Abstract
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been a Public Health Emergency of International Concern, which was officially declared by the World Health Organization. SARS-CoV-2 is a member of the family Coronaviridae that consists of a group of enveloped viruses with single-stranded RNA genome, which cause diseases ranging from common colds to acute respiratory distress syndrome. Although the major transmission routes of SARS-CoV-2 are inhalation of aerosol/droplet and person-to-person contact, currently available evidence indicates that the viral RNA is present in wastewater, suggesting the need to better understand wastewater as potential sources of epidemiological data and human health risks. Here, we review the current knowledge related to the potential of wastewater surveillance to understand the epidemiology of COVID-19, methodologies for the detection and quantification of SARS-CoV-2 in wastewater, and information relevant for human health risk assessment of SARS-CoV-2. There has been growing evidence of gastrointestinal symptoms caused by SARS-CoV-2 infections and the presence of viral RNA not only in feces of infected individuals but also in wastewater. One of the major challenges in SARS-CoV-2 detection/quantification in wastewater samples is the lack of an optimized and standardized protocol. Currently available data are also limited for conducting a quantitative microbial risk assessment (QMRA) for SARS-CoV-2 exposure pathways. However, modeling-based approaches have a potential role to play in reducing the impact of the ongoing COVID-19 outbreak. Furthermore, QMRA parameters obtained from previous studies on relevant respiratory viruses help to inform risk assessments of SARS-CoV-2. Our understanding on the potential role of wastewater in SARS-CoV-2 transmission is largely limited by knowledge gaps in its occurrence, persistence, and removal in wastewater. There is an urgent need for further research to establish methodologies for wastewater surveillance and understand the implications of the presence of SARS-CoV-2 in wastewater.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, Wastewater-based epidemiology (WBE), Virus detection method, Quantitative microbial risk assessment (QMRA)
Graphical abstract
Highlights
-
•
Presence of SARS-CoV-2 RNA in wastewater has been reported.
-
•
SARS-CoV-2 RNA in wastewater can be used to monitor COVID-19 in a community.
-
•
Effective concentration method is needed for recovery of SARS-CoV-2 from wastewater.
-
•
Surrogate coronavirus data help to predict survival of SARS-CoV-2 in wastewater.
-
•
Data on the infectivity of SARS-CoV-2 in wastewater for risk assessment are limited.
1. Introduction
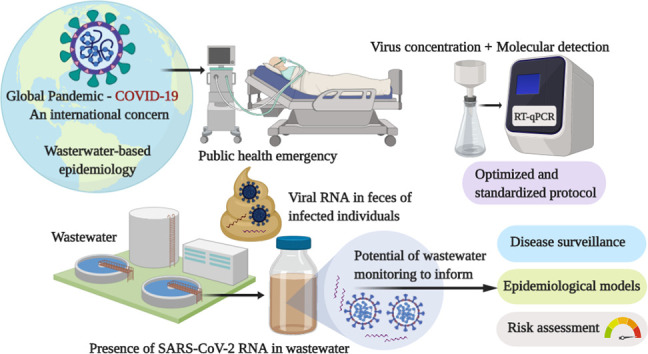
In December 2019, China reported an outbreak of pneumonia of unknown etiology occurring in Wuhan, Central China's Hubei Province to the World Health Organization (WHO) (WHO, 2020a). Shotgun metagenomic sequencing of bronchoalveolar lavage samples indicated that this outbreak was associated with a novel coronavirus (nCoV) (Zhu et al., 2020). The nCoV was confirmed to have 75–80% nucleotide similarity to severe acute respiratory syndrome coronavirus (SARS-CoV) (Zhu et al., 2020) and was officially designated as SARS-CoV-2 after being provisionally named as 2019-nCoV (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). SARS-CoV-2 together with SARS-CoV belong to the species Severe acute respiratory syndrome-related coronavirus in the subgenus Sarbecovirus of the family Coronaviridae that consists of a group of enveloped viruses with a single-stranded, positive-sense RNA genome. SARS-CoV and SARS-CoV-2 are distantly related to Middle East respiratory syndrome coronavirus (MERS-CoV), which belongs to the species Middle East respiratory syndrome-related coronavirus within the genus Betacoronavirus (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). SARS-CoV-2 is also distantly related to ‘classical’ human CoV strains (229E, OC43, NL63, and HKU1) belonging to the genus Alphacoronavirus or Betacoronavirus that have been studied since the 1960s and are estimated to cause 15 to 30% of cases of common colds worldwide (Mesel-Lemoine et al., 2012).
The disease caused by SARS-CoV-2 is referred to as coronavirus disease 2019 (COVID-19). Symptoms of COVID-19 at the onset of illness include fever, myalgia, fatigue, and dry cough, and more than half of patients developed dyspnea (Chen et al., 2020b; Guan et al., 2020; Huang et al., 2020; Wang et al., 2020a, 2020b). On March 11, 2020, WHO declared the current COVID-19 situation a global pandemic on the basis of “alarming levels of spread, severity, and inaction” (Bedford et al., 2020). The WHO then declared the outbreak of COVID-19 a Public Health Emergency of International Concern on January 31, 2020 (WHO, 2020b). As of April 26, 2020, SARS-CoV-2 has further spread to almost all countries and territories around the world with >2,724,809 confirmed cases and 187,847 confirmed deaths, according to WHO (WHO, 2020c). The case fatality rate was estimated as 5.3–8.4% for COVID-19 (Jung et al., 2020), which is lower than SARS (up to 50%) or MERS (34.4–69.2%) (Park et al., 2018;Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c; WHO, 2003). The basic reproduction number (R0) of SARS-CoV-2 was estimated as 1.4–6.5 (Boldog et al., 2020; Jung et al., 2020; Liu et al., 2020; B. Tang et al., 2020; WHO, 2020c), meaning that each infected individual could transmit the virus to another 1.4–6.5 cases - comparable to that of SARS-CoV (R0 of 2 to 5) (Lipsitch et al., 2003; Riley et al., 2003; Wallinga and Teunis, 2004).
Both viable SARS-CoV-2 and viral RNA are shed in bodily excreta, including saliva, sputum, and feces, which are subsequently disposed of in wastewater. Although it is believed that the major transmission route of this virus is inhalation via person-to-person aerosol/droplet transmission, and fomite to hand contamination, currently available evidence indicates the need for better understanding of the role of wastewater as potential sources of epidemiological data and as a factor in public health risk. In this paper, we thoroughly reviewed the current knowledge related to the potential of wastewater surveillance for understanding the epidemiology of COVID-19. Given the rapid emergence of SARS-CoV-2, previous studies on human CoVs, SARS-CoV, MERS-CoV, and surrogate viruses can help to inform predictions of the likely environmental fate and subsequent risks of SARS-CoV-2. We also identified critical research needs that will strengthen our understanding on the occurrence, persistence, and potential public health risks associated with SARS-CoV-2 in wastewater. The synthesis of recent findings highlights that the presence of SARS-CoV-2 RNA in wastewater provides an opportunity to use wastewater as a surveillance tool for the invasion, prevalence, molecular epidemiology, and potential eradication of the virus in a community.
2. Gastrointestinal symptoms in COVID-19 and shedding of SARS-CoV-2 in excreta
Human CoVs, including SARS-CoV and MERS-CoV, are known to cause gastrointestinal symptoms in addition to respiratory symptoms (Leung et al., 2003; Memish et al., 2015). In fact, previous studies demonstrated that these viruses replicate in the gastrointestinal tract (Leung et al., 2003; Zhou et al., 2017). Recent reports revealed that 2–10% of COVID-19 patients had gastrointestinal symptoms, including diarrhea (Chen et al., 2020a, Chen et al., 2020b; Gao et al., 2020; Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c). Although the exact mechanism of COVID-19-induced gastrointestinal symptoms largely remains elusive (Gu et al., 2020), a recent study reported that SARS-CoV-2 infects gastrointestinal glandular epithelial cells (Xiao et al., 2020). Angiotensin-converting enzyme 2 (ACE2) is known to be the cellular receptor for SARS-CoV-2 as well as SARS-CoV (Yan et al., 2020), and the receptor ACE2 is abundantly expressed in the small intestine as well as lung and oral mucosa (Hamming et al., 2004; Xu et al., 2020b). This evidence supports the possibility of SARS-CoV-2 replication in gastrointestinal tract.
Previous studies on SARS-CoV and MERS-CoV detected their viral RNA in feces (Corman et al., 2016; Leung et al., 2003). It has been reported that SARS-CoV RNA loads could be as high as 107 copies/mL in diarrhea and 2.5 × 104 copies/mL in urine (Hung et al., 2004). A number of recent studies reported the presence of SARS-CoV-2 RNA in human feces (Gu et al., 2020; Holshue et al., 2020; Song et al., 2020). As shown in Table 1 , SARS-CoV-2 RNA has been detected in excreta specimens, such as feces and anal/rectal swabs (Gao et al., 2020; Holshue et al., 2020; Jiehao et al., 2020; A. Tang et al., 2020b; Wölfel et al., 2020; Xiao et al., 2020; J. Zhang et al., 2020; W. Zhang et al., 2020b). Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c reported that SARS-CoV-2 RNA was detected by reverse transcription-quantitative PCR (RT-qPCR) in feces in 29% of the patients who were ill, and diarrhea was reported in approximately 2 to 10% of the cases (D. Wang et al., 2020b). Xiao et al. (2020) also found SARS-CoV-2 RNA in 39 (53%) of 73 fecal samples from hospitalized patients. Positive isolation from feces persisted in 23% of the patients even after it disappeared from the respiratory tract (Xiao et al., 2020). Viral RNA concentrations in feces were determined by several studies to be up to 108 copies per gram of feces (Lescure et al., 2020; Pan et al., 2020; Wölfel et al., 2020).
Table 1.
Detection of SARS-CoV-2 in human excreta specimens.
| Specimen | Country | Method | Positive ratea | Remarks | Reference |
|---|---|---|---|---|---|
| Feces or anal/rectal swab | China | qPCR | 14/31 (45%) | (W. Zhang et al., 2020b) | |
| qPCR | 8/22 (36%) | (J. Zhang et al., 2020) | |||
| qPCR | 9/17 (53%) | Day 0–11; 550–1.21 × 105 gene copies/mL | (Pan et al., 2020) | ||
| qPCR | 8/10 (80%) | Pediatric patients; Positive for a mean of 21 (range: 5–28) days | (Y. Xu et al., 2020a) | ||
| qPCR | 5/6 (83%) | Day 3–13 | (Jiehao et al., 2020) | ||
| qPCR | 54/66 (82%) | (Ling et al., 2020) | |||
| qPCR | 39/73 (53%) | (Xiao et al., 2020) | |||
| qPCR | 1/1 (100%) | Asymptomatic | (A. Tang et al., 2020a) | ||
| qPCR | 41/74 (55%) | Positive for a mean of 27.9 days (range: 8–48) | (Y. Wu et al., 2020a) | ||
| qPCR | 12/19 (63%) | (Chen et al., 2020c) | |||
| qPCR | 10/10 (100%) | (Lo et al., 2020) | |||
| Cell culture | 1/1 (100%) | Culturable virus isolated | (Zhang et al., 2020a) | ||
| qPCR | 44/153 (29%) | (W. Wang et al., 2020c) | |||
| Cell culture | 2/4 (50%) | Culturable virus isolated | |||
| USA | qPCR | 1/1 (100%) | Day 7 | (Holshue et al., 2020) | |
| Singapore | qPCR | 4/8 (50%) | (Young et al., 2020) | ||
| Germany | qPCR | 8/9 (89%) | Up to 108 copies/g-feces | (Wölfel et al., 2020) | |
| Cell culture | 0/4 (0%) | No culturable virus isolated | |||
| France | qPCR | 2/5 (40%) | 6.3 × 105–1.3 × 108 gene copies/g-feces | (Lescure et al., 2020) | |
| Urine | China | qPCR | 4/58 (7%) | (Ling et al., 2020) | |
| qPCR | 0/10 (0%) | (Lo et al., 2020) | |||
| Germany | qPCR | 0/9 (0%) | (Wölfel et al., 2020) | ||
| France | qPCR | 0/5 (0%) | (Lescure et al., 2020) |
Based on number of patients tested.
Some clinical studies reported prolonged fecal shedding of SARS-CoV-2 RNA for up to seven weeks after first symptom onset (Jiehao et al., 2020; Y. Wu et al., 2020a; Xiao et al., 2020). Another study reported that viral RNA could be detected in the feces of 81.8% cases even with a negative throat swab result (Ling et al., 2020). Recent reports implied that significant proportions (17.9–30.8%) of infected individuals are asymptomatic (Mizumoto et al., 2020; Nishiura et al., 2020), and SARS-CoV-2 RNA was detected in the feces of asymptomatic individuals as well (Tang et al., 2020a). Although two studies demonstrated the presence of culturable SARS-CoV-2 in fecal samples from COVID-19 patients (Wang et al., 2020b; Zhang et al., 2020a), a more recent study reported that culturable virus was not isolated from feces despite high viral RNA concentrations (Wölfel et al., 2020). This discrepancy could be due to small differences in protocols for virus isolation from feces between laboratories, such as pretreatment method, cell lines, and number of blind passages, as viruses shed in feces are generally fastidious. The use of a recently reported engineered cell line that is highly susceptible to SARS-CoV-2 may enable enhanced virus isolation from feces (Matsuyama et al., 2020). Further research is needed to determine the concentrations of viral RNA and culturable virus particles, if any, in feces of symptomatic and asymptomatic individuals with SARS-CoV-2 infection.
3. Evidence for the presence of SARS-CoV-2 and related CoVs in wastewater
Our knowledge on the presence of CoVs in wastewater is largely limited likely due, at least in part, to the lack of previous environmental investigations focusing on CoVs. As CoVs are an enveloped virus that are thought to be primarily spread via person-to-person contact rather than the fecal-oral route (which has been postulated but not confirmed), their presence in feces requires more nuanced interpretation. In addition, the presence of CoV RNA in wastewater has not garnered widespread use as a disease surveillance tool but is gaining traction in this regard (Ahmed et al., 2020; Lodder and de Roda Husman, 2020). In addition, prior investigations demonstrated that standard virus concentration methods are inefficient to recover enveloped viruses from environmental water samples (Haramoto et al., 2009; Ye et al., 2016). Despite these considerations, one of the first detections of CoVs in wastewater was achieved in 2013 (Wong et al., 2013a). This study reported on detection of DNA and RNA viruses over a 12-month study in the USA and CoVs were found in wastewater in 1 of 12 samples using microarrays. The main focus of the study was on nonenveloped enteric viruses and this detection was not followed up with RT-qPCR to obtain quantitative data. In the same year, a viral metagenomic investigation allowing for untargeted molecular analysis of whole viral community identified the CoV HKU1 genome (a ‘common cold’ CoV) in sewage sludge (Bibby and Peccia, 2013), providing evidence for CoV presence in wastewater. A more recent study also reported the molecular detection of animal CoV belonging to the genus Alphacoronavirus in surface water in Saudi Arabia (Blanco et al., 2019).
