Abstract
Breast cancer is one of the most common life-threatening malignancies and the top cause of cancer deaths in women. Although many conventional therapies exist for its treatment, breast cancer still has many handicaps to overcome. Cancer stem cells (CSCs) are a well-known cause of tumor recurrences due to the ability of CSCs for self-renewal and differentiation into cell subpopulations, similar to stem cells. To fully treat breast cancer, a strategy for the treatment of both cancer cells and CSCs is required. However, current strategies for the eradication of CSCs are non-specific and have low efficacy. Therefore, surface biomarkers to selectively treat CSCs need to be developed. Here, 34 out of 641 surface biomarkers on CSCs were identified by proteomic analysis between the human breast adenocarcinoma cell line MCF-7 and MCF-7-derived CSCs. Among them, carcinoembryonic antigen-related cell adhesion molecules 6 (CEACAM6 or CD66c), a member of the CEA family, was selected as a novel biomarker on the CSC surface. This biomarker was then experimentally validated and evaluated for use as a CSC-specific marker. Its biological effects were assessed by treating breast cancer stem cells (BCSCs) with short hairpin (sh)-RNA under oxidative cellular conditions. This study is the first to evaluate the biological function of CD66c as a novel biomarker on the surface of CSCs. This marker is available as a moiety for use in the development of targeted therapeutic agents against CSCs.
Keywords: apoptosis, breast cancer stem cell, CD66c, surface biomarker
INTRODUCTION
Cancer is a difficult disease to treat and remains a top priority in terms of diseases for which treatments are still lacking. Despite the development of innovative therapeutic methods in recent decades, breast cancer still affects the lives of millions of women around the world. Breast cancer is one of the most common life-threatening malignancies and the leading cause of death by cancer in women. Although breast cancer can be cured after various anti-cancer treatments, it has a high rate of reoccurrence. Cancer stem cells (CSCs) are known to play an important role in the recurrence of cancer, wherein 65% of cancer is caused by CSCs (Rodini et al., 2017; Song and Giovannucci, 2015).
Recurrence and metastasis are caused by CSCs that survive anti-cancer treatment (Baumann et al., 2008; Koch et al., 2010). CSCs are defined as cells that are capable of self-renewal and differentiation, similar to stem cells (Lee et al., 2011; Nassar and Blanpain, 2016; Zhu and Fan, 2018). A small subpopulation of CSCs can initiate the formation of a tumor and is associated with resistance to various anti-tumor drugs (Maugeri-Sacca et al., 2011). CSCs activate various mechanisms via the regulation of cell signals and protein expression for the resistance to anti-cancer treatments, including Wnt/β-catenin, Notch and Hedgehog signaling as reprogramming signals, high drug transporter expression, the activation of efficient DNA repair system, and the expression of anti-apoptotic protein and detoxifying enzymes (Colak and Medema, 2014; Krishnamurthy and Kurzrock, 2018).
Unlike other cancer cells, CSCs are dormant during the cell cycle, a stage known as quiescence, which allows them to evade anti-cancer drugs. After anti-cancer treatment, that is when the environment is once again favorable, these cells become active and multiply rapidly (Nassar and Blanpain, 2016; Zhu and Fan, 2018).
CSCs are able to use their cellular environment intelligently and efficiently, known as the niche environment (Scadden, 2006; Vinogradov and Wei, 2012; Zhu and Fan, 2018). They induce hypoxic stability to enhance tumorigenicity (Vinogradov and Wei, 2012; Scheel et al., 2011). These survival mechanisms allow CSCs to evade the effect of anti-cancer treatments. Due to these characteristics, it is important to know how many CSCs remain after anti-cancer treatment as the presence of CSCs after therapy can be used to assess the effectiveness of treatment. It is, therefore, important to identify reliable and unique CSC biomarkers for each type of tumor (Ahn et al., 2008; Jung, 2017; Luo et al., 2015).
Anti-cancer therapies focusing on CSCs are likely to form the core of future effective anticancer strategies. Many CSC surface markers have been identified and developed that specifically target CSCs. For example, CD24–/CD44+ has been reported to be a representative surface biomarker of breast-derived CSCs (BCSCs) (Crabtree and Miele, 2018; Honeth et al., 2008; Lombardo et al., 2015; Zhu and Fan, 2018). Despite the progress made in these studies, for the identification, isolation, and characterization of CSCs, the research remains controversial and has been challenged by recent studies in terms of its heterogeneity and plasticity, such that targeting CSCs remains a challenge for the treatment of tumors (Jaggupilli and Elkord, 2012; Yang et al., 2017; Zhu and Fan, 2018). The underlying technology for the development of CSC treatments currently focuses on identifying the characteristics of CSCs rather than on targeting them, which is still found in the early stages of research. Moreover, the development of CSC tracking techniques and cell-specific tracking markers is needed in order to maximize the therapeutic effect of the treatment on cancer cells and CSCs, in addition to the functional control of the treatments at the cellular level.
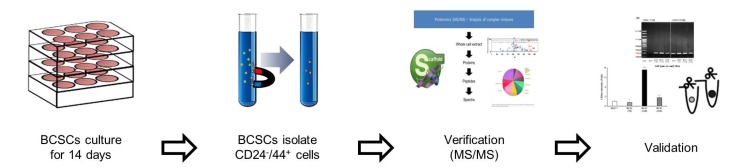
In this study, a mass spectrometry analysis was performed to develop a surface biomarker of BCSCs (Fig. 1). The resulting biomarker was validated and examined for use as a specific marker on the surface CSCs via various experiments. The biological effects of CD66c in BCSCs were also assessed by treating BCSCs with short hairpin (sh)RNA expressing a lentiviral vector against the CD66c gene.
Fig. 1. Scheme 1.
Experimental processing for the identification of novel biomarkers on the surface of breast cancer-derived CSCs.
MATERIALS AND METHODS
Cell culture
MCF-7 (ATCC [American Type Culture Collection], USA) human breast adenocarcinoma cell lines were cultured as adherent cells using DMEM (Hyclone, USA) supplemented with 10% fetal calf serum (Hyclone) and 1% penicillin G-streptomycin solution (Gibco, USA). Cells were maintained in a humidified atmosphere (5% CO2) at 37°C.
Culture of breast cancer stem cells
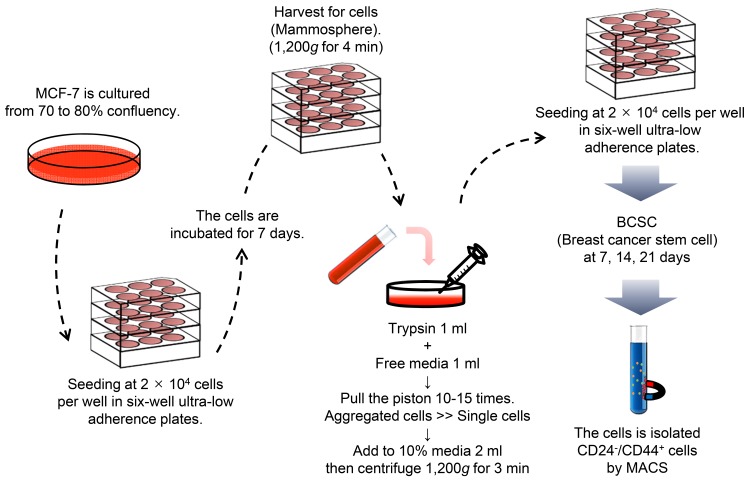
A concentration of 20,000 cells/1.5 ml of culture medium was used to seed the CSCs derived from MCF-7 cells in 6-well ultra-low attachment plates (Corning, USA). Cells possess the capacity for growth and sphere formation in the presence of serum-free Dulbecco’s modified Eagle’s medium/F12 medium (DMEM/F12) (Gibco) and specific supplements including and 10% fetal calf serum (Hyclone), 1% penicillin G-streptomycin solution (Gibco), 5 µg/ml insulin, 20 ng/ml EGF (Gibco), 20 ng/ml b-FGF (Gibco), and B27 (Invitrogen, USA). Every 2-3 days, it was filled 1 ml of the fresh media containing the components in an each-well plate, as mentioned above. Cells grown in these conditions as non-adherent spherical of cells (usually named “mammospheres”) were collected every seven days by gentle centrifugation (Dontu et al., 2003). After centrifugation, the harvested cells were gently suspended in 1 ml of trypsin and 1 ml of serum-free DMEM/F12 medium and then dissociated by pipetting with a 24 gauge needle (Fig. 2). A ZOE fluorescent microscope (Bio-Rad, USA) was used to capture the images.
Fig. 2. Scheme 2.
Manufacturing process of BCSCs derived from MCF-7 cells. MCF-7 was seeded in 6-well ultra-low attachment plates. To expand the CSC populations, the seeded cells were grown on several 6-well plates until mammospheres were formed.
