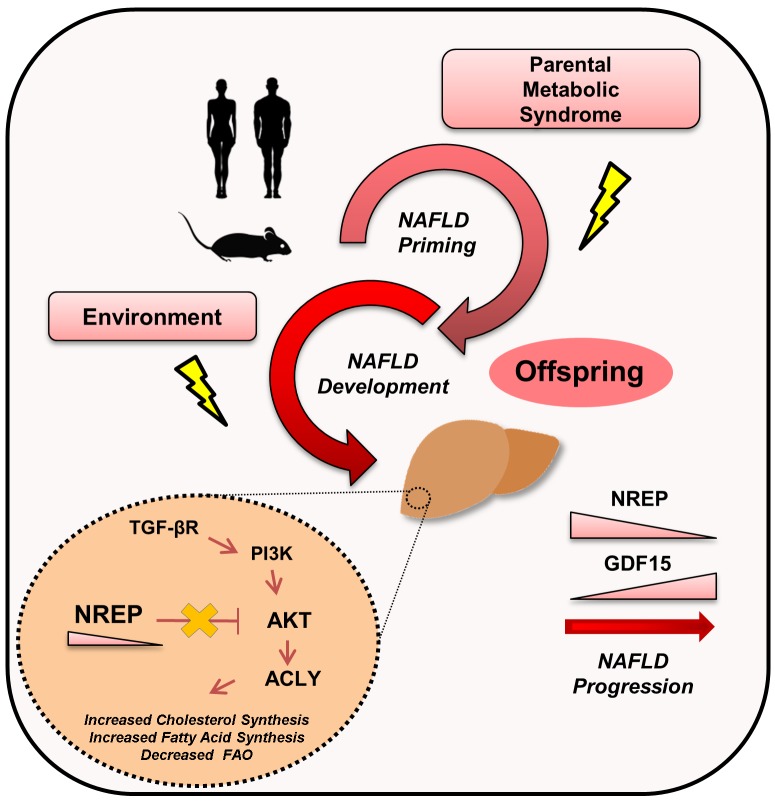
Abstract
The prevalence of nonalcoholic fatty liver disease (NAFLD) is increasing worldwide. Although gene-environment interactions have been implicated in the etiology of several disorders, the impact of paternal and/or maternal metabolic syndrome on the clinical phenotypes of offspring and the underlying genetic and epigenetic contributors of NAFLD have not been fully explored. To this end, we used the liver-specific insulin receptor knockout (LIRKO) mouse, a unique nondietary model manifesting 3 hallmarks that confer high risk for the development of NAFLD: hyperglycemia, insulin resistance, and dyslipidemia. We report that parental metabolic syndrome epigenetically reprograms members of the TGF-β family, including neuronal regeneration–related protein (NREP) and growth differentiation factor 15 (GDF15). NREP and GDF15 modulate the expression of several genes involved in the regulation of hepatic lipid metabolism. In particular, NREP downregulation increases the protein abundance of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and ATP-citrate lyase (ACLY) in a TGF-β receptor/PI3K/protein kinase B–dependent manner, to regulate hepatic acetyl-CoA and cholesterol synthesis. Reduced hepatic expression of NREP in patients with NAFLD and substantial correlations between low serum NREP levels and the presence of steatosis and nonalcoholic steatohepatitis highlight the clinical translational relevance of our findings in the context of recent preclinical trials implicating ACLY in NAFLD progression.
Keywords: Development, Hepatology
Keywords: Diabetes, Embryonic development, Epigenetics
Introduction
The liver receives dietary lipids and fatty acids derived from adipose tissue and hepatocytes, generally retaining no more than about 5% of its weight in lipids in normal physiological conditions in contrast to accumulating lipids in obesity (1–3). Nonalcoholic fatty liver disease (NAFLD) encompasses a heterogeneous set of conditions characterized by an increased hepatic lipid accumulation (steatosis), which may lead to inflammation and fibrosis resulting in nonalcoholic steatohepatitis (NASH), cirrhosis, and end-stage liver disease (1–3).
The global prevalence of NAFLD is estimated to be about 24% and represents a massive economic burden on the health care system (3). Notably, NAFLD in the pediatric community has a prevalence of 3%–12% and can affect up to 80% of the obese subpopulation (2, 4). NAFLD has a strong genetic component, and familial aggregation studies identified that siblings and parents of children with NAFLD were more likely to manifest fatty liver (5). Cross-sectional studies in twins also report a strong positive correlation with the presence of hepatic steatosis among monozygotic twins (6). The identification of several polymorphisms in genes such as APOC3, PNPLA3, TM6SF2, and PPP1R3B that correlate with NAFLD in genome-wide association studies reflects the genetic architecture of the disease (7).
Nevertheless, several studies have underscored the importance of environmental factors in the development of NAFLD early in life via epigenetic mechanisms (2, 3). Such epigenetic factors may prime fetal livers, increasing their susceptibility to NAFLD, and can potentially explain the missing heritability and increasing incidence of NAFLD over the last few decades (2). The importance of environmental factors for reprogramming in offspring is well documented in several animal models designed to study the effects of early-life exposures in the parents on the phenotypes of their offspring (8–13). However, a vast majority of these models represent nutritional interventions that result in sex-specific phenotypes dependent on maternal or paternal transmission and lack human translation.
In humans, dissecting the individual contributors to NAFLD, such as lipids, glucose, hormones, or diet, is challenging. Thus, experimental models continue to provide excellent opportunities to characterize factor(s) that impact NAFLD priming. In this study, we used a unique genetic model of tissue-specific–mediated insulin resistance, characterized by a complete biallelic deletion of exon 4 of the insulin receptor in hepatocytes — the liver-specific insulin receptor knockout (LIRKO) mouse (14, 15) — to identify new genes that contribute to prenatal developmental priming of NAFLD in the wild-type (WT) offspring. At 2 months of age LIRKO mice present hyperglycemia and hyperinsulinemia. Furthermore, LIRKOs have increased levels of hepatic cholesterol (15). Indeed, many changes seen in cholesterol metabolism in LIRKOs are also observed in humans with metabolic syndrome (15). For example, both show decreased levels of HDL and increased secretion of apolipoprotein B and VLDL. These findings make the LIRKO mouse a unique nondietary model of insulin resistance, hyperglycemia, dyslipidemia, and atherosclerosis that resembles several clinical features of the human metabolic syndrome.
We report that members of the TGF-β family are differentially expressed in the offspring of parents with metabolic syndrome, including neuronal regeneration–related protein (NREP) and growth differentiation factor 15 (GDF15). Here, for the first time to our knowledge, we report the role of NREP in mediating NAFLD development by controlling hepatic lipid metabolism. The clinical relevance of these findings is strengthened by the observation of low hepatic expression of NREP in human patients with NAFLD and the negative correlation between serum NREP levels and NAFL activity score in an independent cohort of well-characterized obese NAFLD patients.
Results
Parental genetic insulin resistance impacts body weight trajectories and body composition in offspring.
To determine the effects of parental metabolic syndrome (for breeding scheme, see Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI127502DS1), we compared the WT progeny obtained (a) by breeding of male LIRKO mice with control females (FL, “father LIRKO”) or (b) by breeding of LIRKO females with control males (ML, “mother LIRKO”) with offspring obtained (c) by breeding of control males and females (“control”) (for characteristics of animals used for breeding, see Supplemental Figure 1, B–M, and Supplemental Table 1).
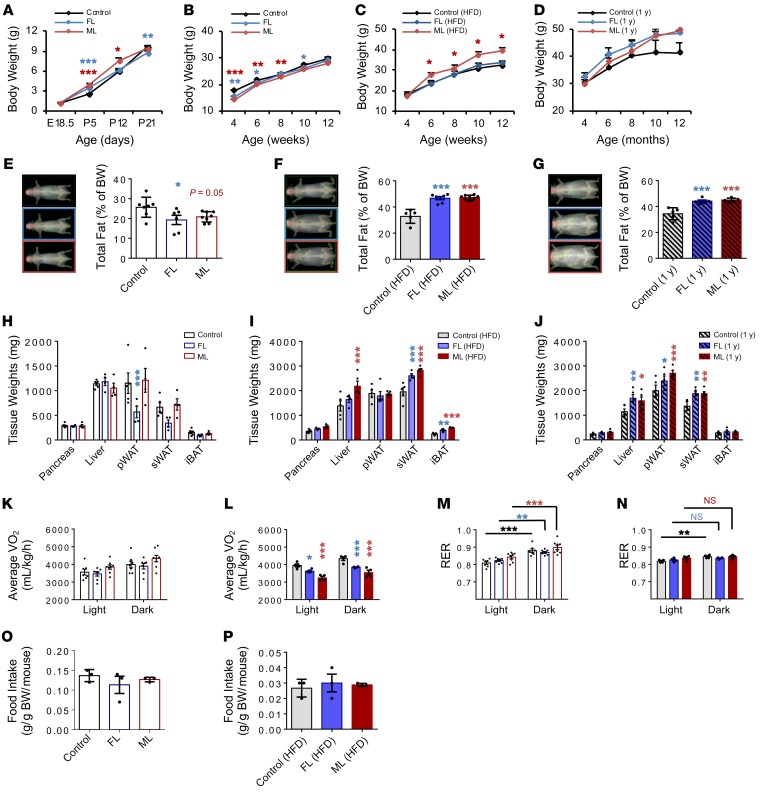
Because altered birth weights are risk factors for the development of NAFLD in children (16), we began by analyzing the body weight trajectories of the offspring. Consistent with our previous observation (10), offspring from insulin-resistant parents at postnatal day 5 presented transient increased body weights compared with controls (Figure 1A). However, from 4 to 8 weeks of age, FL and ML groups preserved low body weights compared with controls (Figure 1B). In fact, challenging FL and ML offspring with a high-fat diet increased the body weight trajectory in the latter from 6 to 12 weeks of age (Figure 1C), and a similar trend was evident in FL and ML groups during aging (Figure 1D).
Figure 1. Body weight trajectories and body composition are altered in FL and ML offspring.
(A) Body weight trajectories in male offspring of parents who are both controls, whose father is LIRKO (FL), or whose mother is LIRKO (ML) from E18.5 to P21; control, n = 27–49, corresponding to 5–6 litters; FL, n = 27–49, 5–6 litters; ML, n = 17–21, 3–4 litters). (B) Body weight on chow diet from 4 to 12 weeks of age (control, n = 9–24, 6 litters; FL, n = 10–18, 5 litters; ML, n = 13–20, 5 litters). (C) Body weight on high-fat diet (HFD) from 4 to 12 weeks of age (control, n = 9–24, 5 litters; FL, n = 6–10, 3 litters; ML, n = 8–13, 3 litters). (D) Body weight on chow diet from 4 to 12 months of age (control, FL, and ML, n = 5 per group). (E–G) Total fat mass measured by DEXA on chow (E) and HFD (F) at 3 months of age, and 1-year-old offspring on chow (G) (chow: control, n = 7; FL, n = 6; ML, n = 8; HFD: control, n = 5; FL, n = 7; ML, n = 8; aging: n = 5). (H–J) Body composition on chow (H) or HFD (I) or in aged (J) mice. (K and L) Energy expenditure (VO2) in 24-hour light/dark cycle measured by CLAMS in chow (K) and HFD (L) groups at 3 months of age (chow: control, n = 7; FL and ML, n = 8; HFD: n = 4). (M and N) Respiratory exchange ratio (RER) measured by CLAMS in control, FL, and ML offspring on chow (M) or HFD (N) (chow: control, n = 7; FL and ML, n = 8; HFD: n = 4 per group). (O and P) Food intake on chow (O) or HFD (P). Significance was determined by 2-way ANOVA with Holm-Šidák test. All data are shown as mean ± SEM and represent 3 or more litters. *P < 0.05; **P < 0.01; ***P < 0.001. pWAT, perigonadal white adipose tissue; sWAT, flank subcutaneous white adipose tissue; iBAT, interscapular brown adipose tissue.