During the SARS outbreak in 2004 in China, SARS-CoV RNA was detected in 100% (10/10) of untreated and 30% (3/10) of disinfected wastewater samples collected from a hospital in Beijing, China receiving SARS patients (Wang et al., 2005). Wastewater was also believed to be at least partly responsible for aprevious SARS outbreak due to a faulty ventilation and plumbing system (McKinney et al., 2006). There have been initial reports of the molecular detection of SARS-CoV-2 in wastewater in the Netherlands, USA, France, and Australia (Ahmed et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Nemudryi et al., 2020; F. Wu et al., 2020b; Wurtzer et al., 2020). These studies reported the detection of SARS-CoV-2 RNA in untreated wastewater with maximum concentrations over 106 copies per liter. The study in France detected SARS-CoV-2 RNA in treated wastewater as well, with concentrations of up to nearly 105 copies per liter (Wurtzer et al., 2020). Details of these reports on molecular detection of SARS-CoV-2 RNA in wastewater are summarized in Table 2 . Beyond these initial reports, continuous monitoring of SARS-CoV-2 in wastewater in multiple geographical regions is underway.
Table 2.
Details of reported molecular detection of SARS-CoV-2 in wastewater.
| Sampling location |
Water type | Virus detection methods |
Detection results |
Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Country | State/city | Virus concentration method | qPCR assaya | Sequence confirmation | Positive rate | Maximum concentration (copies/L) | ||
| Australia | Brisbane, Queensland | Untreated wastewater | Electronegative membrane-direct RNA extraction; ultrafiltration | N_Sarbeco NIID_2019-nCOV |
Direct sequence of qPCR products (Sanger + MiSeq) | 2/9 (22%) | 1.2 × 102 | (Ahmed et al., 2020) |
| The Netherlands | Amsterdam, The Hague, Utrecht, Apeldoorn, Amersfoort, Schiphol, Tilburg | Untreated wastewater | Ultrafiltration | CDC N1, N2, N3 E_Sarbeco |
Not done | 14/24 (58%) | Not available | (Medema et al., 2020) |
| USA | Massachusetts | Untreated wastewater | PEG precipitation | CDC N1, N2, N3 | Direct sequence of qPCR products (Sanger) | 10/14 (71%) | >2 × 105 | (F. Wu et al., 2020b) |
| France | Paris | Untreated wastewater | Ultracentrifugation | E_Sarbeco | Not done | 23/23 (100%) | >106.5 | (Wurtzer et al., 2020) |
| Treated wastewater | Ultracentrifugation | E_Sarbeco | Not done | 6/8 (75%) | ~105 | |||
| USA | Bozeman, Montana | Untreated wastewater | Ultrafiltration | CDC N1, N2 | Re-amplification by regular PCR followed by Sanger sequencing | 7/7 (100%) | >3 × 104 | (Nemudryi et al., 2020) |
See Table 3 for details of each qPCR assay.
4. Understanding COVID-19 epidemiology through wastewater surveillance
Wastewater-based epidemiology (WBE) serves as an important tool to trace the circulation of viruses in a community, providing opportunities to estimate their prevalence, genetic diversity, and geographic distribution (Sinclair et al., 2008; Xagoraraki and O'Brien, 2020). Wastewater systems offer a practical approach to identify viruses excreted in the feces of an entire region (Carducci et al., 2006; La Rosa and Muscillo, 2013). Using this approach, it becomes possible to monitor the epidemiology of virus infections even if they are not evident by clinical surveillance, especially because traditional epidemiological approaches may be limited by the asymptomatic nature of many viral infections and underdiagnosis of clinical cases (Johansson et al., 2014; Qi et al., 2018). These limitations are applicable for not only fecally-shed viruses such as adenovirus, norovirus, sapovirus, enterovirus, rotavirus, and hepatitis A virus (Okabayashi et al., 2008; Rodríguez-Lázaro et al., 2012; Yoshida et al., 2009), but also for other viruses that are rarely or never reported by epidemiological surveillance systems, such as Saffold virus, cosavirus, and salivirus/klassevirus (Bonanno Ferraro et al., 2020; Kitajima et al., 2014, Kitajima et al., 2015; Thongprachum et al., 2018).
SARS-CoV-2 is known to cause asymptomatic or pauci-symptomatic infections (Lai et al., 2020; Mizumoto et al., 2020; Nishiura et al., 2020; A. Tang et al., 2020a) making it difficult to determine the actual degree of viral circulation in a community and in making comparisons among different countries that have different clinical diagnostic testing capabilities with even different diagnostic methods/assays (Ortiz-Ospina and Hasell, 2020). Meanwhile, wastewater surveillance could provide an unbiased method of evaluating the spread of infection in different areas, even where resources for clinical diagnosis are limited and when reporting systems are unavailable or not feasible, such as in developing countries. Moreover, wastewater monitoring can help to detect variations in the circulating strains through phylogenetic analysis, allowing for comparisons between regions and assessment of evolution of the virus genome over time as demonstrated previously for enteric viruses (Bisseux et al., 2018; La Rosa et al., 2014; Lodder et al., 2013), and more recently for SARS-CoV-2 (Nemudryi et al., 2020).
The importance of wastewater surveillance is also highlighted by its ability to detect low levels of viruses; this can happen when the number of infected cases is decreasing following public health interventions, which has been the case for successful examples such as poliovirus eradication programs (Asghar et al., 2014). It is also useful to determine when a new virus is introduced into a population (Savolainen-Kopra et al., 2011; Sinclair et al., 2008) or when fluctuations occur due to changes in seasons or precipitation (Hellmér et al., 2014; Prevost et al., 2015; Sedmak et al., 2003). Thus, such a surveillance strategy can be useful as an “early warning” system (Xagoraraki and O'Brien, 2020) to determine if re-introduction of SARS-CoV-2 had occurred in a community, or conversely as an assessment of whether exposures have decreased sufficiently following public health interventions, such as lockdown, social isolation, and social distancing. Virome analysis of wastewater opens up further possibilities of detecting novel viruses before their clinical recognition in a community, allowing for preventative measures and allocation of resources to potentially affected areas (Bibby, 2013; Bibby et al., 2019; Fernandez-Cassi et al., 2018; Ng et al., 2012). A recent study explored the numbers of SARS-CoV-2 RNA copies observed in untreated wastewater that could estimate the number of infected individuals in the catchment via Monte Carlo simulation. The model estimated a median range of 171 to 1090 infected persons in the catchment, which was in reasonable agreement with clinical observations (Ahmed et al., 2020). The authors identified the need for further methodological and molecular assay validation for enveloped viruses in wastewater in order to enhance the accuracy of wastewater surveillance. While a number of applications of wastewater surveillance are obvious, there are limitations. Correlating levels of viruses with a specific number of cases identified epidemiologically may be challenging because of differences in excretion rates of viruses during the course of infection, temporal delays and the inconsistent capture of spatial variability due to travel and use of multiple wastewater systems in time, and dilution due to precipitation, inactivation during the wastewater transport process, and/or infrequent or absent clinical testing (La Rosa and Muscillo, 2013). Also, stability of the genome in wastewater, low efficiency of virus concentration methods, sampling variability (e.g., grab vs. composite) and lack of sensitive detection assays especially at low virus concentrations may collectively limit virus detection and quantification.
Despite these challenges, multiple efforts are underway to develop environmental surveillance programs for SARS-CoV-2. As noted above, there have been initial reports of the molecular detection of SARS-CoV-2 in wastewater in the Netherlands, USA, France, and Australia (Ahmed et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Nemudryi et al., 2020; F. Wu et al., 2020b; Wurtzer et al., 2020). One of these recent studies carried out in the USA adopted a wastewater surveillance approach to reveal phylogeny of circulating SARS-CoV-2 strains, infer viral ancestry, and observe the efficacy of public health interventions to contain the outbreak (i.e., mandated social isolation) (Nemudryi et al., 2020). These recent studies and other potentially ongoing efforts in many parts of the world may help to inform epidemiological modeling of the prevalence of SARS-CoV-2 in communities, as well as serve as a warning signal to communities attempting to mitigate the spread of the infection. To gain public acceptance of wastewater surveillance, a framework highlighting ethical issues related to basic access to sanitation, privacy, and rights may be required. It should be widely understood that one of the advantages of WBE is that this approach provides epidemiological information on disease prevalence in a community by circumventing individual stigmatization, which often results from clinical diagnosis in the ongoing COVID-19 outbreak (Murakami et al., 2020).
5. Methods for SARS-CoV-2 detection in wastewater
Although viral loads in the feces of COVID-19 patients are variable (Table 1), SARS-CoV-2 RNA can be sometimes detected with comparable concentrations to many enteric viruses (~108 viruses per gram of feces) (Bosch, 1998; Prüss et al., 2002; Wyn-Jones and Sellwood, 2001). Nevertheless, it will likely be necessary to perform a virus concentration step(s) prior to subsequent detection of SARS-CoV-2, even in untreated wastewater, as conducted previously (Ahmed et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Nemudryi et al., 2020; F. Wu et al., 2020b; Wurtzer et al., 2020) (Table 2).
Numerous types of methods have been developed for concentrating viruses in wastewater; however, most of those studies aimed to establish concentration methods for nonenveloped enteric viruses such as norovirus, enterovirus, adenovirus, and hepatitis A virus, using culturable viruses and/or bacteriophages as model viruses (Haramoto et al., 2018). Electropositive or electronegative membranes have been widely used to concentrate enteric viruses in untreated and treated wastewater samples (Cashdollar and Wymer, 2013; Haramoto et al., 2018; Ikner et al., 2012). These methods were developed based on electrostatic interactions between filters and viruses, utilizing the fact that a majority of enteric viruses have a net negative electrostatic charge in environmental water near neutral pH. In this method, negatively charged virus particles directly adsorb onto electropositive filter or adsorb onto electronegative filter via salt-bridging with a multivalent cation (Ikner et al., 2012; Michen and Graule, 2010). Another commonly used membrane-based method for concentrating viruses in environmental water samples is ultrafiltration, which is based on size exclusion (Hill et al., 2005, Hill et al., 2007). Other methods including polyethylene glycol (PEG) precipitation (Lewis and Metcalf, 1988), ultracentrifugation (Fumian et al., 2010), and skimmed-milk flocculation (Calgua et al., 2013) have also been used for concentrating viruses from wastewater samples.
The effectiveness of these virus concentration methods has been well demonstrated by successful detection of various types of indigenous enteric viruses which were not used as a model virus during the method development (Fong and Lipp, 2005; Haramoto et al., 2018). However, limited knowledge is available on recovery efficiencies of enveloped viruses, including CoVs, with the existing virus concentration methods. Ye et al. (2016) reported greater adsorption of enveloped viruses (mouse hepatitis virus [MHV] and Pseudomonas phage Φ6) to the solid fraction of wastewater compared to nonenveloped viruses. Haramoto et al. (2009) reported that enveloped koi herpesvirus showed high enough adsorption efficiency to an electronegative filter (87% or higher). Taken together, these results suggest that virus concentration methods using filters may potentially be used to recover SARS-CoV-2 from water and wastewater and requires further investigation. Even within enteric viruses, recovery efficiencies of viruses can vary greatly depending on virus and water types (Haramoto et al., 2018). Therefore, little scientific evidence is available to inform judgments of the usefulness of these existing virus concentration methods for enveloped SARS-CoV-2, which has quite different characteristics in structural and physical properties from enteric viruses. For example, Wang et al. (Xin Wei Wang et al., 2005b) reported that recovery of SARS-CoV from wastewater was only 1% using an electropositive membrane filter method, a significant decrease in performance compared to that observed for many types of enteroviruses (Li et al., 1998).
Nevertheless, virus concentration will likely be necessary to increase the chance of detection of SARS-CoV-2 in wastewater and research is needed to evaluate the recovery efficiency. Meanwhile, efforts are needed to evaluate the applicability of these existing methods to concentrate SARS-CoV-2. For method evaluation and development, low-pathogenic CoV strains (such as MHV and classical human CoVs) and/or Pseudomonas phage Φ6 may be used as models of SARS-CoV-2 for biosafety reasons. Recent studies in Australia, France, the Netherlands, and USA reported that SARS-CoV-2 RNA was successfully detected in wastewater using different concentration methods, such as ultrafiltration, PEG precipitation, and electronegative membrane adsorption followed by direct RNA extraction (Ahmed et al., 2020; Medema et al., 2020; Nemudryi et al., 2020; F. Wu et al., 2020b; Wurtzer et al., 2020).
Concentration volumes of water are one of the important factors that can affect the results of detection of viruses; normally, concentrating <100 mL of untreated wastewater samples is sufficient to detect enteric viruses (Haramoto et al., 2018). The initial studies reporting molecular detection of SARS-CoV-2 in wastewater concentrated up to 200 mL of raw wastewater samples (Ahmed et al., 2020; Medema et al., 2020; Nemudryi et al., 2020; F. Wu et al., 2020b; Wurtzer et al., 2020). However, a larger volume of wastewater sample may need to be processed for the detection of SARS-CoV-2 in regions where COVID-19 is less prevalent in the community.
Currently, detection of SARS-CoV-2 primarily relies on RT-qPCR or (nested) RT-PCR (CDC, 2020a; China CDC, 2020; Corman et al., 2020; Department of Medical Sciences, Ministry of Public Health, T, 2020; Institut Pasteur, 2020; Poon et al., 2020; Shirato et al., 2020). Currently available RT-qPCR and nested RT-PCR assays for SARS-CoV-2 are summarized in Table 3 . Corman et al. (2020) developed three TaqMan-based qPCR assays targeting RNA-dependent RNA polymerase (RdRp), envelope (E), and nucleocapsid (N) protein genes, with an absolute limit of detection (ALOD) of 3.8, 5.2, and 8.3 RNA copies per reaction, respectively (Corman et al., 2020). The RdRp gene-RT-qPCR assay uses two fluorescent probes to discriminate SARS-CoV-2 from SARS-CoV and bat-SARS-related CoVs, while the E gene-RT-qPCR assay can react with both SARS-CoV-2 and SARS-CoV. Because of its slightly higher ALOD than the other two assays, the performance of the N gene-RT-qPCR assay was not assessed in detail in that study. In contrast, Shirato et al. (2020) reported that among the three assays, only the N gene-RT-qPCR assay, which was specific only for SARS-CoV-2, worked well under their RT-qPCR platform (Shirato et al., 2020). The N protein gene is the most widely used gene target for developing RT-qPCR assays (CDC, 2020a; Chu et al., 2020; Corman et al., 2020; Shirato et al., 2020). An N gene-RT-qPCR assay developed by Shirato et al. (2020) was reported to be able to detect as low as ~5 RNA copies per reaction (Shirato et al., 2020), which is comparable to the assay developed by Corman et al. (2020).
Table 3.
