Abstract
Background
Mesenchymal stem cells (MSCs) selectively differentiate into adipocytes or osteoblasts, and several molecules control the fate determination of MSCs. Understanding these key checkpoints greatly contributes to the ability to induce specific MSC differentiation for clinical applications. In this study, we aimed to explore whether TNF receptor-associated factor 4 (TRAF4) affects MSC adipogenic differentiation, which we previously reported that could positively regulated the osteogenic differentiation.
Methods
Western blotting and Real-time Polymerase Chain Reaction were used to detected the expression pattern of TRAF4 during adipogenic differentiation. Lentivirus was constructed to regulate TRAF4 expression, and oil red O staining and Western blotting were used to assess its role in adipogenesis, which was confirmed in vivo by implanting an MSC-matrigel mixture into nude mice. Western blotting was used to detect the activated signaling pathways, and a specific inhibitor and agonist were used to clear the roles of the key signaling pathways. Additionaly, Co-Immunoprecipitation was conducted to find that Pyruvate kinase isozyme type M2 (PKM2) interacts with TRAF4, and to further explore their binding and functional domains. Finally, an RNA-binding protein immunoprecipitation assay and Western blotting were used to detect whether N6-methyladenosine mediates the decreased TRAF4 expression during adipogenic differentiation.
Findings
The results demonstrated that TRAF4 negatively regulates MSC adipogenesis in vitro and in vivo. Mechanistically, we revealed that TRAF4 binds to PKM2 to activate the kinase activity of PKM2, which subsequently activates β-catenin signaling and then inhibits adipogenesis. Furthermore, TRAF4 downregulation during adipogenesis is regulated by ALKBH5-mediated N6-methyladenosine RNA demethylation.
Interpretation
TRAF4 negatively regulates the adipogenesis of MSCs by activating PKM2 kinase activity, which may act as a checkpoint to fine-tune the balance of adipo-osteogenic differentiation, and suggests that TRAF4 may be a novel target of MSCs in clinical use and may also illuminate the underlying mechanisms of bone metabolic diseases.
Funding
This study was supported by the National Natural Science Foundation of China (81871750 and 81971518) and the Science and Technology Project of Guangdong Province (2019B02023600 and 2017A020215070).
Keywords: TRAF4, Mesenchymal stem cells, Adipogenic differentiation, PKM2
Research in context.
Evidence before this study
TRAF4 is a member of the TRAF family of scaffold proteins, and previous animal study had demonstrated that TRAF4 deficiency can lead to serious skeletal malformation, which suggests that TRAF4 plays a critical role in bone development and metabolism, however, its exact molecular mechanism requires further study. Accumulating studies have revealed that the adipogenic-osteogenic balance plays a critical role in bone metabolism. We previously reported that TRAF4 positively regulates the osteogenic differentiation of MSCs by acting as an E3 ubiquitin ligase to degrade Smurf2. However, whether TRAF4 affects the adipogenic differentiation of MSCs remains unclear.
Added value of this study
We demonstrated that TRAF4 negatively regulates MSC adipogenesis in vitro and in vivo, and we further revealed that TRAF4 binds to PKM2 to activate the kinase activity of PKM2, which subsequently activates β-catenin signaling and then inhibits adipogenesis. Taken together, our results indicate that TRAF4 acts as a fate checkpoint to regulate the adipogenic-osteogenic differentiation of MSCs. Interestingly, TRAF4 expression was decreased in the marrow cavity of rats with osteoporosis. Furthermore, TRAF4 downregulation during adipogenesis was regulated by ALKBH5-mediated m6A RNA demethylation.
Implications of all the available evidence
This study demonstrated that TRAF4 may act as a checkpoint to fine-tune the balance of adipogenic-osteogenic differentiation, and it may be a novel target of MSCs in clinical use and may also illuminate the underlying mechanisms of bone metabolic diseases.
Alt-text: Unlabelled box
1. Introduction
Mesenchymal stem cells (MSCs), which are seed cells with a wide range of clinical applications, can selectively differentiate into adipocytes and osteoblasts under the appropriate conditions [1]. As a common progenitor of adipocytes and osteoblasts, MSCs engage in bone homeostasis via the following two mechanisms after differentiation: MSCs can differentiate into osteoblasts that directly mediate bone development [2] or MSCs can differentiate into adipocytes that regulate the bone marrow microenvironment and subsequently affect bone metabolism [3]. Although adipocytes and osteoblasts are differentiated from MSCs, they drive contrasting metabolic decisions [4]. Thus, tightly controlled MSC differentiation is highly significant for the maintenance of bone homeostasis, which has attracted increasing attention in recent years.
Considerable evidence has shown that the adipogenic and osteogenic differentiation of MSCs is under the control of several key checkpoints [5,6]. These molecules positively or negatively affect downstream signaling pathways, including the peroxisome proliferator-activated receptor-gamma (PPAR-γ) [7], transforming growth factor-beta (TGF-β)/bone morphogenic protein (BMP) [8] and Wnt signaling pathways [9], which eventually affect the MSC differentiation direction. Thus, exploring and identifying these checkpoints could improve the application efficiency of MSCs and illuminate the underlying mechanisms of bone metabolism disorders.
TNF receptor-associated factor 4 (TRAF4) is a member of the TRAF family (TRAF 1 to 7) of seven scaffold proteins, which are involved in various cellular physiological processes [10]. TRAF4, like other TRAFs, mainly consists of three domains. Between the N-terminus and C-terminus, there is a RING finger domain, several continuous zinc finger domains and one TRAF domain. The “E3 ubiquitin ligase” characteristic of TRAF4 is of great importance to TRAF4 functions under most circumstances, promoting the proliferation and metastasis of prostate cancers [11]. Additionally, a recent report revealed that TRAF4 directly binds to the juxtamembrane region of EGFR via the TRAF domain and promotes kinase autophosphorylation and activation [12]. Previous animal studies have demonstrated that TRAF4 is required for embryogenesis and that TRAF4 deficiency can lead to serious skeletal malformation, which suggests that TRAF4 plays a critical role in bone development and metabolism [13]; however, the exact molecular mechanism requires further study. Our previous study demonstrated that TRAF4 positively regulates the osteogenic differentiation of MSCs by acting as an E3 ubiquitin ligase to degrade Smurf2 via the K48-linked ubiquitination of Smurf2 [14]. Accumulating studies have revealed that the adipogenic-osteogenic balance plays a critical role in bone metabolism [15]. However, whether TRAF4 affects the adipogenic differentiation of MSCs remains unknown.
Pyruvate kinase isozyme type M2 (PKM2) is an isozyme of pyruvate kinase that functions as a rate-limiting enzyme in the glycolytic pathway and modulates Wnt/β-catenin signaling [16]. Substantial evidence indicates that PKM2 is important for cell differentiation [17] and metabolism [18]. PKM2 is expressed in MSCs and adipocytes [19], and the regulation of PKM2 expression or kinase activity can affect cell differentiation. Jiang et al. found that PKM2 could regulate adipocyte differentiation via the PI3K-AKT pathway [20]. Moreover, abundant evidence has demonstrated that PKM2 can influence β-catenin nuclear localization and activation [16,21], which has been recognized as the master signaling pathway in adipogenic differentiation [22]. However, the upstream regulatory network that regulates PKM2 or kinase activity during adipogenesis remains unknown.
In this study, we found that TRAF4 negatively regulated the adipogenesis of MSCs both in vitro and in vivo. Mechanistically, we revealed that TRAF4 interacts with PKM2 to activate β-catenin signaling and then inhibits adipogenesis. Finally, we observed that TRAF4 expression was decreased in the marrow cavities of rats with osteoporosis, which is a typical bone metabolism disease. Taken together, our results suggest that TRAF4 acts as a fate checkpoint in regulating the adipogenic differentiation of MSCs. Thus, TRAF4 may be a novel target for the clinical application of MSCs in tissue engineering.
Previous studies have demonstrated that the shift in preferential differentiation of MSCs from osteoblasts to adipocytes plays a role in the pathogenesis of osteoporosis via reducing bone mineral density [23]. We have already demonstrated that TRAF4 positively regulated the osteogenic differentiation of MSCs and TRAF4 was found to be abnormally decreased in osteoporosis specimen [14]. Thus, we hypothesis that TRAF4 not only regulate osteogenic differentiation of MSCs but also modulate the adipogenic capacity of MSCs. In this study, we focus on exploring the effect of TRAF4 on the adipogenic differentiation and further clarify the concrete mechanism. Additionally, our results might help further the understanding of the possible mechanisms of bone metabolism disorders.
2. Materials and methods
2.1. Isolation and culture of MSCs
This study was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital of Sun Yat-sen University (Guangzhou, China). Eighteen healthy donors between the ages of 20 and 30 years were selected for this study and signed the informed consent agreement. Bone marrow samples were extracted from the posterior superior iliac spine under sterile conditions, and MSCs were isolated and expanded according to a previously reported method [24]. MSCs were isolated and cultured using DMEM supplemented with 10% FBS. MSCs from passages 3 to 5 were used for the subsequent experiments.
2.2. Culture of 293T cells
Cells from the human embryonic kidney 293T cell line were cultured in glucose Dulbecco's modified Eagle's medium (DMEM, glucose 4.5 mg/L, GIBCO) containing 10% FBS at 37 °C with 5% CO2. After reaching 80%–90% confluence, the 293T cells were collected for the subsequent experiments or passaged into two new flasks. The 293T cells were used to explore the interaction between TRAF4 and PKM2.
