Abstract
Eukaryotes transport biomolecules between intracellular organelles and between cells and the environment via vesicle trafficking. Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE proteins) play pivotal roles in vesicle and membrane trafficking. These proteins are categorized as Qa, Qb, Qc, and R SNAREs and form a complex that induces vesicle fusion for targeting of vesicle cargos. As the core components of the SNARE complex, the SNAP25 Qbc SNAREs perform various functions related to cellular homeostasis. The Arabidopsis thaliana SNAP25 homolog AtSNAP33 interacts with Qa and R SNAREs and plays a key role in cytokinesis and in triggering innate immune responses. However, other Arabidopsis SNAP25 homologs, such as AtSNAP29 and AtSNAP30, are not well studied; this includes their localization, interactions, structures, and functions. Here, we discuss three biological functions of plant SNAP25 orthologs in the context of AtSNAP33 and highlight recent findings on SNAP25 orthologs in various plants. We propose future directions for determining the roles of the less well-characterized AtSNAP29 and AtSNAP30 proteins.
Keywords: abiotic stress responses, cytokinesis, innate immune response, Qbc SNARE, SNAP25
INTRODUCTION
Vesicle trafficking is a fundamental mechanism for maintaining cellular homeostasis in eukaryotes. Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins play an essential role in vesicle trafficking by participating in vesicle fusion at the membrane. Distinct combinations of SNARE proteins allow for many different types of cargo to travel to specific subcellular locations. SNARE proteins are classified as Q SNAREs or R SNAREs depending on the contributing residue (glutamine or arginine, respectively) in the structurally assembled core SNARE complex. Q SNAREs are further categorized as Qa, Qb, and Qc, depending on their location in the four-helix bundle (Bock et al., 2001), which forms the core SNARE complex. The first identified SNARE complex, which is involved in synaptic vesicle exocytosis, forms a parallel four-stranded coiled-coil structure composed of a helix of syntaxin 1A, two helical domains of SNAP25, and a helix of vesicle-associated membrane protein 2 (VAMP2) (Hayashi et al., 1994; Niemann et al., 1994).
The first cDNA encoding SNAP25 was identified in mouse brain (Oyler et al., 1989). Five years later, human SNAP25 was cloned and sequenced (Zhao et al., 1994), and other SNAP25 homologs, SNAP23, SNAP29, and SNAP47, were subsequently identified (Ravichandran et al., 1996; Steegmaier et al., 1998; Holt et al., 2006). SNAP25 functions in synaptic vesicle trafficking by interacting with syntaxin1A and VAMP2 (Hayashi et al., 1994; Niemann et al., 1994). Moreover, SNAP25 functions in cell division as a subunit of the trafficking machinery along with syntaxin 2 and VAMP8, a component that facilitates abscission of the midbody (Gromley et al., 2005). Enrichment of SNAP25 at the midbody was also observed in a study on zebrafish embryos (Li et al., 2006). These reports indicated that SNAP25 proteins are essential for the final stages of cytokinesis during cell division. SNAP23 proteins play a pivotal role in immune cells by mediating the secretion of cytokines, antibodies, and granules (Pagan et al., 2003; Reales et al., 2005; Mollinedo et al., 2006). SNAP29 interacts with syntaxin17 (STX17) and VAMP8 to form a SNARE complex, which mediates the fusion of autophagosomes and lysosomes (Itakura et al., 2012; Takats et al., 2013). The characterized animal SNAP25 homologs are essential for overall cellular homeostasis.
The Arabidopsis thaliana genome encodes 65 putative SNARE proteins, considerably more than in other eukaryotes (35 in humans, 21 in Saccharomyces cerevisiae, and 20 in Drosophila) (Saito and Ueda, 2009). Similar to Arabidopsis, plants encode multiple SNARE proteins (70 in Oryza sativa, 129 in Brassica rapa, and 151 in Glycine max [Phytozome, Plant Genome Resource]). Arabidopsis SNARE proteins localize in distinct subcellular organelles and are involved in various cellular functions, including development, gravitropism, pathogen responses, and abiotic stress responses (Yano et al., 2003; Heese et al., 2001; Kwon et al., 2008; Pajonk et al., 2008; Zhu et al., 2002). Several Arabidopsis SNARE proteins show functional redundancy but form distinct complexes depending on the cellular environment. For example, VACULOLAR PROTEIN SORTING 10-INTERACTING 11 (VTI11) and VTI12 are involved in trafficking of the marker VAC2 (composed of CLAVATA3 fused to a vacuolar sorting signal from a barley [Hordeum vulgare] lectin); however, VTI11 and VTI12 mediate trafficking to lytic and storage vacuoles, respectively, through their SNARE partners (Surpin et al., 2003; Sanmartin et al., 2007). Therefore, determining the interactions and locations of SNARE proteins is crucial to identifying their functional mechanisms.
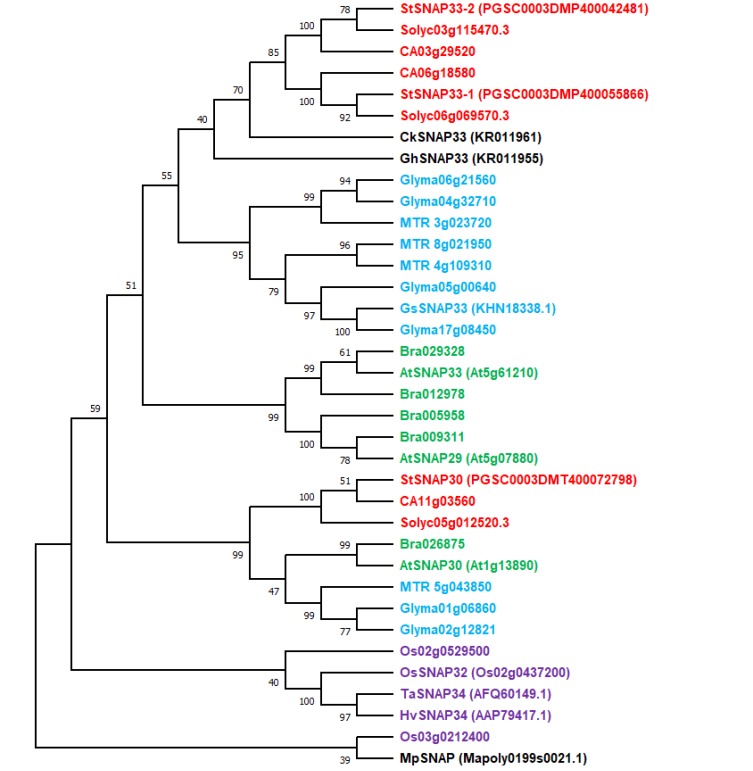
Here, to examine plant SNAP25 proteins, we first generated a phylogenetic tree of plant SNAP25 proteins based on the Qbc SNARE motifs in A. thaliana, B. rapa, Capsicum annuum, Solanum lycopersicum, Solanum tuberosum, G. max, Medicago truncatula, O. sativa, and Marchantia polymorpha. These proteins were analyzed with BLAST using the fulllength amino acid sequence of AtSNAP29/30/33 as a query. SNAP25 proteins from Glycine soja, H. vulgare, Triticum aestivum, Gossypium hirsutum, and Cynanchum komarovii were only listed when functional reports were available in the literature (Fig. 1, Table 1). Using this framework, we discuss recent findings regarding SNAP25 homologs in various plants and their molecular roles in protein–protein or environmental interactions. We also propose future directions for studies on less well-characterized SNAP25 homologs in plants.