Oligonucleotide sequences for selected SARS-CoV-2 RT-qPCR and nested RT-PCR assays.
| Type of PCR | Target gene | Function | Name | Sequence (5′–3′)a | Product length (bp) | Reference |
|---|---|---|---|---|---|---|
| RT-qPCR | RdRp | Forward primer | RdRp_SARSr-F | GTGARATGGTCATGTGTGGCGG | 100 | (Corman et al., 2020) |
| Reverse primer | RdRp_SARSr-R | CARATGTTAAASACACTATTAGCATA | ||||
| TaqMan probe (specific for SARS-CoV-2) | RdRp_SARSr-P2 | FAM-CAGGTGGAACCTCATCAGGAGATGC-BBQ | ||||
| TaqMan probe (reactive with SARS-CoV-2, SARS-CoV, and bat-SARS-related CoVs) | RdRP_SARSr-P1 | FAM-CCAGGTGGWACRTCATCMGGTGATGC-BBQ | ||||
| RdRp | Forward primer | nCoV_IP2-12669Fw | ATGAGCTTAGTCCTGTTG | 108 | (Institut Pasteur, 2020) | |
| Reverse primer | nCoV_IP2-12759Rv | CTCCCTTTGTTGTGTTGT | ||||
| TaqMan probe | nCoV_IP2-12696bProbe(+) | HEX-AGATGTCTTGTGCTGCCGGTA-BHQ1 | ||||
| RdRp | Forward primer | nCoV_IP4-14059Fw | GGTAACTGGTATGATTTCG | 107 | (Institut Pasteur, 2020) | |
| Reverse primer | nCoV_IP4-14146Rv | CTGGTCAAGGTTAATATAGG | ||||
| TaqMan probe | nCoV_IP4-14084Probe(+) | FAM-TCATACAAACCACGCCAGG-BHQ1 | ||||
| ORF1ab | Forward primer | Not provided | CCCTGTGGGTTTTACACTTAA | 119 | (China CDC, 2020) | |
| Reverse primer | Not provided | ACGATTGTGCATCAGCTGA | ||||
| TaqMan probe | Not provided | FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 | ||||
| ORF1b-nonstructural protein 14 | Forward primer | HKU-ORF1b-nsp14F | TGGGGYTTTACRGGTAACCT | 132 | (Poon et al., 2020) | |
| Reverse primer | HKU- ORF1b-nsp14R | AACRCGCTTAACAAAGCACTC | ||||
| TaqMan probe | HKU-ORF1b-nsp141P | FAM-TAGTTGTGATGCWATCATGACTAG-TAMRA | ||||
| E protein | Forward primer | E_Sarbeco_F | ACAGGTACGTTAATAGTTAATAGCGT | 113 | (Corman et al., 2020) | |
| Reverse primer | E_Sarbeco_R | ATATTGCAGCAGTACGCACACA | ||||
| TaqMan probe | E_Sarbeco_P1 | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ | ||||
| N protein | Forward primer | N_Sarbeco_F | CACATTGGCACCCGCAATC | 128 | (Corman et al., 2020) | |
| Reverse primer | N_Sarbeco_R | GAGGAACGAGAAGAGGCTTG | ||||
| TaqMan probe | N_Sarbeco_P | FAM-ACTTCCTCAAGGAACAACATTGCCA-BBQ | ||||
| N protein | Forward primer | 2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | 72 | (CDC, 2020a) | |
| Reverse primer | 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | ||||
| TaqMan probe | 2019-nCoV_N1-P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | ||||
| N protein | Forward primer | 2019-nCoV_N2-F | TTACAAACATTGGCCGCAAA | 67 | (CDC, 2020a) | |
| Reverse primer | 2019-nCoV_N2-R | GCGCGACATTCCGAAGAA | ||||
| TaqMan probe | 2019-nCoV_N2-P | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | ||||
| N protein | Forward primer | 2019-nCoV_N3-F | GGGAGCCTTGAATACACCAAAA | 72 | (CDC, 2020a) | |
| Reverse primer | 2019-nCoV_N3-R | TGTAGCACGATTGCAGCATTG | ||||
| TaqMan probe | 2019-nCoV_N3-P | FAM-AYCACATTGGCACCCGCAATCCTG-BHQ1 | ||||
| N protein | Forward primer | (not provided) | GGGGAACTTCTCCTGCTAGAAT | 99 | (China CDC, 2020) | |
| Reverse primer | (not provided) | CAGACATTTTGCTCTCAAGCTG | ||||
| TaqMan probe | (not provided) | FAM-TTGCTGCTGCTTGACAGATT-TAMRA | ||||
| N protein | Forward primer | HKU-NF | TAATCAGACAAGGAACTGATTA | 110 | (Poon et al., 2020) | |
| Reverse primer | HKU-NR | CGAAGGTGTGACTTCCATG | ||||
| TaqMan probe | HKU-NP | FAM-GCAAATTGTGCAATTTGCGG-TAMRA | ||||
| N protein | Forward primer | WH-NIC N-F | CGTTTGGTGGACCCTCAGAT | 57 | (Department of Medical Sciences, Ministry of Public Health, T, 2020) | |
| Reverse primer | WH-NIC N-R | CCCCACTGCGTTCTCCATT | ||||
| TaqMan probe | WH-NIC N-P | FAM-CAACTGGCAGTAACCABQH1 | ||||
| N protein | Forward primer | NIID_2019-nCOV_N_F2 | AAATTTTGGGGACCAGGAAC | 158 | (Shirato et al., 2020) | |
| Reverse primer | NIID_2019-nCOV_N_R2 | TGGCAGCTGTGTAGGTCAAC | ||||
| Reverse primer (revised version) | NIID_2019-nCOV_N_R2ver3 | TGGCACCTGTGTAGGTCAAC | ||||
| TaqMan probe | NIID_2019-nCOV_N_P2 | FAM-ATGTCGCGCATTGGCATGGA-BHQ1 | ||||
| S protein | Forward primer | RBD-qF1 | CAATGGTTTAACAGGCACAGG | 121 | (Zhou et al., 2020) | |
| Reverse primer | RBD-qR1 | CTCAAGTGTCTGTGGATCACG | ||||
| Nested RT-PCR | ORF1a | 1st PCR forward primer | NIID_WH-1_F501 | TTCGGATGCTCGAACTGCACC | 413 | (Shirato et al., 2020) |
| 1st PCR reverse primer | NIID_WH-1_R913 | CTTTACCAGCACGTGCTAGAAGG | ||||
| 2nd PCR forward primer | NIID_WH-1_F509 | CTCGAACTGCACCTCATGG | 346 | |||
| 2nd PCR reverse primer | NIID_WH-1_R854 | CAGAAGTTGTTATCGACATAGC | ||||
| S protein | 1st PCR forward primer | WuhanCoV-spk1-f | TTGGCAAAATTCAAGACTCACTTT | 547 | (Shirato et al., 2020) | |
| 1st PCR reverse primer | WuhanCoV-spk2-r | TGTGGTTCATAAAAATTCCTTTGTG | ||||
| 2nd PCR forward primer | NIID_WH-1_F24381 | TCAAGACTCACTTTCTTCCAC | 493 | |||
| 2nd PCR reverse primer | NIID_WH-1_R24873 | ATTTGAAACAAAGACACCTTCAC |
Single-letter code: M stands for A or C; R stands for A or G; S stands for C or G; W stands for A or T; and Y stands for C or T. Abbreviations: BBQ, blackberry quencher; BHQ1, black hole quencher 1; FAM, 6-carboxyfluorescein; HEX, hexachloro-6-carboxyfluorescein:TAMRA, 6-carboxytetramethylrhodamine.
Nalla et al. (2020) evaluated the performance of seven RT-qPCR assays targeting RdRp, E, and N genes (CDC, 2020a; Corman et al., 2020), where clinical respiratory and swab samples including SARS-CoV-2 positive samples were tested (Nalla et al., 2020). Based on the results of experiments using dilutions of a SARS-CoV-2-positive sample, the authors found that the N gene- (N2) and E gene-RT-qPCR assays developed by CDC (2020a) and Corman et al. (2020), respectively, showed the highest sensitivity of ~6.3 genomic equivalents per reaction. However, since the limited number of SARS-CoV-2-positive samples collected from a certain region were used in this study, further studies using samples from various locations worldwide are needed to establish a ‘gold standard’ assay.
Unlike clinical samples, a lower ALOD value is required when SARS-CoV-2 is tested in wastewater samples with low virus concentrations due to dilution and a low prevalence of COVID-19. Unfortunately, data regarding ALOD are not available for many of the existing RT-qPCR assays, most likely because these methods were designed for application to rapidly screening clinical samples. It is likely that RT-qPCR assays showing an ALOD of <10 copies per reaction could be useful for screening of wastewater samples for SARS-CoV-2 (Corman et al., 2020; Shirato et al., 2020). A SYBR Green-based qPCR targeting spike (S) protein gene has been also developed, although no ALOD data are provided (Zhou et al., 2020). Digital RT-PCR may enable more sensitive and accurate detection/quantification of SARS-CoV-2 RNA in wastewater samples than RT-qPCR as suggested recently for clinical samples (Dong et al., 2020; Suo et al., 2020).
When detecting SARS-CoV-2 RNA in wastewater by RT-qPCR, confirmation of positive RT-qPCR signals by sequencing analysis is highly recommended until the assay specificities have been validated against environmental samples. This is because the currently available RT-qPCR assays were developed for clinical diagnosis, which may be quite different from environmental applications. Some of recent studies on SARS-CoV-2 detection in wastewater reported sequence confirmation of RT-qPCR positive samples by directly sequencing qPCR products or re-amplification with regular PCR followed by sequencing (Ahmed et al., 2020; Nemudryi et al., 2020; F. Wu et al., 2020b).
Two nested RT-PCR assays targeting open reading frame 1a (ORF1a) and S protein genes are also available (Shirato et al., 2020), which could be used to elucidate the genetic diversity of SARS-CoV-2 circulating in human populations. As a thermal cycler is essential for PCR, novel assays which do not require any thermal cycler, such as loop-mediated isothermal amplification (LAMP) method, are expected to be developed, which will enable detection of SARS-CoV-2 RNA more easily and rapidly, especially in situations where sufficient laboratory equipment is not available. Additional efforts may be made to assess viral infectivity in wastewater using an engineered cell line with high susceptibility to SARS-CoV-2 (Matsuyama et al., 2020) and/or to detect infectious viral particles selectively by utilizing viability qPCR, such as ethidium bromide monoazide (EMA) or propidium monoazide (PMA) treatment followed by RT-qPCR, or integrated cell culture (ICC)-RT-PCR/qPCR (Farkas et al., 2020).
A critical issue in the application of molecular-based methods, including RT-qPCR, to wastewater samples is PCR inhibition during the detection process. It has been recommended that a process control(s) should be included in the analysis to monitor the levels of loss of targets and/or inhibition from the sample concentration to the detection steps (Haramoto et al., 2018). Three types of process controls are proposed, depending on the points of their inoculation: (i) whole process controls, to be inoculated into a water sample before virus concentration; (ii) molecular process controls, to be inoculated into a viral concentrate before nucleic acid extraction; and (iii) RT-qPCR controls, to be inoculated before RT-qPCR. At least one of these process controls is recommended to be included to avoid false-negative results and for concentration methods with low virus recovery efficiencies. Based on the results, samples may need to be reanalyzed (Haramoto et al., 2018). For a reliable process control, it is appropriate to select a virus which is genetically closely related to and/or has similar physical characteristics as the target virus and is expected not to be present in the tested water. Low-pathogenic animal CoVs such as MHV represent ideal process controls for SARS-CoV-2. Nonetheless, the selection of already established process controls for enteric viruses, such as murine norovirus and mengovirus, which are both single-stranded RNA viruses, may be acceptable at this stage.
6. Survival and inactivation of CoVs and enveloped surrogate viruses in water and wastewater matrices
The magnitude of human health risks varies depending on the inactivation of pathogens, including SARS-CoV-2 in water environments. Understanding the inactivation of SARS-CoV-2 and its RNA will ultimately improve control measures and wastewater treatment requirements, but little has been documented on the persistence of CoVs in water and wastewater matrices. Currently available data on survival of coronaviruses and an enveloped phage in water and wastewater are summarized in Table 4 . Wang et al. (2005) investigated the persistence of SARS-CoV, Escherichia coli and f2 phage in hospital wastewater, domestic sewage, tap water, phosphate buffered saline, feces, urine, water, and wastewater with high various concentrations (5, 10, 20 and 40 mg/L) of chlorine and chlorine dioxide. They also investigated the effect of contact time on inactivation of SARS-CoV in wastewater with low (10 mg/L chlorine and chlorine dioxide) and high (20 mg/L of chlorine and 40 mg/L of chlorine dioxide) concentrations. Results indicated that coronavirus persisted longer (inoculated titer of 105 TCID50; detectable with RT-PCR for 14 days) at 4 °C compared to 20 °C (3 days) in hospital wastewater, domestic sewage, and dechlorinated tap water. At 20 °C, SARS-CoV persisted in three fecal samples for 3 days and two urine samples for 17 days (inoculated titer of 105 TCID50). SARS-CoV was more vulnerable to disinfectants compared to E. coli and f2 phage. Free chlorine was more effective in inactivating SARS-CoV than chlorine dioxide. Free residue chlorine of >0.5 mg/L or chlorine dioxide of 2.19 mg/L in wastewater were sufficient for the complete removal of SARS-CoV (Xin Wei Wang et al., 2005b).
Table 4.
Survival of human and animal coronaviruses and an enveloped phage in water and wastewater.
| Virusesa | Water type | Temperature (°C) | Days persisted | Reduction time |
Reference | ||
|---|---|---|---|---|---|---|---|
| T90 | T99 | T99.999 | |||||
| Human coronavirus | Filtered tap water | 23 | - | - | 6.76 day | - | (Gundy et al., 2009) |
| Unfiltered tap water | 23 | - | - | 8.09 day | - | ||
| Filtered tap water | 4 | - | - | 392 day | - | ||
| Filtered primalry effluent | 23 | - | - | 1.57 day | - | ||
| Unfiltered primalry effluent | 23 | - | - | 2.36 day | - | ||
| Unfiltered secondary effluent | 23 | - | - | 1.85 day | - | ||
| Feline coronavirus | Filtered tap water | 23 | - | - | 6.76 day | - | (Gundy et al., 2009) |
| Unfiltered tap water | 23 | - | - | 8.32 day | - | ||
| Filtered tap water | 4 | - | - | 87.0b day | - | ||
| Filtered primalry effluent | 23 | - | - | 1.60 day | - | ||
| Unfiltered Primalry effluent | 23 | - | - | 1.71 day | - | ||
| Unfiltered secondary effluent | 23 | - | - | 1.62 day | - | ||
| TGEV | Reagent grade water | 4 | - | - | 220 day | - | (Casanova et al., 2009) |
| 25 | - | - | 22.0 day | - | |||
| Pasteurized settled sewage | 4 | - | - | 7.00 day | - | ||
| 25 | - | - | 9.00 day | - | |||
| MHV | Reagent grade water | 4 | - | - | >365b day | - | (Casanova et al., 2009) |
| 25 | - | - | 17.0 day | - | |||
| Pasteurized settled sewage | 4 | - | - | 70.0 day | - | ||
| 25 | - | - | 49.0 day | - | |||
| MHV | Unpasteurized wastewater | 10 | - | 36 h | - | - | (Ye et al., 2016) |
| 25 | - | 13 h | - | - | |||
| Pasteurized wastewater | 10 | - | 149 h | - | - | ||
| 25 | - | 19 h | - | - | |||
| Pseudomonas phage Φ6 | Unpasteurized wastewater | 10 | - | 28 h | - | - | |
| 25 | - | 7 h | - | - | |||
| Pasteurized wastewater | 10 | - | 146 h | - | - | ||
| 25 | - | 53 h | - | - | |||
| Pseudomonas phage Φ6 | Primary influent | 22 | - | - | - | 6 day | (Casanova and Weaver, 2015) |
| 30 | - | 1 day | - | - | |||
| Pseudomonas phage Φ6 | Autoclaved river water | 23 | - | 7.1 day | - | - | (Aquino de Carvalho et al., 2017) |
| Nonautoclaved river water | 23 | - | 3.1 day | - | - | ||
| Dechlorinated tap water | 22 | - | 3.1 day | - | - | ||
| Autoclaved wsatewater influent | 22 | - | 2.5 day | - | - | ||
| Deionised water | 4 | - | 66.1 day | - | - | ||
| 25 | - | 1.6 day | - | - | |||
| 37 | - | 0.34 day | - | - | |||
| 45 | - | 0.017 day | - | - | |||
| SARS-CoVc | Hospital wastewater | 20 | 3 | - | - | - | (Xin Wei Wang et al., 2005b) |
| Domestic sewage | 20 | 3 | - | - | - | ||
| Dechlorinated tap water | 20 | 3 | - | - | - | ||
| Phosphate buffer saline | 20 | 14 | - | - | - | ||
| Hospital wastewater | 4 | 14 | - | - | - | ||
| Domestic sewage | 4 | 14 | - | - | - | ||
| Dechlorinated tap water | 4 | 14 | - | - | - | ||
| Phosphate buffer saline | 4 | 14 | - | - | - | ||
| Feces | 20 | 3 | - | - | - | ||
| Urine | 20 | 17 | - | - | - | ||
TGEV, transmissible gastroenteritis virus; MHV, mouse heptitis virus; SARS-CoV, severe acute respiratory syndrome coronavirus.