To further explore the body weight differences, we subjected the offspring to dual-energy x-ray absorptiometry (DEXA; Supplemental Methods). On chow diets, the FL and ML groups presented decreased total fat mass (Figure 1E); however, when challenged with a high-fat diet or subjected to aging, both FL and ML offspring accumulated significantly more fat than controls (Figure 1, F and G). We then manually measured different tissues and identified the liver and subcutaneous white adipose tissue as the most altered among the groups (Figure 1, H–J). We next examined the differences in energy expenditure using a comprehensive laboratory animal monitoring system (CLAMS). Indeed, on a high-fat diet, FL and ML groups presented significantly decreased energy expenditure (Figure 1, K and L) and metabolic inflexibility in shifting from a fatty acid to a carbohydrate energy source compared with controls (Figure 1, M and N). These changes could not be explained by differences in food intake among groups on either the chow (Figure 1O) or the high-fat diet (Figure 1P).
Insulin sensitivity is altered in FL and ML offspring.
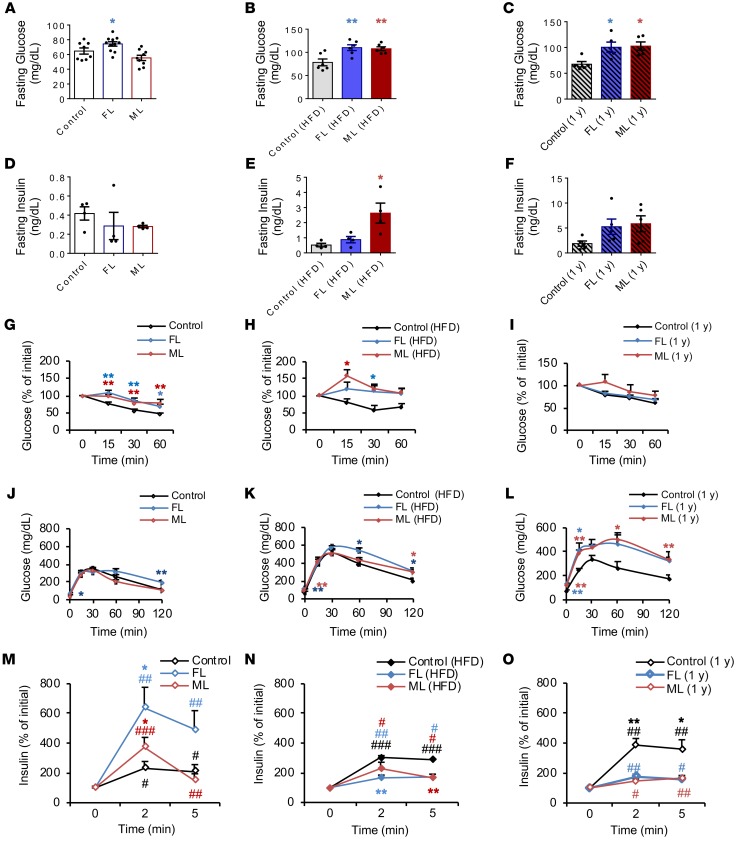
We speculated that the impairments observed in energy expenditure would affect glucose metabolism and insulin sensitivity. Fasting blood glucose levels were increased in FL on chow (Figure 2A) and in FL and ML groups fed a high-fat diet or as a consequence of aging (Figure 2, B and C). On the other hand, fasting serum insulin levels remained unchanged among groups on chow diet and were elevated only in the ML offspring on the high-fat diet (Figure 2, D–F, and Supplemental Table 2). Both ML and FL groups presented a phenotype of insulin resistance on both chow and high-fat diets (Figure 2, G and H). The insulin sensitivity phenotypes in aging were not significantly different between groups, likely because the control animals also developed insulin resistance (Figure 2I). Glucose tolerance was relatively normal in the ML group and mildly impaired in the FL group on chow (Figure 2J). On a high-fat diet, both FL and ML groups presented mildly impaired glucose tolerance (Figure 2K), which worsened significantly in both groups upon aging (Figure 2L). FL and ML offspring showed compensation for the insulin resistance by a significantly greater β cell secretory response to glucose in the chow-fed group (Figure 2M), and this response was blunted in animals on a high-fat diet and with aging (Figure 2, N and O). Female offspring shared many of the metabolic phenotypes with their male siblings (Supplemental Figure 2). Together these data indicate that parental insulin resistance, as exhibited in the LIRKO model, induces insulin resistance even in offspring with a normal complement of hepatic insulin receptors; potentially reprograms their metabolic response, as shown by an inability to adapt to a high-calorie diet; and leads to altered growth and adiposity.
Figure 2. Altered insulin sensitivity in FL and ML offspring.
(A–C) Fasting glucose levels in chow (A) and HFD (B) at 2 months of age and aged offspring (C) (chow: control, n = 8, 3 litters; FL, n = 11, 4 litters; ML, n = 5, 3 litters; HFD: control, FL, and ML, n = 6, 3 litters per group). (D and E) Fasting serum insulin levels in chow (D) and HFD (E) at 2 months of age and aged animals at 12 months of age (F) (chow and HFD: n = 4, 4 litters per group; aged: n = 5, 3 litters per group). (G–I) Insulin tolerance tests in chow (G), HFD (H), and aged (I) (chow: control, n = 10, 4 litters; FL, n = 11, 4 litters; ML, n = 6, 3 litters; HFD: control, n = 3; FL, n = 4; ML, n = 3; 3 litters per group, 2 months of age; aged: n = 5 per group, 1 year old). (J–L) Blood glucose values following an intraperitoneal glucose tolerance test in aged control, FL, or ML offspring. (M–O) Insulin levels plotted as percentage of basal levels after an intraperitoneal glucose injection on chow (M) or HFD (N) diets or in aged offspring (O). All data are based on n = 3–11 per group representing a minimum of 3 independent litters per group; data were analyzed using 1-way ANOVA with Holm-Šidák multiple-comparisons test in A–F and 2-way ANOVA with Holm-Šidák multiple-comparisons test in G–O. *P < 0.05; **P < 0.01. #Statistical comparison between time points 0 and 2 or 5 minutes (M–O). Data are expressed as means ± SEM. #P < 0.05; ##P < 0.01; ###P < 0.001.
FL and ML offspring are primed to develop high-fat diet–induced hepatic steatosis.
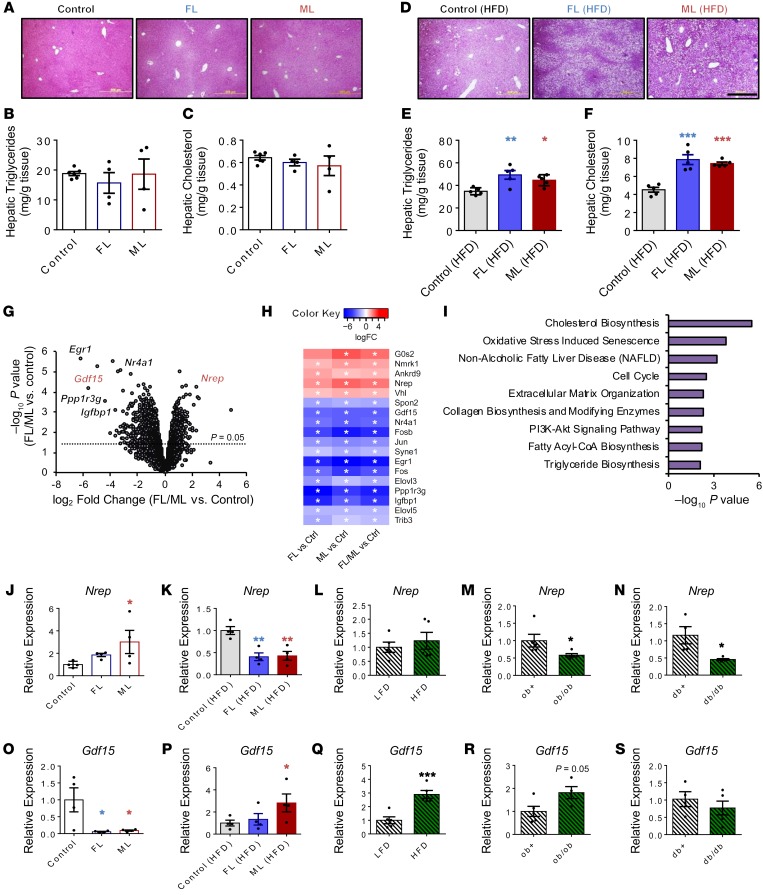
The observation that humans adapt to excess nutrients mostly by storing energy as triglycerides in adipose tissue and in “ectopic sites” such as the liver (17) prompted us to focus on the liver and flank subcutaneous white adipose tissues to further investigate the cause of the metabolic impairment(s). On a chow diet, FL and ML offspring presented normal liver histological architecture (Figure 3A), without significant changes in hepatic triglycerides (Figure 3B) or cholesterol (Figure 3C). Notably, FL and ML offspring presented hepatic steatosis on the high-fat diet compared with controls (Figure 3D and Supplemental Figure 3A). Concordantly, the liver triglyceride (Figure 3E) and cholesterol (Figure 3F) content was increased, and gene expression analyses revealed upregulation of master regulators of lipid metabolism such as Pparγ (Supplemental Figure 3, B–D) in the FL and ML groups. The alterations in hepatic gene expression patterns in FL and ML mice fed a high-fat diet were restricted to lipid metabolism without significantly affecting glycolysis, gluconeogenesis, or glycogenesis (Supplemental Figure 3, E and F). Finally, interrogation of the hepatic insulin signaling cascade in FL and ML groups fed a chow diet revealed an impaired phospho-GSK3β response to insulin consistent with liver insulin resistance (Supplemental Figure 3G, left and right). Flank subcutaneous white adipose tissue presented normal morphology (Supplemental Figure 3H), and the gene expression patterns in adipocytes (Supplemental Figure 3, I and J) were independent of the increase in fat mass observed in FL and ML groups challenged with a high-fat diet.
Figure 3. Hepatic transcriptome of NAFLD-primed offspring reveals Nrep and Gdf15.
