Abstract
Ovarian cancer (OC) is a severe malignancy featuring a poor prognosis due to rapid metastasis and chemotherapy resistance. In this study, we extensively investigated the upstream and downstream mechanisms of miR-548e in regulating OC progression and cisplatin resistance. Our results indicated that ZFAS1 was highly expressed and promoted OC cell proliferation, migration, invasion, and cisplatin resistance by directly suppressing miR-548e expression. ZFAS1 co-localized with miR-548e in the cytosols of OC cells. miR-548e repressed CXCR4 expression, and elevated CXCR4 expression promoted OC cell proliferation, migration, invasion, and cisplatin resistance. Cisplatin resistance induced by ZFAS1 and CXCR4 overexpression in OC cells was mediated by their suppression on let-7a and elevation of BCL-XL/S expression. ZFAS1 knockdown and miR-548e and let-7a overexpression impaired cisplatin resistance and suppressed lung metastatic nodule formation in nude mice. In conclusion, ZFAS1 binds with miR-548e to enhance CXCR4 expression to promote OC cell proliferation and metastasis, which also enhances cisplatin resistance by suppressing let-7a and elevating BCL-XL/S protein expression.
Keywords: ovarian cancer, ZFAS1, miR-548e, CXCR4, Let-7a, metastasis, cisplatin resistance
Introduction
Ovarian cancer (OC) is one severe gynecologic malignancy affecting the female reproductive system that is still listed as leading cause of cancer-related death worldwide, with an estimated 52,100 and 22,240 new cases each year in China and the USA, respectively.1 The pathogenesis of OC has been established as a landscape driven by serial genetic mutations such as TP53 (tumor protein p53), BRCA1 (breast cancer 1), and CCNE1 (cyclin E1), which were explored as targets for drug development.2 However, current clinical treatment has been greatly challenged by the emerging intrinsic and acquired resistances of OC to available chemotherapeutic agents during the past decades.3,4 It is pressingly needed to fully understand the molecular mechanisms underlying OC development and progression, as well as the drug resistances of cancer cells. Hopefully, recent investigations revealed that epigenetic alterations performed critical roles in OC by regulating both the pathogenic processes and multi-drug resistances (MDRs),5,6 which caused extensive concerns in the cancer research community.
Non-coding RNAs such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have been characterized as essential players of epigenetic regulations in various biological and pathogenic processes in cancers.7,8 For instance, the lncRNA ZFAS1 (zinc finger antisense 1), previously shown downregulated in breast cancer and regulated alveolar development and epithelial cell differentiation, has been differentially expressed and associated with many other cancer types such as hepatocellular carcinoma and OC.9 Moreover, the ZFAS1 expression was greatly elevated by cisplatin in OC cells and suggested its potential involvement in chemotherapy resistance, which deserves further investigation.10 In addition, miRNAs also exerted key regulatory effects in OC development.11 Two miRNAs, miR-548a-3p and miR-548c-3p, were specifically expressed in human oocytes,11 and the downregulation of miR-548c-3p in OC tissues was recently reported to closely associate with cancer cell proliferation, migration, and invasion.12 However, little is known about the expressional regulation and roles of other members of this miRNA family, such as miR-548a/e/az-3p (miR-548a/e/az), during OC pathogenesis. Bioinformatic analysis predicted miR-548a/e/az as potential targets of the lncRNA ZFAS1, which was not previously reported and might be an essential signaling axis driving OC development and drug resistance.
Chemokine receptor 4 (CXCR4) acted as a unique receptor of CXC chemokine ligand 12 (CXCL12) and performed key roles in cancer development and metastasis.13,14 Early investigation has characterized CXCR4 as an independent prognostic factor for OC patients, as evidenced by its high expression in refractory and recurrent OC cases, which was closely correlated with lymph node metastasis.15 The CXCL12/CXCR4 axis was shown to be extensively involved in OC development by regulating cancer cell proliferation, migration, invasion, and metastasis, as well as the myeloid-derived suppressor cell accumulation in the cancer environment.16,17 Moreover, highly expressed CXCR4 was also linked with the resistances of OC cells to distinct chemotherapies.18,19 However, the mechanisms elevating CXCR4 expression in OC remain poorly understood. It is also predicted by bioinformatic analysis that CXCR4 could be directly targeted by miR-548a/e/az, which has not been experimentally validated so far.
In addition, the pathogenic roles of elevated CXCR4 expression in tumorigenesis and chemotherapy resistances have been mediated by its suppression of let-7a expression.20,21 In acute myeloid leukemia (AML), CXCR4 activation greatly downregulated the expression of let-7a, and let-7a could enhance AML cells’ sensitivity to cytarabine treatment by suppressing c-Myc and BCL-XL (B-cell lymphoma-extra large).21 Similarly, the let-7a expression was also reported to be repressed by CXCR4 in pancreatic cancer cells (PCCs), thus resulting into progression of PCC tumorigenesis, metastasis, and chemotherapy resistance by targeting and suppressing the expression of high mobility group A2 protein (HMGA2).20 However, the suppression of let-7a expression by CXCR4 in drug resistances of OC still remains unknown.
In this study, we verified our hypothesis that lncRNA ZFAS1 could directly target and suppress miR-548e expression to elevate CXCR4 expression, which promoted OC metastasis and meanwhile altered the downstream let-7a/BCL-XL/S axis to enhance chemotherapy resistance to cisplatin (please see Figure 10). These results unveiled a new epigenetic alteration-mediated signaling underlying OC pathogenesis, which also provided a basis for non-coding-RNA-based OC early diagnosis and novel anti-cancer drug development.
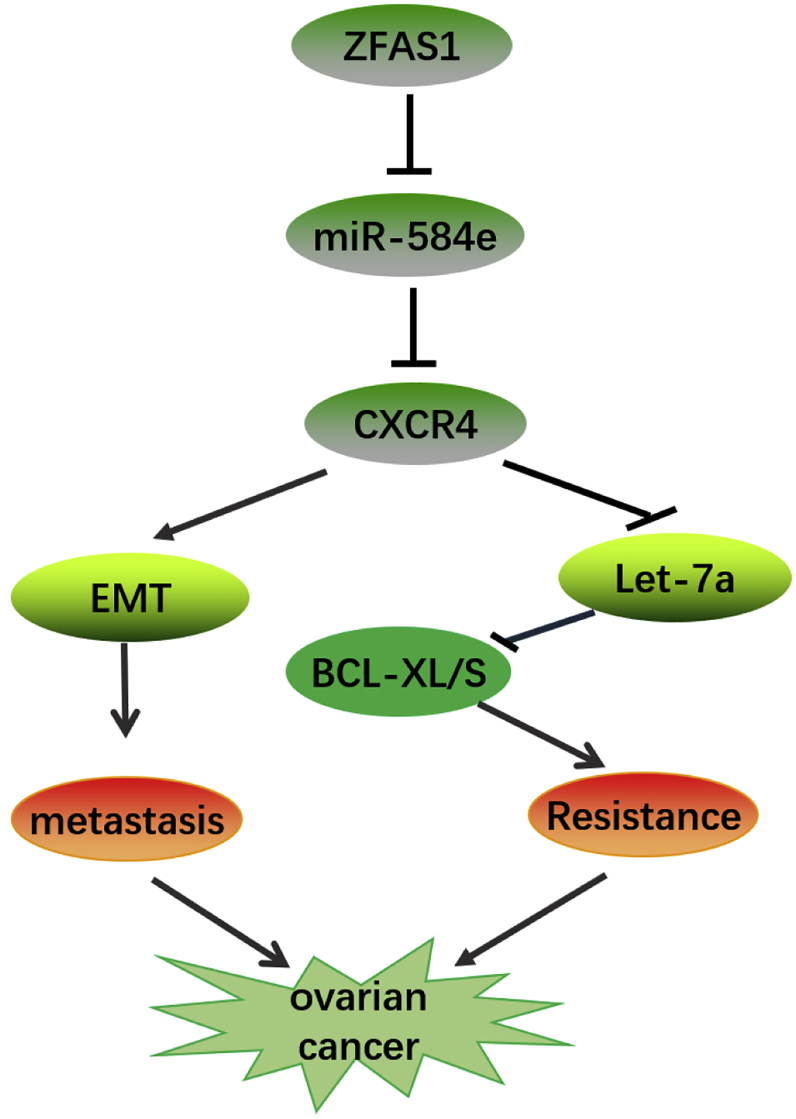
Figure 10.
A Schematic Illustration of ZFAS1/miR-548e/CXCR4 Signaling Axis in Regulating OC Cell Proliferation, Metastasis, and Cisplatin Resistance
In OC cells, ZFAS1 binds with miR-548e to enhance CXCR4 expression to promote OC cell proliferation, metastasis, and invasion, resulting in EMT and OC metastasis. Under cisplatin treatment, the ZFAS1/miR-548e/CXCR4 axis further suppresses let-7a expression and elevates BCL-XL/S expression to enhance OC cell resistance to cisplatin treatment.