Projected value.
Determined by RT-PCR.
Gundy et al. (2009) determined the survival of human CoV 229E and enteric feline CoV (ATCC-990) in water and wastewater using plaque assay or median culture infectious dose (TCID50) technique. The times for 99 and 99.9% inactivation (T 99 and T 99.9, respectively) were determined for filtered and unfiltered tap water at 23 °C, filtered tap water at 4 °C, filtered and unfiltered primary effluent at 23 °C and secondary effluent (activated sludge) at 23 °C. The survival of both human and feline CoVs showed similar patterns and was highly dependent on water temperature, level of organic matter, and biological activity. The T 99 for tap water for both human and feline CoV were faster at 23 °C (7–9 days) than 4 °C (>87 days). The inactivation rates of both CoVs were faster in filtered tap water compared to unfiltered tap water at 23 °C, suggesting increased protection and survival in the presence of organic matter and suspended solids. CoVs were inactivated rapidly in wastewater, with T 99 values of <3 days (Gundy et al., 2009).
Casanova et al. (2009) determined the persistence of two surrogate CoVs, transmissible gastroenteritis virus (TGEV), and MHV in reagent grade water, lake water, and pasteurized settled sewage in North Carolina, USA using quantal assays for cytopathic effect (CPE). In general, both the surrogate viruses persisted for significantly shorter durations at 25 °C compared to 4 °C for all water types. For reagent grade water, TGEV and MHV persisted for shorter durations (T 99 = 22 and 16 days, respectively) at 25 °C than at 4 °C (>220 days for both viruses). For lake water, TGEV and MHV T 99 values were 13 and 10 days, respectively, over a 14-day experiment. However, at 4 °C, one log10 reduction was observed at day 14 for TGEV, while no reduction was observed for MHV up to day 14 at 4 °C. Both viruses persisted shorter in pasteurized settled sewage samples, and T 99 reduction times were nine days for TGEV, and seven days for MHV. At 4 °C, T 99 values of TGEV and MHV were 49 and 70 days, respectively, suggesting surrogate CoVs can remain infectious for long periods in water and pasteurized settled sewage at a lower temperature (Casanova et al., 2009).
A technical brief from WHO suggested that there is no evidence about the survival of SARS-CoV-2 in wastewater or drinking water. It is likely that enveloped CoVs are less stable in the environment and are more susceptible to chlorine, pH, and temperature than most of nonenveloped enteric viruses (WHO, 2020d). Therefore, conventional wastewater treatment processes should inactivate SARS-CoV-2, and multiple barriers used in drinking water treatment plants should suffice to remove SARS-CoV-2 to levels of non-detect and low risks (<10−4 annual risk). However, limited data published to date suggest that asurrogate CoV remain infectious in water environments for days to weeks, depending on temperature and other physico-chemical factors (Pratelli, 2008). Therefore, it is crucial to determine the persistence of SARS-CoV-2 in sewage and environmental waters using molecular and cell culture assays. If biosafety is a concern/limitation, then enveloped surrogate viruses such as low-pathogenic human CoV (e.g., 229E or OC43), feline CoV, MHV, or Pseudomonas phage Φ6 can be used.
Ye et al. (2016) compared the persistence and partitioning behavior of two model enveloped viruses, MHV and Pseudomonas phage Φ6 in raw and pasteurized wastewater samples using cell culture and plaque assays. MHV and Φ6 were seeded into unpasteurized and pasteurized wastewater and incubated at 10 and 25 °C to mimic typical winter and summer temperatures of wastewater. The T 90 values of MHV and Φ6 in unpasteurized wastewater at 25 °C were 13 and 7 h, respectively. In contrast, the T 90 values of MHV and Φ6 were slower in unpasteurized wastewater at 10 °C with T 90 values of 36 and 28 h, respectively. Both viruses persisted relatively longer in pasteurized wastewater than unpasteurized wastewater. Based on the results, the authors concluded that although MHV and Φ6 were inactivated rapidly in wastewater, their persistence could still be of concern for wastewater treatment facilities, stormwater overflows, and wastewater intrusion in drinking water (Ye et al., 2016). Their results on comparative viral persistence in pasteurized and unpasteurized wastewater implied that enhancement of competition and predation contributed by indigenous microbial communities in wastewater could be a potential medium-term strategy to fight against the ongoing and future viral disease outbreaks.
Similarly, Aquino de Carvalho et al. (2017) evaluated the persistence of Φ6 in a variety of matrices, including water and wastewater. The T 90 of Φ6 under these conditions was highly variable, from 24 min to 117 days. Significant factors included temperature, biological activity, and the composition of the test media. Beyond direct study findings, the authors reported that the aqueous stability of enveloped viruses in water matrices was highly variable, and a single surrogate was insufficient to capture the behavior of all enveloped viruses (Aquino de Carvalho et al., 2017).
Given the limited available data on SARS-CoV-2 in water matrices, it may also be informative to consider recent reports of viral persistence on surfaces. van Doremalen et al. (2020) evaluated the surface stability of SARS-CoV-2 compared to SARS-CoV. The half-life of SARS-CoV-2 varied from 0.8 h on copper to 6.8 h on plastic. The authors also identified comparable environmental persistence between the two viruses (van Doremalen et al., 2020). Chin et al. (2020) also reported on the surface persistence and disinfection of SARS-CoV-2. The authors identified a high temperature dependence on the inactivation kinetics, and rapid removal of the virus using bleach, ethanol, benzylalkonium chloride, povidone‑iodine, and chloroxylenol (Chin et al., 2020). Overall, these limited results suggest that previous data on CoVs are likely to be useful for informing the environmental persistence of SARS-CoV-2, and that SARS-CoV-2 is likely rapidly inactivated under increased temperature and by major disinfectants. In fact, a number of the existing disinfectant products have been approved by the United States Environmental Protection Agency (USEPA) for use against SARS-CoV-2 (USEPA, 2020).
7. Respiratory viruses in wastewater and the occupational risk
In the field of environmental virology, the focus on waterborne transmission has been primarily on enteric viruses. However, respiratory viruses including adenoviruses, coxsackieviruses, and indeed CoVs have been known to occur in wastewater (Sinclair et al., 2008; Xin Wei Wang et al., 2005a) and wastewater-polluted waters (Wigginton et al., 2015). Going back to early descriptions, it has been known that these viruses cause diarrheal as well as respiratory diseases (Britton, 1980) but limited study of viral respiratory diseases has been performed in the wastewater context. There are fewer data available on the presence and concentrations of respiratory viruses in wastewater. Fong et al. (2010) found in addition to enteric adenoviruses 40 and 41 (type F) in sewage, combined sewer overflows and rivers receiving these discharges contained respiratory adenoviruses 2 and 3 (types C and B) as well as adenovirus 12 that causes meningoencephalitis with initial replication in the gastrointestinal or respiratory tract (Fong et al., 2010).
There is no doubt that swimming in sewage-contaminated waters is associated with respiratory disease; however, the etiological agent is not frequently identified (Wade et al., 2010). Studies on the Great Lakes suggest this could be due to adenoviruses (Wong et al., 2009). Respiratory disease as an occupational risk for sewage workers has also been studied with mixed results. Four key studies were reviewed as shown in Table 5 . In Switzerland, no health impacts were found in garbage collectors and wastewater treatment plant (WWTP) workers (Tschopp et al., 2011). However, in three other investigations, gastrointestinal effects were observed, and two of the three studies noted respiratory health impacts (Khuder et al., 1998; Lee et al., 2009; Smit et al., 2005).
Table 5.
Epidemiological studies of health effects for wastewater treatment plant workers.
| Study descriptiona | No. of individuals evaluated | Results | Reference |
|---|---|---|---|
| WWTP workers in 11 cities in Northern Ohio were evaluated via questionnaire for a 12-month study; controls were college maintenance and refiner workers. | 150 WWTP workers vs. 54 controls | The WWTP workers had significantly higher gastroenteritis, abdominal pain, and headaches. No significant differences were reported for respiratory and other symptoms. | (Khuder et al., 1998) |
| WWTP workers in 67 plants in the Netherlands were evaluated via questionnaire for a 12-month study; no controls; personal endotoxin exposure was assessed (8 h measurements: n = 460). | 468 WWTP workers | Dose-response relationships were found with endotoxin levels for: “lower respiratory and skin symptoms”, “flu-like and systemic symptoms”, and “upper respiratory symptoms”. | (Smit et al., 2005) |
| WWTP workers in Iowa were evaluated via questionnaire for a 3-year study; controls were workers at water treatment plant (WTP) workers; endotoxins sampled as an exposure indicator. | 93 WWTP workers vs. 54 WTP worker controls | Odds ratios were statistically higher for respiratory, ocular and skin irritation, neurology, and gastrointestinal symptoms in WWTP workers. Tasks related to sludge handling were identified as high-risk. | (Lee et al., 2009) |
| A 5-year study conducted in Switzerland; controls were gardeners, waterway maintenance, public transport, and forestry workers. | 247 WWTP workers; 52 solid waste workers vs. 304 controls | No effects for occupational exposure to bioaerosols were reported. | (Tschopp et al., 2011) |
WWTP, wastewater treatment plant; WTP, water treatment plant.
8. Quantitative microbial risk assessment (QMRA) for respiratory viruses and SARS-CoV-2
Environmental engineering and science, and in particular QMRA have major roles to play in reducing the impact of the current COVID-19 outbreak (Haas, 2020; Wigginton and Boehm, 2020). The process of QMRA involves relating an environmental concentration of an infectious agent to an exposure dose and subsequently a probability of developing an infection or illness (Haas et al., 2014). Gaps needed to fill include characterizing persistence, fate, and transport (including airborne transport and deposition, for example), and exposure to be able to define the risk. In addition to basic questions like “what is the risk” (potentially in relation to some baseline) for a particular context, QMRA can be used to address questions for SARS-CoV-2 such as: (i) What ventilation/air exchange rate is recommended for different settings (e.g. workplace, healthcare facility) to prevent transmission consistent with a risk target?; (ii) Is a 6-ft/2 m “social distance” protective enough?; (iii) What should surface disinfection targets be for different settings and what are the best technologies or disinfectants for achieving these targets (e.g. UV light)?; (iv) What wastewater treatment disinfection targets might be needed?
SARS-CoV-2 transmission is known to occur via hand-to-face (nasal-pharyngeal: eyes, nose, and mouth) contact with contaminated fomites, and inhalation of aqueous aerosols including coughs. The fecal-oral route or aspiration have been postulated as potential exposure routes, although no cases of transmission via the fecal-oral route have been reported to date (CDC, 2020b; Y. Wu et al., 2020a; Yeo et al., 2020). There are some preliminary data to suggest that the virus is shed longer from the digestive tract than the respiratory tract (Hu et al., 2020).
In general for respiratory viruses, a review by Van Leuken et al. (2016) highlighted the ability of bioaerosols, particularly from farming and wastewater exposures, to carry infectious agents, including viruses, and to present disease risks at considerable distances from the source (Van Leuken et al., 2016). Additionally, numerous exposures are possible at close-range, especially for occupational populations (e.g. wastewater workers and nurses). Many QMRAs have also focused on wastewater biosolids applications, addressing adenovirus, astrovirus, coxsackievirus, echovirus, enterovirus, hepatitis A virus, hepatitis E virus, norovirus, and rotavirus, both during the actual period of application as well as at various exposures from farm to fork (Hamilton et al., 2020). SARS-CoV-2 can be spread and persist in aerosols, hence measurement in wastewater aerosols would be informative for risk assessment (Bushmaker et al., 2020; van Doremalen et al., 2020; Guo et al., 2020).
A key component of bioaerosol QMRA is modeling the dispersion of aerosols and/or transfer of microorganisms from water to air. A key study of stormwater reuse for inhalation-ingestion of adenovirus and norovirus provides an example of considering the aerosol size profile of a particular activity in order to calculate the number of viruses aerosolized and the subsequent deposited dose (Lim et al., 2015). Methods utilized for other microbial risk studies are the use of a water-to-air transfer coefficient or computational fluid dynamics approaches (Hamilton and Haas, 2016). The studies of viral aerosols emerging from wastewater facilities have often focused on coliphage as an indicator for human pathogenic viruses, but most studies have not simultaneously sampled the wastewater and the aerosols produced, or identified how the viruses are distributed in aerosols with respect to the aerosol size profile (Table 6 ). Fannin et al. (1985) were not able to detect animal viruses in the aerosols (Fannin et al., 1985). Adenoviruses are known to be quite stable in air and high concentrations were found by qPCR, thus viable viruses were not addressed (Masclaux et al., 2014). The phage data suggest a 10,000-fold level of dilution and inactivation (Brenner et al., 1988). There has been no published study to date testing SARS-CoV-2 in aerosols from wastewater facilities, but a recent laboratory-scale study on persistence of coronaviruses in aerosols revealed that SARS-CoV-2 could maintain its infectivity in aerosols for up to 16 h (Fears et al., 2020), suggesting potential human exposure if wastewater aerosols contain viable SARS-CoV-2. Further investigations are needed to elucidate the presence of SARS-CoV-2 and its viability in wastewater bioaerosols and associated public health risks.
Table 6.
