Abstract
Inflammation is closely related with the progression of cancer and is an indispensable component that orchestrates the tumor microenvironment. Studies suggest that different mediator and cellular effectors, including cytokines (interleukins, tumor necrosis factor-α [TNF-α], transforming growth factor-β [TGF-β], and granulocyte macrophage colony-stimulating factor [GM-CSF]), chemokines, as well as some transcription factors (nuclear factor κB [NF-κB], signal transducer and activator of transcription 3 [STAT3], hypoxia-inducible factor-1α [HIF1α]), play a crucial role during cancer-related inflammation (CRI). MicroRNAs (miRNAs) are the key components of cellular physiology. They play notable roles during posttranscriptional gene regulation and, thus, might have a potential role in controlling the inflammatory cascade during cancer progression. Taking into consideration the role identified for miRNAs in relation to inflammatory cytokines, we have tried to review their participation in neoplastic progression. Additionally, the involvement of miRNAs with some important transcription factors (NF-κB, STAT3, HIF1α) and proteins (cyclooxygenase-2 [COX-2], inducible nitric oxide synthase [iNOS]) closely associated with inflammation during cancer has also been discussed. A clear insight into the responsibility of miRNAs in cytokine signaling and inflammation related to CRI could project them as new therapeutic molecules, which could lead to improved treatment of CRI in the near future.
Keywords: miRNA, cancer, inflammation, cytokines, tumor microenvironment
Graphical Abstract
Cancer is a public health issue of serious concern throughout the world. MicroRNAs (miRNAs) are involved with the regulation of posttranscriptional gene regulation and thus might participate in the neoplastic progression. miRNAs might be involved in the regulation of inflammatory cytokines, transcription factors (NF-κB, STAT3, HIF1α), and inflammatory proteins (COX-2, iNOS), closely associated with inflammation during cancer.
Main Text
Cancer is a public health issue of serious concern, as it represents the major cause of deaths throughout the world. GLOBOCAN 2018 estimated approximately 8.1 million new cancer cases globally.1 Among the various types of cancer, lung, prostate, breast, stomach, liver, pancreatic, and colon cancers are the leading cause of deaths throughout the globe.2 However, studies have shown that chronic inflammation and related infections influence the cause of various types of cancer and are the underlying reason for 15%–20% of deaths from cancer.3,4
Cancer was linked to inflammation for the first time in the 19th century when, in 1863, Rudolf Virchow hypothesized the functional relationship of chronic inflammation to the development of cancer.3 It has been observed that the cause of inflammation may range from microbial infections to exposure to chemicals and allergens to various pathological conditions. Inflammation is a double-edged sword, having a very delicate balance between the development of an anti- or pro-tumorigenic environment. A well-regulated acute inflammation is considered as anti-tumorigenic, whereas unregulated chronic inflammation develops a pro-tumorigenic environment.4,5 It was observed that chronic infection with inflammation develops into ∼25% of all cancer cases throughout the world.6,7 Takahashi et al.8 showed that the presence of high carcinogenic substances is a source of tumor initiation due to their ability to cause chronic inflammation. Tobacco smoke is one such example of a potent tumor promoter with the capability of activating chronic inflammation. These observations have led to the idea that different cancers might develop from inflammation and continual irritation.9 Evidence based on various findings, ranging from molecular studies to epidemiological research, has documented the relationship between cancer and inflammation.
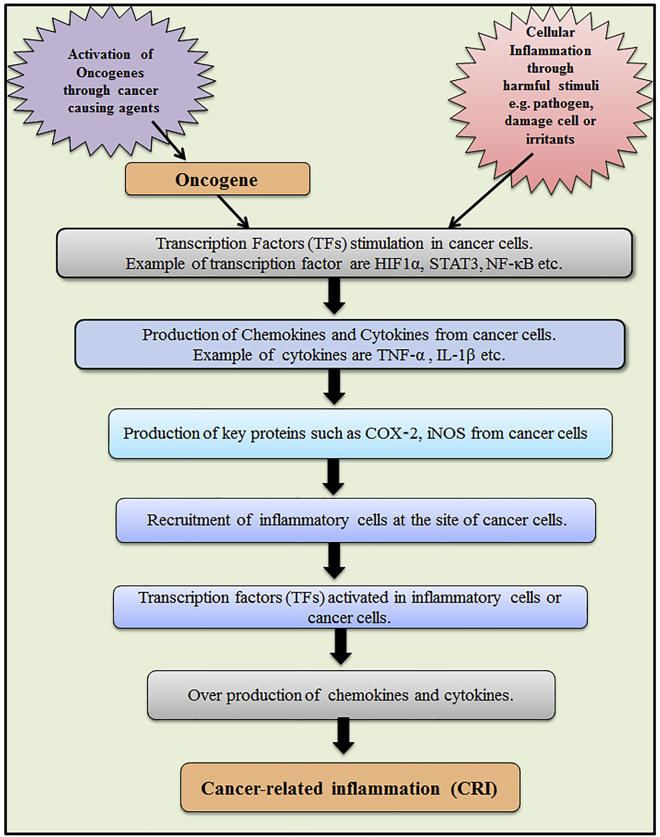
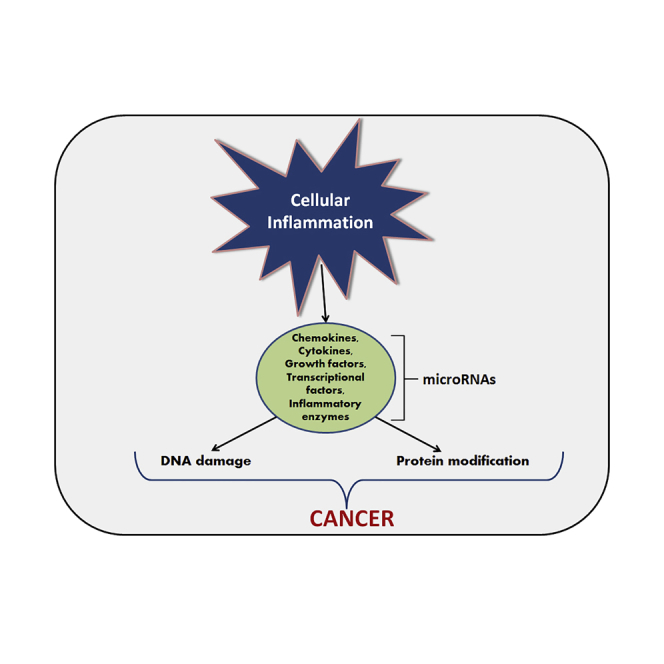
It has been suggested that cancer-related inflammation (CRI) is one of the most important physiological parameters in the diagnosis of malignancy. It is well documented that CRI is triggered by several factors, including chemokines and cytokines (Figure 1).10 Different inflammatory cytokines are dysregulated during cancer that might act as a biomarker for detecting cancer. However, comprehensive studies are requisite to recognize the level of inflammatory cytokines before considering them as biomarkers.
Figure 1.
Pathway Depicting the Process of the Cancer-Related Inflammation and Mediators Involved during This Cascade
MicroRNAs (miRNA) are single-stranded non-coding RNAs that are also called ncRNAs, containing a small number of nucleotides, usually 21–23 nt. The miRNA controls target gene expression, particularly via the cleavage procedure or by modulation of target proteins at the translational level.11, 12, 13 Additionally, it has been observed that miRNAs are involved in gene silencing at the co-translational stage (enrolling protein decay factors such as exosomes) and even at the pre-translational stage (during chromatin remodeling).14 The first miRNA that was discovered is called lin-4, which was found to control the developmental timing in C. elegans (nematodes) by targeting the mRNA of a key developmental protein called lin-14.15,16 Thereafter, an additional miRNA (let-7) was also found to be a contributing factor in the development of C. elegans.17 It was observed that these small miRNAs are not only present in C. elegans but also exist in murine and human cells. The sequences of these miRNAs were found to be highly conserved.18 Studies have revealed that miRNAs encompass approximately 1% of the total human genome, and the miRNA family is among the most crucial gene families.19 Researchers have been depositing miRNA sequences into databases, and these miRNA sequences are available from public databases.20 The name of the miRNA is allocated by following a common nomenclature system after the discovery of new miRNAs.21
Researchers have perceived that miRNAs are potential molecules that might play a crucial role in controlling cellular pathways and might play a noteworthy role in health and diseases.22,23 The role of miRNAs in regulating the mitogen-activated protein kinase (MAPK) signaling pathway during chronic myeloid leukemia (CML) has been evaluated.24,25 Additionally, a link between insulin resistance in type 2 diabetes (T2D) and inflammation has been established.26 Also, miRNAs linked with the key controller proteins of insulin resistance and the insulin signaling pathway have also been assessed.27 Chakraborty et al.28 discussed the accountability of miRNAs associated with key regulator proteins of the insulin signaling pathway and pancreatic cancer development. An emphasis on the role and responsibility of miRNAs in regulating cancer stem cells further demonstrates the significance of miRNAs in carcinogenesis.29 The function of miRNAs in cytokine signaling pathways related to inflammation during rheumatoid arthritis was also reviewed.30 It has been projected that chemotherapeutic agents might exert their anticancer activity by modulating miRNA expression.31 The role of miRNA in CRI has also been reported in esophageal cancer32 and oral squamous cell carcinoma (OSCC).33
Considering the regulatory role of miRNAs in the onset of inflammation and carcinogenesis, we have tried to summarize the regulatory abilities of miRNAs in major cytokine signaling pathways. An insight into the modulatory role of miRNAs in controlling the key inflammatory cytokine signaling pathways could help us to understand CRI. In addition, we have depicted a snapshot about the role of miRNAs in the regulation of some other key protein factors (other than the cytokine) in inflammation related to cancer.
Predisposing Factors, Inflammatory Factors, and Pathways Associated with CRI
Several inflammatory pathologies may lead to increased risk of cancer, such as inflammatory bowel diseases (IBDs), and chronic gastritis caused by H. pylori might lead to gastric cancer, UV irradiation-associated skin inflammation might lead to skin cancer, gall bladder stone-associated chronic cholecystitis might lead to gall bladder cancer, and infection with hepatitis B virus (HBV) and HCV might lead to liver cancer.34 Several pathways linking cancer and inflammation have been recognized and are known as intrinsic or extrinsic pathways depending on the extent of their involvement. The intrinsic pathway is associated with the activation of genetic events that initiate pro-inflammatory programs, which guide the development of the inflammatory microenvironment. The tyrosine kinases pathway and the Ras-Raf pathway are examples of intrinsic pathways associated with inflammation and oncogenes. Conversely, the extrinsic pathway is associated with the inflammatory conditions leading to cancer development.35 In the extrinsic pathway, inflammatory leukocytes and soluble mediators are the driving forces for the establishment of the inflammatory microenvironment. The inflammatory mediators are the key controllers at the junction of intrinsic and extrinsic pathways.
Various types of inflammatory cells are known to be the essential components of the tumor microenvironment participating in CRI. The tumor microenvironment consists of a number of different inflammatory cell types, including cancer-associated fibroblasts (CAFs), stromal cells, tumor-associated macrophages (TAMs), infiltrating immune cells, pericytes, and endothelial cells comprising the tumor vasculature.36 Tumor cells communicate with these inflammatory cells, via secreting cytokines and growth factors, to stimulate tumor growth and develop resistance against chemotherapy.
