Abstract
BACKGROUND
Neurofibroma/schwannoma hybrid nerve sheath tumors (N/S HNSTs) are neoplasms associated with larger nerves that occur sporadically and in the context of schwannomatosis or neurofibromatosis type 2 or 1. Clinical management of N/S HNSTs is challenging, especially for large tumors, and established systemic treatments are lacking.
METHODS
We used next-generation sequencing and array-based DNA methylation profiling to determine the clinically actionable genomic and epigenomic landscapes of N/S HNSTs.
RESULTS
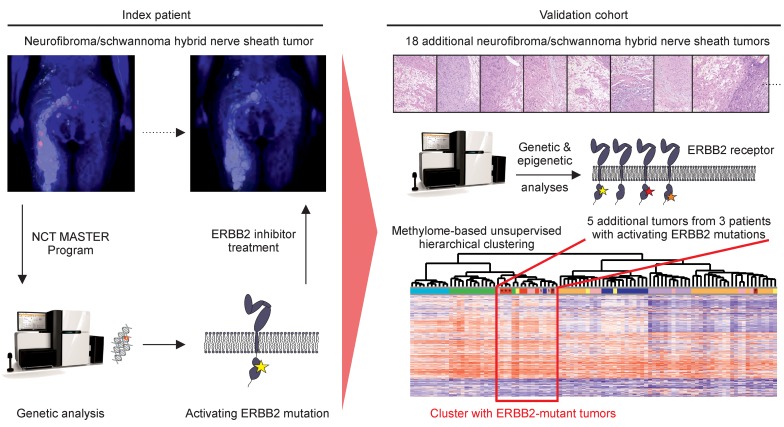
Whole-exome sequencing within a precision oncology program identified an activating mutation (p.Asp769Tyr) in the catalytic domain of the ERBB2 receptor tyrosine kinase in a patient with schwannomatosis-associated N/S HNST, and targeted treatment with the small-molecule ERBB inhibitor lapatinib led to prolonged clinical benefit and a lasting radiographic and metabolic response. Analysis of a multicenter validation cohort revealed recurrent ERBB2 mutations (p.Leu755Ser, p.Asp769Tyr, p.Val777Leu) in N/S HNSTs occurring in patients who met diagnostic criteria for sporadic schwannomatosis (3 of 7 patients), but not in N/S HNSTs arising in the context of neurofibromatosis (6 patients) or outside a tumor syndrome (1 patient), and showed that ERBB2-mutant N/S HNSTs cluster in a distinct subgroup of peripheral nerve sheath tumors based on genome-wide DNA methylation patterns.
CONCLUSION
These findings uncover a key biological feature of N/S HNSTs that may have important diagnostic and therapeutic implications.
FUNDING
This work was supported by grant H021 from DKFZ-HIPO, the University Cancer Center Frankfurt, and the Frankfurt Research Funding Clinician Scientist Program.
Keywords: Genetics, Oncology
Keywords: Molecular diagnosis, Neurological disorders, Oncogenes
Introduction
Neurofibroma/schwannoma hybrid nerve sheath tumors (N/S HNSTs) are peripheral nerve sheath tumors that are recognized as a distinct entity by the 2016 update of the World Health Organization (WHO) Classification of Tumours of the Central Nervous System (1). While schwannomas are solely composed of Schwann cells, neurofibromas are heterogeneous tumors consisting of Schwann cells, axonal processes, perineurial/perineurial-like cells, fibroblasts, and mast cells (2). N/S HNSTs are composed of schwannoma-like nodules within a neurofibroma-like tumor (3), and more than 60% of cases are associated with an underlying tumor predisposition syndrome. In particular, 26%, 17%, and 9% of N/S HNST patients are afflicted with neurofibromatosis type 2 (NF2), schwannomatosis, and neurofibromatosis type (NF1), respectively (4); 71% of patients with schwannomatosis develop N/S HNSTs (4); and 26% of NF2 patients undergoing resection of peripheral nerve sheath tumors have N/S HNSTs (5). Thus, N/S HNSTs may occur in up to 1 in 10,000 persons per year, assuming a birth incidence for schwannomatosis of 1 in 40,000 (6).
Treatment of N/S HNSTs is based on surgical resection. However, due to the association of neurofibromas, schwannomas, and N/S HNSTs with larger nerves as well as the number and extent of manifestations typically seen in cases of NF1-associated plexiform nerve sheath tumors (7), local control is sometimes difficult to achieve. Thus, despite generally being considered benign due to WHO grade I histology in both tumor components, N/S HNSTs can be associated with substantial morbidity, such as motor disability, neuropathic pain, or disfigurement (6). In patients who are not surgical candidates, palliative radiotherapy may be considered depending on the localization and extent of the tumor(s). In contrast, although clinical trials have been conducted in subgroups of schwannomas and neurofibromas (8, 9), there is no established systemic treatment for N/S HNSTs, and molecular drug targets are lacking. Thus, there is an urgent need for novel treatment options to improve clinical outcomes in patients with N/S HNSTs, especially those with unresectable symptomatic tumors.
