Abstract
Pathogenic aminoacyl-tRNA synthetases (ARSs) are attractive targets for anti-infective agents because their catalytic active sites are different from those of human ARSs. Mupirocin is a topical antibiotic that specifically inhibits bacterial isoleucyl-tRNA synthetase (IleRS), resulting in a block to protein synthesis. Previous studies on Thermus thermophilus IleRS indicated that mupirocin-resistance of eukaryotic IleRS is primarily due to differences in two amino acids, His581 and Leu583, in the active site. However, without a eukaryotic IleRS structure, the structural basis for mupirocin-resistance of eukaryotic IleRS remains elusive. Herein, we determined the crystal structure of Candida albicans IleRS complexed with Ile-AMP at 2.9 Å resolution. The largest difference between eukaryotic and prokaryotic IleRS enzymes is closure of the active site pocket by Phe55 in the HIGH loop; Arg410 in the CP core loop; and the second Lys in the KMSKR loop. The Ile-AMP product is lodged in a closed active site, which may restrict its release and thereby enhance catalytic efficiency. The compact active site also prevents the optimal positioning of the 9-hydroxynonanoic acid of mupirocin and plays a critical role in resistance of eukaryotic IleRS to anti-infective agents.
Keywords: active site closure, aminoacyl-tRNA synthetases, anti-infective agents, crystal structure, mupirocin
INTRODUCTION
Developing structure-based anti-infective drugs requires structural information on the active sites of validated target proteins (Kuntz, 1992). These targets must be conserved and essential for the survival of the target organisms, and the active sites must possess structural differences between eukaryotic and pathogenic molecules (Kwon et al., 2019; Yao and Fox, 2013). Aminoacyl-tRNA synthetases (ARSs) catalyze the addition of amino acids to their cognate tRNAs with high fidelity in the initial step of protein synthesis (Delarue, 1995). This canonical function of ARSs is essential and responsible for carrying accurate genetic information in every living organism.
ARSs are grouped into class 1 and class 2, depending on the core structure and oligomeric state (Ribas de Pouplana and Schimmel, 2001). Class 1 ARSs are further divided into three subclasses, 1a, 1b, and 1c, according to sequence homology. Isoleucyl-tRNA synthetase (IleRS) is a multi-domain enzyme with catalytic, editing (connective polypeptide 1 or CP1), and anticodon-binding domains. Together with leucyl-, valyl-, methionyl-, cysteinyl-, and arginyl-tRNA synthetase (LeuRS, ValRS, MetRS, CysRS, and ArgRS), IleRS belongs to class 1a ARSs, which are characterized by a Rossmann fold in the catalytic domain harboring conserved His-Ile-Gly-His (HIGH) and Lys-Met-Ser-Lys-Ser/Arg (KMSKS/R) motifs in the active site. The aminoacylation process requires a two step-reaction: activation of amino acids with ATP to form aminoacyl-AMP; and transfer of amino acids to their cognate tRNA (Antonellis and Green, 2008; Schimmel, 2018). IleRS possesses error-correction activity in the editing domain that enhances the accuracy of aminoacylation and maintains translation fidelity (Ling et al., 2009). In the double-sieve mechanism, larger amino acids are first filtered in the synthetic active site of the catalytic domain, and those of smaller size are removed in the editing active site of the editing domain (Fersht and Dingwall, 1979; Fukai et al., 2000). The misactivated substrate is believed to be hydrolyzed in the editing domain via shuttling of the 3’-acceptor stem of tRNA between the syntheticand editing active sites (Silvian et al., 1999).
Mupirocin (pseudomonic acid A) is a topical antibiotic used to treat infection by Staphylococcus aureus or Streptococcus pyogenes (Sutherland et al., 1985). The compound has been clinically used as an antibiotic against methicillin-resistant S. aureus (MRSA). Mupirocin contains a short fatty acid side chain (9-hydroxynonanoic acid) linked to monic acid by an ester linkage. Mupirocin reversibly binds to the active site of bacterial and archaeal IleRSs, and competes with isoleucine and ATP or Ile-AMP, thereby inhibiting protein synthesis ( and Mellows, 1978). However, mupirocin weakly inhibits IleRS in rat, with a Ki of 20 µM that is 8,000 times greater than the Ki of the Escherichia coli B enzyme (Hughes and Mellows, 1980).
Crystal structures of S. aureus (Sa) and Thermus thermophilus (Tt) IleRS complexed with mupirocin have been reported (Nakama et al., 2001; Silvian et al., 1999). Mupirocin inhibits Tt IleRS with 4-fold less potency than Sa IleRS (Nakama et al., 2001). The moderately conserved His (His581 in Tt IleRS, His585 in Sa IleRS) and Leu/Phe (Leu583/Phe587) in the active site of bacterial IleRSs are replaced with Asn/Ser/Thr and Leu/Ile, respectively, in eukaryotic IleRS. Double substitution of His581 and Leu583 to Leu and His in Tt IleRS increased the Ki value of the mutant toward mupirocin by an order of magnitude compared with wild-type Tt IleRS (Nakama et al., 2001). Based on this analysis, it has been proposed that differences in the two active site residues may confer specificity for mupirocin in bacterial IleRS but not eukaryotic IleRS. However, there are no structures of eukaryotic IleRSs available, making it difficult to elucidate the resistance mechanism of eukaryotic IleRS toward mupirocin.
