Abstract
Age-related cataract patients regularly have hypertension, hyperglycemia, and hyperlipidemia. In oxidative conditions, increased reactive oxygen species can oxidize natural low-density lipoprotein into oxidative low-density lipoprotein (oxLDL). However, the relationship between oxLDL and the occurrence of cataracts is still unclear. In this study, 1515 differentially expressed transcripts were identified by analyzing the results of RNA sequencing in a human lens epithelial cell line (HLEpiC). Compared with control groups, oxLDL-treated HLEpiC had 806 up-regulated transcripts and 709 down-regulated transcripts. Our genome-wide transcriptome results showed that differentially expressed genes, such as Rho signaling (Rho A and Cdc42) and Na+/K+-ATPase family (ATP1B1), are involved in lens epithelial cell differentiation and cell homeostasis. In conclusion, oxLDL greatly influences transcriptional expression and these differentially expressed genes may play an important role in the development of cataracts. Our findings may provide new targets in the treatment for cataracts.
Keywords: OxLDL, RNA sequence, HLEpiC, cataract, cell differentiation
Introduction
Cataracts are one of the leading causes of visual impairment and blindness worldwide [1,2]. A recent World Health Organization survey found that 51% of blindness and 33% of visual impairment cases worldwide are caused by cataract [3]. Despite considerable efforts by the World Health Organization and the Vision 2020 initiative, the global health burden of vision loss caused by cataracts increased from 1990 to 2015 [4]. Currently, cataract surgery combined with intraocular lens implantation is considered the best treatment strategy. Although the continuous improvement of surgical methods and the development of the intraocular lens have greatly improved cataract treatment, the cure rate for cataracts is far lower than the incidence rate. In addition, cataract surgery is accompanied by complications such as retinal detachment, iris prolapse, and blindness [5]. Therefore, clarifying the underlying mechanisms of cataracts will greatly improve treatment outcomes.
Hyperlipidemia is a systemic disease characterized by abnormal lipid metabolism or transport, resulting in increased plasma levels of one or more lipids. Low-density lipoprotein is the main indicator for the diagnosis of hyperlipidemia. Many epidemiological investigations have confirmed that hyperlipidemia is closely related to the occurrence of cataracts. Many mechanisms that may cause cataracts, and animal studies have shown that oxidative damage is closely linked to age related cataracts [6]. Oxidative stress is a phenomenon caused by an imbalance between the production and accumulation of reactive oxygen species (ROS) in cells and tissues, exceeding the ability of biological systems to detoxify these products. Under oxidative stress, increased ROS can oxidize natural low-density lipoprotein into oxidative low-density lipoprotein (oxLDL). Plasma malondialdehyde and oxLDL levels are reportedly higher in patients with cataracts [7]. Recent literature has indicated that oxidative stress should be considered a major or secondary cause of many cardiovascular diseases [8]. The role of stress signaling pathways induced by oxLDL in the lens capsule cells is not well understood, however.
The lens is a transparent crystal structure formed by the lens capsule, lens epithelium, and lens fibers. Transparency and refraction of the lens require that lens epithelial cells undergo coordinated proliferation, migration, and elongation [9]. Lens fibers have very broad cytoskeletons that preserve their precise shape and packaging; the destruction or mutation of certain cytoskeletal elements can result in loss of transparency [10]. The actin cytoskeletal structure plays an important role in lens epithelial cell proliferation and differentiation into mature fibroblasts [11]. Cell cycle exit and perfect implementation of lens fiber end differentiation are prerequisites for lens transparency and refraction, as well [12]. The clarity of the lens depends, in addition to the correct execution of the final differentiation process, on many other factors. These include modulation of the between cell membrane and cytoplasmic protein interactions of the lens crystals, proper maintenance of electrochemical gradients, and overall cellular homeostasis [13]. Many cataract patients have high lens sodium content, reflecting the importance of cellular ion concentrations in the condition [14,15]. Furthermore, any disturbance in these cellular processes can cause opacity of the lens.
Materials and methods
Cell culture
The HLEpiC line was purchased from American Type Culture Collection (HB-8065, Manassas, VA, USA). HLEpiCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gibco, USA). The cells were treated with or without oxLDL (100 μg/ml) and cultured at 37°C in a 5% CO2 incubator for 24 h [16].