Fig. 1. Phylogenetic tree of SNAP25 proteins in plants.
Phylogenetic analysis was performed using MEGAX software (Kumar et al.,2018) with full amino acid sequences of the retrieved SNAP25 proteins from public databases; UniProt, GenBank, Sol Genomics, and Phytozome, and additionally only functionally studied SNAP25 proteins from G. soja, H. vulgare, T. aestivum, G. hirsutum, and C. komarovii. The bootstrap values from 1000 replicates are depicted at the nodes. Different colors indicate the following: green, Brassicaceae; red, Solanaceae; blue, Fabaceae; violet, Poaceae; black, other functionally reported SNAP25 proteins. All information in the tree is described in Table 1
Table 1.
Plant SNAP25 proteins used to construct the phylogenetic tree
| Species | Name (in tree) | Accession No. | Gene ID | ID source |
|---|---|---|---|---|
| Arabidopsis thaliana | AtSNAP29 | Q9SD96 | At5g07880 | UniProt |
| AtSNAP30 | Q9LMG8 | At1g13890 | ||
| AtSNAP33 | Q9S7P9 | At5g61210 | ||
| Brassica rapa | Bra012978 | M4D918 | Bra012978 | UniProt |
| Bra029328 | M4EKL1 | Bra029328 | ||
| Bra005958 | M4CP20 | Bra005958 | ||
| Bra009311 | M4CYL4 | Bra009311 | ||
| Bra026875 | M4EDL5 | Bra026875 | ||
| Glycine max | Glyma04g32710 | C6T803 | Glyma04g32710 | UniProt |
| Glyma06g21560 | I1KCY1 | Glyma06g21560 | ||
| Glyma05g00640 | I1JZK8 | Glyma05g00640 | ||
| Glyma17g08450 | C6TJG5 | Glyma17g08450 | ||
| Glyma02g12821 | K7K7S4 | Glyma02g12821 | ||
| Glyma01g06860 | I1J5Y3 | Glyma01g06860 | ||
| Glycine soja | GsSNAP33 | KHN18338.1 | KHN18338 | GenBank |
| Medicargo trancatula | MTR_3g023720 | KEH33106 | MTR_3g023720 | GenBank |
| MTR_4g109310 | KEH31992 | MTR_4g109310 | ||
| MTR_5g043850 | AES96982 | MTR_5g043850 | ||
| MTR_8g021950 | AET01821 | MTR_8g021950 | ||
| Oriza sativa | OsSNAP32 | Q5EEP3 | AAW82752 | UniProt |
| OsSNAP29 | Q10Q25 | Os03g0212400 | ||
| Hordeum vulgare | HvSNAP34 | AAP79417.1 | AAP79417 | GenBank |
| Triticum aestivum | TaSNAP34 | AFQ60149.1 | AFQ60149 | GenBank |
| Zea mays | Zm00001d019505_P001 | A0A1D6HXY8 | Zm00001d019505_P001 | UniProt |
| Zm00001d016686_P002 | A0A1D6H9U2 | Zm00001d016686_P002 | ||
| Capsicum annuum | CA03g29520 | CA03g29520 | CA03g29520 | Sol Genomics |
| CA06g18580 | CA06g18580 | CA06g18580 | ||
| CA11g03560 | CA11g03560 | CA11g03560 | ||
| Solanum lycopersicum | Solyc06g069570.3 | Solyc06g069570.3 | Solyc06g069570.3 | Sol Genomics |
| Solyc03g115470.3 | Solyc03g115470.3 | Solyc03g115470.3 | ||
| Solyc05g012520.3 | Solyc05g012520.3 | Solyc05g012520.3 | ||
| Solanum tuberosum | PGSC0003DMP400055866 | PGSC0003DMP400055866 | PGSC0003DMP400055866 (StSNAP33-1) | Sol Genomics |
| PGSC0003DMP400042481 | PGSC0003DMP400042481 | PGSC0003DMP400042481 (StSNAP33-2) | ||
| PGSC0003DMP400049245 | PGSC0003DMP400049245 | PGSC0003DMP400049245 | ||
| Gossypium hirsutum | GhSNAP33 | ALD83640.1 | KR011955 | GenBank |
| Cynanchum komarovii | CkSNAP33 | ALH22085.1 | KR011961 | GenBank |
| Marchantia polymorpha | MpSNAP | Mapoly0199s0021.1 | Mapoly0199s0021.1 | Marchantia |
All information regarding SNAP25 protein species, name, accession No., and gene ID was retrieved from publicly available databases; UniProt, GenBank (MpSNAP sequence obtained from Marchandia), Sol Genomics, and Phytozyome. SNAP25 proteins in A. thaliana, B. rapa, C. annuum, S. lycopersicum, S. tuberosum, G. max, M. truncatula, O. sativa, and M. polymorpha were found by BLAST using AtSNAP29/30/33 as a query. The other SNAP25 proteins from G. soja, H. vulgare, T. aestivum, G. hirsutum, and C. komarovii were only listed when a functional report was available in the literature.
STRUCTURE OF THE HUMAN SNAP25 PROTEIN
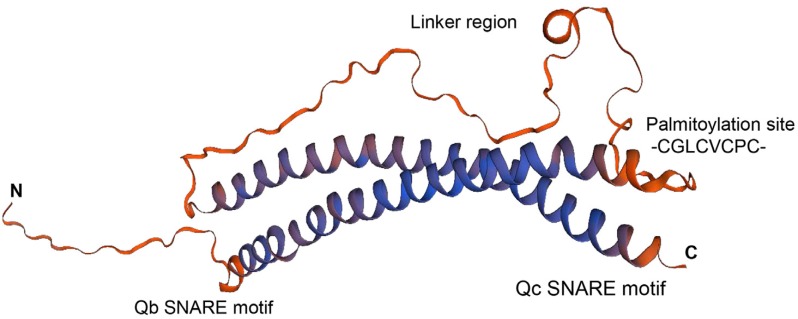
In general, SNARE proteins have a transmembrane domain and an α-helical coiled-coil domain termed the “SNARE domain”, which forms part of the SNARE complex (Weimbs et al., 1997). The coiled-coil domains in SNARE proteins twist together to induce the fusion of vesicles and membranes. Unlike general SNARE proteins, SNAP25 proteins consist of two SNARE domains, namely Qb and Qc, and a linker (Weimbs et al., 1997). The N-terminal Qb SNARE domain is connected via a linker region to the C-terminal Qc SNARE domain of the SNAP25 protein (Fig. 2).