2.3. Adipogenic differentiation induction
MSCs were seeded into 12-well plates at a concentration of 1.5 × 104 cells per cm2. MSCs were induced with adipogenic medium containing DMEM (10% FBS), 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich), 0.2 mM indomethacin (Sigma-Aldrich), 10 μg/mL insulin (Sigma-Aldrich), and 1 μM dexamethasone (Sigma-Aldrich). The medium was replaced every 3 days for 14 days. The cells were stained with oil red O (ORO) at the end of culturing.
2.4. ORO staining and quantification
After specific induction for the indicated days, MSCs were fixed with 4% paraformaldehyde for 20 min and then stained with ORO working solution for 20 min at room temperature. The oil red O was dissolved in isopropyl alcohol at the concentration of 12 mM, and then diluted with deionized water to the concentration of 7.2 mM before staining. After three washes with PBS, the MSCs were observed and recorded immediately under a microscop. The stained cells were destained thoroughly with 600 μl isopropyl alcohol. A 200 μl aliquot was transferred to a 96-well plate, and the absorbance was measured at 520 nm.
2.5. Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
SYBR Premix Ex Taq (TaKaRa) and a LightCycler 480 Real-TimePCR system (Roche) were used to detect the mRNA levels of relative genes in our experiment. Total RNA was extracted from the MSCs using TRIzol (Invitrogen) according to the manufacturer's protocol, and cDNA was then generated using a PrimeScript™ RT Reagent Kit (TaKaRa). The expression level of each gene was normalized to the mRNA level of GAPDH using the 2−△△CT method. The sequences of the specific primers used in this study are listed in Supplemental Table 1.
2.6. Western blot analysis
Western blotting was performed as previously described [25]. Briefly, MSCs were lysed with RIPA buffer (Beyotime), and the proteins were then boiled with loading buffer after quantification. Equal amounts of protein extracts were separated by 10% SDS-PAGE and subsequently transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were blocked with 5% nonfat milk and incubated with primary antibodies against TRAF4 (ab88612), p-CREB (ab32096), CREB (ab32515), PKM2 (ab150377), C/EBP-α (ab40764), fatty-acid-binding protein 4 (FABP4) (ab13979), ALKBH5 (ab195377), FTO (ab92821), METTL3 (ab186002), METTL14 (ab107540), WTAP (ab195380) (the above eleven antibodies are from Abacam), PPAR-γ (2443S), active β-catenin (19807S), β-catenin (8480S), p-Akt (4060S), Akt (9272S), p-p38 (4511S), p38 (8690S), p-ERK (4370S), ERK (4695), p-JNK (4668S), JNK (9252S), p-PKM2 (3827S), Flag (14793S), MYC (2276S), HA (3724S) (the above fifteen antibodies are from Cell Signaling Technology) and GAPDH (AF0006, Beyotime) overnight at 4 °C. All the antibodies were diluted in Tris buffered saline Tween (TBST) consisting of 20 mM Tris base, 150 mM NaCl, 0.1% Tween 20 and deionized water. After washing three times with TBST, the membranes were incubated with HRP-conjugated secondary antibodies (1:3000; Santa Cruz) for 1 h at room temperature. The target protein bands were detected using an Immobilon Western Chemiluminescent HRP Substrate (Millipore) and analyzed using Image J software (version 1.49e).
2.7. Lentivirus construction and infection
The protocols used were reported previously. The TRAF4 overexpression lentiviruses (OE-TRAF4) and vector control (NC1) were synthesized by GenePharma. Lentiviruses encoding short hairpin RNA (shRNA) for TRAF4 were constructed with a target sequence of 5′-GCACTAAGGAGTTCGTCTTTG-3′(sh-TRAF4). The negative control shRNA sequence was 5′-TTCTCCGAACGTGTCACGTTTC-3′ (NC2). All of the above lentiviruses were generated by cotransfecting pGLVH1/GFP/Puro (GenePharma, China) and packaging plasmids (pGag/Pol,pRev, and pVSV-G) into 293T cells. Culture supernatants containing lentiviruses were filtered and concentrated 72 h after transfection. Then, 109 TU/mL lentivirus and 5 μg/mL polybrene were premixed in medium and incubated with MSCs for 24 h at a multiplicity of infection of 30. The related experiments were conducted after the corresponding lentivirus infection and treatments.
2.8. Cell apoptosis and proliferation assay
MSCs were seeded into the indicated plates, and then infected with lentiviruses (NC1, OE-TRAF4, NC2 and sh-TRAF4). MSC apoptosis was detected by using an Annexin V-PE Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer's protocol. After being cultured in adipogenic medium from 1 to 11 days, cell proliferation was analyzed using a Cell Counting Kit-8 (CCK-8) assay (Doindo Molecular Technologies).
2.9. Plasmid construction and transfection
Expression plasmid constructs, including pcDNA3.1(+)-Myc-TRAF4,pcDNA3.1(+)-Myc-TRAF4-RING domain deletion, pcDNA3.1(+)-Myc-TRAF4-TRAF domain deletion, pcDNA3.1(+)-Flag-PKM2, pcDNA3.1(+)-Flag-PKM2-AB/C-domain deletion, pcDNA3.1(+)-Flag-PKM2-C-domain deletion, pcDNA3.1(+)-Flag-PKM2-N/C-domain deletion, pcDNA3.1(+)-Flag-PKM2-N/AB-domain deletion and pcDNA3.1(+)-HA-ubiquitin, were all constructed and purchased from Shanghai (Shanghai). A Lipofectamine 3000 Transfection Kit (Invitrogen, L3000-015) was used for transfection according to the manufacturer's instructions with minor modifications. Briefly, 293T cells were seeded into 6-well plates at a density of 3 × 105 cells/well. One day later, the 293T cells were transfected with 2.5 µg/well plasmid, 5 µl Lipo 3000 and 5 µl P3000 according to the manufacturer's instructions. The total amounts of transfected plasmids in each well were equalized by adding empty pcDNA3.1(+)-vector.
2.10. Adipogenic differentiation of preadipocytes in vivo
This experiment was performed to observe the effect of TRAF4 on the in vivo adipogenic differentiation of preadipocytes derived from MSCs. MSCs were infected with lentiviruses (NC1, OE-TRAF4, NC2 and sh-TRAF4) and then cultured in adipogenic medium for 5 days before being transplanted in vivo. Preadipocytes (1 × 106) were harvested and mixed with 150 μL Matrigel (BD Biosciences) and then injected subcutaneously into two symmetrical sites on the backs of 8-week-old BALB/c-nu/nu female mice (Laboratory Animal Center of Sun Yat-Sen University; n = 9 per group). BALB/c-nu/nu mice were used to assess the potential in vivo effect of TRAF4 on adipogenesis, which complementary verification via in vitro cell experiments. The preadipocytes/Matrigel implants were obtained 8 weeks after transplantation. The implants were fixed in 4% paraformaldehyde and embedded in paraffin for hematoxylin and eosin (H&E) staining and immunohistochemical staining for Perilipin-1, which was used to observe the adipogenic differentiation of preadipocytes in vivo.
2.11. H&E staining and immunohistochemical staining
The sections were deparaffinized in xylene and hydrated with decreasing concentrations of ethanol. For H&E staining, the sections were first incubated with hematoxylin for 5 min and then stained with eosin for 3 min after clearing. Immunohistochemical staining for Perilipin-1 (ab3526, Abcam) was performed to investigate adipogenesis. Briefly, after antigen retrieval, the sections were blocked in 5% serum and then incubated with the primary antibody. Next, the sections were incubated with the appropriate biotin-conjugated secondary antibody and subsequently with a streptavidin solution, which was followed by color development. After staining, allsections were dehydrated with increasing concentrations of ethanol and xylene. All sections were visualized under a light microscope (Nikon).
2.12. Immunofluorescence staining
MSCs were seeded onto sterile glass coverslips, which were induced in adipogenic medium for the indicated days. Then, the MSCs or adipocytes were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 for 15 min at room temperature. After blocking, the cells were incubated with anti-TRAF4 (ab88612, Abcam) and anti-PKM2 (4053, Cell Signaling Technology), Perilipin-1 (ab3526, Cell Signaling Technology), or active β-Catenin (19807S, Cell Signaling Technology) antibodies overnight at 4 °C. The appropriate secondary antibodies were subsequently added for incubation for 1 h at room temperature. Next, the MSCs were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to reveal the nuclei. Thereafter, the glass slides were covered with coverslips after antifade mounting medium (P0126, Beyotime) was applied. Finally, the samples were observed under a laser scanning confocal microscope, and images were obtained using an LSM 5 Exciter confocal imaging system (Carl Zeiss).
2.13. Coimmunoprecipitation and LC-MS/MS
MSCs and 293T cells were seeded into 10-cm dishes, and MSCs were cultured in adipogenic medium, while 293T cells were cotransfected with the indicated plasmids on the second day. All cells were lysed and coimmunoprecipitated using a Dynabeads™ Protein G Immunoprecipitation Kit (10007D, Invitrogen) according to the manufacturer's instructions. The primary antibodies used in this experiment included TRAF4 (sc-390232, Santa Cruz), PKM2 (ab150377, Cell Signaling Technology), Flag-Tag (14793, Cell Signaling Technology), Myc-Tag (2276, Cell Signaling Technology), and their IgG control (3452 or 37988, Cell Signaling Technology). The immunoprecipitates were collected, washed, and boiled in Laemmli sample buffer for 10 min. The samples were separated by SDS-PAGE and subsequently dyed using a Coomassie blue staining kit (Beyotime Institute of Biotechnology). Differential bands were collected for further LC-MS/MS analysis for the TRAF4 interaction proteins in MSCs, and the analysis result and raw data were deposited to the ProteomeXchange with the accession number PXD017524. Western blotting was performed to confirm the interaction between the two proteins, as described above. Notably, a special secondary antibody (ab131366, Abcam) that recognized only native (nonreduced) antibodies was used to minimize the heavy and light chain bands in the WB.