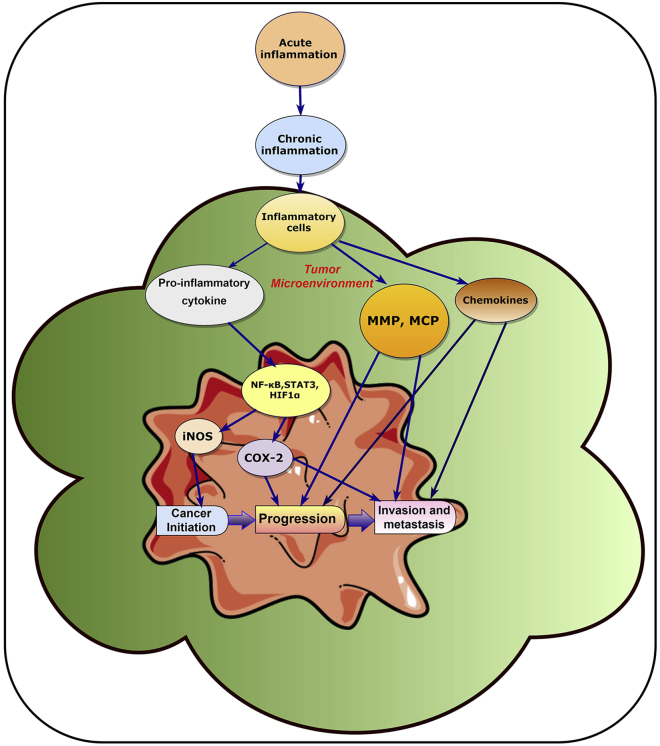
It has been demonstrated that several endogenous and exogenous inflammatory mediators/factors contribute to CRI via crosstalk between the inflammatory cells and tumor cells (Figure 2).10 It is well known that chemokines, cytokines, and several transcription factors act as inflammatory mediators and are important in the development of the inflammatory microenvironment, leading to increased risk of cancer development. Thus, cytokines are among the key machineries of CRI.37 The cytokines include interleukins (ILs), especially IL-1, IL-6, IL-17, and IL-23, tumor necrosis factor (TNF)-α, transforming growth factor (TGF)-β, growth factors, and colony-stimulating factors, which are either membrane-bound or are secreted molecules. These molecules have functional roles in proliferation, activation, and differentiation of cancer and immune cells.34,38 Chemokines are the major family of cytokines that are important in the development of CRI. Based on the relative position of their first two cysteine residues, chemokines can be categorized into four groups (CC, CXC, XC, and CX3C). Among these, the chemokines CCL2 and CXCL8 have a significant role in CRI.10,39 However, in addition to cytokines and chemokines, several other key endogenous factors, such as transcription factors, are also associated in the development of CRI, e.g., nuclear factor-κB (NF-κB), hypoxia-inducible factor-1α (HIF1α) and signal transducer and activator of transcription 3 (STAT3). Hence, intervention in the communication between cells and molecules within the tumor microenvironment can be an effective strategic approach for the development of effective therapies against cancer.
Figure 2.
Summary of the Inflammatory Microenvironment in Cancer Development
The tumor microenvironment plays an important role in the process of cancer development. In this microenvironment, pro-inflammatory cytokine (secreted by the inflammatory cell) modulates NF-κB, STAT3, and HIF1α, which activates iNOS and COX-2. iNOS and COX-2 proteins help in the cancer initiation and progression process. The inflammatory cells present in the inflammatory microenvironment also activates the production of matrix metallopeptidases (MMPs), monocyte chemoattractant protein (MCP), as well as cytokines, which results in cancer progression, invasion, and metastasis.
The Role of Extracellular miRNAs in Establishing Tumor Microenvironment
During inflammation, the expression pattern of miRNAs is found altered, suggesting that miRNAs might offer a link between inflammation and cancer. Moreover, it has been noted that some miRNAs have a prominent role in the regulation of innate immunity.40 Upregulated or downregulated miRNAs can lead to the development of immune tolerance, potential autoimmunity, a hyper-inflammatory phenotype, and cancer commencement and progression. Inflammatory mediators, chemokines, cytokines, and different proteins can regulate miRNA expression, which may contribute to the regulation of multiple genes and the gene regulatory networks associated with inflammation and oncogenesis. Indeed, miRNAs have also been found crucial for the development, differentiation, function, as well as survival of different cell types such as T and B lymphocytes, macrophages, dendritic cells, and other immune cells.41
Several miRNAs are released from cells and have a functional role to establish the tumor microenvironment.42 These extracellular miRNAs can perform their function not only on cancer cells but also on the endothelial cells, immune cells, and fibroblasts present in the tumor microenvironment. It has been noted that extracellular miRNAs have an influence on tumor progression through bidirectional communication such as stromal-to-tumor and tumor-to-stromal communication.43 The route by which miRNAs are released in the extracellular environment is multifarious, including exosomes. Some miRNAs mediate the functioning of the extracellular vesicle in the tumor microenvironment. For example, miR-1 is known to mediate extracellular vesicle function, which is downregulated in glioblastoma multiforme (GBM).44
Extracellular miRNAs can enter tumor cells, immune cells, or stromal cells present within the tumor microenvironment, where they can reprogram the cell transcriptome, resulting in alterations in the growth of tumor and metastasis by affecting cell growth, cell differentiation, migration, and angiogenesis. Some of the miRNAs involved in migration and metastasis are miR-200, miR-10b, and miR-21. miRNAs involved in angiogenesis are miR-16, miR-92a, miR-21, and miR-494.42 In addition, some exosomal miRNA are known to modulate the tumor immune response. For example, IL-4-stimulated macrophages have been found to secrete exosomes containing oncogenic miRNAs (miR-223), which helps in the invasiveness of breast cancer cells.45 It has also been demonstrated that when exosomal miR-21 attaches with Toll-like receptor 7/8 (TLR7/8) on macrophages, it helps to promote the secretion of IL-6, leading to enhancement of the pro-inflammatory response. As reported, certain miRNAs either get upregulated (e.g., Let-7g, miR-26b, miR-101, miR-141, miR-200b, miR-200c, miR-205, and miR-342-3p) or downregulated (e.g., miR-31-3p, miR-221-5p, and miR-221-3p) in CAFs.46 However, further investigations are needed to bring deep insight into the particular involvement of these extracellular miRNAs in the tumor microenvironment.
Role of miRNAs in CRI and Cytokine Signaling
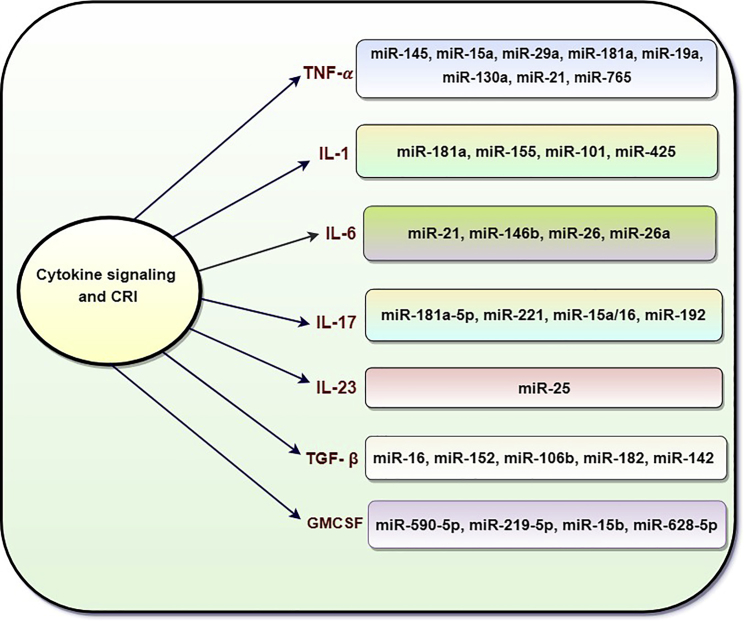
As discussed earlier, cytokines are key players of inflammation that might contribute in acute and chronic inflammation. Cancer progression is being controlled by immune cells, which are regulated by the cytokines in the tumor microenvironment.34,47 Evidence has revealed that miRNAs regulate the genes associated with secretion of different cytokines,48 suggesting that cytokine-mediated CRI is well regulated (Figure 3). Some of the inflammatory cytokines and associated miRNAs responsible for the pathogenesis of CRI are summarized below.
Figure 3.
Schematic Diagram Depicting miRNA-Controlled Cytokine Cascades in Cancer-Related Inflammation (TNF-α, IL-1, IL-6, IL-17, IL-23, TGF-β, GM-CSF)
TNF-α
TNF-α, a potent cytokine, acts as an inflammatory mediator that is secreted by diverse types of cells such as macrophages, T and B cells, fibroblasts, and monocytes. TNF-α was primarily identified as an endotoxin-induced serum factor in the 1970s.49 Thereafter, the TNF-α gene was cloned in a cell line and the protein was characterized after purification. The characterized protein was then made commercially available.50 It has been suggested that TNF-α is an important mediator of inflammation and apoptosis.51 Based on the cell type, TNF-α interacts with either TNF-receptor 1 (TNFR1) or TNFR2 and can stimulate different cell processes such as necrosis, apoptosis, immune cell activation, differentiation, angiogenesis, and cell migration. These cellular activities are of great significance in tumor immune surveillance. When TNF-α binds to TNFR1, it can activate survival of the NF-κB pathway by sequential recruitment of TNFR-associated death domain (TRADD) and TNFR-associated factors and receptor-interacting protein (RIP). On the contrary, TNFR1 may also activate the caspase-dependent apoptotic pathway by stimulating the formation of the TRADD-Fas-associated death domain (FADD)-pro-caspase-8 complex. Therefore, being associated with critical cellular behavior, TNF-α can be held responsible for both pro- and anti-tumoral effects.52,53
miR-145 is assumed to be a tumor suppressor and it is downregulated in multiple cancers.54 Zheng et al.55 showed that miR-145 is downregulated in triple-negative breast cancer (TNBC) tissue and the MDA-MB-231 cell line, while miR-145 is overexpressed in MDA-MB-231 cells treated with TNF-α, resulting in induced cell death and apoptosis. The co-immunoprecipitation assay showed that miR-145 facilitates the formation of the RIP1-FADD-caspase-8 apoptotic complex induced by TNF-α, leading to a caspase-8-mediated apoptotic signaling pathway. They further demonstrated that cellular inhibitor of apoptosis protein-1 (cIAP), an apoptotic inhibitor, is a target for miR-145. cIAP ubiquitinates RIP1 and prevents apoptosis via TAK1 and NF-κB, Thus, miR-145-mediated downregulation of cIAP1 promotes de-ubiquitination of RIP1 and formation of a complex with caspase-8 and FADD, suggesting that miR-145 might regulate TNF-α-induced apoptosis in TNBC.
B cell chronic lymphocytic leukemia (CLL) is a well-known leukemia type, being prevalent in different parts of the world, particularly in the Western world. Higher levels of TNF-α have been observed in CLL patients.56,57 Some miRNAs have the ability to regulate the TNF/TNFR gene superfamily in CLL. Srivastava et al.58 performed in silico target prediction and elucidated that miR-15a, miR-29a, and miR-181a directly target and deregulate members of TNF ligands (i.e., TNF, TNFR1, and LTβR [lymphotoxin β receptor]), a member of the TNFR superfamily (i.e., TNFRSF1A), and adaptor molecules (i.e., TNFR-associated factors [TRAFs] 3–6). These members of the TNF/TNFR superfamily and adaptor molecules are known to activate the NF-κB survival pathway. Therefore, miRNA-mediated targeting of the TNF/TNFR superfamily might be a possible therapy against leukemia.
