Abstract
The pandemic coronavirus infectious disease (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is rapidly spreading across the globe. In this issue of the JCI, Chen and colleagues compared the clinical and immunological characteristics between moderate and severe COVID-19. The authors found that respiratory distress on admission is associated with unfavorable outcomes. Increased cytokine levels (IL-6, IL-10, and TNF-α), lymphopenia (in CD4+ and CD8+ T cells), and decreased IFN-γ expression in CD4+ T cells are associated with severe COVID-19. Overall, this study characterized the cytokine storm in severe COVID-19 and provides insights into immune therapeutics and vaccine design.
Keywords: COVID-19
A rapidly expanding pandemic
In early December 2019, several local health facilities first reported pneumonia cases of unknown origin in Wuhan, China. This new coronavirus infectious disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported on December 1, 2020 and identified as a previously unknown betacoronavirus (1, 2). Since then, studies have increasingly demonstrated that SARS-CoV-2 can be transmitted effectively among humans through aerosol or fomites (3–5). With transmission capabilities even before symptom onset (6, 7), this pandemic is rapidly evolving and expanding. As of April 2, 2020, a total of 962,977 confirmed cases of COVID-19 and 49,180 deaths have been reported across 180 countries and regions. Recent models indicated that as much as 86% of all infections early in the spread were undocumented, suggesting the real number of infections is likely much greater (8). As a public health emergency of international concern, COVID-19 was declared a pandemic by the World Health Organization. Public health authorities around the globe are now racing to contain the spread.
SARS-CoV-2: not a common cold
Coronaviruses are a family of enveloped RNA viruses broadly distributed in humans and animals that cause acute and chronic diseases. Of the six coronavirus species previously known to cause human diseases, four typically cause common cold symptoms and two — SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) — can cause fatal respiratory disease (9, 10). SARS-CoV-2 shares many similarities with SARS-CoV, which caused a global epidemic in 2002–2003, affecting more than 8,000 patients and causing 774 deaths in 26 countries (9). SARS-CoV-2 is closely related to two bat-derived SARS-like coronaviruses and SARS-CoV in phylogenetic distance (11). SARS-CoV-2 uses the same cellular receptor as SARS-CoV, angiotensin-converting enzyme 2 (ACE2) (12), suggesting a similar tropism and route of entry.
Although the majority of SARS-CoV-2–infected individuals may have no or mild symptoms, SARS-CoV-2 infection is not simply a common cold. Despite similarities between SARS-CoV and SARS-CoV-2, SARS-CoV-2 is causing a global pandemic, while SARS-CoV infection was limited to contained geographic regions. There are other notable differences as well. First, SARS-CoV-2 is infectious before symptom onset (6, 7), while SARS-CoV has no reported instance of transmission before the onset of symptoms (9). Second, up to 56% of SARS-CoV-2–infected individuals present without fever even on hospital admission (13), further increasing the difficulty of proactive entry health screening and containment. Additionally, the crude mortality rate of SARS-CoV-2 infection is estimated at 3%–4%, which is remarkably higher than that of influenza infection (typically well below 0.1%) (14). Therefore, drastic quarantine measures have been implemented in multiple countries, including a record-breaking scale of travel restrictions within communities and between countries.
Eight days after symptom onset
Shortly after the outbreak, health care systems in affected areas are experiencing overwhelming demand for patient care. Even after suspending most noncritical medical care, the skyrocketing increase in COVID-19 cases has paralyzed the health care system even in developed countries. As the magnitude of the outbreak exceeds health care capacities, point-of-care indicators that can distinguish severe versus moderate disease for immediate triage and clinical disposition have become critically important to prioritize medical resources.
In this issue of the JCI, Chen et al. (15) compared clinical presentations of a total of 21 patients. These patients are older (average 56 years) with comorbidities (33%) and late presentations (~8 days after symptom onset). Chen et al. (15) categorized the disease severity as severe versus moderate, defined by the presence of respiratory distress at presentation: peripheral capillary oxygen saturation (SpO2) of 93% or lower or respiratory rate of 30 breaths per minute or greater under ambient air, in line with the parameters outlined in the Chinese 6th edition guidelines. Chen et al. (15) found that the presence of respiratory distress on admission is associated with leukocytosis and increased inflammatory indicators (such as D-dimer and procalcitonin). These laboratory changes have been previously reported in COVID-19 patients requiring intensive care unit (ICU) admissions (1). Of note, the clinically confirmed cases presented in Chen et al. (15) had fever, respiratory symptoms, laboratory-confirmed SARS-CoV-2 infection, radiologically confirmed pneumonia (by chest CT) and severity requiring hospital admission. Therefore, the clinical characteristics presented in this study represent the more severe end of confirmed COVID-19 cases with respiratory distress and pneumonia, which are not the same as mild or asymptomatic cases identified through contact tracing. Nevertheless, a simple emergency room triage using respiratory rate and pulse oximetry in high-risk COVID-19 patients approximately 8 days after symptom onset is correlated with an unfavorable outcome that likely requires mechanical ventilation and ICU admission.