2.14. Measurement of pyruvate kinase activity
MSCs were seeded into 6-well plates and transfected with the indicated lentiviruses (NC1, OE-TRAF4, NC2 and sh-TRAF4). After induction in adipogenic medium for three days, the pyruvate kinase activity was measured using a Pyruvate Kinase Assay Kit (ab83432, Abcam) according to the manufacturer's instructions.
2.15. PKM2 inhibitor and activator preparation and MSC treatment
To regulate the PKM2 activity of MSCs, PKM2-IN-1 (inhibitor, MCE) and DASA-58 (activator, MCE) were used in our study. According to the manufacturer's instructions, PKM2-IN-1 (10 mM) and DASA-58 (10 mM) were dissolved in dimethyl sulfoxide (DMSO) and stored at −80 °C for subsequent experiments. MSCs were seeded into a 12-well plate and induced in adipogenic medium containingPKM2-IN-1 (10 μM) or DASA-58 (5 μM). Equal amounts of DMSO were added to the negative control (NC) groups.
2.16. RNA-binding protein immunoprecipitation assay
An RNA immunoprecipitation (RIP) assay was performed using a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer's instructions. In brief, approximately 5 × 106 MSCs or adipogenic differentiation MSCs were lysed via an RIP assay and then incubated with the prepared beads-antibody complex (anti-m6A antibody (202003, Synaptic Systems) or purified rabbit IgG (17-701, Merck Millipore)) in RIP immunoprecipitation buffer. All proteins were digested by proteinase K, and the RNA was extracted using phenol chloroform. Thereafter, the purified RNA was reversed transcribed into cDNA and subjected to quantitative PCR to detect TRAF4 expression.
2.17. Establishment of ovariectomized rats
All female Sprague-Dawley rats were purchased from the Laboratory Animal Center of Sun Yat-Sen University at the age of two months. In this study, all rats were housed in a pathogen-free facility on a 12-hour light/dark cycle with water and food provided ad libitum. Rats were anesthetized with isoflurane and underwent sham (sham) or ovariectomization (OVX) surgery. Then, the animals were observed for general health twice daily for the first week and allowed 3 months for healing. We established the ovariectomized rats to determine whether TRAF4 was involved in the pathogenesis of osteoporosis. After 3 months, the rats were sacrificed for the related assays. Micro-CT (Siemens) was performed to analyzed the cancellous bone mass between the two groups, and the parameters were calculated using an Inveon Research Workplace (Siemens). Moreover, H&E staining was used to evaluate the trabecula and adipocytes, and IHC was performed to detected TRAF4 expression in sections of femur speciments from the two groups.
2.18. Statistical analysis
The experiments were repeated at least three times, and all data are expressed as the means±standard deviations (SDs). Statistical significance was determined by Student's t-test (two groups) and one-way analysis of variance (ANOVA) followed by Bonferroni test (three or more groups), and the Pearson correlation test was performed for correlation analyses, which were performed using SPSS 22.0 software (Chicago, IL, USA). A P-value < 0.05 was considered significant.
3. Results
3.1. TRAF4 negatively correlates with the adipogenic differentiation capacity of MSCs
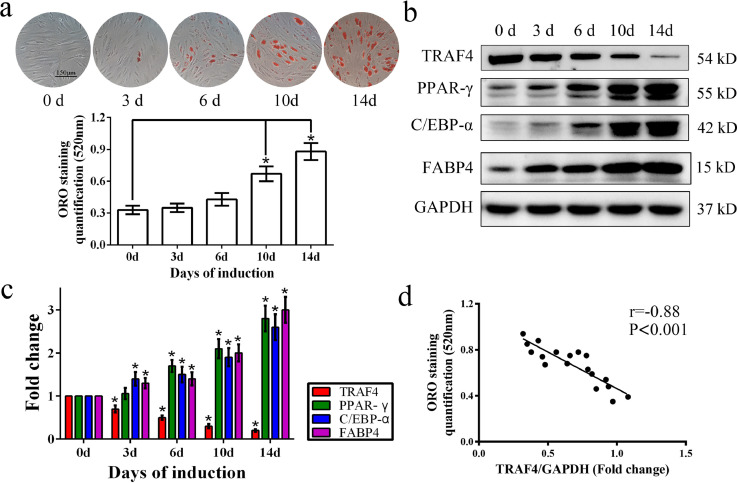
MSCs were isolated, cultured and identified as previously described [25]. All MSCs used in this study conformed to the standard criteria stated by the International Society for Cellular Therapy. MSCs were cultured in adipogenic medium for up to 14 days, and ORO staining and Western blotting were performed to detect the adipogenic differentiation of MSCs. As shown in our results, ORO staining increased gradually from 0 to 14 days (Fig. 1a). Consistent with this result, the protein levels of key adipogenic markers, including PPAR-γ, C/EBP-α and FABP4, were upregulated during adipogenic differentiation within 14 days (Fig. 1b and c). However, TRAF4 expression was decreased during MSC adipogenic differentiation from day 0 to day 14 (Fig. 1b and c). Further analysis demonstrated a strong negative relationship between TRAF4 expression and ORO staining quantification (Fig. 1d). In other words, TRAF4 was negatively associated with MSC adipogenic differentiation.
Fig. 1.
TRAF4 negatively correlated with the adipogenic differentiation of MSCs. (a) ORO staining and quantification gradually increased from 0 to 14 days (scale bar=150 μm). (b, c) Western blotting showed that the protein levels of PPAR-γ, C/EBP-α and FABP4 were gradually increased during adipogenic differentiation (0 d, 3 d, 6d, 10 d and 14d), while TRAF4 decreased after induction toward adipogenic differentiation. (d) A Pearson correlation test revealed a strong negative relationship between TRAF4 expression and ORO staining quantification. All data are presented as the means±SDs. *p < 0.05, according to a one-way analysis of variance (n = 3 independent experiments with three different MSC samples).
3.2. TRAF4 negatively regulated the adipogenic differentiation of MSCs in vitro
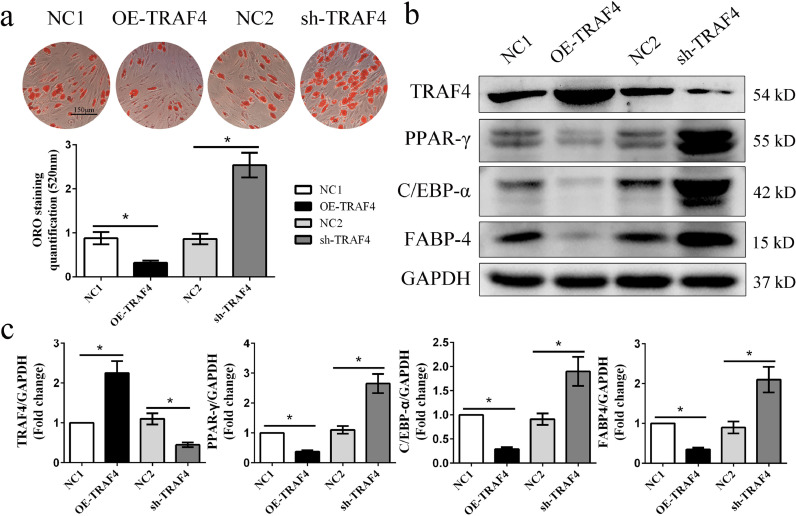
To confirm that TRAF4 is involved in the process of MSC adipogenic differentiation, we constructed a lentivirus to regulate TRAF4 expression and assessed the role of TRAF4 in adipogenesis. The transduction efficiencies of the lentivirus were high and comparable among the different groups (Fig. S1a). The ORO staining results demonstrated that TRAF4 overexpression remarkably decreased the formation of adipocytes, while TRAF4 knockdown promoted the adipogenic differentiation capacity of MSCs in vitro (Fig. 2a). Consistent with the above results, the PPAR-γ, C/EBP-α, and FABP4 protein levels were decreased in TRAF4-overexpressing MSCs but increased in TRAF4-knockdown MSCs (Fig. 2b and c). Moreover, the immunofluorescence result targeting Perilipin-1 was consistent with the ORO and Western blot results, further confirming that TRAF4 negatively regulates the adipogenic differentiation of MSCs (Fig. S3). The differentiation capacity of MSCs might also depend on their state and proliferation. Therefore, we detected MSC apoptosis and proliferation among the different groups. Our results show that MSC apoptosis and proliferation were not influenced by TRAF4 overexpression, as well as not influenced by TRAF4 knockdown (Fig. S1b). Above all, these results indicate that TRAF4 negatively regulates the adipogenic differentiation of MSCs in vitro.
Fig. 2.
