Abstract
Lipid-rich myelin forms electrically insulating, axon-wrapping multilayers that are essential for neural function, and mature myelin is traditionally considered metabolically inert. Surprisingly, we discovered that mature myelin lipids undergo rapid turnover, and quaking (Qki) is a major regulator of myelin lipid homeostasis. Oligodendrocyte-specific Qki depletion, without affecting oligodendrocyte survival, resulted in rapid demyelination, within 1 week, and gradually neurological deficits in adult mice. Myelin lipids, especially the monounsaturated fatty acids and very-long-chain fatty acids, were dramatically reduced by Qki depletion, whereas the major myelin proteins remained intact, and the demyelinating phenotypes of Qki-depleted mice were alleviated by a high-fat diet. Mechanistically, Qki serves as a coactivator of the PPARβ-RXRα complex, which controls the transcription of lipid-metabolism genes, particularly those involved in fatty acid desaturation and elongation. Treatment of Qki-depleted mice with PPARβ/RXR agonists significantly alleviated neurological disability and extended survival durations. Furthermore, a subset of lesions from patients with primary progressive multiple sclerosis were characterized by preferential reductions in myelin lipid contents, activities of various lipid metabolism pathways, and expression level of QKI-5 in human oligodendrocytes. Together, our results demonstrate that continuous lipid synthesis is indispensable for mature myelin maintenance and highlight an underappreciated role of lipid metabolism in demyelinating diseases.
Keywords: Metabolism, Neuroscience
Keywords: Demyelinating disorders, Multiple sclerosis
Introduction
Myelin is a specialized multilayer membrane structure that enables rapid saltatory nerve conduction, and it is assembled by oligodendrocytes in the central nervous system (CNS) (1). To sustain the normal function of neurons, the compact myelin structure needs to persist throughout adult life (1). Many myelin-related CNS diseases, including multiple sclerosis (MS), develop in adulthood, underscoring the importance of active maintenance of the structural integrity of myelin in preventing neurological disability (2). Traditionally, demyelinating processes are thought to be the result of oligodendrocyte death induced by either extrinsic or intrinsic insults (3–5). However, evidence suggests that oligodendrocyte death is not the underlying etiology for all pathological demyelinating processes. More specifically, oligodendrocytes are largely preserved in 30%–60% of early active MS lesions with severe demyelination (6–8), and the turnover rates of oligodendrocytes remain generally low, albeit heterogeneous, across multiple CNS regions during aging despite global myelin volume decline (9–12). These observations suggest that myelin content could be tightly regulated in a manner that is independent of oligodendrocyte survival.
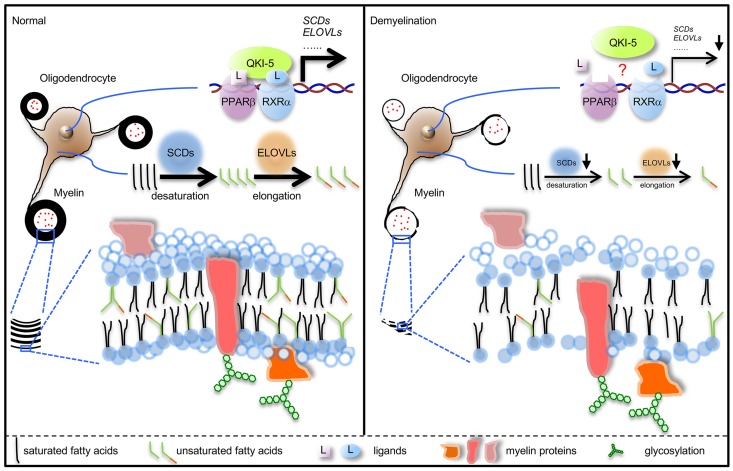
The dry mass of CNS myelin is composed of approximately 70% lipids and 30% proteins — a composition that is characterized by a higher ratio of lipids to proteins than those found in other biological membranes (1). Myelin is distinguished by its high proportion of cholesterol and cerebroside (13). In addition, myelin contains approximately twice as much oleic acid (fatty acid 18:1 cis-9) and more than 5 times the level of long-chain fatty acids (LCFAs) versus levels found in erythrocyte or hepatocyte membranes (13). It has long been observed that major myelin structural proteins and myelin cholesterol exhibit very slow rates of turnover in rodents, leading to the widely believed presumption for decades that CNS myelin is a static material (14–17). Paradoxically, the discoveries of metabolic enzymes in purified myelin indicate that myelin might be a metabolically dynamic substance with rapid turnover of some of its building blocks (18, 19).
The currently available oligodendrocyte-specific genetic demyelinating mouse models are all induced or accompanied by oligodendrocyte apoptosis, rendering these models unsuitable for studying myelin turnover per se (3, 4, 20–22). Unlike those proteins involved in oligodendrocyte survival (20–22), quaking (Qki, encoded by the gene Qk, which undergoes alternative splicing to express Qki-5, Qki-6, and Qki-7), a known regulator of oligodendrocyte differentiation and myelination (23–28), does not regulate cell death (29). In addition, our recent analyses suggested that Qki regulates an array of metabolic pathways in neural stem cells (NSCs) (29), raising the possibility that Qki might be involved in the metabolic turnover of myelin structural components.
In this study, we used an oligodendrocyte-specific Qk conditional knockout mouse model as a tool to address the following questions: (i) Are the structural components of mature myelin static or dynamic? (ii) Is the continuously active metabolism of myelin components required for the maintenance of myelin integrity in adulthood? (iii) Does interruption of myelin metabolic homeostasis play a pathogenic role in demyelinating diseases? Here, we found that structural lipid components of mature myelin are highly dynamic and their turnover is tightly controlled by Qki via coactivation of the peroxisome proliferator-activated receptor β–retinoid X receptor α (PPARβ-RXRα) complex. A continuous supply of the myelin lipids is essential for the maintenance of myelin integrity in adulthood. Our data reveal a previously underappreciated role of lipid metabolism in demyelinating diseases and pave the way for future development of novel disease-modifying therapies that minimize or even reverse myelin loss by modulating lipid biosynthesis in mature oligodendrocytes.
Results
Qki regulates myelin homeostasis independently of oligodendrocyte death.
To determine whether Qki is required for mature oligodendrocyte survival and myelin maintenance, we deleted Qk in mature myelinating oligodendrocytes by injecting Plp-CreERT2 Qkfl/fl mice (hereafter called Qk-iCKO mice) with tamoxifen at age 8 weeks — the time by which the myelin structure is fully established (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI131800DS1) (1). Approximately 2 weeks after tamoxifen injection, Qk-iCKO mice progressively developed tremors, splayed gait, ataxia, and hind limb paresis that culminated in paralysis with hunched posture, hyperpnea, significant weight loss, and early lethality (Supplemental Figure 1, C–F, and Supplemental Video 1). In contrast, both Plp-CreERT2 Qkfl/+ mice and wild-type (WT) mice displayed no neurological signs; therefore, both cohorts were used as controls.
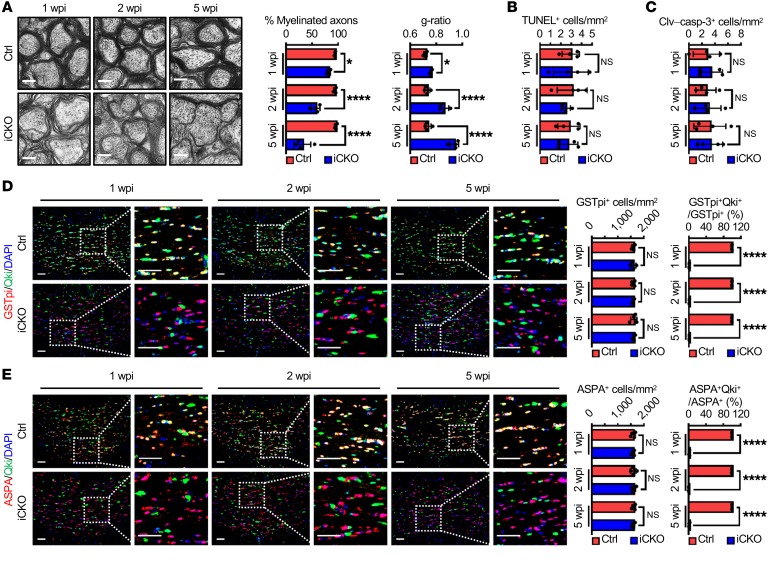
Electron microscopy–based ultrastructural analyses of the optic nerves revealed that demyelination occurred as early as 1 week after Qk deletion. We found that 18.2%, 43.1%, and 68.4% of the axons in Qk-iCKO mice were completely devoid of myelin sheaths 1, 2, and 5 weeks after tamoxifen injection, respectively, whereas about 95% of the axons in controls were wrapped by compact myelin (Figure 1A and Supplemental Figure 1G). Moreover, the myelin sheaths surrounding spared axons in Qk-iCKO mice became gradually thinner than did those in controls (ratio of the inner axonal diameter to the total outer diameter [g-ratio]: 0.76 versus 0.72, 0.86 versus 0.73, and 0.94 versus 0.74 at 1, 2, and 5 weeks after tamoxifen injection, respectively; Figure 1A). Similarly, the percentages of myelinated axons and the thicknesses of myelin sheaths in the corpus callosum and spinal cords of Qk-iCKO mice were also significantly reduced compared with those in the respective CNS regions of control mice (Supplemental Figure 1, H and I), revealing that Qki has a global impact on the mature myelin maintenance across multiple CNS regions. In addition, irregular nodes of Ranvier were frequently observed in the optic nerves of Qk-iCKO mice (Supplemental Figure 1, J and K), and the optic nerves of Qk-iCKO mice had an approximately 54.5% decrease in their conduction velocities compared with the control mice (Supplemental Figure 1, L–N). As the morphological characteristics, diameter, and density of axons remained unaffected in the optic nerves of Qk-iCKO mice (Supplemental Figure 1, O and P), these data suggest that demyelination itself is the cause of the progressive neurological deficits and increased mortality of Qk-iCKO animals. Besides, we also observed demyelination-induced gliosis and inflammation, including upregulation of GFAP+ activated astrocytes and IBA1+ microglia and infiltration of CD3+ T cells, in the corpus callosum of Qk-iCKO mice (Supplemental Figure 1, Q–S).
Figure 1. Qki regulates myelin homeostasis independently of oligodendrocyte death.
(A) Representative electron micrographs and quantification of the optic nerves of Qk-iCKO mice and controls (Ctrl) 1, 2, and 5 weeks post tamoxifen injection (wpi). n = 4 mice/group. Scale bars: 500 nm. (B and C) Quantification of dead cells by TUNEL assay and immunofluorescent staining of cleaved caspase 3 (Clv–casp-3) in the corpus callosum of the experimental mice in A (n = 4 mice/group). (D and E) Representative images and quantification of immunofluorescent staining of GSTpi-Qki (D) and ASPA-Qki (E) in the corpus callosum of the experimental mice in A (n = 4 mice/group). Scale bars: 50 μm. Data are mean ± SD. *P < 0.05; ****P < 0.0001 by 2-way ANOVA with Holm-Sidak multiple-comparisons test. NS, not significant.