TNF-α is also associated with colorectal cancer (CRC) progression. It has been shown that the level of TNF-α is elevated in CRC patients with lymph node metastasis. One of the critical molecular steps during metastasis that facilitates epithelial cells to metastasize to distant sites is the epithelial-to-mesenchymal transition (EMT). Growing evidence suggests that miRNAs might regulate the genes related to EMT. Upregulation of miR-19a in CRC tissue and a relationship between the elevated level of miR-19a expression and lymph node metastasis in CRC cell lines were observed by Huang et al.59 The study also showed that miR-19a expression is increased by TNF-α, while in turn it represses TNF-α formation by a negative feedback loop; however, the exact mechanism is still not clear. This study also found that an increased level of miR-19a is associated with a decreased level of epithelial marker (E-cadherin) and an increased level of mesenchymal markers (N-cadherin, fibronectin, and vimentin) in CRC cells, suggesting induction of EMT.
Zhang et al.60 observed that TNF-α stimulates the nuclear translocation of NF-κB followed by the induction of miR-130a expression and downregulation of TNF-α. However, more mechanistic insight needs to be explored. They suggested a negative feedback loop of NF-κB/miR-130a/TNF-α in cervical cancer cells that might be responsible for the low level of TNF-α, resulting in carcinogenesis by avoiding apoptosis. Another miRNA, miR-21, also serves as a regulator of apoptosis along with cancer cell proliferation and is being suggested as a potential marker for detecting metastasis.61 Inhibition of phosphatase and tensin homology (PTEN) expression could be a plausible mechanism regulated by overexpressed miR-21 that affects cervical cancer cell migration and proliferation.62
Although it has been shown that miR-21-mediated upregulation of TNF-α has a positive effect on HeLa cervical cancer cell proliferation, it exerts no effect on cell apoptosis.63 According to Zheng et al.,64 overexpression of TNF-α increases expression of miR-765, which reduces cancer cell migration, probably by directly repressing EMP3 translation and consequent upregulation of p66Shc. However, note that in Zheng et al.’s work a high dose of TNF-α (100 ng/mL) was used, which demonstrated an inhibitory effect on migration of the OSCC line instead of a stimulatory effect.65 Therefore, as TNF-α is a double-edged sword, understanding the crosstalk between various miRNAs and TNF-α and their roles in inducing different signaling pathways in cancer biology might assist in developing promising cancer therapeutic agents.
IL-1
IL-1 was first discovered at the beginning of the 1940s, and since then it has become one of the significant pro-inflammatory cytokines in medical science. The IL-1 family includes eleven members. Among them, seven members are ligands with agonist activity, i.e., IL-1 (α, β), IL-33, IL-18 and IL-36 (α, β, γ). Another three members (IL-1Ra, IL-38, and IL-36Ra) are receptor antagonists. Only IL-37 is an anti-inflammatory cytokine.66 As reported, the pro-inflammatory cytokine IL-1 has two important forms, i.e., IL-1α and IL-1β.67 The genes of these two cytokines are located on chromosome 2 and are in close vicinity, sharing ∼27% aa sequence homology.68 In comparison to TNF-α, the inflammatory signaling mechanisms of IL-1α and IL-1β are much more complex. It has been noticed that these signaling mechanisms are mediated through cell-surface receptors.37 IL-1 was found to modulate the expression of genes that are known to mediate inflammation and cancer.69 In light of the current therapeutic approaches, a correlation between inflammation and tumorigenesis has also been exploited.
An elevated expression level of NF-κB (Rel A), IL-1β, and miR-181a in colon cancer was noted by Hai Ping et al.70 in colon cancer. It was observed that IL-1β induced the expression of miR-181a via the NF-κB signaling pathway, and miR-181a was able to promote cell proliferation by repressing PTEN, which is an important tumor suppressor and is frequently mutated in various cancers.
In another study, IL-1β upregulated the expression of miR-425, mediated by NF-κB signaling, in gastric cancer cells. Overexpressed miR-425 promoted the growth of gastric cancer cells by negatively regulating PTEN.71 The expression of another miRNA, miR-155, was induced by IL-1β in melanoma cells, where miR-155 mediated the downregulation of MITF-M (microphthalmia-associated transcription factor).72 MITF-M, a key transcription factor, is expressed solely in melanocytes and has the ability to regulate several genes responsible for differentiation, survival, and proliferation.
It was observed that silica particle-induced IL-1β secretion downregulated miR-101 and subsequently increased the expression of enhancers for zeste homolog 2 (EZH2), conferring metastasis and cancer cell proliferation in the Xuan Wei lung cancer cell line. Conversely, miR-101 suppresses translation of EZH2, attenuating cell growth and migration.73 Similarly, in another study, Wang et al.74 observed that the level of IL-1β is highly increased in patients with non-small-cell lung cancer (NSCLC). IL-1β repressed the expression of miR101 and caused an upregulation of the miR-101 target gene, Lin28B, a suppressor of the tumor-suppressive let-7 family of miRNAs. The IL-1β/miR-101/Lin28B pathway is dependent on cyclooxygenase-2 (COX-2) activity and promoted proliferation and migration of NSCLC cells. In conclusion, this pathway links inflammation signaling to cancer cell proliferation and migration in NSCLC and thus may in part explain inflammation-promoted tumorigenesis.
IL-6
IL-6 is a pleiotropic inflammatory cytokine produced by T helper cells, synovial fibroblasts, monocytes, and macrophages.75 IL-6 has various effects on the cells of the immune system and is essential for the regulation of a variety of immune functions such as immunoglobulin G (IgG) production and plasma cell differentiation.76 An elevated IL-6 level has been observed in the serum of patients with cancer inflammation,77 suggesting that overproduction of this protein cascade leads to an inflammatory condition. Besides inflammation, IL-6 is also involved in cancer progression.78,79
Dong et al.80 described the role of IL-6 and miR-21 on programmed cell death 4 (PDCD4) gene expression in the prostate cancer cell lines PC-3 and LNCaP. PDCD4 is a suppressor of tumor progression. Prostate cancer is among the prevalent cancers in the US. According to their study, miR-21 can reduce the expression of PDCD4 by expressing IL-6 in prostate cancer cells.80 PDCD4 binds with translation initiation factors, such as eIF4G and eIF4A, to inhibit the process of translation, resulting in the inhibition of pro-oncogenic factors.
In another study, Xiang et al.81 observed that IL-6 activates STAT3. Constitutive activation of STAT3 in cancer promotes cell proliferation, survival, invasion, and angiogenesis. The study reported that in normal tissues, STAT3 can transcriptionally upregulate miR-146b, which further suppresses the activation of NF-κB by translational repression of the NF-κB activators, i.e., TRAF6 and IL-1R-associated kinase (IRAK1). Consequently, IL-6, an NF-κB target gene, is downregulated, preventing autocrine stimulation of STAT3 in a feedback loop. However, in cancer cells, the tumor-suppressive function is lost by promoter methylation of the miR-146b gene, which may result in persistent inflammation, prolonged NF-κB-IL-6-STAT3 activation, and subsequent tumor progression.81,82
Furthermore, it has been shown that miR-26 might control tumorigenicity and inflammation by downregulating the secretion of IL-6 and NF-κB signaling.83 Chen et al.83 observed that instead of directly targeting IL-6, miR-26 reduces IL-6 transcription triggered by TNF-α through silencing NF-κB signaling related factors high mobility group AT-hook 1 (HMGA1) and mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1) in adenocarcinomic alveolar basal epithelial A549 cells. In contrast, Jones et al.84 observed that miR-26 directly targets IL-6 3′ UTR and silences IL-6 expression in A549 cells. This raises the possibility that miRNAs might act as a cytokine silencer, either by acting indirectly on signaling pathways or by directly acting on cytokine transcripts.
It has been observed that the upregulation of IL-6 results in a decrease of miR-26a expression in hepatocellular carcinoma (HCC) cells.85 In addition, miR-26a was found to repress the tumor growth and metastasis of human HCC in the nude mice model, and the tumor suppresser effect of overexpressed miR-26a is parallel to that of genetic inhibition of IL-6.86 It was suggested that miR-26a targets IL-6 and inhibits IL-6/STAT3 signaling. It is plausible that suppression of STAT might inhibit the transcription of anti-apoptotic genes, such as Bcl-2, Mcl-1, cyclin D1, and matrix metallopeptidase 2 (MMP2). Therefore, miR-26a possibly blocks the G1/S transition and promotes apoptosis in HCC cells. However, the molecular machinery by which miR-26a inhibits HCC remains unclear.86
IL-17
A pro-inflammatory cytokine, IL-17, is also secreted by a diversity of cells, including activated T helper cells, natural killer (NK) cells, macrophages, dendritic cells, γδ-T cells, and lymphoid tissue inducers.87,88 As IL-17 regulates the activities of the NF-κB and MAPK pathway proteins,89 it has been related to the progression of various inflammatory diseases, such as autoimmune disease.90,91 An association between IL-17 and cancer-associated inflammation has also been observed.91 Cao et al.92 suggested a negative correlation between miR-181a-5p and IL-17 in NSCLC and found that IL-17 upregulates vascular cell adhesion molecule 1 (VCAM-1) and downregulates miR-181a-5p via the NF-κB pathway. Furthermore, IL-17-induced VCAM-1 expression was diminished by the transfection of miR-181a-5p mimic. The suppression of VCAM-1 is possibly due to the direct binding of miR-181a-5p to 3′ UTR of VCAM-1. Here, miR-181 functions as a tumor suppressor by reducing tumor cell proliferation and migration.92 VCAM-1 is associated with pathophysiological conditions, such as autoimmune diseases, infection tumor progression, and metastasis. Therefore, miR-181a-5p-mediated targeting of VCAM-1 could be a therapeutic approach against NSCLC progression under the influence of IL-17.
In another study, it was found that the expression levels of IL-17 and miR-221 were increased in papillary thyroid carcinoma (PTC).93 It was found that IL-17 and miR-221 were positively correlated to TNM (tumor, node, metastasis) staging, capsular invasion, and lymph node metastasis. Multiple myeloma (MM) is a type of cancer-related to plasma cells and is a type of white blood cell (WBC) cancer. Bone marrow-derived mononuclear cells (BM-MNCs) isolated from MM-positive patients showed upregulation in vascular endothelial growth factor (VEGF)-A (an angiogenic growth factor) and suppressed miR-15a/16 expression, suggesting the association of miR-15a/16 with the progression of MM.94 Furthermore, it was observed that miR-15a/16 decreased the proliferation of human MM U266 cells by downregulating Bcl-2 (an anti-apoptotic factor). miR-15a/16 was also found to repress VEGF-A at the post-transcriptional level and decrease IL-17 expression, revealing the role of miR-15a/16 against angiogenesis and enhancing antitumor immune response, respectively.94 As both VEGF and Bcl-2 are regulated by the STAT3 signaling pathway, the role of miR-15a/16 in targeting STAT3 cannot be overruled. In accordance, another study demonstrated that IL-17 facilitated cell proliferation and migration and inhibited apoptosis by triggering the regulatory feedback loop involving miR-192-targeted IL-17Rs (IL-17RE and IL-17RA) in MM cells.95 IL-17 is also known to induce NF-κB96 and MAPK97; therefore, it is possible that miR-192 is also regulated by IL-17 via the upstream binding site of NF-κB or MAPK. Another possibility is that IL-17-induced IL-6 activates STAT3, which might subsequently result in cancer pathogenesis.98
IL-23
IL-23 is a heterodimeric pro-inflammatory type 1 cytokine that plays a critical role in tumorigenesis by inducing CRI.99,100 IL-23 is composed of the IL-12/p40 subunit and p19 subunit belonging to the superfamily of IL-6. It has been documented that overexpression of IL-23 was observed during various cancer conditions, and was shown to promote tumor metastasis.101, 102, 103 For example, IL-23 was found to facilitate metastasis in hepatocellular carcinoma via NF-κB-mediated upregulation of MMP9.102 Similarly, IL-23-induced metastatic melanoma brain has also been reported via the upregulation of MMP2.104 Members of the MMP family are associated with cancer invasion and angiogenesis, and thus upregulation of MMPs might be a possible mechanism elicited by IL-6-induced cancer cells for invasion and migration. Suzuki et al.103 showed the role of IL-23 in inducing proliferative and invasive activities of the colorectal carcinoma DLD-1 cell line via TGF-β production.