Results
ZFAS1 Targets and Suppresses miR-548e Expression in OC
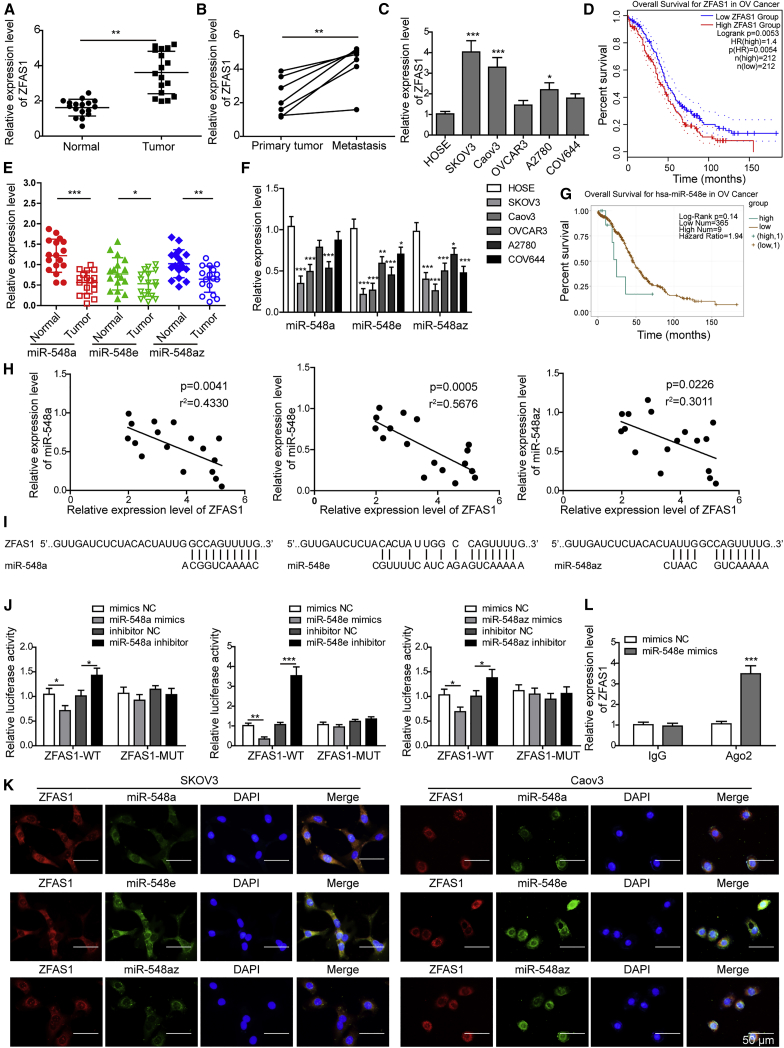
To investigate the association of ZFAS1 and the miR-548 family during OC pathogenesis, we first detected the expression of ZFAS1 and miR-548 members in OC tissues and cell lines. We observed that ZFAS1 expression was significantly increased in OC tissues compared with the adjacent non-cancerous tissues (Figure 1A). Also, ZFAS1 expression in metastatic cancer tissues in lymph nodes from seven metastatic OC patients was remarkably higher than those primary ovarian tumors (Figure 1B). Consistently, we observed that ZFAS1 expression was significantly elevated in OC cell lines SKOV3, Caov3, and A2780 and slightly increased in OC cell lines OVCAR3 and COV644, compared with the human ovarian epithelial (HOSE) cells (Figure 1C). GEPIA (gene expression profiling interactive analysis) data showed that high ZFAS1 expression was correlated with poor survival time in OC patients (Figure 1D). On the contrary, the expression of miR-548a, miR-548e, and miR-548az in both OC tissues and cell lines showed a significant decrease compared with corresponding controls (Figures 1E and 1F). Moreover, low expression of miR-548e in OC patients was correlated with reduced percent survival, compared with those with high miR-548e expression (Figure 1G). Furthermore, we found that ZFAS1 expression showed significantly negative correlation with miR-548a, miR-548e, and miR-548az expression levels in OC tissues (Figure 1H).
Figure 1.
ZFAS1 Targets and Suppresses miR-548e Expression in Ovarian Cancer (OC)
(A) Relative expression of ZFAS1 in OC tissues and adjacent non-cancerous tissues. ZFAS1 expression levels in ovarian tissues from 17 OC patients were determined by qRT-PCR. (B) Relative expression of ZFAS1 in primary and metastatic ovarian tumor tissues from seven metastatic OC patients by qRT-PCR. (C) ZFAS1 expressional alterations in OC cell lines SKOV3, Caov3, OVCAR3, A2780, and COV644. HOSE cells were used as the control. (D) Correlation of ZFAS1 expression levels with OC patient survival time based on the GEPIA database. (E) miR-548 family member expressions in OC tissues and adjacent non-cancerous tissues by qRT-PCR. (F) miR-548 family member expressions in SKOV3, Caov3, OVCAR3, A2780, COV644, and HOSE cells. (G) Correlation between miR-548e expression with OC patient survival time based on TCGA data. (H) Negative correlations between ZFAS1 with miR-548a, miR-548e, or miR-548az in OC tissues. (I) The binding sites of ZFAS1 with miR-548a, miR-548e, or miR-548az as predicted by bioinformatics. (J) Validation of the binding of ZFAS1 with miR-548a, miR-548e, or miR-548az in HEK293T cells by dual-luciferase reporter assay. (K) Co-localization of ZFAS1 with miR-548a, miR-548e, or miR-548az in SKOV3 and Caov3 cells by fluorescence in situ hybridization (FISH). Scale bar: 50 μm. (L) The binding of ZFAS1 with miR-548e in HEK293T cells confirmed by RIP assay. ZFAS1, zinc finger antisense 1; SD, standard deviation; WT, wild-type; MUT, mutant; NC, negative control; Ago2, argonaute 2; shZFAS1, short hairpin RNA targeting ZFAS1; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Bioinformatics analysis predicted that ZFAS1 could directly bind with miR-548 family members (Figure 1I). To test this, we performed dual-luciferase reporter assays to validate their association and found that ZFAS1 could bind with all three miR-548 family members (Figure 1J). Among them, ZFAS1 showed the greatest affinity with miR-548e sequences, as shown by the greatly enhanced or suppressed luciferase signals in HEK293T cells expressing wild-type ZFAS1 (ZFAS1-WT) treated with miR-548e inhibitor or mimics, respectively, which were not observed in HEK293T cells expressing mutant ZFAS1 (ZFAS1-MUT) (Figure 1J). Through a fluorescence in situ hybridization (FISH) assay, we found that ZFAS1 was co-localized with miR-548a, miR-548e, and miR-548az in cytosols of SKOV3 and Caov3 cells, but the most significant co-localization was observed between ZFAS1 and miR-548e (Figure 1K). Our RNA immunoprecipitation (RIP) assay also showed a great increase of co-precipitated ZFAS1 levels after cells being treated with miR-548e mimics compared with negative control (Figure 1L). For further validation, we suppressed and elevated ZFAS1 expression in SKOV3 and Caov3 cells that exhibited the highest ZFAS1 expression level by transfecting them with shZFAS1 and recombinant pcDNA3.1-ZFAS1, respectively (Figure S1A). In these two OC cell lines, ZFAS1 silencing significantly produced the increased miR-548e expression, while ZFAS1 overexpression resulted into greatly repressed miR-548e expression (Figure S1B). These results proved that the highly expressed ZFAS1 suppressed miR-548e expression during OC development through direct binding.
ZFAS1 Promotes OC Progression and Cisplatin Resistance by Suppressing miR-548e Expression
To investigate the cellular functions of ZFAS1 and miR-548e, the OC cell lines with simultaneous silencing of both ZFAS1 and miR-548e were established (Figure S2A). The miR-548e expression levels in SKOV3 and Caov3 cells were elevated by shZFAS1 transfection, which were then significantly downregulated by co-transfection with miR-548e inhibitor (Figure S2B). Importantly, we observed by cell counting kit-8 (CCK-8) clone formation, would healing, and the Transwell system that the proliferation rates, migration, and invasion capacities of SKOV3 and Caov3 cells were significantly suppressed by shZFAS1 transfection, and they were then effectively reversed by treatment the with miR-548e inhibitor (Figures 2A–2D). Consistently, we found that the expression levels of E-cadherin in SKOV3 and Caov3 cells were markedly increased by short hairpin RNA (shRNA)-mediated ZFAS1 silencing, while the expression levels of N-cadherin, vimentin, MMP-2 (matrix metalloproteinases 2), and Slug proteins in SKOV3 and Caov3 cells were significantly downregulated by shZFAS1 transfection (Figure 2E). However, miR-548e inhibitor treatment significantly suppressed E-cadherin and promoted N-cadherin, vimentin, MMP-2, and Slug protein levels in SKOV3 and Caov3 cells induced by shZFAS1 (Figure 2E). These results showed that ZFAS1 promoted OC cell proliferation, migration, and invasion via repressing miR-548e expression.
Figure 2.