We next determined whether demyelination induced by Qki depletion occurs in an oligodendrocyte death–independent manner. Neither a TUNEL assay nor cleaved caspase 3 staining revealed a significant difference in the number of apoptotic cells in Qk-iCKO mice and controls across multiple CNS regions (Figure 1, B and C, and Supplemental Figure 2, A and B), and the numbers of oligodendrocytes in the corpus callosum, optic nerves, and spinal cords of Qk-iCKO mice were similar to those in the respective CNS regions of controls (Figure 1, D and E, and Supplemental Figure 2, C–G). More importantly, more than 97% of the oligodendrocytes still lacked Qki expression 5 weeks after tamoxifen injection (Figure 1, D and E, and Supplemental Figure 2, C–G), indicating that most cells were the resident oligodendrocytes with Qk deletion but not the newly differentiated oligodendrocytes from oligodendrocyte precursor cells (OPCs), which were positive for Qki expression (Supplemental Figure 2H). Therefore, although the number of proliferating OPCs was higher in Qk-iCKO mice 5 weeks after tamoxifen injection than that in controls (Supplemental Figure 2I), it appears that these OPCs had not differentiated into mature oligodendrocytes. In summary, the observation of severe demyelination without overt oligodendrocyte death in Qk-iCKO animals indicates that mature myelin itself is not static, providing what we believe is a unique mouse model to study oligodendrocyte death–independent myelin homeostasis.
Qki governs mature myelin lipid homeostasis.
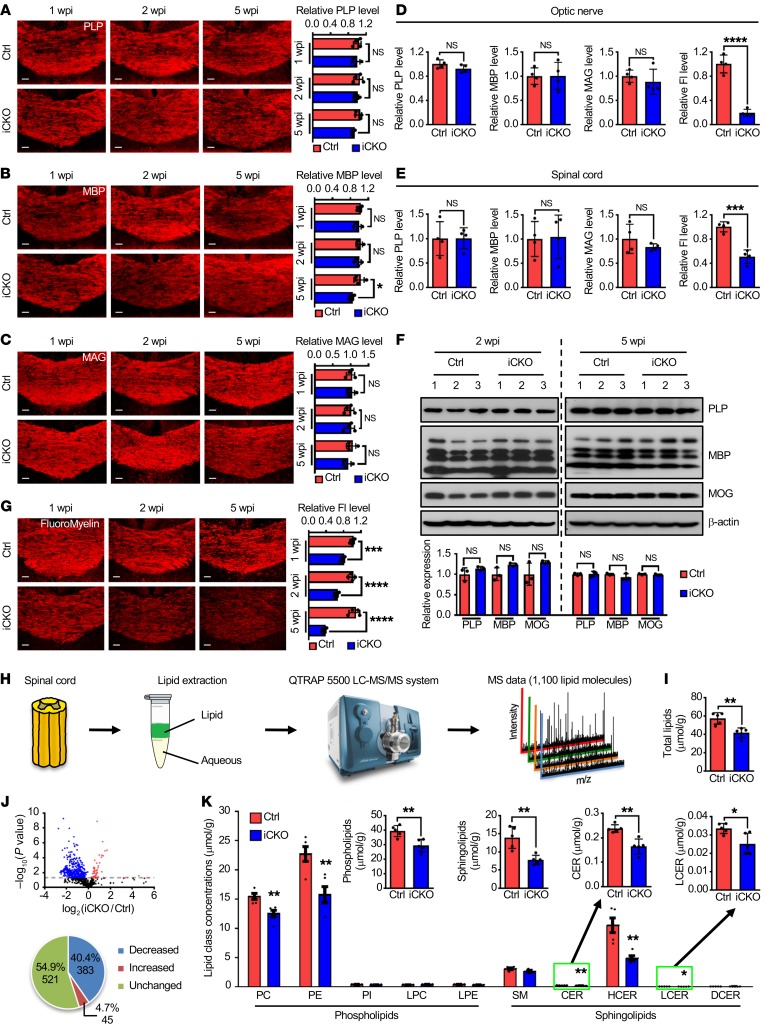
To determine how myelin homeostasis is disturbed in Qk-iCKO mice, we measured the effect of Qki depletion on major myelin structural proteins, including myelin proteolipid protein (PLP), myelin basic protein (MBP), and myelin-associated glycoprotein (MAG). Unexpectedly, immunofluorescent staining revealed only slightly lower levels of these proteins in the corpus callosum of Qk-iCKO mice than in controls (Figure 2, A–C) and unchanged levels in the optic nerves and spinal cords (Figure 2, D and E). Similar results were confirmed by immunoblotting in Qk-iCKO mice and controls 2 and 5 weeks after tamoxifen injection (Figure 2F). However, the degenerative myelin, indicated by staining with the QD-9 antibody, was dramatically increased in the corpus callosum (85.7-fold), optic nerves (39.2-fold), and spinal cords (9.3-fold) of Qk-iCKO mice relative to those in the respective CNS regions of control mice (Supplemental Figure 3). The QD-9 antibody is specific for the exposed epitope of MBP, QDENPVV, which is usually hidden due to the electrostatic interactions of MBP with lipids (30, 31). Therefore, we hypothesized that the drastic epitope unmasking of MBP in Qk-iCKO mice might be due to the dissociation of MBP and myelin lipids. Because 70% of the dry mass of myelin is composed of lipids (1), we next measured total myelin lipid levels using FluoroMyelin, a lipophilic dye with a high selectivity for myelin (32); we observed dramatically lower lipid levels in the corpus callosum (32.7%), optic nerves (19.5%), and spinal cords (50.8%) of Qk-iCKO mice 5 weeks after tamoxifen injection than in controls (Figure 2, D, E, and G). More importantly, the reduction in myelin lipids in Qk-iCKO mice was observed as early as 1 week after tamoxifen injection (Figure 2G), which was much earlier than gliosis, T cell infiltration, and the slight reduction in major myelin structural proteins (Figure 2, A–C, and Supplemental Figure 1, Q–S), suggesting that autonomous myelin lipid reduction is the primary event after Qki depletion and that Qki may control myelin integrity through regulating lipid metabolism.
Figure 2. Loss of Qki interrupts mature myelin lipid homeostasis.
(A–C) Representative images and quantification of PLP (A), MBP (B), and MAG (C) in the corpus callosum of Qk-iCKO mice and controls 1, 2, and 5 weeks post injection (wpi) (n = 4 mice/group). Scale bars: 50 μm. (D and E) Quantification of staining of PLP, MBP, MAG, and FluoroMyelin in the optic nerves (D) and spinal cords (E) of Qk-iCKO mice and controls 5 wpi (n = 4 mice/group). Fl, FluoroMyelin. (F) Immunoblots and quantification showing the expression of PLP, MBP, and myelin oligodendrocyte glycoprotein (MOG) of Qk-iCKO mice and controls 2 and 5 wpi (n = 3 mice/group). (G) Representative images and quantification of staining of FluoroMyelin in the corpus callosum of Qk-iCKO mice and controls 1, 2, and 5 wpi (n = 4 mice/group). Scale bars: 50 μm. (H) Schema depicting the workflow for lipidomic analyses. (I) Quantification of the concentration (μmol/g tissue) of the total lipids measured by mass spectrometry in the spinal cords of Qk-iCKO mice and controls 5 wpi (n = 5 mice/group). (J) Volcano plot (top) illustrating the alterations in the concentrations of each lipid molecule in the samples in I (n = 5 mice/group). The dotted line represents P = 0.05. Pie graph (bottom) illustrating the numbers and percentages of significantly decreased, increased, and unchanged molecules in Qk-iCKO mice relative to controls. (K) Quantification of the concentrations of lipid subclasses in the samples in I (n = 5 mice/group). PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; SM, sphingomyelin; CER, ceramide; HCER, hexosylceramide; LCER, lactosylceramide; DCER, dihydroceramide. Data are mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-way ANOVA with Holm-Sidak multiple-comparisons test (A–C and G) or Student’s t test (D–F, I, and K). NS, not significant.
To identify the lipid species affected by Qki depletion, we quantified lipid concentrations in the spinal cords of Qk-iCKO mice and controls using mass spectrometry–based lipidomic analyses (Figure 2H and Supplemental Tables 1 and 2). Consistent with the results obtained using FluoroMyelin staining, the overall lipid level in the spinal cords of Qk-iCKO mice 5 weeks after tamoxifen treatment was only 72.5% of that detected in controls (Figure 2I). The concentrations of 428 (45.1%) of the 949 lipid molecules detected by mass spectrometry were significantly altered after Qki depletion, and the levels of 383 (40.4%) were significantly reduced after Qki depletion (Figure 2J). Within the entire lipid composition, the total concentration of neutral lipids did not change significantly after Qki depletion (4.14 μmol/g tissue in controls versus 4.48 μmol/g tissue in Qk-iCKO mice; P = 0.68). In contrast, the concentrations of phospholipids and sphingolipids, the 2 major lipid types in myelin, in Qk-iCKO mice were 74.6% and 55.9%, respectively, of the levels in controls (Figure 2K). Taken together, these findings indicate that Qki may control mature myelin maintenance by regulating myelin lipid metabolism.
Qki regulates myelin fatty acid metabolism.
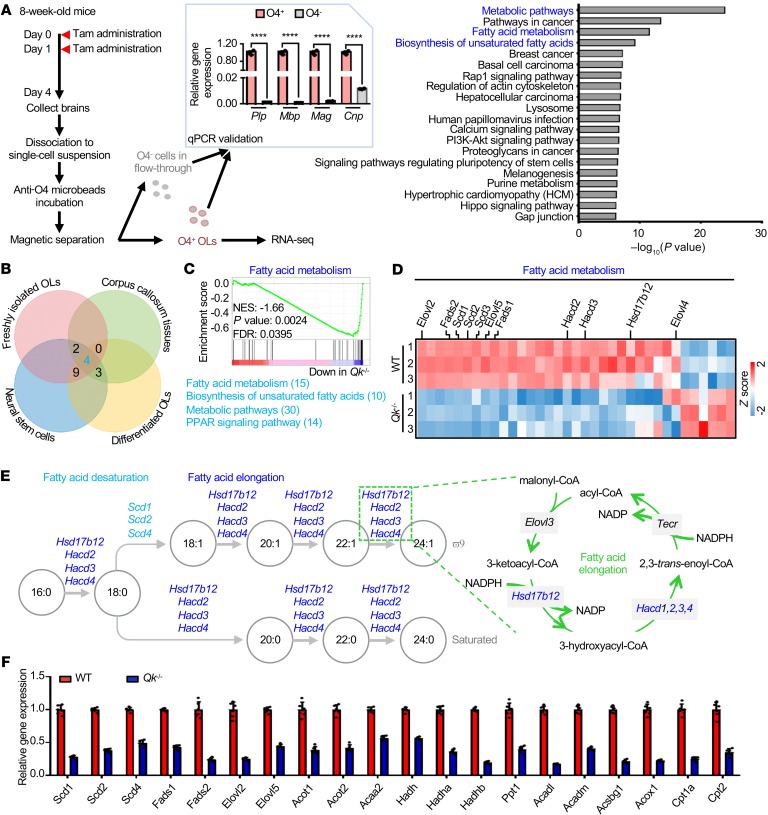
To understand how Qki regulates myelin lipid metabolism, we performed transcriptomic analyses by comparing the differentially expressed genes in freshly isolated oligodendrocytes from Qk-iCKO mice and controls, in the corpus callosum from Qk-iCKO mice and controls, in WT and Qki-depleted oligodendrocytes that differentiated from NSCs, and in WT and Qki-depleted NSCs (Figure 3A and Supplemental Figure 4, A–C). Strikingly, our comparative pathway analyses revealed that the biological processes that were most affected after Qki depletion were involved in fatty acid metabolism, particularly the biosynthesis of unsaturated fatty acids and fatty acid elongation (Figure 3, A–E, and Supplemental Figure 4, A–C). The transcriptional downregulation of these fatty acid metabolism genes after Qki depletion was verified using real-time quantitative PCR (RT-qPCR) (Figure 3F and Supplemental Figure 4D), indicating that the fatty acid metabolism genes are bona fide downstream targets of Qki.
Figure 3. Qki regulates the transcription of genes involved in fatty acid metabolism.