TRAF4 negatively regulated the adipogenic differentiation of MSCs in vitro. (a) MSCs were transduced with lentivirus to regulate TRAF4 expression for 24 h and then cultured in adipogenic medium for 14 days. ORO staining and quantification results show that TRAF4 overexpression remarkably decreased the formation of adipocytes, while TRAF4 knockdown promoted the adipogenic differentiation of MSCs (scale bar = 150 μm). (b, c) Western blotting results demonstrated that the protein levels of PPAR-γ, C/EBP-α, and FABP4 were decreased in TRAF4-overexpressing MSCs but increased in TRAF4-knockdown MSCs. All data are presented as the means±SDs. *p<0.05, according to Student's t-test (n = 3 independent experiments with three different MSC samples).
3.3. TRAF4 impaired the adipogenic differentiation of preadipocytes in vivo
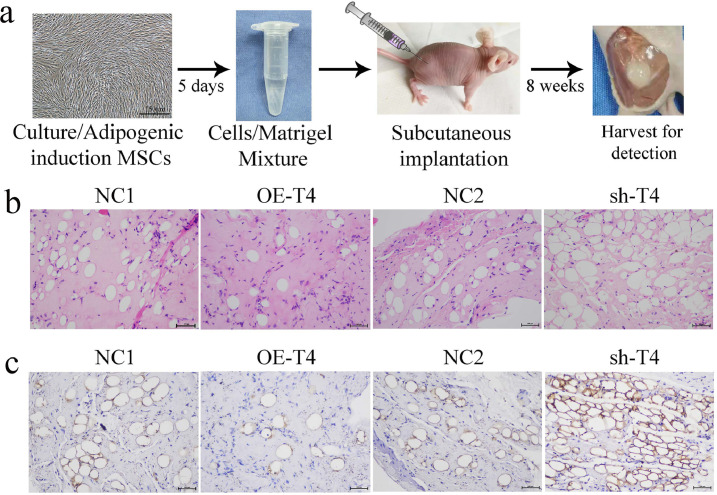
To assess the potential effect on adipogenesis of TRAF4 in vivo, MSCs transfected with a specific lentivirus (NC1, OE-TRAF4, NC2 or sh-TRAF4) were induced in adipogenic medium for 5 days and mixed with Matrigel and then injected subcutaneously into nude mice (Fig. 3a). The preadipocytes/Matrigel plugs were harvested after 8 weeks, and adipocytes were detected using H&E staining and immunohistochemical staining for Perilipin-1. H&E staining showed that there were obviously fewer adipocytes in the OE-TRAF4 group than in the NC1 group, whereas more adipocytes were observed in the sh-TRAF4 group than in the NC2 group (Fig. 3b). Consistent with these results, immunohistochemical staining demonstrated that there were fewer Perilipin-1(+) adipocytes in the overexpression group than in the control group, while more were detected in the knockdown group (Fig. 3c). Collectively, TRAF4 overexpression in preadipocytes derived from MSCs impaired in vivo adipogenesis, while TRAF4 knockdown had the opposite effect, which confirmed the negative adipogenic capacity observed previously in vitro.
Fig. 3.
TRAF4 impaired the adipogenic differentiation of preadipocytes in vivo. (a) Schematic of the in vivo experimental setup. MSCs were infected with lentiviruses (NC1, OE-TRAF4, NC2 and sh-TRAF4), and adipogenesis was induced for 5 days before the cells were transplanted in vivo. Differentiated cells were harvested and mixed with Matrigel, which was implanted in the subcutaneous space of nude mice and detected after 8 weeks. (b) H&E staining showed fewer adipocytes in the TRAF4-overexpressing group than in the NC1 group, whereas more adipocytes were observed in the sh-TRAF4 group (scale bar = 100 μm). (c) The IHC results for Perilipin-1 showed that TRAF4 overexpression inhibited adipogenic differentiation in vivo, while TRAF4 knockdown exerted the opposite effect (scale bar = 100 μm).
3.4. TRAF4 inhibited adipogenic differentiation by activating the β-catenin signaling pathway
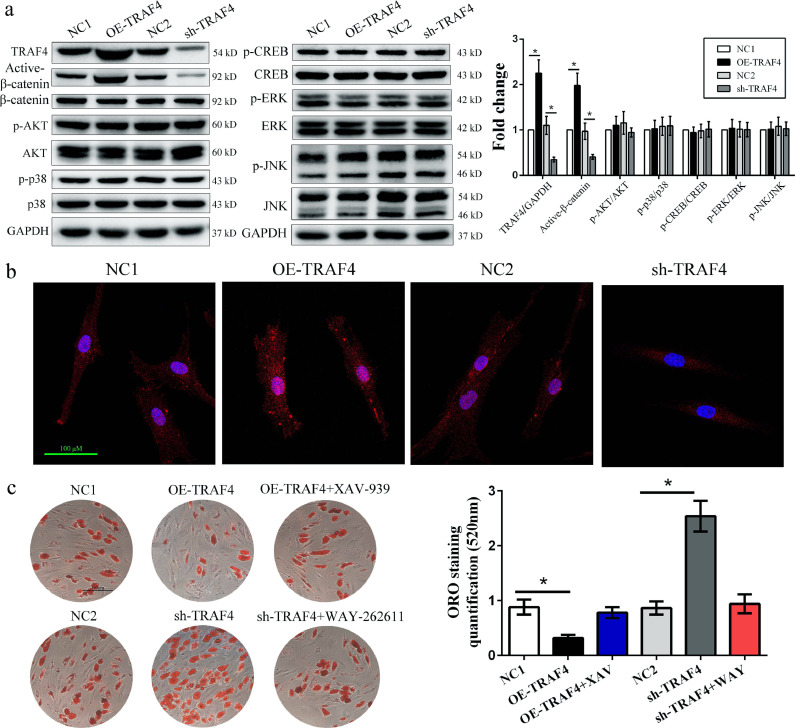
To explore the mechanism by which TRAF4 modulates the adipogenic differentiation of MSCs, we assessed several classical signaling pathways involved in adipogenesis. Western blotting results demonstrated that TRAF4 overexpression promoted the expression of active β-catenin, while TRAF4 knockdown effectively inhibited it. However, there was no difference in the expression of the Smad1/5/8, pERK/ERK, p-p38/p38 and pCREB/CREB signaling pathways (Fig. 4a). In addition, immunofluorescence analyses indicated that MSCs from the OE-TRAF4 group expressed significantly higher levels of active β-catenin than those from the NC1 group, while TRAF4-knockdown MSCs expressed lower levels; These trends were especially visible in the cell nuclei (Fig. 4b). The addition of XAV-939, a specific β-catenin inhibitor, recovered the adipogenic differentiation of MSCs in the OE-TRAF4 group to levels near those in the NC group. Moreover, WAY-262611, a specific β-catenin agonist, reduced the adipogenesis of the sh-TRAF4 group to that of the NC group (Fig. 4c). Taken together, these results indicated that TRAF4 negatively regulated the adipogenic differentiation of MSCs by activating β-catenin signaling.
Fig. 4.
TRAF4 inhibited adipogenic differentiation by activating the β-catenin signaling pathway. (a) Western blotting results of adipogenic differentiation at 10 days indicated that TRAF4 overexpression promoted the expression of active β-catenin, while TRAF4 knockdown effectively inhibited it. (b) Immunofluorescence analyses indicated that active β-catenin expression in the nucleus was increased in TRAF4-overexpressing MSCs but decreased in TRAF4-knockdown MSCs (scale bar = 100 μm). (c) The ORO staining and quantification results on day 14 revealed that XAV-939 (a specific β-catenin inhibitor) recovered the adipogenic differentiation of MSCs in the OE-TRAF4 group to a level near that in the NC group, while WAY-262611 (a specific β-catenin agonist) reduced the adipogenesis of the sh-TRAF4 group to a level similar to that of the NC group (scale bar=150 μm). All data are presented as the means±SDs. *p < 0.05, according to Student's t-test (a) and a one-way analysis of variance (c) (n = 3 independent experiments with three different MSC samples).
3.5. TRAF4 binds with PKM2 through the RING domain and N- and C-terminals on PKM2
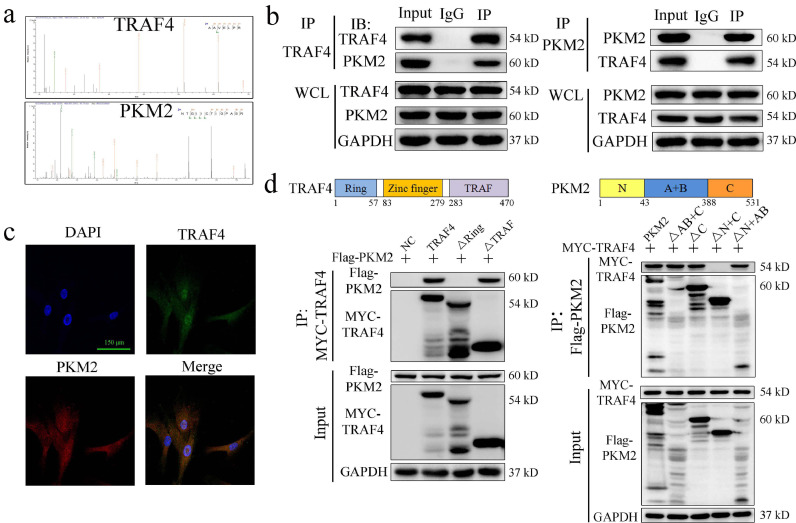
To explore the mechanism underlying the negative regulation of adipogenesis by TRAF4, we performed co-IP and LC-MS/MS experiments to identify the proteins that interact with TRAF4 during adipogenic differentiation. The results revealed that multiple proteins, which are listed in Supplementary Document 1, may interact with TRAF4. The PKM2 protein was detected in the co-IP complex (Fig. 5a), indicating that PKM2 may be a binding partner of TRAF4. Subsequently, the reciprocal co-IP assay results revealed that endogenous TRAF4 could interact with PKM2 in MSCs (Fig. 5b).