(A) H&E-stained liver sections from offspring on chow diet (original magnification, ×200; scale bar: 200 μm). (B) Hepatic triglyceride content in chow-diet offspring. (C) Hepatic cholesterol content in chow-diet offspring. Selected-pathway analyses of differentially expressed genes. (D) H&E-stained liver sections from offspring on HFD (original magnification, ×200; scale bar: 200 μm). (E) Hepatic triglyceride content in HFD offspring. (F) Hepatic cholesterol content in HFD offspring. (G) Volcano plot RNA sequencing representation of differently expressed genes (chow: control, n = 4; FL, n = 3; ML, n = 3). (H) Heatmap representation of the most significantly altered genes. (I) Selected-pathway analyses of altered genes in FL and ML compared with controls. (J and K) Hepatic Nrep gene expression analyses by quantitative PCR (qPCR) on chow (J) and HFD (K) at 12 weeks of age (chow and HFD: n = 4 litters/group). (L) Hepatic Nrep mRNA by qPCR in mice challenged with a 6-week low-fat diet (LFD) and HFD at 12 weeks of age (LFD and HFD: n = 5, diet intervention of 6 weeks). (M and N) Hepatic Nrep mRNA levels by qPCR in ob/ob (M) and db/db mice (N) at 12 weeks of age (n = 5 per group). (O and P) Hepatic Nrep gene expression analyses by qPCR in FL, ML, and controls on chow (O) and HFD (P) (chow and HFD: n = 4 litters/group). (Q) Hepatic Nrep mRNA by qPCR in mice challenged with a 6-week LFD and HFD (LFD and HFD: n = 5, diet intervention of 6 weeks). (R and S) Hepatic Nrep mRNA levels by qPCR in ob/ob (R) and db/db mice (S) at 12 weeks of age (n = 5). Unless otherwise stated: chow: control, n = 8, 3 litters; FL, n = 11, 4 litters; ML, n = 5, 3 litters; HFD: control, FL, and ML, n = 6, 3 litters per group. RNA sequencing data were based on control (n = 4 mice/litters), FL (n = 3 mice/litters), and ML (n = 3 mice/litters). One-way ANOVA with Holm-Šidák multiple-comparisons test; 2-tailed unpaired t test in L–N and Q–S. *P < 0.05; **P < 0.01; ***P < 0.001. In H, asterisk indicates FDR < 0.25 in FL and FDR < 0.10 in ML and FL/ML comparisons.
Hepatic transcriptomic analyses in FL and ML offspring reveal several members of the TGF-β family and identify Nrep and Gdf15.
To explore the presence of a gene signature that could define the metabolic changes, we undertook RNA sequencing in the liver mRNA of FL and ML groups and compared it with that in control offspring on chow diet (Figure 3G). An unbiased global analysis revealed several members of the TGF-β family of genes that were commonly altered between FL and ML compared with controls (Figure 3H), including Nrep and Gdf15. ConsensusPathDB overrepresentation pathway analysis of differentially expressed genes commonly altered between FL and ML revealed enrichment in the cholesterol synthesis, fatty acyl-CoA synthesis, collagen synthesis, triglyceride synthesis, and AKT signaling pathways (Figure 3I, Supplemental Figure 4, A–D, and Supplemental Table 3).
The NAFLD “multiple-hit hypothesis” posits that diverse factors act to trigger disease development on genetically susceptible individuals (18). We, therefore, proceeded to investigate the effects of a metabolic hit, such as a high-calorie diet, on the behavior of metabolic genes reprogrammed by parental metabolic syndrome. First, we validated the RNA sequencing experiment by analyzing the top candidate genes by reverse transcriptase PCR (RT-PCR) in the chow-fed group (Supplemental Figure 5A) and explored changes in the gene expression patterns in the high-fat diet group (Supplemental Figure 5B) as well as in other models such as short-term high-fat feeding (Supplemental Figure 5C), mouse models exhibiting hepatic steatosis and steatohepatitis such as ob/ob (Supplemental Figure 5D), and mice with hepatic steatosis such as db/db (Supplemental Figure 5E). Among several candidates belonging to the TGF-β superfamily, NREP and GDF15 emerged as the most significant and consistently altered genes. Nrep was upregulated in the insulin-resistant FL and ML offspring compared with controls on chow (Figure 3J). However, challenging offspring with a high-fat diet resulted in a significant decrement in Nrep mRNA in the FL and ML groups (Figure 3K). Hepatic Nrep expression was not altered by a short-term high-fat diet (Figure 3L) but was consistently downregulated in ob/ob (Figure 3M) and db/db (Figure 3N) livers, suggesting its involvement in the pathophysiology of NAFLD in rodent models. Interestingly, Nrep and Gdf15, despite belonging to the same family, presented a virtually opposite pattern of expression among the different models. Namely, Gdf15 mRNA levels were downregulated in the FL and ML groups on chow (Figure 3O), while being elevated mainly in ML offspring when challenged with a high-fat diet (Figure 3P). Finally, Gdf15 mRNA levels were upregulated upon short-term high-fat diet (Figure 3Q) but were not altered in ob/ob (Figure 3R) or db/db mice (Figure 3S).
Nrep and Gdf15 are susceptible to DNA methylation modifications.
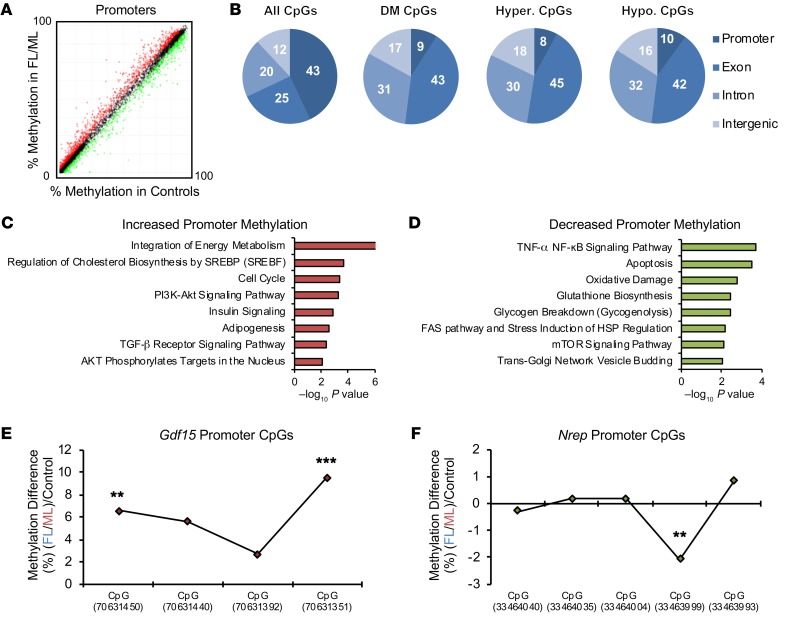
To investigate a potential epigenetic layer of gene regulation, we performed genome-wide DNA methylation analyses by enhanced reduced representation bisulfite sequencing (ERRBS) and compared the controls with a combined (FL plus ML) group on chow diet, since the individual ML and FL groups showed largely similar phenotypes (Figure 4A and Supplemental Methods). ERRBS analyses revealed enrichment for promoter regions — 43% of all CpGs (Figure 4B), as previously reported (19); and these were highly unmethylated, while the hypomethylated CpGs (10% of all CpGs) were more abundant compared with hypermethylated (8% of all CpGs) in FL/ML versus controls (Figure 4B). To determine whether DNA methylation affected the pathways that also showed enrichment at the gene expression level, we performed pathway analyses to confirm that genes with differential DNA methylation were also enriched for cholesterol synthesis, MAPK, AKT, insulin, and TGF-β signaling (Figure 4, C and D, and Supplemental Tables 4 and 5). Next, we assessed the promoter methylation levels of Gdf15 and Nrep. Consistently, with decreased expression, the Gdf15 promoter region revealed several hypermethylated CpGs in FL/ML compared with controls (Figure 4E). On the other hand, consistent with upregulation, the Nrep promoter region presented decreased methylation in the FL/ML group compared with controls (Figure 4F). These results suggest a layer of regulation on gene expression due to DNA methylation that impacts Gdf15 and Nrep.
Figure 4. Hepatic global DNA methylation analyses by ERRBS.
(A) Scatter plot representation of global promoter DNA methylation analyses by enhanced reduced representation bisulfite sequencing ERRBS (n = 4, 4 litters per group). (B) Genomic features of hepatic DNA methylation. All CpGs, all detected cytosines; DM CpGs, differently methylated cytosines; Hyper. CpGs, hypermethylated cytosines; Hypo. CpGs, hypomethylated cytosines. (C) Selected pathways of genes with increased promoter DNA methylation (genes selected for methylation > 0% and FDR < 0.05). (D) Selected pathways of genes with decreased promoter DNA methylation (genes selected for methylation < 0% a/nd FDR < 0.05). (E) DNA methylation in selected covered CpGs spanning the Gdf15 promoter. (F) DNA methylation in selected covered CpGs spanning the Nrep promoter. **FDR < 0.01; ***FDR < 0.001.
NREP is downregulated by palmitate-induced steatosis and modulates fatty acid and cholesterol synthesis.
NREP, also known as P311, is a highly conserved 8-kDa protein belonging to the TGF-β superfamily (20) and has been associated with wound healing (21), nerve and lung regeneration (22), and kidney fibrosis (20). TGF-β signaling is important in the development of fibrosis in advanced NAFLD and is upregulated in NASH (23, 24). It is also shown to reduce β-oxidation and to promote fatty acid synthesis in mouse primary hepatocytes in the presence of palmitate (23). Although the development of steatosis and progression of NAFLD to NASH likely involves inflammation, fibrosis, and fatty acid metabolism, the molecular mechanism(s) underlying these processes are not well understood. We speculated that NREP bridges TGF-β and lipid synthesis pathways to regulate steatosis development by controlling β-oxidation and/or fatty acid and cholesterol synthesis.
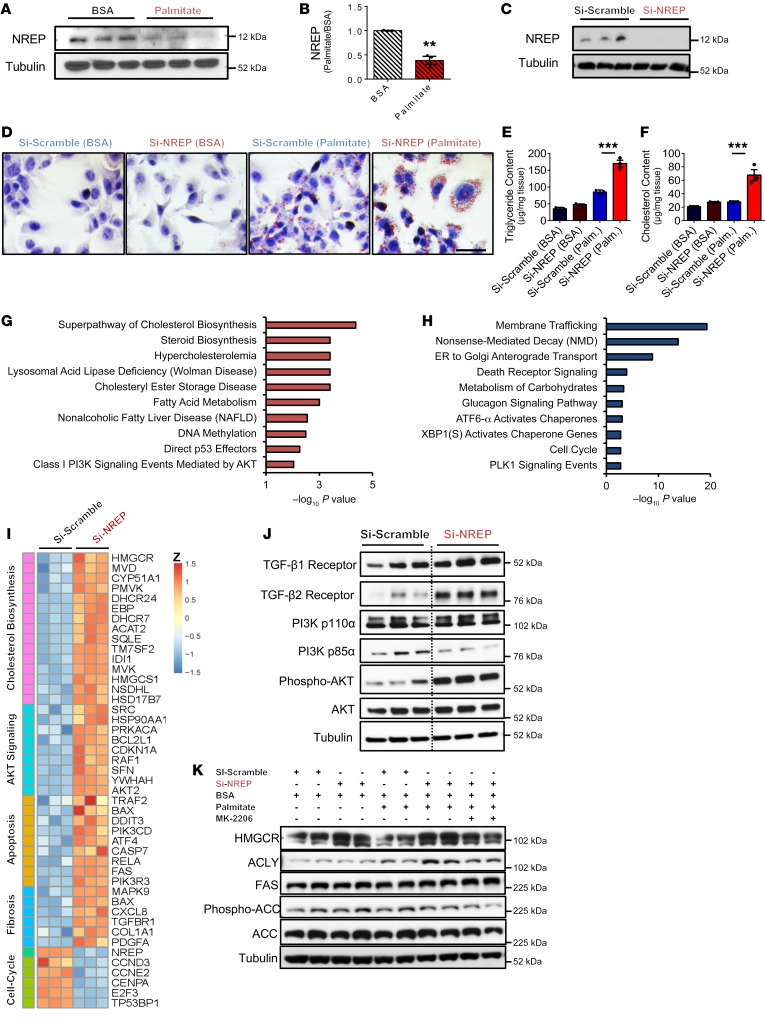
To test this possibility, we first investigated the behavior of candidate genes in a human in vitro model of hepatic steatosis (25–27) by examining HepG2 cells treated with either fatty acid–free BSA or palmitate (Supplemental Figure 6, A and B, and Supplemental Methods). Treatment of HepG2 with palmitate and/or oleate has been used widely to mimic the effects of NAFLD in vitro (27–29). Treatment of HepG2 cells with fatty acids reproduces several clinical aspects of NAFLD, including signaling, apoptosis, and mitochondrial dysfunction (27–29). Palmitate treatment of HepG2 cells mimicked several aspects of hepatic steatosis, and the expression patterns of several candidate genes, including NREP, were similar to those observed in ob/ob and db/db liver samples or in the liver samples from FL and ML groups fed a high-fat diet (Supplemental Figure 6, C and D). Consistently, treatment of HepG2 cells with palmitate induced a decrease in NREP protein levels compared with BSA treatment (Figure 5, A and B).