In the present work, we determined the crystal structure of IleRS from the fungus Candida albicans (Ca) in complex with the product Ile-AMP. In the compact active site, Phe55 from the HIGH loop and Arg410 from the connective polypeptide (CP) core loop enclose the Ile-AMP molecule. Furthermore, the second Lys in the KMSKR loop, Thr57 and Thr59 in the HIGH loop, and Arg410 in the CP core loop form a network to stabilize the closed conformation of the active site, which restricts the release of Ile-AMP. This compact conformation may minimize the use of cellular energy and increase catalytic efficiency. Furthermore, the compact conformation of the active site might be an important feature that confers mupirocin-resistance in eukaryotic IleRS.
MATERIALS AND METHODS
Cloning, expression, and purification
The gene encoding full-length fungal IleRS (residues 1-1088) was amplified by polymerase chain reaction (PCR) from genomic DNA of C. albicans ATCC MYA-2876 and then inserted into the pET28a vector. Two CUG codons for leucine were changed to UCG (serine) in C. albicans by overlap extension PCR. For crystallization, C-terminal domain-truncated Ca IleRS Ca IleRS (residues 1-856) was inserted into the pET28a vector with a His6 tag at the N-terminus. The plasmid containing the IleRS gene was transformed into competent E. coli Rosetta (DE3) cells, and bacteria were cultured in Luria-Bertani (LB) broth medium. Initially, the protein was purified by an Ni2+ column using an imidazole gradient in 20 mM TRISHCl (pH 7.4), 300 mM NaCl, 7 mM 2-mercaptoethanol, and 5% glycerol. The eluted protein was purified further with an ion-exchange column (Resource-Q; GE Healthcare, USA) followed by gel filtration on a Superdex 200 16/600 column equilibrated with 20 mM TRIS-HCl (pH 7.4), 100 mM NaCl, 5 mM dithiothreitol (DTT), and 5% glycerol. The IleRS used for crystallization was concentrated to 30 mg/ml.
For microscale thermophoresis (MST) experiments, Ca IleRS mutants were constructed by overlap extension PCR. Thr599 and Ile601 were simultaneously mutated to histidine and phenylalanine. Human IleRS was expressed in Spodoptera frugiperda (Sf9) cells, and baculoviruses were generated in Sf9 cells with the Bac-to-Bac system (Invitrogen, USA). Cells were infected with baculovirus at a cell density of 2.0 × 106 cells per ml, cultured at 27°C, and harvested after 72 h. Both the Ca IleRS mutant and human IleRSs were purified by the same procedure used for wild-type Ca IleRS.
Crystallization and X-ray diffraction data collection
Crystals of Ca IleRS were grown at 20°C by the sitting-drop vapor-diffusion method. The crystallization buffer contained 0.1 M MES (pH 6.5) and 3% polyethylene glycol (PEG) 8000. The resulting crystals belonged to the P21 space group (a = 56.5 Å, b = 137.6 Å, c = 73.7 Å, β = 106.3°) and contained one IleRS molecule in the asymmetric unit. Diffraction data were collected at –170°C using crystals flash-frozen in crystallization buffer containing 35% glycerol. Diffraction data were collected at 0.9795 Å on Beamline 11C at the Pohang Advanced Light Source. Diffraction data were integrated and scaled using the HKL-2000 package (Otwinowski and Minor, 1997).
Structure determination and refinement
The structure of Ca IleRS was determined by the molecular replacement method. Successive rounds of manual building with COOT (Emsley and Cowtan, 2004) and refinement (rigid body, xyz coordinates, and individual B-factors) with PHENIX (Adams et al., 2010) were employed to build the complete model. The final refined model of Ca IleRS at 2.9 Å resolution (Rwork/Rfree of 17.4%/23.9%) contains 94% of residues in the most favored region and one residue in the disallowed region (Table 1).
Table 1.
Data collection and refinement statistics
| Ca IleRS–Ile-AMP | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 56.5, 137.6, 73.7 |
| α, β, γ (°) | 90, 106, 90 |
| Resolution (Å) | 49.3-2.9 (2.95-2.9)a |
| Measured reflections | 110,098 |
| Unique reflections | 23,327 |
| Completeness (%) | 97.8 (99.2) |
| Average (I/σ) | 11.4 (1.3) |
| R merge (%) | 0.15 (1.16) |
| CC*a | 0.997 (0.862) |
| CC1/2a | 0.987 (0.591) |
| Redundancy | 4.7 (4.9) |
| Wilson B factor (Å2) | 52.3 |
| Refinement statistics | |
| Resolution (Å) | 49.35-2.9 |
| R factor/R free (%)b | 17.4/23.9 |
| No. of atoms | |
| Protein | 6692 |
| Ile-AMP | 31 |
| Water | 4 |
| B-factors | |
| Protein | 53.0 |
| Ile-AMP | 47.3 |
| Water | 44.4 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.137 |
| Clash scorec | 11.0 |
| Ramachandran plotc | |
| Most favored (%) | 94 |
| Allowed (%) | 5.7 |
| Disallowed (%) | 0.3 |
Values in parentheses are for the highest shell.
aKarplus and Diederichs (2012).
bR = |Fobs–Fcalc|/Fobs, where Fobs = Fpi and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections).
cClash score and Ramachandran plot are calculated by Molprobity (Chen et al., 2010).