Fig. 5.
TRAF4 bound with PKM2 through the RING domain and N- and C-terminals on PKM2. (a) The peptide sequences of the PKM2 protein were detected in the co-IP complex. (b)Co-IP assay results revealed that endogenous TRAF4 could interact with PKM2 in MSCs. (c) The immunofluorescence results showed that TRAF4 colocalized with PKM2 and was expressed in both the cytoplasm and nuclei of MSCs (scale bar = 150 μm; green, TRAF4; red, PKM2; blue, DAPI). (d) The structural compositions of TRAF4 and PKM2 are shown. The Western blotting results show that TRAF4 binds to PKM2 through the RING domain and N- and/or C-terminals of PKM2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further confirm the interaction between TRAF4 and PKM2, we detected their locations in MSCs using immunofluorescence staining. The results showed that TRAF4 colocalized with PKM2, and these two proteins were both expressed in the cell cytoplasm and nucleus (Fig. 5c). Altogether, these results suggest that TRAF4 can physically associate with PKM2 in MSCs.
TRAF4 consists of a RING domain, three zinc-finger domains, a coiled-coil domain and a TRAF domain. Previous studies have reported that TRAF4 interacts with other proteins primarily through the RING domain or the TRAF domain [12,26]. Thus, we transfected plasmids encoding Myc-TRAF4 or deletion mutants (TRAF4 ΔRING or TRAF4 ΔTRAF) and Flag-PKM2 into 293T cells, which were then subjected to immunoprecipitation with an anti-Myc antibody and immunoblotting with an anti-Flag antibody. Our results showed that the RING domain of TRAF4 is necessary for the interaction with PKM2. Similarly, we coexpressed Flag-PKM2 or PKM2 mutants along with full-length Myc-tagged TRAF4. The results reveal that both the N- and C-terminals of PKM2 can interact with TRAF4 (Fig. 5d). Taken together, these results indicated that TRAF4 binds to PKM2 through the RING domain and N- and/or C-terminals of PKM2.
3.6. TRAF4 inhibits adipogenic differentiation by activating pyruvate kinase activity
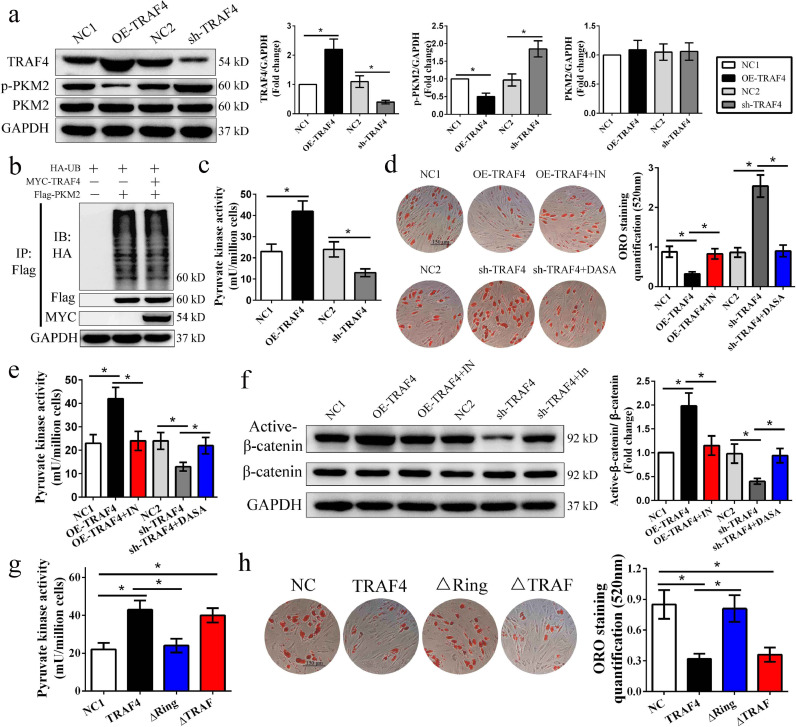
To investigate the potential role of PKM2 in the TRAF4-mediated inhibition of adipogenic differentiation, we detected PKM2 and p-PKM2 expression after modulating TRAF4 expression in MSCs. The Western blotting results indicated that there was no difference in PKM2 expression among the groups. However, our results showed that p-PKM2 (Y105) levels were decreased in the TRAF4-overexpressing group but increased in the TRAF4-knockdown group compared to their corresponding control groups (Fig. 6a). Previous studies indicated that TRAF4 usually acts as an E3 ubiquitin ligase to affect the ubiquitination of proteins that bind with it. However, TRAF4 showed no effect on the ubiquitination of PKM2 in our study (Fig. 6b), which may explain why there was no difference in PKM2 expression among the groups above. Thus, we analyzed the pyruvate kinase activity and found that TRAF4 overexpression increased pyruvate kinase activity, while TRAF4 knockdown decreased pyruvate kinase activity in MSCs during adipogenesis (Fig. 6c). Previous studies demonstrated that PKM2 plays a role in adipogenic differentiation [20]. Thus, we speculated that the activity of PKM2 may also affect adipogenic differentiation in MSCs. To assess this question, a kinase agonist (DASA) and inhibitor (PKM2-IN) of PKM2 were applied during adipogenic differentiation. The results revealed that increasing the concentration of PKM2-IN gradually promoted adipogenesis, while increasing the concentration of DASA exerted the opposite effect (Fig. S2). Further confirming the role of PKM2 activity in the TRAF4-mediated inhibition of adipogenic differentiation, 10 μM PKM2-IN increased adipogenesis in the TRAF4-overexpressing group similar to that in the NC1 group, and 5 μM DASA reduced the adipogenesis of the TRAF4-knockdown group similar to that of the NC2 group (Fig. 6d). Moreover, the results showed that 10 μM PKM2-IN reduced PKM2 activity and active β-catenin expression in the TRAF4-overexpressing group to levels near those of the NC1 group, while 5 μM DASA exerted an inverse effect on the TRAF4 knockdown group (Fig. 6e and f). Altogether, the data indicate that TRAF4 inhibits adipogenic differentiation by increasing the PKM2 activity of MSCs.
Fig. 6.
TRAF4 inhibited adipogenic differentiation by activating pyruvate kinase activity. (a) Western blotting results of adipogenic differentiation at 10 days show that TRAF4 did not affect the expression of PKM2 but inhibited the expression of p-PKM2. (b) MYC-TRAF4 showed no effect on the ubiquitination of Flag-PKM2. (c) TRAF4 overexpression increased pyruvate kinase activity, while TRAF4 knockdown decreased pyruvate kinase activity in MSCs after adipogenic induction for 10 days. (d) ORO staining and quantification demonstrated that 10 μM PKM2-IN (a kinase inhibitor) could rescue the impaired adipogenic differentiation in the TRAF4-overexpressing group, while 5 μM DASA (a kinase agonist) could reduce the enhanced adipogenic differentiation in the TRAF4-knockdown group (scale bar = 150 μm). (e) 10 μM PKM2-IN decreased the pyruvate kinase activity in the TRAF4-overexpressing group to a level near that in the NC1 group, while 5 μM DASA increased the pyruvate kinase activity in the sh-TRAF4 group. (f) 10 μM PKM2-IN decreased the active β-catenin expression in the TRAF4-overexpressing group to a level near that in the NC1 group, while 5 μM DASA exerted an inverse effect on the TRAF4-knockdown group. (g-h) Overexpression of TRAF4 or the mutant retaining the RING domain (ΔTRAF) could increase PKM2 activity and inhibit adipogenesis, whereas the TRAF4 mutant lacking the RING domain (ΔRING) could not. All data are presented as the means±SDs. *p < 0.05, according to Student's t-test (a) and a one-way analysis of variance (c–h) (n = 3 independent experiments with three different MSC samples).
To identify the functional domain of TRAF4 that inhibits the adipogenic differentiation of MSCs, we detected the effect of TRAF4 and mutants (TRAF4 ΔRING or TRAF4 ΔTRAF) on the PKM2 activity and adipogenic differentiationof MSCs. Interestingly, overexpression of TRAF4 or the mutant retaining the RING domain (ΔTRAF) could increase PKM2 activity and inhibit adipogenesis, whereas the TRAF4 mutant lacking the RING domain (ΔRING) could not (Fig. 6g and h). In summary, the RING domain of TRAF4, which binds to PKM2, is required for the TRAF4-mediated inhibition of adipogenic differentiation in MSCs.