Figure 5. NREP is downregulated by palmitate-induced steatosis in HepG2, and NREP regulates hepatic lipid metabolism.
(A) NREP protein levels in HepG2 cells treated with BSA or palmitate for 24 hours (n = 3 independent experiments). (B) Quantification of NREP protein levels. (C) NREP knockdown (KD) in HepG2 cells at protein levels (n = 3). (D) Representative oil red staining in HepG2 cells with NREP KD challenged with palmitate for 24 hours (n = 3 independent experiments; original magnification, ×400; scale bar: 50 μm). (E and F) Triglyceride (E) and cholesterol (F) content quantification in HepG2 cell lysates after stimulation for 24 hours with 500 μM palmitate (n = 3 independent experiments). (G and H) RNA sequencing selected enriched pathway analyses of upregulated (G) and downregulated genes (H) in HepG2 cells with NREP KD compared with scramble (n = 3 per group). (I) Heatmap representation of differently expressed genes involved in cholesterol biosynthesis, AKT signaling, apoptosis, fibrosis, and cell cycle. (J) Basal signaling analyses in lysates from HepG2 cells treated with scramble (left) or NREP KD (right) (n = 3 independent experiments). (K) Protein levels of indicated proteins in HepG2-scramble and NREP KD treated with BSA or palmitate for 24 hours in the presence of AKT inhibitor (MK-2206) or DMSO (n = 2 independent experiments). Significance was determined by 2-tailed unpaired t test in B, and 1-way ANOVA with Holm-Šidák multiple-comparisons test in E and F. **P < 0.01; ***P < 0.001. Data are expressed as means ± SEM.
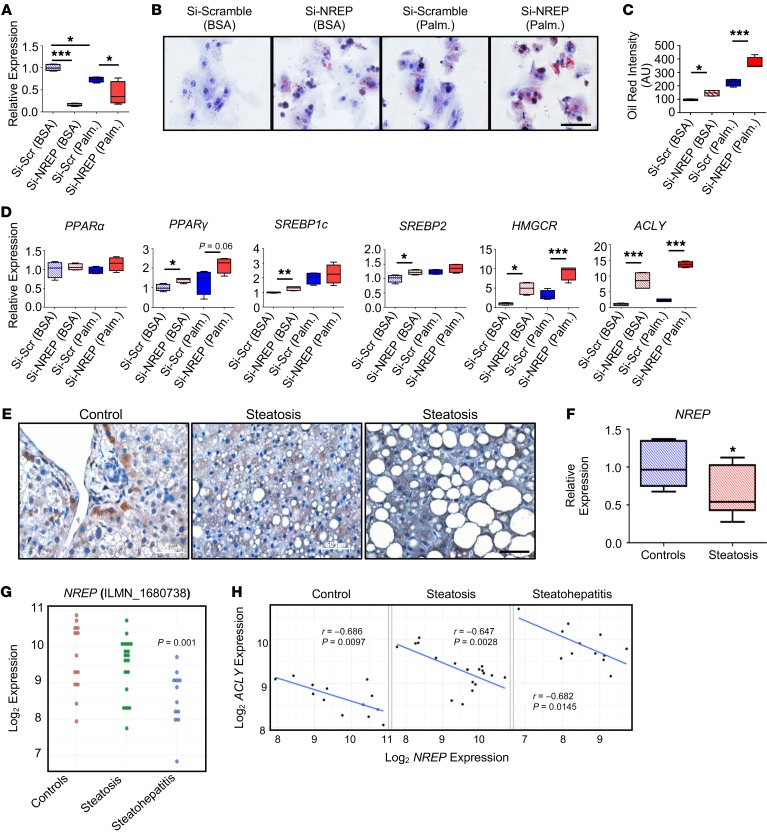
Next, to determine the possible direct role of NREP in the development of hepatic steatosis, we contrasted the effects of knockdown versus overexpression of NREP in HepG2 cells (Supplemental Methods). Treatment of cells with a knockdown of NREP (Figure 5C) with palmitate displayed greater lipid droplet accumulation (Figure 5D) that was consistent with increased triglycerides and cholesterol content compared with scramble (Figure 5, E and F). To determine whether triglyceride accumulation resulted from decreased β-oxidation, we analyzed fatty acid oxidation using the Seahorse XF96 instrument (Supplemental Methods). Cells lacking NREP exhibited decreased basal and maximal respiration in the presence of BSA-palmitate substrate (Supplemental Figure 7, A and B). The impaired fatty acid–dependent respiration and concomitant increase in triglyceride and cholesterol content in HepG2 cells lacking NREP were supported by a decrease in PPARα expression and an increase in the transcriptional network of genes associated with fatty acid (PPARγ, SREBP1c, FAS, ELOVL5), glycerolipid (LPIN1), and cholesterol synthesis (SREBP2, HMGCR, and FDFT1) in palmitate-induced steatosis (Supplemental Figure 7C).
To further evaluate the global transcriptomic changes induced by NREP downregulation, we used RNA sequencing in HepG2 cells lacking NREP. Enriched pathway analyses of upregulated genes revealed pathways involved in cholesterol synthesis, fatty acid metabolism, NAFLD, and PI3K/AKT signaling (Figure 5G). In contrast, enriched downregulated genes included those for membrane trafficking, nonsense-mediated decay, glucagon signaling, and cell cycle progression (Figure 5H). Differentially expressed genes included HMGCR (cholesterol synthesis) and TGFBR1 (TGF-β signaling and fibrosis) (Figure 5I). These results are consistent with a previous study identifying Nrep as the most significantly downregulated hepatic gene in response to an olive oil bolus and showing a negative correlation between Nrep mRNA and hepatic triglycerides and cholesterol content in rats (30). Together these data argue for a potential role for NREP in the development of steatosis in a human in vitro model of hepatic steatosis.
NREP modulates HMGCR and ATP-citrate lyase protein levels via the noncanonical TGF-β pathway.
The signaling pathways that orchestrate the development of NAFLD are not entirely known. TGF-β signaling is important in liver homeostasis, development, and regeneration and is involved in molecular mechanisms that lead to liver fibrosis (31). The role of TGF-β signaling, and in particular, the noncanonical branch TGF-β receptor (TGF-βR)/PI3K/AKT, is virtually not explored in the context of lipid metabolism and NAFLD priming. NREP knockdown increased the abundance of TGF-β1 and TGF-β2 receptor proteins, and this was accompanied by the decrease in the regulatory subunit PI3K p85α (Figure 5J and Supplemental Figure 7D). Consistent with increased TGF-βR/PI3K signaling, NREP knockdown induced a robust increase in AKT phosphorylation (Figure 5J and Supplemental Figure 7D). Interestingly, NREP overexpression (Supplemental Figure 8, A and B) virtually reversed all the effects of the knockdown. Thus, HepG2 cells overexpressing NREP showed less lipid accumulation when stimulated with palmitate (Supplemental Figure 8C) and partially increased basal fatty acid oxidation (Supplemental Figure 8, D and E), which resulted in lower triglyceride and cholesterol content (Supplemental Figure 8, F and G). NREP overexpression increased the gene transcriptional network associated with β-oxidation while reducing the expression of genes related to fatty acid and cholesterol synthesis (Supplemental Figure 8H). Finally, NREP overexpression caused a significant and notable decrease in phospho-AKT and phospho-mTOR (Supplemental Figure 8I). Together, these data prompted us to hypothesize that NREP regulates hepatic metabolism by acting on common nodes of cholesterol and fatty acid synthesis via the noncanonical branch of the TGF-βR/AKT pathway.
ATP-citrate lyase (ACLY) is a cytosolic enzyme responsible for the production of acetyl-CoA — a substrate for de novo cholesterol and fatty acid synthesis (32–35). ACLY is regulated by AKT signaling and has been a focus of recent clinical trials to treat hypercholesterolemia and NAFLD (33, 35). On the other hand, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), a rate-limiting enzyme for cholesterol synthesis and a target of statins (36), is increased in NAFLD and correlates with the histological severity of the disease (37). To investigate whether NREP is linked to ACLY and HMGCR via AKT signaling, we challenged HepG2 cells knocked down for NREP with either BSA or palmitate, in the presence or absence of an AKT inhibitor (MK-2206) or DMSO. Palmitate treatment increased ACLY expression but not HMGCR in HepG2 cells, while cells with a knockdown of NREP exhibited a further increase in ACLY and HMGCR protein levels that was partially blocked by an AKT inhibitor (Figure 5K and Supplemental Figure 8J). Nrep knockdown in AML12 cells (Supplemental Figure 9, A and B) increased AKT phosphorylation in BSA and maintained AKT phosphorylation in palmitate-treated cells (Supplemental Figure 9, C and D), leading to upregulation of ACLY in comparison with scramble cells (Supplemental Figure 9, C and D). Overall, these data support a novel role for NREP in regulating hepatic TGF-βR/PI3K/AKT signaling to modulate ACLY and HMGCR in response to steatosis.
We observed that the effects of GDF15 were opposite to those of NREP. For example, GDF15 knockdown in HepG2 cells (Supplemental Figure 10, A–C) increased fatty acid–dependent maximum respiration (Supplemental Figure 10, D and E) that was potentially secondary to an increase in PPARα and PGC1α mRNA levels in comparison with scramble cells in response to palmitate (Supplemental Figure 10F). This occurred without changes in AKT or mTOR phosphorylation (Supplemental Figure 10G). On the other hand, GDF15 overexpression (Supplemental Figure 11, A–C) resulted in mild alterations in fatty acid–dependent respiration (Supplemental Figure 11, D and E), and an increase in PPARγ and SREBP1c mRNA (Supplemental Figure 11F) in BSA-treated cells. GDF15-deficient cells presented increased phosphorylation of AKT (Supplemental Figure 11G) in the basal state.
NREP is expressed in hepatocytes and its expression is decreased in NAFLD.