Binding affinity measurement by microscale thermophoresis
MST assays were performed with a Monolith NT.115 instrument (NanoTemper Technologies, Germany) (Duhr and Braun, 2006; Seidel et al., 2013). Each titration curve con- tained 16 points prepared by serial dilutions of analytes and a constant concentration of the fluorescein-labeled ligand. To measure the binding affinity between mupirocin and purified human IleRS and Ca IleRSs (wild-type and mutants), proteins were fluorescently labeled using a Monolith His-Tag Labeling Kit RED-TRIS-NTA 2nd Generation (NanoTemper Technolo- gies). A 2 mM sample of mupirocin was serially diluted 2-fold by 15 times and then the 16 different concentrations of mupirocin were incubated with 400 nM of labeled human IleRS or Ca IleRS (wild-type or mutants). Experiments were performed in HEPES buffer supplemented with 0.05% (w/v) Tween-20 and 0.05% bovine serum albumin (BSA). Samples were loaded into high-precision Monolith NT.115 Capillaries (NanoTemper Technologies). MST assays were performed with 60% light-emitting diode (LED) power using a green filter and 60% MST power. Normalized fluorescence read- ings (thermophoresis plus T-jump) were plotted as a function of analyte concentration, and curve fitting and dissociation constant (Kd) calculation were performed with MO.Affinity Analysis (ver. 2.3; NanoTemper Technologies). For each data set, three independent MST measurements were carried out.
Accession numbers
The atomic coordinate has been deposited at the Protein Data Bank, with an accession code 6LDK.
RESULTS AND DISCUSSION
The roles of two mupirocin-binding residues in eukaryotic IleRSs
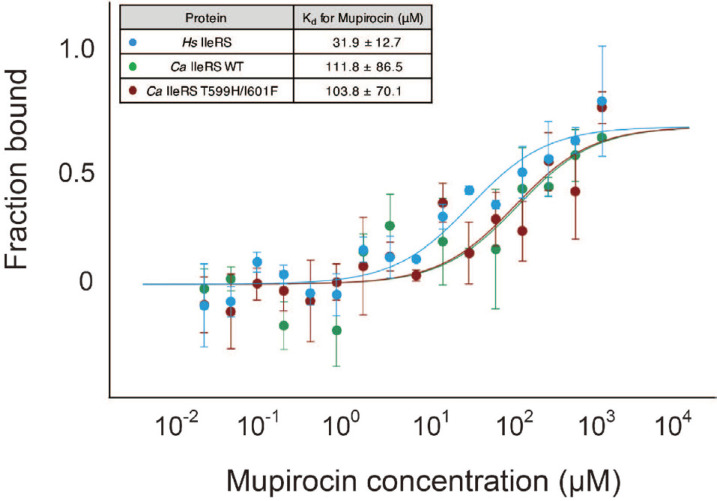
In both Tt IleRS and Sa IleRS structures complexed with mupirocin, His581 (His585 in Sa IleRS) and Leu583 (Phe587 in Sa IleRS), which interact with mupirocin, are not conserved in eukaryotic IleRSs (Nakama et al., 2001; Silvian et al., 1999; Fig. 1). His581 (His585) interacts with the pyran ring of monic acid, and Leu583 (Phe587) is stacked against the C1 to C3 chain of monic acid (PDB 1JZS, 1FFY). Previous studies showed that mutation of these residues to those conserved in eukaryotic IleRS or mupirocin-resistant S. aureus (type II) increased the Ki value for mupirocin, and the authors proposed that these residues are critical in selecting mupirocin (Nakama et al., 2001). The equivalent residues in Ca IleRS are Thr599 and Ile601 (Fig. 1). We therefore mutated Thr599 and Ile601 to His and Phe, respectively, and examined the binding affinity of the Ca IleRS mutant toward mupirocin using MST. Because the active site of Ca IleRS might be pre-occupied with Ile-AMP, we performed extensive dialysis to remove any residual Ile-AMP prior to binding analysis. Both wild-type and mutant Ca IleRS enzymes exhibited similar Kd values (103-111µM) toward mupirocin, suggesting that the mutation did not affect the binding of mupirocin to Ca IleRS (Fig. 2). Thus, our analyses suggest that these two residues are not critical for the mupirocin-resistance of eukaryotic IleRS. The Kd of human IleRS toward mupirocin was ~3.5-fold lower than that of Ca IleRS (Fig. 2).
Fig. 1. Structure-based sequence alignment of IleRS orthologues.
The organisms are Candida albicans (UniProt ID, Q59RI1), Homo sapiens (P41252), Gallus gallus (A0A3Q2UG33), Danio rerio (Q6PGU7), Drosophila melanogaster (Q8MSW0), Caenorhabditis elegans (Q21926), Saccharomyces cerevisiae (P48526), Methanocaldococcus jannaschii (Q58357), Methanothermobacter thermautotrophicus (O27428), Thermus thermophilus (P56690), Escherichia coli (P00956), Staphylococcus aureus (P41972). Strictly conserved and highly conserved residues are highlighted in yellow and pale yellow, respectively. The secondary structure is displayed above the sequence. Every 10th residue is marked with a black dot. Functionally important residues are indicated: the signature motif in the Rossmann fold is indicated by black stars; active site residues are marked with blue circles; The IleRS residues mutated in this study are highlighted with red boxes. Sequence alignment was performed using PROMALS3D (Pei et al., 2008).
Fig. 2. Binding of mupirocin to human IleRS, wild-type and mutant Ca IleRSs measured by MST.
The affinity of mupirocin to the Ca IleRS mutant is similar to that toward wild-type Ca IleRS. The affinity of mupirocin toward human IleRS is ~3.5- fold lower than that of Ca IleRS. Each data set for the binding analysis is derived from three independent experiments, and each experiment contains 17 measurements.