ZFAS1 Enhances OC Cell Proliferation, Migration, and Invasion by Suppressing miR-548e Expression
(A and B) The proliferation rates of SKOV3 and Caov3 cells transfected with shZFAS1 and miR-548e inhibitor. Cell proliferations were analyzed by CCK-8 (A) and clone formation assay (B), respectively. (C) The migration capacities of SKOV3 and Caov3 cells transfected with shZFAS1 and miR-548e inhibitor. Cell migration was analyzed by wound-healing assay. (D) The invasion capacities of SKOV3 and Caov3 cells transfected with shZFAS1 and miR-548e inhibitor. Cell invasion capacity was assessed using the Transwell system. (E) Protein abundances of E-cadherin, N-cadherin, vimentin, MMP-2, and Slug in SKOV3 and Caov3 cells transfected with shZFAS1 and miR-548e inhibitor. Protein levels were determined by western blotting. ZFAS1, zinc finger antisense 1; shZFAS1, short hairpin RNA targeting ZFAS1; NC, negative control; MMP-2, matrix metalloproteinases 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
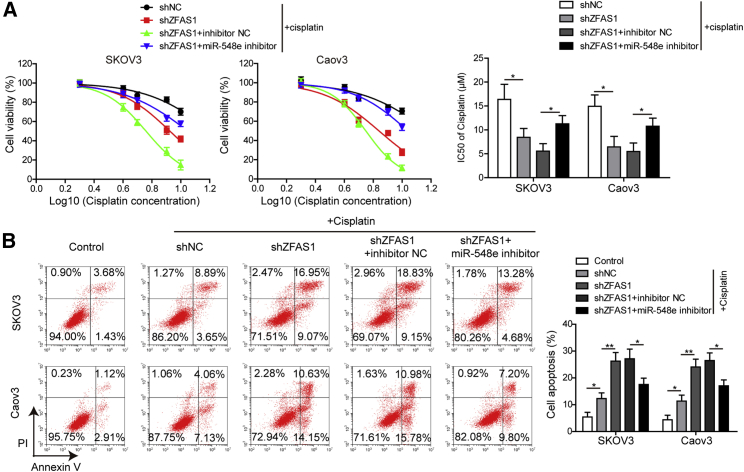
Moreover, we found that the resistances of SKOV3 and Caov3 cells to cisplatin treatment were also greatly impaired by shZFAS1 transfection, but simultaneous treatment with miR-548e inhibitor significantly recovered the cisplatin resistance of both SKOV3 and Caov3 cells transfected with shZFAS1 (Figure 3A). The half-maximal inhibitory concentration (IC50) values of cisplatin in SKOV3 and Caov3 cells showed a significant decrease after shZFAS1 transfection, and the IC50 values were then greatly elevated by co-transfection with miR-548e inhibitor (Figure 3A). In addition, we observed that cisplatin treatment induced apoptosis in SKOV3 and Caov3 cells, and the cisplatin-induced apoptosis of SKOV3 and Caov3 cells was remarkably promoted by shZFAS1 transfection (Figure 3B). But miR-548e inhibitor significantly suppressed the apoptosis of SKOV3 and Caov3 cells transfected with shZFAS1 (Figure 3B). Together, these observations showed that ZFAS1 silencing could effectively inhibit cisplatin resistance through its suppression on miR-548e expression.
Figure 3.
ZFAS1 Promotes OC Cell Resistance to Cisplatin by Inhibiting miR-548e Expression
(A) The cisplatin resistance of SKOV3 and Caov3 cells transfected with shZFAS1 and miR-548e inhibitor. The viability of cells treated with 2, 4, 6, 8, and 10 μM cisplatin were measured by CCK-8 assay for calculate of the IC50 values. (B) The apoptosis of SKOV3 and Caov3 cells transfected with shZFAS1 and miR-548e-3p inhibitor under cisplatin treatment. Cell apoptosis was evaluated by the flow cytometry. ZFAS1, zinc finger antisense 1; shZFAS1, short hairpin RNA targeting ZFAS1; NC, negative control; IC50, half maximal inhibitory concentration; ∗p < 0.05 and ∗∗p < 0.01.
miR-548e Targets and Suppresses CXCR4 Expression in OC
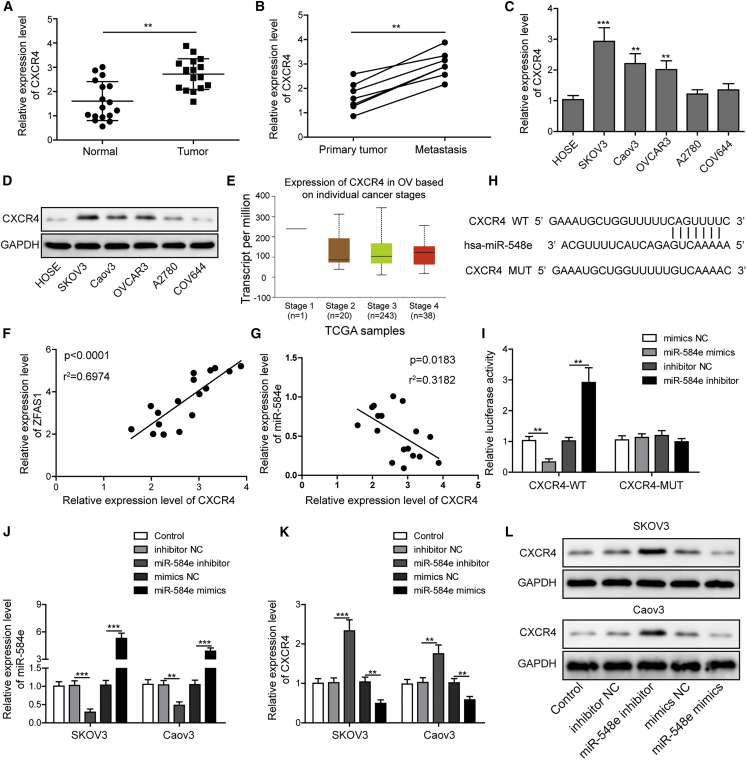
To explore the involvement of CXCR4 and its relationship with miR-548e in OC pathogenesis, we subsequently analyzed the expression of CXCR4 in OC tissues and cell lines. We showed here that CXCR4 expression was significantly elevated in OC tissues compared with the corresponding adjacent ovarian tissues (Figure 4A). Also, CXCR4 expression in metastatic cancer tissues in lymph nodes was significantly higher than that in primary ovarian tumor tissues (Figure 4B). Both the mRNA and protein levels of CXCR4 in OC cell lines SKOV3, Caov3, and OVCAR3 were all significantly higher than the HOSE cells (Figures 4C and 4D). Moreover, The Cancer Genome Atlas (TCGA) data analysis revealed that OC patients at later stages (stage IV) exhibited slightly higher expression of CXCR4 compared with those at earlier stages (stages II and III) (Figure 4E).
Figure 4.
miR-548e Suppresses CXCR4 Expression in OC via Direct Binding
(A) Relative CXCR4 expression in OC tissues and adjacent non-cancerous tissues by qRT-PCR. (B) Relative CXCR4 expression in primary and metastatic ovarian tumor tissues from seven metastatic OC patients by qRT-PCR. (C and D) CXCR4 expressional levels in OC cell lines SKOV3, Caov3, OVCAR3, A2780, COV644, and HOSE cells by qRT-PCR (C) and western blotting (D). HOSE cells were measured as the control. (E) Correlation of CXCR4 expression with OC patient stages based on TCGA data. (F and G) The correlations of CXCR4 expression with ZFAS1 (F) and miR-548e (G) expression in OC tissues. (H) The binding sites of miR-548e with CXCR4 gene 3′ UTR region. (I) The binding validation through dual-luciferase reporter assay in HEK293T cells. (J) Silencing or overexpression of miR-548e expression in both SKOV3 and Caov3 cells with miR-548e inhibitor and mimics, respectively. miR-548e expression was evaluated by qRT-PCR. (K and L) The expression of CXCR4 in SKOV3 and Caov3 cells with silenced or enhanced miR-548e expression. CXCR4 level was detected by qRT-PCR (K) and western blotting (L), respectively. CXCR4, chemokine receptor 4; SD, standard deviation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ZFAS1, zinc finger antisense 1; WT, wild-type; MUT, mutant; UTR, untranslated region; NC, negative control; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Furthermore, we found that CXCR4 expression in OC tissues exhibited significantly positive correlation with ZFAS1 expression but significantly negative correlation with miR-548e expression in OC tissues (Figures 4F and 4G). Our bioinformatics analysis showed that 3′ UTR region of the CXCR4 could be directly targeted by miR-548e, which was then verified by the dual-luciferase reporter assay in HEK293T cells (Figure 4H). The elevated or suppressed luciferase activities in HEK293T cells expressing the WT CXCR4 sequences (CXCR4-WT) were induced by miR-548e mimics or inhibitor, respectively, but not in HEK293T cells expressing MUT CXCR4 sequences (CXCR4-MUT), proving the direct binding of miR-548e with CXCR4 (Figure 4I). In addition, the miR-548e expression in SKOV3 and Caov3 cells were then silenced or enhanced by transfecting with miR-548e inhibitor or mimics, respectively (Figure 4J). In these cells, we observed that miR-548e inhibitor greatly increased CXCR4 mRNA and protein levels, while miR-548e mimics significantly downregulated CXCR4 mRNA and protein levels in both SKOV3 and Caov3 cells (Figures 4K and 4L). These results convincingly validated that the CXCR4 expression in OC cells were effectively suppressed by the binding of miR-548e.
miR-548e Represses OC Progression and Cisplatin Resistance by Targeting CXCR4 Expression
To analyze the pathogenic role of CXCR4 mediated by miR-548e, we established SKOV3 and Caov3 cells with elevated miR-548e expression by mimics and CXCR4 overexpression. The expression levels of miR-548e in SKOV3 and Caov3 cells were not influenced by the simultaneous transfection with recombinant pcDNA3.1-CXCR4 plasmids (Figure S3A). However, the expression levels of CXCR4 in SKOV3 and Caov3 cells were significantly suppressed by miR-548e mimics, which were then partially recovered by transfection with recombinant pcDNA3.1-CXCR4 plasmids (Figure S3B). In the above cells, we observed that miR-548e mimic treatment resulted in significant suppression of the proliferation, migration, and invasion capacities of SKOV3 and Caov3 cells, which were then remarkably mitigated by CXCR4 overexpression (Figures 5A–5D). Consistently, the E-cadherin expressions in both SKOV3 and Caov3 cells were significantly promoted by miR-548e mimics, while the expression levels of N-cadherin, vimentin, MMP-2, and Slug proteins in these two cell lines were greatly downregulated by miR-548e mimics (Figure 5E). On the contrary, CXCR4 overexpression significantly repressed E-cadherin but enhanced N-cadherin, vimentin, MMP-2, and Slug expressions in both SKOV3 and Caov3 cells treated with miR-548e mimics (Figure 5E).