We here report successful targeted treatment of an index patient with schwannomatosis-associated N/S HNST harboring an activating ERBB2 mutation within a genomics-guided precision oncology program, and describe for what we believe is the first time that a substantial proportion of N/S HNSTs are associated with recurrent, pharmacologically tractable ERBB2 mutations.
Results
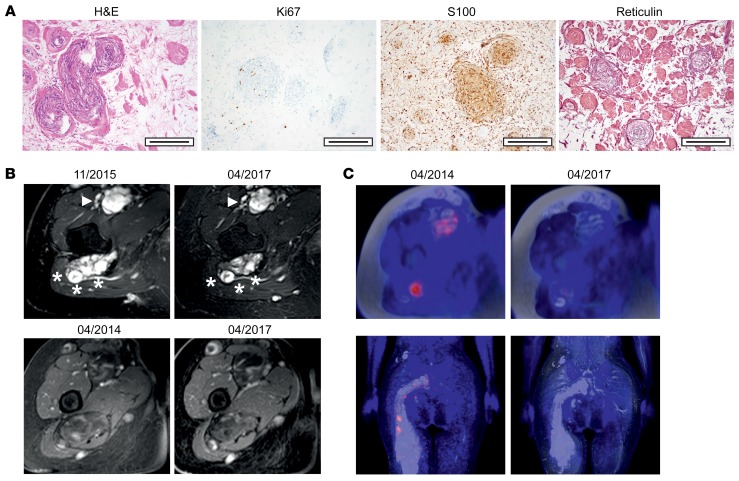
A 30-year-old woman presented with pain in her right leg in 2004. Magnetic resonance imaging (MRI) showed a large tumor mass comprising multiple nodules located around peripheral nerves in the lower right body quadrant, including lumbar and sacral spinal nerve roots, the right lumbosacral plexus, and the right femoral and sciatic nerves. Additional nodules were present in the abdomen and the left leg. Partial resection of nodules in the ventral compartment of the upper right thigh and the right gluteal region was performed in 2005, and a diagnosis of N/S HNST was established. There were no clinical signs of NF2, such as vestibular schwannomas, or dermatologic abnormalities, and the patient fulfilled the clinical criteria of sporadic schwannomatosis (10) with no family history suggestive of a hereditary tumor predisposition syndrome. Over time, the tumor slowly progressed (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI130787DS1), and partial resection of a large tumor mass around the right lateral femoral cutaneous nerve was performed in 2011. Hematoxylin and eosin (H&E) staining showed an encapsulated tumor with compact, cell-dense areas as well as areas with scattered, spindle-shaped cells with filament fragments. Focally, a slight lymphoid infiltrate was detectable in perivascular regions. The tumor cells displayed strong expression of S100 and slight expression of reticulin in a punctuated pattern around balloon-shaped swirls. The proliferation rate was low (average, 1%; range, 0%–3%) (Figure 1A).
Figure 1. Histologic appearance, tumor burden, and assessment of response to lapatinib in a patient with N/S HNST.
(A) H&E, S100, reticulin, and Ki67 staining. Scale bars: 200 μm. (B) Axial MRI studies showing decreased tumor masses over time. Upper panel, T2-weighted and fat-suppressed images of hyperintense tumors along the right sciatic (asterisks) and femoral (arrowhead) nerves; lower panel, T1-weighted, contrast-enhanced, and fat-suppressed images of slightly and inhomogeneously contrast-enhancing tumors along the right sciatic and femoral nerves. (C) FDG-PET/MRI studies showing decreased tracer uptake over time. Upper row, axial images of tumors along the right sciatic and femoral nerves; lower row, coronal images of tumors along the right sciatic nerve.
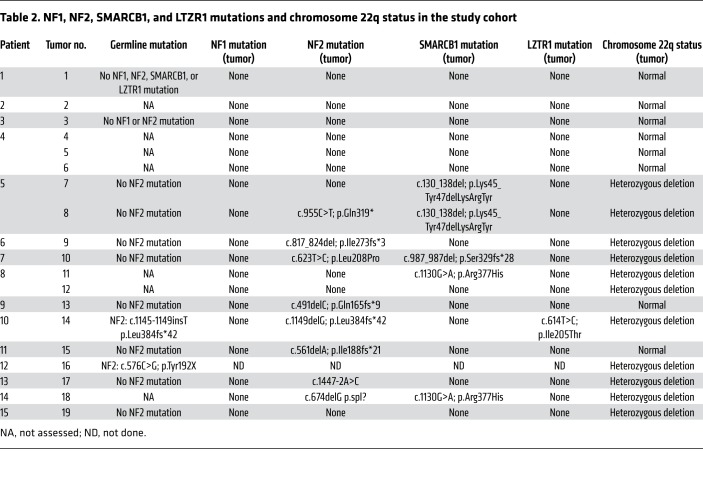
Due to progressive pareses and intense neuropathic pain, and because no further established therapy was available, the patient was enrolled in the NCT MASTER (Molecularly Aided Stratification for Tumor Eradication Research) program, a nationwide registry trial for genomics-guided stratification of younger adults with advanced-stage cancer across all histologies and patients with rare tumors who have exhausted standard treatment options (11). Whole-exome sequencing of tumor tissue and matched peripheral blood mononuclear cells showed a quiet genomic profile with only 2 somatic missense mutations and no DNA copy number alterations. Of particular clinical interest was a heterozygous ERBB2 p.Asp769Tyr mutation (variant allele frequency, 16%), which was confirmed by Sanger sequencing both in the initial specimen and a subsequent biopsy from a different tumor nodule, and whose expression was verified by RNA sequencing. Analysis of germline sequences (Supplemental Table 1) identified no pathogenic variants in established tumor predisposition genes, including NF1, NF2, SMARCB1, and LZTR1.
