al.2011; Hsieh et al.2013). Unsurprisingly, although SecA-only channels can translocate OmpA, proOmpA or unfolded PhoA, proPhoA is translocated only by SecA-SecYEG channels (Zhang et al.2013), consistent with observations that SecYEG-deficient membrane vesicles translocate proOmpA, but not proPhoA (Yang, Lian and Tai 1997a; Yang, Yu and Tai 1997b).
Although the molecular characterizations of the SecYEG protein-conducting channel have been quite extensive, little is known about the SecA-only channel. A model for SecA-only channel has been proposed (You et al.2013, Fig. 1c). In this model, SecA functions as an asymmetric dimer in membranes SecAM and SecAS; (Chen, Brown and Tai 1998; Tang et al.2011; You et al.2013), not a monomer, as has been proposed in the SecA-SecYEG channel (Or et al.2005). SecAM. is permanently embedded into the membrane (Chen, Xu and Tai 1996), providing the structural integrity of the channel and a similar protein translocating functions as the SecYEG channel, but lacks signal-peptide specificity-as in Prl suppressors (Derman et al.1993; Huie and Silhavy 1995; Duong and Wickner 1999). Perhaps, SecAM is a primitive counterpart to the SecYEG protein channel, acting as a protein-conducting core with less efficiency and specificity. SecAS binds precursors and acts as the ATPase motor to drive the proteins through the SecAM channel (Fig. 1c). In summary, SecA is an ATPase, an integral membrane protein capable of forming ring-pore structures, and a protein-conducting channel capable of eliciting ion channel activity and promoting protein translocation. A recent study established these capacities by dissecting various domains for their intrinsic and lipid-stimulated ATPase, ring-like pore structure, ion channel activity, protein translocation activity and interactions with SecYEG-SecDF-YajC (Hsieh et al.2017a). The existence of SecA-only channels partially explains why there are far more SecA proteins than SecYEG complexes in bacteria (Mizushima, Tokuda and Matsuyama 1992; Seoh and Tai 1997).
Development of simple assays for SecA-only channel activity makes it possible to measure the in vitro SecA activity of assorted bacterial species, which otherwise would be difficult to do with non-cogent SecYEG protein translocation assays (Zhang et al.2013). Simplifying SecA-only channel assays also provides opportunities to evaluate for these inhibitors that target SecA and its physiological functions.
INHIBITORS TARGETING SecA
The need for new antibiotics/antimicrobials is widely recognized with the ever-increasing abundance of drug-resistant strains, including methicillin-resistant Staphylococcus aureus (MRSA) and multiple drug resistance (MDR), as reviewed extensively (Levy and Marshall 2004; Nikaido 2009; Rao et al.2014; Chaudhary et al.2015a). Most virulence factors and toxins from pathogenic bacteria are secreted through the Sec-transport system; consequently, inhibitors for their components would be logical antimicrobial candidates (Lanzetta et al.1979; Stephens and Shapiro 1997; Rao et al.2014). However, the inhibitors need to be specific and unique to bacteria to be useful for therapeutic application. Thus, inhibitors/antibiotics that affect the membrane, general proteases (Chen and Tai 1987a, Chen and Tai 1987a, 1989), and proteins with homologs to mammalian translocation systems are not of any practical application. For example, decatransin inhibits both SecYEG translocation and Sec61 translocation (Junne et al.2015). An extensive review of antibiotics/inhibitors targeting bacterial secretory pathway have been presented, including signal peptidase, and other secretory pathways (Rao et al.2014). In recent years SecA has been widely recognized as a druggable target. Several excellent reviews have extensively delineated the needs, assays, chemical synthesis, properties and potency of specific SecA inhibitors (Chen et al.2010; Segers and Anne 2011; Segers et al.2011; Rao et al.2014; Chaudhary et al.2015a). This part of the mini-review on antibiotics/inhibitors will focus on more recent developments, structural classes of potent and selective small-molecule inhibitors, and the perspectives of the inhibitors as antimicrobials.
Different assays have been used for screening SecA inhibitors in the above reviews. To summarize, they are evaluated based on: (1) ATPase activity of various forms of intrinsic, unregulated or mutated SecA (Lanzetta et al.1979; Lill, Dowhan and Wickner 1990; Mitchell and Oliver 1993; Gouridis et al.2010); (2) ion channel activity of SecA-liposomes or membranes in the semi-physiological oocytes -with or without SecYEG (Lin et al.2006; Hsieh et al.2011; Lin et al.2012); (3) protein translocation into membranes vesicles (Huang et al.2012; De Waelheyns et al.2015) and SecA-liposomes with or without SecYEG (Hsieh et al.2011) and (4) efficacy in inhibiting bacterial growth combined with inhibition of the secretion of virulence factors and toxins (Jin et al.2015). The first three assays determine the IC50 (50% inhibition of activity) and differentiate activities of the SecA-only and SecA-SecYEG channels, while the inhibition of bacterial growth is expressed as MIC (minimal inhibitory concentration). It is important to note that the roles of SecA and ATP hydrolysis in bacterial growth are not well understood. For example, a truncation of SecA by deletion of short N-terminal or C-terminal reduces both translocation ATPase and even protein translocation, but does not significantly affect the growth of E. coli (Floyd et al.2014; Na et al.2015). The rationales for these assays and summaries of structures of various SecA inhibitors have been described (Segers and Anne 2011; Rao et al.2014; Chaudhary et al.2015a). This review will briefly summarize the relevant literature, focusing more on biological effects.