It has been speculated that IL-23 regulates miRNA expression. For example, IL-23 induces the expression of the miR-133B/miR-206 cluster in IL-17-producing T cells, contributing to T cell differentiation.105 Indeed, it was shown that miR-133B/miR-206 and IL-17A share regulatory elements and are co-regulated by IL-23 via the STAT3 signaling mechanism. Also, IL-23 was found to upregulate the expression of miR-25 in thyroid cancer cells (TCCs).106 Overexpressed miR-25 inhibits suppressor of cytokine signaling (SOCS) by directly binding to its 3′ UTR region and promotes the invasion and migration of TCCs,106 suggesting SOCS-mediated negative feedback regulation of IL-23-induced cancer progression.
TGF-β
TGF-β is a multifunctional cytokine that signals via protein kinase receptors.107 TGF-β promotes or inhibits tumorigenesis by regulating carcinoma initiation, metastasis, and progression.108 The importance of TGF-β has been established in the inflammation associated with cancer.108 Wang et al.109 showed that miR-16 inhibits TGF-β1-induced EMT through the activation of autophagy in NSCLC cells. Conversely, it has been noted that TGF-β controls human leukocyte antigen (HLA)-G expression in gastric cancer (GC) cells, where miR-152 plays a key role. TGF-β stimulates HLA-G expression in the course of inhibiting miR-152 in GC, and it was suggested that miR-152 might have a therapeutic role in the treatment of GC.110 TGF-β has an effect on cancer cell propagation, especially in breast cancer cells. miR-106b is upregulated by TGF-β1 in highly invasive breast cancer cells. Enhanced levels of miR-106b define the influencing paradox of TGF-β in breast cancer cells.111 In another study, Qiu et al.112 demonstrated that TGF-β plays a noteworthy function in tumor metastasis through the expression of miRNA. miR-182 is a key molecule in the regulation of cancer development, especially gallbladder cancer (GBC) metastasis. It has been found that miR-182 negatively regulates cell adhesion molecule 1 (CADM1) expression in GBC.111,112 It has also been observed that the loss of the function of miRNA, such as miR-142, through the hypermethylation process could cause TGF-β-mediated tumor metastasis and growth in HCC.113
GM-CSF
Granulocyte macrophage colony-stimulating factor (GM-CSF) is a pro-inflammatory cytokine and a WBC growth factor produced by macrophages, T cells, endothelial cells, and NK cells. This cytokine is a monomeric glycoprotein. The gene of this cytokine is located at the chromosome region 5q31. Moreover, GM-CSF is known as a haemopoietic growth factor and can trigger the activation of neutrophils and peripheral monocytes.37,114 GM-CSF is also significant for the development of inflammatory responses during different inflammatory diseases114 as well as in cancer.115,116 A genetically engineered pancreatic ductal adenocarcinoma mouse model showed that GM-CSF secreted from tumors plays an important role in regulating inflammation and immune suppression in the tumor microenvironment.117 Importantly, it has been observed that alterations in GM-CSF-induced signaling pathways resulted in acute myeloid leukemia (AML).118 More importantly, Favreau and Sathyanarayana119 observed that GM-CSF, along with IL-3 and GCSF, regulates (both upregulates and downregulates) the expression of various miRNAs in an AML progenitor cell model (AML-193 cell line). Upregulated miRNAs include miR-219-5p and miR-590-5p, while downregulated miRNAs include miR-15b and miR-628-5p. Among upregulated miRNAs, miR-590-5p targets bone morphogenetic protein receptor type II (BMPR2) and poly(C) binding protein 2 (PCBP2); both of these proteins are known to possess tumor-suppressive properties. Another upregulated miRNA, miR-219-5p, targets TGF-β/BMP-induced signaling protein Smad4, which binds to Hoxa9 and inhibits Hoxa9-Nup98-induced AML. Among downregulated miRNAs, miR-628-5p targets Foxo3/Foxo3a, whereas miR-15b targets Bcl-2. It has been shown that elevated expressions of Foxo3 or phosphorylated (p-)Foxo3 have an adverse effect on AML prognosis.120,121 Therefore, targeting cytokines regulated by miRNAs may be instructive against CRI, and thus may provide therapeutic interventions against cancer progression.
miRNA Mediated Regulation of Other Major Players in CRI
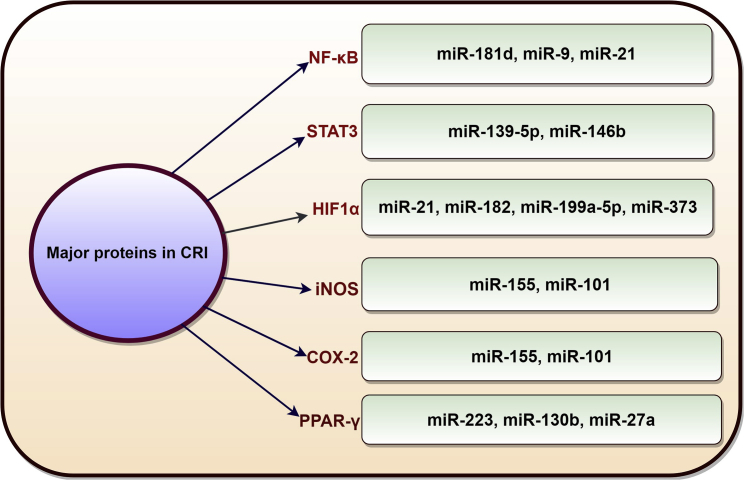
In addition to the major cytokines involved in CRI, there are other significant players that are associated with CRI and are in crosstalk with miRNAs (Figure 4).
Figure 4.
Schematic Diagram Shows miRNAs That Control Other Significant Proteins (Other Than the Cytokines) Associated with CRI (NF-κB, STAT3, HIF1α, iNOS, COX-2, PPARγ)
Transcription Factors
NF-κB. NF-κB is a combined term, which refers to dimeric transcription factors related to an important protein family called the Rel family.122 In the cytoplasm, NF-κB occurs in an inactive form (inactive complex NF-κB-inhibitory NF-κB [IκB]), where the IκB subunit acts as an inhibitor for NF-κB and prevents it from nuclear localization. Upon stimulation by external stimuli, the IκB subunit undergoes rapid phosphorylation, ubiquitination, and, finally, proteolytic degradation. Upon the release of the IκB form, the NF-κB-IκB complex allows NF-κB to translocate into the nucleus and regulate targeted gene transcription. Several studies have established NF-κB as a key molecule for inducing inflammation and as a regulator of innate immunity. Moreover, NF-κB has been implicated as a hallmark of carcinogenesis.123 NF-κB is vital for tumor cell development and also crucial for the inflammatory cells. In the inflammatory cells, NF-κB triggers different genes that encode for inflammatory/pro-inflammatory cytokines and other molecules such as adhesion molecules, inducible nitric oxide synthase (iNOS), and COX-2.123 Some clear evidence indicates that NF-κB is involved in the initiation and progression of tumor cells and tissues, where CRI normally occurs.124 However, the NF-κB pathway is firmly controlled by some inhibitors that function at different phases of the pathway. There is some emerging evidence that miRNAs are examples of these inhibitors and have regulatory control over the NF-κB signaling pathway in cancer. For instance, it has been demonstrated that miR-181d regulates the mesenchymal (MES) phenotype through the NF-κB pathway in GBM. In the absence of miR-181d, the MES subtype of GBM displays a more malignant phenotype.125 In another study, Wang et al.126 observed that epigenetic regulation of the miR-9 family in CLL is due to activation of the NF-κB signaling pathway. Bera et al.127 confirmed that miR-21 regulates NF-κB-mediated cyclin D1 expression in renal cancer cell propagation.
STAT3. Identical to NF-κB, STAT3 is a point of junction for various oncogenic signaling pathways. It is well established that cytokines can activate and regulate STAT family transcription factors, especially STAT3, by the signaling cascade of Janus-activated kinases (JAK).128 The binding of ligands induces conformational/structural changes in cytokine receptors, stimulating the associated JAKs and consequently phosphorylating and triggering STAT signaling. In numerous solid tumors, JAK/STAT activation has been acknowledged as a significant characteristic. However, the mechanism of activation of the pathway is not well understood. However, it is well established that the deregulation of a network of JAK/STAT signaling causes cancer.129 This key signaling cascade is very crucial in immune cells as well as in tumor cells engaged in oncogenesis. The STAT3 protein is known to inhibit apoptosis.130 Promising evidence has proven that miRNAs control the STAT3 pathway in different types of cancer. It has been noted that the deletion of miR-139-5p might stimulate NF-κB, MAPK, and STAT3 signaling and promote colorectal cancer and intestinal inflammation.131 Another miRNA entitled miR-146b has been shown to inhibit NF-κB-mediated production of IL-6 and, consequently, the activation of STAT3 in cancer. A study proposed crosstalk between STAT3 and the NF-κB signaling pathways during the occurrence of inflammation in oncogenesis.81
HIF1α. HIF1α, a subunit of a heterodimeric transcription factor, is a mediator of oxygen homeostasis and has a compelling role in cell survival and tumor invasion.132 HIF1 plays an important role in inflammation and leukocyte adhesion. Increasing evidence has suggested that HIFs, in addition to supervising glycolytic energy production, can act as a governing switch for innate immunity, pro-inflammatory gene expression, and cell migration.133 The HIF2α protein is intensively expressed in tumor-associated macrophages and is in the detectable range for human cancers.134 The status of a cell determines the crosstalk between proteins, NF-κB, and HIF1 during a response. It has been documented that some pro-inflammatory cytokines, such as IL-1β, TNF-α, and others, activate HIF1α in an NF-κB-dependent manner. The activated form of HIF1 then plays a pivotal role in linking the oncogenic and inflammatory pathways.135,136 miRNAs have a role in regulating the expression of HIF1α. miR-21 is an onco-miRNA that influences important targets of different genes of malignant melanoma. Noticeably, this miRNA controls PTEN, HIF1α, and TIMP3 during angiogenesis.137 It has also been noted that another miRNA, miR-182, augments HIF1α signaling in prostate cancer via targeting PHD2 and FIH1.138 In melanoma, miR-199a-5p inhibits tumor proliferation by involving HIF1α.139 miR-373 is associated (positively associated) with HIF1α and TWIST and controls metastasis through miR-373-TXNIP-HIF1α-TWIST signaling in breast cancer.140
Enzymes
iNOS. iNOS is an enzyme-catalyzing NO production and is an important cellular signaling molecule. The gene that codes iNOS is located on chromosome 17(17q11.2-q12) and is overexpressed in different inflammatory diseases and a range of cancers.141 It is well established that NO is a noteworthy regulatory molecule in cancer development and inflammatory responses. Conversely, iNOS is regulated by inflammatory/pro-inflammatory cytokines, such as IL-1β and TNF-α.142 Wang et al.143 found that the upregulation of miR-27a is associated with increased expression of iNOS in thyroid cancer cells.