ERBB2 p.Asp769Tyr has been identified as a rare mutation in a range of carcinomas, in particular breast invasive ductal carcinoma (http://www.cbioportal.org), and studies in MCF10A human mammary epithelial cells have shown that this variant causes constitutive activation of the ERBB2 receptor tyrosine kinase (RTK) and confers sensitivity to ERBB2-directed antibodies and small-molecule tyrosine kinase inhibitors (12). Given these findings and the lack of therapeutic alternatives, treatment with 1500 mg lapatinib daily was initiated in October 2015 and has since then been well tolerated. Prior to lapatinib treatment, the patient had bilateral paresis of foot and toe elevation (2/5 on the right and 4/5 on the left side according to the British Medical Research Council Scale) with corresponding gait impairment. There was numbness of the right foot and lower leg as well as the medial part of the right thigh. Severe neuropathic pain (5–6/10 on a numeric rating scale) was present at all times despite treatment with gabapentin and nortriptyline and caused frequent insomnia. After 6 months of therapy, pareses had improved to 4+/5 on the right and 4+/5 on the left side and gait was no longer impaired. Furthermore, neuropathic pain had improved (1–2/10 on a visual analog scale) and was only present on 1–2 days per week for brief periods, and sleep was rarely affected. In contrast, the numbness of the right leg remained unchanged. As of October 2019, the patient was still on lapatinib, and the improvements of motor strength and neuropathic pain, which had begun as early as 4 weeks after initiation of therapy, were lasting. Interestingly, a 3-week interruption of lapatinib due to paronychia of the right hallux caused an increase in neuropathic pain, which subsided after reintroduction of therapy, indicating continued efficacy of ERBB2 blockade. Clinical improvement was paralleled by a reduction in size of some lesions (Figure 1B and Supplemental Figure 2, A–C) as well as a lasting metabolic response on sequential fluorodeoxyglucose-positron emission tomography (FDG-PET)/MRI studies (Figure 1C and Supplemental Figure 2B). Together, these results uncover mutant ERBB2 as a potentially novel driver of N/S HNST that provides an immediate therapeutic opportunity.
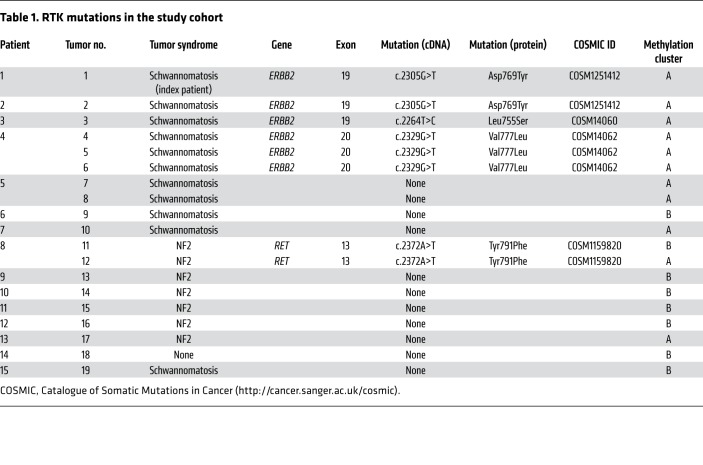
To investigate the frequency of mutations in ERBB2 or other RTKs in N/S HNSTs, we collected 18 additional tumors from 14 additional patients (Supplemental Table 2). This cohort included 7 patients that met clinical diagnostic criteria for sporadic schwannomatosis, 2 of which had unusual clinical characteristics in addition to peripheral nerve sheath tumors (patient 3: marfanoid habitus, scoliosis, bilateral acetabular protrusions, mitral valve prolapse, dilated ascending aorta, history of retrognathia and juvenile glaucoma; patient 4: generalized epileptic seizures, dermal neurofibromas), 6 patients with NF2, and 1 patient without a tumor predisposition syndrome (Table 1).
Table 1. RTK mutations in the study cohort.
Employing targeted next-generation sequencing, we detected ERBB2 mutations (p.Leu755Ser, p.Asp769Tyr, p.Val777Leu) in 3 of 7 (43%) schwannomatosis patients, whereas no ERBB2 mutations were found in NF2 or sporadic N/S HNST cases, resulting in a mutation frequency of 27% (4 of 15 patients) in the entire cohort including the index patient (Table 1). These kinase domain mutations had previously been found in breast cancer and characterized as activating and sensitive to pan-ERBB inhibitors such as neratinib (12). Of the 6 patients with NF2, 1 (17%) had tumors that harbored a heterozygous RET p.Tyr791Phe mutation (Table 1), which has been identified in various cancers and characterized as activating, although its cancer-driving role has been disputed (13). Histologic analysis revealed no phenotypic differences compared with samples without RTK alterations (Supplemental Figure 3).