Azide was the first known inhibitor of SecA, using an azide-resistant mutant of E. coli derived from a specific mutation in SecA (Oliver et al.1990). Indeed, azide (at mM concentrations) does inhibit SecA translocation ATPase, protein translocation and the SecA-dependent ion current activity (Lin et al.2006). Subsequently, however, azide resistant mutants were also found in SecY (PrlA mutants) as well as SecE (PrlG mutants) (Huie and Silhavy 1995; Li et al.2007; Maillard et al.2007). Consequently, azide appears to be more of a SecA-SecYEG translocation inhibitor and a more generic ATPase inhibitor than a specific inhibitor of SecA function (Bowler et al.2006). There are several other inhibitors reported with various activity and MIC (Alksne et al.2000; Parish et al.2009; Jang et al.2011; Segers et al.2011; De Waelheyns et al.2015). One natural fungal product CJ-21 056 has been found to inhibit translocation at 38.4 μM, with an MIC of 12 μM for MRSA and Enterococcus faecalis (Sugie et al.2002). Using an ATPase assay to screen chemical libraries, the best indole derivative has an effective IC50 of 0.25 μM; however, the reported MIC for Agrobacterium tumefaciens is at 0.76 mM (Akula et al.2011). The large discrepancy may be due to its permeability into bacterial membranes, which may be circumvented to test its efficacy.
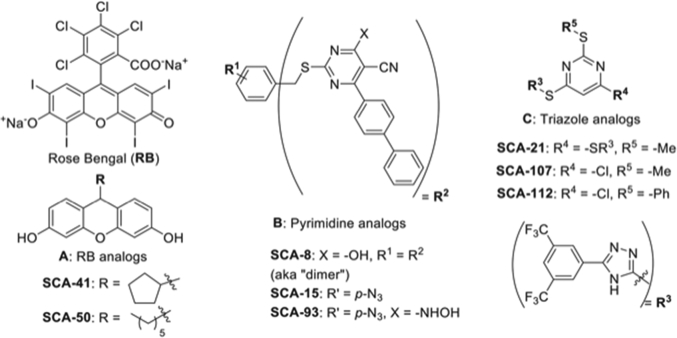
The Tai and Wang labs have undertaken multiple approaches, both in assay development and the design and synthesis of broad-spectrum, small molecule, SecA inhibitors against Gram-positive and Gram-negative bacteria, including MRSA and MDR pathogens. Three structural classes of small molecule compounds have been designed and evaluated (Fig. 2): Rose Bengal (RB)analogs (Huang et al.2012; Cui et al.2013; Jin et al.2015), 5-cyano-6-thiouracil-pyrimidine analogs (Li et al.2008; Chen et al.2010; Chaudhary et al.2015b) and triazole-pyrimidine analogs (Cui et al.2016; Jin et al.2016). Fluorescein dyes RB and erythrosine were found to effectively inhibit SecA activities (Huang et al.2012). RB is the first sub-micromolar inhibitor of ATPase activity in the unregulated E. coli SecA (EcSecA); with EcSecA68N (EcSecA which lacks a C-terminus) having an IC50 of 0.5 μM. In addition, RB showed an IC50 of 25 μM against intrinsic SecA ATPase (Bacillus subtilis SecA at 7 μM), 5 μM against membrane/lipid ATPase, 0.9 μM against translocation ATPase and 0.25 μM against protein translocation. The antibacterial activity against Gram-positive B. subtilis, as measured in MIC, approximates 3.1 μM. Using the NR698 mutant strain of Gram-negative E. coli, which has an outer-membrane deletion and thus increased permeability (Ruiz et al.2005), the MIC is also at 3.1 μM, though the MIC for the wild-type is over 1 mM. Permeability is apparently a major factor for the different sensitivity (see below). Molecular docking studies suggest that RB binds close to the ATP binding site, similar to CJ 21 058 (Huang et al.2012). RB is a competitive inhibitor for ATP at low concentrations, but is a non-competitive inhibitor at high ATP concentrations (Hsieh et al.2015). RB is relatively large (Mr 1017) and known to have other targets, such as F1F0-ATPase (Huang et al.2012), resulting in cytotoxicity. Nevertheless, RB was used as a lead compound for further optimization. The most active derivative of this group, SCA-50 (Mr 243), is a non-competitive inhibitor of ATP, active against SecA1 and SecA2 ATPase from S. aureus with an IC50 of 12 μM for SecA1, and 1 μM against SaSecA1 channel activity. SCA-50 inhibits S. aureus Mu50 toxin secretion, and is also active against E. coli NR698 and B. subtilis (Cui et al.2013). Interestingly, SCA-50 is far more active against several MRSA strains (MIC 4 μM) than RB (MIC 21–59 μM) (Jin et al.2015). In addition to the RB analogs, 5-cyano-6-thiouracil pyrimidine analogs (Li et al.2008; Chen et al.2010; Chaudhary et al.2015b) and triazole-pyrimidine analogs (Cui et al.2016; Jin et al.2016) have also been developed and evaluated with the hope that different scaffolds may target different parts of SecA molecules (Figs. 2 and 3). Using antimicrobial assays as well as SecA-only ion channel and protein translocation assays, SCA-15 was identified as the most active compound from the thiouracil-pyrimidine series, and along with SCA-107 and SCA-112 from the triazole-pyrimidine series. They are all non-competitive inhibitors against ATP in SecA inhibition, and have a relatively low inhibition of ATPase activity, suggesting their effects on ATPase are allosteric for SecA in the lipid environment rather than binding directly to the ATP site (Jin et al.2015; Jin et al.2016). The lack of effects of these SecA inhibitors on other ATPases, such as H+-ATPase and SpuB, indicates a certain selectivity of the inhibitors for SecA functions. This is significant because by not targeting the high-affinity ATP site, it allows further optimization to minimize toxicity issues without concerns for off-target effects on other ATP-binding proteins. Indeed, molecular docking studies show that they bind different part of SecA, close to but not at the ATP binding (Fig. 3). Various SecA from other bacteria has also been evaluated in the SecA-only channel activity; the inhibitors are also very active against SecA from P. aeruginosa, S. typhymurium and S. aureus, and SecA1 from S. pyogenes (Zhang et al.2013; Jin et al.2016). The effects of these inhibitors on SecA-only channel activity, protein translocation and antimicrobial activity are summarized in Tables 1 and 2. SCA-107 is the most active inhibitor among these series of compounds.
Figure 2.
Three classes of small molecule SecA inhibitors: Class A, RB analogs; Class B, Thiouracil-pyrimidine analogs and Class C, Triazole-pyrimidine analogs (From Jin et al.2016).