Overall structure of Ca IleRS
To understand the basis for the mupirocin-resistance of eukaryotic IleRS, we attempted to determine the structure of eukaryotic IleRS. We initially crystallized full-length Ca IleRS (residues 1-1088, 125 kDa), which is composed of the amino-terminal, catalytic, editing, anticodon-binding, and carboxyl-terminal domains. However, the crystals diffracted weakly to 7 Å. The structure of full-length Tt IleRS (residues 1-1043, PDB 1ILE) revealed that the C-terminal 217 residues are partially disordered in the absence of tRNA, which suggests that the C-terminal domain might be flexible. Thus, we removed the C-terminal domain by aligning the sequence with Tt IleRS to obtain crystals with better diffraction quality.
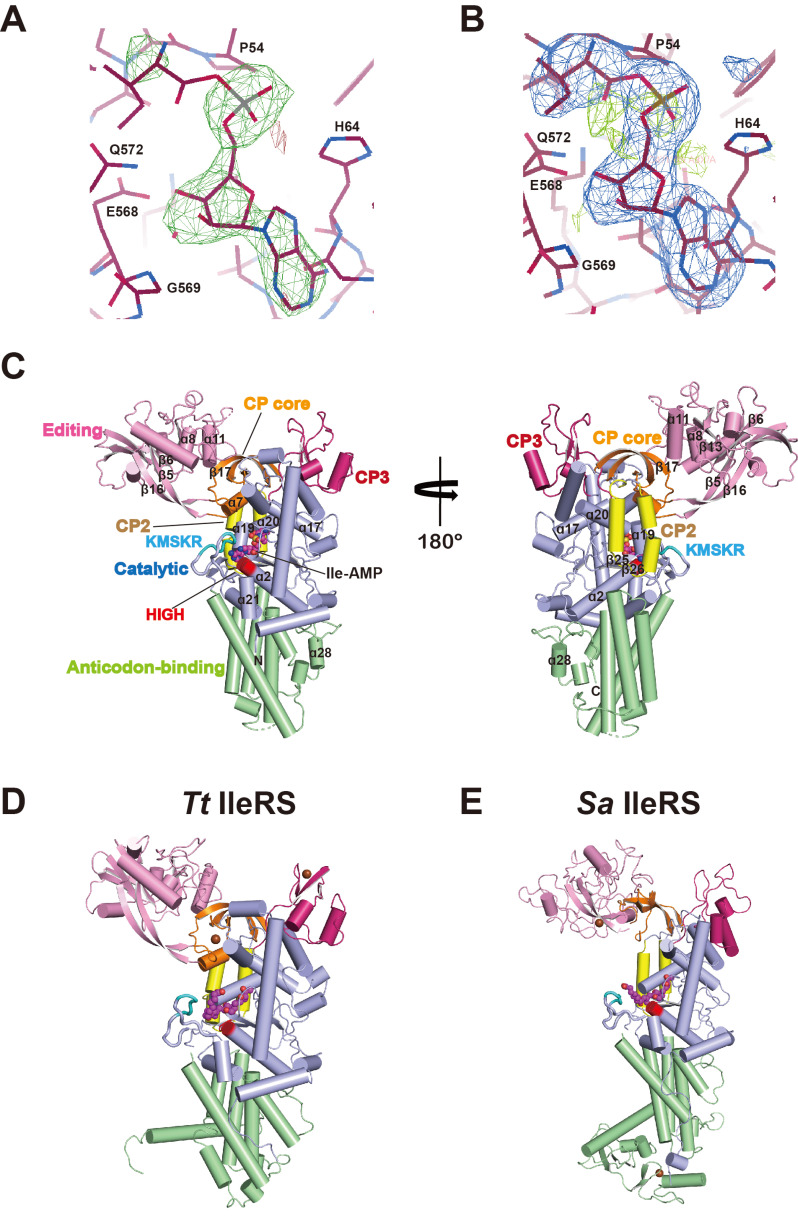
We determined the crystal structure of the C-terminal truncated form of Ca IleRS (residues 1-856) at 2.9 Å resolution by molecular replacement using the Tt IleRS structure as a search model (Nureki et al. 1998; Table 1). Although we did not add any ATP, amino acids, or analogues, the structure clearly revealed electron density for an unknown ligand at the active site, into which an Ile-AMP molecule fitted well (Figs. 3A and 3B). We presumed that Ile-AMP is formed as a consequence of the reaction between Ile and ATP in the active site during or prior to purification or crystallization of IleRS.
Fig. 3. Comparison of the overall structures of prokaryotic and eukaryotic IleRSs.
(A and B) Electron density map of the Ca IleRS active site. (A) The 2.9 Å |Fo|-|Fc| map contoured at 3.0 σ is generated with phases from molecular replacement. Ile- AMP was fitted into the electron density at the active site. The final model is modeled onto the map. (B) A composite omit map around the ligand binding site. (C) Overall structure of Ca IleRS complexed with Ile-AMP in two different orientations. The catalytic domain (light blue), the CP core (orange), the editing domain (CP1, pink), and the CP2 (yellow), CP3 (magenta), and anticodon-binding domains (pale green) are shown. The HIGH and KMSKR motifs are colored in red and cyan, respectively. The bound Ile-AMP molecule is shown in sphere representation. (D and E) Ribbon representation of the structures of the bacterial IleRS-mupirocin complexes Tt IleRS (PDB 1JZS, left; D) and Sa IleRS (PDB 1FFY, right; E). Each domain is colored as in Fig. 3C. The mupirocin molecule is shown in spheres. The Sa IleRS structure is from the Sa IleRS- tRNAIle complex with the tRNA omitted.