RNA Sequence and data analysis
Total RNA was extracted from cell lysates using an RNA extraction kit according to the manufacturer’s instructions (Hefei Nuower Biotechnology Co., Ltd, China). The high-throughput Illumina HiSeq 2500 was used to sequence the samples. We obtained RNA-seq FastQ raw data and used Trimmomatic to remove lower mass readings [17]. The quality of clean data was assessed using FastQC software [18]. HISAT2 (v2.0.13) software was used to map the approved data to the reference genome of the human (National Center for Biotechnology Information [NCBI] genome assembly version Rnor_6.0) and the data were annotated with the annotation file (.gtf) NCBI Rnor_6.0 [19,20]. The transcript expression level was determined by Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) [21]. Kallisto software was used to obtain a count of known mRNAs and edgR package was used to find the differences in the number of reads [22]. The differentially expressed genes (DEGs) were determined using RNA Sequence data as significantly up-regulated or down-regulated genes with a statistical significance (two-sided P value < 0.05). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to determine the function of DEGs [23].
Protein-protein interaction (PPI) network integration
PPI analysis reveals the general organization principles of functional cellular networks to find new protein function. The STRING is an online search tool (http://string.embl.de/) used to find the interaction and functional relationships between proteins. Cytoscape software (http://www.cytoscape.org/) is widely used to construct biomolecular interaction networks and make the models [24]. We used the molecular complex detection (MCODE) function in Cytoscape software (version 3.6.0) to find modules with MCODE scores ≥ 3 and nodes ≥ 3 in the PPI network. We used topology analysis to analyze the connectivity of nodes in the PPI network and find higher degrees of important nodes (central proteins). The top 10 hub genes were selected for analysis. The analysis of individual modules was performed using Metascape with a statistical significance (P < 0.05).
Results
Quality of RNA sequencing
We sequenced and obtained raw data from six HLEpiC samples through the Illumina platform. After trimming and filtering, the quality of data was good. The median score of the reads in all samples was above 30 and all samples passed the quality check (Figure 1).
Figure 1.
Sequencing quality. Plot showing the sequencing quality of the six sample reads. The X-axis is the base position for each reading (bp indicates base pair). The Y-axis is the sequence quality score.
Identification of DEGs
We investigated the overall transcript expression levels of HLEpiC in the control and oxLDL-treated groups by using log2 (FPKM+1) values and found that the overall changes were not significantly different from the control groups. Cluster analysis of differentially expressed transcripts showed significant differences between the groups (Figure 2). A total of 1,515 gene expression differences were significant, of which 806 genes were up-regulated and 709 genes were down-regulated. The results of differential expression of transcripts are shown as volcano map in Figure 3A. Under oxLDL treatment, the top ten up-regulated DEGs were ATP1B1, AC03426.1, AC005912.1, AC005000.1, AC113935.1, RPL26P19, EEF1A1P6, NORAD, ALDOA and AL080243.2. The top ten down-regulated DEGs were ATP1B1, AC03426.1, AC005912.1, AC005000.1, AC113935.1, RPL26P19, EEF1A1P6, NORAD, ALDOA (Figure 3B).
Figure 2.
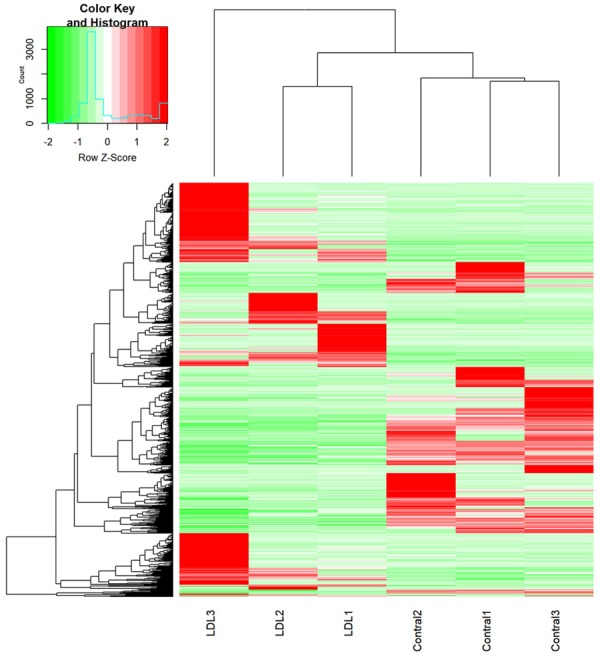
Overview of expression profiles of transcripts. Transcript-based hierarchical clustering analysis combining tissues with similar natures. Scaled to log2 (FPKM+1) expression values by different color intensities, the Z scores of each transcript are normalized, and green and red represent low and high expression levels, respectively.