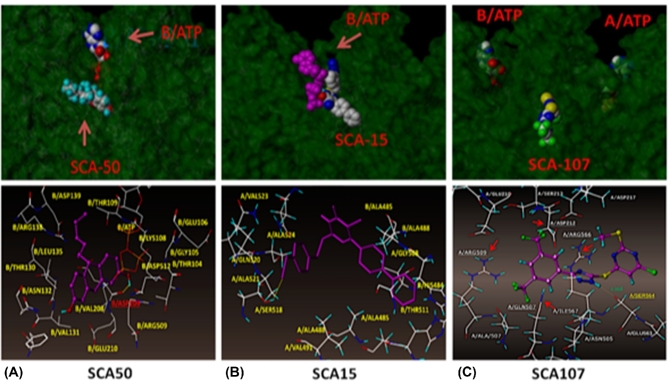
Figure 3.
Molecular modeling of three classes of small molecule SecA inhibitors. The program used is SYBYL 2.0 for file PDB ID- 2FSG with SecA dimers A and B. (A) Class A inhibitor (see Fig. 2), SCA-50, binds close to the ATP site of monomer B; (B) Class B inhibitor, SCA-15, binds at the interface of A and B; close to the ATP site of the B momoner and (C) Class C inhibitor, SCA-107, binds to the interface of A and B monomers (slightly closer to the ATP site of A, partially blocking the entrance to this ATP site). SCA-15 binds relatively closer to ATP site of the B monomer, followed by SCA-50, then the SCA-107. At the interface, SCA-15 binds to the B monomer, facing the A monomer. Lower panels: Inhibitors bindings relative to surrounding amino acid residues. (SCA-50 modified from Jin et al.2015, SCA-15 from Chaudhary et al.2015a, SCA-107, from Cui et al.2016).
Table 1.
Summary of inhibition of SecA-only liposomes activities by SecA inhibitors. SecA of E. coli (EcSecA), P. aeruginosa (PaSecA), S. aureus (SaSecA1) and B. anthracis (BaSecA1) were tested. A. IC50 for ion channel activity of SecA-liposome determined in the oocytes; B. ProOmpA translocation into SecA-liposomes (From Hsieh et al.2013; Jin et al.2016. * indicates unpublished data; ND, not determined).
| A. Inhibition on liposome ion-channel activity, IC50 (μM) | ||||
|---|---|---|---|---|
| SecA protein | RB | SCA-50 | SCA-15 | SCA-107 |
| M r: 1017 | M r: 298 | M r: 436 | M r: 472 | |
| EcSecA | 0.4 | 2.3 | *4.2 | 1.6 |
| PaSecA | 0.3 | 3.0 | *3.2 | 1.3 |
| SaSecA1 | 0.4 | 1.0 | *2.3 | 0.6 |
| BaSecA1 | 0.3 | *1.0 | *2.8 | 0.7 |
| B. Inhibition on liposomes translocation activity, IC50 (μM) | ||||
| EcSecA | PaSecA | SaSecA1 | BaSecA1 | |
| SCA-107 | 3.5 | 3.0 | *2.0 | *2.8 |
| Rose Bengal | 1.0 | 1.0 | *1.0 | ND |
Table 2.
MIC of SecA inhibitors to various bacterial strains, including some Gram-positive and Gram-negative MRSA and MDR pathogens. *With (+ ) or without (-) PMBN (Polymxin B nanopeptide) which alone had no inhibition on growth ND, not determined (From Jin et al., 2015, 2016;).
| Strains, MIC (μM) | PMBN* | RB | SCA-50 | SCA-15 | SCA-107 | Vancomycin |
|---|---|---|---|---|---|---|
| Mr: 1017 | Mr: 298 | Mr: 436 | Mr: 472 | Mr: 1446 | ||
| Gram-positive bacteria | ||||||
| S. aureus Mu50 | - | 50.0 | 12.5 | *5.0 | 1.6 | 5.5 |
| S. aureus Newman | - | 50.0 | 12.5 | ND | 1.6 | ND |
| B. anthracis Sterling | - | ND | *6.3 | 6.3 | 3.1 | ND |
| Gram-negative bacteria | ||||||
| E. coli NR698 | - | 4.4 | 15.8 | 12.5 | 5.2 | ND |
| E. coli MC4100 | - | >1250 | >1250 | >1250 | >1250 | >150 |
| E. coli MC4100 | + | 15.6 | 3.0 | 15.6 | 1.5 | 21.6 |
| P. aeruginosa T15464 | + | 0.4 | 6.3 | ND | 0.4 | 21.6 |
| A. baumannii ATCC9955 | + | 79.2 | 25.0 | ND | 2.1 | 172.5 |
| S. flexneri ATCC12022 | + | 7.4 | 5.8 | 25.0 | 4.7 | 43.1 |
The outer membrane of Gram-negative bacteria is a major permeability barrier, hindering the effectiveness of many antibiotis/antimicrobials. This obstacle can be circumvented by two outer membrane permeabilizers, PMBN and NAB7061. These compounds are polymyxin B derivatives that lack the fatty acid tail, have minimal toxicity, and allow antimicrobials to enter Gram-negative bacteria (Vaara et al.2008; Vaara et al.2010). In the presence of these permeabilizers, all classes of SecA inhibitors are active against several Gram-negative pathogens, including P. aeruginosa, S. typhymurium, Shigella flexneli, A. baumannii and Klebsiella pneumonia with low MIC (Table 2). These SecA inhibitors are broad spectrum bactericidal inhibitors against Gram-positive and Gram-negative pathogens, and are more effective than the last resort antibiotic, vancomycin (Jin et al.2015; Jin et al.2016). The results from the SecA-only ion channel activity and protein translocation assays parallel most closely those of the antimicrobial assays.