To delineate potential additional steps of tumorigenesis and corroborate the clinical diagnoses, we analyzed the sequencing data and genome-wide DNA methylation profiles of all tumors for mutations in NF1, NF2, SMARCB1, or LZTR1, and for loss of heterozygosity of chromosome 22q, where NF2 is located, respectively (Table 2), and observed that ERBB2 mutations were mutually exclusive with alterations of NF2, SMARCB1, or LZTR1 and loss of heterozygosity of chromosome 22q. No NF1 mutation was detected in the study cohort (Table 2). Of note, both RET-mutant tumors from a patient who had been diagnosed with NF2 lacked NF2 alterations, whereas the mutational profiles of all other tumors were consistent with the respective clinical diagnoses. Collectively, these results identify activating ERRB2 mutations as recurrent events in N/S HNST arising in schwannomatosis patients, and suggest that this tumor entity may be driven by aberrant ERBB2 signaling in a substantial proportion of cases.
Table 2. NF1, NF2, SMARCB1, and LTZR1 mutations and chromosome 22q status in the study cohort.
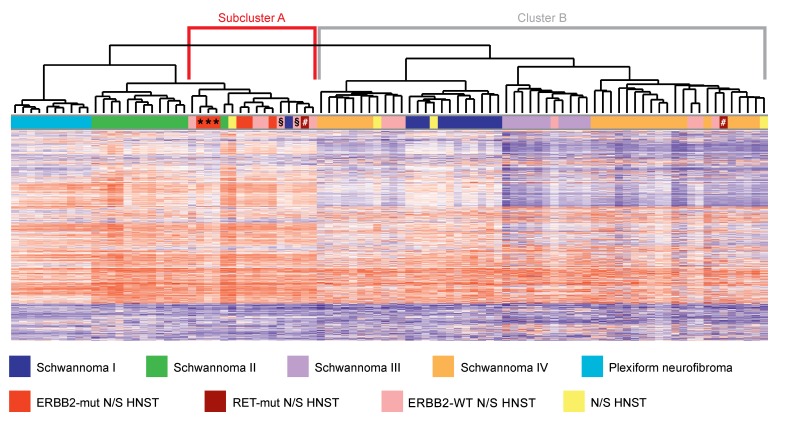
DNA methylation profiling has emerged as a powerful tool to distinguish biologically distinct tumor entities (14), and a recent analysis of peripheral nerve sheath tumors revealed several clinically important methylation subgroups (15). DNA methylation-based classification of the index tumor yielded high classifier scores for the subgroups “melanocytic schwannoma” and “regular schwannoma”, indicating a potential relationship with these entities. Unsupervised hierarchical clustering of the DNA methylation profiles of our cohort of 19 N/S HNSTs and 80 benign Schwann cell tumors from previously described methylation categories (15) identified a distinct subcluster of tumors (Figure 2, Subcluster A) that contained the majority of N/S HNSTs associated with sporadic schwannomatosis (7 of 9 tumors), including all ERBB2-mutant cases, whereas most N/S HNSTs associated with NF2 (5 of 7 tumors), which lack activating ERBB2 mutations, lay outside this cluster (Figure 2, Cluster B).
Figure 2. DNA methylation profiling of peripheral nerve sheath tumors.
Dendrogram showing the results of unsupervised hierarchical clustering of the DNA methylation profiles of N/S HNSTs and 80 benign Schwann cell tumors. “Schwannoma I-IV” refers to previously described methylation subgroups (15). DNA methylation levels (beta values) are represented as heatmap. mut, mutant; WT, wild-type. *, §, and # identify tumors from the same patient.
Discussion
We here report for the first time that a substantial proportion of N/S HNSTs harbor ERBB2 kinase domain mutations that have been shown in other tumor entities to induce constitutive ERBB2 signaling, promote oncogenesis, and confer sensitivity to pharmacologic ERBB2 inhibition (12, 16). Furthermore, N/S HNSTs that had occurred in a patient with NF2 harbored a RET p.Tyr791Phe variant, which has been described as activating mutation in familial medullary thyroid carcinoma (17). Together, these findings indicate that N/S HNSTs are frequently driven by aberrant RTK signaling, and consistent with the hypothesis of a distinct molecular pathogenesis, we observed that ERBB2-mutant N/S HNSTs cluster in a separate subgroup of peripheral nerve sheath tumors based on genome-wide DNA methylation analysis (Figure 2). The notion that these tumors might define a separate subgroup of N/S HNSTs is further supported by the mutual exclusivity of activating ERBB2 variants and mutations known to drive tumorigenesis in patients with schwannomatosis, NF2, and NF1 (Table 2).