The overall structure of Ca IleRS can be divided into six regions: the catalytic domain (Rossmann fold, residues 15-176 and 535-649, light blue), the CP core (residues 177-202 and 408-426, orange), the editing domain (CP1, residues 203-407, pink), the CP2 (427-461, yellow) and CP3 (473-534, magenta) subdomains, and the anticodon-binding domain (residues 650-828, pale green; Figs. 1 and 3C). The catalytic domain occupies the core of the molecule and possesses a central deep cleft with two signature motifs, 61HIGH and 609KMSKR, and one Ile-AMP molecule. The HIGH motif is located at the beginning of the long α2 helix that lies beneath the Rossmann fold core strands. The KMSKR loop connects the β26 strand and the α21 helix near the α2 helix containing the HIGH motif. The four CP subdomains are inserted within the catalytic domain, and the CP core with the β-β-α-β fold is located on the top of the catalytic domain. The editing domain containing the binding site for Val or Ile is inserted between the β7 helix and the β17 strand of the CP core through its first and last strands (β5 and β16). The binding site for Val is surrounded by the β6 strand, the α8 helix, the β13 strand, and the α11 helix (Supplementary Fig. S1; Fukunaga and Yokoyama, 2006). The CP2 subdomain is formed from a pair of anti-parallel α-helices followed by the β17 strand of the CP core. The CP3 subdomain, composed of two anti-parallel strands and two α-helices, is located on the opposite side of the CP2 domain, and connects the CP2 subdomain and second half of the catalytic domain. The anticodon-binding domain containing a five-helix bundle occupies the bottom part of the enzyme.
Comparison with bacterial IleRSs
At present, the crystal structures of two bacterial IleRSs, Tt IleRS and Sa IleRS, have been reported in both ligand-free and ligand-bound forms (Nakama et al., 2001; Nureki et al., 1998; Silvian et al., 1999). Ca IleRS shares 38% and 25% sequence identity with Tt IleRS and Sa IleRS, respectively. The overall structure of Ca IleRS superimposes onto Tt IleRSs and Sa IleRSs with a root mean square deviation (rmsd) value of 2.1 Å for 706 Cα atoms and 2.5 Å for 486 Cα atoms, respectively. The individual domains share more similar structures (rmsd value of 1.4 Å for 167-437 Cα atoms), with the largest difference in the anticodon-binding domains, which aligned with an rmsd value of 2.3 Å for 80 Cα atoms.
When the catalytic domains of IleRSs are superimposed, the editing domain of Ca IleRS is rotated by 5.7° and shifted by 5.3 Å toward the KMSKR loop of the catalytic domain relative to Tt IleRS, which induces a more closed conformation (Fukunaga and Yokoyama, 2006 Figs. 3C and 3D). Structural alignment between Ca IleRSand tRNAIle-bound Sa IleRS revealed that the editing domain undergoes a dramatic movement to accommodate the cognate tRNA (Silvian et al., 1999). The editing domain of Sa IleRS complexed in tRNAIle rotates by 11.5° relative to the rest of the molecules due to interactions with the tRNA. The editing active sites between Ca IleRS and Sa IleRS are separated by 11.2 Å (Figs. 3C and 3E).
The structures of the CP insertions and anticodon-binding domains are clearly different from those in bacterial IleRSs (Figs. 3C-3E). The CP cores of both Ca IleRS and Tt IleRS share the same β-β-α-β topology, whereas Sa IleRS lacks the α-helix. The zinc-binding motif in the CP core of Tt IleRS is absent in Ca IleRS. While the CP3 domains of Ca IleRS and Sa IleRS lack a zinc-binding motif, the equivalent domain of Tt IleRS contains a zinc-binding motif. The β-β-α-α topology of the CP3 subdomain of Ca IleRS differs from the β-α-α-β-β topology in Tt IleRS and the three α-helical fold in Sa IleRS. The C-terminal end of the anticodon-binding domain of Ca IleRS has a unique short α-helix (α28) insertion that is not present in other IleRSs.
Binding of Ile-AMP at the active site
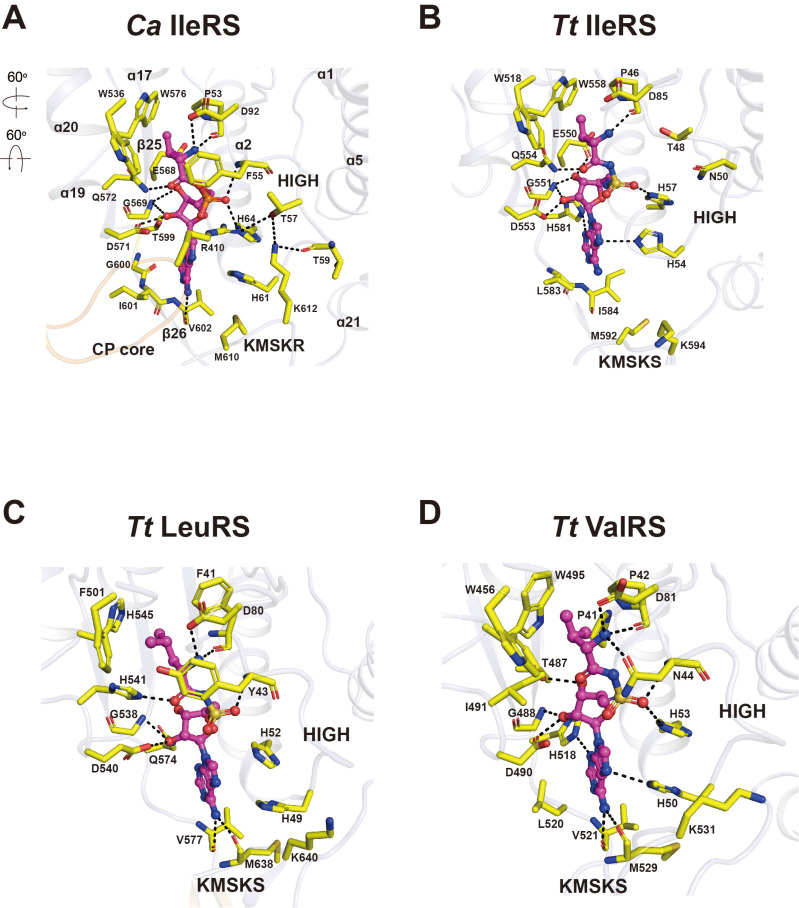
The crystal structure of Ca IleRS revealed clear electron density for Ile-AMP at the synthetic active site, although no ATP or amino acids were added during purification and crystallization (Silvian et al., 1999 Figs. 3A and 3B). The Ile-AMP molecule is bound at the central cleft between the first and second half of the catalytic domain, with the CP core positioned on top. The active site adopts a very compact conformation (Fig. 4A). The side chain of the Ile moiety is lodged at the hydrophobic pocket formed by the HIGH loop and α17 and α20 helices through van der Waals interactions. Ile-AMP resides on top of the α2 helix and the β25 and β26 strands. The backbone amide of Ile forms H-bonds with the carbonyl oxygen of Pro53 and the side chain of Asp92, and the carbonyl group of Ile forms an H-bond with Gln572.