Figure 3.
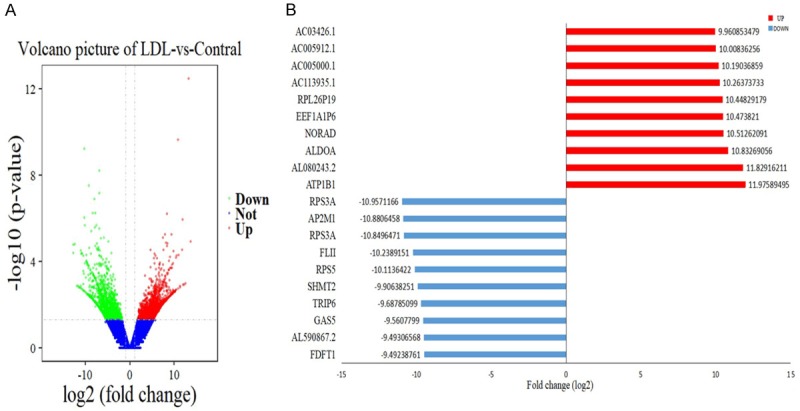
Differential expression of data between two sets of samples. A. Volcano map. Red points represent up-regulated transcripts screened on the basis of absolute fold change ≥ 2.0 and a corrected P value of < 0.05. Green points represent the expression of transcripts that were down-regulated, screened on the basis of absolute fold change ≥ 2.0 and a corrected P value of < 0.05. The black points represent transcripts with no significant change. B. The most substantially (P < 0.05) up-regulated and down-regulated transcripts based on oxLDL treatment of HLEpiC (n = 3).
DEGs reflect changes in biological function
We classified DEGs according to their respective biological processes by analysis with GO function annotation. Differential genes of transcripts were involved in cell cycle phase transition: cell proliferation-related processes and apoptotic signaling pathways (Figure 4A). In the cellular component category, differential genes of transcripts are mainly involved in cell cycle regulation, chromosomal regions, nuclear chromosomes, microtubule tissue centers, biofilm-related components, mitochondrial envelopes, Golgi membranes, and vesicle membranes (Figure 4B). Differential genes of transcripts related to molecular function are mainly involved in ribonucleic acid-related components, transcriptional community activity, translation factor activity, RNA binding, transcriptional repressor activity, cadherin binding, protein serine/threonine kinase activity, protein heterodimerization activity, Ral GTPase binding, oxidoreductase activity, peptidase activator activity, actin binding, NEDD8 transferase activity, and ubiquitin-like protein ligase binding (Figure 4C). Interestingly, several important pathways, including the neurotrophin signaling pathway, ubiquitin-mediated proteolysis, and the MAPK signaling pathway were found in the top twenty pathways rich with transcriptional differences (Figure 5).
Figure 4.
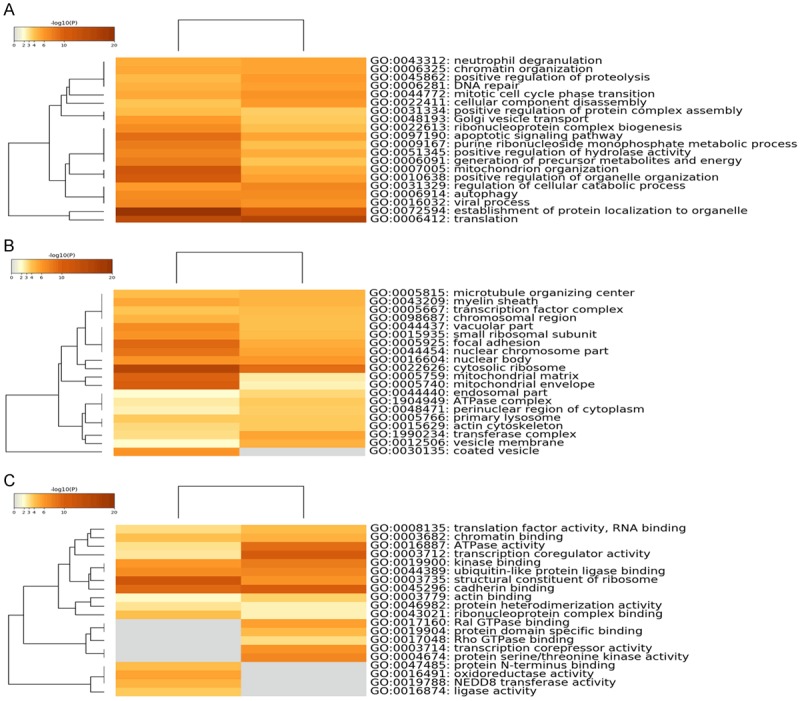
GO functional category enrichment following oxLDL treatment of HLEpiC. Heat maps of the top 20 enriched (A) biologic processes, (B) cell composition, and (C) molecular functions following oxLDL treatment of HLEpiC. The result on the left is based on the down-regulated gene list, and the right is based on the up-regulated gene list. Colors represent P values.