Fig. 2. Structure of SNAP25 in humans.
SNAP25 proteins comprise a coiled-coil of the Qb domain at the N-terminus, a linker region, and a coiled-coil of the Qc domain at the C-terminus. Images were prepared by SWISS-MODEL, an automated protein structure homology-modeling server, using SNAP25 protein from humans (UniProtKB - P60880) (Waterhouse et al., 2018).
SNAP25 homologs have no transmembrane domains; therefore, it is unclear how SNAP25 homologs localize to the cellular membranes. One possibility is that lipid modifications allow the SNAP25 proteins to associate with membranes. For example, PtSNAP25 from Paramecium tetraurelia contains a myristoylation site for membrane attachment (Schilde et al., 2008). Moreover, mammalian membrane-targeted SNAP25 is palmitoylated at a cysteine residue in the linker region (Gonzalo and Linder, 1998; Gonzalo et al., 1999). A snap25 mutant lacking the cysteine site showed a slower rate of synaptic vesicle fusion in mouse cells than the wild-type protein (Nagy et al., 2008). Recovery of the cysteine residue in the linker region of SNAP23 complemented the snap25 mutant with respect to the rate of fusion in synaptic vesicles (Nagy et al., 2008). However, the mammalian SNAP25 homologs SNAP29 and SNAP47 localize to subcellular membranes even without the cysteine residue; thus, these homologs cannot complement the function of SNAP25 in snap25 mutants (Arora et al., 2017).
Similar to SNAP29 and SNAP47, plant SNAP25 homologs have a linker region lacking the cysteine residue. This indicates that plant SNAP25 homologs use another mechanism, possibly fatty acid modification, for cell membrane attachment. However, to our knowledge, no structural studies on plant SNAP25 homologs have been reported and further studies are needed to determine how plant SNAP25 localizes to the cellular membrane, and how it forms complex structures with other SNARE proteins.
MOLECULAR FUNCTIONS OF SNAP25 FAMILY PROTEINS IN PLANTS
Cytokinesis
The mechanism of cytokinesis in plants differs from that in animals. An animal cell is divided by cytoplasmic abscission through the formation of a cleavage furrow (Cao and Wang, 1990; Mierzwa and Gerlich, 2014), whereas a plant cell is divided by the formation of a cell plate through vesicle fusion and concomitant formation of vesicular-tubular structures (Ahn et al., 2017). Plants contain a Qa SNARE, a cytokinesis-specific syntaxin termed SYP111 (SYNTAXIN OF PLANTS 111)/KNOLLE (meaning tuber-shaped in German), which localizes to the Golgi stacks and plasma membranes, and is involved in the fusion of vesicles from the Golgi to the center of the dividing cell (Lauber et al., 1997; Volker et al., 2001). The delivered vesicles and de novo-synthesized tubular structures fuse and expand toward the parental plasma membrane to form the cell plate. The Arabidopsis SNAP25 homolog AtSNAP33 is the principal interacting partner of KNOLLE/ SYP111 together with AtVAMP721 (Heese et al., 2001). Arabidopsis snap33 mutants show severe necrotic cotyledons and a seedling-lethal phenotype (Heese et al., 2001). A genetic study on snap33 npsn11 (NPSN11, Novel Plant SNARE11) and snap33 syp71 double mutants, along with mutants of other cytokinesis-specific SNARE proteins, revealed a major cytokinetic defect with dividing cells showing abnormal morphologies (El Kasmi et al., 2013). Seminal studies showed that AtSNAP33 plays a critical role in cytokinesis. Heterologously expressed CkSNAP33 from C. komarovii in Arabidopsis transgenic plants promotes growth thereby increasing root length and leaf area (Wang et al., 2017). Direct evidence of increased cell numbers of the root and leaf in CkSNAP33-expressing lines suggests that heterologous expression of CkSNAP33 is associated with cytokinesis.
Other SNAP25 homologs, such as the Qbc SNAREs AtSNAP29 and AtSNAP30, also interact with KNOLLE/ SYP111 in vitro (Heese et al., 2001). AtSNAP33 is expressed throughout the plant, whereas AtSNAP29 and AtSNAP30 are rather highly expressed in the roots and flowers, respectively (Lipka et al., 2007). However, the functions of AtSNAP29 and AtSNAP30 have not been characterized. AtSYP132 is a Qa SNARE, which interacts with VAMP721/722/724 to form a SNARE complex in growing root hairs, and is involved in cytokinesis in flowering plants (Ichikawa et al., 2014; Park et al., 2018). AtSNAP29 and AtSNAP30, may be involved in the development of specific tissues by interacting with other AtSYPs.
Members of Fabaceae, such as G. max, M. truncatula, and Lotus japonicus, contain SYP132 orthologs and various VAMP72 family members, which are involved in symbiosis as well as in delivering cargos to the cell plate in dividing cells (Gavrin et al., 2016; Ivanov et al., 2012; Sogawa et al., 2018). G. max has six putative SNAP25 homologs, which are likely components of the SNARE complex and are involved in symbiosis and cytokinesis (Sharma et al., 2016). Analysis of the G. max genome suggests that six members of the GlymaSNAP25 protein family have evolved to have novel functions in symbiosis with environmental microbes and cellular trafficking, in addition to their roles in cytokinesis.
Innate immune responses
Unlike animals, plants do not have an adaptive immune system, but depend on their innate immunity against various pathogens. Secreted defense molecules are crucial for defense against non-host-adapted pathogens and plants primarily secrete compounds associated with defense, such as pathogenesis-related (PR) proteins and secondary metabolites, via SNARE complex-mediated vesicle trafficking. PENETRATION RESISTANCE1 (PEN1)/AtSYP121 is the main syntaxin involved in mediating resistance responses (Collins et al., 2003). Additionally, the PEN2/PEN3 pathway transports phytoalexins to the outside of the cell via a membrane transporter (Lipka et al., 2005; Stein et al., 2006). In response to pathogen infection, Arabidopsis increases the expression of AtSNAP33, which is regulated by the salicylic acid (SA) pathway (Wick et al., 2003). AtSNAP33 forms SNARE complexes with PEN1/AtSYP121 and VAMP721/722, which deliver undetermined defense cargos to the penetration sites of fungal pathogens (Kim et al., 2014; Kwon et al., 2008; Pajonk et al., 2008). Arabidopsis pen1 mutants and VAMP721/722-silenced mutants were vulnerable to infection by the plant pathogenic fungus Blumeria graminis (Kwon et al., 2008; Pajonk et al., 2008).