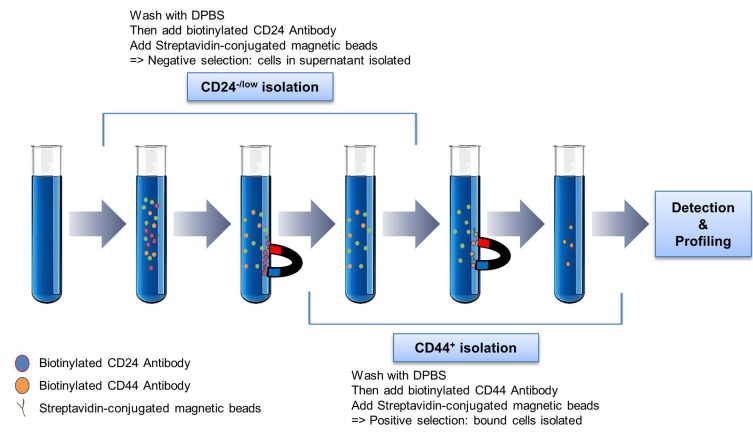
Isolation of CD24–/CD44+ breast cancer stem cells
Breast cancer stem cells (BCSCs) after 7, 14, and 21 days were harvested in 2 ml of 1× plus buffer (R&D Systems, USA). A single-cell suspension of the BCSCs was isolated by human CD24-/CD44+ BCSCs isolation kit (R&D Systems) according to the manufacturer’s instructions. From whole cells, CD24-/ CD44+ cells were sorted using a biotinylated human antibody and streptavidin-conjugated magnetic beads. The CD24-/ CD44+ cells were selected as a positive BCSC model. To identify the characteristics of BCSCs, we were confirmed via flow cytometry analysis (FACS) analysis by staining obtained cells with fluorochrome-conjugated anti-human CD24- and CD44+ antibodies. Flow cytometry was used to confirm whether or not the isolated cells are CSCs (Fig. 3).
Fig. 3. Scheme 3.
Isolation of BCSCs formed from mammospheres by magnetic activated cell sorting (MACS).
Mass spectrometry analysis
BCSCs and MCF-7 cells were lysed 30 µl of lysis buffer (20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1% NP-40 and 1× protease phosphatase inhibitor), and maintain constant agitation for 30 min at 4°C. The lysed samples were centrifuged at 12,000 rpm for 15 min and the supernatant was transferred to another vial for further tryptic digestion. To check protein concentration, the samples quantified using the Coomassie Plus (Bradford, UK) Assay Reagent (Thermo Fisher Scientific).
Lysate protein (100 µg) were washed in 10 mM DTT solution and incubated for 1 h at 37°C. The samples were added 50 mM IAA solution and incubated for 1 h at room temperature in the dark. We added 50 mM ABC (ammonium bicarbonate) to reducing the DTT concentration to the final 1 mM. The protein that lysed BCSCs and MCF-7 cells were digested by trypsin solution (proteins:trypsin = 40:1) incubation at 37° C overnight. Finally, we added up to 1% TFA (trifluoroacetic acid) to stop the trypsin digestion.
Also, to clean up the sample using MCX cartridge, the following process was carried out. We equilibrated the column by adding 1 ml of methanol and washed by 1 ml of 0.1% TFA. The column was added 500 to 1,000 µl of the acidified sample (< pH 2.0) and washed with 1 ml of 0.1% TFA and then with 1 ml of methanol. It added 500 µl of elution buffer which contains 50% ACN (acetonitrile) and 5% ammonium hydroxide. After passing the elution buffer through the column, the elute was dried in a speed vacuum to complete dryness. The peptides were extracted with 0.1% formic acid for LC injection, or store at –20°C before the analysis.
Samples are subsequently separated by online reversed-phase chromatography for each run using a Thermo Scientific Easy nano LC II autosampler with a reversed-phase peptide trap EASY-Column (100 µm inner diameter, 2 cm length) and a reversed-phase analytical EASY-Column (75 µm inner diameter, 10 cm length, 3 µm particle size; both Thermo Scientific, USA), and electrospray ionization was subsequently performed using a 30 µm (inner diameter) nano-bore stainless steel online emitter (Thermo Scientific) and a voltage set at 2.6 kV, at a flow rate of 300 nl/min. The chromatography system was coupled on-line with an LTQ VelosOrbitrap mass spectrometer equipped with an ETD source. To improve peptide fragmentation of phosphopeptides, we applied a data-dependent switching mode to select for collision-induced dissociation. Protein identification was accomplished utilizing the Sorcerer 2 and searches were performed against the 2014 UniProt human DB. A fragment mass tolerance of 1.0 Da, peptide mass tolerance of 25 ppm, and maximum missed cleavage of 2 was set. Result filters were performed with peptides number per protein (minimal number of peptides: 1). The carbamidomethylation (+57 Da) of cysteine (C) is set as a static modification, and the following variable modification were allowed: Glu→pyro-Glu of the n-terminus (–18 Da), ammonia-loss of the n-terminus (–17 Da), Gln→pyro-Glu of the n-terminus (–17 Da), and oxidation of methionine (+16 Da). Each processed data was subsequently transformed to.sf file with Scaffold 3 program (Lee et al., 2014). We technically performed triplicates.
Flow cytometry analysis
After the appropriate preparation of samples, 100 µl of cell suspensions were incubated with appropriate amounts of each CEACAM6 monoclonal antibody (CD66c, 5 µg/ml final concentration; Thermo Scientific) for 90 min at 4°C. The cells were then washed once with 500 µl Dulbecco’s phosphate-buffered saline (DPBS), incubated in 100 µl DPBS with secondary antibodies: Alexa Fluor 488-conjugated goat anti-mouse antibody (4 µg/ml final concentration; Thermo scientific) at 4°C for 30 min. Cells were analyzed using flow cytometry (NovoCyte; ACEA Biosciences, USA) after antibody staining for the analysis.
Reverse transcription-polymerase chain reaction
MCF-7 and BCSCs during 7, 14, 21 days were harvested, and total RNA was prepared using TRIzol (Invitrogen) following the manufacturer’s manual. Then RNA samples were converted into cDNA by DiaStar 2× RT Pre-Mix (Solgent, Korea). Synthesized cDNA was subjected to conventional polymerase chain reaction (PCR) using a Solg 2× Taq PCR Pre-Mix (Solgent) according to the manufacturer’s protocols and performed on T100 (Bio-Rad). Fold changes in relative gene expression were calculated by the expression of GAPDH as an internal control. The primers are used in this study are as follows: CD66c-F, 5’-GATCTTGTGAATGAAGA AGCAAC-3’, CD66c-R, 5’-GTCAT GTTGCCATTGGACAG-3’ and GAPDH-F, 5’-AGGGCTGCTTTTAACTCTGGT-3’, GAPDH-R, 5’-CCCCACTTGATTTTGGAGGGA-3’.
Western blot analyses
For the western blot analysis, cells were lysed using a RIPA buffer (ATTO, Japan). Protein concentration was measured using a BCA kit (Thermo Scientific). Equal amounts of protein (20 µg) were resolved with 10% SDS-PAGE and transferred to nitrocellulose (NC) membranes (Bio-Rad). Membranes were probed with primary antibodies for 16-18 h at 4˚C and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000, 31430; Invitrogen) for 1 h at room temperature. CEACAM6 (1:1,250, MA5-24164; Thermo Scientific) antibodies were used as a target marker, and β-actin (1:5,000, SC-47778; Santa Cruz Biotechnology, USA) antibody was used as an internal control. Electrochemiluminescence was performed with a Fusion SL (Vilber Lourmat, France).
On-cell binding assays
LICOR Odyssey Imaging system (LI-COR Biosciences, USA) allows for the quantitative determination of extracellular protein expression levels of cells growing in 96 well culture plates. For the on-cell binding assays, this plate was incubated with a blocking buffer for 30 min on ice, and then this was incubated overnight at 4˚C with CEACAM6 (mouse monoclonal, 1:500; Thermo Scientific) and β-actin (mouse monoclonal, 1:1,200; Santa Cruz Biotechnology) antibodies in blocking buffer. After being washed in blocking buffer Tween-20 (TBST; 3×, 5 min each time), this was treated with IRDye 800CW goat anti-mouse secondary antibody (1:15,000, 926-32210; LI-COR Biosciences). Densitometric analysis of IRDye signals was carried out using Odyssey Image system at 800 nm (green intensity). The protein levels were normalized to β-actin.
Immunocytochemistry
Cultures were grown in 4-well removable-chamber slides (Thermo Scientific). This slide was fixed directly using 4% paraformaldehyde for 5 min at 37°C and 100% methanol for 10 min at –20°C and then blocked by phosphate-buffered saline (PBS) with bovine serum albumin. It was incubated overnight at 4˚C with the primary antibody for CEACAM6 (1:500; Thermo Scientific). It was followed by an Alexa Fluor 488-conjugated goat anti-mouse antibody (4 µg/ml final concentration, A32723; Invitrogen) and PE-conjugated mouse anti-human CD44 antibody (7:100, 860217; R&D Systems) at 4°C for 30 min. After rinsing, the slides were mounted with a mounting solution (Fluoroshield Mounting Medium with DAPI; Abcam, UK) and observed using a digital fluorescence microscope (Nikon, USA).
Knockdown of CD66c in BCSCs by shRNA
A total of 1 × 105 BCSCs sorted by the isolation kit were transfected with shRNA-CD66c in lentiviral GFP vector (Origene, USA) or shRNA-scrambled (SC) in lentiviral GFP vector (Origene) using TransIT-X2 (Mirus Bio, USA), following the manufacturer’s instructions. The same plasmid without any insert, as well as a plasmid with a scrambled vector, was used as negative controls. GFP expressing BCSCs were observed after 48 h of incubation. Cell viability was assessed by crystal violet staining and MTT assay.