(A) Schema depicting the workflow for the fresh isolation of oligodendrocytes from Qk-iCKO mice and controls for RNA-seq (left). n = 3 mice/group. After being validated by RT-qPCR (middle), the O4+ oligodendrocytes (OLs) were used for RNA-seq. Bar graph (right) shows the top 20 enriched pathways. Tam, tamoxifen. (B) Venn diagram depicting the overlap among the top 50 enriched pathways identified from the RNA-seq of freshly isolated mouse OLs, differentiated OLs, NSCs, and adult corpus callosum. The numbers of common genes overlapped in more than 3 cell/tissue types are shown next to individual pathways. (C) GSEA shows the fatty acid metabolism signature in freshly isolated mouse oligodendrocytes. NES, normalized enrichment score; FDR, false discovery rate. (D) Heatmap representing the differentially expressed genes involved in fatty acid metabolism in freshly isolated OLs. (E) Schema depicting desaturating and elongating reactions of fatty acids with the corresponding enzymes. The reduction in fatty acid desaturases is shown in light blue, and the reduction in enzymes of the fatty acid elongation cycle is shown in dark blue. (F) RT-qPCR validation of the differentially expressed genes involved in fatty acid metabolism in WT and Qki-depleted (Qk–/–) differentiated oligodendrocytes. Data are representative of 3 independent experiments (F). Data are mean ± SD. ****P < 0.0001 by Student’s t test.
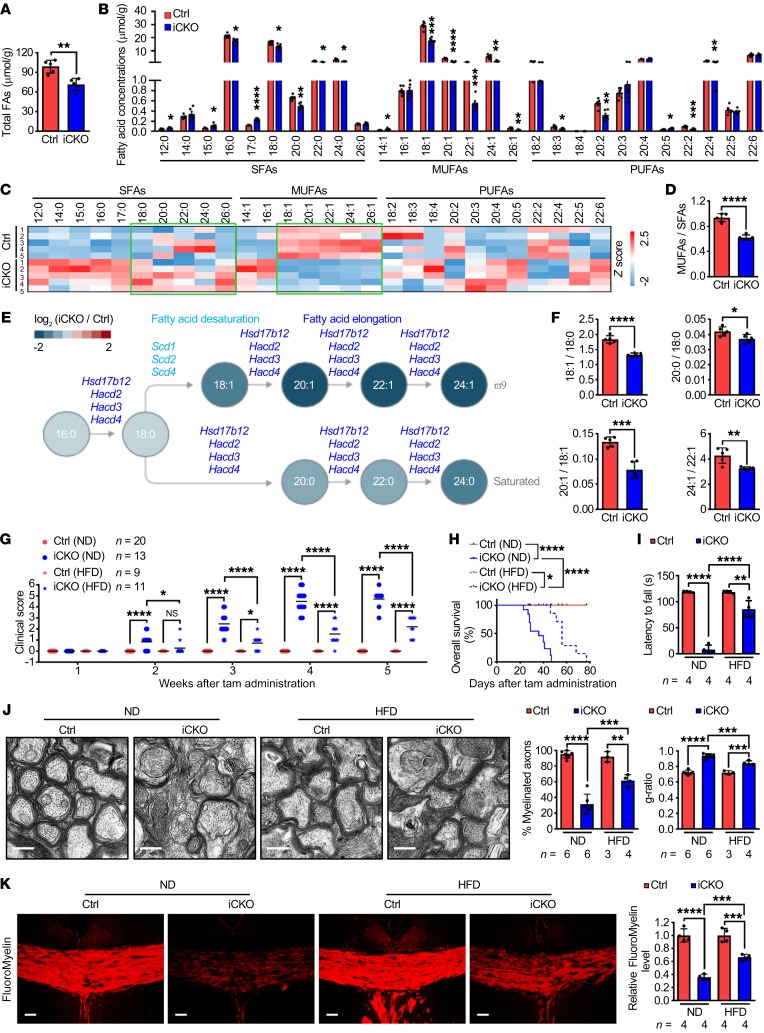
Fatty acid moieties are the major contributors to lipid molecule complexity. Indeed, consistent with the transcriptional regulation of fatty acid metabolism pathways by Qki, further analysis of the lipidomic data revealed that the overall fatty acid level in the spinal cords of Qk-iCKO mice was only 72.2% of that in controls (Figure 4A). Fatty acids can be classified by saturation status and by carbon chain length. We identified the fatty acids whose concentrations were the most substantially reduced by Qki depletion using each of these 2 classifications.
Figure 4. Qki controls fatty acid desaturation and elongation and HFD alleviates Qki deficiency–induced demyelination.
(A and B) Quantification of the concentration of total and each species of fatty acids (FAs) in the spinal cords of Qk-iCKO mice and controls 5 weeks post injection (wpi) (n = 5 mice/group). (C) Heatmap plotting the compositions of each fatty acid species in the samples in A (n = 5 mice/group). The green boxes highlight the saturated and monounsaturated LCFAs and VLCFAs. (D) Quantification of the MUFA/SFA ratios in the samples in A (n = 5 mice/group). (E) Schema showing the desaturating and elongating reactions of fatty acids with the reduction in specific fatty acid molecules (from lipidomic data) and the downregulated corresponding enzymes (from RNA-seq data) in Qk-iCKO mice relative to controls. (F) Quantification of the product/substrate ratios of representative fatty acid desaturating and elongating reactions in the samples in A (n = 5 mice/group). (G and H) The clinical scores (G) and Kaplan-Meier overall survival curves (log-rank test; H) of Qk-iCKO mice and controls fed a normal diet (ND) or HFD. The experimental mouse number is indicated in G. (I) Latency to fall (in seconds) off the rotarod (5 rpm) for Qk-iCKO mice and controls fed an ND or HFD for 5 wpi. (J) Representative electron micrographs and quantification of the percentage of myelinated axons and g-ratio of the optic nerves of Qk-iCKO mice and controls fed an ND or HFD for 5 wpi. Scale bars: 500 nm. (K) Representative images and quantification of FluoroMyelin level in the corpus callosum of Qk-iCKO mice and controls fed an ND or HFD for 5 wpi. Scale bars: 50 μm. Data are mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by Student’s t test (A, B, D, and F) or 1-way ANOVA followed by Bonferroni’s post hoc test (G and I–K). NS, not significant.
First, the fatty acids were sorted based on their saturation status into 3 groups: saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs), and polyunsaturated fatty acids (PUFAs); MUFA levels decreased the most after Qki depletion (to 54.2% of levels in controls; Figure 4B and Supplemental Figure 5, A–D). Because fatty acid composition is crucial for the fluidity and rigidity of the membrane structure of myelin (33), we then determined whether the fatty acid composition was altered in the spinal cords of Qk-iCKO mice (Figure 4C and Supplemental Figure 5E). The composition analysis revealed that the percentage of MUFAs among all fatty acids in Qk-iCKO mice was lower than that in controls (41.0% in controls versus 30.8% in Qk-iCKO mice; P = 1.16 × 10–5), and the percentage of SFAs among all fatty acids in Qk-iCKO mice was higher than that in controls (43.8% in controls versus 49.5% in Qk-iCKO mice; P = 1.21 × 10–5). Accordingly, the ratio of MUFAs to SFAs in Qk-iCKO mice was significantly lower than that in controls (0.94 in controls versus 0.62 in Qk-iCKO mice; P = 1.26 × 10–5), indicating that the desaturating reactions in converting SFAs to MUFAs are impaired in Qk-iCKO mice (Figure 4, D and E). For example, the ratio of oleic acid (18:1) to stearic acid (18:0) in Qk-iCKO mice was significantly lower than that in controls (1.84 in controls versus 1.32 in Qk-iCKO mice; P = 0.00003; Figure 4F), indicating that this critical desaturating reaction is compromised in Qk-iCKO mice. This finding is consistent with the aforementioned transcriptional downregulation of stearoyl-CoA desaturases 1, 2, 3, and 4 (Scd1, -2, -3, and -4), genes that encode the enzymes that catalyze the desaturation of stearic acid, after Qki depletion (Figure 3D and Figure 4E).
Second, an analysis of the fatty acids based on their carbon chain lengths revealed that the levels of LCFAs and very-long-chain fatty acids (VLCFAs), but not medium-chain fatty acids (MCFAs), were significantly diminished after Qki depletion (Supplemental Figure 5, F–H). The composition analysis further indicated that the percentage of VLCFAs among the total fatty acids was the most diminished after Qki depletion (8.6% in controls versus 5.3% in Qk-iCKO mice; P = 0.0049). Accordingly, the fatty acid ratios of 20:0/18:0, 20:1/18:1, 24:1/22:1, 22:5/20:5, and 22:4/20:4 were all significantly reduced after Qki depletion (Figure 4, E and F, and Supplemental Figure 5, I and J), suggesting that these elongating reactions are impaired. These results are consistent with the aforementioned transcriptional downregulation of hydroxysteroid 17-β dehydrogenase 12 (Hsd17b12) and 3-hydroxyacyl-CoA dehydratase 2, 3, and 4 (Hacd2, -3, and -4), genes that encode the enzymes responsible for the elongation of these fatty acids, after Qki depletion (Figure 3D, Figure 4E, and Supplemental Figure 5I).
In summary, complex lipidomic analyses revealed that the most markedly reduced fatty acids after Qki depletion were the SFAs, MUFAs, LCFAs, and VLCFAs (Supplemental Figure 5, B, C, G, and H). Our finding that Qki is a transcriptional regulator of fatty acid metabolism raised the strong possibility that impaired lipid metabolism is a cause of the Qki depletion–induced demyelinating phenotype. To test this hypothesis, we determined whether dietary supplementation of fatty acids alleviated the severe demyelinating symptoms of Qk-iCKO animals. Indeed, compared with the Qk-iCKO mice that were fed a normal diet, the mice that received a high-fat diet (HFD) enriched with various fatty acids had a markedly longer time to the onset of neurological signs, lower mean clinical scores, better coordinate movement on rotarod tests, and longer median overall survival durations (56 days versus 36 days; Figure 4, G–I, Supplemental Figure 5K, and Supplemental Video 2). Ultrastructural analyses confirmed that Qk-iCKO mice fed the HFD had a significantly higher percentage of myelinated axons (31.7% in the normal diet group versus 61.5% in the HFD group; P < 0.0001) and thicker myelin sheaths (g-ratio: 0.94 in the normal diet group versus 0.84 in the HFD group; P = 0.0001) than did mice fed a normal diet (Figure 4J). Moreover, Qk-iCKO mice fed the HFD had 85.8% higher myelin lipid levels than did mice fed a normal diet (Figure 4K and Supplemental Figure 5L). Together, these results demonstrate that a Qki-mediated continuous fatty acid supply is required for mature myelin maintenance and that an HFD can partially rescue Qki loss–induced demyelination.
Qki regulates fatty acid metabolism via PPARβ-RXRα.