Associations with disease severity
The two coronaviruses that have been known to cause fatal pneumonia, SARS-CoV and MERS-CoV, are associated with rapid virus replication, an elevation of proinflammatory cytokines, and inflammatory cell infiltration (16). In SARS-CoV infection, it was immune dysregulation, rather than the level of peak viremia, that induced an insufficient type I interferon (IFN) response (too little and too late), aberrant proinflammatory cytokine secretion by alveolar macrophages, and subsequent CD4+ and CD8+ T cell dysfunction (17). Little is known about the determinants of disease severity and immune dysregulation in SARS-CoV-2 infection in humans. The study by Chen et al. (15) provides a window for us to examine the immune dysregulation at 7–10 days of onset, when the cytokine storm rages and the respiratory distress peaks.
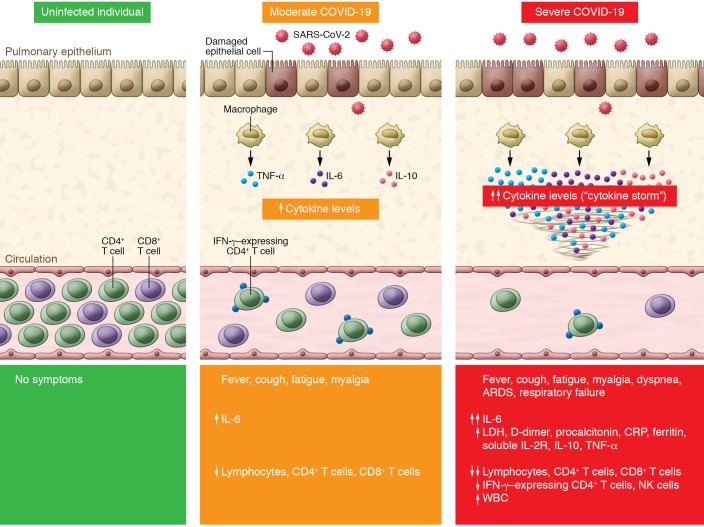
Cytokines and chemokines have long been understood to have an important role in immunity and immunopathology, but dysregulated and exuberant immune responses have been shown to potentially cause lung damage and diminished survival. In SARS-CoV-2–infected individuals, interleukin 6 (IL-6), IL-10, and tumor necrosis factor α (TNF-α) surge during illness and decline during recovery. Patients requiring ICU admission have significantly higher levels of IL-6, IL-10, and TNF-α and fewer CD4+ and CD8+ T cells (18). Further, the level of IL-6, IL-10, and TNF-α inversely correlates with CD4+ and CD8+ T cell count (18), confirming previous animal studies showing that it is the cytokine storm that dampens adaptive immunity against SARS-CoV infection (17). In this issue of the JCI, Chen and colleagues (15) present one of the first studies comparing the immunological characteristics between severe and moderate COVID-19. Despite an increase in white blood cell count, CD4+ and CD8+ T cell counts were significantly decreased (P = 0.018 and 0.035, respectively) in severe COVID-19 patients. Serum cytokine levels and analysis of lymphocyte composition on admission suggest that SARS-CoV-2 infection is associated with lymphopenia (particularly in CD4+ T cells and CD8+ T cells but not in B cells), overproduction of cytokines (IL-6, soluble IL-2 receptor [IL-2R], IL-10, and TNF-α), and decreased IFN-γ expression in CD4+ T cells in severe COVID-19, which correlated with disease severity of COVID-19 (Figure 1). Levels of IL-6, IL-2R, IL-10, and TNF-α were mildly elevated or within the normal range in moderate cases, but markedly elevated in most of the severe cases. These cytokines are likely produced by highly inflammatory macrophages that have been implicated in cytokine storm (19). The total lymphocyte counts, and specifically CD4+ T cells and CD8+ T cells, were slightly lower in moderate cases and significantly decreased in severe COVID-19 (15). Most importantly, the authors found that IFN-γ expression in CD4+ T cells was lower (although not statistically significant, P = 0.063) in patients with severe versus moderate COVID-19. This study demonstrates that cytokine storm is associated with COVID-19 disease severity, likely through increased pulmonary pathology, T cell depletion, and CD4+ T cell dysfunction. These findings are in line with those of Diao and colleagues, which suggested that in addition to reduction in T cell counts, surviving T cells appear functionally exhausted (18).