Crystal violet assay for assessing the viability of cultured cells
After transfection, this plate was stained with 0.5% crystal violet (Sigma, USA). After 4 h of incubation, the excess crystal violet was washed out with PBS, and the stained cells were dissolved methanol for 30 min (Shi et al., 2015). The absorbance from each sample was measured in duplicate using a spectropho-tometric microplate reader at a wavelength of 450 nm (Versamax Microplate Reader; Molecular Devices, USA).
MTT assay
BCSCs transfected with lentivirus shRNA-CD66c, lentivirus shRNA-scrambled, and negative control was treated with 5 mM H2O2 for 4 h, and then recovered for 1 day. A cell viability assay was analyzed by MTT (3-(4,5-dimethylthiazol2-yl)-2,5-diphenyltetrazolium bromide) (Sigma). MTT (2.5 mg/ml in PBS) was added to each well and incubated for an additional 3 h at 37˚C. The formazan crystals were dissolved in DMSO, and the absorbance was read at 570 nm by a microplate reader (Versamax Microplate Reader). Triplecate wells were measured in each treatment group
Apoptosis induction analysis of BCSCs treated with H2O2 under oxidative stress condition
For an apoptosis assay, the supernatant was aspirated, and cells were resuspended in 150 µl binding buffer, before staining with 5 µl Annexin V-FITC and 5 µl PI at room temperature for 15 min in the dark. After incubation, cells were processed as directed by the kit instructions (Annexin V-FITC Apoptosis Detection Kit I; BD Biosciences, USA) and analyzed using NovoCyte Flow Cytometer (ACEA Biosciences).
Statistical analysis
Statistical significance of data was determined by applying the paired t-test and one-way ANOVA. Significance of analysis of variance is indicated in the figures; *P < 0.05, **P < 0.02, and ***P < 0.01. Statistical analyses were performed with the Prism software for the Windows (ver. 5.01; GraphPad Software, USA).
RESULTS
Isolation and identification of BCSCs
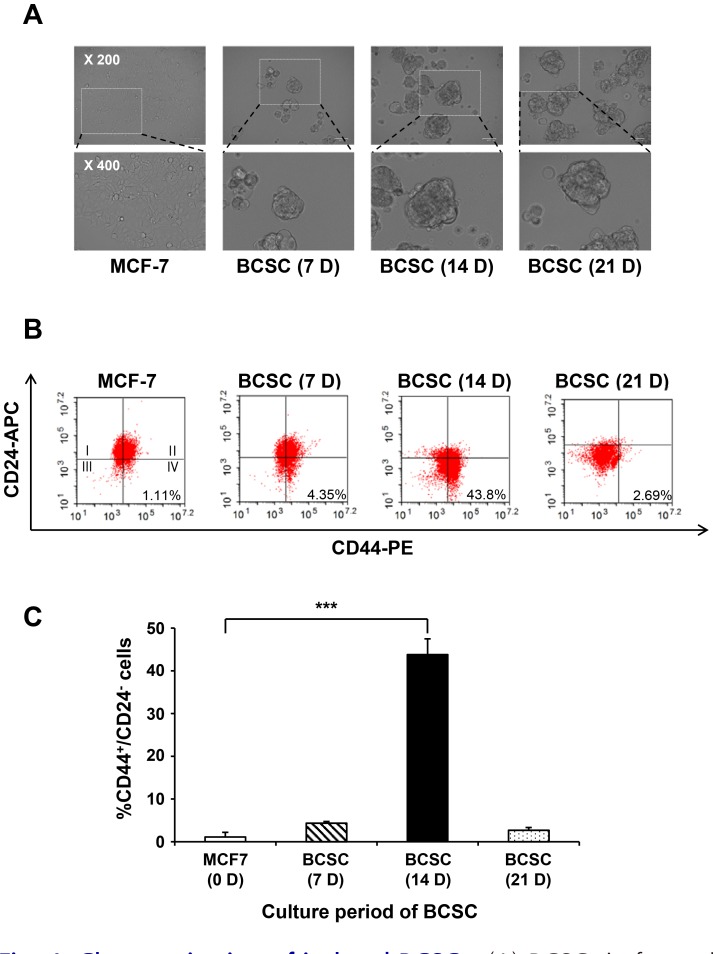
MCF-7 breast cancer cells were cultivated as parental cells for the proteomic analysis of BCSCs. The cells grew as adherent epithelial-like monolayer cells with a polygonal shape and clear, sharp boundaries between them. Under mammosphere culture conditions, MCF-7 cells were cultured for 7, 14, and 21 days in the non-adherent surface compared to their parental counterparts. These cells formed mammosphere starting from the third day of cell culture. It appeared that the size of the BCSCs increased in a time-dependent manner, in contrast to MCF-7 cells (Fig. 4A).
Fig. 4. Characterization of isolated BCSCs.
(A) BCSCs in formed mammospheres were observed using an optical microscope on days 7, 14, and 21. The size of the cells increased in a time-dependent manner. (B) Flow cytometric analysis of cells for CD24–/CD44+. To identify the characteristics of BCSCs, the cell population expressing CD44+ and CD24– were analyzed by flow cytometry. After 3 weeks, the highest level of CD24–/CD44+ CSC marker expression was observed after 14 days. (C) Quantitative data of BCSCs expressing CD24–/CD44+ in a time-dependent manner. Data are expressed as the mean ± SEM. ***P < 0.01.
At each culture time, CD24–/CD44+ markers were used to determine whether the characteristics of the cultured cells represented those of BCSCs. MCF-7 cells and MCF-7-derived CSCs were analyzed by flow cytometry at 7, 14, and 21 days post-culture (Fig. 4B). The population of cells expressing CD24–/CD44+ after 7, 14, and 21 day of culturing increased on was average by 4.35%, 43.80%, and 2.69%, respectively, compared to MCF-7 cells (1.11%). The expression of the CD24–/CD44+ marker was highest after 14 days of culture and was found to best represent the characteristics of BCSCs (Figs. 4B and 4C). Therefore, the cells at 14 days post-culture were used as BCSCs for future experiments.
Identification of novel biomarkers on the surface of BCSCs via proteomic analysis
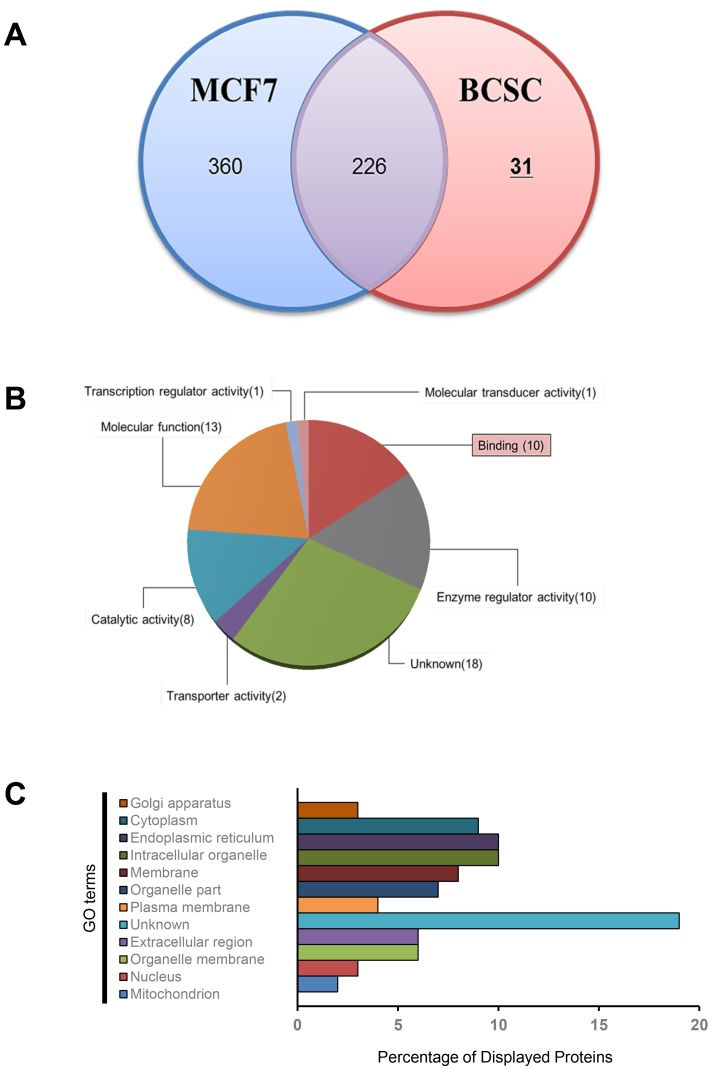
To compare the expression of surface proteins between BCSCs and MCF-7 cells, liquid chromatography-mass spectrometry (LC-MS) was performed, and the results were analyzed. After comparative proteomics, a total of 617 proteins were analyzed using a Venn diagram (Fig. 5A, Supplementary Files S1 and S2). Among the 617 proteins, 31 candidates were identified in BCSCs. The proteins expressed in the BCSCs were re-analyzed for statistical over-representation of the Gene Ontology (GO) category. Using charts divided by category, the 31 candidates in the BCSCs were found to overlap in each category (Figs. 5B and 5C). Four candidate groups were identified within the plasma protein category, namely carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6 or CD66c), ATP synthase subunit gamma, mitochondrial (ATP5C1), guanylate-binding protein 1 (GBP1), and serine/threonine-protein kinase (PAK4). Among these, CD66c was ultimately selected as the novel surface biomarker of BCSCs. Anchored cell surface glycoproteins are known to be responsible for cellular adhesion and typically exert anti-apoptosis functions (Cameron et al., 2012; Hong et al., 2015; Johnson and Mahadevan, 2015; Rizeq et al., 2018).