To investigate the translational relevance of our findings, we first validated the effects of NREP knockdown in primary human hepatocytes. Concordant with previous results in HepG2 cells (Figure 5, A and B), palmitate treatment decreased NREP mRNA levels by 30% (Figure 6A). Although we achieved about 70%–80% silencing of NREP expression in primary human hepatocytes in the absence of palmitate, treatment with the fatty acids likely induced toxicity in the hepatocytes as evidenced by a variation in the ability to knock down NREP (~30%) (Figure 6A). Finally, NREP abrogation in primary human hepatocytes increased lipid droplet accumulation in BSA- and palmitate-treated cells (Figure 6, B and C). NREP knockdown in primary human hepatocytes cultured in BSA increased expression of PPARγ, SREBP1c, SREBP2, HMGCR, and ACLY mRNA (Figure 6D). Palmitate treatment further increased HMGCR and ACLY mRNA levels in primary human hepatocytes bearing an NREP knockdown (Figure 6D), consistent with our findings in HepG2 cells. Next, we sought to analyze NREP expression in human liver samples from patients with histologically defined hepatic steatosis (sample information in Supplemental Table 6). Immunohistological analyses revealed virtually undetectable NREP protein in hepatocytes from patients with hepatic steatosis in contrast to the presence of NREP-positive hepatocytes in the controls (Figure 6E). Consistent with our hypothesis, hepatic NREP mRNA was decreased by 40% in patients manifesting hepatic steatosis (Figure 6F). Next, we validated our results by reanalyzing publicly available data sets comparing the transcriptomic signatures of biopsy-obtained samples from controls and steatosis and NASH patients. Reanalyses of the GSE33814 data set from a previous study comparing the gene expression signatures between steatohepatitis versus steatosis and controls (38) were congruent with our observations of NREP mRNA levels tending to be lower in steatosis and dramatically downregulated in steatohepatitis (Figure 6G). In the same study, reexamination of the relationship between NREP and ACLY mRNA levels showed a strong opposite correlation between them (Figure 6H).
Figure 6. NREP relevance in human hepatic steatosis.
(A) NREP knockdown in human primary hepatocytes (n = 3 independent experiments, hepatocytes from 5 pooled healthy donors). (B) Representative oil red staining showing lipid droplets in human primary hepatocytes treated with BSA or challenged with palmitate for 24 hours (n = 3 independent experiments; original magnification, ×400; scale bar: 50 μm). (C) Quantification of oil red staining intensity (n = 3 independent experiments). (D) RT-PCR analyses of genes involved in β-oxidation (PPARα), transcriptional regulation of fatty acid (PPARγ, SREBP1c) and cholesterol (SREBP2) metabolism, and acyl-CoA (ACLY) and cholesterol synthesis (HMGCR) (n = 3 independent experiments, hepatocytes from 5 pooled healthy donors). (E and F) NREP protein (E) and mRNA (F) levels in human liver samples from controls and patients with steatosis (control, n = 7; steatosis, n = 8; Supplemental Table 6). (G) Hepatic mRNA levels in controls and steatosis and steatohepatitis patients by microarrays (GSE33814). (H) Pearson’s correlations between NREP and ACLY mRNA levels in all groups in controls, steatosis, and steatohepatitis. Significance in comparisons between 2 groups was determined by 2-way ANOVA with Holm-Šidák multiple-comparisons test in A, 1-way ANOVA with Holm-Šidák multiple-comparisons test in C and D, 2-tailed unpaired t test in F, Benjamini-Hochberg method (see Methods) in G, and Pearson’s correlations in H. *P < 0.05; **P < 0.01; ***P < 0.001. All data are shown as mean ± SEM.
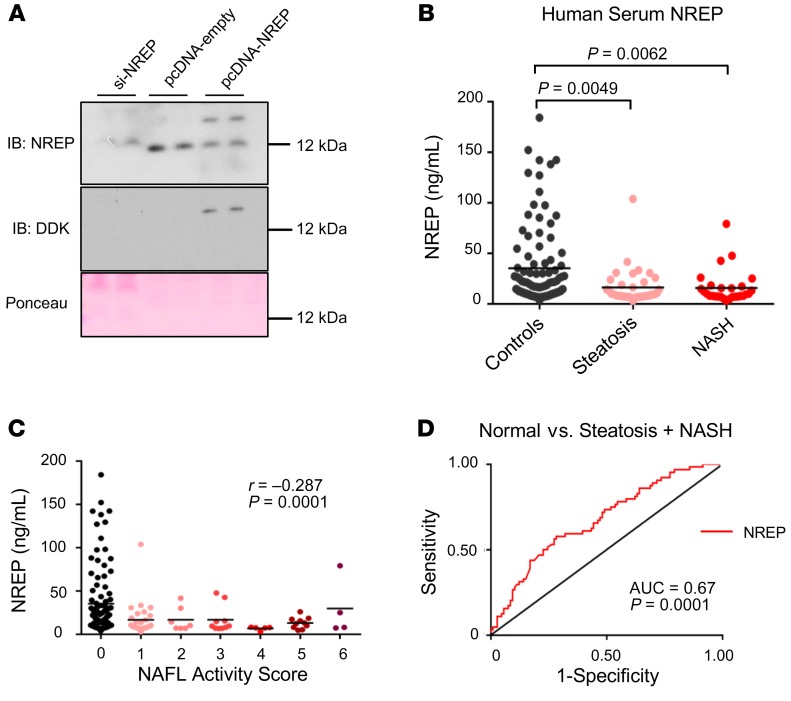
NREP circulating levels are decreased in NAFLD and correlate with steatosis grade and NAFL activity score.
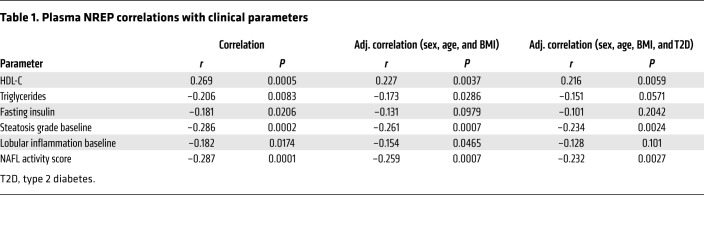
The last decade has revealed the significance of TGF-β and TGF-β–related proteins, including cytokines and secreted growth factors, in modulating diverse signaling pathways (39). Therefore, to explore whether NREP is secreted by hepatocytes, we collected cell- and FBS-free culture medium supernatants from HepG2 cells with a knockdown of NREP, scramble, or NREP overexpression. NREP was easily detected in the culture medium from scramble and NREP-overexpressing HepG2 cultured cells, in contrast to culture medium from HepG2 cells with an NREP knockdown (Figure 7A). Next, to examine whether circulating NREP levels reflect changes in hepatic mRNA and protein levels in patients with NAFLD, we analyzed plasma levels from controls (n = 106) and patients with simple steatosis (n = 36) or NASH (n = 28) in an extended and comprehensive obese liver biopsy-proven cohort (i.e., the Kuopio cohort; Supplemental Table 7). Consistent with our hypothesis, plasma NREP levels were significantly decreased in both steatosis and NASH compared with controls (Figure 7B). Indeed, several metabolic parameters had significant correlations with NREP (Table 1). For example, serum NREP levels correlated positively with HDL cholesterol (r = 0.27) and negatively with serum triglycerides (r = –0.21), reflecting its involvement in cholesterol and triglyceride metabolism. Furthermore, NREP correlated strongly with steatosis grade (Table 1) and NAFL activity score (Table 1 and Figure 7C). Estimation of the area under a receiver operating characteristic curve (AUROC) as a diagnostic tool revealed a moderate sensitivity/specificity for NREP for the prediction of steatosis and NASH (AUC, 0.67; P = 0.0001; Figure 7D).
Figure 7. NREP is secreted by hepatocytes and its plasma levels reflect the changes in hepatic NREP mRNA seen in NAFLD.
(A) NREP protein levels in the supernatant of HepG2 cells with knockdown of NREP, scramble, or NREP overexpression, cultured for 48 hours in FBS-free medium. (B) NREP plasma levels in obese control, steatosis, and NASH patients (control, n = 106; steatosis, n = 36; NASH, n = 28). (C) Correlation between plasma NREP and NAFL activity score. (D) Receiver operating characteristic curves of NREP in controls versus steatosis plus NASH. Significance was determined by 1-way ANOVA with Kruskal-Wallis test with Dunn’s multiple-comparisons test in B, and adjusted Spearman’s correlations in C and D. Data are expressed as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Table 1. Plasma NREP correlations with clinical parameters.
Finally, we measured GDF15 levels from controls and patients with steatosis or NASH in our cohort (Supplemental Figure 12A). GDF15 plasma levels were increased in steatosis and NASH, consistent with recent work reporting that GDF15 is a biomarker of advanced fibrosis (40). GDF15 correlated strongly with several clinical features important in NAFLD pathophysiology, including inflammation and NAFLD activity score (Supplemental Figure 12B). Estimation of AUROC as a diagnostic tool revealed very high specificity for GDF15 as a biomarker for steatosis and NASH (Supplemental Figure 12C). Consistent with a previous report (40), we were not able to detect GDF15-positive hepatocytes (data not shown) in our sample cohort (Supplemental Table 6), and while our in vitro studies in HepG2 cells suggest that GDF15 silencing increases fatty acid oxidation concomitant with increased expression of PPARα and PGC-1α, a recent study has reported that Gdf15-KO mice present aggravated NASH (41). These discrepancies led us to focus on NREP not only as an early biomarker of NAFLD but also as a potential therapeutic target.
To test this, we conducted a pilot experiment in mice fed either a low-fat diet or a high-fat diet for about 1 year. Mice receiving PBS or 20 ng of human recombinant NREP protein delivered by an osmotic pump per day per mouse (Supplemental Figure 13A) were followed for 12 days. NREP administration decreased body weights of high-fat diet mice compared with PBS (Supplemental Figure 13A). Furthermore, NREP in vivo treatment decreased blood glucose levels in high-fat diet NREP-treated mice (Supplemental Figure 13B), without changes in C-peptide levels (Supplemental Figure 13C), in comparison with the high-fat diet mice on PBS pumps. NREP infusion improved glucose excursions during a glucose tolerance test (Supplemental Figure 13D) without changes in insulin sensitivity (Supplemental Figure 13E). Finally, hepatic gene expression analyses (Supplemental Figure 13F) revealed that NREP administration reduced the expression of Pparγ and Acly, consistent with our previous findings pointing to an important relationship between NREP and genes involved in the regulation of lipid metabolism. NREP administration also decreased the expression of the fibrotic markers Col1a1 and Acta2 (Supplemental Figure 13F), pointing to NREP as a potential therapeutic agent to counter hepatic fibrosis.
Discussion
There is increasing evidence that epigenetic mechanisms may contribute to the development of NAFLD (42). Contrary to simple steatosis, which is generally benign, NASH is strongly associated with comorbidities and reduced life span. Follow-up studies have demonstrated that the progression of steatosis to NASH and consequently hepatic fibrosis is not simply linear and may be influenced by genetic and/or environmental mechanisms (3). This provides a strong rationale for the identification of new early responsive genes triggered by the parental metabolic syndrome and of mechanisms underlying NAFLD progression that are translatable to clinical therapy.