3.7. Excessive adipogenesis and decreased TRAF4 were observed in the marrow cavity of osteoporotic rats
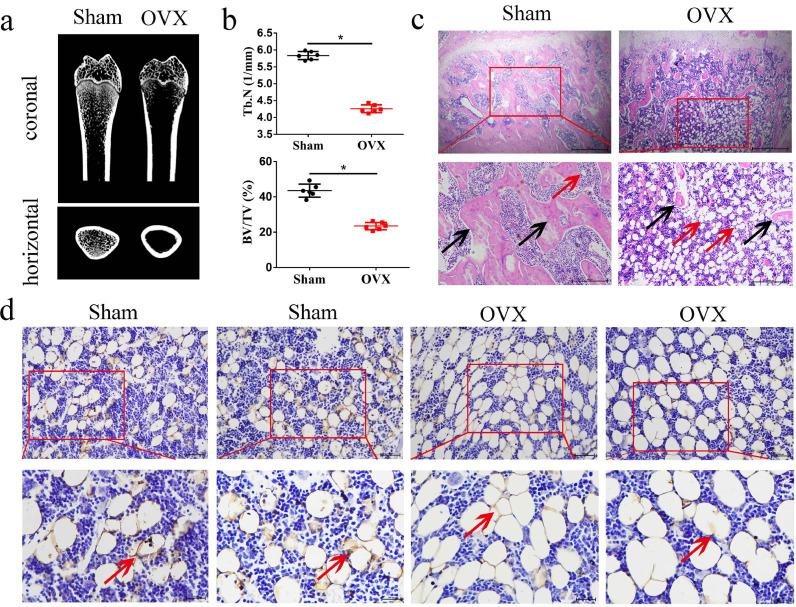
We previously reported that TRAF4 positively regulates the osteogenic differentiation of MSCs and might be involved in the pathogenesis of osteoporosis [26]. Substantial evidence suggests that the balance between osteogenesis and adipogenesis plays an important role in osteoporosis [27,28]. We constructed a rat model of postmenopausal osteoporosis using ovariectomy surgery (OVX). The CT results demonstrated that the cancellous bone mass in the proximal femur was significantly decreased in the OVX groups compared with the sham group (Fig. 7a), and further analysis indicated that OVX significantly decreased trabecular Tb.N and BV/TV in the OVX groups compared to the sham group (Fig. 7b). Moreover, these results were confirmed by H&E staining (Fig. 7c, black arrows). Interestingly, the H&E staining results revealed that excessive adipogenesis was found in OVX rats compared with sham-operated rats (Fig. 7c, red arrows). Furthermore, the immunohistochemistry (IHC) results demonstrated that TRAF4 is decreased in the marrow cavity, especially in the adipocytes of osteoporotic rats (Fig. 7d, red arrows). Above all, these results indicated that TRAF4 is decreased, with excessive adipogenesis in osteoporosis. Thus, we speculated that TRAF4 might affect the adipogenesis of MSCs and is involved in the pathogenesis of osteoporosis.
Fig. 7.
Excessive adipogenesis but decreased TRAF4 was observed in the marrow cavities of osteoporotic rats. We constructed a rat model using ovariectomy surgery (OVX) to explore the pathogenesis of postmenopausal osteoporosis. (a) Representative micro-CT images demonstrated that the cancellous bone mass was significantly decreased in the OVX group compared with the sham group. (b) Micro-CT analysis indicated that the bone mineral density, bone volume, and trabecular number were reduced in OVX rats. (c) H&E staining results showed that excess adipocytes (red arrows) and fewer trabecula (black arrows) were observed in OVX rats (scale bar = 500 μm). (d) The IHC results demonstrated that TRAF4 was decreased in the marrow cavity, especially in the adipocytes of osteoporotic rats (red arrows, scale bar = 100 μm). All data are presented as the means±SDs. *p < 0.05, according to Student's t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.8. ALKBH5-mediated m6A RNA demethylation regulates the mRNA expression of TRAF4 during adipogenesis
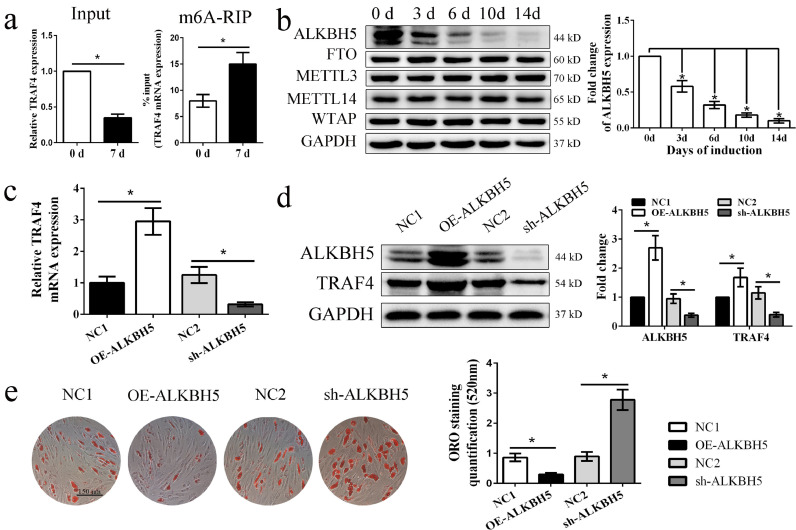
Why TRAF4 expression was downregulated during MSC adipogenic differentiation remains unknown. Previous studies demonstrated that N(6)-methyladenosine (m6A) methylation plays an important role in regulating posttranscriptional gene expression [29]. Thus, we wondered whether m6A methylation modifications were involved in TRAF4 mRNA expression regulation during the adipogenic differentiation of MSCs. First, m6A-IP-qPCR indicated that the m6A abundance of TRAF4 was markedly increased on day 7 compared to day 0, while TRAF4 was decreased on day 7 in the input groups (Fig. 8a). These results suggest that TRAF4 may be regulated by m6A methylation. Second, we examined the expression patterns of known m6A writers (Mettl3, Mettl14 and Wtap) and erasers (ALKBH5 and FTO) during the adipogenic differentiation of MSCs. The Western blotting results indicated that ALKBH5 gradually decreased during adipogenic differentiation, while the others did not (Fig. 8b).
Fig. 8.
ALKBH5-mediated m6A RNA demethylation regulated TRAF4 mRNA expression during adipogenesis. (a) The m6A-IP-qPCR results demonstrated that the m6A abundance of TRAF4 was markedly increased on day 7 after adipogenic induction, while TRAF4 was decreased on day 7 in the input samples. (b) Western blotting demonstrated that ALKBH5 gradually decreased during adipogenic differentiation. (c, d) The TRAF4 mRNA and protein expression levels were increased in the ALKBH5-overexpressing group and decreased in the ALKBH5-knockdown group. (e) ORO staining and quantification results demonstrated that ALKBH5 negatively regulated the adipogenic differentiation capacity of MSCs in vitro (scale bar = 150 μm). All data are presented as the means±SDs. *p<0.05, according to Student's t-test (a, c–e) and a one-way analysis of variance (b) (n = 3 independent experiments with three different MSC samples).
Furthermore, we constructed a lentivirus to regulate ALKBH5 expression to confirm this speculation. ALKBH5 expression was efficiently increased in the overexpression group and decreased in the knockdown group, as shown byWestern blotting. Next, we detected TRAF4 expression using qPCR and Western blotting. Our results showed that the TRAF4 mRNA and protein expression levels were increased in the ALKBH5-overexpressing group but decreased in the ALKBH5-knockdown group compared to the corresponding negative groups (Fig. 8c and d). In addition, the ORO staining results demonstrated that ALKBH5 overexpression decreased the formation of adipocytes, while ALKBH5 knockdown promoted the adipogenic differentiation capacity of MSCs in vitro (Fig. 8e). Taken together, these results indicated that TRAF4 downregulation during adipogenesis was regulated by m6A RNA demethylation.
4. Discussion
Previously, we demonstrated that TRAF4 positively regulates the osteogenic process of MSCs both in vitro and in vivo [26]. In this study, we identified a downregulated TRAF4 expression pattern during the adipogenic differentiation process of MSCs and confirmed that TRAF4 negatively regulated the adipogenesis of MSCs both in vitro and in vivo. Further mechanistic investigations revealed that TRAF4 interacts with PKM2, activating its kinase activity and then activating β-catenin signaling to regulate the adipogenesis of MSCs. Surprisingly, TRAF4 downregulation during adipogenesis was regulated by m6A RNA demethylation (Fig. S4). In addition, we observed that TRAF4 was decreased in the bone marrow cavities of femurs in osteoporotic rats. These results suggest that TRAF4 is a differentiation checkpoint in MSCs that conversely regulates the adipogenic and osteogenic differentiation of MSCs. Moreover, TRAF4 could be a promising target of MSC-based tissue engineering and may be involved in the pathogenesis of some bone metabolism diseases, such as osteoporosis.
MSCs, which are also known as multipotent stromal cells, are extensively applied in tissue engineering due to their self-renewal and multiple differentiation abilities [30]. However, the clinical application of MSCs poses a potential problem due to the possibility of differentiation into unwanted tissues [7]. Moreover, disruption of the balance during MSC differentiation may lead to metabolic diseases, such as skeletal fragility and osteoporosis [31]. Therefore, controlling the specific differentiation of MSCs, especially osteogenesis or adipogenesis, is quite important. Accumulating studies have demonstrated that there is an inverse and competitive balance between adipogenesis and osteogenesis [32,33], and these processes are recognized as the crossroads of MSC differentiation. MSC differentiation contains two steps: lineage commitment and maturation. The first step determines the lineage-specific differentiation, which is usually irreversible once the first step has begun [34]. Recently, studies have indicated that some molecules are involved in regulating the first step of MSC differentiation, which plays a critical role in regulating adipo-osteogenic differentiation as a molecular checkpoint [35,36]. For example, Runx2 can promote the MSC differentiation potential toward osteoblasts while inhibiting the lineage commitment to adipocytes [37]. Although great progress has been made over the past decades in exploring these regulators regarding adipo-osteogenic differentiation, further studies are required to identify more and better checkpoints. We previously reported that TRAF4 positively regulated the osteogenic capacity of MSCs by acting as an E3 ubiquitin ligase to degrade Smurf2 via the K48-linked ubiquitination of Smurf2 [26]. However, whether TRAF4 affects the adipogenic differentiation of MSCs remains unknown.