Fig. 5. Comparative proteome analysis of isolated BCSCs and MCF-7 cells.
(A) Venn diagram of isolated proteins of BCSCs by mass spectrometry (MS). The 31 proteins indicated on the right were upregulated in BCSCs compared to MCF-7 cells. (B) The classification according to molecular function of the 31 proteins represented and (C) GO analysis of in various biological processes in the plasma membrane of BCSCs.
CD66c as a novel biomarker on the surface of BCSCs at the transcriptomic and proteomic level
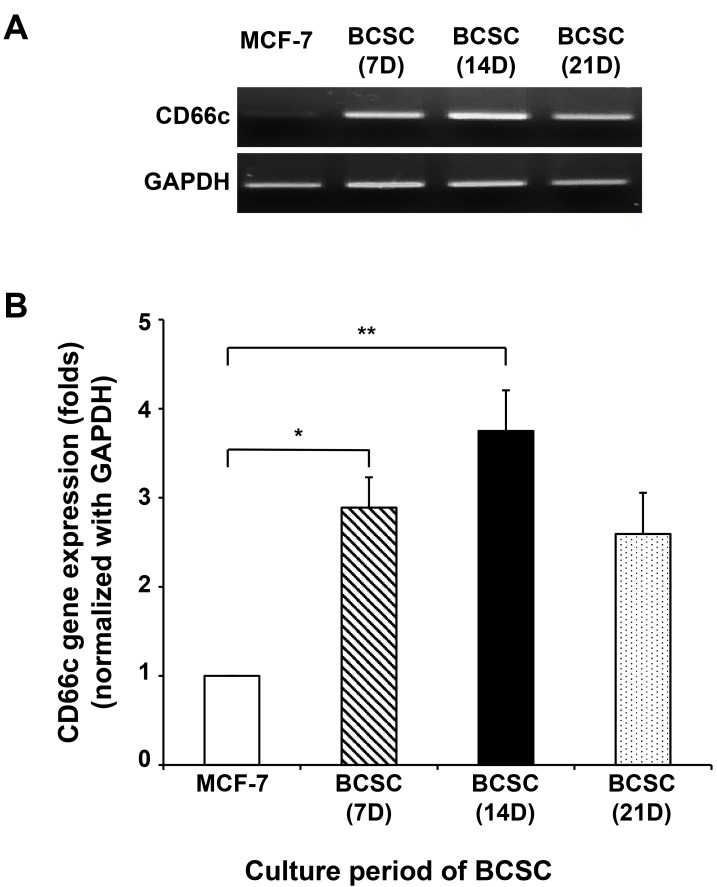
The changes in the expression of CD66c, a surface biomarker of BCSCs, at the transcriptomic level between breast cancer cells and CSCs were investigated. RNA extraction and cDNA synthesis by reverse transcription-polymerase chain reaction (RT-PCR) were used to confirm the existence of the CD66c gene in the BCSCs. The isolated total RNA from these cells was normalized to GAPDH. As shown Fig. 6, the expression of CD66c was significantly higher in BCSCs cultured for 14 days compared to MCF-7 cells. CD66c expression was found to gradually decrease after 14 days. This result indicates that CD66c expression tended to increase when cells were characterized as CSCs, leading to the induction of their expression in BCSCs.
Fig. 6. The expression of the CD66c gene at the transcriptomic level in cultured BCSCs in a time-dependent manner.
(A) The expression of CD66c in BCSCs was confirmed by RT-PCR using mRNA extracted from BCSCs at the transcriptomic level. (B) Quantitative data of the expressed CD66c gene normalized by GAPDH in BCSCs. Data are expressed as the mean ± SEM. *P < 0.05 or **P < 0.02 versus MCF-7 group.
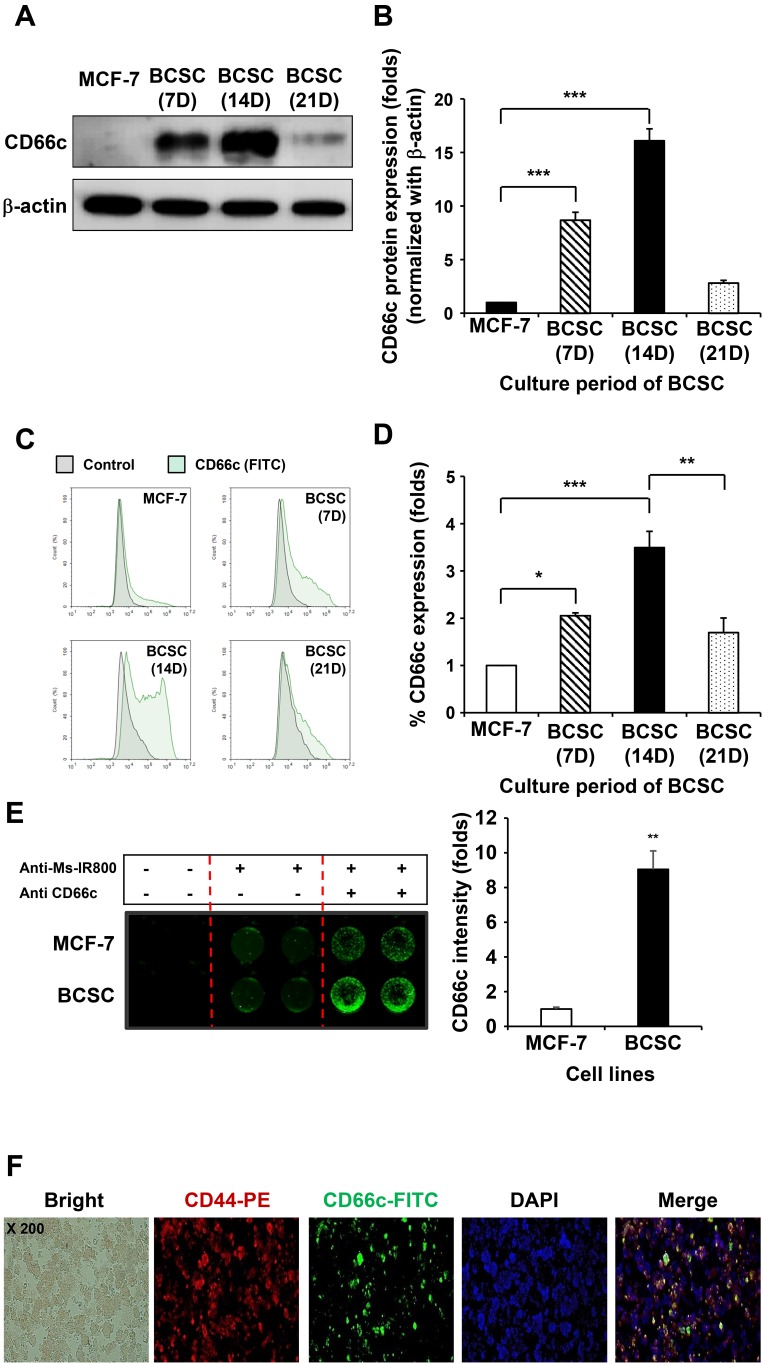
Next, in order to confirm whether CD66c expression at the genome occurs at the protein level, breast cancer cells and BCSCs were cultured and checked for CD66c protein expression using western blotting and flow cytometry. As shown in Fig. 7A, CD66c protein expression increased up to day 14, before subsequently decreasing. As such, CD66c expression was highest on day 14. The quantitative data of the CD66c band showed a similar tendency (Fig. 7B).
Fig. 7. The expression of CD66c at the protein level in cultured BCSCs in a time-dependent manner.
The expression of CD66c protein increased in a time-dependent manner in BSCSs compared to MCF-7 cells, in agreement with the transcriptomic results. These results indicate that the expression of CD66c is related to the maintenance period in CSCs, but not in cancer cells. (A) Confirmation of the expression of CD66c protein by western blotting. (B) Quantitative graph of the western blot. Data are expressed as the mean ± SEM. ***P < 0.01 versus MCF-7 group. (C) Resulting peaks of the CD66c protein levels as a factor of time expressed on the surface of BCSCs in the form of a histogram according to FACS data. (D) Quantitative graph of the shifted peak in the FACS data. Data are expressed as the mean ± SEM. *P < 0.05 and ***P < 0.01 for MCF-7 versus BCSC (7 days) or BCSC (14 days); **P < 0.02 for BCSC (14 days) versus BCSC (21 days). (E) The images of CD66c expressed on the surface of the BCSCs according to cell binding assay. Data are expressed as the mean ± SEM. **P < 0.02 versus MCF-7 group. (F) Identification of the characteristics of CSCs in BCSCs expressing CD66c by immunocytochemistry.
FACS was performed to assess whether CD66c is expressed on the surface of the cell (Fig. 7C). As shown in the FACS result, the expression peak of CD66c in each group was found to shift to the right. Its expression increased in BCSCs cultured for 7 and 14 days compared to MCF-7 cells, while its expression decreased in BCSCs cultured for 21 days (Fig. 7D). This correlated with the verification results for CD66c protein expression (Figs. 7A and 7B).
The surface expression of CD66c was then verified using the Odyssey Image system. MCF-7 cells and BCSCs were seeded onto plates and analyzed using a one-color IR-based quantitative-on-cell binding assay. β-Actin was used to ensure equal protein loading. As shown in Fig. 7E, the surface fluorescence of CD66c on the BCSCs was significantly greater than that of MCF-7 cells.