SNAP25 orthologs play a role in the innate immune response against fungal pathogens in Arabidopsis and in monocots, such as H. vulgare, O. sativa, and T. aestivum (Collins et al., 2003; Bao et al., 2008; Luo et al., 2016; Chandra et al., 2017). HvSNAP34 is required for the development of resistance against B. graminis penetration by forming ternary SNARE complexes with ROR2 (REQUIRED FOR mlo SPECIFIED RESISTANCE 2), a PEN1 ortholog, and HvVAMP721, an AtVAMP721 ortholog (Collins et al., 2003; Kwon et al., 2008). The expression of OsSNAP32, an O. sativa SNAP25 ortholog, increases upon infection with the blast fungus Magnaporthe oryzae. The O. sativa cultivar Suyunuo (a glutinous rice variety) lacks resistance to M. oryzae. Overexpression of OsSNAP32 in blast-susceptible Suyunuo plants increased resistance to the blast fungus (Luo et al., 2016). Reciprocally, RNA interference (RNAi) lines of the blast-resistant O. sativa landrace Heikezijing, with reduced expression of OsSNAP32 produced more lesions, as observed for the susceptible plants (Luo et al., 2016). T. aestivum SNAP25 homologs are also involved in the defense against leaf rust (Chandra et al., 2017). The expression levels of GhSNAP33 in G. hirsutum and CkSNAP33 in C. komarovii increased upon inoculation with the fungal pathogen Verticillium dahlia, which shows a broad host range (Wang et al., 2017; 2018). Ectopic expression of CkSNAP33 and GhSNAP33 in transgenic Arabidopsis plants resulted in increased resistance to V. dahlia compared to that observed in wild-type Arabidopsis (Wang et al., 2017; 2018). SNAP25 orthologs also participate in defense against bacterial and oomycete pathogens. For example, the pathogens Pseudomonas syringae pv. maculicola and Phytophthora infestans increase the expression of StSNAP33 in S. tuberosum (Eschen-Lippold et al., 2012). StSNAP33-silenced plants exhibited a chlorotic phenotype, enhanced SA concentrations, increased StPR1 gene expression, and enhanced callose deposition. However, StSNAP33-silenced plants did not show greater resistance when inoculated with P. infestans and the necrotrophic pathogen Botrytis cinerea compared to StSYP1-RNAi plants. In contrast, StSNAP33-silenced plants showed hypersensitive responses to Agrobacterium tumefaciens and Escherichia coli (Eschen-Lippold et al., 2012). These results indicate that StSNAP33 uses different modes of action for modulating innate immune responses based on the interacting microbes in the environment.
SNAP25 homologs in G. max may have specific roles in the defense against nematodes in roots. Overexpression of Glyma17g08450 (SNAP25-3) in G. max increases the resistance to nematodes, whereas RNA interference of Glyma17g08450 in plants allowed for greater nematode invasion (Sharma et al., 2016).
The AtSNAP33 orthologs in dicots as well as in monocots have conserved functions as a component of a ternary SNARE complex in innate immune responses against environmental pathogens. However, plants have other SNAP25 homologs, such as AtSNAP29 and AtSNAP30, which are differentially expressed throughout the plant and may have evolved to counter distinct pathogens or to trigger immune responses via different SNARE interactions to enable plant survival in varied environments.
Abiotic stress responses
Plants have evolved high-order trafficking mechanisms via numerous SNAREs to maintain cellular homeostasis against environmental stresses. Plant vacuoles and the trans-Golgi network function in plant responses to stressful environ ments. Various endosomal-specific SNARE proteins such as VTI11, SYP22, and SYP51 interact with multiple members in the VAMP71 family, which are involved in directing the vesi cle transport to the vacuole and plasma membrane (Ebine et al., 2008; Leshem et al., 2010). These SNARE proteins, such as Qa SNAREs and R SNAREs, are widely involved in endoso mal trafficking by carrying cargos containing storage proteins, reactive oxygen species (ROS), and unknown materials to respond to abiotic stresses (Ebine et al., 2008; Leshem et al., 2010). A study on the extreme halophyte Salicornia brachiate suggested that a novel salt-inducible gene SbSLSP (S. brachi ata SNARE-like superfamily protein), confers salt and drought tolerance by maintaining membrane stability, and reduces the accumulation of Na+ ion and ROS (Singh et al., 2016).
Plants have evolved high-order trafficking mechanisms via numerous SNAREs to maintain cellular homeostasis against environmental stresses. Plant vacuoles and the trans-Golgi network function in plant responses to stressful environ ments. Various endosomal-specific SNARE proteins such as VTI11, SYP22, and SYP51 interact with multiple members in the VAMP71 family, which are involved in directing the vesi cle transport to the vacuole and plasma membrane (Ebine et al., 2008; Leshem et al., 2010). These SNARE proteins, such as Qa SNAREs and R SNAREs, are widely involved in endoso mal trafficking by carrying cargos containing storage proteins, reactive oxygen species (ROS), and unknown materials to respond to abiotic stresses (Ebine et al., 2008; Leshem et al., 2010). A study on the extreme halophyte Salicornia brachiate suggested that a novel salt-inducible gene SbSLSP (S. brachi ata SNARE-like superfamily protein), confers salt and drought tolerance by maintaining membrane stability, and reduces the accumulation of Na+ ion and ROS (Singh et al., 2016).
How SNARE and SNAP25 proteins mechanistically func tion in abiotic stress responses remains unclear; however, several studies have suggested that they have important roles in mediating these responses. For example, AtSYP61 and AtSYP121 are involved in aquaporin distribution, and Arabidopsis SNAP25 proteins may interact with these SNARE proteins during abiotic stress responses (Hachez et al., 2014). AtVAMP71 suppression in Arabidopsis increased water loss and altered the control of stomatal opening/closing via inappropriate ROS localization under drought stress (Lesh em et al., 2010). Therefore, SNARE proteins in endosomal compartments may function in mediating resistance to abi otic stresses (Leshem et al., 2010). Exposure to mechanical stresses such as wind and wounding increased the expression of AtSNAP33 (Wick et al., 2003). Moreover, the expression of OsSNAP32 increased under drought and cold stress in O. sativa (Bao et al., 2008). Arabidopsis plants heterologously overexpressing GhSNAP33 or GsSNAP33 showed increased drought tolerance (Nisa et al., 2017; Wang et al., 2018). Moreover, the SNARE proteins interacting with SNAP25 ho mologs in plants such as AtVAMP721/722 and NtSYP121 are regulated by abscisic acid (ABA) for abiotic stress responses. AtVAMP721/722-deficient plants are sensitive to ABA, and the PEN1/AtSYP121 ortholog in Nicotiana tabacum, NtS YP121, is regulated by ABA (Kargul et al., 2001; Yi et al., 2013).