Although the association of obesity and diabetes with NAFLD development is well documented, there is global consensus regarding the roles of the environment during preconception, such as diet, body composition, metabolism, smoking, and stress, in modulating chronic disease risk (43). Models of maternal overnutrition, such as high-fat diet–induced obesity, are among the most extensively studied paradigms and have been reported to promote hepatic triglyceride accumulation and lead to NAFLD in nonhuman primates (13). However, adoption of a diet-induced obesity model to mimic metabolic syndrome in humans is limited by the difficulty in distinguishing the contributions of diverse metabolic phenotypes observed in obesity and type 2 diabetes versus those directly linked to high-fat consumption.
Previous studies have used genetic mouse models to modulate the effects of either paternal and maternal hyperglycemia (Akita mice) (44) or maternal hyperinsulinemia (IRS-1–heterozygous mice) (45) on their respective offspring; however, virtually none have investigated the combined contributions of hyperglycemia, hyperinsulinemia, and dyslipidemia. We used the LIRKO mouse, a unique model of genetic insulin resistance, to study the effects of metabolic syndrome in NAFLD priming (14, 46). In the Akita model, paternal and maternal hyperglycemia resulted in decreased body weights, elevated fasted blood glucose levels, and mild glucose intolerance in male offspring (44); however, no phenotype was described regarding the development of hepatic steatosis. On the other hand, the effects of maternal hyperinsulinemia have been shown to induce hepatic steatosis in male offspring at 6 months of age despite an absence of changes in body composition or energy expenditure, suggesting that the development of steatosis is driven by increased circulating serum insulin levels (45). We used an unbiased, transversal, and original approach to identify new genes and mechanisms underlying NAFLD priming in a genetic model of metabolic syndrome and used only WT offspring to exclude effects secondary to insulin receptor ablation. We focused on common genes and pathways affecting both sexes and common to paternal and maternal contributions that exclude confounding effects of in utero exposures and/or lactation. Our results point to parental metabolic syndrome acting concomitantly to impact body weight trajectories and body composition in the offspring secondary to decreased energy expenditure and metabolic inflexibility leading to hepatic lipid accumulation and insulin resistance. Analyses of the hepatic transcriptomic data sets in the offspring allowed us to identify a subset of genes that are transversely affected by paternal and maternal metabolic syndrome.
We report that maternal and paternal genetic insulin resistance epigenetically reprograms NREP in hepatocytes to reflect a novel molecular bridge between TGF-β signaling and hepatic lipid metabolism that is highly susceptible to environmental triggers such as a high-calorie diet. A notable observation is the reduced expression of NREP in hepatocytes in rodent models of steatosis and steatohepatitis, human in vitro models of steatosis, and human liver samples from patients with hepatic steatosis. NREP belongs to the TGF-β superfamily, which encompasses a group of regulatory proteins that control several aspects of cell biology (47). TGF-β1, a cytokine, was the first protein to be identified in this family (48) and is known to be involved in the fibrogenic response by stellate cell activation (23) and in the adaptive response seen in the F1 and F2 offspring of fathers with a history of liver damage (11). Although the canonical branch of TGF-β signaling modulates hepatic lipid metabolism by regulating lipogenesis and β-oxidation, its exact role in NAFLD development and, in particular, the involvement of the noncanonical branch of TGF-β signaling have not been explored (23).
Our mechanistic data suggest that NREP is epigenetically modified by DNA methylation and controls hepatic lipid content (e.g., cholesterol and triglycerides) by modulating fatty acid oxidation and regulating the expression of master regulators of fatty acid synthesis such as PPARγ and SREBP1c, while controlling ACLY and HMGCR levels in an AKT-dependent manner. ACLY is an enzyme highly expressed in liver that is responsible for the production of acetyl-CoA necessary for the synthesis of cholesterol and fatty acids and for protein acetylation (33, 34). It is strategically positioned at the intersection of these pathways and may work as a nutrient sensor (33). Silencing of hepatic ACLY protects against NAFLD development in the db/db mouse (34), and recent studies performed in humans reported that ACLY expression is increased in NAFLD patients (49). Although all these data point to ACLY as an attractive target, a suitable pharmacological inhibitor is lacking. On the other hand, HMGCR catalyzes the rate-limiting step in cholesterol biosynthesis and has been an attractive clinical target of lipid-lowering drugs — statins. HMGCR expression is increased in NAFLD and positively correlates with the severity of the disease, partially explaining the increased cholesterol synthesis in NAFLD patients (37). Interestingly, our RNA sequencing data revealed that NREP ablation induces the expression of several fibrotic genes and may explain the concomitant increase in lipid synthesis and fibrosis seen in human NAFLD progression.
In contrast to the expression patterns in NREP, we noted that GDF15 expression is increased in in vivo models of steatosis and steatohepatitis and by in vitro modeling of hepatic steatosis in HepG2 cells. Consistently, knockdown of GDF15 in the latter increased the expression of PPARα and PGC1α mRNA while increasing the maximal respiration capacity on a BSA-palmitate substrate. However, we did not detect changes in fatty acid oxidation when we overexpressed GDF15 in HepG2 cells. These data suggest that the upregulation of GDF15 in NAFLD is pathological rather than compensatory. Furthermore, GDF15 plasma levels are increased in steatosis and NASH patients as compared with controls, which is consistent with Koo et al., who suggested that circulating GDF15 serves as a new biomarker for advanced fibrosis in NAFLD (40). Given the lack of expression of GDF15 in hepatocytes (40), further studies are necessary to understand the biology of GDF15 in the pathophysiology of NAFLD.
Our data are consistent with previous studies involving AKT signaling in the regulation of ACLY (50). However, the precise role of the PI3K/AKT pathway in the development of NAFLD is complex (51). Excessive AKT activation leads to NAFLD development by maturation of the transcription factor SREBP1c, leading to increased transcription of several lipogenic enzymes (52, 53). PTEN is a negative regulator of AKT and is frequently mutated in several cancers. PTEN-deficient mice develop severe NAFLD as a result of increased phospho-AKT and lipid synthesis (51, 54). Recently, Hippo signaling was reported to prevent NAFLD development by inhibiting IRS-2/AKT signaling (55). Interestingly, in a recent study on fibroblasts, receptor-interacting protein kinase-3 (RIPK3) was shown to promote fibrinogenesis in an AKT-dependent activation of ACLY in response to TGF-β1 (56). These results, in the light of previous reports that NREP overexpression inhibits TGF-β1 and TGF-β2 receptor levels (21) and that NREP can bind to the latency-associated protein (LAP) of TGF-β1 and TGF-β2 to decrease TGF-β autoinduction (57), indicate that NREP regulates AKT signaling and ACLY and HMGCR levels via the noncanonical branch of the TGF-β pathway.
Four observations from our studies support the concept that NREP is regulating NAFLD development. First, reanalysis of a previously published data set (38) demonstrated decreased hepatic NREP mRNA in patients with steatohepatitis and a strong negative correlation between hepatic NREP and ACLY mRNA levels. Second, plasma NREP levels are decreased in patients with simple steatosis and NASH. Third, plasma NREP is strongly negatively correlated with steatosis grade and NAFL activity score in the Kuopio cohort. Finally, administration of recombinant NREP to mice with diet-induced obesity improves their hepatic lipid and fibrotic gene expression signatures.
In summary, the present study identifies NREP as a novel molecular mediator of NAFLD development that further elucidates the role of TGF-β signaling in mediating hepatic lipid accumulation and fibrosis development. Further studies in diverse ethnic cohorts will strengthen the possible utility of NREP as a steatosis or NASH biomarker. Interestingly, several obese control patients presented decreased serum NREP levels. Future longitudinal studies will reveal whether this heterogeneity seen in controls represents differing susceptibilities to progress toward NAFLD and whether NREP is a predictor of NAFLD progression. We propose that recombinant NREP or gene therapy–mediated approaches targeting hepatic NREP can be harnessed to improve NAFLD and its associated complications in the clinic.
Methods
Mice and diets.
Liver-specific insulin receptor knockout (LIRKO) mice (insulin receptor–IRlox/lox albumin-Cre+/–) were generated as previously described (14). In brief, the control offspring group consisted of F1 offspring from a control male and female (insulin receptor–IRlox/lox albumin-Cre–/–). Control parents were crossed for 4 generations to minimize any epigenetic memory from the presence of Cre. Father LIRKO (FL) offspring resulted from the crossing of a male LIRKO (insulin receptor–IRlox/lox albumin-Cre+/–) with a control female. Mother LIRKO (ML) offspring resulted from the breeding of a control male with a LIRKO female (see Supplemental Methods for further detail).
Study subjects.
The present study contains data from a total of 170 obese individuals selected from an ongoing Kuopio Obesity Surgery Study (ref. 60; for clinical characteristics see Supplemental Table 6). All patients with alcohol consumption of more than 2 doses per day and patients with previously diagnosed liver diseases, not related to obesity, were excluded from the study. Clinical parameters were assessed before surgery, after 4 weeks of a very-low-calorie diet. Liver biopsies were collected as a wedge biopsy during elective Roux-en-Y gastric bypass surgery at Kuopio University Hospital. The histological assessment of liver biopsy samples was performed by a pathologist according to the standard criteria (1). Subjects were divided into 3 categories based on the liver phenotype: normal, simple steatosis, or NASH (as described previously; ref. 59).
Intraperitoneal glucose tolerance test, insulin tolerance test, and in vivo glucose-stimulated insulin secretion.
Glucose tolerance tests (GTTs) and insulin tolerance tests (ITTs) were performed as previously reported (60–62). Briefly, for GTT, animals were fasted for 16 hours overnight, and 20% (vol/vol) dextrose was given through an i.p. injection in a 2 g/kg body weight proportion. Blood glucose levels were measured by tail punch immediately before and at 15, 30, 60, and 120 minutes after injection. ITTs were performed after fasting of the animals for 5 hours (10–15 hours), and 0.5 U/kg body weight insulin was administered by i.p. injection. Glucose levels were measured by tail punch at time points 0, 15, 30, and 60 minutes after injection and were plotted to the initial glucose levels. Briefly, for glucose-stimulated insulin secretion (GSIS), animals were fasted for 16 hours overnight, and 20% (vol/vol) dextrose was given through an i.p. injection in a 3 g/kg body weight proportion. Serum was collected at 0, 2, and 5 minutes after glucose injection.
Lipid isolation and measurements.
Livers and HepG2 lysates were homogenized for 10 minutes in an ice-cold chloroform/methanol (2:1) mixture. Neutral lipid extraction was performed overnight at room temperature. For phase separation, distilled water was added, samples were centrifuged, and the organic bottom layer was collected. The organic phase was dried using a SpeedVac (Thermo Fisher Scientific) and redissolved in chloroform. Triglyceride and cholesterol content of each sample was measured after evaporation of the organic phase using colorimetric kits according to manufacturer protocol (Stanbio LiquiColor Triglycerides and Stanbio Cholesterol LiquiColor).
RNA isolation and RT-PCR.