Fig. 4. Comparison of the Ile-AMP binding in prokaryotic and eukaryotic IleRSs and class ARSs.
(A) Close-up view of the interactions between Ile-AMP and the active site residues of Ca IleRS. Phe55 of the HIGH loop covers the top of the Ile moiety. Arg410 of the CP core loop interacts with the phosphate moiety of Ile-AMP and Thr57. Lys612 of the KMSKR loop interacts with Thr57 and Thr59 of the HIGH loop. Arrows indicate the orientation of the active site relative to that of the overall structure in Fig. 3C. (B-D) Close-up view of the interaction between the cognate aa-AMP analogues and bacterial class 1a ARSs Tt IleRS (1JZQ; B), Tt LeuRS (1H3N; C), and Tt ValRS (1GAX; D). The figures are in the same orientation as in Fig. 4A.
Phe55 encloses the backbone of Ile, further stabilizing ligand binding. The space between Trp576 and the terminal methyl of Ile is optimal, preventing unfavorable interactions with Leu or Val. The phosphate oxygens of Ile-AMP are surrounded by the HIGH loop and the CP core loop linking the β16 and β17 strands. The backbone amide of Phe55 and the side chain of His64 in the HIGH loop form H-bonds with the phosphate oxygens of Ile-AMP. Arg410 in the CP core loop interacts with the phosphate oxygen and covers the ribose group. The ribose ring of Ile-AMP is recognized by the β25 strand and the α19 helix from the second half of the catalytic domain. The O2’ atom of the ribose ring are stabilized by H-bonds with Asp571 (α19) and the backbone amide of Gly569 (β25), and the O3’ atom forms an H-bond with Glu568. The adenine base of Ile-AMP is surrounded by the three regions of the catalytic domain: His61 and His64 in the HIGH loop; the backbone of Gly600 to Val602 of theβ26 strand; and Met610 of the KMSKR loop that stabilizes the adenine ring via van der Waals interactions. Lys612 (the second Lys of the 609KMSKR motif) is directed toward to the HIGH loop, and engages in H-bonds with the main chain of Thr59 and the side chain of Thr57. These interactions restrict the KMSKR loop of Ca IleRS to the closed conformation.
Comparison of synthetic active sites in eukaryotic and bacterial IleRSs
There are three notable differences in the synthetic active sites between Ca- and Tt IleRS complexed with aminoacyl-AMP: the HIGH loop; the CP core loop; and the conformation of the KMSKR loop. While the aromatic group of Phe55 covers the top of the Ile moiety in Ca IleRS, the equivalent Thr48 is far away from Ile in Tt IleRS (Fig. 4B). The highly conserved Arg410 interacts with the phosphate moiety of Ile-AMP and Thr57 in the HIGH loop, thereby shifting the CP core loop to cover the top of the active site. No such movement is ob- served in Tt IleRS, suggesting that this interaction is specific to eukaryotic IleRSs. The conformation of the KMSKR loop differs the most between eukaryotic and bacterial IleRSs. In Ca IleRS, the KMSKR loop is directed toward the adenine base, and the HIGH loop is stabilized by H-bonds, contributing to the binding of Ile-AMP, whereas in Tt IleRS, the correspond- ing Lys594 is exposed on the surface and does not interact with other residues. These key differences might uniquely contribute to the binding of Ile-AMP in the active site of eukaryotic IleRSs. There are several additional protein-ligand interactions in Ca IleRS. For example, the phosphate moiety of Ile-AMP is recognized by the main chain amide of Phe55 in Ca IleRS. Also, the carbonyl oxygen of Val602 of Ca IleRS forms a H-bond with the N6 atom of the adenine base.