How SNAP25 homologs are maintained at appropriate levels in various organelles, such as the trans-Golgi network, vacuole, or plasma membrane, during abiotic stress respons es is unclear. Additional studies will provide insight into how plants adapt to varied environments with the involvement of their highly expanded families of SNARE proteins.
UNDETERMINED ASPECTS OF PLANT SNAP25
Interacting partners
Work on animal SNAP25s has identified interactions with proteins lacking SNARE motifs; however, whether plant SNAP25s interact with other non-SNARE proteins remains to be determined. For example, during mitosis, the C-terminal SNARE domain of SNAP29 directly recruits kinetochores and supports tissue development in Drosophila (Morelli et al., 2016). The eye disks in the Drosophila snap29 mutant showed defects in epithelial architecture and increased apoptosis (Morelli et al., 2014; 2016).
Similarities in domain structure can help identify candidate interactors. Unlike other SNAP25 homologs in mammals, human SNAP29 harbors an NPF (Asn-Pro-Phe) motif, which interacts with the Eps15-Homology (EH) domain-containing protein1, EHD1 (Rotem-Yehudar et al., 2001; Rapaport et al., 2010). EHD1 is required for the redistribution of the endocytic recycling compartment (Lin et al., 2001). EHD1 and SNAP29 directly interact and form a complex with insulin-like growth factor 1 receptor, IGF-1R (Rotem-Yehudar et al., 2001; Rapaport et al., 2010). Based on these interactions in animals, we predicted that AtSNAP29 and AtSNAP33, which harbor the NPF motif, can interact with AtEHD1, a regulator of endocytosis. It has been shown that down-regulation of AtEHD1 delays the internalization of the styryl dye FM4-64, an endocytosis marker (Bar et al., 2008).
Studies on SNAP29 and SNAP47 in animals suggested that plant SNAP25 homologs could interact with other proteins in autophagy or endocytosis, or other functions, in addition to their role in regulating SNARE complexes. As an autophagy-regulating Qbc SNARE in animals, SNAP29 was identified together with STX17 on autophagosomes (Diao et al., 2015). Autophagy Related 14 (ATG14), an essential autophagy-specific regulator, directly binds to the STX17–SNAP29 binary SNARE complex on autophagosomes, thus priming the SNAP29 complex to interact with VAMP8 and stimulating autophagosome–endolysosome fusion (Diao et al., 2015). Additionally, animal SNAP47 interacts with STX16 and VAMP7, which localize in ATG9a-resident vesicles from recycling endosomes (Aoyagi et al., 2018). The autophagic trafficking of SNAP29 and SNAP47 proteins was hijacked by coxsackievirus B3 (CVB3) and enterovirus D68 (EV-D68), respectively, to enhance viral replication (Mohamud et al., 2018; Corona et al., 2018).
A recent report identified QUIRKY, a member of the family of multiple C2 domain and transmembrane region proteins, as interacting with an Arabidopsis Qa SNARE, and suggested that PEN1/AtSYP121 is engaged in the export of florigen from phloem companion cells to sieve elements through its interaction with QUIRKY in the induction of flowering (Liu et al., 2019). Based on the study of QUIRKY, a Qbc SNARE partner of PEN1/AtSYP121 may be involved in regulating developmental phases by exporting florigen.
Post-translational modifications
Animal SNAP25 proteins undergo several post-translational modifications to modulate their functions. In animals, O-GlcNAc-modification of SNAP29 exacerbates the dysfunction of autophagy induced by arsenic and diabetes (Dodson et al., 2018; Huang et al., 2018). NEK3 (NIMA-never in mitosis gene A-related kinase 3)-mediated serine 105 (S105) phosphorylation of SNAP29 is important for its membrane association (Rapaport et al., 2018). A serine 105 to alanine (S105A) mutant of SNAP29 could not localize to the Golgi or rescue the CEDNIK (cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma) syndrome that occurs because of an early stop codon in SNAP29 (Rapaport et al., 2018). These reports indicate that post-translational modifications of mammalian SNAP25 proteins are important for their cellular functions.
The NetOGlyc 4.0 Server (http://www.cbs.dtu.dk/services/NetOGlyc/) predicts distinct O-GlcNAc-modification sites on Arabidopsis SNAP25 proteins, with 17 residues in AtSNAP29, 18 residues in AtSANP30, and 27 residues in AtSNAP33. However, there is no clear evidence of the function of O-GlcNAc-modification in plants. Phosphoproteome profiling of Arabidopsis seedlings revealed that AtSNAP33 in the plasma membrane has one phosphoserine, although AtSNAP29 and AtSNAP30 were not identified (Reiland et al., 2009). The role of phosphoserine in AtSNAP33 has not been characterized.
Numerous tools for predicting post-translational modifications based on large-scale proteome profiling are available, but the predictions require additional experimental validation. For example, regarding the phosphorylation site in AtSNAP33, one prediction tool (Functional Analysis Tools for Post-Translational Modifications, FAT-PTM; https://bioinformatics.cse.unr.edu/fat-ptm/proteins/) identified 12 residues and another prediction tool (a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor, PhosPhAt 4.0; http://phosphat.uni-hohenheim.de/) identified 20 residues. However, only 9 residues were identified by both prediction tools and whether the predicted residues are functionally meaningful, such as S105 of SNAP25 in CEDNIK syndrome, requires further validation.
AtSNAP29 proteins contain K119 as a potential ubiq-uitination modification site without a phosphorylation site from FAT-PTM tool and 13 phosphorylation sites from PhosPhAt4.0 tool. AtSNAP30 proteins contain S183 as a matched phosphorylation site from both 1 residue in FAT-PTM and 14 residues in PhosPhAt4.0. Interestingly, phosphorylated AtSNAP30 proteins were enriched in pollen (Mayank et al., 2012), where AtSNAP30 proteins are specifically and highly expressed. However, which residues contribute to post-translational modifications in specific cellular functions require further analysis.
CONCLUSION AND PERSPECTIVES
Based on genetic, biochemical, and cell biological studies in Arabidopsis, the unique Qbc SNAREs, which are SNAP25 homologs, function in several vesicle-trafficking processes, such as cytokinesis, innate immune responses, and abiotic stress responses. The results of various studies of plant SNAP25 proteins support the idea that the characterized biological roles of SNAP25 proteins are fairly conserved in monocots and dicots (Table 2). The recently identified interactors of SNAP25 homologs have novel functions, such as the regulation of development of specific tissues (Ichikawa et al., 2014; Liu et al., 2019; Park et al., 2018). Additionally, in the Fabaceae, potential SNAP25 interactors, such as G. max VAMP721a and VAMP721d, are required for symbiotic interactions. GlymaSNAP25 homologs are involved in suppressing nematodes, which are deadly parasites in the roots of legumes and L. japonicus LjVAMP72a and LjVAMP72b are required for root symbiosis and root hair formation (Gavrin et al., 2016; Ivanov et al., 2012; Sharma et al., 2016; Sogawa et al., 2018). These SNAP25 homologs may function as targets for interacting with other SNARE proteins in different organisms to affect survival.