RNA isolation and RT-PCR were performed as previously reported (63). In brief, total RNA (>200 nt) was extracted using standard Trizol reagent (Invitrogen) according to manufacturer instructions. The resultant aqueous phase was mixed (1:1) with 70% RNA-free ethanol and added to Qiagen RNeasy Mini Kit columns (Qiagen), and the kit protocol was followed. RNA quality and quantity were analyzed using NanoDrop 1000 and used for the reserve transcription step using the high-capacity cDNA synthesis kit (Applied Biosciences). cDNA was analyzed using the ABI 7900HT system (Applied Biosciences), and gene expression was calculated using the ΔΔCt method. Data were normalized to β-actin. See Supplemental Table 8 for primers.
Western blotting.
Total proteins were harvested from tissue and cell line lysates using RIPA buffer and M-PER protein extraction reagent (Thermo Fisher Scientific), respectively, supplemented with proteinase and phosphatase inhibitors (MilliporeSigma) according to the manufacturer’s guidelines. Protein concentrations were determined using the BCA standard protocol followed by standard Western immunoblotting protocol of proteins. The blots were developed using chemiluminescent substrate ECL (Thermo Fisher Scientific) and quantified using Image studio Lite version 5.2 software (LI-COR). See Supplemental Table 9 for antibody details and online supplemental material for uncut original blots.
Fatty acid oxidation by Seahorse.
The Seahorse XF96 (Agilent) was used to measure oxygen consumption rate (OCR) according to the manufacturer’s protocol. Cells were seeded into XF96 cell culture microplates at a density of 50,000 cells per well. To determine fatty acid oxidation (FAO), cells were incubated with a substrate-limited medium 1 day before, then FAO running medium plus BSA-palmitate substrate was added with or without etomoxir (ETO), an inhibitor of carnitine palmitoyltransferase-1 (CPT1) (Agilent). After measurement of the basal FAO respiration, 1 μM oligomycin, 0.5 μM FCCP, and 2 μM antimycin were subsequently injected to determine ATP-coupled, maximal, nonmitochondrial FAO, and FAO-contributed OCR was calculated by subtraction of the non-FAO respiration (with ETO) from the total respiration (without ETO).
Cell culture and treatments.
HepG2 cells were obtained from ATCC and were cultured in DMEM supplemented with 10% (vol/vol) FBS and nonessential amino acids. For in vitro modeling of hepatic steatosis in HepG2, cells were treated with fatty acid–free BSA (MilliporeSigma) or 500 μM of albumin-conjugated palmitate in 25 mM glucose DMEM for 24 hours. Albumin-conjugated palmitate was prepared by dissolving of sodium palmitate (MilliporeSigma) with distilled water and NaOH at 70°C and then conjugated with fatty acid–free albumin (BSA; MilliporeSigma). Human plateable primary hepatocytes were cultured in William’s E medium supplemented with primary hepatocyte supplements (Thermo Fisher Scientific) and HepExtend supplement (Thermo Fisher Scientific). For the analyses of HepG2 supernatant, 12 mL of HepG2 FBS-free culture medium was collected after 72 hours of knockdown or NREP overexpression and passed through a 0.22-μm filter. The supernatant was further centrifuged at maximum speed for 10 minutes. A total of 10 mL of cell culture supernatant was purified and concentrated into 100 μL using StrataResin (Agilent) according to the manufacturer protocol. Forty microliters of the resulting medium was run on a 20% SDS-PAGE gel according to Western blot standard protocol.
siRNA gene silencing.
HepG2 cells were trypsinized at 70% confluence, and reverse transfection was performed using 30 nM of genome SMARTpool Non-targeting siRNA (scramble), siNREP, or siGDF15 targeting 4 different sequences (Dharmacon). Lipofectamine RNAiMA reagent (Invitrogen) in Opti-MEM (Invitrogen) was used, and medium was exchanged after 8 hours of transfection. After 48 hours, cells were treated with DMEM containing BSA or BSA-palmitate for 24 hours. Human plateable primary hepatocytes were seeded on collagen-treated plates containing William’s E medium supplemented with primary hepatocyte supplements (Thermo Fisher Scientific) and HepExtend supplement (Thermo Fisher Scientific), and left for 6 hours to allow attachment. Cells were forward-transfected with 100 nM of nontargeting siRNA or siNREP, and medium was exchanged after 12 hours. At 24 hours, cells were treated with William’s E supplemented with BSA or BSA-palmitate for 24 hours. See Supplemental Table 9 for siRNA information.
Plasmid transfections.
HepG2 cells at 70% confluence were forward-transfected with pCMV-empty, pCMV-Myc-NREP, or pCMV-Myc-GDF15 (Origene) using Lipofectamine 3000 (Invitrogen) and Opti-MEM (Invitrogen) according to manufacturer protocols. Medium was exchanged with fresh DMEM after 8 hours of transfection, and at 48 hours cells were treated with DMEM containing BSA or BSA-palmitate for 24 hours. See Supplemental Table 9 for plasmid information.
Analytical methods used in human studies.
Plasma glucose, insulin, serum lipids, and lipoproteins were measured from fasting venous blood samples. Plasma glucose was measured by enzymatic hexokinase photometric assay (Konelab Systems Reagents, Thermo Fisher Scientific, Vantaa, Finland). Plasma insulin was determined by immunoassay (ADVIA Centaur Insulin IRI, no. 02230141, Siemens Medical Solutions Diagnostics). Cholesterol, HDL cholesterol, and triglyceride concentrations were assayed by standard automated enzymatic methods (Roche Diagnostics). Plasma NREP levels were measured with a quantitative sandwich ELISA using a capture mouse monoclonal antibody whose immunogen is a recombinant partial protein corresponding to amino acids 1–68 of human NREP and a detection rabbit polyclonal whose immunogen is a synthetic peptide corresponding to human NREP. Standards consisted of recombinant full-length human NREP protein expressed by E. coli (MBS9323406, MyBioSource). Plasma GDF15 levels were measured using a commercially available solid-phase sandwich ELISA (DGD150, R&D Systems).
Bioinformatics.
RNA sequencing data were aligned to the genome using STAR (64), mapped reads were assigned to genomic features using featureCounts (65), the counts were normalized using trimmed mean of M values (TMM) (66), and the normalized counts were incorporated into linear modeling using limma voom (67). DNA methylation data were analyzed with methylKit (68). FDRs were calculated using the Benjamini-Hochberg method. Bioinformatic analyses were done with R software. Pathway analyses were done in ConsensusPathDB (69).
Data availability.
Data from RNA sequencing performed on livers of control, FL, and ML offspring were deposited with the NCBI’s Gene Expression Omnibus under accession code GSE141077. ERRBS data from livers of control, FL, and ML offspring were deposited under accession code GSE141379. RNA sequencing data from HepG2 cells were deposited under accession code GSE141078.
Statistics.
Analyses were performed with GraphPad Prism 6.0. Normal distribution was calculated using the D’Agostino and Pearson omnibus normality test followed by unpaired, 2-tailed Student’s t test; 1-way ANOVA; or 2-way ANOVA followed by Dunn’s, Kruskal-Wallis, Holm-Šidák, or least significant difference multiple-comparisons tests, as appropriate. Spearman’s rank correlation was used for correlation analysis. Correlation analyses were conducted with SPSS version 25 (IBM SPSS Statistics). P values less than 0.05 were considered statistically significant.
Study approval.
All animal experiments were conducted in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care. All protocols were approved by the Institutional Animal Care and Use Committee of the Joslin Diabetes Center in accordance with NIH guidelines. All human studies and protocols used were approved by the Joslin Diabetes Center’s Committee on Human Studies (CHS#5-05). The study protocol of the Kuopio cohort was approved by the Ethics Committee of Northern Savo Hospital District (54/2005, 104/2008, and 27/2010), and carried out in accordance with the Helsinki Declaration. Informed written consent was obtained from all participants.
Author contributions
DFDJ conceived the idea, designed and performed the experiments, analyzed the data, and wrote the manuscript. KO assisted with overexpression experiments. TK assisted with the experiments on AML12 cells and human primary hepatocytes. GB assisted with the implantation of the osmotic pumps. CHW performed Seahorse FAO. NKB assisted with RT-PCR experiments. JH performed immunohistochemistry. ED assisted with in vivo GSIS. LH and RR assisted with DNA methylation experiments. AMS, YHT, and TH researched data and assisted with technical support and/or critical reading of the manuscript. DK, VM, and JP assisted with human serum samples and correlation experiments. JP is the principal investigator of the Kuopio Obesity Surgery Study. RNK contributed to conceptual discussions, designed the experiments, supervised the project, and wrote the manuscript. All the authors reviewed, commented and edited the manuscript.
Supplementary Material
Acknowledgments
LIRKO mice were provided by C. Ronald Kahn (Joslin Diabetes Center). We thank Ines Sousa-Lima (CEDOC), Mary Elizabeth Patti (Joslin Diabetes Center), and Stephan Kissler (Joslin Diabetes Center) for advice. We thank the Joslin Specialized Assay Core, Joslin Physiology Core, and Joslin Bioinformatics Core (NIH grant P30 DK36836). We thank the Epigenomics Core at Weill Cornell Medicine for assistance with ERRBS. We thank the Genomic Services Laboratory at HudsonAlpha Institute for Biotechnology for assistance with RNA sequencing. DK was supported by the Academy of Finland (contract 316458). DFDJ was supported by the Portuguese Foundation for Science and Technology (FCT) (SFRH/BD/51699/2011), Albert Ronald Travel Fellowship 2015, and 2016 FLAD R&D@PhD Internship grant. AMS acknowledges support by the FCT under project UID/AGR/04033/2019 (CITAB). YHT is supported by NIH grants R01 DK077097 and R01 DK102898. JP and the Kuopio Obesity Study were supported by grants from the Academy of Finland (contract 138006), Finnish Diabetes Research Foundation, Kuopio University Hospital Project (NUDROBE), and Finnish Cultural Foundation (to JP). RNK is supported by NIH grants R01 DK067536, R01 DK103215, and R01 DK117639.
Version 1. 04/06/2020
Electronic publication
Version 2. 05/01/2020
Print issue publication
Funding Statement
Y-HT is supported by NIH grants R01 DK077097 and R01 DK102898.
RNK is supported by NIH grants R01 DK067536, R01 DK103215 and R01 DK117639.
DFDJ was supported by the Portuguese Foundation for Science and Technology (FCT) – SFRH/BD/51699/2011, Albert Ronald Travel Fellowship 2015 and 2016 FLAD R&D@PhD Internship grant.
AMS acknowledges support by FCT under the project UID/AGR/04033/2019 (CITAB).
JP and the Kuopio Obesity Study were supported by the grants from the Academy of Finland (contract number 138006), the Finnish Diabetes Research Foundation, Kuopio University Hospital Project grant (NUDROBE) and the Finnish Cultural Foundation (JPI).
Footnotes
ED’s present address is: Department of Pharmacology, School of Medicine, New York Medical College, Valhalla, New York, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(5):2391–2407.https://doi.org/10.1172/JCI127502.
Contributor Information
Dario F. De Jesus, Email: Dario.DeJesus@joslin.harvard.edu.
Kazuki Orime, Email: Kazuki.Orime@joslin.harvard.edu.
Dorota Kaminska, Email: dorota.kaminska@uef.fi.
Tomohiko Kimura, Email: Tomohiko.Kimura@joslin.harvard.edu.
Giorgio Basile, Email: Giorgio.Basile@joslin.harvard.edu.
Chih-Hao Wang, Email: Chih-Hao.wang@joslin.harvard.edu.
Larissa Haertle, Email: larissa.haertle@uni-wuerzburg.de.