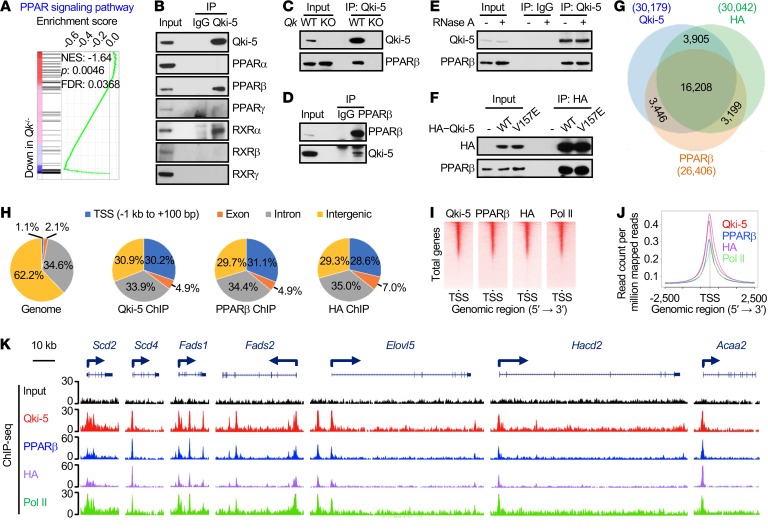
The PPAR signaling pathway is a major regulator of lipid metabolism, adipogenesis, and energy homeostasis (34). Interestingly, pathway analyses with transcriptomic data revealed that the PPAR signaling pathway was one of the cascades that was most profoundly affected by Qki depletion (Figure 3B and Figure 5A), suggesting that Qki may regulate lipid metabolism through this pathway. Based on the observations that the expression of all homologs of PPAR in the corpus callosum was comparable in Qk-iCKO mice and controls and that most Qki-5 was localized on chromatin (Supplemental Figure 6, A and B), we hypothesized that Qki-5 interacts with the PPAR-RXR complex to modulate the transcription of fatty acid metabolism genes. Indeed, coimmunoprecipitation (co-IP) experiments revealed that PPARβ and RXRα, but not other PPAR and RXR homologs, specifically interacted with Qki-5 (Figure 5, B–D).
Figure 5. Qki-5 interacts with PPARβ-RXRα to regulate the transcription of genes involved in fatty acid metabolism.
(A) GSEA shows the PPAR signaling pathway signature in freshly isolated mouse oligodendrocytes. (B) Co-IP using an anti–Qki-5 antibody in differentiated oligodendrocytes, followed by the detection of homologs of PPAR and RXR via immunoblotting. (C) Co-IP using an anti–Qki-5 antibody in differentiated oligodendrocytes, followed by detection of PPARβ via immunoblotting. Qk-knockout (Qk-KO) cells served as a negative control to exclude nonspecific immunoprecipitation from the anti–Qki-5 antibody. (D) Reciprocal co-IP using an anti-PPARβ antibody, blotted with an anti–Qki-5 antibody. An IgG antibody served as a negative control. (E) Co-IP of Qki-5 and PPARβ in differentiated oligodendrocytes that were treated with RNase A while alive. (F) Co-IP of HA–Qki-5 (WT or V157E mutant) and PPARβ in differentiated oligodendrocytes. (G) Overlap of ChIP-seq peak sets of Qki-5, PPARβ, and HA in differentiated oligodendrocytes with ectopic expression of HA-PPARβ. (H) Genomic global distribution of the ChIP-seq peaks of Qki-5, PPARβ, and HA. (I) ChIP-seq density heatmaps of Qki-5, PPARβ, HA, and Pol II within ± 1 kb of the transcription start site (TSS). All peaks are rank ordered from high to low Qki-5 occupancy. (J) Average genome-wide occupancies of Qki-5, PPARβ, HA, and Pol II within ± 2.5 kb of the TSS. (K) Representative ChIP-seq binding peaks of Qki-5, PPARβ, HA, and Pol II on gene loci associated with fatty acid metabolism. Data are representative of 3 independent experiments (B–F).
Although Qki-5 is an RNA-binding protein (23), the interaction between Qki-5 and PPARβ was not affected by either RNase A treatment or mutation of Qki-5 (V157E), which disrupts the RNA-binding ability (27), indicating that RNA was not essential for the formation of the Qki-5–PPARβ–RXRα complex (Figure 5, E and F, and Supplemental Figure 6C). In fact, approximately 96.5% of the differentially expressed genes after Qki depletion do not contain a Qki response element (QRE) (Supplemental Figure 6D). In support of this, the transcription of fatty acid metabolism genes was similarly upregulated by ectopically expressed Qki-5WT and Qki-5V157E (Supplemental Figure 6E), suggesting that Qki regulates gene expression independently of its RNA-binding ability.
To uncover the molecular mechanism by which the Qki-PPARβ-RXRα complex regulates the transcription of genes involved in lipid metabolism, differentiated oligodendrocytes with ectopic expression of hemagglutinin-tagged PPARβ (HA-PPARβ) were analyzed by chromatin immunoprecipitation and sequencing (ChIP-seq) using anti–Qki-5, anti-PPARβ, and anti-HA antibodies, and 30,179 Qki-5–binding peaks, 26,406 PPARβ-binding peaks, and 30,042 HA-binding peaks were identified (Figure 5G). Consensus PPAR response element (PPRE) motifs were significantly enriched in both PPARβ- and HA-binding events (Supplemental Figure 6F), confirming the specificity of the ChIP. Strikingly, we found that 74.4% (19,654 of 26,406) of PPARβ sites and 66.9% (20,113 of 30,042) of HA sites were cobound by Qki-5 (Figure 5G). Moreover, genomic distribution analyses of Qki-5– and PPARβ-binding events showed similarly high enrichment at promoter/transcription start site regions (30.2%, 31.1%, and 28.6% of Qki-5–, PPARβ-, and HA-binding events, respectively, whereas only 1.1% of regions are considered promoter regions across the whole genome; Figure 5, H–J). Reinforcing the notion that Qki-5 and PPARβ coregulate the transcription of fatty acid metabolism genes, Qki-5, PPARβ, HA, and Pol II occupancies were codetected at the promoter regions of 45 genes involved in fatty acid metabolism, such as Scd2, Scd4, fatty acid desaturase 1 (Fads1), Fads2, ELOVL fatty acid elongase 5 (Elovl5), and Hacd2 (Figure 5K and Supplemental Figure 6G), and the mRNA levels of majority of these genes (73.3%; 33 of 45) were significantly downregulated in Qk–/– oligodendrocytes relative to WT oligodendrocytes (Figure 3, D–F). Consistently, Qki-5– and PPARβ-binding events showed similarly high enrichment at promoter/transcription start site regions in freshly isolated mouse oligodendrocytes (Supplemental Figure 6, H–L). Importantly, we found that the promoter occupancies of PPARβ at Scd2 and carnitine palmitoyltransferase 1c (Cpt1c) were greatly diminished by Qki depletion (Supplemental Figure 6M), suggesting that Qki either recruits or stabilizes PPARβ at the promoters. In addition, we found that the PPARβ-specific agonist GW501516 stimulated the transcription of fatty acid metabolism genes with comparable potency in WT and Qk–/– oligodendrocytes (Supplemental Figure 6N), indicating that Qki-5 might serve as a transcriptional coactivator instead of an essential component of the PPARβ-RXRα complex. In support of this, ectopically expressed Qki-5 promoted the transcription of a PPAR-responsive luciferase reporter (Supplemental Figure 6O). Together, these findings strongly suggest that Qki-5 functions as a coactivator of PPARβ-RXRα to regulate the expression of fatty acid metabolism genes that are essential for maintaining mature myelin homeostasis.
PPARβ and RXR agonists alleviate Qki deficiency–induced demyelination.
A previous study showed that PPARβ-null mice had altered myelin in the corpus callosum, yet it was unclear whether the effect was cell autonomous and whether PPARβ regulates myelin lipid homeostasis (35). To address these questions, we deleted Ppard (gene that encodes PPARβ) in mature myelinating oligodendrocytes by injecting Plp-CreERT2 Ppardfl/fl mice (hereafter called Ppard-iCKO mice) with tamoxifen at age 8 weeks (Supplemental Figure 7, A and B). Compared with control mice, at 5 weeks after tamoxifen injection Ppard-iCKO mice displayed significantly reduced motor coordinative movement (Supplemental Figure 7C), completely devoid of myelin sheaths in 36.7% of the axons, and significant reduction in the thickness of myelin sheaths surrounding the remaining axons (g-ratio: 0.83 versus 0.72; P = 0.0066; Supplemental Figure 7, D–G), yet a similar number of oligodendrocytes in the corpus callosum (Supplemental Figure 7, H and I). These results indicate that, similarly to Qki, PPARβ is critical for the maintenance of oligodendrocyte death–independent myelin homeostasis. Consistently, depletion of PPARβ in mature oligodendrocytes resulted in a significant reduction in myelin lipids but not major myelin structural proteins in the corpus callosum (Supplemental Figure 7, J and K). Together with the data indicating that Qki interacts with PPARβ to transcriptionally control myelin lipid metabolism (Figure 5 and Supplemental Figure 6), the similar demyelinating phenotypes of Ppard-iCKO mice and Qk-iCKO mice support the model that Qki and PPARβ work together in regulating myelin lipid metabolism and controlling myelin homeostasis.
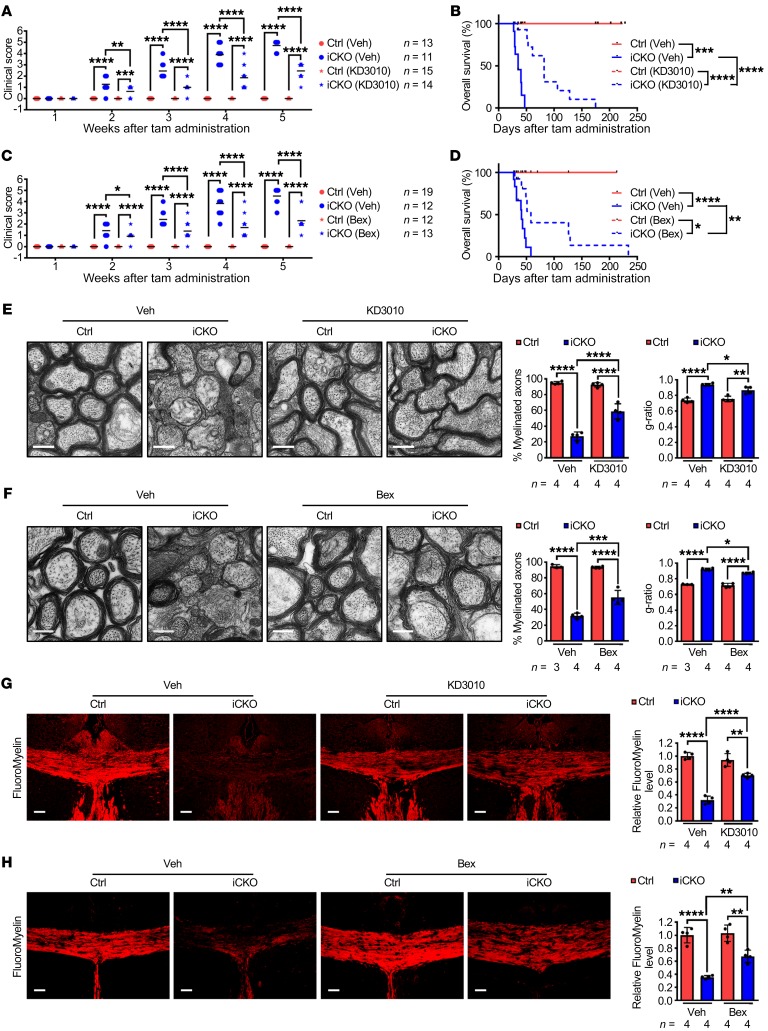
Because loss of Qki diminished but not totally abolished PPARβ-RXRα transcriptional activity (Supplemental Figure 6N), we next determined whether the CNS-permeant PPARβ agonist KD3010 (36) and RXR agonist bexarotene (37) could alleviate Qki depletion–induced demyelinating symptoms. Oral administration of KD3010 or bexarotene in Qk-iCKO mice significantly delayed the onset of neurological deficits, lowered clinical scores, and prolonged median survival times (2.34- or 1.41-fold, respectively) compared with treatment with vehicle (Figure 6, A–D, Supplemental Figure 8, A and B, and Supplemental Videos 3 and 4). Strikingly, 23.1% (3 of 13) of Qk-iCKO mice treated with KD3010 or bexarotene lived for more than 100 days, with the maximum survival duration increased by 3.72- or 4.03-fold compared with that of Qk-iCKO mice treated with vehicle, respectively (Figure 6, B and D). Consistent with their clinical improvement, Qk-iCKO mice treated with KD3010 or bexarotene had 115.3% or 73.7% more myelinated axons, with thicker myelin sheaths, than did Qk-iCKO mice treated with vehicle (Figure 6, E and F). In support of this, Qk-iCKO mice treated with KD3010 or bexarotene had 119.4% or 89.5% higher myelin lipid levels than did Qk-iCKO mice treated with vehicle (Figure 6, G and H, and Supplemental Figure 8, C and D). Additionally, the reduction in expression of Scd1, Scd2, Scd4, and Elovl5 in Qk-iCKO mice was significantly alleviated by treatment with KD3010 (Supplemental Figure 8E). These significant alleviations of Qki depletion–induced demyelinating symptoms and enhancement of myelin lipid biosynthesis by PPARβ and RXR agonists confirm that PPARβ-RXRα–dependent fatty acid metabolism is a key downstream target of Qki in maintaining mature myelin homeostasis and provide justification for the use of PPARβ/RXRα agonists in future clinical trials for patients with demyelinating diseases.