Table 2.
Reported functions of plant SNAP25 proteins
| Name | Expression | Interaction | Reference | Function |
|---|---|---|---|---|
| AtSNAP33 | Whole plant | KNOLLE, VAMP721/722 | (Heese et al., 2001; El Kasmi et al., 2013) | Cell division |
| SYP123, VAMP721/722/724 | (Ichikawa et al., 2014) | |||
| SYP132, VAMP721/722 | (Park et al., 2018) | |||
| PEN1, VAMP721/722 | (Kwon et al., 2008; Pajonk et al., 2008) | Biotic stress | ||
| AtSNAP29 | Root, whole plant | KNOLLE/SYP111 | (Heese et al., 2001) | ND |
| AtSNAP30 | Flower | KNOLLE/SYP111 | (Heese et al., 2001) | ND |
| Glyma17g08450 | Root | ND | (Sharma et al., 2016) | Biotic stress |
| GsSNAP33 | Pod, root, seed, stem | ND | (Nisa et al., 2017) | Abiotic stress |
| OsSNAP32 | Leaf, root, flowering panicle | ND | (Luo et al., 2016) | Biotic stress |
| (Bao et al., 2008; Luo et al., 2016) | Abiotic stress | |||
| HvSNAP34 | ND | ROR2, HvVAMP721 | (Collins et al., 2003) | Biotic stress |
| GhSNAP33 | Leaf, root, stem | ND | (Wang et al., 2018) | Biotic stress |
| Abiotic stress | ||||
| StSNAP33-1 | ND | StSYP1 | (Eschen-Lippold et al., 2012) | Biotic stress |
| CkSNAP33 | Root, stem, leaf | ND | (Wang et al., 2017) | Biotic stress |
ND, not determined.
AtSNAP29 and AtSNAP30 have distinct expression profiles throughout development of Arabidopsis and in different tissues. They also respond differently to environmental stresses; therefore, their roles likely reflect their spatiotemporal effects. AtSNAP29 or AtSNAP30 may interact independently with Qa SNAREs and R SNAREs, whose expression is synchronized. Studies on the subcellular localization, post-translational modification, identification of interacting Qa SNAREs or R SNAREs for the SNAP25 proteins, and structural analysis of new SNARE complexes at the cellular membrane are essential for exploring the mechanisms and roles of Qbc SNARE SNAP25 proteins in plants. snap29 and snap30 mutants of Arabidopsis and snap25 mutants of other crops can be generated using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome editing tools to study the developmental phenotypes and tissue-specific biological roles of SNAP25 proteins in plants.
ACKNOWLEDGMENTS
This work was supported by funds from the Basic Science Research Program of National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology (grant No. 2018R1A2B6006233). Both authors appreciate the PCGE lab members.
Footnotes
AUTHOR CONTRIBUTIONS
K.H.W. and H.K. wrote and approved the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahn G., Kim H., Kim D.H., Hanh H., Yoon Y., Singaram I., Wijesinghe K.J., Johnson K.A., Zhuang X., Liang Z., et al. SH3 Domain-Containing Protein 2 plays a crucial role at the step of membrane tubulation during cell plate formation. Plant Cell. 2017;29:1388–1405. doi: 10.1105/tpc.17.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi K., Itakura M., Fukutomi T., Nishiwaki C., Nakamichi Y., Torii S., Makiyama T., Harada A., Ohara-Imaizumi M. VAMP7 regulates autophagosome formation by supporting Atg9a functions in pancreatic beta-cells from male mice. Endocrinology. 2018;159:3674–3688. doi: 10.1210/en.2018-00447. [DOI] [PubMed] [Google Scholar]
- Arora S., Saarloos I., Kooistra R., van de Bospoort R., Verhage M., Toonen R.F. SNAP-25 gene family members differentially support secretory vesicle fusion. J. Cell Sci. 2017;130:1877–1889. doi: 10.1242/jcs.201889. [DOI] [PubMed] [Google Scholar]
- Bao Y.M., Wang J.F., Huang J., Zhang H.S. Molecular cloning and characterization of a novel SNAP25-type protein gene OsSNAP32 in rice (Oryza sativa L. ). Mol. Biol. Rep. 2008;35:145–152. doi: 10.1007/s11033-007-9064-8. [DOI] [PubMed] [Google Scholar]
- Bar M., Aharon M., Benjamin S., Rotblat B., Horowitz M., Avni A. AtEHDs, novel Arabidopsis EH-domain-containing proteins involved in endocytosis. Plant J. 2008;55:1025–1038. doi: 10.1111/j.1365-313X.2008.03571.x. [DOI] [PubMed] [Google Scholar]
- Bock J.B., Matern H.T., Peden A.A., Scheller R.H. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Cao L.G., Wang Y.L. Mechanism of the formation of contractile ring in dividing cultured animal cells. I. Recruitment of preexisting actin filaments into the cleavage furrow. J. Cell Biol. 1990;110:1089–1095. doi: 10.1083/jcb.110.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S., Halder P., Kumar M., Mukhopadhyay K. Genome-wide identification, cloning and characterization of SNARE genes in bread wheat (Triticum aestivum L) and their response to leaf rust. Agri Gene. 2017;3:12–20. doi: 10.1016/j.aggene.2016.11.002. [DOI] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Huckelhoven R., Stein M., Freialdenhoven A., Somerville S.C., et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Corona A.K., Saulsbery H.M., Corona Velazquez A.F., Jackson W.T. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018;22:3304–3314. doi: 10.1016/j.celrep.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L.M., Li J., Vivona S., et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Liu P., Jiang T., Ambrose A.J., Luo G., Rojo de la Vega M., Cholanians A.B., Wong P.K., Chapman E., Zhang D.D. Increased O-GlcNAcylation of SNAP29 drives arsenic-induced autophagic dysfunction. Mol. Cell. Biol. 2018;38:e00595–17. doi: 10.1128/MCB.00595-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell. 2008;20:3006–3021. doi: 10.1105/tpc.107.057711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi F., Krause C., Hiller U., Stierhof Y.D., Mayer U., Conner L., Kong L., Reichardt I., Sanderfoot A.A., Jurgens G. SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell. 2013;24:1593–1601. doi: 10.1091/mbc.e13-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschen-Lippold L., Landgraf R., Smolka U., Schulze S., Heilmann M., Heilmann I., Hause G., Rosahl S. Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 2012;193:985–996. doi: 10.1111/j.1469-8137.2011.04024.x. [DOI] [PubMed] [Google Scholar]
- Gavrin A., Chiasson D., Ovchinnikova E., Kaiser B.N., Bisseling T., Fedorova E.E. VAMP721a and VAMP721d are important for pectin dynamics and release of bacteria in soybean nodules. New Phytol. 2016;210:1011–1021. doi: 10.1111/nph.13837. [DOI] [PubMed] [Google Scholar]