SecA functions as a membrane protein that spans the cytoplasmic membrane and forms a channel (Wang et al.2003; You et al.2013) (Fig. 1c). As a result, SecA inhibitors may access SecA directly from the extracellular matrix and exert their effects without entering inside cells (alternatively, SecA inhibitors may be poor substrates for efflux pumps). About 50% of E. coli SecA is in the cytoplasmic membrane, while all SecA1 of S. pyogenes is in the membranes (Rosch and Caparon 2004; Rosch and Caparon 2005). Consequently, targeting SecA may allow the possibility of avoiding the effect of bacterial efflux transporters, which are responsible for MDR in highly pathogenic strains of bacteria including MRSA (Nikaido 2009; Jin et al.2015; Jin et al.2016). Indeed, SecA is required for the proper assembly of many efflux pumps with or without signal peptides. Even though S. aureus strains Mu50 and N315 are resistant to QacA efflux-mediated antiseptics, the levels of efflux pumps have no effects on the MIC and bactericidal nature of SecA inhibitors (Jin et al.2015). Similar results were obtained with MDR strains of P. aeruginosa with MexJK-Opr-H pumps and E. coli with AcrAB pumps (Jin et al.2016). These findings show that differing activities of the efflux pumps have no effect on the MIC of the SecA inhibitors, validating the ability for SecA inhibitors to abrogate the effects of efflux pumps in either Gram-positive or Gram-negative bacteria.
The biochemical specificity of inhibitors targeting SecA has been shown by the binding assays and immunoprecipitation pull-down study for S. aureus SecA1 and E. coli SecA (Jin et al.2015; Jin et al.2016). The critical genetic evidence is the identification of an E. coli azide-resistant strain carrying a single seca-azi-9 mutation that is resistant to SCA-107 (Jin et al.2016). This strain has an increased MIC of about 10–20 fold. The mutated ecsecA gene has been cloned, and identified as having single mutation resulting in SecAL515F. Complementation of the azide resistance of seca-azi-9 in E. coli with B. subtilis SecA and subsequent increase in MIC further verifying the seca-azi-9 resistance to SCA-107. Purified SecA-L515F protein, when assayed for channel activity and protein translocation, further confirms that the resistance is due to the singular SecA mutation (Jin et al.2016), providing important genetic evidence of SecA being a key target for SCA-107 achieving its antimicrobial effect. Interestingly, this mutant is also partially resistant to SCA-50. Two recent articles that evaluated the effects of SecA inhibitors, based on thiouracil derivatives containing acyl thiourea or triazol-thiadiazole, reported that these inhibitors were also active against Gram-positive bacteria (Cui et al.2017a, b). However, there are limited data on the correlation between SecA inhibition assays and MIC determination. Thus, it is hard to compare directly with other published SecA inhibitors.
The ultimate goal in searching for SecA inhibitors would be to test their efficacy in protecting the host from infection by bacterial pathogens. Indeed, SCA-107 protects mice against lethal infection by the S. aureus Newman strain (Eichenbaum, Tai, Wang, manuscript in preparation). This is encouraging and warrants further investigation of these inhibitors. Further work in this field also needs to improve the solubility and the delivery of these inhibitors (Hu, Akula and Wang 2016), as well as overcoming the permeability hindrance of the outer membrane of Gram-negative bacteria. So far the best inhibitor is effective at high nM concentrations, which is better than many existing antibiotics, but may be optimized further. However, direct quantitative comparisons in efficacy of the different inhibitors are difficult to make when one is reminded of the fact that the SecA concentration in bacterial cells is around the 5–8 μM range.
The presence of SecYEG alters the sensitivity of these inhibitors in both ion channel activity and protein translocation (Hsieh et al.2013; Hsieh et al.2017b). The IC50 values of RB and SCA-107 are higher in the presence of SecYEG, and in membrane with wild-type SecYEG (Table 3A). This difference was observed in liposome reconstitution with or without SecYEG, or in membrane with or without SecYEG (Table 3A). Many prlD/secA, prlA/secY and prlG/secE suppressors are resistant to azide (Oliver et al.1990; Osborne and Silhavy 1993; Huie and Silhavy 1995; Maillard et al.2007). Their membrane vesicles exhibit a biphasic mode of inhibition in ion channel activity and in protein translocation, one with an IC50 similar to wild-type, the other with a much higher IC50 (Hsieh et al.2017b). However, all bacterial strains have similar MIC despite vast differences in their IC50 (Table 3B). It appears that SecA-only channels, which are more sensitive to SCA-107, have specific physiological functions in the cells that are distinct from those of SecA-SecYEG channels.
Table 3A.
Changes of sensitivity to SecA inhibitors by SecYEG for ion channel activuity in the oocytes. SecA-liposomes reconstituted with equal amount of SecYEG as in the MC4100 membranes (Memb). The SecYEG in MC4100 or PrlA4* membranes was removed by cholate (Watanabe, Nicchitta and Blobel 1990) to yield reconstituted membrane (RE-4100 Memb); and then similarly reconstituted with SecYEG-DFC (Re-MC4100 + SecYEGDFC) or RE-PrlA4 Memb. (Hsieh et al.2011, 2017a).
| A. Changes of ion channel sensitivity to SecA inhibitors by SecYEG | |||||||
|---|---|---|---|---|---|---|---|
| Channel activity IC50 (μM) | SecA-liposomes | SecA-lipo + YEGDFC | MC4100 Memb | RE-MC4100 Memb | RE-MC4100 + YEGDFC | PrlA4 Memb | RE- PrlA4 Memb |
| Rose Bengal | 0.4 | 3.8 | 4.7 | 0.4 | 4.4 | 73 | 0.6 |
| SCA-107 | 1.1 | 3.3 | 2.9 | 0.8 | 2.1 | >90 | 2.5 |
Table 3B.