Figure 1. Cytokine storm and T cell lymphopenia is associated with COVID-19 severity.
SARS-CoV-2 infection causes COVID-19. Compared with uninfected individuals (left panel), moderate COVID-19 cases exhibit an increase in IL-6 and a decrease in total T lymphocyte counts, particularly CD4+ T cells and CD8+ T cells (middle panel). Severe COVID-19 cases have further increased production of IL-6, IL-2R, IL-10, and TNF-α, while total T lymphocytes, particularly CD4+ T cells and CD8+ T cells, and IFN-γ–expressing CD4+ T cells markedly decrease (right panel). The level of cytokine storm and T cell lymphopenia is associated with pulmonary damage, respiratory distress, and unfavorable outcome. ARDS, acute respiratory distress syndrome; CRP, C-reactive protein; LDH, lactate dehydrogenase.
Study limitations
Studying the immunological features of COVID-19 presents new challenges and limitations. The statistical power of this study is severely hindered by the small sample size. Further, this study compares clinical presentations in moderate versus severe cases in high-risk populations and fails to present the clinical spectrum of the majority of SARS-CoV-2 infections, which is mild or asymptomatic, especially in young and healthy populations (15). Prospective studies that examine both innate and adaptive immune cell subsets and functions at different time points, such as early during the asymptomatic viremic stage, during respiratory distress, and after recovery, may identify mechanisms driving immune dysregulation in patients who have different disease susceptibilities, such as children versus the elderly.
Taken together, the results show that SARS-CoV-2–induced cytokine storm is associated with disease severity and outcome. Understanding the immune dysregulation in patients with COVID-19 not only provides a greater understanding of SARS-CoV-2 pathogenesis but also identifies targets for immune therapeutics. Although antiviral agents are currently being explored, the use of antiviral agents alone may not be sufficient to stop the cytokine storm, pulmonary destruction, and respiratory distress in patients who presented late after infection. Targeted immunomodulation that reduces the cytokine storm may ameliorate pulmonary inflammation and hopefully reduce mortality. Further studies on viral factors driving immune dysregulation may provide insights into shaping vaccine responses toward protective immunity.
Acknowledgments
The authors are supported by research funding from Yale Top Scholar, a Rudolf J. Anderson Fellowship, NIH grants R01 AI141009, R61 DA047037, R01 AI147868, UM1 AI126620 BEAT-HIV Delaney Collaboratory, and P50 AI150464 Center for the Structural Biology of Cellular Host Elements in Egress, Trafficking, and Assembly of HIV (CHEETAH), a Gilead HIV Research Scholar Grant, and the American Foundation for AIDS Research (amfAR).
Version 1. 03/27/2020
In-Press Preview
Version 2. 04/13/2020
Electronic publication
Version 3. 05/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(5):2202–2205. https://doi.org/10.1172/JCI137647.
See the related article at Clinical and immunological features of severe and moderate coronavirus disease 2019.
Contributor Information
Savannah F. Pedersen, Email: savannah.pedersen@yale.edu.
Ya-Chi Ho, Email: ya-chi.ho@yale.edu.
References
- 1.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. doi: 10.1056/nejmc2004973. van Doremalen N, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1 [published online March 17, 2020]. N Engl J Med . [DOI] [PMC free article] [PubMed]
- 6.Rothe C, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. doi: 10.1001/jama.2020.2565. Bai Y, et al. Presumed asymptomatic carrier transmission of COVID-19 [published online February 21, 2020]. Jama . [DOI] [PMC free article] [PubMed]
- 8. doi: 10.1126/science.abb3221. Li R, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) [published online March 16, 2020]. Science . [DOI] [PMC free article] [PubMed]
- 9.Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349(25):2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 10.Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. doi: 10.1126/science.abb2762. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2 [published online March 4, 2020]. Science . [DOI] [PMC free article] [PubMed]
- 13. doi: 10.1056/nejmoa2002032. Guan WJ, et al. Clinical characteristics of coronavirus disease 2019 in China [published online February 28, 2020]. N Engl J Med . [DOI] [PMC free article] [PubMed]
- 14. WHO. Coronavirus disease 2019 (COVID-19) Situation Report – 46. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200306-sitrep-46-covid-19.pdf Updated March 6, 2020. Accessed March 27, 2020.
- 15.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Channappanavar R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. doi: 10.1101/2020.02.18.20024364. Diao B, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). medRxiv. 2020:2020.02.18.20024364. Posted on medRxiv February 20, 2020. [DOI] [PMC free article] [PubMed]
- 19. doi: 10.1101/2020.02.23.20026690. Liao M, et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing [preprint]. Posted on medRxiv February 26, 2020. [DOI]