Figure 5.
miR-548e Inhibits OC Cell Proliferation, Migration, and Invasion by Suppressing CXCR4 Expression
(A and B) The proliferation rates of SKOV3 and Caov3 cells with enhanced miR-548e and CXCR4 expression levels. Both the CCK-8 (A) and clone-formation assay (B) were used to detect cell proliferation. (C) The migration capacities of SKOV3 and Caov3 cells transfected with miR-548e mimics and CXCR4-overexpressing plasmids. Cell migration was measured through the wound-healing assay. (D) The invasion capacities of SKOV3 and Caov3 cells with enhanced miR-548e and CXCR4 expression levels. Cell invasion capacity was analyzed with the Transwell system. (E) E-cadherin, N-cadherin, vimentin, MMP-2, and Slug protein levels in SKOV3 and Caov3 cells transfected with miR-548e mimics and CXCR4-overexpressing plasmids. Protein abundance was measured by western blotting. NC, negative control; CXCR4, chemokine receptor 4; MMP-2, matrix metalloproteinases 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
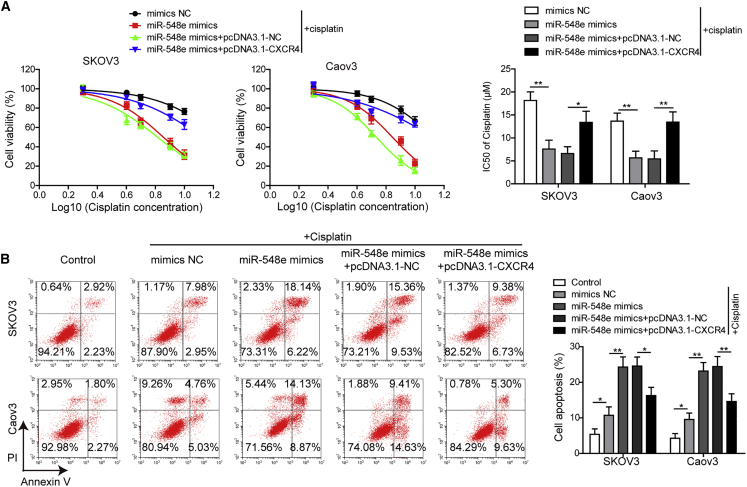
Also, cisplatin resistances of SKOV3 and Caov3 cells were greatly suppressed by miR-548e mimics, which were then enhanced by CXCR4 overexpression in SKOV3 and Caov3 cells transfected with miR-548e mimics (Figure 6A). Specifically, miR-548e mimics significantly downregulated the IC50 values of cisplatin in SKOV3 and Caov3 cells, which were then greatly increased by co-transfection with CXCR4-overexpressing vectors in both cell lines (Figure 6A). Finally, we found that cisplatin-induced SKOV3 and Caov3 cell apoptosis was greatly enhanced by miR-548e mimics, but CXCR4 overexpression significantly inhibited the apoptosis of SKOV3 and Caov3 cells transfected with miR-548e mimics (Figure 6B). These results suggested that miR-548e overexpression could effectively inhibit OC cell proliferation, invasion, and cisplatin resistance, which was mediated by its suppression on CXCR4 expression.
Figure 6.
miR-548e Impairs OC Cell Resistance to Cisplatin by Repressing CXCR4 Expression
(A) Effects of miR-548e mimics and CXCR4 overexpression on resistance of SKOV3 and Caov3 cells to cisplatin treatments. Viabilities of cells treated with 2, 4, 6, 8, or 10 μM cisplatin were detected using the CCK-8 assay, and IC50 values were calculated to evaluate drug resistance. (B) The cisplatin-induced SKOV3 and Caov3 cell apoptosis after being transfected with miR-548e mimics and CXCR4 overexpressing plasmids. Flow cytometry was used to detect cell apoptosis. NC, negative control; CXCR4, chemokine receptor 4; IC50, half maximal inhibitory concentration; ∗p < 0.05 and ∗∗p < 0.01.
CXCR4 Enhances OC Cell Resistance to Cisplatin by Regulating the let-7a/BCL-XL/S Axis
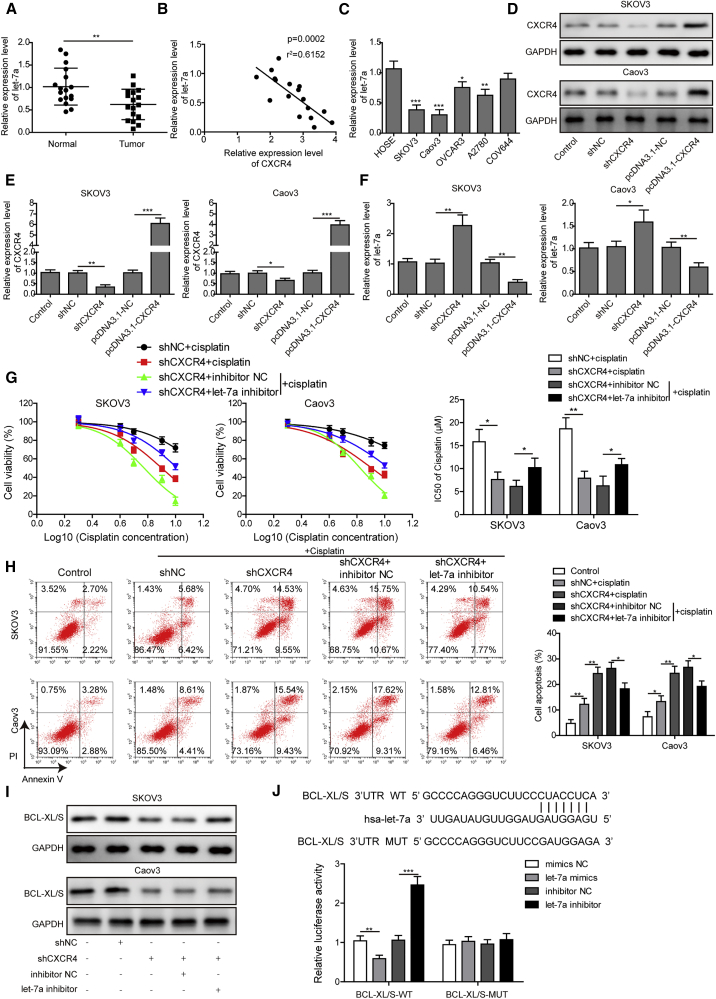
For more insights into the roles of CXCR4 in regulating cisplatin resistance, we further analyzed the involvement of the let-7a in these processes. First, we observed that let-7a expression levels in OC tissues were significantly lower than these adjacent non-cancerous ovarian tissues (Figure 7A). Moreover, we found that let-7a expression showed a significantly negative correlation with CXCR4 expression in OC tissues (Figure 7B). In these OC cell lines, the expression levels of let-7a were also greatly lowered compared with the HOSE cells (Figure 7C). Next, we separately suppressed and elevated CXCR4 expressions in SKOV3 and Caov3 cells by transfecting with shCXCR4 or the recombinant pcDNA3.1-CXCR4 plasmids, respectively (Figures 7D and 7E). Moreover, we showed that let-7a expression levels in both the SKOV3 and Caov3 cells were significantly elevated by transfection with shCXCR4 but remarkably suppressed by transfection with the pcDNA3.1-CXCR4 plasmids (Figure 7F), indicative of the inhibitory roles of CXCR4 on let-7a expression in OC cells.
Figure 7.
CXCR4 Promotes Cisplatin Resistance of OC Cells through Suppressing the let-7a/BCL-XL/S Axis
(A) Relative let-7a expression levels in OC tissues and adjacent non-cancerous ovarian tissues. Expressional changes of let-7a in OC tissues were analyzed by qRT-PCR. (B) Negative correlation of let-7a with CXCR4 expression in OC tissues. (C) Relative let-7a expression in SKOV3, Caov3, OVCAR3, A2780, and CAV644 cells. Its expression in HOSE cells was detected as the control. (D and E) Silencing and elevation of CXCR4 expression in SKOV3 and Caov3 cells with shCXCR4 and pcDNA3.1-CXCR4 plasmids, respectively. Expression levels of CXCR4 in SKOV3 and Caov3 cells were detected by qRT-PCR (D) and western blotting (E). (F) Relative expression of let-7a in SKOV3 and Caov3 cells with silenced or elevated CXCR4 expression by qRT-PCR. (G) Influences of shCXCR4 and let-7a inhibitor on cisplatin resistance of SKOV3 and Caov3 cells. Cell viabilities were detected by the CCK-8 method under treatments with 2, 4, 6, 8, or 10 μM cisplatin, and the resistances were assessed by the IC50 values. (H) Effects of shCXCR4 and let-7a inhibitor on cisplatin-induced SKOV3 and Caov3 cell apoptosis. Flow cytometry was performed to detect cell apoptosis. (I) Abundances of BCL-XL/S proteins in SKOV3 and Caov3 cells transfected with shCXCR4 and let-7a inhibitor. Protein levels were analyzed by western blotting. (J) The binding of let-7a with BCL-XL/S gene sequences predicted by informatics (top) and confirmed by dual-luciferase reporter assay (bottom). NC, negative control; CXCR4, chemokine receptor 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IC50, half maximal inhibitory concentration; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
More importantly, we found that the cisplatin resistances of both SKOV3 and Caov3 cells were greatly impaired by transfection with shCXCR4, and IC50 values were then significantly recovered by let-7a inhibitor in SKOV3 and Caov3 cells transfected with shCXCR4 (Figure 7G). The IC50 values of cisplatin in SKOV3 and Caov3 cells were significantly decreased by shCXCR4 but effectively elevated by the let-7a inhibitor (Figure 7G). Consistently, shCXCR4 induced the marked increase of SKOV3 and Caov3 cell apoptosis under cisplatin treatment but the let-7a inhibitor significantly suppressed the apoptosis of SKOV3 and Caov3 cells transfected with shCXCR4 under cisplatin treatment (Figure 7H). Furthermore, we also showed that shCXCR4 induced the significant decrease of BCL-XL/S (Bcl-2-like protein 1) expression in SKOV3 and Caov3 cells, which was mitigated by simultaneous transfection with let-7a inhibitor (Figure 7I). Finally, our bioinformatics analysis predicted that let-7a could directly bind with the 3′ UTR region of the BCL-XL/S gene (Figure 7J). In HEK293T cells expressing the WT BCL-XL/S, the luciferase activity was significantly suppressed by treatment with let-7a mimics but was greatly elevated by the let-7a inhibitor (Figure 7J), showing the direct binding of let-7a with BCL-XL/S. Together, these observations indicated that CXCR4 promoted the cisplatin resistance of OC cells by negatively regulating the let-7a/BCL-XL/S axis.