Biphasic responses of PrlA/SecY membranes to inhibitor SCA-107. The IC50 (μM) inhibition of PrlA membranes were extrapolated for low IC50L and high IC50H: Ion channel activity in oocytes, and in vitro pOmpA translocation into membrane vesicles (From Hsieh et al.2017a). All suppressor strains are derivatives of MC4100 (Osborne and Silhavy 1993).
| B. Biphasic responses of PrlA mutant membranes and MIC to SecA inhibitor SCA-107 | |||||||
|---|---|---|---|---|---|---|---|
| MC4100 | PrlA4 | PrlA666 | Other PrlA, PrlG1 | ||||
| Prl membranes | IC50 | IC50L | IC50H | IC50L | IC50H | IC50L | IC50H |
| IC50 on channel activity (μM) | 3.9 | 3.4 | 125 | 1.7 | 154 | 1.4–6.1 | >100 |
| IC50 on translocation activity (μM) | 1.5 | 2.6 | >50 | 2.7 | >50 | 2.1–3.1 | >50 |
| MIC of SCA-107 (μM) | 1.5–5.5 | 3.1 | 0.8 | 1.6–3.1 | |||
Along with all the rapid advances in molecular analysis of post-translational translocation, an understanding of co-translational secretion has not been pursued vigorously, even though extracellular labeling of nascent peptides on membrane-bound ribosomes, and the nascent peptides as the sole attachment of ribosome to membranes were reported early (Smith et al.1977; Smith, Tai and Davis 1978; Davis and Tai 1980). Importantly, ATP is required, in addition to the translationally-specific GTP, for bacterial co-translational translocation, implying the involvement of SecA with membrane-bound ribosomes (Chen and Tai 1987b). Indeed, SecA is involved in co-translational translocation with or without signal peptides (Karamyshev and Johnson 2005; Huber et al.2011) as well as post-translational translocation (Huber et al.2011). Additionally, SecA binds ribosomes (Fig. 1e) and covers the nascent peptide emerging site with its N-terminal helix that is required for stable ribosome association (Kramer et al.2009; Singh et al.2014). SecA has also been shown to interact with ribosomes in a mutually exclusive manner to SecYEG (Wu et al.2012). Moreover, while membrane proteins may insert spontaneously into or across membranes (Engelman and Steitz 1981), the insertion of some proteins that require signal recognition particles has been demonstrated to be mediated by SecA (Qi and Bernstein 1999; Neumann-Haefelin et al.2000), as have the co-translational insertion of membrane proteins without signal-peptides (Herskovits and Bibi 2000; Halbedel et al.2014; Rawat et al.2015). SecA mediates co-translational targeting and translocation of an inner membrane protein (Wang, Yang and Shan 2017), implicating a role for SecA-bound ribosomes in co-translational translocation (Fig. 1f). One intriguing question emerges: whether the ‘efficient’ biogenesis and integration of signal-peptide-less SecYEG into membranes also involve SecA. Energetically, the SecA-only channel is less efficient than the SecA-SecYEG channel, and extensive ATP hydrolysis is required for post-translational protein translocation, even through the SecB-SecA-SecYEG channels (Mao, Hardy and Randall 2009). It is possible that these post-translational pathways are less efficient than their co-translational counterparts, which perhaps can push nascent peptides by protein synthesis, or minimally save energy by translocating partially folded nascent peptides.
CONCLUSIONS AND PERSPECTIVES
Effective broad-spectrum inhibitors for SecA-dependent protein translocation have been developed, and may have therapeutic applications. These inhibitors are less effective against SecA-SecYEG channels than against the SecA-only channels, which translocate proteins with or without signal-peptides albeit with lower efficiency. It is interesting to note that when bacterial cells enter stationary phase, the amounts of SecA increase while the SecYEG levels decrease (Yang, Lu and Tai 2013), suggesting an important role for these SecA-only-channels in the secretion of proteins without signal-peptides (Tanji et al.1991; Yang et al.2011). Since ribosomes can only attach to membranes via nascent peptides (Smith, Tai and Davis 1978), we propose that the interaction of SecA with membrane-ribosomes (Fig. 1f) may have special importance in the fundamental mechanism(s) of bacterial cell physiology, which SecA inhibitors may help to unravel. The interactions of SecA with membrane-bound ribosomes in co-translational secretion raise the intriguing prospect of combined application of both SecA inhibitors and ribosome antibiotics (Davis, Chen and Tai 1986).
Acknowledgements
We thank Drs. Tom Silhavy and Don Oliver for strains, and John Ingraham for comments and coworkers over the years for their contributions in the studies. The corresponding author (PCT) would like to dedicate the article to the memory of late Bernard Davis who worked extensively on the antibiotic actions on ribosomes, and the biochemical mechanisms of bacterial protein secretion.
FUNDING
JSJ, HSH and ASC were fellows of Molecular Basis of Diseases Program at Georgia State University. Most of the experimental work was supported in parts by National Institute of Health grants GM34766 (PCT) and AI104168 (PCT, BW), and Chinese National Science Foundation Grant 31 230 016 (SFS).
Conflict of interest. None declared.