Figure 5.
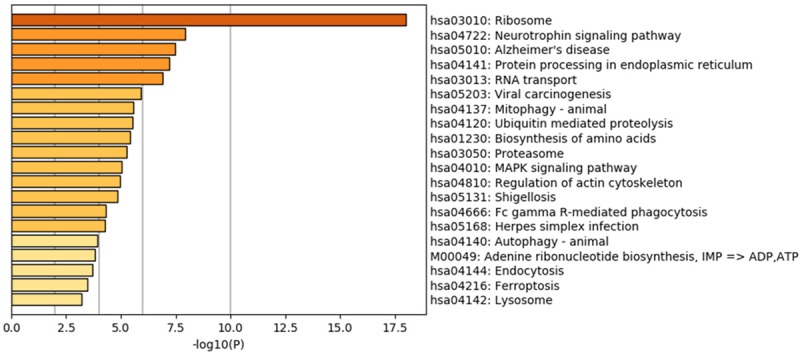
KEGG enrichment results. Scatterplot of the top 20 KEGG enrichment results of differentially expressed transcripts in each pairwise comparison that were annotated in the specific pathway term. A high RichFator represents greater intensiveness. A lower P value represents greater intensity.
PPI network integration
To further identify the biologic relevance of differentially expressed genes after oxLDL treatment, we constructed a PPI network encoded by DEGs (Figure 6A). We analyzed the PPI network of significantly differentially expressed transcripts and obtained seven distinct modules. The main pathways involved in these genes include the following cellular structure and assembly processes: amino acid biosynthesis, proteasome, ribosome, cell adhesion, actin cytoskeleton regulation, adherens junction formation, and related signaling pathways. These related pathways included B cell receptor signaling pathway, T cell receptor signaling pathway and cAMP signaling pathway (Figure 6B). Moreover, the ten most significant interaction genes were Cdc42, Rhog, Cdkn1a, Ccl2, Ngf, Prkar2b, Hif1a, Bub3, Vegfa and Plk4 (Figure 7).
Figure 6.
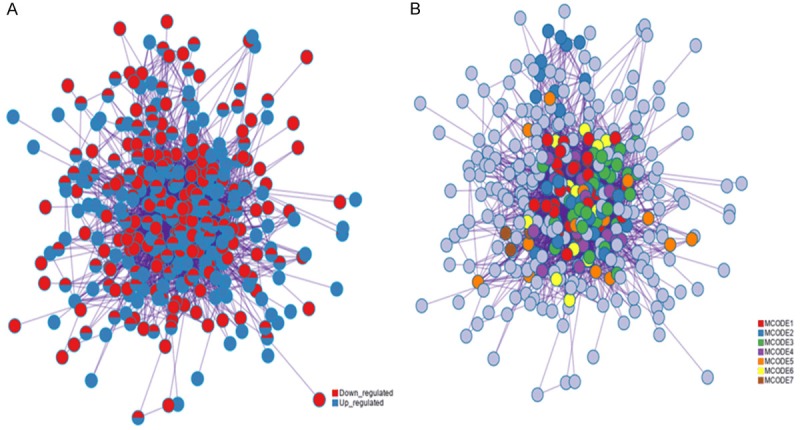
Protein-protein interaction. (A) Interaction network and (B) Modules. Circles represent genes, lines represent protein interactions between genes, and line colors represent evidence of interactions between proteins.
Figure 7.