COX-2. COX-2 is an enzyme, and the expression of COX-2 might be regulated by a variety of stimuli (such as pro-inflammatory cytokines, including TNF and IL-1). It is also overexpressed in several types of cancer. COX-2 has been proposed as a linker between inflammation and cancer. Nevertheless, the exact role of COX-2 in CRI is not fully known.144 It has been reported that miRNAs can enhance COX-2 expression during inflammation. miR-155 is shown to enhance COX-2 expression in inflammation, and its inhibition displayed a deleterious effect on tumorigenicity. Therefore, miR-155 might be considered a promising target for antagonizing COX-2 expression in colorectal and other cancers.145 It has been found that miR-101 regulates prostate cancer cells through COX-2 modulation in vivo. Therefore, more detailed studies on miR-101-mediated COX-2 modulation might offer new cancer therapy.146
Receptors
PPARγ. Peroxisome proliferator-activated receptor γ (PPARγ), a type II nuclear receptor, is involved in inflammation, cell differentiation, as well as cell proliferation. miRNAs regulate this nuclear receptor, which also participates in the regulation of transcription factors such as NF-κB. PPARγ has a tumor-suppressive function. In addition, several studies also suggested the implication of PPARs in inflammatory processes and specific cancers.147,148 During inflammation, the expression of miR-223 is controlled through direct binding of PPARγ with PPAR response elements (PPREs) within the promoter of pre-miR-223. For macrophage polarization, it has been noted that miR-223 and its target genes (such as Rasa1 and genuine) are significant effectors.149 Both miR-130b and miR-27a also have the ability to target PPARγ and reduce its expression, resulting in an enhanced cancer cells growth and aggressiveness during inflammation in cancer.150
Therapeutic Possibilities of miRNA Against CRI
Presently, miRNAs are known to regulate critical cellular processes by simultaneously modulating multiple targets and have been regarded as a possible therapeutic tool.151 It has been observed that oncogenic miRNAs are overexpressed in different human cancers or CRI. Thus, the inhibition or downregulation of these miRNAs might re-establish the normal functioning of a gene. There are several strategies to inhibit the miRNAs, such as the use of antisense anti-miR oligonucleotides (AMOs), locked nucleic acid (LNA) anti-miRNAs, miRNA sponges, antagomirs, and miRNA masks. Another effective alternative strategy to re-establish the normal function of a gene is the use of “miRNA mimics.” Moreover, SMIRs (small molecule inhibitors of miRNAs) can actively block miRNA-target interaction or can inhibit miRNA biogenesis. Another method used is to agitate the mode of transport and to block extracellular miRNAs in exosomes. It has been observed that GW4869, a small molecule, is an inhibitor of neutral sphingomyelinase and can inhibit miRNA and exosome secretion.152 It may be concluded that miRNA-based therapeutics have great promise for the treatment of CRI, as they are highly specific and perfect candidates for targeted therapies.
Challenges for miRNA-Based Therapeutics
The development of miRNA-based therapeutic systems may encounter various challenges. As it is well known that the efficacy of miRNA-based therapeutics depends on the successful delivery of miRNA into the target site, finding a suitable delivery system is one of the major challenges.153 Various factors might affect the successful delivery of miRNAs into the target cancer site. These include the leaky structure of tumor vessels, slow diffusion of miRNAs in the solid tumor due to the complex or higher interstitial fluid pressure, and non-specific uptake of miRNAs by non-malignant cells.154 However, even if some miRNAs reach the tumor site, they may get degraded by the endosomes or lysosomes in the cancer cells.155 Therefore, measures for endosomal escape and the release of therapeutic miRNA cargo into the cytoplasm must be taken into account.
The short half-life and instability of naked miRNAs, as they are prone to degradation by nucleases present in the blood, are other issues to be resolved before using miRNAs in cancer therapy.156 Moreover, systemic miRNA delivery might trigger the secretion of undesirable inflammatory cytokines and interferons,157 leading to the activation of the innate immune system that might induce side effects and toxicities, such as neurotoxicity.158 Besides immunogenicity, off-target gene silencing is also one of the biggest hurdles, as miRNAs can interact with complementary 3′ UTRs of non-target genes regulating multiple pathways.159 Thus, strategies should be utilized that must minimize the side effects and toxicities mediated by therapeutic miRNAs by reducing off-target gene silencing.
Conclusion
CRI appears to be one of the greatest challenges faced by medical scientists who are performing research in the area of current medicines and new drug discovery. It is very apparent from the research data that miRNA-mediated regulation might play an important role in the development of CRI. The identified miRNA candidates associated with CRI have already offered an indication that key molecular mechanisms are involved with the inflammation and cytokine signaling pathways (Table 1). Moreover, several miRNAs associated with CRI that are involved in regulating the key factors (NF-κB, STAT3, HIF1α) of molecular pathways during the course of inflammation have also been acknowledged (Table 2). However, studies on the roles of miRNAs in CRI are still in the early stages, and several questions remain to be answered about the key regulatory events mediated by miRNAs such as (1) how a single miRNA or miRNA family regulates CRI, (2) whether a single miRNA can perform different functions during the regulation of CRI, and (3) clarifying the complex network between the miRNA, cytokines, and CRI. miRNAs that regulate CRI might have the potential to prevent migration of the cells toward the tumor site. This knowledge might re-educate us about tumor cell inflammatory infiltrates. We hypothesize that a better understanding of the miRNA-mediated regulation mechanisms linking inflammation and cancers will be beneficial to the development of efficient prevention and therapies for CRI. In-depth knowledge about the miRNA controlled CRI might provide a potential approach for reversing tumor-related inflammation, which could be the start of a new era for anticancer therapies, especially for CRI.
Table 1.
miRNAs Related to Major Cytokines Signaling and CRI
| Cytokine Cascades | Pro-inflammatory/Inflammatory Role | miRNAs | Remark | References |
|---|---|---|---|---|
| TNF-α | pro-inflammatory role | miR-145 | supposed to be a tumor suppressor, and it is downregulated in multiple cancers | 54 |
| miR-145 | this miRNA supports TNF-α-induced apoptosis in triple-negative breast cancer | 55 | ||
| miR-15a, miR-29a, miR-181a | miRNAs regulate the TNF/TNFR gene superfamily in chronic lymphocytic leukemia | 58 | ||
| miR-19a | connected with lymph node metastasis | 59 | ||
| miR-130a | this miRNA regulates cervical cancer cells; TNF-α may induce NF-κB activity, which may activate miR-130a | 60 | ||
| miR-21 | cell propagation in cervical cancer | 63 | ||
| miR-765 | TNF-α inhibited the metastasis of OSCC through the miR-765-EMP3-p66Shc axis | 64 | ||
| IL-1 | pro-inflammatory role | miR-181a | IL-1β stimulated the expression of miR-181a, which regulates colon cancer | 70 |
| miR-155 | induced by IL-1β in melanoma cells | 72 | ||
| miR-101 | IL-1β mediated suppression of this miRNA in particle-induced lung cancer | 73 | ||
| miR-425 | IL-1β induces the upregulation of this miRNA in gastric cancer | 71 | ||
| miR-101 | vital for inflammation formation, especially for lung cancer tumorigenesis | 74 | ||
| IL-6 | inflammatory role | miR-21 | this miRNA regulates the PDCD4 gene, and IL-6 inhibits expression of PDCD4 in prostate cancer cells | 76 |
| miR-146b | inhibits NF-κB-mediated production of IL-6 and consequent activation of STAT3 in breast cancer cells | 77 | ||
| miR-26 | role in the regulation of IL-6 in tumorigenicity and inflammation | 83 | ||
| miR-26a | represses metastasis and tumor growth of HCC through the IL-6/STAT3 pathway | 86 | ||
| miR-26a | upregulation of IL-6 provides the decrease of this miRNA expression in HCC | 85 | ||
| IL-17 | pro-inflammatory role | miR-181a-5p | VCAM-1 expression by this miRNA under IL-17 introduction; this miRNA causes tumor cell migration and cell proliferation | 92 |
| miR-221 | role in papillary thyroid carcinoma | 93 | ||
| miR-15a/16 | this miRNA reduces IL-17 and VEGF-A levels where it acts as a tumor suppressor in multiple myeloma (MM) | 94 | ||
| miR-192 | regulatory feedback loop related to IL-17/miR-192/IL-17Rs in MM progression | 95 | ||
| IL-23 | pro-inflammatory role | miR-25 | this miRNA is overexpressed in cancer cells and is involved in IL-23-associated cell migration and invasion in thyroid cancer cells | 106 |
| TGF-β | pro-inflammatory role | miR-16 | this miRNA inhibits TGF-β1, which controls epithelial-to-mesenchymal transition through the creation of autophagy in NSCLC cells | 109 |
| miR-152 | TGF-β stimulates HLA-G expression in the course of inhibiting this miRNA in gastric cancer | 111 | ||
| miR-106b | this miRNA is unregulated by TGF-β1 during breast cancer proliferation | 111 | ||
| miR-182 | this miRNA regulates gallbladder cancer (GBC) metastasis development through TGF-β induction | 112 | ||
| miR-142 | TGF-β-related tumor metastasis and growth and in HCC | 113 | ||
| GM-CSF | pro-inflammatory role | miR-590-5p, miR-219-5p, miR-15b, miR-628-5p | expression of these miRNAs control by GM-CSF and G-CSF in acute myeloid leukemia | 114 |
TNF-α, tumor necrosis factor α; TNFR, tumor necrosis factor receptor; NF-κB, nuclear factor κB; OSCC, oral squamous cell carcinoma; EMP3, epithelial membrane protein 3; IL-1β, interleukin 1β; PDCD4, programmed cell death protein 4; STAT3, signal transducer and activator of transcription 3; HCC, hepatocellular carcinoma; VCAM-1, cascular cell adhesion protein 1; TGF-β1, transforming growth factor β1; NSCLC, non-small-cell lung carcinoma; HLA-G, human leukocyte antigen G; GM-CSF, granulocyte macrophage colony-stimulating factor; G-CSF, granulocyte colony stimulating factor.
Table 2.
miRNAs Related to Other Significant Players (Other than Cytokines) Associated with CRI
| Other Significant Players | Related miRNA | Remark | References |
|---|---|---|---|
| NF-κB | miR-181d | regulates the mesenchymal phenotype through a pathway called NF-κB pathways in glioblastoma | 125 |
| miR-9 | inactivation of this miRNA family in chronic lymphocytic leukemia (CLL) triggers a pathway called the NF-κB pathway | 126 | |
| miR-21 | NF-κB-mediated cyclin D1 expression in renal cancer cell propagation | 127 | |
| STAT3 | miR-139-5p | regulates NF-κB, MAPK, and STAT3 signaling, and it promotes colorectal cancer and intestinal inflammation | 131 |
| miR-146b | inhibited NF-κB-mediated production of IL-6 and consequent activation of STAT3 in cancer | 83 | |
| miR-146b | regulated the cancer-associated inflammation in different cancers | 82 | |
| HIF1α | miR-21 | this miRNA affects the different genes of malignant melanoma | 137 |
| miR-182 | this miRNA augments HIF1α signaling in prostate cancer | 138 | |
| miR-199a-5p | this miRNA inhibits tumor proliferation mediated by HIF1α | 139 | |
| miR-373 | this miRNA is associated with HIF1α and controls metastasis in breast cancer | 140 | |
| iNOS | miR-27a | upregulation of this miRNA has been noted for thyroid cancer cells | 134 |
| COX-2 | miR-155 | enhances COX-2 expression in inflammation in colorectal and other cancers | 136 |
| miR-101 | regulates prostate cancer through COX-2 modulation | 137 | |
| PPARγ | miR-223 | PPARγ directly enhanced miR-223 expression and controls macrophage polarization | 149 |
| miR-130b, miR-27a | both miRNAs target PPARγ and promote cancer cell growth and aggressiveness | 150 |
NF-κB, nuclear factor κB; STAT3, signal transducer and activator of transcription 3; MAPK, mitogen-activated protein kinase; HIF1α, hypoxia-inducible factor 1α; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2; PPARγ, peroxisome proliferator-activated receptor-γ.
Author Contributions
C.C. and A.R.S researched data for the article, substantively contributed to the discussion of content, and contributed to writing the article. G.S. significantly contributed to the discussion of content, generated the figures, and reviewed the manuscript. S.-S.L. reviewed and edited the manuscript before submission.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1A2B4012944).