To explore this question, we assessed the adipogenic differentiation of MSCs after knocking down and overexpressing TRAF4. Our results revealed that TRAF4 could inhibit the adipogenesis of MSCs both in vitro and in vivo. These results, along with our previous results [26], suggested that TRAF4 is one of the key checkpoints regulating the adipo-osteogenic differentiation of MSCs. Surprisingly, TRAF4 expression in MSCs gradually decreased during adipogenic differentiation. Similarly, Hong et al. [5] demonstrated that TAZ, an adipocyte differentiation inhibitor, was decreased during differentiation. We conclude that TRAF4 is a checkpoint for MSC differentiation and may function as a “clutch” during adipogenesis, so gradually decreased TRAF4 levels will promote the adipogenic differentiation ability of MSCs. Therefore, decreased TRAF4 was observed after adipogenic induction, and TRAF4 downregulation may be a necessary process to initiate adipogenesis.
Adipogenic differentiation, just one of the multiple differentiation of MSCs. Thus, maintaining MSC differentiation toward desired cells, such as adipocytes, requires a complex regulatory process. Many studies have indicated that cellular/molecular signaling pathways and microenvironmental changes can affect adipogenic differentiation. Cao et al. demonstrated that PPAR-γ plays an important role in promoting adipogenic differentiation [1,5]. Similarly, some other proteins have also been reported to increase the adipognesis of MSCs, such as CEBPB, Twist-1, and Oct4. By constrast, Foxa 1, HOXC8 and GATA have been found to exert an inhibitory effect on adipogenic differentiation. Moreover, most of the above molecules might regulate adipogenic differentiation via the Wnt/β-catenin, PI3K/Akt or BMPR-1A/TGF-β signaling pathways.
Some questions remain concerning how TRAF4 inhibits the adipogenic differentiation of MSCs. We previously reported that TRAF4 promoted osteogenesis by acting as an E3 ubiquitin ligase to degrade Smurf2 [26]. Interestingly, the mechanism by which TRAF4 inhibits adipogenesis is completely different from that involved in the osteogenesis of MSCs. Large studies have indicated that TRAF4 acts as a classical ubiquitin enzyme [11], so we speculated that it potentially interacts with a key molecule directly to regulate adipogenesis. Our results revealed that TRAF4 binds to PKM2 and inhibits adipogenesis through the RING domain, while TRAF4 interacts with Smurf2 through the TRAF domain, and the promotion to osteogenesis relies on both the RING and TRAF domains. Large previous studies demonstrated that the RING domain plays a critical role in TRAF4-mediated ubiquitination [38], which always affects protein degradation, while the TRAF domain can mediate protein kinase activation [39]. However, our results revealed that TRAF4 did not affect the ubiquitination of PKM2 but affected its kinase activity, while the RING domain was the functional domain in our study. These results suggested that the RING domain could also exert an effect on kinase activity in addition to ubiquitination. Similarly, some RING finger proteins without the TRAF domain were also reported to activate the kinase pathway [40]. These results not only revealed that different domains showed different biological functions but also indicated that the same domain of a protein may exert diverse functions under different circumstances. Moreover, we found that TRAF4 regulates adipogenesis and osteogenesis by respectively modulating β-catenin and Smad1 signaling. Much evidence has confirmed that PKM2 is involved in the regulation of β-catenin signaling. Yang et al. demonstrated that PKM2 regulates β-catenin transactivation via c-Src-mediated β-catenin Y333 phosphorylation and EGFR activation [16]. Zheng et al. indicated that PKM2 reduction inhibits β-catenin activity [21,41]. In brief, PKM2 might regulate the β-catenin signaling pathway via affecting β-catenin nuclear localization or activating its expression. Moreover, studies have reported that the activation of β-catenin signaling inhibited the adipogenic differentiation of MSCs [36,42]. Above all, TRAF4 might regulate adipogenic differentiation through the PKM2-catenin axis, independently of its osteogenic regulatory mechanism.
Although adipogenic and osteogenic differentiation have been well explained, the fate decision and balance of adipo-osteogenic differentiation are of great importance. The most critical problem is how to maintain the balance when MSCs initiate their differentiation into osteoblasts or adipoblasts. The present study demonstrated that the microenvironment in vivo and a variety of external cues, including chemical, physical and biological factors, contribute to the adipo-osteogenic differentiation balance of MSCs [43,44]. All of these factors could activate various molecules and trigger different signaling pathways that guide MSCs to differentiate into either lineage. Thus, checkpoints that can regulate both adipogenic and osteogenic differentiation are of great significance because they determine the fate of MSCs. MSC-based skeletal therapies have attracted increasing attention in recent years [45], but their clinical efficiency is limited due to the possibility of differentiation into unwanted tissues [7]. Checkpoints that can drive MSCs to differentiate into the indicated lineage will improve the clinical application of MSCs. In addition, dysregulation of the adipo-osteogenic balance has been linked to several diseases, such as osteoporosis and osteopenia [15]. MSCs can selectively differentiate into adipocytes and osteoblasts, and the prevailing though is that there is an opposite relationship between the two differentiation. This opposing phenomenon and adipo-osteogenic differentiation imbalance can also be observed in osteoporosis. Moerman et al. have demonstrated that adipocytes were increased while osteoblasts were decreased during osteoporosis, which was accompanied by a reduced bone mineral density [46]. Further studies revealed that MSCs from old humans or mice had a high ability to differentiate into adipocytes rather than osteoblasts via multiple mechanisms, which might help to explain osteoporosis [23]. Our results, along with our previous study [26], demonstrated that TRAF4 could increase the osteogenic differentiation and decrease the adipogenic differentiation of MSCs and that TRAF4 was decreased in osteoporotic rats. The decreased-TRAF4-mediated adipo-osteogenic differentiation imbalance might help explain the reduced bone trabecula and increased adipocytes in marrow cavity of osteoporosis. Additionally, these results suggested that TRAF4 may be a potential therapeutic target and diagnostic marker for osteoporosis. Thus, elucidating the roles of the checkpoints in the adipo-osteogenic balance contributes to our understanding of the mechanisms of some bone metabolic diseases.
As the adipocyte-or-osteoblast differentiation checkpoint is of great medical importance, finding the pathway upstream of TRAF4 that regulates TRAF4 expression is also very important. In our study, we demonstrated that both TRAF4 mRNA and protein levels were decreased during MSC adipogenic differentiation from day 0 to day 14. In recent years, abundant research has indicated that N6-methyladenosine (m6A) is the most prevalent internal modification in eukaryotic mRNA [47,48]. Therefore, we speculated that decreased TRAF4 expression may be regulated by m6A. Interestingly, we found that ALKBH5-dependent demethylation of m6A could regulate TRAF4 mRNA expression and is required for adipogenic differentiation. Zhao et al. [49] previously reported that FTO (another m6A demethylase) depletion blocked adipogenesis by controlling exonic splicing of the adipogenic regulatory factor RUNX1T1. However, these results seem controversial. After careful analysis, we used cells harvested from human bone marrow in our study, whereas in their study, the cells were from a mouse 3T3-L1 preadipocyte cell line. Thus, the above factor might have contributed to this “discrepancy”. However, the cells used in our study were harvested from humans rather than from mice; therefore, our study is more suitable for life science applications in the human body. Similarly, this “discrepancy” has been reported in other bioprocesses [50,51]. Despite the “discrepancy” between these studies, both suggest that m6A plays a critical role in cell biology, such as in adipogenic differentiation, and further investigations are required to address this “discrepancy”.
In conclusion, we demonstrated that TRAF4 negatively regulated the adipogenesis of MSCs by activating PKM2 kinase activity, which may act as a checkpoint to fine-tune the balance between adipogenic and osteogenic differentiation. These results suggest that TRAF4 may be a novel therapeutic target to improve the clinical applications of MSCs but may also illuminate the underlying mechanism of bone metabolism diseases, such as osteoporosis. However, there are still some limitations to this study. To further explore the specific role of TRAF4 in adipogenic-osteogenic differentiation, transgenic mice in which TRAF4 is targeted toward MSCs are necessary, and we are currently developing these transgenic mice for future studies.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
The authors thank American Journal Experts for providing English language editing of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81871750 and 81971518) and the Science and Technology Project of Guangdong Province (2019B02023600 and 2017A020215070).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102722.
Contributor Information
Zhongyu Xie, Email: xiezhy23@mail.sysu.edu.cn.
Peng Wang, Email: wangp57@mail.sysu.edu.cn.
Huiyong Shen, Email: shenhuiy@mail.sysu.edu.cn.