In addition to providing new insights into underlying pathogenetic mechanisms, our findings may broaden the therapeutic armamentarium against N/S HNSTs. Pharmacologic treatment options for schwannomas or neurofibromas have been explored previously in the context of NF2 and NF1. In patients with NF2-associated vestibular schwannomas, both the dual EGFR/ERBB2 inhibitor lapatinib and the anti-VEGF antibody bevacizumab induced transient responses in a subset of tumors; in patients with NF1-associated plexiform neurofibromas, the MEK inhibitor selumetinib caused a decrease in tumor volume in 70% of cases (8, 9, 18, 19). The rationale to investigate the activity of lapatinib and selumetinib, respectively, was that loss of the Merlin tumor suppressor encoded by NF2 is frequently associated with activation of EGFR or ERBB2 signaling (9), and that inactivation of the RAS GTPase-activating protein encoded by NF1 stimulates the RAS-MEK-ERK pathway (18). Our finding of recurrent ERBB2 kinase domain mutations now also suggests a targeted therapeutic strategy for N/S HNSTs occurring in non-NF patients. Importantly, the efficacy of ERBB2 kinase inhibition in ERBB2-mutant cancers varies across tumor entities and mutation types, with the greatest activity seen in breast, cervical, and biliary cancers, and in tumors that contain missense variants in the catalytic domain (16). The prolonged response to lapatinib we observed in a patient with ERBB2-mutant N/S HNST is encouraging and supports prospective mutational profiling in N/S HNST patients with unresectable tumors as well as a molecularly guided clinical trial of ERBB2 inhibitors such as lapatinib, afatinib, or neratinib to establish mutant ERBB2 as valid therapeutic target in this disease and to identify modifiers of response.
Our findings may also have diagnostic implications as they indicate that the heterogeneous group of N/S HNSTs, which is currently defined based on histologic criteria alone (1), may be deciphered further using molecular analyses. ERBB2-mutant N/S HNSTs were exclusively associated with schwannomatosis, and future diagnostic algorithms may therefore incorporate the presence of an ERBB2 mutation as supportive or even core criterion (6, 10). Such additional parameters are urgently needed given that the differential diagnosis of tumor syndromes can be challenging due to overlap between entities, diverse mutations causing common phenotypes, and continuously evolving clinical criteria.
Finally, our results add to an emerging body of evidence indicating that oncogenic driver mutations can also be found in benign conditions and in premalignant lesions (20), and it will be interesting to study the impact of mutant ERBB2 signaling on the course of disease in N/S HNST patients and to identify individuals who may be candidates for preemptive therapeutic strategies.
Methods
Tumor samples.
For whole-exome and RNA sequencing of the index case, a fresh-frozen tumor specimen and matched peripheral blood mononuclear cells were collected. Samples were pseudonymized, and tumor histology and cellularity were assessed at the Institute of Pathology, Heidelberg University Hospital, prior to further processing. For targeted next-generation sequencing, sections of 18 formalin-fixed paraffin-embedded (FFPE) tumors from 14 patients were provided by the Institute of Pathology and Neuropathology, University Hospital Tübingen, and the Institute of Neurology, University Hospital Frankfurt. These tumors were evaluated by a board-certified neuropathologist at the Institute of Pathology and Neuropathology, University Hospital Tübingen, and original diagnoses were independently confirmed by another board-certified neuropathologist at the Institute of Neurology, University Hospital Frankfurt. Tumor tissue was microdissected to achieve a tumor cell content of at least 15%.
Whole-exome and RNA sequencing.
DNA and RNA from the tumor specimen and DNA from the blood sample were isolated using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen), followed by quality control and quantification using a Qubit 2.0 Fluorometer (Life Technologies), a 2200 TapeStation system (Agilent), and a 2100 Bioanalyzer system (Agilent).
Exome capturing was performed using SureSelect Human All Exon V5 in-solution capture reagents (Agilent). Briefly, 1.5 μg genomic DNA was fragmented to 150–200 bp (paired-end) insert size with a Covaris S2 device, and 250 ng Illumina adapter-containing libraries were hybridized with exome baits at 65°C for 16 hours. Paired-end sequencing (101 bp) was carried out with a HiSeq 2500 instrument (Illumina) in rapid mode. RNA sequencing libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina). Briefly, mRNA was purified from 1 μg total RNA using oligo(dT) beads, poly(A)+ RNA was fragmented to 150 bp and converted into cDNA, and cDNA fragments were end-repaired, adenylated on the 3′ end, adapter-ligated, and amplified with 12 cycles of PCR. The final libraries were validated using a Qubit 2.0 Fluorometer (Life Technologies) and a Bioanalyzer 2100 system (Agilent). Paired-end sequencing (2 × 101 bp) was carried out with a HiSeq 2500 instrument (Illumina) in rapid mode.