Figure 6. PPARβ and RXR agonists alleviate Qki deficiency–induced demyelination.
(A–D) The clinical scores and Kaplan-Meier overall survival curves (log-rank test) of Qk-iCKO mice and controls receiving daily oral administration of KD3010, bexarotene (Bex), or vehicle (Veh). The experimental mouse number is indicated in A and C. (E and F) Representative electron micrographs and quantification of the percentage of myelinated axons and g-ratio of the optic nerves of the experimental mice after 5 weeks of treatment. Scale bars: 500 nm. (G and H) Representative images and quantification of FluoroMyelin level in the corpus callosum of the experimental mice after 5 weeks of treatment. Scale bars: 50 μm. Data are mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA followed by Bonferroni’s post hoc test.
Myelin lipids are preferentially reduced in MS.
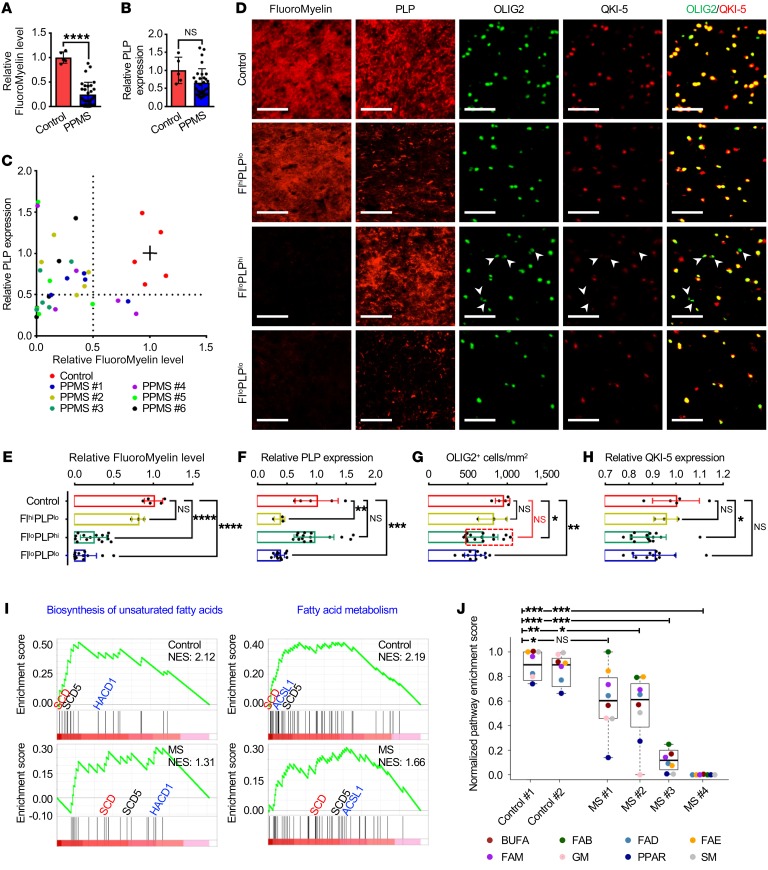
Experimental autoimmune encephalomyelitis manifests as an acute ascending paralysis that either remits or plateaus, and is widely used as an animal model of relapsing-remitting MS (38). In contrast, the clinical course of our Qk-iCKO mice is characterized by a chronic accumulation of neurological disability that is more reminiscent of the primary progressive subset of MS. To investigate the potential role of myelin lipid dysregulation in the pathogenesis of human demyelinating diseases, we obtained 30 individual lesions that were characterized following the Lassmann/Brück method (39) from the brains of 6 patients with primary progressive MS (PPMS) (Supplemental Figure 9A). Frontal lobe white matter tissues from 5 donors without neurological disease were analyzed in parallel as controls. The average level of myelin lipids in these MS lesions, as measured by FluoroMyelin staining, was 24.8% of that in the control tissues (Figure 7A). In contrast, despite marked variations among individual lesions, the average levels of the major myelin structural proteins (PLP, MBP, and MAG) were not significantly different in the MS lesions compared with the controls (Figure 7B and Supplemental Figure 9, B and C). By grouping these MS lesions on the basis of their myelin lipid and protein levels, we found that 50% (15 of 30) of the lesions exhibited low lipid levels but normal protein levels (FlloPLPhi), 10% exhibited normal lipid levels but low protein levels (FlhiPLPlo), and the remaining 40% exhibited low levels of both lipids and proteins (FlloPLPlo) (Figure 7, C–F). These data suggest that myelin lipids are preferentially reduced in a subset of PPMS lesions.
Figure 7. Lipids are more vulnerable than proteins in PPMS lesions.
(A and B) Quantification of staining of FluoroMyelin (A) and PLP (B) in the frontal lobes from 5 human brains without neurological disease (control) and 30 lesions from 6 patients with PPMS. (C) Scatter plot of FluoroMyelin intensity versus PLP intensity in the samples in A. The solid lines among red dots represent the average levels of FluoroMyelin and PLP in controls. (D–H) Representative images (D) and quantification of FluoroMyelin level (E), PLP level (F), OLIG2+ cell number (G), and QKI-5 level (H) in controls and 3 subtypes of PPMS lesions, which were divided according to the level of FluoroMyelin and PLP in C. n = 5 in the control group; n = 3 in the FlhiPLPlo group; n = 15 in the FlloPLPhi group; n = 12 in the FlloPLPlo group. Scale bars: 50 μm. (I and J) Comparison of GSEA of fatty acid metabolism pathways (I) and the NES (J) between MS lesions and the controls. Box plots indicate medians, interquartile values, ranges, and individual pathways. BUFA, biosynthesis of unsaturated fatty acids; FAB, fatty acid biosynthesis; FAD, fatty acid degradation; FAE, fatty acid elongation; FAM, fatty acid metabolism; GM, glycerophospholipid metabolism; SM, sphingolipid metabolism. Data are mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by Student’s t test (A and B), 1-way ANOVA followed by Bonferroni’s post hoc test (E–H), or Wilcoxon’s signed-rank test (J). NS, not significant.
Oligodendrocyte death is often considered a primary pathogenic event in MS (2–4). To determine whether the observed lower levels of myelin lipids in MS lesions were correlated with a lower density of oligodendrocytes, we counted the number of OLIG2+ oligodendrocytes in each MS lesion (Supplemental Figure 9D). We found that FlloPLPlo lesions had significantly lower numbers of oligodendrocytes than did control tissues (P < 0.01), suggesting that oligodendrocyte death was the major cause of the demyelination and decreased levels of myelin lipids and proteins that were observed in this subgroup of lesions (Figure 7, D and G). In contrast, 80% (12 of 15) of the FlloPLPhi lesions (indicated by the red-dashed box in Figure 7G) had normal oligodendrocyte densities, suggesting that diminished myelin lipid levels, rather than oligodendrocyte death, might be responsible for demyelination in this subgroup of lesions (Figure 7, D and G). The remaining 20% (3 of 15) of the FlloPLPhi lesions had relatively low oligodendrocyte density but normal myelin protein staining. One explanation for this discrepancy is the long half-life of these myelin structural proteins (14, 15). Interestingly, although the number of samples was limited, the FlhiPLPlo lesions did not exhibit significantly lower oligodendrocyte numbers or lipid levels than did control tissues, suggesting the existence of additional demyelinating mechanisms that were independent of oligodendrocyte death or lipid reduction (Figure 7, D and G).
In support of the important role of the Qki-PPARβ-RXRα complex in the pathogenesis of PPMS, we found that QKI-5 was reduced specifically in the FlloPLPhi group for which myelin lipid reduction is the major pathogenic alteration (Figure 7, D and H). We next determined the expression levels of the genes (normalized with the PLP expression level) of 8 Qki-PPARβ-RXRα–associated lipid metabolism pathways, including fatty acid metabolism, fatty acid biosynthesis, biosynthesis of unsaturated fatty acids, fatty acid elongation, fatty acid degradation, PPAR signaling, glycerophospholipid metabolism, and sphingolipid metabolism, from published microarray data (40); a gene set enrichment analysis (GSEA) revealed that the activities of these pathways were significantly downregulated in the MS lesions relative to controls (Figure 7, I and J, and Supplemental Figure 9E). Notably, the levels of SCD, SCD5, HACD1, ELOVL1, and ELOVL5, the known Qki-PPARβ-RXRα target genes that encode key enzymes for fatty acid desaturation and elongation, were all significantly lower in the MS lesions than controls (Figure 7I and Supplemental Figure 9E). Together, our data indicate that disruption of the Qki-PPARβ-RXRα complex–regulated lipid metabolism may play an important role in pathological demyelination of subsets of patients with MS.
Discussion
Myelin has long been considered to be an inert material (14–17), but our studies with Qk-iCKO and Ppard-iCKO genetic mouse models reveal that myelin itself, independent of oligodendrocyte survival, is a highly dynamic substance. We found that mature myelin homeostasis requires a continuous Qki-PPARβ-RXRα–dependent biosynthesis of the structural lipid components, particularly unsaturated and elongated fatty acids (Supplemental Figure 9F), probably owing to their high vulnerability to exogenous insults (5). An intriguing observation in our study is that depletion of Qki or PPARβ did not affect oligodendrocyte survival yet had detrimental effects on myelin integrity. We postulate that because myelin sheaths are such extraordinarily large extensions of membrane structure, they have a tremendously higher demand for continuous lipid synthesis than do other cell membranes; therefore, myelin membranes should be particularly sensitive to loss of Qki or PPARβ. Supporting the notion that Qki-dependent lipid metabolism is not essential for general cell survival, our previous work showed that, consistent with Qki’s role as a tumor suppressor (41), Qki depletion in Pten/Trp53-deficient NSCs greatly promoted cell growth and gliomagenesis (29).
Distinct from the widely held dogma that Qki functions primarily as an RNA-binding protein in oligodendrocyte differentiation and myelination (23–28), we uncovered what we believe is a previously unknown function of Qki-5 — as a transcriptional coactivator of the PPARβ-RXRα complex to regulate fatty acid metabolism during myelin homeostasis. Although previous studies have already shown reduced myelin lipid content in the qkv mouse (42, 43), which carries a germline mutation resulting in reduced expression of Qki protein (along with 2 other proteins, Pacrg and Park2) from birth (23), it is worth noting that the reduction in all myelin structural components, including myelin lipids and myelin proteins, is secondary to the impaired oligodendrocyte differentiation and subsequent failure of myelin formation in qkv mice during development (23–28).