ZFAS1 Promotes OC Cisplatin Resistance by Regulating the let-7a/BCL-XL/S Signaling Axis
To further analyze the roles of the let-7a/BCL-XL/S axis in ZFAS1-regulated OC chemotherapy resistance, the SKOV3 and Caov3 cells were co-transfected with pcDNA3.1-ZFAS1 plasmids and let-7a mimics. We observed that ZFAS1 expression in both SKOV3 and Caov3 cells were greatly increased by pcDNA3.1-ZFAS1 plasmids but not altered by the combined transfection with let-7a mimics (Figure 8A). However, the expression levels of let-7a in SKOV3 and Caov3 cells were greatly suppressed by ZFAS1 overexpression, while let-7a expression levels were then remarkably promoted by let-7a mimics (Figure 8B). Also, we found that the BCL-XL/S protein levels were markedly upregulated by ZFAS1 overexpression but were then significantly downregulated by let-7a mimics in SKOV3 and Caov3 cells (Figure 8C). More importantly, we also showed that the enhanced cisplatin resistance of SKOV3 and Caov3 cells by ZFAS1 overexpression was also significantly suppressed by simultaneous transfection with let-7a mimics (Figure 8D). The IC50 values of cisplatin in SKOV3 and Caov3 cells were increased by ZFAS1 overexpression but greatly downregulated by co-transfection with let-7a mimics (Figure 8D). Consistently, ZFAS1 overexpression reduced the apoptotic SKOV3 and Caov3 cells under cisplatin treatment, which were also significantly abrogated by the transfection with let-7a mimics (Figure 8E).
Figure 8.
ZFAS1 Promotes OC Cisplatin Resistance by Suppressing the let-7a/BCL-XL/S Signaling Axis
(A) ZFAS1 relative expression levels in SKOV3 and Caov3 cells with ZFAS1 overexpression and transfected with let-7a mimics. ZFAS1 expression was detected by the qRT-PCR method. (B) Relative let-7a expression levels in SKOV3 and Caov3 cells with ZFAS1 overexpression and transfected with let-7a mimics by the qRT-PCR method. (C) BCL-XL/S protein abundances in SKOV3 and Caov3 cells with elevated ZFAS1 and let-7a expression by western blotting. GAPDH was included as the internal standard. (D) Cisplatin resistances of SKOV3 and Caov3 cells with ZFAS1 overexpression and transfected with let-7a mimics. Cell viabilities measured by the CCK-8 method under cisplatin treatments were used to calculate the IC50 values. (E) The apoptosis of SKOV3 and Caov3 cells with elevated ZFAS1 and let-7a expression by flow cytometry. (F) Tumors generated from SKOV3 cells transfected let-7a mimics in nude mice under cisplatin treatment. Cell line-based xenograft (CDX) models were established using nude mice to evaluated cisplatin resistance in vivo. (G) The sizes and weights of tumors developed from SKOV3 cells transfected let-7a mimics in nude mice under cisplatin treatment. (H) Ki-67 protein levels in tumors generated from SKOV3 cells transfected let-7a mimics in nude mice under cisplatin treatment. Scale bar: 50 μm. (I) BCL-XL/S mRNA levels in tumors developed from SKOV3 cells transfected let-7a mimics in nude mice under cisplatin treatment. ZFAS1, Zinc finger antisense 1; NC, negative control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IC50, half maximal inhibitory concentration; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
To provide in vivo evidence, we further investigated the effects of let-7a on cisplatin resistance using the cell-line-based xenograft models (Figure 8F). After transplanting SKOV3 cells transfected with let-7a mimics into nude mice, we found that the sizes and weights of tumors generated from SKOV3 cells transfected with let-7a mimics under cisplatin treatment were significantly lower than those from the negative controls (Figures 8F and 8G). Consistently, we demonstrated that the Ki-67 expression in tumors from SKOV3 cells under cisplatin treatment were greatly suppressed by let-7a mimics, compared with that in tumors from negative control cells, indicating the suppressed cancer cell proliferation by let-7a in in vivo tumorigenesis model (Figure 8H). Moreover, the mRNA levels of BCL-XL/S in tumors derived from SKOV3 cells transfected with let-7a mimics in nude mice under cisplatin treatment were also significantly lower than those from negative control cancer cells (Figure 8I). Together, these results indicated that cisplatin resistance enhanced by ZFAS1 in OC was mediated by the suppression of let-7a expression and the resultant alteration of BCL-XL/S expression.
ZFAS1 Knockdown or miR-548e Overexpression Suppresses Cisplatin Resistance and Lung Metastasis of OC in Animal Models
For further verification of ZFAS1 and miR-548e’s pathogenic roles, we finally investigated the influences of their expressional alterations on OC cisplatin resistance and lung metastasis using in vivo tumorigenesis models (Figure 9A). We found that tumors developed in nude mice injected with SKOV3 cells transfected with shZFAS1 or miR-548e mimics under cisplatin treatment exhibited remarkably reduced sizes and weights, compared with those from negative control cells in nude mice (Figures 9A and 9B). Consistently, the Ki-67 assay showed that proliferation rates of cells in tumors derived from SKOV3 cells with shZFAS1 under cisplatin treatment were greatly suppressed compared with the negative controls (Figure 9C). Also, the expression of BCL-XL/S in tumor tissues showed similar decrease induced by shZFAS1 or miR-548e mimics (Figure 9D). These analyses based on an in vivo model further validated the regulating roles of ZFAS1 and miR-548e in cisplatin resistance of OC.
Figure 9.
ZFAS1 Knockdown or miR-548e Overexpression Represses OC Cisplatin Resistance and Lung Metastasis in Models of In Vivo Tumorigenesis
(A) Tumors formed in nude mice injected with SKOV3 cells transfected with shZFAS1 or miR-548e mimics under cisplatin treatment. (B) The sizes and weights of tumors formed in nude mice injected with SKOV3 cells transfected with shZFAS1 or miR-548e mimics under cisplatin treatment. (C) Cell proliferation rates in tumors developed from SKOV3 cells transfected with shZFAS1 in nude mice under cisplatin treatment. Ki-67 expression was detected by immunohistochemistry to evaluate cell proliferation. Scale bar: 50 μm. (D) BCL-XL/S abundances in tumors generated in nude mice injected with SKOV3 cells transfected with shZFAS1 or miR-548e mimics under cisplatin treatment. (E) Metastatic lung nodules developed in nude mice injected with SKOV3 cells transfected with shZFAS1. (F and G) Relative expression of E-cadherin (F) and N-cadherin (G) in metastatic lung nodules developed from SKOV3 cells transfected with shZFAS1 or miR-548e mimics. (H) ZFAS1 expression in metastatic lung nodules derived from SKOV3 cells transfected with shZFAS1 by qRT-PCR. (I and J) Relative miR-548e (I) and CXCR4 (J) expression in metastatic lung nodule tissues generated in nude mice injected with SKOV3 cells transfected with shZFAS1 or miR-548e mimics. ZFAS1, zinc finger antisense 1; NC, negative control; shZFAS1, short hairpin RNA targeting ZFAS1; CXCR4, chemokine receptor 4; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
In addition, we revealed using the in vivo model that the metastatic lung nodule formation in nude mice injected with SKOV3 cells transfected with shZFAS1 was significantly suppressed, in comparison with those injected with negative control cells (Figure 9E). Consistently, we also found that E-cadherin expression in metastatic lung nodules formed in nude mice injected with SKOV3 cells with shZFAS1 or miR-548e mimics were significantly elevated compared with the negative controls (Figure 9F). On the contrary, the expression of N-cadherin showed opposite alterations in metastatic lung nodules formed in nude mice (Figure 9G). The decreased ZFAS1 expression was also detected in metastatic lung nodules developed in nude mice injected with SKOV3 cells transfected with shZFAS1 (Figure 9H). Also in metastatic lung nudes formed in nude mice, the expression of miR-548e was markedly elevated by SKOV3 cells with both shZFAS1 and miR-548e mimics (Figure 9I). Contrarily, the expression levels of CXCR4 in metastatic lung nodules developed in nude mice were significantly downregulated by pre-injecting SKOV3 cells with shZFAS1 or miR-548e mimics (Figure 9J). Together, these results from the in vivo animal model proved that ZFAS1 knockdown or miR-548e overexpression could effectively repress cisplatin resistance and lung metastasis of OC.