Exome sequencing reads were mapped to the 1000 Genomes Phase 2 assembly of the human reference genome (NCBI build 37.1) using BWA (version 0.6.2) with default parameters and maximum insert size set to 1000 bp (21). BAM files were sorted with SAMtools (version 0.1.19) (22), and duplicates were marked with Picard tools (version 1.90). Average target coverage was ×180 for the tumor and ×160 for the control. For the detection of single-nucleotide variants (SNVs), we applied an in-house analysis pipeline based on SAMtools mpileup and bcftools with parameter adjustments to allow for calling of somatic variants with heuristic filtering as previously described (23–25). After annotation with RefSeq (version September 2013) using ANNOVAR (26), somatic, nonsilent coding variants of high confidence were selected. Short insertions and deletions (indels) were identified using Platypus (version 0.5.2; parameters genIndels = 1, genSNPs = 0, ploidy = 2, nIndividuals = 2) by providing the tumor and control BAM files (27). To be of high confidence, somatic calls (control genotype 0/0) were required to either have the Platypus filter flag PASS or pass custom filters allowing for low variant frequency using a scoring scheme. Candidates with the badReads flag, alleleBias, or strandBias if the variant allele frequency was less than 10%, or more than 2 of the remaining flags were discarded. Additionally, combinations of Platypus non-PASS filter flags, bad quality values, low genotype quality, very low variant counts in the tumor, and presence of variant reads in the control were not tolerated. Indels were annotated with ANNOVAR, and somatic high-confidence indels falling into a coding sequence or splice site were extracted. Copy number variants were analyzed by read-depth plots and an in-house pipeline using the VarScan2 copynumber and copyCaller modules (28). Regions were filtered for unmappable genomic stretches, merged by requiring at least 70 markers per called copy number event, and annotated with RefSeq genes using BEDtools (29).
RNA sequencing reads were mapped with STAR (version 2.3.0e) (30). For building the index, the 1000 Genomes reference sequence with GENCODE version 17 transcript annotations were used. For alignment, the following parameters were used: alignIntronMax 500000, alignMatesGapMax 500000, outSAMunmapped Within, outFilterMultimapNmax 1, outFilterMismatchNmax 3, outFilterMismatchNoverLmax 0.3, sjdbOverhang 50, chimSegmentMin 15, chimScoreMin 1, chimScoreJunctionNonGTAG 0, chimJunctionOverhangMin 15. The output was converted to sorted BAM files with SAMtools, and duplicates were marked with Picard tools (version 1.90).
Expression levels were determined per gene and sample as reads per kilobase (kb) of exon model per million mapped reads, and RefSeq was used as gene model. For each gene, overlapping annotated exons from all transcript variants were merged into nonredundant exon units with a custom Perl script. Nonduplicate reads with mapping quality above zero were counted for all exon units with coverageBed from the BEDtools package. Read counts were summarized per gene and divided by the combined length of its exon units (in kb), and the total number of reads (in millions) was counted by coverageBed. SNVs and indels were annotated with RNA information by generating a pileup of the DNA variant position in the RNA BAM file with SAMtools.
Hybrid capture-based panel sequencing.
DNA was extracted using a Maxwell 16 Research System (Promega), followed by quantification using the QuBit 2.0 DNA High Sensitivity Kit (Thermo Fisher Scientific). Library preparation for the capture-based TruSight Oncology 500 panel (Illumina) (Supplemental Table 3)was performed as previously described (31). DNA integrity was assessed using the Genomic DNA ScreenTape Analysis on a 4150 TapeStation System (Agilent). To fragment DNA to a length of 90–250 bp, 80 ng DNA was sheared for 50–78 seconds using an ME220 Focused-Ultrasonicator (Covaris). Following target capture and purification steps, enriched libraries were amplified by 15 cycles of PCR and subsequently quality controlled using the KAPA SYBR Library Quantification Kit (Thermo Fisher Scientific) on a StepOnePlus qPCR system (Thermo Fisher Scientific). Libraries were sequenced on a NextSeq 500 instrument (Illumina) to a mean coverage of ×1096 using high-output cartridge and v2 chemistry. All assays were performed according to the manufacturers’ protocols.
Processing of raw sequencing data and variant calling was carried out using the TruSight Oncology 500 Local App (version 1.3.0.39). Called variants were verified by visual inspection in the Integrative Genomics Viewer (32). Only variants with an allele frequency above 5% and a minimum coverage of greater than ×100 were taken into account.
DNA methylation profiling.
Tumor DNA was analyzed using Infinium Human Methylation 450K BeadChip or Infinium MethylationEPIC BeadChip arrays (Illumina) at the DKFZ Genomics and Proteomics Core Facility. Data were processed with the R/Bioconductor package minfi (version 1.20).
Immunohistochemistry.
Immunohistochemical stainings were performed on FFPE whole-tissue slides using the following primary antibodies: S100 (polyclonal rabbit, 1:5000, Dako), Ki67 (monoclonal mouse, clone MIB-1, 1:200, Dako) and reticulin (Reticulum II Staining Kit, Ventana).
Sequencing data deposition.
Sequencing data were deposited in the European Genome-Phenome Archive under accession EGAS00001003776.
Statistics.
For unsupervised hierarchical clustering, the 25,000 probes with the highest median absolute deviation (MAD) across the beta values of 80 benign Schwann cell tumors (15) were selected. Samples were hierarchically clustered using Euclidean distance and Ward’s linkage method. Filtering and genome-wide DNA copy number analyses were performed using the R/Bioconductor package conumee as described (33).
Study approval.