REFERENCES
- Akula N, Zheng H, Han FQ et al. Discovery of novel SecA inhibitors of Candidatus Liberibacter asiaticus by structure based design. Bioorg Med Chem Lett 2011;21:4183–8. [DOI] [PubMed] [Google Scholar]
- Alksne LE, Burgio P, Hu W et al. Identification and analysis of bacterial protein secretion inhibitors utilizing a SecA-LacZ reporter fusion system. Antimicrob Agents Chemother 2000;44:1418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JJ, Oxender DL. Genetic separation of high- and low-affinity transport systems for branched-chain amino acids in Escherichia coli K-12. J Bacteriol 1978;136:168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baars L, Wagner S, Wickstrom D et al. Effects of SecE depletion on the inner and outer membrane proteomes of Escherichia coli. J Bacteriol 2008;190:3505–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer BW, Shemesh T, Chen Y et al. A “push and slide mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell 2014;157:1416–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler MW, Montgomery MG, Leslie AG et al. How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci 2006;103:8646–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelli RJ, Chen L, Tai PC et al. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell 1988;55:683–92. [DOI] [PubMed] [Google Scholar]
- Chatzi KE, Sardis MF, Economou A et al. SecA-mediated targeting and translocation of secretory proteins. Biochim Biophys Acta 2014;1843:1466–74. [DOI] [PubMed] [Google Scholar]
- Chaudhary AS, Chen W, Jin J et al. SecA: a potential antimicrobial target. Future Med Chem 2015a;7:989–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary AS, Jin J, Chen W et al. Design, syntheses and evaluation of 4-oxo-5-cyano thiouracils as SecA inhibitors. Bioorg Med Chem 2015b;23:105–17. [DOI] [PubMed] [Google Scholar]
- Chen L, Rhoads D, Tai PC. Alkaline phosphatase and OmpA protein can be translocated posttranslationally into membrane vesicles of Escherichia coli. J Bacteriol 1985;161:973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tai PC. ATP is essential for protein translocation into Escherichia coli membrane vesicles. Proc Natl Acad Sci 1985;82:4384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tai PC. Effects of antibiotics and other inhibitors on ATP-dependent protein translocation into membrane vesicles. J Bacteriol 1987;169:2373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tai PC. Effects of inhibitors of membrane signal peptide peptidase on protein translocation into membrane vesicles. Arch Microbiol 1989;153:90–94. [DOI] [PubMed] [Google Scholar]
- Chen LL, Tai PC. Evidence for the involvement of ATP in co-translational protein translocation. Nature 1987;328:164–6. [DOI] [PubMed] [Google Scholar]
- Chen W, Huang YJ, Gundala SR et al. The first low ?M SecA inhibitors. Bioorg Med Chem 2010;18:1617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Brown T, Tai PC. Identification and characterization of protease-resistant SecA fragments: secA has two membrane-integral forms. J Bacteriol 1998;180:527–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Tai PC. A Significant Fraction of Functional SecA Is Permanently Embedded in the Membrane. J Biol Chem 1996;271:29698–706. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pan X, Tang Y et al. Full-length Escherichia coli SecA dimerizes in a closed conformation in solution as determined by cryo-electron microscopy. J Biol Chem 2008;283:28783–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tai PC, Sui SF. The active ring-like structure of SecA revealed by electron crystallography: conformational change upon interaction with SecB. J Struct Biol 2007;159:149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DB, Smith VF, Crane JM et al. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol 2008;382:74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Jin J, Chaudhary AS et al. Design, synthesis and evaluation of Triazole-Pyrimidine analogues as SecA inhibitors. ChemMedChem 2016;11:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Jin J, Hsieh YH et al. Design, synthesis and biological evaluation of rose bengal analogues as SecA inhibitors. ChemMedChem 2013;8:1384–93. [DOI] [PubMed] [Google Scholar]
- Cui P, Li X, Zhu M et al. Design, synthesis and antibacterial activities of thiouracil derivatives containing acyl thiourea as SecA inhibitors. Bioorg Med Chem Letters 2017a;27:2234–7. [DOI] [PubMed] [Google Scholar]
- Cui P, Li X, Zhu M et al. Design, synthesis and antimicrobial activities of thiouracil derivatives containing triazolo-thiadiazole as SecA inhibitors. Eur J Med Chem 2017b;127:159–65. [DOI] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet 1998;32:59–94. [DOI] [PubMed] [Google Scholar]
- Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci 1986;83:6164–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD, Tai PC. The mechanism of protein secretion across membranes. Nature 1980;283:433–8. [DOI] [PubMed] [Google Scholar]
- De Waelheyns E, Segers K, Sardis MF et al. Identification of small-molecule inhibitors against SecA by structure-based virtual ligand screening. J Antibiot 2015;68:666–73. [DOI] [PubMed] [Google Scholar]
- Derman AI, Puziss JW, Bassford PJ Jr et al. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. Embo J 1993;12:879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 2008;77:643–67. [DOI] [PubMed] [Google Scholar]
- Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. Embo J 1999;18:3263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 1994;78:835–43. [DOI] [PubMed] [Google Scholar]
- Eichler J, Wickner W. The SecA subunit of Escherichia coli preprotein translocase is exposed to the periplasm. J Bacteriol 1998;180:5776–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: The helical hairpin hypothesis. Cell 1981;23:411–22. [DOI] [PubMed] [Google Scholar]
- Floyd JH, You Z, Hsieh YH et al. The dispensability and requirement of SecA N-terminal aminoacyl residues for complementation, membrane binding, lipid-specific domains and channel activities. Biochem Biophys Res Commun 2014;453:138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouridis G, Karamanou S, Koukaki M et al. In vitro assays to analyze translocation of the model secretory preprotein alkaline phosphatase. Methods Mol Biol 2010;619:157–72. [DOI] [PubMed] [Google Scholar]
- Halbedel S, Kawai M, Breitling R et al. SecA is required for membrane targeting of the cell division protein DivIVA in vivo. Front Microbiol 2014;5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits AA, Bibi E. Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc Natl Acad Sci 2000;97:4621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YH, Huang YJ, Zhang H et al. Dissecting structures and functions of SecA-only protein-conducting channels: ATPase, pore structure, ion channel activity, protein translocation, and interaction with SecYEG/SecDF*YajC. PLoS One 2017a;12:e0178307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YH, Zhang H, Jin J et al. Biphasic actions of SecA inhibitors on Prl/Sec suppressors: Possible physiological roles of SecA-only channels. Biochem Biophys Res Commun 2017;482:296–300. [DOI] [PubMed] [Google Scholar]