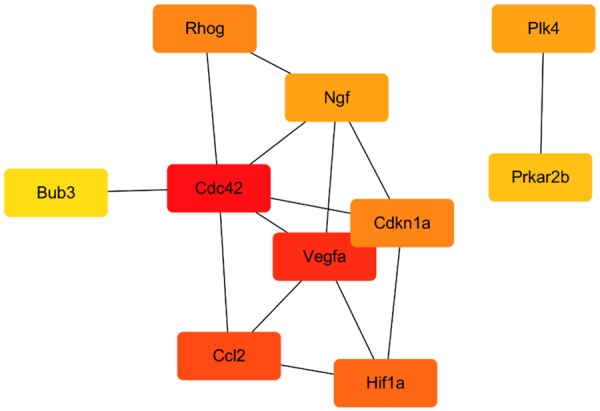
Top 10 hub genes. Red indicates genes with high scores. Yellow and orange indicate genes with middle scores, increasing respectively.
OxLDL treatment changes expression of Rho A GTPases and Na+/K+-ATPase
Rho GTPases are critical to actin organization and include Rho, Rac, and Cdc42. Three families regulate Rho GTPase activity: guanine-nucleotide exchange factor (GEF), GTPase activating protein (GAP), and purine-nucleoside dissociation inhibitor (GDI). RNA sequence and IPA data indicate that Rho A and Cdc42 are significantly up-regulated and that all three families of Rho GTPase regulatory factors are also significantly up-regulated. Expression of ATP1B1 was also up-regulated significantly after oxLDL treatment. The protein encoded by ATP1B1 belongs to the Na+/K+ and H+/K+ ATPase β chain protein family and plays a critical role in osmoregulation and cellular homeostasis.
Discussion
In the present study, we found that DEGs treated with oxLDL are associated with lens epithelial cell (HLEpiC) differentiation and maintenance of cellular homeostasis. Among the top ten up-regulated transcripts ATP1B1 was up-regulated most significantly. The protein encoded by this gene is a subunit of the Na+/K+-ATPase, which is a membrane protein responsible for establishing and maintaining an electrochemical balance across the plasma membrane by pumping Na+ and K+ against the gradient [25]. These salt gradients are essential for osmotic adjustment, sodium-coupled transport of various organic and inorganic molecules, and electrical excitability of nerves and muscles. After oxLDL treatment, oxidation may damage the cell membrane of HLEpiC, significantly changing the function of the Na+/K+-ATPase pump and leading to increased membrane permeability to Na+. This may result in an increased lens Na+ concentration and cause the lens to over-hydrate, starting the process of cataract. The degree of abnormal lens Na+ distribution is reportedly related to the degree of cortical opacity. Furthermore, ATP1B1 is involved in cell adhesion and establishes epithelial cell polarity [26]. It has previously been reported that endogenous biocurrents in the equator of the lens promote lens differentiation in mammals through Na+/K+-ATPase (ATP1A1 and ATP1B1) and result in excessive lens hydration and loss of protein [27].
Among the top ten down-regulated transcripts we found FLII, also known as actin remodeling protein. FLII may act as a transcriptional coactivator, working through hormone-activated nuclear receptors and synergizing with NCOA2 and CARM1 [28]. FLII can inhibit the formation of cell processes by inhibiting Cdc42 [29]. Therefore, we hypothesize that FLII may play a role in regulating the cytoskeletal rearrangements involved in cell division and cell migration. The partial secondary RNA structure of the transcript encoded by the GAS5 gene mimics the glucocorticoid response element (GRE) [30], meaning that it can bind to the DNA binding domain of the glucocorticoid receptor. This action prevents the glucocorticoid receptor from being activated, thereby preventing it from regulating the transcription of its target gene. This transcript is also thought to regulate the transcriptional activity of other receptors, such as androgens, progesterone and mineralocorticoid receptors, which may bind to its GRE mimetic region. Multiple functions are associated with this process, including cell growth, arrest and apoptosis. Cdkn1a encodes a potent cyclin-dependent kinase inhibitor that may contribute to the implementation of apoptosis after caspase activation [31]. At the same time, the apoptotic gene BCL2L12 is significantly up-regulated, which encodes a member of the protein family containing Bcl-2 homology domain 2, regulating apoptosis [32].