- Gonzalo S., Linder M.E. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol. Biol. Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Greentree W.K., Linder M.E. SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J. Biol. Chem. 1999;274:21313–21318. doi: 10.1074/jbc.274.30.21313. [DOI] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C.T., Mirabelle S., Guha M., Sillibourne J., Doxsey S.J. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Hachez C., Laloux T., Reinhardt H., Cavez D., Degand H., Grefen C., De Rycke R., Inzé D., Blatt M.R., Russinova E., et al. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell. 2014;26:3132–3147. doi: 10.1105/tpc.114.127159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., McMahon H., Yamasaki S., Binz T., Hata Y., Sudhof T.C., Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese M., Gansel X., Sticher L., Wick P., Grebe M., Granier F., Jurgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J. Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M., Varoqueaux F., Wiederhold K., Takamori S., Urlaub H., Fasshauer D., Jahn R. Identification of SNAP-47, a novel Qbc-SNARE with ubiquitous expression. J. Biol. Chem. 2006;281:17076–17083. doi: 10.1074/jbc.M513838200. [DOI] [PubMed] [Google Scholar]
- Huang L., Yuan P., Yu P., Kong Q., Xu Z., Yan X., Shen Y., Yang J., Wan R., Hong K., et al. O-GlcNAc-modified SNAP29 inhibits autophagy-mediated degradation via the disturbed SNAP29-STX17-VAMP8 complex and exacerbates myocardial injury in type I diabetic rats. Int. J. Mol. Med. 2018;42:3278–3290. doi: 10.3892/ijmm.2018.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Hirano T., Enami K., Fuselier T., Kato N., Kwon C., Voigt B., Schulze-Lefert P., Baluska F., Sato M.H. Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:790–800. doi: 10.1093/pcp/pcu048. [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Ivanov S., Fedorova E.E., Limpens E., De Mita S., Genre A., Bonfante P., Bisseling T. Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8316–8321. doi: 10.1073/pnas.1200407109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargul J., Gansel X., Tyrrell M., Sticher L., Blatt M.R. Protein-binding partners of the tobacco syntaxin NtSyr1. FEBS Lett. 2001;508:253–258. doi: 10.1016/S0014-5793(01)03089-7. [DOI] [PubMed] [Google Scholar]
- Kim H., O'Connell R., Maekawa-Yoshikawa M., Uemura T., Neumann U., Schulze-Lefert P. The Powdery Mildew Resistance Protein RPW8.2 Is Carried on VAMP721/722 Vesicles to the Extrahaustorial Membrane of Haustorial Complexes. Plant J. 2014;79:835–847. doi: 10.1111/tpj.12591. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H.S., Lipka U., Humphry M., Bau S., Straus M., Kwaaitaal M., Rampelt H., et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jurgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y., Golani Y., Kaye Y., Levine A. Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 2010;61:2615–2622. doi: 10.1093/jxb/erq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.M., Webb S.E., Lee K.W., Miller A.L. Recruitment and SNARE-mediated fusion of vesicles in furrow membrane remodeling during cytokinesis in zebrafish embryos. Exp. Cell Res. 2006;312:3260–3275. doi: 10.1016/j.yexcr.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Lin S.X., Grant B., Hirsh D., Maxfield F.R. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat. Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- Lipka V., Dittgen J., Bednarek P., Bhat R., Wiermer M., Stein M., Landtag J., Brandt W., Rosahl S., Scheel D., et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- Lipka V., Kwon C., Panstruga R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 2007;23:147–174. doi: 10.1146/annurev.cellbio.23.090506.123529. [DOI] [PubMed] [Google Scholar]
- Liu L., Li C., Teo Z.W.N., Zhang B., Yu H. The MCTP-SNARE complex regulates florigen transport in Arabidopsis. Plant Cell. 2019;31:2475–2490. doi: 10.1105/tpc.18.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Zhang H., He W., Zhang Y., Cao W., Zhang H., Bao Y. OsSNAP32, a SNAP25-type SNARE protein-encoding gene from rice, enhanced resistance to blast fungus. Plant Growth Regul. 2016;80:37–45. doi: 10.1007/s10725-016-0152-4. [DOI] [Google Scholar]
- Mayank P., Grossman J., Wuest S., Boisson-Dernier A., Roschitzki B., Nanni P., Nühse T., Grossniklaus U. Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 2012;72:89–101. doi: 10.1111/j.1365-313X.2012.05061.x. [DOI] [PubMed] [Google Scholar]
- Mierzwa B., Gerlich D.W. Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Mohamud Y., Shi J., Qu J., Poon T., Xue Y.C., Deng H., Zhang J., Luo H. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication. Cell Rep. 2018;22:3292–3303. doi: 10.1016/j.celrep.2018.02.090. [DOI] [PubMed] [Google Scholar]
- Mollinedo F., Calafat J., Janssen H., Martin-Martin B., Canchado J., Nabokina S.M., Gajate C. Combinatorial SNARE complexes modulate the secretion of cytoplasmic granules in human neutrophils. J. Immunol. 2006;177:2831–2841. doi: 10.4049/jimmunol.177.5.2831. [DOI] [PubMed] [Google Scholar]
- Morelli E., Ginefra P., Mastrodonato V., Beznoussenko G.V., Rusten T.E., Bilder D., Stenmark H., Mironov A.A., Vaccari T. Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy. 2014;10:2251–2268. doi: 10.4161/15548627.2014.981913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli E., Mastrodonato V., Beznoussenko G.V., Mironov A.A., Tognon E., Vaccari T. An essential step of kinetochore formation controlled by the SNARE protein Snap29. EMBO J. 2016;35:2223–2237. doi: 10.15252/embj.201693991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G., Milosevic I., Mohrmann R., Wiederhold K., Walter A.M., Sorensen J.B. The SNAP-25 linker as an adaptation toward fast exocytosis. Mol. Biol. Cell. 2008;19:3769–3781. doi: 10.1091/mbc.e07-12-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H., Blasi J., Jahn R. Clostridial neurotoxins: new tools for dissecting exocytosis. Trends Cell Biol. 1994;4:179–185. doi: 10.1016/0962-8924(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Nisa Z.U., Mallano A.I., Yu Y., Chen C., Duan X., Amanullah S., Kousar A., Baloch A.W., Sun X., Tabys D., et al. GsSNAP33, a novel Glycine soja SNAP25-type protein gene: improvement of plant salt and drought tolerances in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2017;119:9–20. doi: 10.1016/j.plaphy.2017.07.029. [DOI] [PubMed] [Google Scholar]
- Oyler G.A., Higgins G.A., Hart R.A., Battenberg E., Billingsley M., Bloom F.E., Wilson M.C. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J. Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan J.K., Wylie F.G., Joseph S., Widberg C., Bryant N.J., James D.E., Stow J.L. The t-SNARE syntaxin 4 is regulated during macrophage activation to function in membrane traffic and cytokine secretion. Curr. Biol. 2003;13:156–160. doi: 10.1016/S0960-9822(03)00006-X. [DOI] [PubMed] [Google Scholar]