Comparison of the recognition of aa-AMP between Ca IleRS and bacterial class 1a ARSs
Among the class 1a ARSs, IleRS, LeuRS, and ValRS share particularly high levels of sequence identity and are therefore thought to have evolved from a common ancestor (Brown and Doolittle, 1995). The crystal structures of Tt LeuRS and Tt ValRS have been reported in aminoacyladenylate-bound forms (Cusack et al., 2000; Fukai et al., 2000). The aminoacyl moiety binding sites in Tt LeuRS and Tt ValRS are surrounded by aromatic side chain residues that form a hydrophobic pocket, similar to that of Ca IleRS (Figs. 4C and 4D). The backbone of the Leu moiety in Tt LeuRS is covered by the aromatic group of Tyr43, similar to Ca IleRS, whereas Asn44 occupies the same position and engages in H-bonding with the amino group of the Val moiety in Tt ValRS. Although Arg410 is a highly conserved residue, the conformation of Arg410 enclosing the active site is only observed in Ca IleRS.
The conformation of the KMSKS motif in Tt LeuRS and Tt ValRS is significantly different from that in Ca IleRS (Figs. 4A, 4C, and 4D). The second Lys of the KMSKS motif is exposed to the surface in Tt LeuRS and Tt ValRS, and this loop adopts an open conformation. Nevertheless, Met (Met638 in Tt LeuRS and Met529 in Tt ValRS) and the neighboring Val residue form H-bonds with the N6 atom of adenine, partly closing the loop. Overall, eukaryotic IleRS has a much more compact active site conformation than its prokaryotic counterpart, which might explain why the reaction intermediate is trapped in the active site of Ca IleRS.
The closed conformation of the KMSKR loop contributes to the binding of mupirocin in Ca IleRS
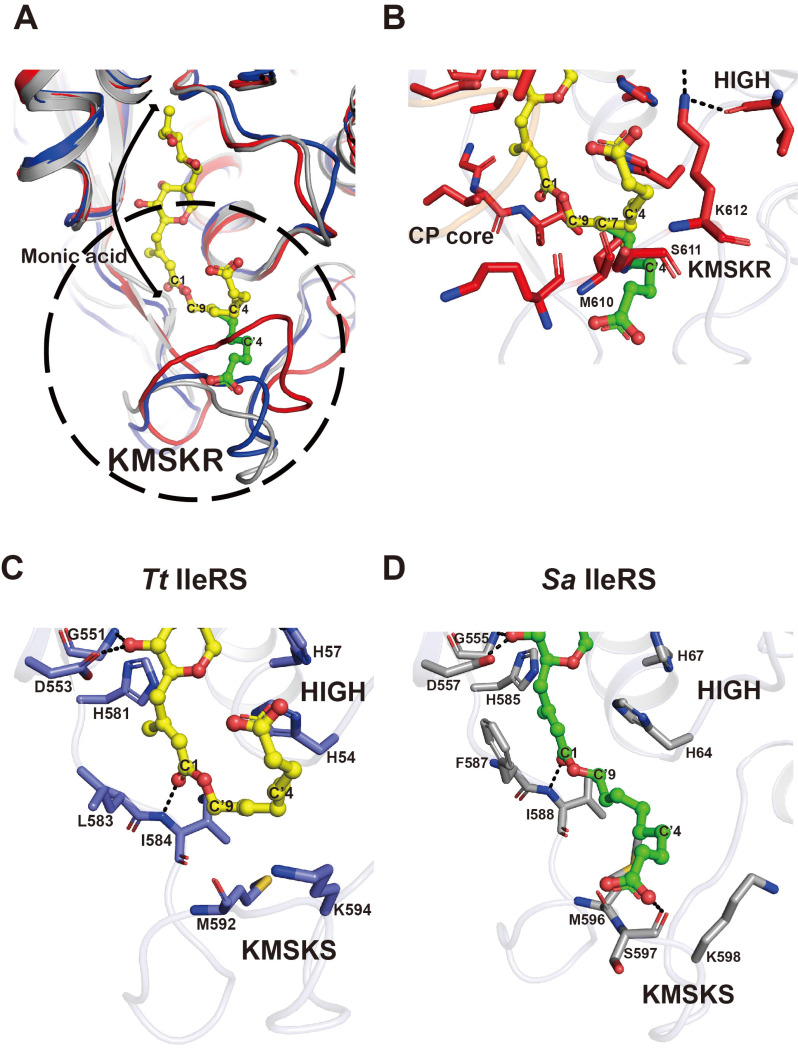
Although the catalytic domains are highly conserved among all bacterial and eukaryotic IleRSs, mupirocin selectively inhibits bacterial and archaeal IleRSs but not eukaryotic enzymes (Hughes and Mellows, 1980). The crystal structures of two bacterial IleRSs, Tt IleRS and Sa IleRS, have been reported in their mupirocin-bound forms (Nakama et al., 2001; Silvian et al., 1999). In both structures, the mupirocin molecule is tightly bound to the synthetic active site, but the conformation of the 9-hydroxynonanoic acid differs slightly, indicating that mupirocin-binding may be dynamic in nature.
The structures of apoand mupirocin-bound Tt IleRS have virtually identical open KMSKS loop conformations (Nakama et al., 2001). In both Tt and Sa enzymes, the monic acid moiety of mupirocin is recognized by an Ile-specific pocket, similar to the ribose moiety of Ile-AMS (Fig. 5A). The 9-hydroxynonanoic acid (C’2 to C’9 and the carboxyl group) of mupirocin is bent toward the opposite side of the KMSKS loop, and sandwiched between the HIGH and KMSKS motifs (Fig. 5C). By contrast, the 9-hydroxynonanoic acid of mupirocin is extended toward the KMSKS loop in Sa IleRS, and the carboxyl group of the nonanoic acid moiety is recognized by the carbonyl oxygen of Ser597 (the first Ser of the KMSKS motif; Fig. 5D). In both structures, the second Lys of the KMSKS motif is exposed to the surface, creating a space to accommodate the nonanoic acid moiety.