Contributor Information
Chiranjib Chakraborty, Email: drchiranjib@yahoo.com.
Sang-Soo Lee, Email: 123sslee@gmail.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 4.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mantovani A., Allavena P., Sica A., Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 6.Eiró N., Vizoso F.J. Inflammation and cancer. World J. Gastrointest. Surg. 2012;4:62–72. doi: 10.4240/wjgs.v4.i3.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diakos C.I., Charles K.A., McMillan D.C., Clarke S.J. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi H., Ogata H., Nishigaki R., Broide D.H., Karin M. Tobacco smoke promotes lung tumorigenesis by triggering IKKβ- and JNK1-dependent inflammation. Cancer Cell. 2010;17:89–97. doi: 10.1016/j.ccr.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philip M., Rowley D.A., Schreiber H. Inflammation as a tumor promoter in cancer induction. Semin. Cancer Biol. 2004;14:433–439. doi: 10.1016/j.semcancer.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Colotta F., Allavena P., Sica A., Garlanda C., Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 11.Shukla G.C., Singh J., Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol. Cell. Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Cho W.C. Grand challenges and opportunities in deciphering the role of non-coding RNAs in human diseases. Front. Genet. 2011;2:1. doi: 10.3389/fgene.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valinezhad Orang A., Safaralizadeh R., Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 16.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 17.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 18.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 19.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinske L.C., Heyn J., Galante P.A., Ohno-Machado L., Kreth S. Setting up an intronic miRNA database. Methods Mol. Biol. 2013;936:69–76. doi: 10.1007/978-1-62703-083-0_5. [DOI] [PubMed] [Google Scholar]
- 21.Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho W.C. MicroRNAs in cancer—from research to therapy. Biochim. Biophys. Acta. 2010;1805:209–217. doi: 10.1016/j.bbcan.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Felekkis K., Touvana E., Stefanou Ch., Deltas C. MicroRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- 24.Huang P., Han J., Hui L. MAPK signaling in inflammation-associated cancer development. Protein Cell. 2010;1:218–226. doi: 10.1007/s13238-010-0019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakraborty C., Sharma A.R., Patra B.C., Bhattacharya M., Sharma G., Lee S.S. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget. 2016;7:42683–42697. doi: 10.18632/oncotarget.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoelson S.E., Lee J., Goldfine A.B. Inflammation and insulin resistance. J. Clin. Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty C., Doss C.G.P., Bandyopadhyay S., Agoramoorthy G. Influence of miRNA in insulin signaling pathway and insulin resistance: micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip. Rev. RNA. 2014;5:697–712. doi: 10.1002/wrna.1240. [DOI] [PubMed] [Google Scholar]
- 28.Chakraborty C., George Priya Doss C., Bandyopadhyay S. miRNAs in insulin resistance and diabetes-associated pancreatic cancer: the “minute and miracle” molecule moving as a monitor in the ‘genomic galaxy’. Curr. Drug Targets. 2013;14:1110–1117. doi: 10.2174/13894501113149990182. [DOI] [PubMed] [Google Scholar]
- 29.Chakraborty C., Chin K.Y., Das S. miRNA-regulated cancer stem cells: understanding the property and the role of miRNA in carcinogenesis. Tumour Biol. 2016;37:13039–13048. doi: 10.1007/s13277-016-5156-1. [DOI] [PubMed] [Google Scholar]
- 30.Sharma A.R., Sharma G., Lee S.S., Chakraborty C. miRNA-regulated key components of cytokine signaling pathways and inflammation in rheumatoid arthritis. Med. Res. Rev. 2016;36:425–439. doi: 10.1002/med.21384. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty C., Doss C.G.P., Sarin R., Hsu M.J., Agoramoorthy G. Can the chemotherapeutic agents perform anticancer activity through miRNA expression regulation? Proposing a new hypothesis [corrected] Protoplasma. 2015;252:1603–1610. doi: 10.1007/s00709-015-0776-7. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M., Zhang L., Cui M., Ye W., Zhang P., Zhou S., Wang J. miR-302b inhibits cancer-related inflammation by targeting ERBB4, IRF2 and CXCR4 in esophageal cancer. Oncotarget. 2017;8:49053–49063. doi: 10.18632/oncotarget.17041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christopher A.F., Gupta M., Bansal P. Micronome revealed miR-19a/b as key regulator of SOCS3 during cancer related inflammation of oral squamous cell carcinoma. Gene. 2016;594:30–40. doi: 10.1016/j.gene.2016.08.044. [DOI] [PubMed] [Google Scholar]
- 34.Landskron G., De la Fuente M., Thuwajit P., Thuwajit C., Hermoso M.A. Chronic inflammation and cytokines in the tumor microenvironment. J. Immunol. Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Prete A., Allavena P., Santoro G., Fumarulo R., Corsi M.M., Mantovani A. Molecular pathways in cancer-related inflammation. Biochem. Med. (Zagreb) 2011;21:264–275. doi: 10.11613/bm.2011.036. [DOI] [PubMed] [Google Scholar]
- 36.Comen E.A., Bowman R.L., Kleppe M. Underlying causes and therapeutic targeting of the inflammatory tumor microenvironment. Front. Cell Dev. Biol. 2018;6:56. doi: 10.3389/fcell.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germano G., Allavena P., Mantovani A. Cytokines as a key component of cancer-related inflammation. Cytokine. 2008;43:374–379. doi: 10.1016/j.cyto.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- 39.Turner M.D., Nedjai B., Hurst T., Pennington D.J. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Momen-Heravi F., Bala S. miRNA regulation of innate immunity. J. Leukoc. Biol. 2018;103:1205–1217. doi: 10.1002/JLB.3MIR1117-459R. [DOI] [PubMed] [Google Scholar]
- 41.Gracias D.T., Katsikis P.D. MicroRNAs: key components of immune regulation. Adv. Exp. Med. Biol. 2011;780:15–26. doi: 10.1007/978-1-4419-5632-3_2. [DOI] [PubMed] [Google Scholar]
- 42.Bell E., Taylor M.A. Functional roles for exosomal microRNAs in the tumour microenvironment. Comput. Struct. Biotechnol. J. 2016;15:8–13. doi: 10.1016/j.csbj.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki H.I., Katsura A., Matsuyama H., Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene. 2015;34:3085–3094. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bronisz A., Wang Y., Nowicki M.O., Peruzzi P., Ansari K., Ogawa D., Balaj L., De Rienzo G., Mineo M., Nakano I. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014;74:738–750. doi: 10.1158/0008-5472.CAN-13-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M., Chen J., Su F., Yu B., Su F., Lin L., Liu Y., Huang J.D., Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rupaimoole R., Calin G.A., Lopez-Berestein G., Sood A.K. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budhu A., Wang X.W. The role of cytokines in hepatocellular carcinoma. J. Leukoc. Biol. 2006;80:1197–1213. doi: 10.1189/jlb.0506297. [DOI] [PubMed] [Google Scholar]
- 48.Asirvatham A.J., Magner W.J., Tomasi T.B. miRNA regulation of cytokine genes. Cytokine. 2009;45:58–69. doi: 10.1016/j.cyto.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carswell E.A., Old L.J., Kassel R.L., Green S., Fiore N., Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aggarwal B.B., Kohr W.J., Hass P.E., Moffat B., Spencer S.A., Henzel W.J., Bringman T.S., Nedwin G.E., Goeddel D.V., Harkins R.N. Human tumor necrosis factor. Production, purification, and characterization. J. Biol. Chem. 1985;260:2345–2354. [PubMed] [Google Scholar]
- 51.Balkwill F. TNF-α in promotion and progression of cancer. Cancer Metastasis Rev. 2006;25:409–416. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 52.Wajant H. The role of TNF in cancer. Results Probl. Cell Differ. 2009;49:1–15. doi: 10.1007/400_2008_26. [DOI] [PubMed] [Google Scholar]
- 53.Kubota T., Miyagishima M., Frye C.S., Alber S.M., Bounoutas G.S., Kadokami T., Watkins S.C., McTiernan C.F., Feldman A.M. Overexpression of tumor necrosis factor-α activates both anti- and pro-apoptotic pathways in the myocardium. J. Mol. Cell. Cardiol. 2001;33:1331–1344. doi: 10.1006/jmcc.2001.1393. [DOI] [PubMed] [Google Scholar]
- 54.Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., Elble R., Watabe K., Mo Y.Y. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng M., Wu Z., Wu A., Huang Z., He N., Xie X. miR-145 promotes TNF-α-induced apoptosis by facilitating the formation of RIP1-FADDcaspase-8 complex in triple-negative breast cancer. Tumour Biol. 2016;37:8599–8607. doi: 10.1007/s13277-015-4631-4. [DOI] [PubMed] [Google Scholar]
- 56.Ferrajoli A., Keating M.J., Manshouri T., Giles F.J., Dey A., Estrov Z., Koller C.A., Kurzrock R., Thomas D.A., Faderl S. The clinical significance of tumor necrosis factor-alpha plasma level in patients having chronic lymphocytic leukemia. Blood. 2002;100:1215–1219. [PubMed] [Google Scholar]
- 57.Bojarska-Junak A., Rolinski J., Wasik-Szczepaneko E., Kaluzny Z., Dmoszynska A. Intracellular tumor necrosis factor production by T- and B-cells in B-cell chronic lymphocytic leukemia. Haematologica. 2002;87:490–499. [PubMed] [Google Scholar]
- 58.Srivastava S., Tsongalis G.J., Kaur P. Role of microRNAs in regulation of the TNF/TNFR gene superfamily in chronic lymphocytic leukemia. Clin. Biochem. 2016;49:1307–1310. doi: 10.1016/j.clinbiochem.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 59.Huang L., Wang X., Wen C., Yang X., Song M., Chen J., Wang C., Zhang B., Wang L., Iwamoto A. hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Sci. Rep. 2015;5:13350. doi: 10.1038/srep13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J., Wu H., Li P., Zhao Y., Liu M., Tang H. NF-κB-modulated miR-130a targets TNF-α in cervical cancer cells. J. Transl. Med. 2014;12:155. doi: 10.1186/1479-5876-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Zhan X., Yan D., Wang Z. Circulating microRNA-21 is involved in lymph node metastasis in cervical cancer by targeting RASA1. Int. J. Gynecol. Cancer. 2016;26:810–816. doi: 10.1097/IGC.0000000000000694. [DOI] [PubMed] [Google Scholar]
- 62.Xu J., Zhang W., Lv Q., Zhu D. Overexpression of miR-21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN. Oncol. Rep. 2015;33:3108–3116. doi: 10.3892/or.2015.3931. [DOI] [PubMed] [Google Scholar]