Discussion
OC is a severe human malignancy featured by hidden symptoms at an early stage, rapid progression, metastasis, and poor prognosis.22 Even worse, current therapies for OC have usually been limited by multiple intrinsic and acquired resistances to available chemotherapy drugs.3,4 Hence, it is of immense clinical significance to investigate the molecular mechanisms underlying OC pathogenesis and drug resistances. In the present study, we extensively investigated the pathogenic roles of lncRNA ZFAS1 and its interaction with the miR-548e in OC cell proliferation, migration, invasion, and resistance to cisplatin treatment. We revealed for the first time that the high expression of ZFAS1 in OC cells bound with miR-548e to promote CXCR4 expression, thus regulating OC cell proliferation, migration, invasion, and cisplatin resistance. Furthermore, we proved that CXCR4 exerted its cisplatin-resistance-promoting roles by negatively regulating let-7a, which suppressed BCL-XL/S expression. The roles of ZFAS1, miR-548e, and let-7a on cisplatin resistance and lung metastasis of OC were finally confirmed by in vivo tumorigenesis model by injecting nude mice with modified OC cells. Together, we revealed a new ZFAS1/miR-548e/CXCR4 axis driving OC metastasis and promoted OC cisplatin resistance by modulating the let-7a/BCL-XL/S signaling (Figure 10), which also provided novel targets for OC early diagnosis and new treatment development.
lncRNAs are a large group of non-coding transcripts with a length of over 200 nucleotides that post-transcriptionally modulate gene expression mainly by acting as sponges of target miRNAs.23 It has been well documented during the past decades that lncRNA-miRNA networks performed key regulatory roles in the pathogenesis, metastasis, and multiple drug resistance of various human cancers.24,25 Among a large quantity of lncRNAs associated with tumorigenesis, ZFAS1 is a lncRNA originally characterized as mammary development regulator, which was later reported to be closely associated with a number of human cancers.26 Although ZFAS1 was shown to regulate OC malignancy and elevated in cisplatin-treated OC cell lines,10,27 the downstream signaling cascades mediating its roles in OC pathogenesis, metastasis, and cisplatin resistance still remain unknown. In the present study, we confirmed that ZFAS1 was highly expressed in OC tissues and cell lines, which was negatively correlated with the expression of miR-548e and other miR-548 family members in OC. ZFAS1 and miR-548e showed negative and positive correlations with OC patient survival time, respectively. Based on bioinformatics prediction, we validated for the first time that ZFAS1 could suppress miR-548e expression, which was verified by combination of the dual-luciferase reporter assay and RIP methods in this study. We also indicated that ZFAS1 and miR-548 family members were mainly co-localized in the cytosols of OC cells by FISH assay. Through modulating ZFAS1 and miR-548e expression in OC cells, we persuasively demonstrated that the regulation of OC cell proliferation, migration, invasion, and cisplatin resistance by ZFAS1 was mediated by its suppression on miR-548e expression. These results provided novel insights into the molecular pathogenesis of OC, as well as the non-coding RNA-mediated anti-ovarian-cancer drug resistance development. Of note, we showed that two other members of the miR-548 family could also be targeted by ZFAS1, and their involvements in OC pathogenesis and cisplatin resistance deserve further investigations.
As introduced above, it was already known that elevated CXCR4 expression was closely associated with OC cell proliferation, migration, invasion, and metastasis, as well as anti-cancer drug resistance.16,18,19 In this study, we also verified the significant increase of CXCR4 expression in OC tissues and cell lines. Furthermore, we showed that CXCR4 expression was positively correlated with ZFAS1 but negatively correlated with miR-548e expression in OC tissues. However, little is known about the mechanisms responsible for significantly increased CXCR4 expression in OC cells. Based on our bioinformatics, we convincingly proved that miR-548e could directly bind with CXCR4 3′ UTR regions by dual-luciferase reporter assay, thus resulting into altered CXCR4 expression in OC cells. This is the first evidence of molecular interaction between miR-548e and CXCR4. More importantly, we clearly demonstrated that the effects of miR-548e on OC cell proliferation, migration, invasion, and cisplatin resistance were mediated by its inhibition on CXCR4 expression. At this point, we unveiled for the first time that ZFAS1 promoted OC cell proliferation, migration, invasion, and cisplatin resistance through its binding with and suppression on miR-548e, which further resulted into elevated CXCR4 expression in OC cells. In light of the oncogene roles of CXCR4 in other cancers such as melanoma, gastric cancer, and hepatocellular carcinoma,26 further investigations of the ZFAS1/miR-548e/CXCR4 axis in these malignancies would facilitate full understanding of epigenetic regulation of human tumorigenesis.
Cisplatin has been applied as a potent anti-cancer agent for several decades due to its great capability of blocking DNA synthesis during cell proliferation.28 Like other widely used chemotherapy reagents, the application of cisplatin has been greatly impaired by frequent resistance among various cancer types.28,29 It has been recently established that the evolution of cisplatin resistance in cancer cells was closely linked with alteration of miRNA expressional profiles.30 For instance, the cisplatin resistance of nasopharyngeal carcinoma cells was associated with suppression of the let-7a expression by lncRNA NEAT1 (nuclear paraspeckle assembly transcript 1).31 Moreover, the let-7a expression could also be suppressed by CXCR4, which was involved in chemoresistance formation in leukemia and PCCs.20,21 However, the suppression of let-7a expression by CXCR4 associated with cisplatin resistance in OC cells has not been previously explored. Here, we showed that let-7a expression in OC cells could also be negatively modulated by CXCR4, which mediated the CXCR4-induced cisplatin resistance. The anti-apoptotic protein BCL-XL/S was previously identified as an enhancer of cisplatin resistance in ovarian and lung cancers.32,33 In this study, we also showed that the suppression of let-7a expression by CXCR4 caused elevated BCL-XL/S expression, which was involved in ZFAS1-induced cisplatin resistance in OC cells. Finally, the roles of ZFAS1 and let-7a in promoting OC cell cisplatin resistance and lung metastasis were validated by our in vivo tumorigenesis assay using mouse models. Recently, targeting lncRNAs has been suggested as a novel strategy for treating various human cancers,34,35 and the anti-OC roles of shZFAS1 shown in this study suggested a promising option for developing new treating regiments.
In summary, we reported in this study that the lncRNA ZFAS1 targeted miR-548e to enhance CXCR4 expression in OC cells, which effectively promoted OC cell proliferation, metastasis, and resistance to cisplatin treatment. Furthermore, the cisplatin resistance-promoting role of the ZFAS1/miR-548e/CXCR4 axis in o OC was mediated by suppressing let-7a and elevating BCL-XL/S expression (Figure 10). These results revealed a novel non-coding RNA-mediated OC pathogenesis and chemotherapy resistance, which also provided a basis for developing new anti-cancer treatments and overcoming resistances of OC to available therapy modules.
Material and Methods
Bioinformatics Analysis
The correlation between lncRNA ZFAS1 or miR-548e with OC patient survival time was analyzed using the starBase platform through the pan-cancer module as previously introduced36 and the GEPIA database (http://gepia.cancer-pku.cn/). Specifically, the correlation between ZFAS1 expression and survival analysis was performed based on RNA sequencing (RNA-seq) data of totally 374 ovarian serous cystadenocarcinoma patients downloaded from TCGA project, which was scaled with log2 (Fregments Per Kilobase per Million [FPKM] + 0.01). The correlation between miR-548e expression and survival analysis was performed based on RNA-seq data of totally 424 OC patients. The expression of CXCR4 in OC based on individual cancer stages also downloaded from TCGA project through the UALCAN portal (http://ualcan.path.uab.edu/index.html).37 The target miRNAs of lncRNA ZFAS1 and their binding sites were predicted by the lncBase Predicted v.2 tool (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted).38 The binding sites of mRNAs with miR-548e or let-7a were predicted through the TargetScan software (release 7.2; http://www.targetscan.org/vert_72/).39
Clinical Tissue Collection
The OC tissues and adjacent non-cancerous ovarian tissues were surgically collected from 17 OC patients registered at the Department of Obstetrics and Gynecology, Affiliated Haikou Hospital of Xiangya Medical College, Central South University (Haikou, Hainan, China) between August 2017 and July 2018. Detailed clinical data of OC patients in this study including disease stages, metastasis status, and other clinical pathogenic information were provided in Table 1. Written informed consents were obtained from each OC patient. The research including tissue collection was carried out with approval from the Ethical Committee of the Affiliated Haikou Hospital of Xiangya Medical College, Central South University (Haikou, Hainan, China). Samples were frozen immediately after being surgically resected and were stored in liquid nitrogen until further assays.
Table 1.
Clinicopathological Characteristics in Patients with OC
| Clinical Parameters | Cases (n) |
|---|---|
| Age (Years) | |
| <60 years | 12 |
| ≥60 years | 5 |
| Histology | |
| Serous carcinoma | 11 |
| Mucinous adenocarcinoma + others | 6 |
| Differentiation | |
| High grade | 9 |
| Low grade | 8 |
| Figo Stage | |
| I | 5 |
| II-IV | 12 |
| Lymph Node Metastasis | |
| Yes | 7 |
| No | 10 |
Cell Culture and Treatment
OC cell lines SKOV3, Caov3, and OVCAR3 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Other OC cell lines (A2780 and COV644), human embryonic kidney cells (HEK293T), and HOSE cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin at 37°C in a humidified culture chamber with supply of 5% CO2. For analysis of cell chemotherapy resistance, cultured cells after specified transfections were cultured in standard DMEM containing 2, 4, 6, 8, or 10 μM cisplatin (Selleck Chemicals, Houston, TX, USA) for 24 h, the IC50 values of cisplatin on cells were determined by measuring cell viability using the CCK-8 method as shown below.
Cell Transfections
The shZFAS1 for silencing of ZFAS1 expression and the negative control shNC, the shCXCR4 for silencing of CXCR4 expression and the negative control shNC, the miR-548a/e/az inhibitor for silencing of miR-548a/e/az expression and the inhibitor NC, the miR-548a/e/az mimics and mimics NC, and the let-7a inhibitor/mimics and negative controls NC, were all synthesized by GenePharma (Shanghai, China).
For establishment of recombinant pcDNA3.1-ZFAS1 vectors, the ZFAS1-encoding genomic sequences were amplified by the PCR method, then were used to ligate with the pcDNA3.1 vectors (Thermo Fisher Scientific, Waltham, MA, USA). The recombinant pcDNA3.1-CXCR4 vectors were established through ligating pcDNA3.1 plasmid with CXCR4 CDSs (coding sequences). Above lncRNA and mRNA shRNAs, miRNA inhibitors and mimics, and their corresponding negative controls, as well as recombinant and empty vectors, were introduced into cultured cells using the Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Madison, WI, USA) as instructed by the manufacture.