Presence of viruses in aerosols at wastewater treatment plants.
| Virus | Detection method | Plant descriptionc | Country | Wastewater levelsd | Aerosol levels | Remark | Reference |
|---|---|---|---|---|---|---|---|
| Coliphages | Plaque assaya | Aeration basin - lagoons | USA | 3.25 × 103 to 5.53 × 105 PFU/L | <1 to 9 PFU/m3 | (Brenner et al., 1988) | |
| Coliphages | Plaque assay-MPNb | Two-stage aeration basins | USA | Not reported | 5.0 × 10−3 to 7.6 × 10−2 MPN PFU/m3 | (Fannin et al., 1985) | |
| Somatic coliphages | Plaque assay | 7 WWTPs | Finland | Not reported | Up to 380 PFU/m3 | (Heinonen-Tanski et al., 2009) | |
| F-specific coliphages | Plaque assay | 7 WWTPs | Finland | Not reported | Up to 70 PFU/m3 | (Heinonen-Tanski et al., 2009) | |
| Adenoviruses | qPCR | 79 WWTPs | Switzerland | Not reported | Up to 2.27 × 106 copies/m3 | 104/123 (84%) air samples positive | (Masclaux et al., 2014) |
| Adenoviruses, norovirus GI and GII, FRNA bacteriophages GIII, enteroviruses | qPCR | 1 WWTP activated sludge chamber, exhaust duct, and treated air | Japan | Up to 2.5 × 107 copies/L (norovirus GII) | Up to 3.2 × 103 copies/m3 | Air samples positive for adenovirus (4/16), norovirus GI (6/16), FRNA bacteriophages GIII (3/16), and enteroviruses (3/16) | (Matsubara and Katayama, 2019) |
Three different E. coli strains (ATCC 13706, 15,597, and 11,303) were used as hosts.
MPN, most probable number.
WWTP, wastewater treatment plant.
PFU, plaque-forming unit.
A recent QMRA study for enteric viruses via exposure to wastewater bioaerosols focused on adenoviruses as the hazards (Carducci et al., 2018). The analysis highlighted the following: (i) workers at highest risk were related to exposures at the influent and biological oxidation tanks for >3 min; (ii) adenovirus concentrations drove the risk; (iii) risks of 10−2, 10−3, 10−4, and 10−5 were related to levels of 565, 170, 54 and 6 copies/m3 in the bioaerosol, respectively; (iv) this relates to an estimated level of approximately 104 to 106 copies/L in the oxidation tank. Similarly, a recent study focusing on rotavirus and norovirus bioaerosol exposures to WWTP workers noted risks that exceeded common public health risk benchmarks of 10−4 infections or 10−6 disability adjusted life years (DALY) per person per year for airborne concentrations above the aeration tank of 27 and 3,099 viruses/m3-h, for rotavirus and norovirus, respectively (Pasalari et al., 2019). Taken together, these data help to provide a comparison of relative risks and put concentrations of viruses into perspective.
To date, few QMRAs for CoVs have been conducted including one for MERS-CoV in a hospital setting (Adhikari et al., 2019) and another for a residential bathroom exposure (Watanabe et al., 2010). Adhikari et al. (2019) made use of a SARS-CoV dose-response model to examine the impact of a coughing patient and resulting exposures to nurses, healthcare workers, and family visits. Mean daily risks were on the order of ~10−4 and were highest for healthcare workers and nurses compared to family visitors and patients in the same room. The concentration of viruses in saliva were a driving variable in the Monte Carlo analysis, and respiratory masks were a more effective intervention than increasing the air exchange rate. Watanabe et al. (2010) developed new SARS-CoV dose-response models and reconstructed a dose observed during a 2003 Hong Kong apartment complex outbreak due to exposures to sewage via sewer gas entering through bathroom floor drains and U-traps after a toilet flush under negative pressure by high-capacity bathroom fans. The authors made use of epidemiological attack rate data (ranging from 0.038 to 0.144) by floor of a building where 99 cases of SARS were reported (Li et al., 2005) to arrive at estimates of 16–49 plaque-forming units (PFU) on the first floor, 63–160 PFU for residents on a middle floor, and 42–117 PFU for residents on upper floors, with higher attack rates on higher floors likely due to air flow in the building. Tabulating attack rates for SARS-CoV-2 could provide similarly useful information for estimating doses and/or risks in retrospect; however such an exercise requires information on an environmental measurement coupled with the total number of persons exposed in addition to the number of individuals infected and/or ill. Given the parameters developed in the existing QMRA models, a summary of parameters for potential use in QMRA models and data gaps are summarized in Table 7 .
Table 7.
QMRA parameters of SARS-CoV-2 and relevant respiratory viruses (SARS-CoV, MERS-CoV, and influenza viruses).
| Parameter | SARS-CoV-2 | SARS-CoV | MERS-CoV | Influenza virus | Reference |
|---|---|---|---|---|---|
| Dose response (see Table 8) | Not available | Available | Available | Available | (Huang et al., 2018; Kitajima et al., 2011; Lunn et al., 2019; QMRA Wiki, 2020a, QMRA Wiki, 2020b; Watanabe et al., 2010) |
| Excretion in saliva (copies/mL) | 9.9 × 102–1.2 × 108 | 7.08 × 103–6.38 × 108 | 7–2.02 × 105 (Avg.: 4.17 × 104) | 101–107 | (Adhikari et al., 2019; Sueki et al., 2016; To et al., 2020a, To et al., 2020b; Wang et al., 2004) |
| Feces (copies/g-feces) | Up to 108 copies/swab | 5.1 × 101–107 | Up to 103 | 103.7–106 | (Chan et al., 2011; Cheng et al., 2004; Drosten et al., 2013; Hung et al., 2004; Isakbaeva et al., 2004; D. Wang et al., 2020; Wigginton and Boehm, 2020; Wölfel et al., 2020) |
| Urine (copies/mL) | ND | Up to 104.4 | Up to 2.69 × 103 | Not available | (Drosten et al., 2013; Hung et al., 2004; W. Wang et al., 2020c) |
| Nasal swabs (copies/mL) | 1.4 × 106–1.5 × 107 | 2.47 × 104–6.97 × 107 | Up to 103.7 | 102.7–109.3 (varies by strain) | (Drosten et al., 2013; Lee et al., 2009; Ngaosuwankul et al., 2010; D. Wang et al., 2020b; Wong et al., 2005; Zou et al., 2020) |
| Attack rate (%) | Up to 80 | <1–100 depending on scenario | 0.42–15.8 | 5–30 | (Burke et al., 2020; Park et al., 2018; Verity et al., 2020; WHO, 2011, WHO, 2019) |
| Case fatality rate (CFR) (%) | 5.3–8.4 | Up to 50; most estimates ~9–17% | 34.4–69.2 | <1 (seasonal flu)–60 (H5N1) | (Jung et al., 2020; Li et al., 2008; Park et al., 2018; C. Wang et al., 2020a; WHO, 2003, WHO, 2011; J. Y. Wong et al., 2013b) |
| Basic reproduction number (R0) | 1.4–6.5 | 2–5 | 0.45–8.1 | 1.7–2.8 (varies by strain) | (Boldog et al., 2020; Cheng and Shan, 2020; Coburn et al., 2009; Jung et al., 2020; Lai et al., 2020; Liu et al., 2020; Park et al., 2018; B. Tang et al., 2020; Wallinga and Teunis, 2004; WHO, 2020e) |
| Incubation period (days) | 2–14 | 2–10 | 4.5–7.8 | 1–4 | (CDC, 2004; Lessler et al., 2009; Linton et al., 2020; Park et al., 2018; WHO, 2018) |
9. Dose-response of SARS-CoV-2 and relevant respiratory viruses
To conduct a QMRA, a dose-response model is required providing the relationship between exposure and health outcome or endpoint. To date, no quantitative dose-response model is available for SARS-CoV-2. This is partially due to the absence of an appropriate animal model for pathogen dosing (Gralinski and Menachery, 2020). Contributing factors to the lack of appropriate animal model include the inability to cause disease without passaging the virus through a mouse host and milder disease in primates compared to humans (Gralinski and Menachery, 2020). The search for an appropriate animal model for various applications (treatments and vaccines) is underway and several models are being explored, placing strains on the supply of transgenic laboratory mice (Boodman, 2020; Callaway, 2020; Warren, 2020).
Existing animal models for SARS-CoV and MERS-CoV include non-human primates (macaques, cynomolgus monkeys, African green monkeys, rhesus macaques, and common marmosets), hamsters, ferrets, and transgenic mice (Gretebeck and Subbarao, 2015). A recent study by Rockx et al. (2020) indicated that SARS-CoV-2 results in a severity of infection that is intermediate between that of SARS-CoV and MERS-CoV based on a direct comparison of the three viruses in a combined intratracheal and intranasal dosing study of female adult cynomolgus macaques with a dose of 106 TCID50 for all viruses. Shedding varied depending on the age of the animal, with higher levels detected in nasal swabs of aged animals compared to younger animals (Rockx et al., 2020).
The reported viral doses in current animal model experiments underway for SARS-CoV-2 are 104 PFU − 106.5 TCID50 in ferrets (Blanco-Melo et al., 2020; Kim et al., 2020; Richard et al., 2020), 102–105 TCID50 in mice (Bao et al., 2020b; S. Xia et al., 2020a), 105 PFU in hamsters (Chan et al., 2020), and 105–106 TCID50 in macaques (Bao et al., 2020a; Deng et al., 2020; Munster et al., 2020). These studies generally indicated 100% infection or isolation of viral RNA from animals at the inoculated dose and therefore it is not possible to designate a median infectious dose (ID50) or median lethal dose (LD50). Xia et al. (2020a) reported 100% mortality in 12 newborn mice challenged intranasally with 102 TCID50 (J. Xia et al., 2020b). Deng et al. (2020) reported that macaques infected with 106 via an ocular (2/3) or intratracheal (1/3) route of exposure had a positive viral load in nose and throat swabs from 1 to 7 days post inoculation, supporting reports of ocular transmission reported for a healthcare worker infected with SARS-CoV-2 while working with a patient without eye protection (Lu et al., 2020; S. Xia et al., 2020a). Previously infected rhesus macaques challenged intratracheally with SARS-CoV-2 at 106 TCID50 did not display reinfection characteristics when challenged again with the same dose, indicating some immunity conferred from an initial infection (Bao et al., 2020a).
Sufficient data are not available for modeling or pooling dose groups from multiple studies at this time that meets the criteria of (i) >3 unique dose groups and (ii) at least three unique responses. A lack of a dose-response model for SARS-CoV-2 is a critical gap for conducting QMRA for this pathogen. Emerging areas for dose-response testing include dosing of organoids to represent aspects of specific pathogenesis processes such as liver damage (Zhao et al., 2020); however, these approaches have not been reconciled with existing quantitative dose-response modeling calculations as they do not fully encapsulate the ability to represent host immune processes (Haas, 2015).
Existing dose-response models are available for SARS-CoV (QMRA Wiki, 2020a; Watanabe et al., 2010), MERS-CoV (Lunn et al., 2019), and various influenza virus strains (Huang et al., 2018) including H5N1 (Kitajima et al., 2011; QMRA Wiki, 2020b), H1N1 (QMRA Wiki, 2020b; Watanabe et al., 2012), and H3N2 (QMRA Wiki, 2020b; Watanabe et al., 2012) subtypes, but not SARS-CoV-2 (Table 8 ). Available health endpoints (infection or death) varied for these models with infection endpoints available for influenza virus but not MERS-CoV or SARS-CoV, and death endpoints available for influenza virus and SARS-CoV. Modelled LD50 for SARS-CoV ranged from 233 to 324 PFU, compared to estimates ranging from <32–107 PFU for H5N1 highly pathogenic avian influenza virus (Kitajima et al., 2011; QMRA Wiki, 2020b). Modelled ID50 for influenza virus ranged from 6.66 × 105 to 1.25 × 106 TCID50 (QMRA Wiki, 2020b). The MERS-CoV median infectious/lethal dose calculated from Lunn et al. (2019) by solving the exponential model using -ln(0.5/k) (authors specified a pooled analysis with both endpoints) would be approximately 121 PFU.
Table 8.
Dose-response parameters available and sporadic dose-response data where there are limited models and gaps.
| Virus | Dose unitsa | Host type | Exposure route | No. doses | Modelb | Parametersc, d | Health endpoint/ response | N50 | Remarks | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | - | - | Intranasal (Bao et al., 2020a, Bao et al., 2020b, Blanco-Melo et al. 2020), ocular inoculation and intratracheal (Deng et al. 2020), intratracheal and intranasal (Rockx et al. 2020) | - | - | - | - | - | Existing dosing experiments designed to infect all animals ranged from 102 TCID50 (mice)–106 TCID50 (macaques) | (Bao et al., 2020a, Bao et al., 2020b; Blanco-Melo et al., 2020; Deng et al., 2020; Rockx et al., 2020) |
| SARS-CoV | PFU | Pooled transgenic mice, non-transgenic mice | Intranasal | 8 | E | k = 2.45×10-3 | Deathe | 280 | (QMRA Wiki, 2020a; Watanabe et al., 2010) | |
| SARS-CoV | PFU | Transgenic mice | Intranasal | 4 | E | k = 2.97×10-3 | Death | 233 | (QMRA Wiki, 2020a; Watanabe et al., 2010) | |
| SARS-CoV | PFU | Mice | Intranasal | 4 | E | k= 2.14×10-3 | Death | 324 | (QMRA Wiki, 2020a; Watanabe et al., 2010) | |
| SARS-CoV | PFU | Rhesus macaques | Intratracheal | 2 | - | - | Infection | - | Monkeys: 2/2 infected at 108 PFU | (Zhou et al., 2005) |
| MERS-CoV | TCID50 | Mice | Intranasal | 6 | E | k ≅ 5.71×10-3 | Shedding/ mortality | ≅ 121 | Pooled endpoint | (Lunn et al., 2019) |
| MERS-CoV | PFU | Mice | Intranasal | 3 | - | - | Infection/ death | All animals infected: LD50 ~1–2×104 | (Douglas et al., 2018) | |
| MERS-CoV | PFU | Mice | Intranasal | 2 | - | - | Infection/ death | - | Authors stated “sublethal” 5×103 and “lethal” 5×105 doses (no deaths in test animals observed; all sacrificed 4 days post inoculation | (Leist et al., 2019) |
| MERS-CoV | TCID50 | Rhesus macaques | Intratracheal | 1 | - | - | Death | - | 0/4 died at 6.5×107 | (Yao et al., 2014) |
| MERS-CoV | TCID50 | Rhesus macaques | Intratracheal | 1 | - | - | Infection | - | 4/4 infected at 6.5×107 | (Yao et al., 2014) |
| Human coronavirus 229E | TCID50 | Humans | Intranasal | 4 | E | k = 5.39 ×10-2 | Illness (cold) | 13 | (Watanabe et al., 2010) | |
| Animal coronaviruses (MHV-S, HEV-67N, IBVA-5968) | PFU or CD50 | Mice, rats, chicks | Intranasal | 3-6 | E | k = 8.78×10-5 - 9.16×10-2 | Death | ~8 - 5.95×105 | Various coronavirus models fit for comparison with SARS | (Watanabe et al., 2010) |
| Influenza virus (H5N1) | PFU, TCID50 | Mice | Intranasal | 6 | T | α= 4.640×10-1; J0=3.015×102; J1 = 1.000; J2 =1.793 | Infection | <101.5–>107 (depending on strain) | (Kitajima et al., 2011) | |
| Influenza virus (H5N1) | PFU, TCID50 | Mice | Intranasal | 7 | T | α= 2.730×10-1; J0=9.617×104; J1 = 2.7082; J2 =4.666 | Infection | <101.5–>107 (depending on strain) | (Kitajima et al., 2011) | |
| Influenza virus (H5N1) | PFU, TCID50 | Ferrets | Intranasal | 2 | T | k0= -1.707×10-1; k1=-1.502×10-1 | Infection | <101.5–>107 (depending on strain) | (Kitajima et al., 2011) | |
| Influenza virus (H5N1) | PFU, TCID50 | Ferrets | Intratracheal | 2 | T | k0 = -1.480×101; k1 = -7.092 | Infection | <101.5–>107 (depending on strain) | (Kitajima et al., 2011) | |
| Influenza virus (H1N1) | TCID50 | Human | Intranasal | 4 | B | ɑ = 9.04×101 | Infection | 1.25×106 | (QMRA Wiki, 2020b) | |
| Influenza virus (H1N1) | TCID50 | Human | Intranasal | 9 | B | ɑ = 5.81×101 | Infection | 9.45×105 | (QMRA Wiki, 2020b) | |
| Influenza virus (H3N2) | TCID50 | Human | Intranasal | 5 | B | ɑ = 4.29×101 | Infection | 6.66×105 | (QMRA Wiki, 2020b) | |
| Influenza virus (H5N1) | EID50 | Mice | Intranasal | 6 | E | k = 1.09×10-2, | Death | 6.38×101 | (QMRA Wiki, 2020b) | |
| Influenza virus (H1N1, H3N2) | TCID50 | Human | Intranasal | 4-5 | B | ɑ= 1.57×10-1-9.05×10-1; for fixed parameters (α=2.95×10-1; N50=4.42×105) attenuation tion parameter γ=1.07e×10-3 | Infection | Children 3.3×102-1.2×105; Adults 2.7×104-1.2×106 | Pooled data analysis from 11 datasets with respect to virus subtype (H1N1 or H3N2), attenuation method (cold-adapted or avian-human gene reassortment), and human age (adults or children | (Watanabe et al., 2012) |
| Influenza virus (H3N1, H1N1, influenza A, influenza B) | HI titer | Various | Various | Various | B-HI | λ= 0.002- 0.245 | Various | Depends on HI titer | Authors developed a relationship between HI titer and protection against influenza virus | (Huang et al., 2018) |
PFU, plaque forming unit; TCID50, median tissue culture infectious dose; EID50, median egg infectious dose, HI, hemagglutination inhibition.