- 63.Xu L., Xu Q., Li X., Zhang X. MicroRNA-21 regulates the proliferation and apoptosis of cervical cancer cells via tumor necrosis factor-α. Mol. Med. Rep. 2017;16:4659–4663. doi: 10.3892/mmr.2017.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng Z., Luan X., Zha J., Li Z., Wu L., Yan Y., Wang H., Hou D., Huang L., Huang F. TNF-α inhibits the migration of oral squamous cancer cells mediated by miR-765-EMP3-p66Shc axis. Cell. Signal. 2017;34:102–109. doi: 10.1016/j.cellsig.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 65.Tang D., Tao D., Fang Y., Deng C., Xu Q., Zhou J. TNF-alpha promotes invasion and metastasis via NF-kappa B pathway in oral squamous cell carcinoma. Med. Sci. Monit. Basic Res. 2017;23:141–149. doi: 10.12659/MSMBR.903910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garlanda C., Dinarello C.A., Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.England H., Summersgill H.R., Edye M.E., Rothwell N.J., Brough D. Release of interleukin-1α or interleukin-1β depends on mechanism of cell death. J. Biol. Chem. 2014;289:15942–15950. doi: 10.1074/jbc.M114.557561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nicklin M.J., Barton J.L., Nguyen M., FitzGerald M.G., Duff G.W., Kornman K. A sequence-based map of the nine genes of the human interleukin-1 cluster. Genomics. 2002;79:718–725. doi: 10.1006/geno.2002.6751. [DOI] [PubMed] [Google Scholar]
- 69.Kasza A. IL-1 and EGF regulate expression of genes important in inflammation and cancer. Cytokine. 2013;62:22–33. doi: 10.1016/j.cyto.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 70.Hai Ping P., Feng Bo T., Li L., Nan Hui Y., Hong Z. IL-1β/NF-kb signaling promotes colorectal cancer cell growth through miR-181a/PTEN axis. Arch. Biochem. Biophys. 2016;604:20–26. doi: 10.1016/j.abb.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 71.Ma J., Liu J., Wang Z., Gu X., Fan Y., Zhang W., Xu L., Zhang J., Cai D. NF-kappaB-dependent microRNA-425 upregulation promotes gastric cancer cell growth by targeting PTEN upon IL-1β induction. Mol. Cancer. 2014;13:40. doi: 10.1186/1476-4598-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arts N., Cané S., Hennequart M., Lamy J., Bommer G., Van den Eynde B., De Plaen E. MicroRNA-155, induced by interleukin-1ß, represses the expression of microphthalmia-associated transcription factor (MITF-M) in melanoma cells. PLoS ONE. 2015;10:e0122517. doi: 10.1371/journal.pone.0122517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lei Y.M., Zu Y.F., Wang J., Bai S., Shi Y.F., Shi R., Duan J., Cui D., Chen J., Xiang Y., Dong J. Interleukin-1β-mediated suppression of microRNA-101 and upregulation of enhancer of zeste homolog 2 is involved in particle-induced lung cancer. Med. Oncol. 2015;32:387. doi: 10.1007/s12032-014-0387-8. [DOI] [PubMed] [Google Scholar]
- 74.Wang L., Zhang L.F., Wu J., Xu S.J., Xu Y.Y., Li D., Lou J.T., Liu M.F. IL-1β-mediated repression of microRNA-101 is crucial for inflammation-promoted lung tumorigenesis. Cancer Res. 2014;74:4720–4730. doi: 10.1158/0008-5472.CAN-14-0960. [DOI] [PubMed] [Google Scholar]
- 75.Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res. Ther. 2006;8(Suppl 2):S2. doi: 10.1186/ar1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 77.Mitsunaga S., Ikeda M., Shimizu S., Ohno I., Furuse J., Inagaki M., Higashi S., Kato H., Terao K., Ochiai A. Serum levels of IL-6 and IL-1β can predict the efficacy of gemcitabine in patients with advanced pancreatic cancer. Br. J. Cancer. 2013;108:2063–2069. doi: 10.1038/bjc.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tanaka T., Kishimoto T. The biology and medical implications of interleukin-6. Cancer Immunol. Res. 2014;2:288–294. doi: 10.1158/2326-6066.CIR-14-0022. [DOI] [PubMed] [Google Scholar]
- 79.Taniguchi K., Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 2014;26:54–74. doi: 10.1016/j.smim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 80.Dong B., Shi Z., Wang J., Wu J., Yang Z., Fang K. IL-6 inhibits the targeted modulation of PDCD4 by miR-21 in prostate cancer. PLoS ONE. 2015;10:e0134366. doi: 10.1371/journal.pone.0134366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiang M., Birkbak N.J., Vafaizadeh V., Walker S.R., Yeh J.E., Liu S., Kroll Y., Boldin M., Taganov K., Groner B. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci. Signal. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iliopoulos D. MicroRNA circuits regulate the cancer-inflammation link. Sci. Signal. 2014;7:pe8. doi: 10.1126/scisignal.2005053. [DOI] [PubMed] [Google Scholar]
- 83.Chen C.Y., Chang J.T., Ho Y.F., Shyu A.B. miR-26 down-regulates TNF-α/NF-κB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucleic Acids Res. 2016;44:3772–3787. doi: 10.1093/nar/gkw205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones M.R., Quinton L.J., Blahna M.T., Neilson J.R., Fu S., Ivanov A.R., Wolf D.A., Mizgerd J.P. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y., Zhang B., Zhang A., Li X., Liu J., Zhao J., Zhao Y., Gao J., Fang D., Rao Z. IL-6 upregulation contributes to the reduction of miR-26a expression in hepatocellular carcinoma cells. Braz. J. Med. Biol. Res. 2013;46:32–38. doi: 10.1590/S0100-879X2012007500155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang X., Liang L., Zhang X.F., Jia H.L., Qin Y., Zhu X.C., Gao X.M., Qiao P., Zheng Y., Sheng Y.Y. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 87.Fossiez F., Djossou O., Chomarat P., Flores-Romo L., Ait-Yahia S., Maat C., Pin J.J., Garrone P., Garcia E., Saeland S. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J. Exp. Med. 1996;183:2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaffen S.L. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shalom-Barak T., Quach J., Lotz M. Interleukin-17-induced gene expression in articular chondrocytes is associated with activation of mitogen-activated protein kinases and NF-κB. J. Biol. Chem. 1998;273:27467–27473. doi: 10.1074/jbc.273.42.27467. [DOI] [PubMed] [Google Scholar]
- 90.Kuwabara T., Ishikawa F., Kondo M., Kakiuchi T. The role of IL-17 and related cytokines in inflammatory autoimmune diseases. Mediators Inflamm. 2017;2017:3908061. doi: 10.1155/2017/3908061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tabarkiewicz J., Pogoda K., Karczmarczyk A., Pozarowski P., Giannopoulos K. The role of IL-17 and Th17 lymphocytes in autoimmune diseases. Arch. Immunol. Ther. Exp. (Warsz.) 2015;63:435–449. doi: 10.1007/s00005-015-0344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao Y., Zhao D., Li P., Wang L., Qiao B., Qin X., Li L., Wang Y. MicroRNA-181a-5p impedes IL-17-induced nonsmall cell lung cancer proliferation and migration through targeting VCAM-1. Cell. Physiol. Biochem. 2017;42:346–356. doi: 10.1159/000477389. [DOI] [PubMed] [Google Scholar]
- 93.Jiang X.L., Zhang H., Chen Y.L., Peng L. [Expression of microRNA-221 and IL-17 in papillary thyroid carcinoma and correlation with clinicopathologic features] Zhonghua Bing Li Xue Za Zhi. 2017;46:160–165. doi: 10.3760/cma.j.issn.0529-5807.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 94.Li Y., Zhang B., Li W., Wang L., Yan Z., Li H., Yao Y., Yao R., Xu K., Li Z. miR-15a/16 regulates the growth of myeloma cells, angiogenesis and antitumor immunity by inhibiting Bcl-2, VEGF-A and IL-17 expression in multiple myeloma. Leuk. Res. 2016;49:73–79. doi: 10.1016/j.leukres.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 95.Sun Y., Pan J., Mao S., Jin J. IL-17/miR-192/IL-17Rs regulatory feedback loop facilitates multiple myeloma progression. PLoS ONE. 2014;9:e114647. doi: 10.1371/journal.pone.0114647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruddy M.J., Wong G.C., Liu X.K., Yamamoto H., Kasayama S., Kirkwood K.L., Gaffen S.L. Functional cooperation between interleukin-17 and tumor necrosis factor-α is mediated by CCAAT/enhancer-binding protein family members. J. Biol. Chem. 2004;279:2559–2567. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 97.Shen F., Hu Z., Goswami J., Gaffen S.L. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J. Biol. Chem. 2006;281:24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 98.Yu H., Jove R. The STATs of cancer—new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 99.Duvallet E., Semerano L., Assier E., Falgarone G., Boissier M.C. Interleukin-23: a key cytokine in inflammatory diseases. Ann. Med. 2011;43:503–511. doi: 10.3109/07853890.2011.577093. [DOI] [PubMed] [Google Scholar]
- 100.Martin-Orozco N., Dong C. The IL-17/IL-23 axis of inflammation in cancer: friend or foe? Curr. Opin. Investig. Drugs. 2009;10:543–549. [PubMed] [Google Scholar]
- 101.Chen D., Li W., Liu S., Su Y., Han G., Xu C., Liu H., Zheng T., Zhou Y., Mao C. Interleukin-23 promotes the epithelial-mesenchymal transition of oesophageal carcinoma cells via the Wnt/β-catenin pathway. Sci. Rep. 2015;5:8604. doi: 10.1038/srep08604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li J., Lau G., Chen L., Yuan Y.F., Huang J., Luk J.M., Xie D., Guan X.Y. Interleukin 23 promotes hepatocellular carcinoma metastasis via NF-kappa B induced matrix metalloproteinase 9 expression. PLoS ONE. 2012;7:e46264. doi: 10.1371/journal.pone.0046264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suzuki H., Ogawa H., Miura K., Haneda S., Watanabe K., Ohnuma S., Sasaki H., Sase T., Kimura S., Kajiwara T. IL-23 directly enhances the proliferative and invasive activities of colorectal carcinoma. Oncol. Lett. 2012;4:199–204. doi: 10.3892/ol.2012.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klein A., Schwartz H., Sagi-Assif O., Meshel T., Izraely S., Ben Menachem S., Bengaiev R., Ben-Shmuel A., Nahmias C., Couraud P.O. Astrocytes facilitate melanoma brain metastasis via secretion of IL-23. J. Pathol. 2015;236:116–127. doi: 10.1002/path.4509. [DOI] [PubMed] [Google Scholar]
- 105.Haas J.D., Nistala K., Petermann F., Saran N., Chennupati V., Schmitz S., Korn T., Wedderburn L.R., Förster R., Krueger A., Prinz I. Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in αβ and γδ T cells. PLoS ONE. 2011;6:e20171. doi: 10.1371/journal.pone.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mei Z., Chen S., Chen C., Xiao B., Li F., Wang Y., Tao Z. Interleukin-23 facilitates thyroid cancer cell migration and invasion by inhibiting SOCS4 expression via microRNA-25. PLoS ONE. 2015;10:e0139456. doi: 10.1371/journal.pone.0139456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santibañez J.F., Quintanilla M., Bernabeu C. TGF-β/TGF-β receptor system and its role in physiological and pathological conditions. Clin. Sci. (Lond.) 2011;121:233–251. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- 108.Liu S., Chen S., Zeng J. TGF-β signaling: a complex role in tumorigenesis (Review) Mol. Med. Rep. 2018;17:699–704. doi: 10.3892/mmr.2017.7970. [DOI] [PubMed] [Google Scholar]
- 109.Wang H., Zhang Y., Wu Q., Wang Y.B., Wang W. miR-16 mimics inhibit TGF-β1-induced epithelial-to-mesenchymal transition via activation of autophagy in non-small cell lung carcinoma cells. Oncol. Rep. 2018;39:247–254. doi: 10.3892/or.2017.6088. [DOI] [PubMed] [Google Scholar]
- 110.Guan Z., Song B., Liu F., Sun D., Wang K., Qu H. TGF-β induces HLA-G expression through inhibiting miR-152 in gastric cancer cells. J. Biomed. Sci. 2015;22:107. doi: 10.1186/s12929-015-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gong C., Qu S., Liu B., Pan S., Jiao Y., Nie Y., Su F., Liu Q., Song E. miR-106b expression determines the proliferation paradox of TGF-β in breast cancer cells. Oncogene. 2015;34:84–93. doi: 10.1038/onc.2013.525. [DOI] [PubMed] [Google Scholar]