Next, we examined whether the characteristics of CSC are maintained during the expression of CD66c. The expression of the representative CSC marker and CD66c were evaluated by immunocytochemistry (Fig. 7F). Immunofluorescence images showed that the co-expression of CD44 and CD66c was significantly observed in the spheroids generated from MCF-7 cells. Taken together, these results demonstrate that the biomarker CD66c on the surface of BCSCs is unique to BCSC.
Assessment of the biological function of CD66c by silencing in BCSCs
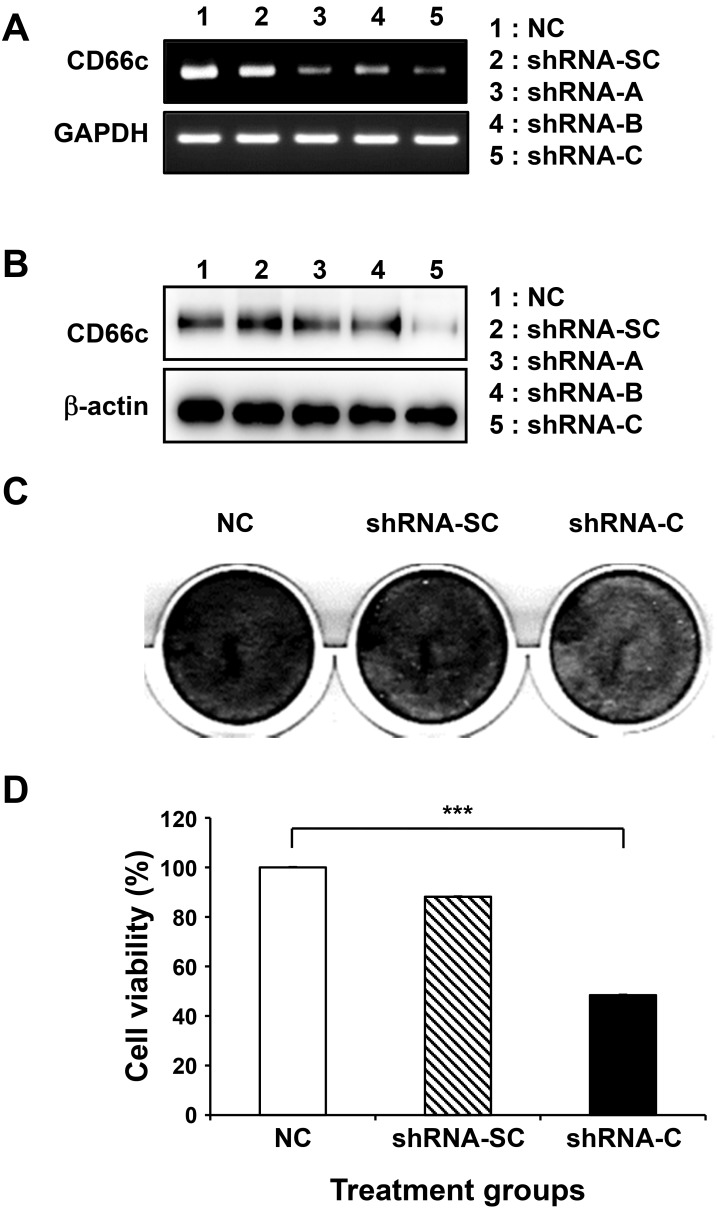
CD66c is known to enhance cell-cell interactions, resistance to apoptosis, and cell invasion (Johnson and Mahadevan, 2015; Lin et al., 2015; Rizeq et al., 2018). Next, we investigated whether CD66c performs these functions in BCSCs, using various shRNA to induce the knock-down of CD66c. Among the shRNA used, shRNA-C was the most effective for CD66c knock-down (Fig. 8A). For the following experiment, we researched after BCSCs seeded at the adherent condition. During the transfected with shRNA that knockdown CD66c in BCSCs, it was confirmed that the CD24–/CD44+ properties of the BCSCs remained unchanged. (Supplementary Fig. S1).
Fig. 8. Biological effects of CD66c in BCSCs by CD66c gene silencing using shRNA.
To determine the biological function of CD66c in BCSCs, BCSCs were treated with several shRNAs. The knock-down of the CD66c gene by shRNA at the transcriptomic (A) and protein (B) level. (C) Cell death and adhesion in BCSCs treated with shRNA-C against CD66c by crystal violet staining. (D) Cell viability in BCSCs treated with shRNA-C. Scrambled shRNA (SC) was used as a negative control here. ***P < 0.01 for NC group versus shRNA-C treated group.
The expression levels of CD66c protein in BCSCs after CD66c knock-down fell by about 2-fold compared to BCSCs treated with other shRNA groups, due to the silencing of the CD66c gene (Fig. 8B). Next, we evaluated the effect on cell proliferation in CSCs when the CD66c gene was knockdown by transfected with shRNA. As shown in Fig. 8C, BCSCs were stained with crystal violet post-shRNA transfection. The results showed that the cells treated with shRNA-C were dyed lightly, resulting in the induction of cell death by shRNA-C. To further clarify this, an MTT assay was conducted. Similar to the results obtained from the crystal violet staining experiment, the MTT results indicated that shRNA-C-treated cells had the lowest cell survival rate (Fig. 8D). MCF-7 cells that did not present CD66c were then evaluated. Unlike the results obtained for the BCSCs, the effects of shRNA-c were not observable in MCF-7 cells (Supplementary Fig. S2), indicating that the effects of shRNA against CD66c are dependent on CD66c expression.
Increased cell death by knock-down of CD66c gene under oxidative stress
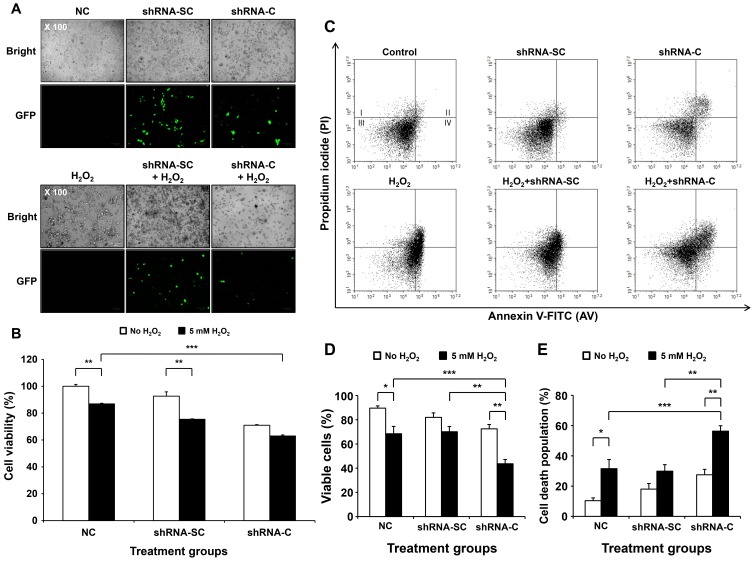
One of the many biological functions of CD66c is the inhibition of apoptosis under hypoxic conditions. To mimic the in vivo environment under in vitro conditions, cells were treated with hydrogen peroxide (H2O2) for 4 h after shRNA-SC and shRNA-C transfection. The shRNA-C expressing vector had inserted with the GFP gene as an indicator of transfection efficiency. We assessed the ability of cells to induce cytotoxicity according to the presence or absence of the CD66c gene under oxidative stress. First, the efficiency of the transfection was investigated based on the expression of GFP vector inserted with shRNA-c against the CD66c gene. As shown in Fig. 9A, the expression of GFP in the cells treated with shRNA-SC was higher compared to cells treated with shRNA-C, due to the decrease in the levels of GFP-expressing cells resulting from the increased cell killing effect, observed in the bright images of the cells treated with shRNA-C (Figs. 9A and 9B). To assess the cytotoxicity by shRNA-C under oxidative stress conditions, an MTT assay was performed. Cells were transduced with shRNA-SC or shRNA-C for 6 h, then treated with H2O2 for 4 h at 48 h post-transfection. Anti-proliferation was found to be induced in BCSCs transfected with shRNA compared to the untreated cells (NC) (100%, 78%, and 64% in NC, shRNA-SC, and shRNA-C for cell viability, respectively). shRNA-C-treated cells showed considerable cytotoxicity compared to the control (NC) or shRNA-SC groups (Fig. 9B). In particular, an apparent increase in cytotoxicity was observed in all groups treated with H2O2. A significant difference (2.6-fold increase in cytotoxicity) was observed between cells treated with shRNA-C and H2O2 and cells in the NC group. These results imply that CSC viability is increased under conditions of oxidative cellular damage in the absence of the anti-apoptosis effect of CD66c, resulting in an increased cell killing effect.
Fig. 9. Enhanced cell death in BCSCs treated with shRNA-C under oxidative stress conditions.