- Hsieh YH, Zhang H, Lin BR et al. SecA alone can promote protein translocation and ion channel activity: SecYEG increases efficiency and signal peptide specificity. J Biol Chem 2011;286:44702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YH, Zhang H, Wang H et al. Reconstitution of functionally efficient SecA-dependent protein-conducting channels: Transformation of low-affinity SecA-liposome channels to high-affinity SecA-SecYEG-SecDF·YajC channels. Biochem Biophys Res Commun 2013;431:388–92. [DOI] [PubMed] [Google Scholar]
- Hsieh YH, Zou J, Jin JS et al. Monitoring channel activities of proteoliposomes with SecA and Cx26 gap junction in single oocytes. Anal Biochem 2015;480:58–66. [DOI] [PubMed] [Google Scholar]
- Hu HJ, Holley J, He J et al. To be or not to be: predicting soluble SecAs as membrane proteins. IEEE Transon Nanobioscience 2007;6:168–79. [DOI] [PubMed] [Google Scholar]
- Hu J, Akula N, Wang N. Development of a microemulsion formulation for antimicrobial SecA inhibitors. PLoS One 2016;11:e0150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Wang H, Gao FB et al. Fluorescein analogues inhibit SecA ATPase: the first sub-micromolar inhibitor of bacterial protein translocation. ChemMedChem 2012;7:571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Rajagopalan N, Preissler S et al. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell 2011;41:343–53. [DOI] [PubMed] [Google Scholar]
- Huie JL, Silhavy TJ. Suppression of signal sequence defects and azide resistance in Escherichia coli commonly result from the same mutations in secA. J Bacteriol 1995;177:3518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JF, Weinkauf S, Henry L et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 2002;297:2018–26. [DOI] [PubMed] [Google Scholar]
- Jang MY, De Jonghe S, Segers K et al. Synthesis of novel 5-amino-thiazolo[4,5-d]pyrimidines as E. coli and S. aureus SecA inhibitors. Bioorg Med Chem 2011;19:702–14. [DOI] [PubMed] [Google Scholar]
- Jilaveanu LB, Oliver DB. In vivo membrane topology of Escherichia coli SecA ATPase reveals extensive periplasmic exposure of multiple functionally important domains clustering on one face of SecA. J Biol Chem 2007;282:4661–8. [DOI] [PubMed] [Google Scholar]
- Jin J, Cui J, Chaudhary AS et al. Evaluation of small molecule SecA inhibitors against methicillin-resistant Staphylococcus aureus. Bioorg Med Chem 2015;23:7061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Hsieh YH, Cui J et al. Using chemical probes to assess the feasibility of targeting SecA for developing antimicrobial agents against Gram-negative bacteria. ChemMedChem 2016;11:2511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junne T, Wong J, Studer C et al. Decatransin, a new natural product inhibiting protein translocation at the Sec61/SecYEG translocon. J Cell Sci 2015;128:1217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanou S, Vrontou E, Sianidis G et al. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol 1999;34:1133–45. [DOI] [PubMed] [Google Scholar]
- Karamyshev AL, Johnson AE. Selective SecA association with signal sequences in ribosome-bound nascent chains: a potential role for SecA in ribosome targeting to the bacterial membrane. J Biol Chem 2005;280:37930–40. [DOI] [PubMed] [Google Scholar]
- Keller RC. The prediction of novel multiple lipid-binding regions in protein translocation motor proteins: a possible general feature. Cell Mol Biol Lett 2011;16:40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N et al. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 2009;16:589–97. [DOI] [PubMed] [Google Scholar]
- Kusters I, Driessen AJ. SecA, a remarkable nanomachine. Cell Mol Life Sci 2011;68:2053–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta PA, Alvarez LJ, Reinach PS et al. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 1979;100:95–97. [DOI] [PubMed] [Google Scholar]
- Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 2004;10:S122–9. [DOI] [PubMed] [Google Scholar]
- Li M, Huang YJ, Tai PC et al. Discovery of the first SecA inhibitors using structure-based virtual screening. Biochem Biophys Res Commun 2008;368:839–45. [DOI] [PubMed] [Google Scholar]
- Li W, Schulman S, Boyd D et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell 2007;26:511–21. [DOI] [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 1990;60:271–80. [DOI] [PubMed] [Google Scholar]
- Lin BR, Gierasch LM, Jiang C et al. Electrophysiological studies in Xenopus oocytes for the opening of Escherichia coli SecA-dependent protein-conducting channels. J Membrane Biol 2006;214:103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BR, Hsieh YH, Jiang C et al. Escherichia coli membranes depleted of SecYEG elicit SecA-dependent ion-channel activity but lose signal peptide specificity. J Membrane Biol 2012;245:747–57. [DOI] [PubMed] [Google Scholar]
- Lycklama ANJA, Driessen AJ. The bacterial Sec-translocase: structure and mechanism. Philos Trans R Soc Lond B Biol Sci 2012;367:1016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard AP, Lalani S, Silva F et al. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J Biol Chem 2007;282:1281–7. [DOI] [PubMed] [Google Scholar]
- Mao C, Hardy SJ, Randall LL. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J Bacteriol 2009;191:978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol 1993;10:483–97. [DOI] [PubMed] [Google Scholar]
- Mizushima S, Tokuda H, Matsuyama S-I. Molecular characterization of Sec proteins comprising the protein secretory machinery of Escherichia coli. In: Walter N, Roland L (eds) New Comprehensive Biochemistry. Elsevier, 1992, 21–32. [Google Scholar]
- Moran U, Phillips R, Milo R. SnapShot: key numbers in biology. Cell 2010;141:1262–1262.e1. [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K. The Sec protein-translocation pathway. Trends Microbiol 2001;9:494–500. [DOI] [PubMed] [Google Scholar]
- Na B, You Z, Yang H et al. Characterization of the minimal length of functional SecA in Escherichia coli. Biochem Biophys Res Commun 2015;456:213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin C, Schafer U, Muller M et al. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. Embo J 2000;19:6419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem 2009;78:119–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DB, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 1981;25:765–72. [DOI] [PubMed] [Google Scholar]
- Oliver DB, Cabelli RJ, Dolan KM et al. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci 1990;87:8227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E, Boyd D, Gon S et al. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem 2005;280:9097–105. [DOI] [PubMed] [Google Scholar]
- Osborne RS, Silhavy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. Embo J 1993;12:3391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolau Y, Papadovasilaki M, Ravelli RB et al. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol 2007;366:1545–57. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol 2007;5:839–51. [DOI] [PubMed] [Google Scholar]