Appendix. Supplementary materials
References
- 1.Cao Y., Gomes S.A., Rangel E.B., Paulino E.C., Fonseca T.L., Li J. S-nitrosoglutathione reductase-dependent PPARgamma denitrosylation participates in MSC-derived adipogenesis and osteogenesis. J Clin Invest. 2015;125(4):1679–1691. doi: 10.1172/JCI73780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan M., Yao W., Liu R., Lam K.S., Nolta J., Jia J. Directing mesenchymal stem cells to bone to augment bone formation and increase bone mass. Nat Med. 2012;18(3):456–462. doi: 10.1038/nm.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jing H., Liao L., An Y., Su X., Liu S., Shuai Y. Suppression of EZH2 prevents the shift of osteoporotic MSC fate to adipocyte and enhances bone formation during osteoporosis. Mol Ther J Am Soc Gene Ther. 2016;24(2):217–229. doi: 10.1038/mt.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdallah B.M., Jafari A., Zaher W., Qiu W., Kassem M. Skeletal (stromal) stem cells: an update on intracellular signaling pathways controlling osteoblast differentiation. Bone. 2015;70:28–36. doi: 10.1016/j.bone.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Hong J.H., Hwang E.S., McManus M.T., Amsterdam A., Tian Y., Kalmukova R. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M., Zheng C., Shou P., Li N., Cao G., Chen Q. SHP1 regulates bone mass by directing mesenchymal stem cell differentiation. Cell Rep. 2016;17(8):2161. doi: 10.1016/j.celrep.2016.10.069. [DOI] [PubMed] [Google Scholar]
- 7.Kim J., Ko J. A novel PPARgamma2 modulator sLZIP controls the balance between adipogenesis and osteogenesis during mesenchymal stem cell differentiation. Cell Death Differ. 2014;21(10):1642–1655. doi: 10.1038/cdd.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Q., Song W.X., Luo Q., Tang N., Luo J., Luo X. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009;18(4):545–559. doi: 10.1089/scd.2008.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett C.N., Ross S.E., Longo K.A., Bajnok L., Hemati N., Johnson K.W. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277(34):30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 10.Inoue J., Ishida T., Tsukamoto N., Kobayashi N., Naito A., Azuma S. Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp Cell Res. 2000;254(1):14–24. doi: 10.1006/excr.1999.4733. [DOI] [PubMed] [Google Scholar]
- 11.Singh R., Karri D., Shen H., Shao J., Dasgupta S., Huang S. TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J Clin Invest. 2018;128(7):3129–3143. doi: 10.1172/JCI96060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai G., Zhu L., Chen X., Sun K., Liu C., Sen G.C. TRAF4 binds to the juxtamembrane region of EGFR directly and promotes kinase activation. Proc Natl Acad Sci U S A. 2018;115(45):11531–11536. doi: 10.1073/pnas.1809599115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regnier C.H., Masson R., Kedinger V., Textoris J., Stoll I., Chenard M.P. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc Natl Acad Sci U S A. 2002;99(8):5585–5590. doi: 10.1073/pnas.052124799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Wang P., Xie Z., Wang S., Cen S., Li M. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2019;26(12):2652–2666. doi: 10.1038/s41418-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q., Shou P., Zheng C., Jiang M., Cao G., Yang Q. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23(7):1128–1139. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spoden G.A., Rostek U., Lechner S., Mitterberger M., Mazurek S., Zwerschke W. Pyruvate kinase isoenzyme M2 is a glycolytic sensor differentially regulating cell proliferation, cell size and apoptotic cell death dependent on glucose supply. Exp Cell Res. 2009;315(16):2765–2774. doi: 10.1016/j.yexcr.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 18.Dayton T.L., Jacks T., Heiden M.G.V. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17(12):1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isidor M.S., Winther S., Markussen L.K., Basse A.L., Quistorff B., Nedergaard J. Pyruvate kinase M2 represses thermogenic gene expression in brown adipocytes. FEBS Lett. 2019 doi: 10.1002/1873-3468.13716. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y., Guo L., Xie L.Q., Zhang Y.Y., Liu X.H., Zhang Y. Proteome profiling of mitotic clonal expansion during 3T3-L1 adipocyte differentiation using iTRAQ-2DLC-MS/MS. J Proteome Res. 2014;13(3):1307–1314. doi: 10.1021/pr401292p. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Q., Lin Z., Xu J., Lu Y., Meng Q., Wang C. Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting beta-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 2018;9(3):253. doi: 10.1038/s41419-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross S.E., Hemati N., Longo K.A., Bennett C.N., Lucas P.C., Erickson R.L. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 23.Singh L., Brennan T.A., Russell E., Kim J.H., Chen Q., Brad Johnson F. Aging alters bone-fat reciprocity by shifting in vivo mesenchymal precursor cell fate towards an adipogenic lineage. Bone. 2016;85:29–36. doi: 10.1016/j.bone.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colter D.C., Class R., DiGirolamo C.M., Prockop D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci U S A. 2000;97(7):3213–3218. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z., Wang P., Li Y., Deng W., Zhang X., Su H. Imbalance between bone morphogenetic protein 2 and noggin induces abnormal osteogenic differentiation of mesenchymal stem cells in ankylosing spondylitis. Arthritis Rheumatol. 2016;68(2):430–440. doi: 10.1002/art.39433. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Wang P., Xie Z., Wang S., Cen S., Li M. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2019;26:2652–2666. doi: 10.1038/s41418-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y., Xie L., Wang M., Xiong Q., Guo Y., Liang Y. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9(1):4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Infante A., Rodriguez C.I. Osteogenesis and aging: lessons from mesenchymal stem cells. Stem Cell Res Ther. 2018;9(1):244. doi: 10.1186/s13287-018-0995-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29(13):1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satija N.K., Gurudutta G.U., Sharma S., Afrin F., Gupta P., Verma Y.K. Mesenchymal stem cells: molecular targets for tissue engineering. Stem Cells Dev. 2007;16(1):7–23. doi: 10.1089/scd.2006.9998. [DOI] [PubMed] [Google Scholar]
- 31.Uccelli A., Moretta L., Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8(9):726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J., Wang S., Qi Q., Yang X., Zhu E., Yuan H. Nuclear factor I-C reciprocally regulates adipocyte and osteoblast differentiation via control of canonical Wnt signaling. FASEB J Off Publ Fed Am Soc Exp Biol. 2017;31(5):1939–1952. doi: 10.1096/fj.201600975RR. [DOI] [PubMed] [Google Scholar]
- 33.Cheng H., Qiu L., Zhang H., Cheng M., Li W., Zhao X. Arsenic trioxide promotes senescence and regulates the balance of adipogenic and osteogenic differentiation in human mesenchymal stem cells. Acta Biochim Biophys Sin. 2011;43(3):204–209. doi: 10.1093/abbs/gmq130. [DOI] [PubMed] [Google Scholar]
- 34.Hemming S., Cakouros D., Isenmann S., Cooper L., Menicanin D., Zannettino A. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32(3):802–815. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- 35.Li X., Huynh H., Zuo H., Salminen M., Wan Y. Gata2 is a rheostat for mesenchymal stem cell fate in male mice. Endocrinology. 2016;157(3):1021–1028. doi: 10.1210/en.2015-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Z., Li Q., Luo S., Liu Z., Luo D., Zhang B. PPARgamma and wnt signaling in adipogenic and osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther. 2016;11(3):216–225. doi: 10.2174/1574888x10666150519093429. [DOI] [PubMed] [Google Scholar]
- 37.Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 38.Bradley J.R., Pober J.S. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20(44):6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 39.Song H.Y., Regnier C.H., Kirschning C.J., Goeddel D.V., Rothe M. Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-kappaB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc Natl Acad Sci U S A. 1997;94(18):9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arshad M., Ye Z., Gu X., Wong C.K., Liu Y., Li D. RNF13, a RING finger protein, mediates endoplasmic reticulum stress-induced apoptosis through the inositol-requiring enzyme (IRE1alpha)/c-Jun NH2-terminal kinase pathway. J Biol Chem. 2013;288(12):8726–8736. doi: 10.1074/jbc.M112.368829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y., Meng F., Lu Z., Chen K., Tao Y., Ouyang Y. Knockdown of PKM2 suppresses tumor progression in human cervical cancer by modulating epithelial-mesenchymal transition via Wnt/beta-catenin signaling. Cancer Manag Res. 2018;10:4191–4202. doi: 10.2147/CMAR.S178219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prestwich T.C., Macdougald O.A. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19(6):612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehmann J.M., Lenhard J.M., Oliver B.B., Ringold G.M., Kliewer S.A. Peroxisome proliferator-activated receptors alpha and gamma are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J Biol Chem. 1997;272(6):3406–3410. doi: 10.1074/jbc.272.6.3406. [DOI] [PubMed] [Google Scholar]
- 44.da Silva M.L., Chagastelles P.C., Nardi N.B. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119(Pt 11):2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 45.Parekkadan B., Milwid J.M... Mesenchymal stem cells as therapeutics. Annu Rev Biomed Eng. 2010;12:87–117. doi: 10.1146/annurev-bioeng-070909-105309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moerman E.J., Teng K., Lipschitz D.A., Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3(6):379–389. doi: 10.1111/j.1474-9728.2004.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F., Yi Y., Miao Y., Long W., Long T., Chen S. N6-methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 2019;15:5785–5798. doi: 10.1158/0008-5472.CAN-18-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lockhart J. The story continues: following the fate of m6A marks in the eukaryotic transcriptome. Plant Cell. 2018;30(7):1385–1386. doi: 10.1105/tpc.18.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X., Yang Y., Sun B.F., Shi Y., Yang X., Xiao W. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary microRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 51.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.