We noticed that the demyelinating phenotypes of Ppard-iCKO mice were not as strong as those of Qk-iCKO mice, and the Qk-deletion-induced demyelinating phenotype was not completely rescued by the HFD, KD3010, or bexarotene (Figures 4 and 6). There are 3 potential explanations for this. First, although the biosynthesis of unsaturated fatty acids, fatty acid metabolism, and fatty acid elongation (which are all downstream of PPARβ signaling) are the pathways most profoundly affected by Qki depletion (Figure 3 and Supplemental Figure 4), we acknowledge the potential contributions of Qki to other PPARβ-independent pathways. In fact, 34.9% (10,525 of 30,179) of Qki-5 DNA binding sites identified by ChIP-seq did not overlap with PPARβ DNA binding sites (Figure 5G), suggesting that PPARβ-independent Qki-5 DNA binding sites might also be involved in demyelination in Qk-iCKO mice. Second, in investigating the transcriptional downregulation of the fatty acid metabolism genes, we focused on Qki-5, because we found that Qki-5 was located on chromatin and interacted with the PPARβ-RXRα complex. However, we cannot rule out the possibility that the cytoplasmic Qki-6 and Qki-7 might also contribute to the demyelinating phenotype of Qk-iCKO mice in a PPARβ-independent manner. Lastly, we found that PPARα could also interact with Qki-5, although this interaction is not as strong as that between PPARβ and Qki-5, and it could be disrupted by sonication (data not shown). Therefore, it is possible that PPARα could compensate for the loss of PPARβ activity in Ppard-iCKO mice. Nevertheless, because no myelin defects have been reported in Ppara-KO mice (44), our data suggested that PPARβ is a more important mediator of Qki’s function in myelin maintenance than PPARα.
Various studies, by taking advantage of ChIP-seq, showed that between 4% and 49% of PPARβ-binding sites resided in promoter regions (45–48). One explanation for this large variation is that these studies (as well as ours) were performed in different cell types. Of note, other than a 16-bp consensus sequence PPRE, NFIA/NFIX, Rfx, NF1, Sp1, NRF1, and Sox9 motifs were also identified as highly enriched motifs by both our PPARβ ChIP and HA ChIP (data not shown), indicating that various cofactors might codetermine where PPARβ interacts on the chromatin. In line with our finding, other studies have also found that PPARβ cooperated with other transcription factors to specifically regulate subsets of genes, and these factors could be tissue or cell type dependent (46).
The great variability in MS genetics, clinical course, appearance on imaging, pathology, and response to therapy suggests that this chronic demyelinating disorder has multiple etiologies (2). Autoimmune attack (49) and autonomous oligodendrocyte death (3) are thought to be the primary pathogenic events in MS. Contrary to these hypotheses, which both imply a major role for oligodendrocyte death in MS, other studies have noted that 30%–60% of early active lesions exhibit no apparent oligodendrocyte death (6–8). Our research found that dysregulation of the myelin lipid metabolism (in both Qk-iCKO and Ppard-iCKO mice) is sufficient to drive a demyelinating syndrome without affecting oligodendrocyte viability, indicating that defective lipid metabolism may play an important role in the formation of some MS lesions.
Most disease-modifying therapies that are used to manage relapsing-remitting MS have been found to be ineffective in progressive forms of the disease (50). There is a dire need for interventions that slow or block the chronic, progressive component of MS. Excitingly, the therapeutic benefits of the HFD, KD3010, and bexarotene in Qk-iCKO mice raise the possibility that similar approaches would be efficacious in a subset of patients with progressive MS with reduced lipid synthesis in oligodendrocytes and/or in individuals with leukodystrophies such as diffusely abnormal white matter, in which the myelin lipid level is greatly reduced yet the myelin protein level is relatively unchanged (51).
Methods
Mice.
Mice bearing the Plp-CreERT2 allele (C57BL/6) (52) and mice bearing the Ppard-loxP allele (C57BL/6) (53) were obtained from The Jackson Laboratory. Mice with the Nestin-CreERT2 allele (C57BL/6) were provided by Ryoichiro Kageyama (Kyoto University, Kyoto, Japan) (54). Mice with the Qk-loxP allele were described previously (29). Mice bearing the Qk-loxP allele were crossed with mice bearing the Plp-CreERT2 allele, in which the expression of tamoxifen-inducible Cre is under the control of the Plp promoter; the Plp-CreERT2 Qkfl/fl, Plp-CreERT2 Qkfl/+, or WT mice were generated and used in further experiments. Mice bearing the Qk-loxP allele were crossed with mice bearing the Nestin-CreERT2 allele, in which the expression of tamoxifen-inducible Cre is under the control of the Nestin promoter, and Nestin-CreERT2 Qkfl/fl pups were used for NSC isolation. All mouse work was performed under pathogen-free conditions at MD Anderson’s animal facility. Mice were fed a standard chow unless otherwise specified, maintained under a 12-hour light/dark cycle, housed in groups, and monitored for signs of ill health every other day.
When the Plp-CreERT2 Qkfl/fl mice were 8 weeks old, 100 μL of tamoxifen (10 mg/mL in corn oil; Sigma-Aldrich) was injected subcutaneously on 2 consecutive days to induce the deletion of Qk in oligodendrocytes. Littermates of both sexes with the same genetic backgrounds and of the same ages were used for all experiments. For the high fatty acid diet rescue experiment, 8-week-old mice were fed either a normal chow or chow supplemented with 13.68% SFAs and 14% MUFAs (58Y1; TestDiet). For the KD3010 rescue experiment, 8-week-old mice were orally administered 50 mg/kg/day KD3010 or vehicle (0.5% carboxymethyl cellulose containing 12.5% ethanol). For the bexarotene rescue experiment, 8-week-old mice were given 100 mg/kg/day oral bexarotene (Cayman Chemical) or vehicle (0.5% carboxymethyl cellulose). For the in vivo Brdu incorporation assay, 50 mg/kg BrdU (Sigma-Aldrich) in phosphate-buffered saline (PBS) was injected into the peritoneal cavity every 8 hours for 4 days before scarification.
The tamoxifen-injected Qk-iCKO mice and controls were monitored daily, and their neurological deficits were assessed weekly using a modified experimental autoimmune encephalomyelitis scoring system to characterize the disease phenotype and severity. In brief, 3 types of neurological deficits, including tremor, hind limb weakness, and paresis, were scored on a scale from 0 to 2. Tremor was scored as T0 (no tremor), T1 (tremor when the tail was holding), or T2 (tremor when walking or standing). Hind limb weakness was rated as W0 (normal), W1 (an angle between the hind leg and paw of around 90°), or W2 (an angle between the hind leg and paw of around 180°). Hind limb paresis was categorized as P0 (normal), P1 (splayed gait and ataxia), or P2 (hind limb paresis or paralysis). The clinical score was the sum of these 3 ratings, ranging from 0 (T0W0P0) to 6 (T2W2P2). When the clinical score reached 5 or 6, most of the mice showed a hunched posture, immobility, and loss of body weight and were moribund. Thus, they were considered to have reached the clinical endpoint and were euthanized if they were not found dead. Euthanized mice were considered in the analyses in the same way as those that died, with the time of death or euthanasia plotted accordingly on the survival curves.
Mice bearing the Ppard-loxP allele were crossed with mice bearing the Plp-CreERT2 allele. The 8-week-old Plp-CreERT2 Ppardfl/fl mice were subcutaneously injected with 100 μL of tamoxifen (10 mg/mL) on 2 consecutive days to induce the deletion of Ppard in oligodendrocytes specifically. To assess motor coordination and balance, the experimental mice were tested on a rotarod apparatus (Harvard Bioscience, Inc.). After 3 days of training, mice were tested in a 2-minute trial on the rotarod apparatus rotating at 20 rpm. If a mouse stayed on the rotarod longer than 2 minutes, a time of 120 seconds was recorded. Mice were tested 4 times, with half-hour intervals between each trial. For statistical analyses, a mean latency of falling from the rotarod of 4 trials of each mouse was used.
Human tissue specimens.
Fresh-frozen tissue specimens were obtained from the Rocky Mountain MS Center Tissue Bank and stored at –80°C until use. In total, 11 postmortem brain slices were analyzed, including 5 control brain slices from donors without neurological disease and 6 from patients with PPMS. The patients’ age at the time of death, sex, postmortem interval, pathological diagnosis, cause of death, disease duration, and disease-modifying therapies are shown in Supplemental Table 3. Lesions were recognized grossly, dissected, and embedded in optimal cutting temperature compound. Twenty-micrometer-thick frozen sections were sliced and stored at –80°C. Histological classification of PPMS lesions (5 from each patient) was characterized following the Lassmann/Brück method (39). Briefly, reduction in the intensity of Luxol fast blue stain and/or immunohistochemistry of MBP was used to indicate the demyelinating activity; the presence/absence and distribution of macrophages/microglia (immunohistochemistry of CD68) was used to indicate inflammatory activity (39). The frontal lobe white matter from neurological disease–free human brains was used as control tissue. The intensities of PLP and FluoroMyelin (Invitrogen) were quantified and normalized with the average level of those in control tissue. The levels of FluoroMyelin and PLP that were lower than 50% of the average level of those in control tissues were defined as Fllo and PLPlo, respectively; all of the lesions were grouped into 3 groups based on their myelin lipid and protein levels: FlloPLPhi, FlhiPLPlo, and FlloPLPlo.
NSC isolation and oligodendrocyte differentiation.
Mouse NSCs were isolated using neural tissue dissociation kits according to the manufacturer’s instructions (Miltenyi Biotec). In brief, whole Nestin-CreERT2 Qkfl/fl mouse forebrains from postnatal day 1 pups (both sexes) were dissociated enzymatically and the single-cell suspension was cultured in NeuroCult Basal Medium (Stemcell Technologies), supplemented with 20 ng/mL epidermal growth factor (ProteinTech), 10 ng/mL basic fibroblast growth factor (ProteinTech), NeuroCult Proliferation Supplement (Stemcell Technologies), 50 units/mL penicillin G, and 50 μg/mL streptomycin in a humidified 37°C incubator with an atmosphere containing 5% CO2. The NSCs were exposed to 100 nM 4-hydroxytamoxifen (Sigma-Aldrich) twice at an interval of 2 days to knockout Qk. To induce oligodendrocyte differentiation, NSCs were cultured on dishes coated with poly-L-ornithine (Sigma-Aldrich) and laminin (Thermo Fisher Scientific) and maintained in neurobasal medium (Thermo Fisher Scientific) containing B-27 (Thermo Fisher Scientific), 2 mM GlutaMAX-I (Thermo Fisher Scientific), 100 ng/mL insulin-like growth factor 1 (IGF-1) (Peprotech), 50 ng/mL 3,3′,5-triiodo-L-thyronine (T3) (Cayman Chemical), 50 units/mL penicillin G, and 50 μg/mL streptomycin. IGF-1 and T3 were added daily, and the differentiation medium was replaced with fresh medium every other day.
Primary oligodendrocyte isolation.
Tamoxifen was administered subcutaneously to 8-week-old Qk-iCKO mice and controls on 2 consecutive days. Four days later, brains were dissociated enzymatically according to the instructions of the Adult Brain Dissociation Kit (Stemcell Technologies). After the removal of debris and red blood cells, the single-cell suspension was incubated with anti-O4 microbeads, followed by magnetic separation. The O4+ oligodendrocytes and the O4– flow-through were collected separately for further RT-qPCR validation. The O4+ oligodendrocytes from Qk-iCKO mice and controls were used for RNA sequencing.
Tissue preparation and immunofluorescence.