- 112.Qiu Y., Luo X., Kan T., Zhang Y., Yu W., Wei Y., Shen N., Yi B., Jiang X. TGF-β upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting CADM1. Mol. Biosyst. 2014;10:679–685. doi: 10.1039/c3mb70479c. [DOI] [PubMed] [Google Scholar]
- 113.Yu Q., Xiang L., Yin L., Liu X., Yang D., Zhou J. Loss-of-function of miR-142 by hypermethylation promotes TGF-β-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 2017;50:e12384. doi: 10.1111/cpr.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Francisco-Cruz A., Aguilar-Santelises M., Ramos-Espinosa O., Mata-Espinosa D., Marquina-Castillo B., Barrios-Payan J., Hernandez-Pando R. Granulocyte-macrophage colony-stimulating factor: not just another haematopoietic growth factor. Med. Oncol. 2014;31:774. doi: 10.1007/s12032-013-0774-6. [DOI] [PubMed] [Google Scholar]
- 115.Ushach I., Zlotnik A. Biological role of granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) on cells of the myeloid lineage. J. Leukoc. Biol. 2016;100:481–489. doi: 10.1189/jlb.3RU0316-144R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi Y., Liu C.H., Roberts A.I., Das J., Xu G., Ren G., Zhang Y., Zhang L., Yuan Z.R., Tan H.S.W. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell Res. 2006;16:126–133. doi: 10.1038/sj.cr.7310017. [DOI] [PubMed] [Google Scholar]
- 117.Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z., Vonderheide R.H. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Testa U., Riccioni R., Militi S., Coccia E., Stellacci E., Samoggia P., Latagliata R., Mariani G., Rossini A., Battistini A. Elevated expression of IL-3Rα in acute myelogenous leukemia is associated with enhanced blast proliferation, increased cellularity, and poor prognosis. Blood. 2002;100:2980–2988. doi: 10.1182/blood-2002-03-0852. [DOI] [PubMed] [Google Scholar]
- 119.Favreau A.J., Sathyanarayana P. miR-590-5p, miR-219-5p, miR-15b and miR-628-5p are commonly regulated by IL-3, GM-CSF and G-CSF in acute myeloid leukemia. Leuk. Res. 2012;36:334–341. doi: 10.1016/j.leukres.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Santamaría C.M., Chillón M.C., García-Sanz R., Pérez C., Caballero M.D., Ramos F., de Coca A.G., Alonso J.M., Giraldo P., Bernal T., Queizán J.A. High FOXO3a expression is associated with a poorer prognosis in AML with normal cytogenetics. Leuk. Res. 2009;33:1706–1709. doi: 10.1016/j.leukres.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 121.Kornblau S.M., Singh N., Qiu Y., Chen W., Zhang N., Coombes K.R. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin. Cancer Res. 2010;16:1865–1874. doi: 10.1158/1078-0432.CCR-09-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 123.Viatour P., Merville M.P., Bours V., Chariot A. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 124.Pikarsky E., Porat R.M., Stein I., Abramovitch R., Amit S., Kasem S., Gutkovich-Pyest E., Urieli-Shoval S., Galun E., Ben-Neriah Y. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 125.Yang F., Liu X., Liu Y., Liu Y., Zhang C., Wang Z., Jiang T., Wang Y. miR-181d/MALT1 regulatory axis attenuates mesenchymal phenotype through NF-κB pathways in glioblastoma. Cancer Lett. 2017;396:1–9. doi: 10.1016/j.canlet.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 126.Wang L.Q., Kwong Y.L., Kho C.S.B., Wong K.F., Wong K.Y., Ferracin M., Calin G.A., Chim C.S. Epigenetic inactivation of miR-9 family microRNAs in chronic lymphocytic leukemia—implications on constitutive activation of NFκB pathway. Mol. Cancer. 2013;12:173. doi: 10.1186/1476-4598-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bera A., Ghosh-Choudhury N., Dey N., Das F., Kasinath B.S., Abboud H.E., Choudhury G.G. NFκB-mediated cyclin D1 expression by microRNA-21 influences renal cancer cell proliferation. Cell. Signal. 2013;25:2575–2586. doi: 10.1016/j.cellsig.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hodge D.R., Hurt E.M., Farrar W.L. The role of IL-6 and STAT3 in inflammation and cancer. Eur. J. Cancer. 2005;41:2502–2512. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 129.Thomas S.J., Snowden J.A., Zeidler M.P., Danson S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer. 2015;113:365–371. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bromberg J.F., Wrzeszczynska M.H., Devgan G., Zhao Y., Pestell R.G., Albanese C., Darnell J.E., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 131.Zou F., Mao R., Yang L., Lin S., Lei K., Zheng Y., Ding Y., Zhang P., Cai G., Liang X., Liu J. Targeted deletion of miR-139-5p activates MAPK, NF-κB and STAT3 signaling and promotes intestinal inflammation and colorectal cancer. FEBS J. 2016;283:1438–1452. doi: 10.1111/febs.13678. [DOI] [PubMed] [Google Scholar]
- 132.Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 133.Imtiyaz H.Z., Simon M.C. Hypoxia-inducible factors as essential regulators of inflammation. Curr. Top. Microbiol. Immunol. 2010;345:105–120. doi: 10.1007/82_2010_74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Talks K.L., Turley H., Gatter K.C., Maxwell P.H., Pugh C.W., Ratcliffe P.J., Harris A.L. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jung Y.J., Isaacs J.S., Lee S., Trepel J., Neckers L. IL-1β-mediated up-regulation of HIF-1α via an NFκB/COX-2 pathway identifies HIF-1 as a critical link between inflammation and oncogenesis. FASEB J. 2003;17:2115–2117. doi: 10.1096/fj.03-0329fje. [DOI] [PubMed] [Google Scholar]
- 136.Zhou J., Schmid T., Brüne B. Tumor necrosis factor-α causes accumulation of a ubiquitinated form of hypoxia inducible factor-1α through a nuclear factor-κB-dependent pathway. Mol. Biol. Cell. 2003;14:2216–2225. doi: 10.1091/mbc.E02-09-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Melnik B.C. miR-21: an environmental driver of malignant melanoma? J. Transl. Med. 2015;13:202. doi: 10.1186/s12967-015-0570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li Y., Zhang D., Wang X., Yao X., Ye C., Zhang S., Wang H., Chang C., Xia H., Wang Y.C. Hypoxia-inducible miR-182 enhances HIF1α signaling via targeting PHD2 and FIH1 in prostate cancer. Sci. Rep. 2015;5:12495. doi: 10.1038/srep12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang X., Lei S., Long J., Liu X., Wu Q. MicroRNA-199a-5p inhibits tumor proliferation in melanoma by mediating HIF-1α. Mol. Med. Rep. 2016;13:5241–5247. doi: 10.3892/mmr.2016.5202. [DOI] [PubMed] [Google Scholar]
- 140.Chen D., Dang B.L., Huang J.Z., Chen M., Wu D., Xu M.L., Li R., Yan G.R. miR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer. Oncotarget. 2015;6:32701–32712. doi: 10.18632/oncotarget.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim Y.H., Woo K.J., Lim J.H., Kim S., Lee T.J., Jung E.M., Lee J.M., Park J.W., Kwon T.K. 8-Hydroxyquinoline inhibits iNOS expression and nitric oxide production by down-regulating LPS-induced activity of NF-κB and C/EBPβ in Raw 264.7 cells. Biochem. Biophys. Res. Commun. 2005;329:591–597. doi: 10.1016/j.bbrc.2005.01.159. [DOI] [PubMed] [Google Scholar]
- 142.Hussain S.P., Trivers G.E., Hofseth L.J., He P., Shaikh I., Mechanic L.E., Doja S., Jiang W., Subleski J., Shorts L. Nitric oxide, a mediator of inflammation, suppresses tumorigenesis. Cancer Res. 2004;64:6849–6853. doi: 10.1158/0008-5472.CAN-04-2201. [DOI] [PubMed] [Google Scholar]
- 143.Wang Y.L., Gong W.G., Yuan Q.L. Effects of miR-27a upregulation on thyroid cancer cells migration, invasion, and angiogenesis. Genet. Mol. Res. 2016;15:1–10. doi: 10.4238/gmr15049070. [DOI] [PubMed] [Google Scholar]
- 144.Kuwano T., Nakao S., Yamamoto H., Tsuneyoshi M., Yamamoto T., Kuwano M., Ono M. Cyclooxygenase 2 is a key enzyme for inflammatory cytokine-induced angiogenesis. FASEB J. 2004;18:300–310. doi: 10.1096/fj.03-0473com. [DOI] [PubMed] [Google Scholar]
- 145.Comer B.S. Does miRNA-155 promote cyclooxygenase-2 expression in cancer? Drug Dev. Res. 2015;76:354–356. doi: 10.1002/ddr.21276. [DOI] [PubMed] [Google Scholar]
- 146.Hao Y., Gu X., Zhao Y., Greene S., Sha W., Smoot D.T., Califano J., Wu T.C., Pang X. Enforced expression of miR-101 inhibits prostate cancer cell growth by modulating the COX-2 pathway in vivo. Cancer Prev. Res. (Phila.) 2011;4:1073–1083. doi: 10.1158/1940-6207.CAPR-10-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Laganà A.S., Vitale S.G., Nigro A., Sofo V., Salmeri F.M., Rossetti P., Rapisarda A.M.C., La Vignera S., Condorelli R.A., Rizzo G., Buscema M. Pleiotropic actions of peroxisome proliferator-activated receptors (PPARs) in dysregulated metabolic homeostasis, inflammation and cancer: current evidence and future perspectives. Int. J. Mol. Sci. 2016;17:999. doi: 10.3390/ijms17070999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fuentes E., Guzmán-Jofre L., Moore-Carrasco R., Palomo I. Role of PPARs in inflammatory processes associated with metabolic syndrome (Review) Mol. Med. Rep. 2013;8:1611–1616. doi: 10.3892/mmr.2013.1714. [DOI] [PubMed] [Google Scholar]
- 149.Ying W., Tseng A., Chang R.C.A., Morin A., Brehm T., Triff K., Nair V., Zhuang G., Song H., Kanameni S. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Invest. 2015;125:4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Portius D., Sobolewski C., Foti M. MicroRNAs-dependent regulation of PPARs in metabolic diseases and cancers. PPAR Res. 2017;2017:7058424. doi: 10.1155/2017/7058424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shah M.Y., Ferrajoli A., Sood A.K., Lopez-Berestein G., Calin G.A. MicroRNA therapeutics in cancer—an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cui M., Wang H., Yao X., Zhang D., Xie Y., Cui R., Zhang X. Circulating microRNAs in cancer: potential and challenge. Front. Genet. 2019;10:626. doi: 10.3389/fgene.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen Y., Gao D.Y., Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Orellana E.A., Abdelaal A.M., Rangasamy L., Tenneti S., Myoung S., Low P.S., Kasinski A.L. Enhancing microRNA activity through increased endosomal release mediated by nigericin. Mol. Ther. Nucleic Acids. 2019;16:505–518. doi: 10.1016/j.omtn.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Y., Wang Z., Gemeinhart R.A. Progress in microRNA delivery. J. Control. Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kabilova T.O., Meschaninova M.I., Venyaminova A.G., Nikolin V.P., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Short double-stranded RNA with immunostimulatory activity: sequence dependence. Nucleic Acid Ther. 2012;22:196–204. doi: 10.1089/nat.2011.0328. [DOI] [PubMed] [Google Scholar]
- 158.Lehmann S.M., Krüger C., Park B., Derkow K., Rosenberger K., Baumgart J., Trimbuch T., Eom G., Hinz M., Kaul D. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 159.Singh S., Narang A.S., Mahato R.I. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011;28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]