E, exponential model; B, Beta-Poisson model; T, dose response time model; B-HI, modified Beta-Poisson model to include a parameter for hemagglutination inhibition titer.
Best fit dose response model parameters are given in table (where a model was not available, available information relating dose to an outcome in an animal or human model is provided); ID50, median infectious dose; LD50, median lethal dose.
The N50 represents the median dose associated with a particular health endpoint.
Watanabe et al. (2010) considered the animal death endpoint to be representative of a SARS-CoV human illness in the dose response model.
Previously, “sublethal”, “moderate”, and “lethal” concentrations of MERS-CoV of 5 × 103, 5 × 104, and 5 × 105 PFU, respectively, have been used for a mouse animal model (Leist et al., 2019), but concentrations of 6.5 × 107 TCID50 or 108 PFU were not lethal in macaques (Yao et al., 2014; Zhou et al., 2005). The lowest dose used in SARS-CoV-2 experiments designed to infect all animals was 102 TCID50, indicating the ID50 might be lower than that value, although this cannot be determined with certainty at this time. Chen et al., 2020a, Chen et al., 2020b indicated that in a placebo (unvaccinated) group of SARS-CoV-dosed macaques, 3/4 dosed animals at 105 TCID50 displayed respiratory distress or alveolar damage after virus inoculation and 3/4, 4/4, and 2/4 had quantifiable viral RNA in pharyngeal swab samples at day 2, 5, and 7, respectively (Y. Chen et al., 2020a). These modeling efforts and sparse animal model experimental data that could not be modelled generally indicated that SARS-CoV and SARS-CoV-2 are more infectious than influenza virus, with SARS-CoV-2 potentially more infectious than SARS-CoV. MERS-CoV had uncertain infectivity but overlapped with ranges for SARS-CoV and SARS-CoV-2. For a death endpoint, LD50 values were higher for influenza virus compared to SARS-CoV (indicating SARS-CoV is more deadly), and MERS-CoV estimates were again overlapping the influenza virus and SARS-CoV values. This is roughly comparable with the severity of infection ranking provided by Rockx et al. (2020) of SARS-CoV > SARS-CoV-2 > MERS-CoV.
10. Knowledge gaps and research needs
At present, significant knowledge gaps exist on the potential role of wastewater in the transmission of SARS-CoV-2. Survival of SARS-CoV-2 in environmental media, including wastewater and water, remains mostly unknown. Recent data indicate that the stability of SARS-CoV-2 is similar to that of SARS-CoV in aerosols and on surfaces (van Doremalen et al., 2020). Using a similar approach, the stability of SARS-CoV-2 in various water matrices should be investigated. The persistence of SARS-CoV-2 needs to be determined in wastewater and environmental water for tropical, sub-tropical and temperate climatic zones as the persistence may be highly variable in different temperatures, as demonstrated in a recent study (Hart and Halden, 2020). Moreover, the persistence of SARS-CoV-2 in wastewater and receiving waters and inactivation mechanisms, such as predation, UV, sunlight, and disinfection should be investigated. Data on SARS-CoV-2 removal and/or inactivation by wastewater and water treatment processes, such as activated sludge, membrane filtration, coagulation-sedimentation, and disinfection (chlorine, chloramine, UV, ozone, etc.) is scarce. If it is difficult to determine log10 reduction values of SARS-CoV-2 itself due to availability of the virus and/or biosafety restrictions, model enveloped virus such as human CoVs, MHV, or Pseudomonas phage Φ6 can be used for laboratory- or pilot-scale experiments.
Currently RT-qPCR assays developed for clinical specimen testing are being used for SARS-CoV-2 RNA detection in environmental water samples. Recent environmental studies reported that different assays might produce conflicting results (Ahmed et al., 2020; Medema et al., 2020). Moreover, the false-negative rates (due to improperly designed primers/probe or virus mutation in the targeted genome region) of these assays need to be assessed by multiple laboratories. For environmental application, the sensitivities of these RT-qPCR methods need to be evaluated. The major limitation of qPCR is that it does not provide information on viability. When viability needs to be assessed, cell culture infectivity assay, EMA/PMA-RT-qPCR, and ICC-RT-qPCR may provide useful information. A standardized protocol to recover and detect SARS-CoV-2 from environmental water samples, including concentration method, PCR assay, and process controls, should be established.
Wastewater surveillance is critical as WBE may provide valuable information on the prevalence of infections in the community. Continuous and systematic monitoring of wastewater may provide early warning signs and will potentially identify undiagnosed or successive disease at the population level, thus alerting public health officials on the ongoing or future viral disease outbreaks. Nationwide and international wastewater surveillance campaigns should be carried out to better understand temporal and spatial dynamics of disease prevalence, molecular epidemiology and evolution of the virus, and efficacy of public health interventions.
QMRA has a potential role to play in reducing the impact of the ongoing COVID-19 outbreak. However, currently available QMRA parameters for SARS-CoV-2 are limited, although previous studies on relevant respiratory viruses (SARS-CoV, MERS-CoV, and influenza viruses) help to assess the likely risks of SARS-CoV-2. Our understanding on the potential role of wastewater in SARS-CoV-2 transmission is largely limited by knowledge gaps in its occurrence and survival in wastewater and environmental waters and removal by wastewater treatment processes. There is an urgent need for collecting these pieces of information to understand and mitigate the human health risks associated with exposure to wastewater and environmental waters potentially contaminated with SARS-CoV-2.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was partly supported by the Japan Society for the Promotion of Science (JSPS) through the Promotion of Joint International Research (Fostering Joint International Research (B)) Grant Number JP18KK0270 and United States National Science Foundation award 2027752. The authors thank Sayalee Joshi from Arizona State University for her technical assistance in drawing the graphical abstract.
Editor: Damia Barcelo
References
- Adhikari U., Chabrelie A., Weir M., Boehnke K., McKenzie E., Ikner L., Wang M., Wang Q., Young K., Haas C.N., Rose J., Mitchell J. A case study evaluating the risk of infection from Middle Eastern respiratory syndrome coronavirus (MERS-CoV) in a hospital setting through bioaerosols. Risk Anal. 2019;39:2608–2624. doi: 10.1111/risa.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., Brien J.W.O., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino de Carvalho N., Stachler E.N., Cimabue N., Bibby K. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environ. Sci. Technol. 2017;51:8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., Bassioni L. El, Akande A.O., Maamoun E. Al, Zaidi S., Adeniji A.J., Burns C.C., Deshpande J., Oberste M.S., Lowther S.A. Environmental surveillance for polioviruses in the global polio eradication initiative. J. Infect. Dis. 2014;210:S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Gao H., Xiao C., Jiayi Liu, Xue J., Lv Q., Jiangning Liu, Yu P., Xu Y., Qi F., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C. 2020. Reinfection Could Not Occur in SARS-CoV-2 Infected Rhesus Macaques. bioRxiv 2020.03.13.990226. [DOI] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao Linna, Liu P., Zhao Li, Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. bioRxiv. 2020 doi: 10.1101/2020.02.07.939389. [DOI] [PubMed] [Google Scholar]
- Bedford J., Enria D., Giesecke J., Heymann D., Ihekweazu C., Kobinger G., Lane H., Memish Z., Oh M., Sall A., Schuchat A., Ungchusak K., Wieler L. COVID-19: towards controlling of a pandemic. Lancet. 2020;6736:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K. Metagenomic identification of viral pathogens. Trends Biotechnol. 2013;31(5):275–279. doi: 10.1016/j.tibtech.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby Kyle. Metagenomics and the development of viral water quality tools. npj Clean Water. 2019;2(1):1–13. [Google Scholar]
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.L., Henquell C. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in Central France, 2014 to 2015. Eurosurveillance. 2018;23:1–11. doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F.J., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ. Virol. 2019;11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B., Liu W.-C., Moeller R., Panis M., Sachs D., Albrecht R., tenOever B.R. 2020. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv 2020.03.24.004655. [DOI] [Google Scholar]
- Boldog P., Tekeli T., Vizi Z., Dénes A., Bartha F.A., Röst G. Risk assessment of novel coronavirus COVID-19 outbreaks outside China. J. Clin. Med. 2020;9:571. doi: 10.3390/jcm9020571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno Ferraro G., Mancini P., Veneri C., Iaconelli M., Suffredini E., Brandtner D., La Rosa G. Evidence of Saffold virus circulation in Italy provided through environmental surveillance. Lett. Appl. Microbiol. 2020;70:102–108. doi: 10.1111/lam.13249. [DOI] [PubMed] [Google Scholar]
- Boodman, E., 2020. From ferrets to mice and marmosets, labs scramble to find right animals for coronavirus studies [WWW document]. URL https://www.statnews.com/2020/03/05/coronavirus-labs-scramble-to-find-right-animals-for-covid-19-studies/.
- Bosch A. Human enteric viruses in the water environment: a minireview. Int. Microbiol. 1998;1:191–196. doi: 10.2436/im.v1i3.39. [DOI] [PubMed] [Google Scholar]
- Brenner K., Scarpino P., Clark C. Animal viruses, coliphages, and bacteria in aerosols and wastewater at a spray irrigation site. Appl. Environ. Microbiol. 1988;54:409–415. doi: 10.1128/aem.54.2.409-415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G. John Wiley & Sons Canada, Ltd; 1980. Introduction to Environmental Virology. [Google Scholar]
- Burke R.M., Midgley C.M., Dratch A., Fenstersheib M., Haupt T., Holshue M., Ghinai I., Jarashow M.C., Lo J., McPherson T.D., Rudman S., Scott S., Hall A.J., Fry A.M., Rolfes M.A. Active monitoring of persons exposed to patients with confirmed COVID-19 - United States, January-February 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:245–246. doi: 10.15585/mmwr.mm6909e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., Munster V.J. erosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B., Rodriguez-Manzano J., Hundesa A., Suñen E., Calvo M., Bofill-Mas S., Girones R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J. Virol. Methods. 2013;187:215–221. doi: 10.1016/j.jviromet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Callaway E. Labs rush to study coronavirus in transgenic animals--some are in short supply [WWW document]. Nat. News. 2020. https://www.nature.com/articles/d41586-020-00698-x#ref-CR1 URL. [DOI] [PubMed]
- Carducci A., Verani M., Battistini R., Pizzi F., Rovini E., Andreoli E., Casini B. 2006. Epidemiological Surveillance of Human Enteric Viruses by Monitoring of Different Environmental Matrices; pp. 239–244. [DOI] [PubMed] [Google Scholar]
- Carducci A., Donzelli G., Cioni L., Federigi I., Lombardi R., Verani M. Quantitative microbial risk assessment for workers exposed to bioaerosol in wastewater treatment plants aimed at the choice and setup of safety measures. Int. J. Environ. Res. Public Health. 2018;15 doi: 10.3390/ijerph15071490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environ. Sci. Technol. Lett. 2015;2:76–78. doi: 10.1021/acs.estlett.5b00029. [DOI] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdollar J.L., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol. 2013;115:1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- CDC Severe acute respiratory syndrome (SARS) [WWW document] 2004. https://www.cdc.gov/sars/about/faq.html URL.
- CDC . Centers Dis. Control Prev; 2020. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes [WWW Document] [Google Scholar]
- CDC How COVID-19 spreads [WWW document] 2020. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fprepare%2Ftransmission.html URL.