Jiang Hu, Email: Jiang.Hu@joslin.harvard.edu.
Ville Männistö, Email: ville.mannisto@kuh.fi.
Ercument Dirice, Email: ercument.dirice@joslin.harvard.edu.
Yu-Hua Tseng, Email: yu-hua.tseng@joslin.harvard.edu.
Thomas Haaf, Email: thomas.haaf@uni-wuerzburg.de.
Jussi Pihlajamäki, Email: jussi.pihlajamaki@uef.fi.
References
- 1.Brunt EM, Tiniakos DG. Histopathology of nonalcoholic fatty liver disease. World J Gastroenterol. 2010;16(42):5286–5296. doi: 10.3748/wjg.v16.i42.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumbaugh DE, Friedman JE. Developmental origins of nonalcoholic fatty liver disease. Pediatr Res. 2014;75(1-2):140–147. doi: 10.1038/pr.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi Z, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 4.Bush H, Golabi P, Younossi ZM. Pediatric non-alcoholic fatty liver disease. Children (Basel) 2017;4(6):E48. doi: 10.3390/children4060048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwimmer JB, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136(5):1585–1592. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loomba R, et al. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149(7):1784–1793. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wruck W, et al. Multi-omic profiles of human non-alcoholic fatty liver disease tissue highlight heterogenic phenotypes. Sci Data. 2015;2:150068. doi: 10.1038/sdata.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jesus DF, Kulkarni RN. Epigenetic modifiers of islet function and mass. Trends Endocrinol Metab. 2014;25(12):628–636. doi: 10.1016/j.tem.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Sales VM, Ferguson-Smith AC, Patti ME. Epigenetic mechanisms of transmission of metabolic disease across generations. Cell Metab. 2017;25(3):559–571. doi: 10.1016/j.cmet.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahraman S, Dirice E, De Jesus DF, Hu J, Kulkarni RN. Maternal insulin resistance and transient hyperglycemia impact the metabolic and endocrine phenotypes of offspring. Am J Physiol Endocrinol Metab. 2014;307(10):E906–E918. doi: 10.1152/ajpendo.00210.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeybel M, et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat Med. 2012;18(9):1369–1377. doi: 10.1038/nm.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCurdy CE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119(2):323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorn SR, et al. Early life exposure to maternal insulin resistance has persistent effects on hepatic NAFLD in juvenile nonhuman primates. Diabetes. 2014;63(8):2702–2713. doi: 10.2337/db14-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael MD, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6(1):87–97. doi: 10.1016/S1097-2765(05)00015-8. [DOI] [PubMed] [Google Scholar]
- 15.Biddinger SB, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7(2):125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton KP, et al. Low and high birth weights are risk factors for nonalcoholic fatty liver disease in children. J Pediatr. 2017;187:141–146.e1. doi: 10.1016/j.jpeds.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savage DB, Petersen KF, Shulman GI. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev. 2007;87(2):507–520. doi: 10.1152/physrev.00024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metab Clin Exp. 2016;65(8):1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Garrett-Bakelman FE, et al. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. J Vis Exp. 2015;(96):e52246. doi: 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao Z, et al. P311 promotes renal fibrosis via TGFβ1/Smad signaling. Sci Rep. 2015;5:17032. doi: 10.1038/srep17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan D, Zhe X, Jakkaraju S, Taylor GA, Schuger L. P311 induces a TGF-beta1-independent, nonfibrogenic myofibroblast phenotype. J Clin Invest. 2002;110(9):1349–1358. doi: 10.1172/JCI15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L, Leung JK, Yamamoto H, Goswami S, Kheradmand F, Vu TH. Identification of P311 as a potential gene regulating alveolar generation. Am J Respir Cell Mol Biol. 2006;35(1):48–54. doi: 10.1165/rcmb.2005-0475OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, et al. Transforming growth factor beta signaling in hepatocytes participates in steatohepatitis through regulation of cell death and lipid metabolism in mice. Hepatology. 2014;59(2):483–495. doi: 10.1002/hep.26698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61(3):1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep. 2016;6:22640. doi: 10.1038/srep22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi-Barve S, et al. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology. 2007;46(3):823–830. doi: 10.1002/hep.21752. [DOI] [PubMed] [Google Scholar]
- 27.Gao D, et al. The effects of palmitate on hepatic insulin resistance are mediated by NADPH oxidase 3-derived reactive oxygen species through JNK and p38MAPK pathways. J Biol Chem. 2010;285(39):29965–29973. doi: 10.1074/jbc.M110.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Ruiz I, Solís-Muñoz P, Fernández-Moreira D, Muñoz-Yagüe T, Solís-Herruzo JA. In vitro treatment of HepG2 cells with saturated fatty acids reproduces mitochondrial dysfunction found in nonalcoholic steatohepatitis. Dis Model Mech. 2015;8(2):183–191. doi: 10.1242/dmm.018234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harasim-Symbor E, Konstantynowicz-Nowicka K, Chabowski A. Additive effects of dexamethasone and palmitate on hepatic lipid accumulation and secretion. J Mol Endocrinol. 2016;57(4):261–273. doi: 10.1530/JME-16-0108. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Beamonte R, et al. Postprandial transcriptome associated with virgin olive oil intake in rat liver. Front Biosci (Elite Ed) 2011;3:11–21. doi: 10.2741/e215. [DOI] [PubMed] [Google Scholar]
- 31.Fabregat I, et al. TGF-β signalling and liver disease. FEBS J. 2016;283(12):2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- 32.Pinkosky SL, et al. Liver-specific ATP-citrate lyase inhibition by bempedoic acid decreases LDL-C and attenuates atherosclerosis. Nat Commun. 2016;7:13457. doi: 10.1038/ncomms13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinkosky SL, Groot PHE, Lalwani ND, Steinberg GR. Targeting ATP-citrate lyase in hyperlipidemia and metabolic disorders. Trends Mol Med. 2017;23(11):1047–1063. doi: 10.1016/j.molmed.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, et al. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. 2009;49(4):1166–1175. doi: 10.1002/hep.22774. [DOI] [PubMed] [Google Scholar]
- 35.Burke AC, Huff MW. ATP-citrate lyase: genetics, molecular biology and therapeutic target for dyslipidemia. Curr Opin Lipidol. 2017;28(2):193–200. doi: 10.1097/MOL.0000000000000390. [DOI] [PubMed] [Google Scholar]
- 36.Sharpe LJ, Brown AJ. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) J Biol Chem. 2013;288(26):18707–18715. doi: 10.1074/jbc.R113.479808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min HK, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15(5):665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starmann J, et al. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One. 2012;7(10):e46584. doi: 10.1371/journal.pone.0046584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson IB, Rifkin DB. Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb Perspect Biol. 2016;8(6):a021907. doi: 10.1101/cshperspect.a021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koo BK, et al. Growth differentiation factor 15 predicts advanced fibrosis in biopsy-proven non-alcoholic fatty liver disease. Liver Int. 2018;38(4):695–705. doi: 10.1111/liv.13587. [DOI] [PubMed] [Google Scholar]
- 41.Kim KH, Kim SH, Han DH, Jo YS, Lee YH, Lee MS. Growth differentiation factor 15 ameliorates nonalcoholic steatohepatitis and related metabolic disorders in mice. Sci Rep. 2018;8(1):6789. doi: 10.1038/s41598-018-25098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JH, Friso S, Choi SW. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients. 2014;6(8):3303–3325. doi: 10.3390/nu6083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleming TP, et al. Origins of lifetime health around the time of conception: causes and consequences. Lancet. 2018;391(10132):1842–1852. doi: 10.1016/S0140-6736(18)30312-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grasemann C, et al. Parental diabetes: the Akita mouse as a model of the effects of maternal and paternal hyperglycemia in wildtype offspring. PLoS One. 2012;7(11):e50210. doi: 10.1371/journal.pone.0050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isganaitis E, et al. Developmental programming by maternal insulin resistance: hyperinsulinemia, glucose intolerance, and dysregulated lipid metabolism in male offspring of insulin-resistant mice. Diabetes. 2014;63(2):688–700. doi: 10.2337/db13-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen SE, et al. High circulating leptin receptors with normal leptin sensitivity in liver-specific insulin receptor knock-out (LIRKO) mice. J Biol Chem. 2007;282(32):23672–23678. doi: 10.1074/jbc.M704053200. [DOI] [PubMed] [Google Scholar]
- 47.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB. Transforming growth factor-beta in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem. 1983;258(11):7155–7160. [PubMed] [Google Scholar]
- 49.Ahrens M, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277(37):33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 51.Matsuda S, Kobayashi M, Kitagishi Y. Roles for PI3K/AKT/PTEN pathway in cell signaling of nonalcoholic fatty liver disease. ISRN Endocrinol. 2013;2013:472432. doi: 10.1155/2013/472432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8(3):224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krycer JR, Sharpe LJ, Luu W, Brown AJ. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol Metab. 2010;21(5):268–276. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 54.He L, et al. The critical role of AKT2 in hepatic steatosis induced by PTEN loss. Am J Pathol. 2010;176(5):2302–2308. doi: 10.2353/ajpath.2010.090931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong SH, et al. Hippo-mediated suppression of IRS2/AKT signaling prevents hepatic steatosis and liver cancer. J Clin Invest. 2018;128(3):1010–1025. doi: 10.1172/JCI95802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imamura M, et al. RIPK3 promotes kidney fibrosis via AKT-dependent ATP citrate lyase. JCI Insight. 2018;3(3):e94979. doi: 10.1172/jci.insight.94979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paliwal S, Shi J, Dhru U, Zhou Y, Schuger L. P311 binds to the latency associated protein and downregulates the expression of TGF-beta1 and TGF-beta2. Biochem Biophys Res Commun. 2004;315(4):1104–1109. doi: 10.1016/j.bbrc.2004.01.171. [DOI] [PubMed] [Google Scholar]
- 58.Walle P, et al. Fatty acid metabolism is altered in non-alcoholic steatohepatitis independent of obesity. Metab Clin Exp. 2016;65(5):655–666. doi: 10.1016/j.metabol.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Männistö VT, et al. Ketone body production is differentially altered in steatosis and non-alcoholic steatohepatitis in obese humans. Liver Int. 2015;35(7):1853–1861. doi: 10.1111/liv.12769. [DOI] [PubMed] [Google Scholar]
- 60.De Jesus DF, et al. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat Metab. 2019;1(8):765–774. doi: 10.1038/s42255-019-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dirice E, et al. Human duct cells contribute to β cell compensation in insulin resistance. JCI Insight. 2019;4(8):99576. doi: 10.1172/jci.insight.99576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dirice E, et al. Increased β-cell proliferation before immune cell invasion prevents progression of type 1 diabetes. Nat Metab. 2019;1(5):509–518. doi: 10.1038/s42255-019-0061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta MK, et al. Insulin receptor-mediated signaling regulates pluripotency markers and lineage differentiation. Mol Metab. 2018;18:153–163. doi: 10.1016/j.molmet.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 66.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11(10):1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from RNA sequencing performed on livers of control, FL, and ML offspring were deposited with the NCBI’s Gene Expression Omnibus under accession code GSE141077. ERRBS data from livers of control, FL, and ML offspring were deposited under accession code GSE141379. RNA sequencing data from HepG2 cells were deposited under accession code GSE141078.