Dual-Luciferase Reporter Assay
The dual-luciferase reporter assay in this study was performed using the Nano-Glo dual-luciferase reporter system (Promega, Madison, WI, USA) following the manufacturer’s instruction, combined with the pmirGLO dual-luciferase miRNA target expression vector. For validation of the binding between lncRNA ZFAS1 or miR-548a/e/az, HEK293T cells expressing the ZFAS1-WT or ZFAS1-MUT were transfected with miR-548a/e/az mimics or inhibitors. To test miR-548e binding with the CXCR4 3′ UTR region, HEK293T cells expressing the CXCR4-WT or CXCR4-MUT were transfected with miR-548e mimics or inhibitors. To verify the binding of let-7a with BCL-XL/S the 3′ UTR region, HEK293T cells expressing the WT BCL-XL/S sequences (BCL-XL/S-WT) or BCL-XL/S MUT sequences (BCL-XL/S-MUT) were transfected with let-7a mimics or inhibitors. Cell transfection was performed as introduced above. 48 h later, cells were lysed and subjected to luciferase activity measurement with a GloMax 20/20 luminometer, using the dual-luciferase reporter assay kit (Promega, Madison, WI, USA) as instructed by the manufacturer. The binding was finally evaluated by relative luciferase activities.
FISH
Subcellular distribution of ZFAS1 and miR-548a/e/az in SKOV3 and Caov3 cells were assessed by FISH method using the FISH tag RNA multicolor kit (#F32956; Thermo Fishier Scientific) as instructed by the manufacturer. In brief, probes targeting ZFAS1, miR-548a, miR-548e, and miR-548az were synthesized by the GenePharma company (Shanghai, China). After being labeled with Alexa Fluor 488 or 594 dyes, these probes (1 μg/mL) were separately incubated with SKOV3 and Caov3 cells at 55°C for 18 h, and cell nuclei were visualized by staining with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) for 5 min. Subcellular localizations of non-coding RNAs were finally observed using a fluorescence microscope.
RIP Assay
The RNA immunoprecipitation (RIP) assay was carried out using the EZ-Magna RIP RNA binding protein immunoprecipitation kit (Merck, Darmstadt, Germany) according to the manufacturer’s instructions. In brief, HEK293T cells transfected with miR-548e mimics or negative controls were collected and lysed in RIP lysis buffer. Cell lysates (100 μL) were then incubated with magnetic beads conjugated with antibodies targeting human Ago2 proteins. Cell lysates incubated with magnetic beads conjugated with antibodies targeting human immunoglobulin G (IgG) were used as the negative control. After being incubated with proteinase K, the RNA samples immunoprecipitated with beads were isolated and quantified. Finally, the relative ZFAS1 levels in collected RNA samples were determined by the quantitative RT-PCR (qRT-PCR) method to validate its binding with miR-548e.
Cell Proliferation Analysis
Cell proliferation rates were evaluated in this study by the CCK-8 and clone-formation assays. The CCK-8 assay was performed using CCK-8 (Solarbio, Beijing, China) following the manufacturer’s instructions. In brief, cells cultured in a 96-well plate were incubated with CCK-8 reagent (15 μL/well) for 2 h, and the absorbances at 490 nm (optical density 490 [OD490]) were measured using a multimode plate reader. Cell proliferation analysis by colony-formation assay was done as previously described.40 In brief, cells were seeded (600 cells/well) and cultured in 6-well plates for 2 weeks under specified treatments, and formed clones were finally stained with 0.5% crystal violet and observed under microscopy (Zeiss, Germany).
Cell Migration and Invasion Analysis
A wound-healing assay was carried out to detect cell migration capability following designated transfection and treatment. In brief, cultured cells were seeded (4 × 105 cells/well) and cultured in 24-well plates for 24 h under standard conditions, then a straight scratch was made in the middle of the cell monolayer with a sterile pipette tip, and wound-healing status was finally observed under microscopy 24 h later. Cell invasion capability was analyzed using the Transwell culture system. In brief, Matrigel basement membrane matrix (BD Biosciences, San Jose, CA, USA) was pre-coated on the inner side of the Transwell chamber, and cells (1 × 106/mL) in serum-free DMEM were added in upper chamber, with the lower chamber filled with DMEM containing 10% FBS. 24 h later, cells that invaded the lower chambers were fixed in 4% paraformaldehyde, stained with crystal violet, and quantified.
Cell Apoptosis Analysis
The apoptosis of cultured cells was evaluated by flow cytometry using the annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. In brief, cells were seeded and cultured in 24-well plates for about 24 h, until a cell confluence of nearly 80% was reached. Then, cells were stained with the annexin V-FITC and PI solution for 15 min, which was finally analyzed using the FACS cytometer (BD Biosciences, San Jose, CA, USA) for determination of apoptotic cell percentages.
Lentivirus-Mediated Cell Transfection
SKOV3/DDP (cisplatin) cells with ZFAS1 silencing and miR-548e or let-7a overexpression for in vivo tumorigenesis assay were established using the lentivirus transfection system as previously introduced with minor modifications.41 The ZFAS1 shRNA, miR-548e mimics, or let-7a mimics as well as their corresponding negative controls synthesized as introduced above were integrated into the GV113 lentivirus (HU6-MCS-CMV-RFP) provided by the Shanghai Genechem (Shanghai, China), which were used to infect the SKOV3/DDP cells with enhanced infection solution containing polybrene as instructed by the manufacturer. Cells with stable expression of target sequences were selected with puromycin (Sigma-Aldrich, St. Louis, MO, USA) for three successive weeks.
In Vivo Tumorigenesis Model
The cell-line-based xenograft models were established for in vivo evaluation of the tumorigenesis and metastasis capacities of cancer cells as previously described with minor modifications.42 Six-week-old female BALB/c nude (nu/nu) mice were obtained from Shanghai SLAC Laboratory Animal Center (Shanghai, China), and they were bred and sustained under standard pathogen-free conditions. This animal experiment was approved by the Animal Care and Use Committee of the Affiliated Haikou Hospital of Xiangya Medical College, Central South University (Haikou, Hainan China).
For tumorigenesis evaluation, approximately 3 × 106 SKOV3 cells in PBS solution after designated transfection and treatments were subcutaneously injected into mice, which were then sustained under standard conditions for 30 days, during which each mouse was orally given cisplatin (2 mg/kg per day). Subsequently, mice were euthanized and subcutaneously implanted tumors were excised, followed by volume and weight measurement. For metastasis analysis, approximately 3 × 106 cells were injected into mice through tail-vein injection. After 30 days with oral cisplatin administration, mice were sacrificed, and the metastatic lung nodules were excised and analyzed. Cancer cell proliferation in tumor tissues were evaluated by Ki-67 expression using a Ki-67 cell proliferation kit (Sangon Biotech, Shanghai, China) following the manufacturer’s instructions.
qRT-PCR
Total RNA samples in cultured cells or tumor tissues were extracted using the Trizol reagent (Beyotime, Beijing, China) following the manufacturer’s instructions. The cDNA was synthesized from 2-μg RNA samples using the miScript II RT kit (QIAGEN, Valencia, VA, USA) as instructed by the manufacturer. Fast SYBR green master mix (Thermo Fisher Scientific, Madison, WI, USA) was used to detect relative expressional levels through quantitative real-time PCR methods following the manufacturer’s instructions. The expressional levels were finally calculated by the standard 2−ΔΔCt method. GAPDH and U6 small nuclear RNA (U6 snRNA) were used as internal references for mRNA and miRNA, respectively. Primers for ZFAS1, miR-548a/e/az, CXCR4, and let-7a were purchased from Sangon Biotech (Shanghai, China).
Western Blotting Assay
Total proteins were extracted from cultured cells using mammalian cell total protein lysis buffer (Sangon Biotech, Shanghai, China) following the manufacturer’s instructions. After protein concentration quantitation using the BCA (bicinchoninic acid) protein assay kit (Sangon Biotech, Shanghai, China), proteins were separated by SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). Following blocking with 5% BSA solution for 2 h at room temperature, membranes with proteins were incubated with primary and secondary antibodies and finally developed with the Pierce enhanced chemiluminescence (ECL) western blotting substrates (Thermo Fisher Scientific, Madison, WI, USA). Primary antibodies used for western blotting were as follows: anti-E-cadherin (#ab15148; Abcam), anti-N-cadherin (#ab18203; Abcam, Cambridge, UK), anti-vimentin (#5741; CST, Framingham, MA, USA), anti-MMP-2 (#ab37150; Abcam), anti-Slug (#ab27568; Abcam), anti-CXCR4 (#97680; CST), anti-BCL-XL/S (#ab32370; Abcam), and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase, #5174; CST). Secondary antibodies targeting rabbit IgG (#ab205718; Abcam) were used in this study. Relative protein levels were determined by band intensities quantitated with the ImageJ software and normalized to GAPDH levels.
Statistical Analysis
The SPSS 20.0 software was used to analyze data obtained in this study from at least three biological replicates, which were presented as mean ± standard deviation (SD). Comparison between two groups was performed using Student’s t test. Comparison among three or more groups was conducted using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. Spearman correlation analysis was performed to analyze the correlation between ZFAS1, miR-548a/e/az, CXCR4, and let-7a in OC tissues. It was considered to be statistically significant when the p value was less than 0.05.