(A) Transfection efficiency images of shRNA-C expressing a lentiviral vector for the knock-down of the CD66c gene. (B) Cell viability of BCSCs treated with shRNA-C under oxidative stress conditions. Data are expressed as the mean ± SEM. **P < 0.02 for in the absence or presence of H2O2. ***P < 0.01 for H2O2-only group versus H2O2treated with shRNA-C group. (C) FACS analysis of cell death induced by treatment with H2O2 post-shRNA-C transfection in BCSCs. (D) Quantitative viable cell population of BCSCs treated with shRNA-C and H2O2 by FACS analysis. Data are expressed as the mean ± SEM. *P < 0.05 or **P < 0.02 for the comparison of no H2O2 treated groups and H2O2 treated groups or **P < 0.02 for shRNA-SC versus shRNA-C groups in the presence of H2O2. ***P < 0.01 for the comparison of NC and shRNA-C groups in the presence of H2O2. (E) Quantitative death cell population induced by shRNA-C treatment with H2O2. Data are expressed as the mean ± SEM. *P < 0.05 or **P < 0.02 for in the absence or presence of H2O2. **P < 0.02 for the comparison of shRNA-SC with H2O2 group and shRNA-C with H2O2 group. ***P < 0.01 for the comparison of H2O2 only group and shRNA-C with H2O2 group.
The enhancement of the CSC killing effect may be caused by apoptosis, due to the absence of the apoptosis inhibition function of the CD66c gene. To address this, BCSCs were evaluated by double staining FACS using FITC-annexin-V and propidium iodide (PI). The cell death population (annexin-V positive and PI negative, 41.16%; annexin-V positive and PI positive, 5.86%) was found increased in the BCSCs treated with shRNA-C and H2O2 compared to those treated with shRNA-SC and H2O2 (10.55% and both positive 3.71%) or the other groups (Figs. 9D and 9E). These results demonstrate that shRNA-C elicits enhanced cell death compared to shRNA-SC, as shown by a decrease in the induction of cell viability.
DISCUSSION
There is currently a high rate of relapse in many types of cancer despite the development of improved therapeutic treatments (Rodini et al., 2017; Song and Giovannucci, 2015). Although many conventional therapies have been used for the treatment of cancer, many handicaps remain to be overcome. Several studies have suggested that the recurrence of cancer is caused by CSCs (Nassar and Blanpain, 2016; Zhu and Fan, 2018). CSCs, also known as tumor initiating cells, are defined as cancer cells with the ability to self-renew, similar to stem cells, and are associated with drug resistance and the formation of a tumorigenic core for tumor formation (Maugeri-Sacca et al., 2011; Nilendu et al., 2018; Zhu and Fan, 2018). Therefore, to avoid relapse after the treatment of cancer, CSCs must be eliminated. To this end, many researchers have studied a variety of treatments targeting CSCs. As a result, many biomarkers of CSCs have been identified (Ahn et al., 2008; Hamam et al., 2017; Luo et al., 2015; Skvortsov et al., 2014). However, the targeting of CSCs for their elimination involves many obstacles (Chen et al., 2012; Deonarain et al., 2009; Huang and Rofstad, 2017). CSC biomarkers have heterogeneous properties due to their genetic characteristics and heterogeneity, making them difficult to selectively recognize and utilize (Meacham and Morrison, 2013). Therefore, CSCs need to be distinguished before they can be used in anti-cancer treatments. As such, there is a need to find novel surface biomarkers for CSCs (Kim and Ryu, 2017).
Here, we attempted to identify a novel biomarker on the surface of certain breast cancer-derived CSCs through the development of CSC biomarkers in order to develop a novel selective anti-cancer treatment (Jung, 2017; Luo et al., 2015; Morrison et al., 2012; Nie et al., 2015) and resolve the issues related to the heterogeneity and differences between cells. First, we isolated and characterized breast cancer-derived CSCs (Bailey et al., 2018; Luo et al., 2015). As CSCs exist in small populations in tumors, they are very difficult to isolate and study. In the present study, several procedures were used to obtain CD24–/CD44+ markers expressing cell-enriched mammospheres from cultured MCF-7 cells (Calvet et al., 2014; Wang et al., 2014) (Figs. 1 and 2). In previous studies, CSCs have been reported to be capable of sustained growth as spheres, wherein sphere formation is enriched in later passage spheres compared to early passage spheres (Kim et al., 2017). In our study, CD24–/CD44+-expressing BCSCs were found to be enriched in 14-day mammospheres compared to 7-day mammospheres; however, after 21 days, their expression decreased (Fig. 4).
Proteomic analysis is a leading research methodology for understanding protein expression in tissues. One theoretical drawback of proteomic research is that the identification of plasma membrane proteins is often hindered due to several reasons: (i) plasma membrane proteins are often more hydrophobic and therefore less soluble than cytosolic proteins (Pogozheva et al., 2013); (ii) they often have significant post-translational modifications (such as glycosylation, phosphorylation, and lipid moieties), making their identification difficult (Diaz-Fernandez et al., 2018); (iii) they are not as relatively abundant as other proteins (Leth-Larsen et al., 2009). Despite these shortcomings in identifying cell surface markers, proteomics still offers a powerful high-throughput technique for the collection of a large dataset of proteins (Chen et al., 2012; Diaz-Fernandez et al., 2018; Rodini et al., 2017). Therefore, we used a proteomic analyzer to identify candidates for surface biomarkers through the comparative analysis of human breast adenocarcinoma cell line MCF-7 and MCF-7-derived CSCs (mammospheres derived from MCF-7 cells) (Abboud et al., 2019; Hertz et al., 2015; Morrison et al., 2012; Nie et al., 2015). Comparing BCSCs and MCF-7 cells, the proteomic analysis identified only 31 of the total 617 proteins for BCSCs. Among them, the 4 strongest candidates for use as biomarkers consisted of several proteins associated with the plasma membrane: CD66c, ATP5C1, GBP1, and PAK4. Plasma membrane proteins were first selected as they can be readily used for further validation and as tools for targeting BCSCs. CD66c, also known as CEACAM6, is an intercellular adhesion molecule that is overexpressed in a wide variety of human cancers and is associated with tumorigenesis, tumor cell adhesion, cancer cell invasion, and metastasis. ATP5C1, also known as ATP Synthase F1 Subunit Gamma, is a mitochondrial membrane ATP synthase (Pecina et al., 2018). GBP1 is a member of the large GTPase family and is induced by interferons and inflammatory cytokines (Britzen-Laurent et al., 2013; Elias et al., 2015; Qiu et al., 2018; Quintero et al., 2017). PAK4, also known as P21 (RAC1) Activated Kinase 4, is part of a conserved family of serine/threonine kinases, originally described as downstream effectors of small Rho GTPases, Rac, and Cdc42, and is crucial for cytoskeletal dynamics, survival, proliferation, metabolism, and invasion (Santiago-Gomez et al., 2019).
Among these, CD66c was selected as the BCSC surface marker since it was perfectly co-expressed with CD24–/CD44+, located across the outer cell membrane, and associated with various biological functions (Crabtree and Miele, 2018; Henderson et al., 2018; Jaggupilli and Elkord, 2012; Kim et al., 2016; Ricardo et al., 2011; Yan et al., 2015) (Fig. 7F). CD66c is part of the carcinoembryonic antigen (CEA) family and is a multifunctional glycoprotein that mediates homotypic binding with other CEA family members, as well as heterotypic binding with integrin receptors. CD66c functions by organizing tissue architecture and regulating signal transduction (Johnson and Mahadevan, 2015; Rizeq et al., 2018). It has been found to have biologically significant roles in cellular invasiveness, resistance to apoptosis, and the metastatic potential of tumor cells (Hong et al., 2015; Lin et al., 2015). CD66c over-expression has been previously reported in mucinous adenocarcinoma in various organs, including the colon, pancreas, breast, ovary, and lung, where it has been frequently correlated with a poor prognosis (Blumenthal et al., 2005; Johnson and Mahadevan, 2015; Lin et al., 2015). As such, the use of CD66c as a novel biomarker on the surface of BCSCs was validated at the transcriptomic and proteomic levels (Figs. 6 and 7). In relation to the expression pattern of the CD24–/CD44+ marker for CSCs, the expression of CD66c was found to increase in BCSCs cultured for 14 days, compared to MCF-7 cells.
High levels of CD66c expression have been associated with a variety of malignancies, including breast cancer (Johnson and Mahadevan, 2015). CD66c has been found to play roles associated with: (i) decreased chemosensitivity; (ii) increased cell adhesion; (iii) improved cell invasion; (iv) metastasis induction; (v) the inhibition of anoikis (Balk-Moller et al., 2014; Blumenthal et al., 2005; 2007; Hong et al., 2015; Johnson and Mahadevan, 2015; Lin et al., 2015; Pańczyszyn and Wieczorek, 2012; Rizeq et al., 2018). Unlike attached cells, one of the many characteristics of CSC culturing is that the cells die early via cell killing during the formation of the spheres. Mitogen-responsive anoikis resistant cells (putative CSCs) proliferate and form new spheres (Cao et al., 2016). Thus, when forming the spheres, the expression of this protein seems to increase, as shown in the results of Fig. 7. The increased expression of CD66c could be the result of the spheres being enriched for anoikis-resistant CSCs (Lee et al., 2018).
CD66c has several functions, including roles in cell-cell binding, anti-apoptosis/anoikis activity, and tumor invasion, and metastasis. Among its functions, CD66c acts as a factor for the regulation of anti-apoptosis/anoikis related to the death of BCSCs (Gemei et al., 2013; Hong et al., 2015; Johnson and Mahadevan, 2015; Lee et al., 2018; Lin et al., 2015). To prove this, the CD66c gene was knocked down in BCSCs using shRNA to suppress the effects of the CD66c expression. As a result, CD66c knockdown was found to affect the growth of BCSCs. The levels of crystal violet staining (for the examination of cell adhesion ability) and the MTT assay results (for cell survival) were found to be reduced in cells transfected with shRNA-C against CD66c, in agreement with the transcriptomic and proteomic results (Fig. 8).