Mice were anesthetized with isoflurane and perfused transcardially with 4% formaldehyde. Their brains, spinal cords, and optic nerves were removed, postfixed in formalin at room temperature for 2 days, and embedded in paraffin. Five-micrometer-thick paraffin-embedded sections were deparaffinized and subjected to heat-induced antigen retrieval in citrate buffer (Poly Scientific R&D Corp.). The sections were blocked with 10% horse serum and incubated with primary antibodies overnight at 4°C in the blocking solution. The following antibodies were used for staining, according to their manufacturer’s directions: anti-Olig2 (catalog AB9610), anti-Olig2 (catalog MABN50), anti-aspartoacylase (anti-ASPA) (catalog ABN1698), and anti-MBP (QD-9; catalog AB5864) were from Merck Millipore; anti-IBA1 (catalog ab107159), anti-Brdu (catalog ab6326), anti-PLP (catalog ab105784), anti-PPARβ (catalog ab137724), and anti-CD68 (catalog ab955) were from Abcam; anti-MAG (catalog 9043), anti-PDGFRα (catalog 3174), and anti–cleaved caspase 3 (catalog 9661) were from Cell Signaling Technology; anti-QKI (catalog SAB5201536) was from Sigma-Aldrich; anti-GFAP (catalog 556330) was from BD Biosciences; anti-GSTpi (catalog 311) was from MBL International; anti-MBP (catalog SMI-94R) was from Covance; anti–QKI-5 (catalog IHC-00574) was from Bethyl Laboratories; anti–QKI-6 (catalog 75-190) and anti-AnkG (catalog MABN466) were from NeuroMab; anti–QKI-7 was from a rabbit immunized with a short synthetic peptide (KGGPIEPSGVLEWIEMPVMPDISAH); anti-CD3 (catalog A0452) was from DAKO; and anti-Caspr was a gift from Matthew N. Rasband (Baylor College of Medicine, Houston, Texas, USA). After 3 washes with Tris-buffered saline containing 0.1% Tween 20, the sections were incubated with species-appropriate secondary antibodies conjugated with Alexa Fluor dyes for 1 hour. To mount the stained sections, mounting medium with DAPI (Vector Laboratories) was used. All stained sections were visualized using a Leica DMi8 microscope, and images were obtained using a Leica DFC345 FX digital monochrome camera.
Frozen sections were thawed at room temperature for 30 minutes and fixed in ice-cold formalin for 10 minutes. After being rehydrated with cold PBS, sections were blocked with 10% horse serum and incubated with primary antibodies and secondary antibodies, as described above. For FluoroMyelin staining, the rehydrated slides were stained directly for 20 minutes at room temperature.
Strong autofluorescence was stimulated in the human brain sections by multiple wavelength channels of the microscope; it was removed in 2 ways. For staining of OLIG2, autofluorescence was removed using the TrueBlack Lipofuscin Autofluorescence Quencher, according to the manufacturer’s instructions (Biotium); the fluorescence images were obtained normally. For FluoroMyelin, PLP, MBP, and MAG, immunofluorescent staining was performed normally using secondary antibodies that were specifically stimulated by the red wavelength channel. The sections were imaged using both the red channel and the green channel, and signals that appeared in both channels were considered autofluorescent noise. Using this information, we devised an algorithm for enhancing the signal-to-noise ratio of the images. The intensities of the green-filtered image were mean-centered. This mean was subtracted from the red image to remove strong background noise signals. The green image was then converted to a binary (black-and-white) format by setting the threshold at the intensity returned by the Greythresh function in Matlab software. The Matlab Regionprops function was used to obtain bounding boxes for all spots observed in the black-and-white–converted green image. The corresponding spots in the red-filtered image were substituted with the mean intensity of the red image. Pixels within the bounding boxes of the red image, corresponding to all spots identified in the green image, were then subjected to Gaussian filtering to blur these regions and thereby minimize noise.
Electron microscopy.
Tamoxifen was administered to 8-week-old Qk-iCKO mice and controls. After 1, 2, and 5 weeks, the mice were anesthetized with isoflurane and transcardially perfused with 2% paraformaldehyde. The corpus callosum, optic nerves, and spinal cords were removed and postfixed in a solution containing 2% paraformaldehyde and 3% glutaraldehyde in PBS at 4°C. Fixed samples were processed at the High Resolution Electron Microscopy Facility at MD Anderson. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques Corp.).
Electrophysiology.
Compound action potential (CAP) recordings in optic nerves were performed as described elsewhere (55, 56). Briefly, optic nerves were dissected and immediately placed in a continuously perfused, oxygenated, and temperature-regulated (25°C) chamber containing standard Locke’s solution (154 mM NaCl, 5.6 mM KCl, 2 mM CaCl2, 5 mM D-glucose, and 10 mM HEPES [pH 7.4]). The ends of the nerve were drawn orthodromically into suction electrodes, with the retinal side in the stimulating electrode and the chasmic side in the recording electrode. Nerves were then stimulated and the responses to a depolarizing current were amplified, digitized, recorded, and analyzed using Molecular Devices Axon pClamp, MultiClamp, and Clampfit software. Nerve conduction velocity was calculated by dividing the length of the optic nerve by the time from stimulation to the peak of the CAP.
Complex lipidomic analyses.
Tamoxifen was administered to 8-week-old Qk-iCKO mice and controls. Each spinal cord was collected, weighed, and lipids were extracted for further mass spectrometric analyses to yield data from each mouse (n = 5 mice/group). Lipidomic analyses were performed by Metabolon, Inc. Lipids were extracted from the spinal cords using dichloromethane and methanol in a modified Bligh-Dyer extraction procedure, in the presence of internal standards, with the lower organic phase used for analysis. The extracts were concentrated under nitrogen and reconstituted in 0.25 mL of dichloromethane/methanol (50:50) containing 10 mM ammonium acetate. The extracts were placed in vials for ion-mobility spectrometry analyses, performed on a SelexION-equipped SCIEX 5500 QTRAP mass spectrometer with both positive- and negative-mode electrospray. Each sample was analyzed twice, with the ion-mobility spectrometry–mass spectrometry conditions optimized for the lipid classes monitored in each analysis. The 5500 QTRAP mass spectrometer was operated in multiple reaction monitoring mode to monitor the transitions for over 1,100 lipids from up to 14 lipid classes, including neutral lipids (cholesteryl ester, triacylglycerol, diacylglycerol, and free fatty acid), phospholipids (phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, lysophosphatidylcholine, and lysophosphatidylethanolamine), and sphingolipids (sphingomyelin, ceramide, hexosylceramide, lactosylceramide, and dihydroceramide). Individual lipid species were quantified based on the ratio of the signal intensity for the target compound to the signal intensity for an assigned internal standard of known concentration. Lipid class concentrations (μmol/g spinal cord tissue) were calculated by summing the quantities of all molecular species within a class, and fatty acid compositions were determined by calculating the proportion of individual fatty acids within each class.
Stable cells.
The coding DNA sequence region of WT or mutant Qki-5 with an HA tag at the C-terminus was subcloned into the lentiviral vector pInducer20 (57), and the lentiviruses packaged in HEK293T cells were used to infect Qki-depleted NSCs. After selection with G418 (Thermo Fisher Scientific), the surviving cells were treated with doxycycline (Sigma-Aldrich) to induce the expression of HA–Qki-5 (WT or mutant). Similarly, the coding DNA sequence region of Ppard gene was subcloned into pInducer20, and doxycycline-inducible PPARβ-expressing stable cells were generated in NSCs.
Dual-luciferase reporter assay.
HEK293T cells were seeded in 24-well plates. A mixture of a PPAR-responsive firefly luciferase reporter, a constitutively expressing Renilla element, a pInducer20-PPARβ construct, and a pInducer20–Qki-5 construct (or the empty vector) was cotransfected. Forty-eight hours after cells were transfected and treated with doxycycline, they were lysed, and luciferase activity was measured using a dual-luciferase reporter assay system (Promega), following the manufacturer’s instructions. Transfection efficiency was normalized to Renilla luciferase activity.
RNA sequencing and pathway enrichment analyses.
Total RNA was isolated as described in Supplemental Methods and treated with DNase (RNase-free, Qiagen). Sequencing was performed by the Illumina HiSeq/MiSeq sequencing service at the Sequencing and Microarray Facility at MD Anderson. Raw reads were aligned with the mouse reference genome (mm10 version) using the TopHat program, allowing 2 mismatches. The DEseq2 package for R was used to perform normalization and create a gene expression matrix. All RNA sequencing data were analyzed from at least 3 biological replicates. The fold-change threshold at which genes were considered to be differentially expressed was set at 1.2-fold with P less than 0.05. Significantly affected pathways were analyzed using Advaita Bio’s iPathwayGuide (http://www.advaitabio.com/ipathwayguide). In addition, GSEA was carried out as described previously to identify functional enrichment profiles (58).
Data availability.
The RNA sequencing data and ChIP sequencing data reported in this study have been deposited in the NCBI’s Gene Expression Omnibus (GEO) under ID codes GSE84134, GSE106468, GSE125581, and GSE126577.
Statistics.
The numerical results are presented as mean ± standard deviation (SD) and were compared using a 2-tailed Student’s t test (unpaired), 2-way ANOVA with Holm-Sidak multiple-comparisons test, or 1-way ANOVA followed by Bonferroni’s post hoc test, as indicated. Animal survival durations were analyzed using the log-rank test. Statistical analyses were conducted using GraphPad Prism 6. The numbers of animals or cells used to perform the experiments are indicated in the figure legends. For immunoblotting and RT-qPCR, representative data from multiple independent replicates are shown. No statistical methods were used to predetermine sample size. P values of less than 0.05 were considered to be statistically significant in all cases. There were no randomization or blinding events during the experiments.
All other methods are detailed in Supplemental Methods.
Study approval.
All mouse work was conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center (Houston, Texas). Informed consent and ethical approval for the use of human samples were collected by the Rocky Mountain MS Center Tissue Bank. MD Anderson provided guidelines for human tissue study procedures.
Author contributions
JH and XZ designed the study and analyzed the data. XZ conducted the experiments. CH and FL performed ChIP and the bioinformatics analysis for ChIP-seq. JR and QW performed the GSEA. JR, CD, TS, and LY assisted in sample preparation for RNA-seq. SRS and MNR performed electrophysiology. DZ, YW, FZ, and YC performed the bioinformatics analysis for RNA-seq. CRC performed mouse genotyping. VR and AUKR designed the algorithm for removing autofluorescence. HZ, ABH, and BMS helped in the analyses of human tissue specimens. JH and XZ wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Shan Jiang, Kun Zhao, and Yanping Cao for mouse husbandry and care and all members of the Hu laboratory for helpful discussions. We thank Kenneth Dunner, Jr. for performing electron microscopy studies. We thank John R. Corboy and Caroline Miller for providing the human brain specimens from the Rocky Mountain Multiple Sclerosis Center Tissue Bank, University of Colorado. We thank Albert R. La Spada for providing KD3010 as the positive control for our own KD3010 synthesis. We also thank David M. Wildrick and the Department of Scientific Publications of MD Anderson for providing editorial assistance. This investigation was supported in part by a grant from the National Multiple Sclerosis Society and a Cancer Prevention and Research Institute of Texas grant (RP120348 and RP170002). JH is supported by the University of Texas Rising STARs Award, the Sidney Kimmel Scholar Award, the Sontag Foundation Distinguished Scientist Award, NIH grant R37CA214800, and the Brockman Foundation.
Version 1. 03/23/2020
Electronic publication
Version 2. 05/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(5):2220–2236.https://doi.org/10.1172/JCI131800.
Contributor Information
Xin Zhou, Email: xzhou8@mdanderson.org.
Chenxi He, Email: hnalice@126.com.
Jiangong Ren, Email: jren1@mdanderson.org.