- Chan M.C.W., Lee N., Chan P.K.S., To K.F., Wong R.Y.K., Ho W.S., Ngai K.L.K., Sung J.J.Y. Seasonal influenza a virus in feces of hospitalized adults. Emerg. Infect. Dis. 2011;17:2038–2042. doi: 10.3201/eid1711.110205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.-P., Kok K.-H., Chu H., Chan K.-H., Sridhar S., Chen Z., Chen H., To K.K.-W., Yuen K.-Y. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Qin C., Wei Q., Li R., Gao H., Zhu H., Deng W., Bao L., Wei T. 2020. Protection of Rhesus Macaque from SARS-Coronavirus Challenge by Recombinant Adenovirus Vaccine. bioRxiv 2020.02.17.951939. [DOI] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Gao G., Xu Y., Pu L., Wang Q., Liming Wang, Wang W., Song Y., Chen M., Linghang Wang, Yu F., Yang S., Tang Y., Zhao L., Wang H., Wang Y., Zeng H., Zhang F. SARS-CoV-2–positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann. Intern. Med. 2020;170:1–3. doi: 10.7326/AITC201903050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Shan J. 2019 novel coronavirus: where we are and what we know. Infection. 2020;48:155–163. doi: 10.1007/s15010-020-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.K.C., Wong D.A., Tong L.K.L., Ip S.M., Lo A.C.T., Lau C.S., Yeung E.Y.H., Lim W.W.L. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. 2020. Stability of SARS-CoV-2 in Different Environmental Conditions; pp. 2–5. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China CDC . China CDC; 2020. Specific Primers and Probes for Detection 2019 Novel Coronavirus [WWW Document] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;7:1–7. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B.J., Wagner B.G., Blower S. Modeling influenza epidemics and pandemics: insights into the future of swine flu (H1N1) BMC Med. 2009;7:1–8. doi: 10.1186/1741-7015-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., Muth D., Sieberg A., Meyer B., Assiri A.M., Binger T., Steinhagen K., Lattwein E., Al-Tawfiq J., Müller M.A., Drosten C., Memish Z.A. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Bao L., Gao H., Xiang Z., Qu Y., Song Z., Gong S., Jiayi Liu, Jiangning Liu, Yu P., Qi F., Xu Y., Li F., Xiao C., Lv Q., Xue J., Wei Q., Liu M., Wang G., Wang S., Yu H., Liu X., Zhao W., Han Y., Qin C. 2020. Ocular Conjunctival Inoculation of SARS-CoV-2 Can Cause Mild COVID-19 in Rhesus Macaques. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Medical Sciences, Ministry of Public Health, T . 2020. Diagnostic Detection of Novel Coronavirus 2019 by Real Time RT-PCR [WWW Document] [Google Scholar]
- Dong L., Zhou J., Niu C., Wang Q., Pan Y., Wang X., Zhang Y., Yang J., Liu M., Yang Zhao, Peng T., Xie J., Gao Y., Wang D., Yun Zhao, Dai X., Fang X. 2020. Highly Accurate and Sensitive Diagnostic Detection of SARS-CoV-2 by Digital PCR. medRxiv 2020.03.14.20036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M.G., Kocher J.F., Scobey T., Baric R.S., Cockrell A.S. Adaptive evolution influences the infectious dose of MERS-CoV necessary to achieve severe respiratory disease. Virology. 2018;517:98–107. doi: 10.1016/j.virol.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D., Junglen S., Müller M.A., Haas W., Guberina H., Röhnisch T., Schmid-Wendtner M., Aldabbagh S., Dittmer U., Gold H., Graf P., Bonin F., Rambaut A., Wendtner C.M. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannin K.F., Vana S.C., Jakubowski W. Effect of an activated sludge wastewater treatment plant on ambient air densities of aerosols containing bacteria and viruses. Appl. Environ. Microbiol. 1985;49:1191–1196. doi: 10.1128/aem.49.5.1191-1196.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Mannion F., Hillary L.S., Malham S.K., Walker D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Heal. 2020;16:1–6. doi: 10.1016/j.coesh.2020.01.007. [DOI] [Google Scholar]
- Fears A.C., Klimstra W.B., Duprex P., Hartman A., Weaver S.C., Plante K.C., Mirchandani D., Plante J.A., Aguilar P.V., Fernández D., Nalca A., Totura A., Dyer D., Kearney B., Lackemeyer M., Bohannon J.K., Johnson R., Garry R.F., Reed D.S., Roy C.J. 2020. Comparative Dynamic Aerosol Efficiencies of Three Emergent Coronaviruses and the Unusual Persistence of SARS-CoV-2 in Aerosol Suspensions. medRxiv. [DOI] [Google Scholar]
- Fernandez-Cassi X., Timoneda N., Martínez-Puchol S., Rusiñol M., Rodriguez-Manzano J., Figuerola N., Bofill-Mas S., Abril J.F., Girones R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2018;618:870–880. doi: 10.1016/j.scitotenv.2017.08.249. [DOI] [PubMed] [Google Scholar]
- Fong T.-T., Lipp E.K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69:357–371. doi: 10.1128/MMBR.69.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumian T.M., Leite J.P.G., Castello A.A., Gaggero A., Caillou M.S.L.d., Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170:42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Gao Q.Y., Chen Y.X., Fang J.Y. 2019 novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020;1(2) doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:1–8. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretebeck L.M., Subbarao K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015;13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D., Du B., Li L., Zeng G., Yuen K., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10–14. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Guo Z.D., Wang Z.Y., Zhang S.F., Li X., Li L., Li C., Cui Y., Fu R.B., Dong Y.Z., Chi X.Y., Zhang M.Y. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas C.N. Microbial dose response modeling: past, present, and future. Environ. Sci. Technol. 2015;49:1245–1259. doi: 10.1021/es504422q. [DOI] [PubMed] [Google Scholar]
- Haas C.N. Coronavirus and environmental engineering science. Environ. Eng. Sci. 2020;37:1–2. [Google Scholar]
- Haas C., Rose J., Gerba C. Second edi. John Wiley & Sons, Inc; 2014. Quantitative Microbial Risk Assessment. [Google Scholar]
- Hamilton K.A., Haas C.N. Critical review of mathematical approaches for quantitative microbial risk assessment (QMRA) of Legionella in engineered water systems: research gaps and a new framework. Environ. Sci. Water Res. Technol. 2016;2:599–613. doi: 10.1039/c6ew00023a. [DOI] [Google Scholar]
- Hamilton K.A., Ahmed W., Rauh E., Rock C., McLain J., Muenich R.L. Comparing microbial risks from multiple sustainable waste streams applied for agricultural use: biosolids, manure, and diverted urine. Curr. Opin. Environ. Sci. Heal. 2020;14:37–50. doi: 10.1016/j.coesh.2020.01.003. [DOI] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ito T., Ohgaki S. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 2009;32:297–300. doi: 10.1111/j.1365-2761.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen-Tanski H., Reponen T., Koivunen J. Airborne enteric coliphages and bacteria in sewage treatment plants. Water Res. 2009;43:2558–2566. doi: 10.1016/j.watres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V.R., Polaczyk A.L., Hahn D., Narayanan J., Cromeans T.L., Roberts J.M., Amburgey J.E. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 2005;71:6878–6884. doi: 10.1128/AEM.71.11.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V.R., Kahler A.M., Jothikumar N., Johnson T.B., Hahn D., Cromeans T.L. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 2007;73:4218–4225. doi: 10.1128/AEM.02713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M., DeBolt C., Lindquist S., Lofy K., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M., Weldon W., Biggs H., Uyeki T., Pillai S. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Shen L., Xu Z., Zhou J., Zhou H. 2020. SARS-CoV-2 May Persist in Digestive Tract Longer Than Respiratory Tract. Preprints. [DOI] [Google Scholar]
- Huang Y., Anderson S.A., Forshee R.A., Yang H. A modified dose–response model that describes the relationship between haemagglutination inhibition titre and protection against influenza infection. J. Appl. Microbiol. 2018;124:294–301. doi: 10.1111/jam.13628. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.N., Cheng V.C.C., Wu A.K.L., Tang B.S.F., Chan K.H., Chu C.M. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4:41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Institut Pasteur P. Inst. Pasteur; Paris: 2020. Protocol: Real-time RT-PCR assays for the detection of SARS-CoV-2.https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2 [WWW Document] (accessed 3.31.20) [Google Scholar]
- Isakbaeva E.T., Khetsuriani N., Beard R.S., Peck A., Erdman D., Monroe S.S., Tong S., Ksiazek T.G., Lowther S., Pandya-Smith I., Anderson L.J., Lingappa J., Widdowson M.A., McLaughlin J., Romney M., Kimura A., Dassey D., Lash B., Terashita D., Klish S., Cody S., Farley S., Lea S., Sanderson R., Wolthuis J., Allard C., Albanese B., Nivin B., McCall P., Davies M., Murphy M., Koch E., Weltman A., Brumund H., Barton C., Whetstone K., Bellini W.J., Bialek S., Comer J.A., Emery S., Helfand R., Hennessy T., James A., LaMonte A., Newbern E.C., Scott S., Simpson L., Siwek A., Smelser C., Stockman L., Lu X., White D. SARS-associated coronavirus transmission, United States. Emerg. Infect. Dis. 2004;10:225–231. doi: 10.3201/eid1002.030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiehao C., Jing X., Daojiong L., Lei X., Zhenghai Q., Yuehua Z., Hua Z., Xiangshi W., Yanling G., Aimei X., He T., Hailing C., Chuning W., Jingjing L., Jianshe W., Mei Z., Children N., Women H., Central S., Zeng M. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.A., Vasconcelos P.F.C., Staples J.E. The whole iceberg: estimating the incidence of yellow fever virus infection from the number of severe cases. Trans. R. Soc. Trop. Med. Hyg. 2014;108:482–487. doi: 10.1093/trstmh/tru092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.-M., Akhmetzhanov A.R., Hayashi K., Linton N.M., Yang Y., Yuan B., Kobayashi T., Kinoshita R., Nishiura H. Real-time estimation of the risk of death from novel coronavirus (COVID-19) infection: inference using exported cases. J. Clin. Med. 2020;9 doi: 10.3390/jcm9020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuder S.A., Arthur T., Bisesi M.S., Schaub E.A. Prevalence of infectious diseases and associated symptoms in wastewater treatment workers. Am. J. Ind. Med. 1998;33:571–577. doi: 10.1002/(SICI)1097-0274(199806)33:6<571::AID-AJIM8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Kim Young-il, Kim Seong-gyu, Kim Se-mi, Webby R.J., Jung J.U., Choi Y.K., Kim Young-il, Kim Seong-gyu, Kim Se-mi, Kim E., Park S., Yu K., Chang J., Kim E.J., Lee S., Casel M.A.B., Um J., Song M.-S., Jeong H.W., Lai V.D., Kim Yeonjae, Chin B.S., Park J.-S., Chung K.-H., Foo S.-S., Poo H., Mo I.-P., Lee O.-J., Webby R.J., Jung J.U., Choi Y.K. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020:1–6. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Huang Y., Watanabe T., Katayama H., Haas C.N. Dose-response time modelling for highly pathogenic avian influenza A (H5N1) virus infection. Lett. Appl. Microbiol. 2011;53:438–444. doi: 10.1111/j.1472-765X.2011.03128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Rachmadi A.T., Haramoto E., Gerba C.P. Quantification and genetic analysis of salivirus/klassevirus in wastewater in Arizona, USA. Food Environ. Virol. 2014;6:213–216. doi: 10.1007/s12560-014-9148-2. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Pepper I.L., Gerba C.P. Occurrence and genetic diversity of human cosavirus in influent and effluent of wastewater treatment plants in Arizona, United States. Arch. Virol. 2015;160:1775–1779. doi: 10.1007/s00705-015-2435-x. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Muscillo M. Molecular detection of viruses in water and sewage. In: Cook N., editor. Viruses in Food and Water: Risks, Surveillance and Control. Woodhead Publishing Limited; Cambridge, UK: 2013. pp. 97–125. [DOI] [Google Scholar]
- La Rosa G., Della Libera S., Iaconelli M., Ciccaglione A.R., Bruni R., Taffon S., Equestre M., Alfonsi V., Rizzo C., Tosti M.E., Chironna M., Romanò L., Zanetti A.R., Muscillo M. Surveillance of hepatitis A virus in urban sewages and comparison with cases notified in the course of an outbreak, Italy 2013. BMC Infect. Dis. 2014;14:1–11. doi: 10.1186/1471-2334-14-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Liu Y.H., Wang C.-Y., Wang Y.-H., Hsueh S.-C., Yen M.-Y., Ko W.-C., Hsueh P.-R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J. Microbiol. Immunol. Infect. 2020;2 doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Chan P.K.S., Hui D.S.C., Rainer T.H., Wong E., Choi K., Lui G.C.Y., Wong B.C.K., Wong R.Y.K., Lam W., Chu I.M.T., Lai R.W.M., Cockram C.S., Sung J.J.Y. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J. Infect. Dis. 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., Jensen K.L., Baric R.S., Sheahan T.P. Increasing the translation of mouse models of MERS coronavirus pathogenesis through kinetic hematological analysis. PLoS One. 2019;14:1–14. doi: 10.1371/journal.pone.0220126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.-C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.-F., Lina B., van-der-Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;2:1–10. doi: 10.1016/s1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J., Reich N.G., Brookmeyer R., Perl T.M., Nelson K.E., Cummings D.A. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect. Dis. 2009;9:291–300. doi: 10.1016/S1473-3099(09)70069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.F., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., Yuen K.Y., Sung J.J.Y. Enteric involvement of severe acute respiratory syndrome - associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.W., Wang X.W., Rui Q.Y., Song N., Zhang F.G., Ou Y.C., Chao F.H. A new and simple method for concentration of enteric viruses from water. J. Virol. Methods. 1998;74:99–108. doi: 10.1016/S0166-0934(98)00078-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Duan S., Yu I.T.S., Wong T.W. Multi-zone modeling of probable SARS virus transmission by airflow between flats in Block E, Amoy Gardens. Indoor Air. 2005;15:96–111. doi: 10.1111/j.1600-0668.2004.00318.x. [DOI] [PubMed] [Google Scholar]
- Li F.C.K., Choi B.C.K., Sly T., Pak A.W.P. Finding the real case-fatality rate of H5N1 avian influenza. J. Epidemiol. Community Health. 2008;62:555–559. doi: 10.1136/jech.2007.064030. [DOI] [PubMed] [Google Scholar]
- Lim K.Y., Hamilton A.J., Jiang S.C. Assessment of public health risk associated with viral contamination in harvested urban stormwater for domestic applications. Sci. Total Environ. 2015;523:95–108. doi: 10.1016/j.scitotenv.2015.03.077. [DOI] [PubMed] [Google Scholar]
- Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song, Gang Z., Huang W., Chen J., Hu B.J., Wang S., Mao E.Q., Zhu L., Zhang W.H., Lu H.Z. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton N.M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A.R., Jung S.-M., Yuan B., Kinoshita R., Nishiura H. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J. Clin. Med. 2020;9:538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L., Gopalakrishna G., Chew S.K., Tan C.C., Samore M.H., Fisman D., Murray M. Transmission dynamics and control of severe acute respiratory syndrome. Science (80-. ) 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., Tian Y., Sin N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;1253:30087. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Rutjes S.A., Takumi K., de Roda Husman A.M. Aichi virus in sewage and surface water, the Netherlands. Emerg. Infect. Dis. 2013;19:1222–1230. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. Wei, Liu X. Fen, Jia Z. Fang. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn T.J., Restif O., Peel A.J., Munster V.J., De Wit E., Sokolow S., Van Doremalen N., Hudson P., McCallum H. Dose-response and transmission: the nexus between reservoir hosts, environment and recipient hosts. Philos. Trans. R. Soc. B Biol. Sci. 2019;374 doi: 10.1098/rstb.2019.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux F.G., Hotz P., Gashi D., Savova-Bianchi D., Oppliger A. Assessment of airborne virus contamination in wastewater treatment plants. Environ. Res. 2014;133:260–265. doi: 10.1016/j.envres.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Katayama H. Development of a portable detection method for enteric viruses from ambient air and its application to a wastewater treatment plant. Pathogens. 2019;8 doi: 10.3390/pathogens8030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K.R., Gong Y.Y., Lewis T.G. Environmental transmission of SARS at Amoy Gardens. J. Environ. Health. 2006;68:26–30. [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R. 2020. Presence of SARS-Coronavirus-2 in Sewage. medRxiv. [DOI] [PubMed] [Google Scholar]
- Memish Z., Perlman S., Van Kerkhove M., Zumla A. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M., Millet J., Vidalain P.-O., Law H., Vabret A., Lorin V., Escriou N., Albert M.L., Nal B., Tangy F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012;86:7577–7587. doi: 10.1128/jvi.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010;109:388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:1–5. doi: 10.2807/1560-7917.es.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., Avanzato V.A., Rosenke R., Hanley P.W., Saturday G., Scott D., Fischer E.R., de Wit E. Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.21.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Hata A., Honda R., Watanabe T. Wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ. Sci. Technol. 2020:20045880. doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer/probe sets and one assay kit. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. medRxiv. 2020 doi: 10.1101/2020.04.15.20066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngaosuwankul N., Noisumdaeng P., Komolsiri P., Pooruk P., Chokephaibulkit K., Chotpitayasunondh T., Sangsajja C., Chuchottaworn C., Farrar J., Puthavathana P. Research influenza A viral loads in respiratory samples collected from patients infected with pandemic H1N1, seasonal H1N1 and H3N2 viruses. Virol. J. 2010;7:1–7. doi: 10.1186/1743-422X-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Suzuki A., Jung S.-M., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R., Linton N.M., Miyama T. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayashi T., Yokota S.I., Ohkoshi Y., Ohuchi H., Yoshida Y., Kikuchi M., Yano K., Fujii N. Occurrence of norovirus infections unrelated to norovirus outbreaks in an asymptomatic food handler population. J. Clin. Microbiol. 2008;46:1985–1988. doi: 10.1128/JCM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ospina E., Hasell J. Our World Data; 2020. How Many Tests for COVID-19 Are Being Performed around the World? [WWW Document] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Jung S., Kim A. MERS transmission and risk factors: a systematic review. BMC Public Health. 2018;18 doi: 10.1186/s12889-018-5484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasalari H., Ataei-Pirkooh A., Aminikhah M., Jafari A.J., Farzadkia M. Assessment of airborne enteric viruses emitted from wastewater treatment plant: atmospheric dispersion model, quantitative microbial risk assessment, disease burden. Environ. Pollut. 2019;253:464–473. doi: 10.1016/j.envpol.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Poon L., Chu D., Peiris M. Detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR [WWW document] 2020. https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn=af1aac73_4 URL.