Tissue samples were used in accordance with the regulations of the respective tissue bank and after approval by the ethics committees of Heidelberg University (Ethik-Kommission, Medical Faculty Heidelberg, Heidelberg, Germany) and Frankfurt University (Ethik-Kommission, University Hospital Frankfurt, Goethe University, Frankfurt, Germany) as well as the Neuro-Oncology Biobank Board Tübingen (Tübingen, Germany) (protocol numbers S-206/2011, SNO-05-2017, and ZNO32, respectively). Written informed consent was obtained prior to the study. Research was conducted in accordance with the Declaration of Helsinki.
Author contributions
MWR, PNH, JPS, DER, AS, and SF conceived and designed the study. MWR, PNH, MK, CH, BH, LG, CHB, ES, WW, BB, AVD, JPS, DER, HG, AS, and SF developed methodology. MWR, PNH, MK, CH, VE, WW, DER, HG, AS, and SF acquired data. MWR, PNH, MK, CH, BH, LG, JS, MUS, MT, GM, MW, VFM, ES, WW, BB, AVD, MM, DER, HG, AS, and SF analyzed and interpreted data. MWR, AS, and SF wrote the manuscript with approval from all other authors.
Supplementary Material
Acknowledgments
The authors thank the DKFZ-HIPO Sample Processing Laboratory and the DKFZ Genomics and Proteomics Core Facility for technical support. We also thank Katja Beck, Viktoria Fischer, Karolin Willmund, and Peter Lichter for infrastructure and program development within DKFZ-HIPO. FDG-PET/MRI studies were performed at the Department of Diagnostic and Interventional Radiology, University Hospital Tübingen, Tübingen, Germany. The authors thank Sergios Gatidis for critical review of the manuscript. MM is supported by grant FNR PEARL P16/BM/11192868 from the Luxembourg National Research Fond. This work was supported by grant H021 from DKFZ-HIPO. MWR and PNH received fellowships from University Cancer Center Frankfurt, and MWR received funding from the Frankfurt Research Funding Clinician Scientist Program.
Version 1. 02/04/2020
In-Press Preview
Version 2. 04/13/2020
Electronic publication
Version 3. 05/01/2020
Print issue publication
Funding Statement
This work was supported by grant H021 from DKFZ-HIPO (Heidelberg Center for Personalized Oncology).
Michael W. Ronellenfitsch and Patrick N. Harter have received fellowships from UCT Frankfurt
Michael W. Ronellenfitsch has received support from the Clinician Scientist Program.
Footnotes
Conflict of interest: MWR has received research funding from UCB. VE has collected personal fees from AstraZeneca, Merck Sharp & Dohme, Novartis, and Thermo Fisher Scientific, with other support from Illumina. WW has collected personal fees from Amgen, Astellas, AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Eli Lilly, Merck, Merck Sharp & Dohme, Novartis, Pfizer, Roche, and Takeda, as well as research funding from Bristol-Myers Squibb, Bruker, Merck Sharp & Dohme, and Roche. AVD and DR have licensed an NF1 antibody to Cell Marque. JPS has a consulting or advisory board membership with, or has received honoraria or travel or accommodation expenses from Abbvie, Medac, Novocure, Roche, and UCB. AS has consulting or advisory board membership with AstraZeneca, Bayer, Bristol-Myers Squibb, Illumina, Novartis, Seattle Genomics, Takeda, and Thermo Fisher Scientific; received honoraria from AstraZeneca, Bayer, Bristol-Myers Squibb, Illumina, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Seattle Genomics, Takeda, and Thermo Fisher Scientific; and received research funding from Bayer, Bristol-Myers Squibb, and Chugai. SF has a consulting or advisory board membership with Bayer and Roche; received honoraria from Amgen, Eli Lilly, PharmaMar, and Roche; received research funding from AstraZeneca, Pfizer, and PharmaMar; and received travel or accommodation expenses from Amgen, Eli Lilly, PharmaMar, and Roche.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(5):2488–2495.https://doi.org/10.1172/JCI130787.
Contributor Information
Michael W. Ronellenfitsch, Email: m.ronellenfitsch@gmx.net.
Patrick N. Harter, Email: patrick.harter@kgu.de.
Martina Kirchner, Email: Martina.Kirchner@med.uni-heidelberg.de.
Christoph Heining, Email: christoph.heining@nct-dresden.de.
Barbara Hutter, Email: b.hutter@dkfz-heidelberg.de.
Laura Gieldon, Email: Laura.Gieldon@uniklinikum-dresden.de.
Jens Schittenhelm, Email: Jens.Schittenhelm@med.uni-tuebingen.de.
Martin U. Schuhmann, Email: martin.schuhmann@med.uni-tuebingen.de.
Marcos Tatagiba, Email: marcos.tatagiba@med.uni-tuebingen.de.
Gerhard Marquardt, Email: Gerhard.Marquardt@kgu.de.
Marlies Wagner, Email: Marlies.Wagner@kgu.de.
Volker Endris, Email: Volker.Endris@med.uni-heidelberg.de.
Christian H. Brandts, Email: Christian.Brandts@kgu.de.
Victor-Felix Mautner, Email: v.mautner@uke.de.