The decreased in cell viability may have been caused by the inhibition of CD66c for anti-apoptosis/anoikis. To mimic the oxidative stress conditions of CSCs in found in the in vivo environment (Zhong et al., 2019), BCSCs were treated with H2O2. Many studies have shown that H2O2-mediated oxidative stress can induce premature senescence in various types of cells. However, it is largely unknown how BCSCs respond to H2O2-mediated oxidative stress in the absence of CD66c protein. Our results showed that H2O2-mediated oxidative stress decreased the number of BCSCs. When CD66c was not expressed (as a result of shRNA treatment), the killing effects of oxidative stress were enhanced in cells treated with H2O2 (Fig. 9B).
One of the major cell death mechanisms is apoptosis. Double-staining FACS using annexin-V and PI was carried out to investigate the mechanisms of increased cell death after shRNA and H2O2 treatment. The knockdown of CD66c after treatment with shRNA-C was found to induce increased levels of apoptosis and necrosis, resulting an enhanced cell death of BCSCs (Figs. 9C and 9D). These results demonstrate that CD66c expression in BCSCs is related to enhanced apoptosis in response to oxidative stress (Zhong et al., 2019). Taken together, our results indicate that CD66c is a novel BCSC surface biomarker that could be used for the targeted therapeutic treatment in clinical research studies.
In conclusion, we attempted to develop a novel biomarker on the surface of BCSCs via proteomic analysis. Identified as a common molecule using proteomic data between MCF-7 breast cancer cells and BCSCs, CD66c was investigated as a potential biomarker for BCSCs. CD66c was validated in BCSCs at the transcriptomic and proteomic level. Compared to MCF-7 cells, CD66c showed a high level of expression on the 14th day in BCSCs, with distinctive features of a CD24–/CD44+ (CSC biomarker). The biological function of CD66c in BCSCs was assessed by shRNA treatment. The down-regulation of the CD66c gene by shRNA induced a decrease in cell proliferation. After the treatment of BCSCs with shRNA under oxidative stress conditions, an increased cytotoxicity was observed. In addition, increased levels of cell death were found as a result of enhanced apoptosis.
This study is the first to evaluate the biological function of CD66c as a novel biomarker on the surfaces of breast cancer-derived CSCs. Taken together, our results indicate that CD66c expressed on the surface of BCSCs may be used as a novel biomarker and a potential molecular target for the treatment of breast cancer.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This research was supported by grants from the National Research Foundation (NRF) funded by the Ministry of Education (2016R1D1A1B03935498).
Footnotes
AUTHOR CONTRIBUTIONS
P.H.K. designed experiments, analyzed data, and helped in manuscript preparation and writing. E.Y.K., J.E.Y., and S.H.J. performed experiments. E.Y.K. wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Abboud M.M., Al Awaida W., Alkhateeb H.H., Abu-Ayyad A.N. Antitumor action of amygdalin on human breast cancer cells by selective sensitization to oxidative stress. Nutr. Cancer. 2019;71:483–490. doi: 10.1080/01635581.2018.1508731. [DOI] [PubMed] [Google Scholar]
- Ahn S.M., Goode R.J., Simpson R.J. Stem cell markers: insights from membrane proteomics? Proteomics 8, 4946-4957. 2008 doi: 10.1002/pmic.200800312. [DOI] [PubMed] [Google Scholar]
- Bailey P.C., Lee R.M., Vitolo M.I., Pratt S.J.P., Ory E., Chakrabarti K., Lee C.J., Thompson K.N., Martin S.S. Single-cell tracking of breast cancer cells enables prediction of sphere formation from early cell divisions. iScience. 2018;8:29–39. doi: 10.1016/j.isci.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk-Moller E., Kim J., Hopkinson B., Timmermans-Wielenga V., Petersen O.W., Villadsen R. A marker of endocrine receptor-positive cells, CEACAM6, is shared by two major classes of breast cancer: luminal and HER2-enriched. Am. J. Pathol. 2014;184:1198–1208. doi: 10.1016/j.ajpath.2013.12.013. [DOI] [PubMed] [Google Scholar]
- Baumann M., Krause M., Hill R. Exploring the role of cancer stem cells in radioresistance. Nat. Rev. Cancer. 2008;8:545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- Blumenthal R.D., Hansen H.J., Goldenberg D.M. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen) Cancer Res. 2005;65:8809–8817. doi: 10.1158/0008-5472.CAN-05-0420. [DOI] [PubMed] [Google Scholar]
- Blumenthal R.D., Leon E., Hansen H.J., Goldenberg D.M. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britzen-Laurent N., Lipnik K., Ocker M., Naschberger E., Schellerer V.S., Croner R.S., Vieth M., Waldner M., Steinberg P., Hohenadl C., et al. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis. 2013;34:153–162. doi: 10.1093/carcin/bgs310. [DOI] [PubMed] [Google Scholar]
- Calvet C.Y., Andre F.M., Mir L.M. The culture of cancer cell lines as tumorspheres does not systematically result in cancer stem cell enrichment. PLoS One. 2014;9:e89644. doi: 10.1371/journal.pone.0089644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S., de Long L.M., Hazar-Rethinam M., Topkas E., Endo-Munoz L., Cumming A., Gannon O., Guminski A., Saunders N. Focal overexpression of CEACAM6 contributes to enhanced tumorigenesis in head and neck cancer via suppression of apoptosis. Mol. Cancer. 2012;11:74. doi: 10.1186/1476-4598-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Livas T., Kyprianou N. Anoikis and EMT: lethal "liaisons" during cancer progression. Crit. Rev. Oncog. 2016;21:155–168. doi: 10.1615/CritRevOncog.2016016955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.S., Wang A.X., Dong B., Pu K.F., Yuan L.H., Zhu Y.M. A new prospect in cancer therapy: targeting cancer stem cells to eradicate cancer. Chin. J. Cancer. 2012;31:564–572. doi: 10.5732/cjc.011.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak S., Medema J.P. Cancer stem cells--important players in tumor therapy resistance. FEBS J. 2014;281:4779–4791. doi: 10.1111/febs.13023. [DOI] [PubMed] [Google Scholar]
- Crabtree J.S., Miele L. Breast cancer stem cells. Biomedicines. 2018;6:77. doi: 10.3390/biomedicines6030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deonarain M.P., Kousparou C.A., Epenetos A.A. Antibodies targeting cancer stem cells: a new paradigm in immunotherapy? MAbs 1, 12-25. 2009 doi: 10.4161/mabs.1.1.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Fernandez A., Miranda-Castro R., de-Los-Santos-Alvarez N., Lobo-Castanon M.J. Post-translational modifications in tumor biomarkers: the next challenge for aptamers? Anal. Bioanal. Chem. 2018;410:2059–2065. doi: 10.1007/s00216-018-0861-9. [DOI] [PubMed] [Google Scholar]
- Dontu G., Abdallah W.M., Foley J.M., Jackson K.W., Clarke M.F., Kawamura M.J., Wicha M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias D., Vever H., Laenkholm A.V., Gjerstorff M.F., Yde C.W., Lykkesfeldt A.E., Ditzel H.J. Gene expression profiling identifies FYN as an important molecule in tamoxifen resistance and a predictor of early recurrence in patients treated with endocrine therapy. Oncogene. 2015;34:1919–1927. doi: 10.1038/onc.2014.138. [DOI] [PubMed] [Google Scholar]
- Gemei M., Mirabelli P., Di Noto R., Corbo C., Iaccarino A., Zamboli A., Troncone G., Galizia G., Lieto E., Del Vecchio L., et al. CD66c is a novel marker for colorectal cancer stem cell isolation, and its silencing halts tumor growth in vivo. Cancer. 2013;119:729–738. doi: 10.1002/cncr.27794. [DOI] [PubMed] [Google Scholar]
- Hamam R., Hamam D., Alsaleh K.A., Kassem M., Zaher W., Alfayez M., Aldahmash A., Alajez N.M. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson T., Chen M., Darrow M.A., Li C.S., Chiu C.L., Monjazeb A.M., Murphy W.J., Canter R.J. Alterations in cancer stem-cell marker CD44 expression predict oncologic outcome in soft-tissue sarcomas. J. Surg. Res. 2018;223:207–214. doi: 10.1016/j.jss.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz E., Cadona F.C., Machado A.K., Azzolin V., Holmrich S., Assmann C., Ledur P., Ribeiro E.E., DE Souza Filho O.C., Manica-Cattani M.F., et al. Effect of Paullinia cupana on MCF-7 breast cancer cell response to chemotherapeutic drugs. Mol. Clin. Oncol. 2015;3:37–43. doi: 10.3892/mco.2014.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeth G., Bendahl P.O., Ringner M., Saal L.H., Gruvberger-Saal S.K., Lovgren K., Grabau D., Ferno M., Borg A., Hegardt C. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K.P., Shin M.H., Yoon S., Ji G.Y., Moon Y.R., Lee O.J., Choi S.Y., Lee Y.M., Koo J.H., Lee H.C., et al. Therapeutic effect of anti CEACAM6 monoclonal antibody against lung adenocarcinoma by enhancing anoikis sensitivity. Biomaterials. 2015;67:32–41. doi: 10.1016/j.biomaterials.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Huang R., Rofstad E.K. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget. 2017;8:35351–35367. doi: 10.18632/oncotarget.10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggupilli A., Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin. Dev. Immunol. 2012;2012:708036. doi: 10.1155/2012/708036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B., Mahadevan D. Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin. Cancer Drugs. 2015;2:100–111. doi: 10.2174/2212697X02666150602215823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.J. Chemical proteomic approaches targeting cancer stem cells: a review of current literature. Cancer Genomics Proteomics. 2017;14:315–327. doi: 10.21873/cgp.20042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Kim M.H., Kim K.S., Park M.J., Jeong J.H., Park S.W., Ji Y.H., Kim K.I., Lee T.S., Ryu P.Y., et al. In vivo monitoring of CD44+ cancer stem-like cells by gamma-irradiation in breast cancer. Int. J. Oncol. 2016;48:2277–2286. doi: 10.3892/ijo.2016.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.T., Ryu C.J. Cancer stem cell surface markers on normal stem cells. BMB Reports. 2017;50:285. doi: 10.5483/BMBRep.2017.50.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Park H.B., Kim P.H., Park J.S., Kim K.S. Enhanced anti-cancer efficacy in MCF-7 breast cancer cells by combined drugs of metformin and sodium salicylate. Biomed. Sci. Lett. 2017;23:290–294. doi: 10.15616/BSL.2017.23.3.290. [DOI] [Google Scholar]