Author Contributions
Guarantor of integrity of the entire study, J.Z., S.-G.H.; Study concepts, J.Z., L.-N.Q.,S.-G.H.; Study design, J.Z., L.-N.Q., S.-G.H.; Definition of intellectual content, J.Z., L.-N.Q., S.-G.H.; Literature research, Q.M., H.-Y.W., J.W., P.Y., J.-T.F., Y.-J.L.; Clinical studies, J.C., H.C., Q.-P.W., X.-R.Y., H.-Y.Y.; Experimental studies, J.Z., L.-N.Q.; Data acquisition, Q.M., H.-Y.W., J.W.; Data analysis, Q.M., H.-Y.W., J.W., P.Y., J.-T.F.; Statistical analysis, Q.M., H.-Y.W., J.W.; Manuscript preparation, J.Z., L.-N.Q.; Manuscript editing, J.Z., L.-N.Q.; Manuscript review, S.-G.H.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by Special Projects for Research and Development of Applied Technology and Demonstration and Promotion in Hainan Province.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.03.013.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Kroeger P.T., Jr., Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Curr. Opin. Obstet. Gynecol. 2017;29:26–34. doi: 10.1097/GCO.0000000000000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson S.W., Ozols R.F., Hamilton T.C. Mechanisms of drug resistance in ovarian cancer. Cancer. 1993;71(2, Suppl):644–649. doi: 10.1002/cncr.2820710224. [DOI] [PubMed] [Google Scholar]
- 4.Mihanfar A., Fattahi A., Nejabati H.R. MicroRNA-mediated drug resistance in ovarian cancer. J. Cell. Physiol. 2017;234:3180–3191. doi: 10.1002/jcp.26060. [DOI] [PubMed] [Google Scholar]
- 5.Van Heetvelde M., Van Bockstal M., Poppe B., Lambein K., Rosseel T., Atanesyan L., Deforce D., Van Den Berghe I., De Leeneer K., Van Dorpe J. Accurate detection and quantification of epigenetic and genetic second hits in BRCA1 and BRCA2-associated hereditary breast and ovarian cancer reveals multiple co-acting second hits. Cancer Lett. 2018;425:125–133. doi: 10.1016/j.canlet.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Matei D., Fang F., Shen C., Schilder J., Arnold A., Zeng Y., Berry W.A., Huang T., Nephew K.P. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Jiang X., Su Z., Li J., Kang P., Li C., Cui Y. Current insight into a cancer-implicated long noncoding RNA ZFAS1 and correlative functional mechanisms involved. Pathol. Res. Pract. 2018;214:1517–1523. doi: 10.1016/j.prp.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Liu R., Zeng Y., Zhou C.F., Wang Y., Li X., Liu Z.Q., Chen X.P., Zhang W., Zhou H.H. Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients. Sci. Rep. 2017;7:18. doi: 10.1038/s41598-017-00050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battaglia R., Vento M.E., Borzì P., Ragusa M., Barbagallo D., Arena D., Purrello M., Di Pietro C. Non-coding RNAs in the Ovarian Follicle. Front. Genet. 2017;8:57. doi: 10.3389/fgene.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X., Cui M., Zhang A., Tong L., Wang K., Li K., Wang X., Sun Z., Zhang H. miR-548c impairs migration and invasion of endometrial and ovarian cancer cells via downregulation of Twist. J. Exp. Clin. Cancer Res. 2016;35:10. doi: 10.1186/s13046-016-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagasawa T. CXC chemokine ligand 12 (CXCL12) and its receptor CXCR4. J. Mol. Med. (Berl.) 2014;92:433–439. doi: 10.1007/s00109-014-1123-8. [DOI] [PubMed] [Google Scholar]
- 14.Smith M.C., Luker K.E., Garbow J.R., Prior J.L., Jackson E., Piwnica-Worms D., Luker G.D. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y.P., Wu X.H., Shi B., Wu W.X., Yin G.R. Expression of chemokine CXCL12 and its receptor CXCR4 in human epithelial ovarian cancer: an independent prognostic factor for tumor progression. Gynecol. Oncol. 2006;103:226–233. doi: 10.1016/j.ygyno.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 16.Guo Q., Gao B.L., Zhang X.J., Liu G.C., Xu F., Fan Q.Y., Zhang S.J., Yang B., Wu X.H. CXCL12-CXCR4 Axis Promotes Proliferation, Migration, Invasion, and Metastasis of Ovarian Cancer. Oncol. Res. 2014;22:247–258. doi: 10.3727/096504015X14343704124430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obermajer N., Muthuswamy R., Odunsi K., Edwards R.P., Kalinski P. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011;71:7463–7470. doi: 10.1158/0008-5472.CAN-11-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H.H., Bellat V., Law B. Chemotherapy induces adaptive drug resistance and metastatic potentials via phenotypic CXCR4-expressing cell state transition in ovarian cancer. PLoS ONE. 2017;12:e0171044. doi: 10.1371/journal.pone.0171044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Jiang K., Qiu X., Li M., Hao Q., Wei L., Zhang W., Chen B., Xin X. Overexpression of CXCR4 is significantly associated with cisplatin-based chemotherapy resistance and can be a prognostic factor in epithelial ovarian cancer. BMB Rep. 2014;47:33–38. doi: 10.5483/BMBRep.2014.47.1.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao G., Wang X., Yu Y. CXCR4/Let-7a Axis Regulates Metastasis and Chemoresistance of Pancreatic Cancer Cells Through Targeting HMGA2. Cell. Physiol. Biochem. 2017;43:840–851. doi: 10.1159/000481610. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y., Jacamo R., Konopleva M., Garzon R., Croce C., Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J. Clin. Invest. 2013;123:2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ataseven B., Grimm C., Harter P., Heikaus S., Heitz F., Traut A., Prader S., Kahl A., Schneider S., Kurzeder C., du Bois A. Prognostic Impact of Port-Site Metastasis After Diagnostic Laparoscopy for Epithelial Ovarian Cancer. Ann. Surg. Oncol. 2016;23(Suppl 5):834–840. doi: 10.1245/s10434-016-5415-9. [DOI] [PubMed] [Google Scholar]
- 23.Furió-Tarí P., Tarazona S., Gabaldón T., Enright A.J., Conesa A. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw443. W176-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao M.X., Jiang Y.P., Tang Y.L., Liang X.H. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8:12472–12483. doi: 10.18632/oncotarget.13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayers D., Vandesompele J. Influence of microRNAs and Long Non-Coding RNAs in Cancer Chemoresistance. Genes (Basel) 2017;8:95. doi: 10.3390/genes8030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong D., Mu Z., Zhao C., Sun M. ZFAS1: a novel tumor-related long non-coding RNA. Cancer Cell Int. 2018;18:125. doi: 10.1186/s12935-018-0623-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia B., Hou Y., Chen H., Yang S., Liu T., Lin M., Lou G. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget. 2017;8:19534–19546. doi: 10.18632/oncotarget.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira J.A., Peixoto A., Neves M., Gaiteiro C., Reis C.A., Assaraf Y.G., Santos L.L. Mechanisms of cisplatin resistance and targeting of cancer stem cells: Adding glycosylation to the equation. Drug Resist. Updat. 2016;24:34–54. doi: 10.1016/j.drup.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal R., Kaye S.B. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer. 2003;3:502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 30.Boac B.M., Xiong Y., Marchion D.C., Abbasi F., Bush S.H., Ramirez I.J., Khulpateea B.R., Clair McClung E., Berry A.L., Bou Zgheib N. Micro-RNAs associated with the evolution of ovarian cancer cisplatin resistance. Gynecol. Oncol. 2016;140:259–263. doi: 10.1016/j.ygyno.2015.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F., Tai Y., Ma J. LncRNA NEAT1/let-7a-5p axis regulates the cisplatin resistance in nasopharyngeal carcinoma by targeting Rsf-1 and modulating the Ras-MAPK pathway. Cancer Biol. Ther. 2018;141:1–9. doi: 10.1080/15384047.2018.1450119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei X., Huang Z., Zhong M., Zhu B., Tang S., Liao D. Bcl-XL small interfering RNA sensitizes cisplatin-resistant human lung adenocarcinoma cells. Acta Biochim. Biophys. Sin. (Shanghai) 2007;39:344–350. doi: 10.1111/j.1745-7270.2007.00286.x. [DOI] [PubMed] [Google Scholar]
- 33.Villedieu M., Louis M.H., Dutoit S., Brotin E., Lincet H., Duigou F., Staedel C., Gauduchon P., Poulain L. Absence of Bcl-xL down-regulation in response to cisplatin is associated with chemoresistance in ovarian carcinoma cells. Gynecol. Oncol. 2007;105:31–44. doi: 10.1016/j.ygyno.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Li C.H., Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J. Biochem. Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 35.Malissovas N., Ninou E., Michail A., Politis P.K. Targeting Long Non-Coding RNAs in Nervous System Cancers: New Insights in Prognosis, Diagnosis and Therapy. Curr. Med. Chem. 2019;26:5649–5663. doi: 10.2174/0929867325666180831170227. [DOI] [PubMed] [Google Scholar]
- 36.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T., Zagganas K., Tsanakas P., Floros E., Dalamagas T., Hatzigeorgiou A.G. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 40.Ying H., Xu Z., Chen M., Zhou S., Liang X., Cai X. Overexpression of Zwint predicts poor prognosis and promotes the proliferation of hepatocellular carcinoma by regulating cell-cycle-related proteins. OncoTargets Ther. 2018;11:689–702. doi: 10.2147/OTT.S152138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Jiang H., Zhou H., Ying X., Wang Z., Yang Y., Xu W., He X., Li Y. Lentivirus-mediated silencing of HOTAIR lncRNA restores gefitinib sensitivity by activating Bax/Caspase-3 and suppressing TGF-α/EGFR signaling in lung adenocarcinoma. Oncol. Lett. 2018;15:2829–2838. doi: 10.3892/ol.2017.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu D.D., Chen X., Sun K.X., Wang L.L., Chen S., Zhao Y. Role of the lncRNA ABHD11-AS1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol. Cancer. 2017;16:138. doi: 10.1186/s12943-017-0709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.