Rhog encodes a member of the Rho family of small GTPases that circulate between an inactive GDP binding state and an active GTP binding state and acts as a molecular switch in the signal transduction cascade. Rho GTPases include Rho, Rac and Cdc42 [33]. Previous studies have reported that Rho GTPase has the role of maintaining lens transparency and structural integrity. Defects in Rho GTPase lead to lens aberrations, including lens epithelial proliferation, differentiation and elongation, polarity, shape and packaging defects [34]. Rho proteins promote reorganization of the actin cytoskeleton and regulate cell shape, attachment and movement. Our genome-wide transcriptome results indicate that the Rho signaling pathway is significantly affected by oxLDL treatment: Rho A and Cdc42 are significantly up-regulated, as are the three families that regulate Rho GTPase activity. Up-regulation of Rho GTPase inhibitor (GDI) can lead to fibroblast maturation disorders, resulting in terminal differentiation of lens epithelial cells in the equatorial region [35]. During fibroblast maturation, the cells undergo a complex process of degradation that clears of all intracellular organelles, including the nucleus. Fibroblast maturation disorders lead to incomplete removal of organelles, affecting the transparency of the entire lens.
Our GO analysis showed that oxLDL is involved in the positive regulation of organelle tissue and the cell cycle. OxLDL treatment affects cell morphology, enhances apoptosis, and blocks normal cell cycle. These processes affect lens epithelial cell function and are associated with the epithelial dysfunction caused by hyperlipidemia. The DEGs-enriched KEGG pathway includes regulation of the actin cytoskeleton, MAPK signaling pathway, protein processing in the endoplasmic reticulum, amino acid biosynthesis, proteasome, endocytosis, and neurotrophin signaling pathways. We constructed and analyzed a PPI network record encoded with differential expression of oxLDL treated HLEpiC and identified ten closely related genes: Rhog, Cdc42, Cdkn1a, Ccl2, Ngf, Prkar2b, Hif1a, Bub3, Vegfa, and Plk4. Of these differentially expressed genes, Rhog, Cdc4, Cdkn1a, Vegfa, Ccl2, and Bub3 were up-regulated. VEGF is a key node in the PPI network and may be the main protein associated with the development of cataracts. This gene is a member of the growth factor family of PDGF/VEGF, which induces proliferation and migration of vascular endothelial cells. VEGF also has anti-angiogenic and vascular permeability-inducing effects [36]. We believe that the up-regulation of VEGFA protein may result in increased HLEpiC permeability.
In conclusion, we used next-generation sequencing technology to determine the expression profile of transcriptomes in HLEpiC after oxLDL treatment to explore the mechanism by which oxLDL induces HLEpiC injury. Bioinformatic analysis revealed several specific genes that are new potential targets for the molecular mechanisms of cataracts with hyperlipidemia.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of Anhui Provincial [Grant No. 1408085MH158]; the National Natural Science Foundation of China [Grant Nos. 81570403, U1732157]; the Outstanding Young Investigator Award of Anhui Medical University; and Scientific Research Foundation of the Institute for Translational Medicine of Anhui Province (SRFITMAP 2017zhyx30).
Disclosure of conflict of interest
None.
References
- 1.Abraham AG, Condon NG, West Gower E. The new epidemiology of cataract. Ophthalmol Clin North Am. 2006;19:415–425. doi: 10.1016/j.ohc.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96:614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Yan W, Muller A, He M. A global view on output and outcomes of cataract surgery with national indices of socioeconomic development. Invest Ophthalmol Vis Sci. 2017;58:3669–3676. doi: 10.1167/iovs.17-21489. [DOI] [PubMed] [Google Scholar]
- 4.He M, Wang W, Huang W. Variations and trends in health burden of visual impairment due to cataract: a global analysis. Invest Ophthalmol Vis Sci. 2017;58:4299–4306. doi: 10.1167/iovs.17-21459. [DOI] [PubMed] [Google Scholar]
- 5.Llop SM, Papaliodis GN. Cataract surgery complications in uveitis patients: a review article. Semin Ophthalmol. 2018;33:64–69. doi: 10.1080/08820538.2017.1353815. [DOI] [PubMed] [Google Scholar]
- 6.Ottonello S, Foroni C, Carta A, Petrucco S, Maraini G. Oxidative stress and age-related cataract. Ophthalmologica. 2000;214:78–85. doi: 10.1159/000027474. [DOI] [PubMed] [Google Scholar]