- Pajonk S., Kwon C., Clemens N., Panstruga R., Schulze-lefert P. Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J. Biol. Chem. 2008;283:26974–26984. doi: 10.1074/jbc.M805236200. [DOI] [PubMed] [Google Scholar]
- Park M., Krause C., Karnahl M., Reichardt I., El Kasmi F., Mayer U., Stierhof Y.D., Hiller U., Strompen G., Bayer M., et al. Concerted action of evolutionarily ancient and novel SNARE complexes in flowering-plant cytokinesis. Dev. Cell. 2018;44:500–511.:e4. doi: 10.1016/j.devcel.2017.12.027. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Lugassy Y., Sprecher E., Horowitz M. Loss of SNAP29 impairs endocytic recycling and cell motility. PLoS One. 2010;5:e9759. doi: 10.1371/journal.pone.0009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Fichtman B., Weidberg H., Sprecher E., Horowitz M. NEK3-mediated SNAP29 phosphorylation modulates its membrane association and SNARE fusion dependent processes. Biochem. Biophys. Res. Commun. 2018;497:605–611. doi: 10.1016/j.bbrc.2018.02.116. [DOI] [PubMed] [Google Scholar]
- Ravichandran V., Chawla A., Roche P.A. Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J. Biol. Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- Reales E., Mora-Lopez F., Rivas V., Garcia-Poley A., Brieva J.A., Campos-Caro A. Identification of soluble N-ethylmaleimide-sensitive factor attachment protein receptor exocytotic machinery in human plasma cells: SNAP-23 is essential for antibody secretion. J. Immunol. 2005;175:6686–6693. doi: 10.4049/jimmunol.175.10.6686. [DOI] [PubMed] [Google Scholar]
- Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., Gruissem W., Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem-Yehudar R., Galperin E., Horowitz M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J. Biol. Chem. 2001;276:33054–33060. doi: 10.1074/jbc.M009913200. [DOI] [PubMed] [Google Scholar]
- Saito C., Ueda T. Chapter 4 Functions of RAB and SNARE proteins in plant life. In: K.W. Jeon., editor. International Review of Cell and Molecular Biology. Academic Press; Amsterdam, The Netherlands: 2009. pp. 183–233. [DOI] [PubMed] [Google Scholar]
- Sanmartin M., Ordonez A., Sohn E.J., Robert S., Sanchez-Serrano J.J., Surpin M.A., Raikhel N.V., Rojo E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3645–3650. doi: 10.1073/pnas.0611147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C., Lutter K., Kissmehl R., Plattner H. Molecular identification of a SNAP-25-like SNARE protein in Paramecium. Eukaryot. Cell. 2008;7:1387–1402. doi: 10.1128/EC.00012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Pant S.R., McNeece B.T., Lawrence G.W., Klink V.P. Co-regulation of the Glycine max soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE)-containing regulon occurs during defense to a root pathogen. J. Plant Interact. 2016;11:74–93. doi: 10.1080/17429145.2016.1195891. [DOI] [Google Scholar]
- Singh D., Yadav N.S., Tiwari V., Agarwal P.K., Jha B. A SNARE-like superfamily protein SbSLSP from the halophyte Salicornia brachiata confers salt and drought tolerance by maintaining membrane stability, K+/Na+ ratio, and antioxidant machinery. Front. Plant Sci. 2016;7:737. doi: 10.3389/fpls.2016.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa A., Yamazaki A., Yamasaki H., Komi M., Manabe T., Tajima S., Hayashi M., Nomura M. SNARE proteins LjVAMP72a and LjVAMP72b are required for root symbiosis and root hair formation in Lotus japonicus. Front. Plant Sci. 2018;9:1992. doi: 10.3389/fpls.2018.01992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M., Yang B., Yoo J.S., Huang B., Shen M., Yu S., Luo Y., Scheller R.H. Three novel proteins of the syntaxin/SNAP-25 family. J. Biol. Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- Stein M., Dittgen J., Sanchez-Rodriguez C., Hou B.H., Molina A., Schulze-Lefert P., Lipka V., Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., et al. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;15:2885–2899. doi: 10.1105/tpc.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S., Nagy P., Varga A., Pircs K., Karpati M., Varga K., Kovacs A.L., Hegedus K., Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker A., Stierhof Y.D., Jurgens G. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J. Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- Wang P., Sun Y., Pei Y., Li X., Zhang X., Li F., Hou Y. GhSNAP33, a t-SNARE protein from Gossypium hirsutum, mediates resistance to Verticillium dahliae infection and tolerance to drought stress. Front. Plant Sci. 2018;9:896. doi: 10.3389/fpls.2018.00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang X., Ma X., Sun Y., Liu N., Li F., Hou Y. Identification of CkSNAP33, a gene encoding synaptosomal-associated protein from Cynanchum komarovii, that enhances Arabidopsis resistance to Verticillium dahliae. PLoS One. 2017;12:e0178101. doi: 10.1371/journal.pone.0178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimbs T., Low S.H., Chapin S.J., Mostov K.E., Bucher P., Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick P., Gansel X., Oulevey C., Page V., Studer I., Durst M., Sticher L. The expression of the t-SNARE AtSNAP33 is induced by pathogens and mechanical stimulation. Plant Physiol. 2003;132:343–351. doi: 10.1104/pp.102.012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano D., Sato M., Saito C., Sato M.H., Morita M.T., Tasaka M. A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8589–8594. doi: 10.1073/pnas.1430749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., Park S., Yun H.S., Kwon C. Vesicle-associated membrane proteins 721 and 722 are required for unimpeded growth of Arabidopsis under ABA application. J. Plant Physiol. 2013;170:529–533. doi: 10.1016/j.jplph.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Zhao N., Hashida H., Takahashi N., Sakaki Y. Cloning and sequence analysis of the human SNAP25 cDNA. Gene. 1994;145:313–314. doi: 10.1016/0378-1119(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Zhu J., Gong Z., Zhang C., Song C.P., Damsz B., Inan G., Koiwa H., Zhu J.K., Hasegawa P.M., Bressan R.A. OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell. 2002;14:3009–3028. doi: 10.1105/tpc.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]