Evelin Schröck, Email: evelin.schrock@tu-dresden.de.
Wilko Weichert, Email: wilko.weichert@tum.de.
Benedikt Brors, Email: b.brors@dkfz.de.
Andreas von Deimling, Email: andreas.vondeimling@med.uni-heidelberg.de.
Michel Mittelbronn, Email: Michel.MITTELBRONN@lns.etat.lu.
Joachim P. Steinbach, Email: joachim.steinbach@med.uni-frankfurt.de.
David E. Reuss, Email: david.reuss@med.uni-heidelberg.de.
Hanno Glimm, Email: hanno.glimm@nct-dresden.de.
Albrecht Stenzinger, Email: Albrecht.Stenzinger@med.uni-heidelberg.de.
Stefan Fröhling, Email: stefan.froehling@nct-heidelberg.de.
References
- 1.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.McClatchey AI. Neurofibromatosis. Annu Rev Pathol. 2007;2:191–216. doi: 10.1146/annurev.pathol.2.010506.091940. [DOI] [PubMed] [Google Scholar]
- 3.Feany MB, Anthony DC, Fletcher CD. Nerve sheath tumours with hybrid features of neurofibroma and schwannoma: a conceptual challenge. Histopathology. 1998;32(5):405–410. doi: 10.1046/j.1365-2559.1998.00419.x. [DOI] [PubMed] [Google Scholar]
- 4.Harder A, et al. Hybrid neurofibroma/schwannoma is overrepresented among schwannomatosis and neurofibromatosis patients. Am J Surg Pathol. 2012;36(5):702–709. doi: 10.1097/PAS.0b013e31824d3155. [DOI] [PubMed] [Google Scholar]
- 5.Montgomery BK, et al. Tumors displaying hybrid schwannoma and neurofibroma features in patients with neurofibromatosis type 2. Clin Neuropathol. 2016;35(2):78–83. doi: 10.5414/NP300895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plotkin SR, Wick A. Neurofibromatosis and Schwannomatosis. Semin Neurol. 2018;38(1):73–85. doi: 10.1055/s-0038-1627471. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich RE, Diekmeier C. Peripheral nerve sheath tumors of the upper extremity and hand in patients with neurofibromatosis type 1: topography of tumors and evaluation of surgical treatment in 62 patients. GMS Interdiscip Plast Reconstr Surg DGPW. 2017;6:Doc15. doi: 10.3205/iprs000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mautner VF, et al. Bevacizumab induces regression of vestibular schwannomas in patients with neurofibromatosis type 2. Neuro-oncology. 2010;12(1):14–18. doi: 10.1093/neuonc/nop010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karajannis MA, et al. Phase II trial of lapatinib in adult and pediatric patients with neurofibromatosis type 2 and progressive vestibular schwannomas. Neuro-oncology. 2012;14(9):1163–1170. doi: 10.1093/neuonc/nos146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plotkin SR, et al. Update from the 2011 International Schwannomatosis Workshop: From genetics to diagnostic criteria. Am J Med Genet A. 2013;161A(3):405–416. doi: 10.1002/ajmg.a.35760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horak P, et al. Precision oncology based on omics data: The NCT Heidelberg experience. Int J Cancer. 2017;141(5):877–886. doi: 10.1002/ijc.30828. [DOI] [PubMed] [Google Scholar]
- 12.Bose R, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvorakova S, et al. Somatic mutations in the RET proto-oncogene in sporadic medullary thyroid carcinomas. Mol Cell Endocrinol. 2008;284(1–2):21–27. doi: 10.1016/j.mce.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Capper D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Röhrich M, et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 2016;131(6):877–887. doi: 10.1007/s00401-016-1540-6. [DOI] [PubMed] [Google Scholar]
- 16.Hyman DM, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554(7691):189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plaza Menacho I, et al. RET-familial medullary thyroid carcinoma mutants Y791F and S891A activate a Src/JAK/STAT3 pathway, independent of glial cell line-derived neurotrophic factor. Cancer Res. 2005;65(5):1729–1737. doi: 10.1158/0008-5472.CAN-04-2363. [DOI] [PubMed] [Google Scholar]
- 18.Dombi E, et al. Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N Engl J Med. 2016;375(26):2550–2560. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plotkin SR, et al. Hearing improvement after bevacizumab in patients with neurofibromatosis type 2. N Engl J Med. 2009;361(4):358–367. doi: 10.1056/NEJMoa0902579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato S, Lippman SM, Flaherty KT, Kurzrock R. The conundrum of genetic “drivers” in benign conditions. J Natl Cancer Inst. 2016;108(8):djw036. doi: 10.1093/jnci/djw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, et al. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaktapour N, et al. BRAF inhibitor-associated ERK activation drives development of chronic lymphocytic leukemia. J Clin Invest. 2014;124(11):5074–5084. doi: 10.1172/JCI76539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DT, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46(8):912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazdal D, et al. Spatial and temporal heterogeneity of panel-based tumor mutational burden in pulmonary adenocarcinoma: separating biology from technical artifacts. J Thorac Oncol. 2019;14(11):1935–1947. doi: 10.1016/j.jtho.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.