Congxin Dai, Email: daicongxin@163.com.
Sharon R. Stevens, Email: Sharon.Stevens@bcm.edu.
Qianghu Wang, Email: wangqh@njmu.edu.cn.
Takashi Shingu, Email: tshingu@mdanderson.org.
Liang Yuan, Email: Liang.Yuan02@tufts.edu.
Yunfei Wang, Email: ywang52@mdanderson.org.
Visweswaran Ravikumar, Email: vravik@umich.edu.
Feng Zhou, Email: zhou_feng@fudan.edu.cn.
Hongwu Zheng, Email: hoz4001@med.cornell.edu.
Matthew N. Rasband, Email: rasband@bcm.edu.
Yiwen Chen, Email: YChen26@mdanderson.org.
Fei Lan, Email: fei_lan@fudan.edu.cn.
Benjamin M. Segal, Email: Benjamin.Segal@osumc.edu.
Jian Hu, Email: jhu3@mdanderson.org.
References
- 1. Armati PJ, Mathey EK. The Biology of Oligodendrocytes. Cambridge, UK: Cambridge University Press; 2010. [Google Scholar]
- 2.Lassmann H, Brück W, Lucchinetti C. Heterogeneity of multiple sclerosis pathogenesis: implications for diagnosis and therapy. Trends Mol Med. 2001;7(3):115–121. doi: 10.1016/S1471-4914(00)01909-2. [DOI] [PubMed] [Google Scholar]
- 3.Traka M, Podojil JR, McCarthy DP, Miller SD, Popko B. Oligodendrocyte death results in immune-mediated CNS demyelination. Nat Neurosci. 2016;19(1):65–74. doi: 10.1038/nn.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locatelli G, et al. Primary oligodendrocyte death does not elicit anti-CNS immunity. Nat Neurosci. 2012;15(4):543–550. doi: 10.1038/nn.3062. [DOI] [PubMed] [Google Scholar]
- 5.Bartzokis G. Age-related myelin breakdown: a developmental model of cognitive decline and Alzheimer’s disease. Neurobiol Aging. 2004;25(1):5–18; author reply 49. doi: 10.1016/j.neurobiolaging.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Brück W, et al. Oligodendrocytes in the early course of multiple sclerosis. Ann Neurol. 1994;35(1):65–73. doi: 10.1002/ana.410350111. [DOI] [PubMed] [Google Scholar]
- 7.Ozawa K, et al. Patterns of oligodendroglia pathology in multiple sclerosis. Brain. 1994;117(pt 6):1311–1322. doi: 10.1093/brain/117.6.1311. [DOI] [PubMed] [Google Scholar]
- 8.Lucchinetti C, Brück W, Parisi J, Scheithauer B, Rodriguez M, Lassmann H. A quantitative analysis of oligodendrocytes in multiple sclerosis lesions. A study of 113 cases. Brain. 1999;122(pt 12):2279–2295. doi: 10.1093/brain/122.12.2279. [DOI] [PubMed] [Google Scholar]
- 9.Yeung MS, et al. Dynamics of oligodendrocyte generation and myelination in the human brain. Cell. 2014;159(4):766–774. doi: 10.1016/j.cell.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Peters A. The effects of normal aging on myelinated nerve fibers in monkey central nervous system. Front Neuroanat. 2009;3:11. doi: 10.3389/neuro.05.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartzokis G, Beckson M, Lu PH, Nuechterlein KH, Edwards N, Mintz J. Age-related changes in frontal and temporal lobe volumes in men: a magnetic resonance imaging study. Arch Gen Psychiatry. 2001;58(5):461–465. doi: 10.1001/archpsyc.58.5.461. [DOI] [PubMed] [Google Scholar]
- 12.Tripathi RB, Jackiewicz M, McKenzie IA, Kougioumtzidou E, Grist M, Richardson WD. Remarkable stability of myelinating oligodendrocytes in mice. Cell Rep. 2017;21(2):316–323. doi: 10.1016/j.celrep.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chrast R, Saher G, Nave KA, Verheijen MH. Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J Lipid Res. 2011;52(3):419–434. doi: 10.1194/jlr.R009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci U S A. 2010;107(32):14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyama BH, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154(5):971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith ME. The turnover of myelin in the adult rat. Biochim Biophys Acta. 1968;164(2):285–293. doi: 10.1016/0005-2760(68)90154-9. [DOI] [PubMed] [Google Scholar]
- 17.Ando S, Tanaka Y, Toyoda Y, Kon K. Turnover of myelin lipids in aging brain. Neurochem Res. 2003;28(1):5–13. doi: 10.1023/A:1021635826032. [DOI] [PubMed] [Google Scholar]
- 18.Vaswani KK, Ledeen RW. Long-chain acyl-coenzyme A synthetase in rat brain myelin. J Neurosci Res. 1987;17(1):65–70. doi: 10.1002/jnr.490170110. [DOI] [PubMed] [Google Scholar]
- 19.Koul O, Chou KH, Jungalwala FB. UDP-galactose-ceramide galactosyltransferase in rat brain myelin subfractions during development. Biochem J. 1980;186(3):959–969. doi: 10.1042/bj1860959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koenning M, et al. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32(36):12528–12542. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii A, Furusho M, Dupree JL, Bansal R. Role of ERK1/2 MAPK signaling in the maintenance of myelin and axonal integrity in the adult CNS. J Neurosci. 2014;34(48):16031–16045. doi: 10.1523/JNEUROSCI.3360-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussien Y, Podojil JR, Robinson AP, Lee AS, Miller SD, Popko B. ER chaperone BiP/GRP78 is required for myelinating cell survival and provides protection during experimental autoimmune encephalomyelitis. J Neurosci. 2015;35(48):15921–15933. doi: 10.1523/JNEUROSCI.0693-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chénard CA, Richard S. New implications for the QUAKING RNA binding protein in human disease. J Neurosci Res. 2008;86(2):233–242. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- 24.Sidman RL, Dickie MM, Appel SH. Mutant mice (Quaking and Jimpy) with deficient myelination in the central nervous system. Science. 1964;144(3616):309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- 25.Darbelli L, Vogel G, Almazan G, Richard S. Quaking regulates neurofascin 155 expression for myelin and axoglial junction maintenance. J Neurosci. 2016;36(14):4106–4120. doi: 10.1523/JNEUROSCI.3529-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L, Tian D, Xia M, Macklin WB, Feng Y. Rescuing qkV dysmyelination by a single isoform of the selective RNA-binding protein QKI. J Neurosci. 2006;26(44):11278–11286. doi: 10.1523/JNEUROSCI.2677-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larocque D, et al. Nuclear retention of MBP mRNAs in the quaking viable mice. Neuron. 2002;36(5):815–829. doi: 10.1016/S0896-6273(02)01055-3. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Zhang Y, Li D, Feng Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J Neurosci. 2000;20(13):4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shingu T, et al. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat Genet. 2017;49(1):75–86. doi: 10.1038/ng.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo A, Akiguchi I, Lee GC, McGeer EG, McGeer PL, Kimura J. Myelin degeneration in multiple system atrophy detected by unique antibodies. Am J Pathol. 1998;153(3):735–744. doi: 10.1016/S0002-9440(10)65617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weil MT, et al. Loss of myelin basic protein function triggers myelin breakdown in models of demyelinating diseases. Cell Rep. 2016;16(2):314–322. doi: 10.1016/j.celrep.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monsma PC, Brown A. FluoroMyelin™ Red is a bright, photostable and non-toxic fluorescent stain for live imaging of myelin. J Neurosci Methods. 2012;209(2):344–350. doi: 10.1016/j.jneumeth.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubbs CD, Smith AD. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim Biophys Acta. 1984;779(1):89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 34.Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications--a review. Nutr J. 2014;13:17. doi: 10.1186/1475-2891-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters JM, et al. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20(14):5119–5128. doi: 10.1128/MCB.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickey AS, et al. PPAR-δ is repressed in Huntington’s disease, is required for normal neuronal function and can be targeted therapeutically. Nat Med. 2016;22(1):37–45. doi: 10.1038/nm.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cramer PE, et al. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science. 2012;335(6075):1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kipp M, Nyamoya S, Hochstrasser T, Amor S. Multiple sclerosis animal models: a clinical and histopathological perspective. Brain Pathol. 2017;27(2):123–137. doi: 10.1111/bpa.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhlmann T, Ludwin S, Prat A, Antel J, Brück W, Lassmann H. An updated histological classification system for multiple sclerosis lesions. Acta Neuropathol. 2017;133(1):13–24. doi: 10.1007/s00401-016-1653-y. [DOI] [PubMed] [Google Scholar]
- 40.Han MH, et al. Janus-like opposing roles of CD47 in autoimmune brain inflammation in humans and mice. J Exp Med. 2012;209(7):1325–1334. doi: 10.1084/jem.20101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darbelli L, Richard S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip Rev RNA. 2016;7(3):399–412. doi: 10.1002/wrna.1344. [DOI] [PubMed] [Google Scholar]
- 42.Baumann NA, Jacque CM, Pollet SA, Harpin ML. Fatty acid and lipid composition of the brain of a myelin deficient mutant, the “quaking” mouse. Eur J Biochem. 1968;4(3):340–344. doi: 10.1111/j.1432-1033.1968.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 43.Baumann N, Bourre JM, Jacque C, Harpin ML. Lipid composition of Quaking mouse myelin: comparison with normal mouse myelin in the adult and during development. J Neurochem. 1973;20(3):753–759. doi: 10.1111/j.1471-4159.1973.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee SS, et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15(6):3012–3022. doi: 10.1128/MCB.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adhikary T, et al. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) PLoS ONE. 2011;6(1):e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khozoie C, et al. Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4. BMC Genomics. 2012;13:665. doi: 10.1186/1471-2164-13-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adhikary T, et al. The transcriptional PPARβ/δ network in human macrophages defines a unique agonist-induced activation state. Nucleic Acids Res. 2015;43(10):5033–5051. doi: 10.1093/nar/gkv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan W, et al. PPARδ promotes running endurance by preserving glucose. Cell Metab. 2017;25(5):1186–1193.e4. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McFarland HF, Martin R. Multiple sclerosis: a complicated picture of autoimmunity. Nat Immunol. 2007;8(9):913–919. doi: 10.1038/ni1507. [DOI] [PubMed] [Google Scholar]
- 50.Segal BM, Stüve O. Primary progressive multiple sclerosis--why we are failing. Lancet. 2016;387(10023):1032–1034. doi: 10.1016/S0140-6736(16)00158-6. [DOI] [PubMed] [Google Scholar]
- 51.Laule C, et al. Diffusely abnormal white matter in multiple sclerosis: further histologic studies provide evidence for a primary lipid abnormality with neurodegeneration. J Neuropathol Exp Neurol. 2013;72(1):42–52. doi: 10.1097/NEN.0b013e31827bced3. [DOI] [PubMed] [Google Scholar]
- 52.Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35(1):63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- 53.Barak Y, et al. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci U S A. 2002;99(1):303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imayoshi I, et al. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nat Neurosci. 2008;11(10):1153–1161. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- 55.Rasband MN, Peles E, Trimmer JS, Levinson SR, Lux SE, Shrager P. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J Neurosci. 1999;19(17):7516–7528. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Susuki K, et al. Gangliosides contribute to stability of paranodal junctions and ion channel clusters in myelinated nerve fibers. Glia. 2007;55(7):746–757. doi: 10.1002/glia.20503. [DOI] [PubMed] [Google Scholar]
- 57.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data and ChIP sequencing data reported in this study have been deposited in the NCBI’s Gene Expression Omnibus (GEO) under ID codes GSE84134, GSE106468, GSE125581, and GSE126577.