- Parish CA, de la Cruz M, Smith SK et al. Antisense-guided isolation and structure elucidation of pannomycin, a substituted cis-decalin from Geomyces pannorum. J Nat Prod 2009;72:59–62. [DOI] [PubMed] [Google Scholar]
- Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys 2012;41:21–40. [DOI] [PubMed] [Google Scholar]
- Qi HY, Bernstein HD. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J Biol Chem 1999;274:8993–7. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V, Oliver D. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J Biol Chem 1997;272:23239–46. [DOI] [PubMed] [Google Scholar]
- Rao CVS, De Waelheyns E, Economou A et al. Antibiotic targeting of the bacterial secretory pathway. Biochim Biophys Acta 2014;1843:1762–83. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007;450:663–9. [DOI] [PubMed] [Google Scholar]
- Rawat S, Zhu L, Lindner E et al. SecA drives transmembrane insertion of RodZ, an unusual single-span membrane protein. J Mol Biol 2015;427:1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science 2004;304:1513–5. [DOI] [PubMed] [Google Scholar]
- Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol 2005;58:959–68. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Falcone B, Kahne D et al. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 2005;121:307–17. [DOI] [PubMed] [Google Scholar]
- Schmidt MG, Rollo EE, Grodberg J et al. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J Bacteriol 1988;170:3404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers K, Anne J. Traffic jam at the bacterial sec translocase: targeting the SecA nanomotor by small-molecule inhibitors. Chem Biol 2011;18:685–98. [DOI] [PubMed] [Google Scholar]
- Segers K, Klaassen H, Economou A et al. Development of a high-throughput screening assay for the discovery of small-molecule SecA inhibitors. Anal Biochem 2011;413:90–96. [DOI] [PubMed] [Google Scholar]
- Seoh HK, Tai PC. Carbon source-dependent synthesis of SecB, a cytosolic chaperone involved in protein translocation across Escherichia coli membranes. J Bacteriol 1997;179:1077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SM, Blobel G. Signal peptides open protein-conducting channels in E. coli. Cell 1992;69:677–84. [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G, Zimmerberg J. Large aqueous channels in membrane vesicles derived from the rough endoplasmic reticulum of canine pancreas or the plasma membrane of Escherichia coli. Proc Natl Acad Sci 1989;86:6176–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kraft C, Jaiswal R et al. Cryo-electron microscopic structure of SecA protein bound to the 70S ribosome. J Biol Chem 2014;289:7190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WP, Tai PC, Davis BD. Nascent peptide as sole attachment of polysomes to membranes in bacteria. Proc Natl Acad Sci 1978;75:814–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WP, Tai PC, Thompson RC et al. Extracellular labeling of nascent polypeptides traversing the membrane of Escherichia coli. Proc Natl Acad Sci 1977;74:2830–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Shapiro L. Bacterial protein secretion–a target for new antibiotics? Chem Biol 1997;4:637–41. [DOI] [PubMed] [Google Scholar]
- Sugie Y, Inagaki S, Kato Y et al. CJ-21,058, a new SecA inhibitor isolated from a fungus. J Antibiot 2002;55:25–29. [DOI] [PubMed] [Google Scholar]
- Tang Y, Pan X, Chen Y et al. Dimeric SecA couples the preprotein translocation in an asymmetric manner. PLoS One 2011;6:e16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Pan X, Tai PC et al. Electron microscopic visualization of asymmetric precursor translocation intermediates: SecA functions as a dimer. Sci China Life Sci 2010;53:1049–56. [DOI] [PubMed] [Google Scholar]
- Tanji Y, Gennity J, Pollitt S et al. Effect of OmpA signal peptide mutations on OmpA secretion, synthesis, and assembly. J Bacteriol 1991;173:1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M, Fox J, Loidl G et al. Novel polymyxin derivatives carrying only three positive charges are effective antibacterial agents. Antimicrob Agents Chemother 2008;52:3229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M, Siikanen O, Apajalahti J et al. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob Agents Chemother 2010;54:3341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Chen Y, Yang H et al. Ring-like pore structures of SecA: implication for bacterial protein-conducting channels. Proc Natl Acad Sci 2003;100:4221–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yang CI, Shan SO. SecA mediates cotranslational targeting and translocation of an inner membrane protein. J Cell Biol 2017;216:3639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Blobel G. SecA protein is required for translocation of a model precursor protein into inverted vesicles of Escherichia coli plasma membrane. Proc Natl Acad Sci 1993;90:9011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Nicchitta CV, Blobel G. Reconstitution of protein translocation from detergent-solubilized Escherichia coli inverted vesicles: PrlA protein-deficient vesicles efficiently translocate precursor proteins. Proc Natl Acad Sci 1990;87:1960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci 2002;11:875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZC, de Keyzer J, Kedrov A et al. Competitive binding of the SecA ATPase and ribosomes to the SecYEG translocon. J Biol Chem 2012;287:7885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CK, Ewis HE, Zhang X et al. Nonclassical protein secretion by Bacillus subtilis in the stationary phase is not due to cell lysis. J Bacteriol 2011;193:5607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CK, Lu CD, Tai PC. Differential expression of secretion machinery during bacterial growth: SecY and SecF decrease while SecA increases during transition from exponential phase to stationary phase. Curr Microbiol 2013;67:682–7. [DOI] [PubMed] [Google Scholar]
- Yang YB, Lian J, Tai PC. Differential translocation of protein precursors across SecY-deficient membranes of Escherichia coli: SecY is not obligatorily required for translocation of certain secretory proteins in vitro. J Bacteriol 1997;179:7386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YB, Yu N, Tai PC. SecE-depleted membranes of Escherichia coli are active. SecE is not obligatorily required for the in vitro translocation of certain protein precursors. J Biol Chem 1997;272:13660–5. [DOI] [PubMed] [Google Scholar]
- You Z, Liao M, Zhang H et al. Phospholipids induce conformational changes of SecA to form membrane-specific domains: AFM structures and implication on protein-conducting channels. PLoS One 2013;8:e72560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hsieh YH, Lin BR et al. Specificity of SecYEG for PhoA precursors and SecA homologs on SecA protein-conducting channels. Biochem Biophys Res Commun 2013;437:212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 2008;455:936–43. [DOI] [PMC free article] [PubMed] [Google Scholar]