- Koch U., Krause M., Baumann M. Cancer stem cells at the crossroads of current cancer therapy failures--radiation oncology perspective. Semin. Cancer Biol. 2010;20:116–124. doi: 10.1016/j.semcancer.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat. Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.C., Fitzgerald M., Bannerman B., Donelan J., Bano K., Terkelsen J., Bradley D.P., Subakan O., Silva M.D., Liu R., et al. Antitumor activity of the investigational proteasome inhibitor MLN9708 in mouse models of B-cell and plasma cell malignancies. Clin. Cancer Res. 2011;17:7313–7323. doi: 10.1158/1078-0432.CCR-11-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Jang Y., Park S., Jang H., Park E.J., Kim H.J., Kim H. Development and evaluation of a CEACAM6-targeting theranostic nanomedicine for photoacoustic-based diagnosis and chemotherapy of metastatic cancer. Theranostics. 2018;8:4247–4261. doi: 10.7150/thno.25131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Kim D.G., Kim B.G., Yang W.S., Hong J., Kang T., Oh Y.S., Kim K.R., Han B.W., Hwang B.J., et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci. 2014;127:4234–4245. doi: 10.1242/jcs.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leth-Larsen R., Lund R., Hansen H.V., Laenkholm A.V., Tarin D., Jensen O.N., Ditzel H.J. Metastasis-related plasma membrane proteins of human breast cancer cells identified by comparative quantitative mass spectrometry. Mol. Cell. Proteomics. 2009;8:1436–1449. doi: 10.1074/mcp.M800061-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.E., Barrette A.M., Chapin C., Gonzales L.W., Gonzalez R.F., Dobbs L.G., Ballard P.L. Expression of human carcinoembryonic antigen-related cell adhesion molecule 6 and alveolar progenitor cells in normal and injured lungs of transgenic mice. Physiol. Rep. 2015;3:e12657. doi: 10.14814/phy2.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo Y., de Giorgio A., Coombes C.R., Stebbing J., Castellano L. Mammosphere formation assay from human breast cancer tissues and cell lines. J. Vis. Exp. 2015;97:e52671. doi: 10.3791/52671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Clouthier S.G., Deol Y., Liu S., Nagrath S., Azizi E., Wicha M.S. Breast cancer stem cells: current advances and clinical implications. Methods Mol. Biol. 2015;1293:1–49. doi: 10.1007/978-1-4939-2519-3_1. [DOI] [PubMed] [Google Scholar]
- Maugeri-Sacca M., Vigneri P., De Maria R. Cancer stem cells and chemosensitivity. Clin. Cancer Res. 2011;17:4942–4947. doi: 10.1158/1078-0432.CCR-10-2538. [DOI] [PubMed] [Google Scholar]
- Meacham C.E., Morrison S.J. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison B.J., Hastie M.L., Grewal Y.S., Bruce Z.C., Schmidt C., Reynolds B.A., Gorman J.J., Lopez J.A. Proteomic comparison of mcf-7 tumoursphere and monolayer cultures. PLoS One. 2012;7:e52692. doi: 10.1371/journal.pone.0052692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar D., Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- Nie S., McDermott S.P., Deol Y., Tan Z., Wicha M.S., Lubman D.M. A quantitative proteomics analysis of MCF7 breast cancer stem and progenitor cell populations. Proteomics. 2015;15:3772–3783. doi: 10.1002/pmic.201500002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilendu P., Kumar A., Kumar A., Pal J.K., Sharma N.K. Breast cancer stem cells as last soldiers eluding therapeutic burn: a hard nut to crack. Int. J. Cancer. 2018;142:7–17. doi: 10.1002/ijc.30898. [DOI] [PubMed] [Google Scholar]
- Pańczyszyn A., Wieczorek M. [Role of CEACAM in neutrophil activation] Postepy Hig. Med. Dosw. (Online) 2012;66:574–582. doi: 10.5604/17322693.1008194. Polish. [DOI] [PubMed] [Google Scholar]
- Pecina P., Nuskova H., Karbanova V., Kaplanova V., Mracek T., Houstek J. Role of the mitochondrial ATP synthase central stalk subunits gamma and delta in the activity and assembly of the mammalian enzyme. Biochim. Biophys. Acta Bioenerg. 2018;1859:374–381. doi: 10.1016/j.bbabio.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Pogozheva I.D., Tristram-Nagle S., Mosberg H.I., Lomize A.L. Structural adaptations of proteins to different biological membranes. Biochim. Biophys. Acta. 2013;1828:2592–2608. doi: 10.1016/j.bbamem.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Guo H., Yang J., Ji Y., Wu C.S., Chen X. Down-regulation of guanylate binding protein 1 causes mitochondrial dysfunction and cellular senescence in macrophages. Sci. Rep. 2018;8:1679. doi: 10.1038/s41598-018-19828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero M., Adamoski D., Reis L.M.D., Ascencao C.F.R., Oliveira K.R.S., Goncalves K.A., Dias M.M., Carazzolle M.F., Dias S.M.G. Guanylate-binding protein-1 is a potential new therapeutic target for triple-negative breast cancer. BMC Cancer. 2017;17:727. doi: 10.1186/s12885-017-3726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo S., Vieira A.F., Gerhard R., Leitao D., Pinto R., Cameselle-Teijeiro J.F., Milanezi F., Schmitt F., Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- Rizeq B., Zakaria Z., Ouhtit A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci. 2018;109:33–42. doi: 10.1111/cas.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodini C.O., Lopes N.M., Lara V.S., Mackenzie I.C. Oral cancer stem cells - properties and consequences. J. Appl. Oral Sci. 2017;25:708–715. doi: 10.1590/1678-7757-2016-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Gomez A., Kedward T., Simoes B.M., Dragoni I., NicAmhlaoibh R., Trivier E., Sabin V., Gee J.M., Sims A.H., Howell S.J., et al. PAK4 regulates stemness and progression in endocrine resistant ER-positive metastatic breast cancer. Cancer Lett. 2019;458:66–75. doi: 10.1016/j.canlet.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Scadden D.T. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Scheel C., Eaton E.N., Li S.H., Chaffer C.L., Reinhardt F., Kah K.J., Bell G., Guo W., Rubin J., Richardson A.L., et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Chen G., Liu X., Qiu Y., Yang S., Zhang Y., Fang X., Zhang C., Liu X. Scutellarein inhibits cancer cell metastasis in vitro and attenuates the development of fibrosarcoma in vivo. Int. J. Mol. Med. 2015;35:31–38. doi: 10.3892/ijmm.2014.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsov S., Debbage P., Skvortsova I. Proteomics of cancer stem cells. Int. J. Radiat. Biol. 2014;90:653–658. doi: 10.3109/09553002.2013.873559. [DOI] [PubMed] [Google Scholar]
- Song M., Giovannucci E.L. Cancer risk: many factors contribute. Science. 2015;347:728–729. doi: 10.1126/science.aaa6094. [DOI] [PubMed] [Google Scholar]
- Vinogradov S., Wei X. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine (Lond.) 2012;7:597–615. doi: 10.2217/nnm.12.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Lv Q., Meng W., Tan Q., Zhang S., Mo X., Yang X. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J. Thorac. Dis. 2014;6:829–837. doi: 10.3978/j.issn.2072-1439.2014.03.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Zuo X., Wei D. Concise review: emerging role of CD44 in cancer stem cells: a promising biomarker and therapeutic target. Stem Cells Transl. Med. 2015;4:1033–1043. doi: 10.5966/sctm.2015-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Xu J., Tang L., Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell. Mol. Life Sci. 2017;74:951–966. doi: 10.1007/s00018-016-2334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G., Qin S., Townsend D., Schulte B.A., Tew K.D., Wang G.Y. Oxidative stress induces senescence in breast cancer stem cells. Biochem. Biophys. Res. Commun. 2019;514:1204–1209. doi: 10.1016/j.bbrc.2019.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Fan Z. Cancer stem cells and tumorigenesis. Biophys. Rep. 2018;4:178–188. doi: 10.1007/s41048-018-0062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.