Fig. 5. Structural basis for the mupirocin-resistance of eukaryotic IleRS.
(A) A model of the mupirocin molecule in the active site of Ca IleRS (red). The model was generated by aligning the catalytic domain of Ca IleRS onto that of Tt IleRS (PDB 1JZS, blue) or Sa IleRS (PDB 1FFY, grey) bound to mupirocin. Two different conformations of the 9-hydroxynonanoic acid moiety are shown in yellow (a bent form from Tt IleRS) and green (an extended form from Sa IleRS). The conformation of the KMSKR loop of Ca IleRS leads to a steric clash with either model of the 9-hydroxynonanoic acid moiety of mupirocin. The region within the dotted circle is highlighted in Figs. 5B-5D. (B) Close-up view of the binding of the 9-hydroxynonanoic acid of mupirocin in the active site of Ca IleRS. (C and D) Close-up view of the interaction between mupirocin and the active site residues of bacterial IleRSs, Tt IleRS (C), and Sa IleRS (D). The figures are shown in the same orientation as in Fig. 5B.
When we modeled mupirocin in Ca IleRS by aligning the catalytic domain of Ca IleRS onto the mupirocin-bound Tt IleRS and Sa IleRS structures (rmsd values of 1.4 Å and 2.5 Å, respectively, for Cα atoms), the KMSKR loop of Ca IleRS (red) was too close, which would limit the efficient binding of mupirocin (Fig. 5A). The monic acid moiety of mupirocin that resembles the Ile and ribose moieties of Ile-AMP can be accommodated in the synthetic active site of Ca IleRS (Fig. 5A). However, despite the flexibility of the 9-hydroxynonanoic acid of mupirocin, it would be difficult for this fragment to adopt an optimal position in the active site of Ca IleRS (Fig. 5B). The C’7 atom of the nonanoic acid moiety is only 1.8 Å away from the carbonyl oxygen and 2.7 Å away from the Cα atom of Met610 in Ca IleRS, and Ser611 and Lys612 of the KMSKR motif of Ca IleRS are also too close to the C’4 atom of the nonanoic acid moiety. Thus, the closed conformation of the KMSKR loop of Ca IleRS might explain the relatively weak affinity of mupirocin toward eukaryotic IleRSs, and thus provide the basis for mupirocin-resistance in eukaryotic IleRSs.
In summary, we determined the structure of fungal IleRS and analyzed structural differences between prokaryotic and eukaryotic IleRSs. We showed that three key features cause the active site to adopt a compact conformation and thereby restrict the release of the Ile-AMP reaction intermediate. Binding of tRNA at the catalytic and editing domains might rearrange the orientation of the editing domain, resulting in the opening the active site conformation to allow ligation of an aminoacyl group to the 3’ acceptor of tRNA (Supplementary Fig. S2). Tight binding of Ile-AMP at the compact active site would limit the release of Ile-ATP, and thus allowing eukaryotic IleRSs to use ATP energy more efficiently in aminoacylation and tRNA charging processes. In addition, the structural basis for mupirocin action presented herein provides new insights that could be used to develop improved anti-infective and anti-fungal drugs.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korea government (MEST, No. 2015R1A2A1A05001694, 2017M3A9F6029733, and NRF-2013M3A6A4044580), and BK21 program (Ministry of Education) to YC.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A., Green E.D. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- Brown J.R., Doolittle W.F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk A., Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 1995;5:48–55. doi: 10.1016/0959-440X(95)80008-O. [DOI] [PubMed] [Google Scholar]
- Duhr S., Braun D. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19678–19682. doi: 10.1073/pnas.0603873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fersht A.R., Dingwall C. Evidence for the Double-Sieve Editing Mechanism in Protein Synthesis. Steric Exclusion of Isoleucine by valyl-tRNA Synthetases. Biochemistry. 1979;18:2627–2631. doi: 10.1021/bi00579a030. [DOI] [PubMed] [Google Scholar]
- Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D.G., Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/S0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Fukunaga R., Yokoyama S. Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J. Mol. Biol. 2006;359:901–912. doi: 10.1016/j.jmb.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Hughes J., Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem. J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., Mellows G. Interaction of pseudomonic acid A with Escherichia coli B isoleucyl-tRNA synthetase. Biochem. J. 1980;191:209–219. doi: 10.1042/bj1910209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus P.A., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz I.D. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- Kwon N.H., Fox P.L., Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019;18:629–650. doi: 10.1038/s41573-019-0026-3. [DOI] [PubMed] [Google Scholar]
- Ling J., Reynolds N., Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- Nakama T., Nureki O., Yokoyama S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem. 2001;276:47387–47393. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- Nureki O., Vassylyev D.G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T.L., Schimmel P., Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pei J., Kim B.H., Grishin N.V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas de Pouplana L., Schimmel P. Aminoacyl-tRNA synthetases: potential markers of genetic code development. Trends Biochem. Sci. 2001;26:591–596. doi: 10.1016/S0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- Seidel S.A., Dijkman P.M., Lea W.A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., Joseph J.S., Srinivasan P., Baaske P., Simeonov A., et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvian L.F., Wang J., Steitz T.A. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. doi: 10.1126/science.285.5430.1074. [DOI] [PubMed] [Google Scholar]
- Sutherland R., Boon R.J., Griffin K.E., Masters P.J., Slocombe B., White A.R. Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob. Agents Chemother. 1985;27:495–498. doi: 10.1128/AAC.27.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P., Fox P.L. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 2013;5:332–343. doi: 10.1002/emmm.201100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.