- 7.Hashim Z, Zarina S. Assessment of paraoxonase activity and lipid peroxidation levels in diabetic and senile subjects suffering from cataract. Clin Biochem. 2007;40:705–709. doi: 10.1016/j.clinbiochem.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piatigorsky J. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation. 1981;19:134–153. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 10.Wride MA. Lens fibre cell differentiation and organelle loss: many paths lead to clarity. Philos Trans R Soc Lond B Biol Sci. 2011;366:1219–1233. doi: 10.1098/rstb.2010.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Cheng C, Nowak RB, Fowler VM. The lens actin filament cytoskeleton: diverse structures for complex functions. Exp Eye Res. 2017;156:58–71. doi: 10.1016/j.exer.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cvekl A, McGreal R, Liu W. Lens development and crystallin gene expression. Prog Mol Biol Transl Sci. 2015;134:129–167. doi: 10.1016/bs.pmbts.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Duncan G, Bushell AR. Ion analyses of human cataractous lenses. Exp Eye Res. 1975;20:223–230. doi: 10.1016/0014-4835(75)90136-0. [DOI] [PubMed] [Google Scholar]
- 15.Yu XS, Jiang JX. Interaction of major intrinsic protein (aquaporin-0) with fiber connexins in lens development. J Cell Sci. 2004;117:871–880. doi: 10.1242/jcs.00945. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y, Ruan Y, Wang Y, Wu S. Polydatin inhibits cell proliferation and expressions of inflammatory cytokines in THP-1 cells induced by ox-LDL via up-regulating SIRT1. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2018;34:193–198. [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang SG, Kim KH, Lee BM, Moon JC. Transcriptome analysis for identifying possible gene regulations during maize root emergence and formation at the initial growth stage. Genes Genomics. 2018;40:755–766. doi: 10.1007/s13258-018-0687-z. [DOI] [PubMed] [Google Scholar]
- 19.Peterson CS, Huang S, Lee SA, Ferguson AV, Fry WM. The transcriptome of the rat subfornical organ is altered in response to early postnatal overnutrition. IBRO Rep. 2018;5:17–23. doi: 10.1016/j.ibror.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 23.Park BJ, Lord D, Hart JD. Bias properties of Bayesian statistics in finite mixture of negative binomial regression models in crash data analysis. Accid Anal Prev. 2010;42:741–749. doi: 10.1016/j.aap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taub M, Springate JE, Cutuli F. Targeting of renal proximal tubule Na,K-ATPase by salt-inducible kinase. Biochem Biophys Res Commun. 2010;393:339–344. doi: 10.1016/j.bbrc.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson PJ, Musil LS, Mathias RT. Point: a critical appraisal of the lens circulation model--an experimental paradigm for understanding the maintenance of lens transparency? Invest Ophthalmol Vis Sci. 2010;51:2303–2306. doi: 10.1167/iovs.10-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao L, Liu J, Pu J, Collinson JM, Forrester JV, McCaig CD. Endogenous bioelectric currents promote differentiation of the mammalian lens. J Cell Physiol. 2018;233:2202–2212. doi: 10.1002/jcp.26074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103:13351–13356. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto Y, Tanaka K, Harimaya K, Nakatani F, Matsuda S, Iwamoto Y. Small GTP-binding protein, Rho, both increased and decreased cellular motility, activation of matrix metalloproteinase 2 and invasion of human osteosarcoma cells. Jpn J Cancer Res. 2001;92:429–438. doi: 10.1111/j.1349-7006.2001.tb01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreis NN, Friemel A, Zimmer B, Roth S, Rieger MA, Rolle U, Louwen F, Yuan J. Mitotic p21Cip1/CDKN1A is regulated by cyclin-dependent kinase 1 phosphorylation. Oncotarget. 2016;7:50215–50228. doi: 10.18632/oncotarget.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MT, Ho SM, Tarapore P, Chung I, Leung YK. Estrogen receptor beta isoform 5 confers sensitivity of breast cancer cell lines to chemotherapeutic agent-induced apoptosis through interaction with Bcl2L12. Neoplasia. 2013;15:1262–1271. doi: 10.1593/neo.131184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narumiya S. The small GTPase Rho: cellular functions and signal transduction. J Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- 34.Rao V, Wawrousek E, Tamm ER, Zigler S Jr. Rho GTPase inactivation impairs lens growth and integrity. Lab Invest. 2002;82:231–239. doi: 10.1038/labinvest.3780415. [DOI] [PubMed] [Google Scholar]
- 35.Maddala R, Reneker LW, Pendurthi B, Rao PV. Rho GDP dissociation inhibitor-mediated disruption of Rho GTPase activity impairs lens fiber cell migration, elongation and survival. Dev Biol. 2008;315:217–231. doi: 10.1016/j.ydbio.2007.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]