- Pratelli A. Canine coronavirus inactivation with physical and chemical agents. Vet. J. 2008;177:71–79. doi: 10.1016/j.tvjl.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Prüss A., Kay D., Fewtrell L., Bartram J. Estimating the burden of disease from water, sanitation, and hygiene at a global level. Environ. Health Perspect. 2002;110:537–542. doi: 10.1289/ehp.02110537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R., Huang Y. ting, Liu J. wei, Sun Y., Sun X. feng, Han H.J., Qin X.R., Zhao M., Wang L. jun, Li W., Li J. hong, Chen C., Yu X.J. Global prevalence of asymptomatic norovirus infection: a meta-analysis. EClinicalMedicine. 2018;(2–3):50–58. doi: 10.1016/j.eclinm.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QMRA Wiki SARS dose response experiments [WWW Document] 2020. http://qmrawiki.org/experiments/sars URL.
- QMRA Wiki Influenza dose response experiments [WWW Document] 2020. http://qmrawiki.org/experiments/influenza/104 URL.
- Richard M., Kok A., de Meulder D., Bestebroer T.M., Lamers M.M., Okba N.M., van Vlissingen M.F., Rockx B., Haagmans B.L., Koopmans M.P., Fouchier R.A. SARS-CoV-2 is transmitted via contact and via the air between ferrets. bioRxiv. 2020 doi: 10.1038/s41467-020-17367-2. https://www.biorxiv.org/content/10.1101/2020.04.16.044503v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J., Leung G.M., Ho L.M., Lam T.H., Thach T.Q., Chau P., Chan K.P., Lo S.V., Leung P.Y., Tsang T., Ho W., Lee K.H., Lau E.M.C., Ferguson N.M., Anderson R.M. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science (80-. ) 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M., Meulder D. de, Amerongen G. van, Brand J. van de, Okba N., Schipper D., Run P. van, Leijten L., Verschoor E., Verstrepen B., Langermans J., Drosten C., Vlissingen M.F. van, Fouchier R., Swart R.L. de, Koopmans M., Haagmans B. 2020. Comparative Pathogenesis of COVID-19, MERS and SARS in a Non-human Primate Model. bioRxiv 2020.03.17.995639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Lázaro D., Cook N., Ruggeri F.M., Sellwood J., Nasser A., Nascimento M.S.J., D’Agostino M., Santos R., Saiz J.C., Rzezutka A., Bosch A., Gironés R., Carducci A., Muscillo M., Kovač K., Diez-Valcarce M., Vantarakis A., von Bonsdorff C.H., de Roda Husman A.M., Hernández M., van der Poel W.H.M. Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 2012;36:786–814. doi: 10.1111/j.1574-6976.2011.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen-Kopra C., Paananen A., Blomqvist S., Klemola P., Simonen M.L., Lappalainen M., Vuorinen T., Kuusi M., Lemey P., Roivainen M. A large Finnish echovirus 30 outbreak was preceded by silent circulation of the same genotype. Virus Genes. 2011;42:28–36. doi: 10.1007/s11262-010-0536-x. [DOI] [PubMed] [Google Scholar]
- Sedmak G., Bina D., MacDonald J. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 2003;69:7181–7187. doi: 10.1128/AEM.69.12.7181-7187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y., Kageyama T., Matsuyama S., Takeda M. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Sinclair R.G., Choi C.Y., Riley M.R., Gerba C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008;65:249–269. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit L.A.M., Spaan S., Heederik D. Endotoxin exposure and symptoms in wastewater treatment workers. Am. J. Ind. Med. 2005;48:30–39. doi: 10.1002/ajim.20176. [DOI] [PubMed] [Google Scholar]
- Song Y., Liu P., Shi X., Chu Y., Zhang J., Xia J., Gao X., Qu T., Wang M. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69(6):1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- Sueki A., Matsuda K., Yamaguchi A., Uehara M., Sugano M., Uehara T., Honda T. Evaluation of saliva as diagnostic materials for influenza virus infection by PCR-based assays. Clin. Chim. Acta. 2016;453:71–74. doi: 10.1016/j.cca.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Suo T., Liu X., Guo M., Feng J., Hu W., Yang Y., Qiuhan Zhang, Wang X., Sajid M., Guo D., Huang Z., Deng L., Chen T., Liu F., Xu K., Yuan Liu, Qi Zhang, Yingle Liu, Xiong Y., Guo G., Chen Y., Lan K. 2020. ddPCR: A More Sensitive and Accurate Tool for SARS-CoV-2 Detection in Low Viral Load Specimens. medRxiv 2020.02.29.20029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Wang X., Li Q., Bragazzi N., Tang S., Xiao Y., Wu J. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J. Clin. Med. 2020;9:462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.-D., Wang H.-L., Dai Y.-X., Li K.-F., Liu J.-N., Wu W.-J., Yuan C., Yu M.-L., Li P., Yan J.-B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26:1–5. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. J. 2020;26 doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongprachum A., Fujimoto T., Takanashi S., Saito H., Okitsu S., Shimizu H., Khamrin P., Maneekarn N., Hayakawa S., Ushijima H. Detection of nineteen enteric viruses in raw sewage in Japan. Infect. Genet. Evol. 2018;63:17–23. doi: 10.1016/j.meegid.2018.05.006. [DOI] [PubMed] [Google Scholar]
- To K.K., Tak O., Tsang Y., Leung W., Tam A.R., Wu T., Lung D.C., Yip C.C., Cai J., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L., Chan W., Chan K., Ip J.D., Ng A.C., Poon R.W., Luo C., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K., Richard F., Yu C., Tam M., Mei M., Shaw T., Hong F., Tong M., Lee M. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;3099:1–10. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Chik-Yan Yip C., Chan K.-H., Wu T.-C., Chan J.M.C., Leung W.-S., Chik T.S.-H., Choi C.Y.-C., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W.-S., Fung A.Y.-F., Hung I.F.-N., Cheng V.C.-C., Chan J.F.-W., Yuen K.-Y. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020:4–6. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp A., Bernard A., Thommen A.M., Jeggli S., Dumont X., Oppliger A., Hotz P. Exposure to bioaerosols, respiratory health and lung-specific proteins: a prospective study in garbage and wastewater workers. Occup. Environ. Med. 2011;68:856–859. doi: 10.1136/oem.2010.060178. [DOI] [PubMed] [Google Scholar]
- USEPA List N: disinfectants for use against SARS-CoV-2 [WWW document] 2020. https://www.epa.gov/pesticide-registration/list-n-disinfectants-use-against-sars-cov-2 URL.
- van Doremalen N., Bushmaker T., Morris D., Holbrook M., Gamble A., Williamson B., Tamin A., Harcourt J., Thornburg N., Gerber S., Lloyd-Smith J., de Wit E., Munster V. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuken J.P.G., Swart A.N., Havelaar A.H., Van Pul A., Van der Hoek W., Heederik D. Atmospheric dispersion modelling of bioaerosols that are pathogenic to humans and livestock - a review to inform risk assessment studies. Microb. Risk Anal. 2016;1:19–39. doi: 10.1016/j.mran.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-dannenburg G., Thompson H., Walker P.G.T., Fu H., Dighe A., Griffin J.T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunubá Z., Fitzjohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-gilani G., Riley S., Elsland S. Van, Volz E., Wang H., Wang Y., Xi X., Donnelly C.A., Ghani A.C., Ferguson N.M. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;3099:1–9. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade T., Sams E., Brenner K., Haugland R., Chern E., Beach M., Wymer L., Rankin C., Love D., Li Q., Noble R., Dufour A. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health. 2010;9:66. doi: 10.1093/ije/27.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J., Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160:509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.K., Chen S.Y., Liu I.J., Chen Y.C., Chen H.L., Yang C.F., Chen P.J., Yeh S.H., Kao C.L., Huang L.M., Hsueh P.R., Wang Jann Tay, Sheng W.H., Fang C.T., Hung C.C., Hsieh S.M., Su C.P., Chiang W.C., Yang J.Y., Lin J.H., Hsieh S.C., Hu H.P., Chiang Y.P., Wang Jin Town, Yang P.C., Chang S.C. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg. Infect. Dis. 2004;10:1213–1219. doi: 10.3201/eid1007.031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J., Guo T., Zhen B., Kong Q., Yi B., Li Z., Song N., Jin M., Xiao W., Zhu X., Gu C., Yin J., Wei W., Yao W., Liu C., Li J., Ou G., Wang M., Fang T., Wang G., Qiu Y., Wu H., Chao F., Li J. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan hospital and the 309th Hospital of the Chinese People’s liberation Army. Water Sci. Technol. 2005;52:213–221. doi: 10.2166/wst.2005.0266. [DOI] [PubMed] [Google Scholar]
- Wang Xin Wei, Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., Li Z., Song N., Jin M., Xiao W.J., Zhu X.M., Gu C.Q., Yin J., Wei W., Yao W., Liu C., Li J.F., Ou G.R., Wang M.N., Fang T.Y., Wang G.J., Qiu Y.H., Wu H.H., Chao F.H., Li J.W. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th hospital. J. Virol. Methods. 2005;128:156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xin Wei, Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B.Y., Guo B.Z., Liu C., Ou G.R., Wang M.N., Fang T.Y., Chao F.H., Li J.W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020:3–4. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren S. Bloom. News; 2020. Texas baboon troop enlisted in humankind's war on coronavirus [WWW document] https://www.bloomberg.com/news/articles/2020-03-07/texas-baboon-troop-enlisted-in-humankind-s-war-on-coronavirus URL.
- Watanabe T., Bartrand T.A., Weir M.H., Omura T., Haas C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010;30:1129–1138. doi: 10.1111/j.1539-6924.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Bartrand T.A., Omura T., Haas C.N. Dose-response assessment for influenza A virus based on data sets of infection with its live attenuated reassortants. Risk Anal. 2012;32:555–565. doi: 10.1111/j.1539-6924.2011.01680.x. [DOI] [PubMed] [Google Scholar]
- WHO Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) [WWW document] 2003. https://www.who.int/csr/sars/en/WHOconsensus.pd URL.
- WHO FAQs: H5N1 influenza [WWW Document] 2011. https://www.who.int/influenza/human_animal_interface/avian_influenza/h5n1_research/faqs/en/ URL.
- WHO, 2018. Influenza (seasonal) [WWW Document]. URL https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed 4.4.20).
- WHO . 2019. Influenza [WWW Document] [Google Scholar]
- WHO Pneumonia of unknown cause – China [WWW document] 2020. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ URL.
- WHO . 2020. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV) [WWW Document] [Google Scholar]
- WHO Coronavirus disease (COVID-19) pandemic [WWW document] 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 URL.
- WHO . 2020. Water, Sanitation, Hygiene and Waste Management for COVID-19. [Google Scholar]
- WHO, 2020e. Statement on the meeting of the international health regulations (2005) emergency committee regarding the outbreak of novel coronavirus (2019-nCoV) [WWW document]. URL https://www.who.int/news-room/detail/23-01-2020-statement-on-the-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) (accessed 4.5.20).
- Wigginton K.R., Boehm A.B. Environmental engineers and scientists have important roles to play in stemming outbreaks and pandemics caused by enveloped viruses. Environ. Sci. Technol. 2020;54:3736–3739. doi: 10.1021/acs.est.0c01476. [DOI] [PubMed] [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi: 10.1039/C5EW00125K. [DOI] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Bruenink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wong S.C.C., Chan J.K.C., Lee K.C., Lo E.S.F., Tsang D.N.C. Development of a quantitative assay for SARS coronavirus and correlation or GAPDH mRNA with SARS coronavirus in clinical specimens. J. Clin. Pathol. 2005;58:276–280. doi: 10.1136/jcp.2004.016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., Kumar L., Jenkins T.M., Xagoraraki I., Phanikumar M.S., Rose J.B. Evaluation of public health risks at recreational beaches in Lake Michigan via detection of enteric viruses and a human-specific bacteriological marker. Water Res. 2009;43:1137–1149. doi: 10.1016/j.watres.2008.11.051. [DOI] [PubMed] [Google Scholar]
- Wong M.V.M., Hashsham S.A., Gulari E., Rouillard J.M., Aw T.G., Rose J.B. Detection and characterization of human pathogenic viruses circulating in community wastewater using multi target microarrays and polymerase chain reaction. J. Water Health. 2013;11:659–670. doi: 10.2166/wh.2013.322. [DOI] [PubMed] [Google Scholar]
- Wong J.Y., Kelly H., Ip D.K.M., Wu J.T., Leung G.M., Cowling B.J. Case fatality risk of influenza a (H1N1pdm09): a systematic review. Epidemiology. 2013;24:830–841. doi: 10.1097/EDE.0b013e3182a67448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;1253:20–21. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Xiao, A., Zhang, J., Gu, X., Lee, W., Kauffman, K., Hanage, W., Matus, M., Ghaeli, N., Endo, N., Duvallet, C., Moniz, K., Erickson, T., Chai, P., Thompson, J., Alm, E., 2020b. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed]
- Wurtzer S., Marechal V., Mouchel J., Moulin L. 2020. Time Course Quantitative Detection of SARS-CoV-2 in Parisian Wastewaters Correlates with COVID-19 Confirmed Cases; pp. 10–13. medRxiv. [DOI] [Google Scholar]
- Wyn-Jones A.P., Sellwood J. A review: enteric viruses in the aquatic environment. J. Appl. Microbiol. 2001;91:945–962. doi: 10.1046/j.1365-2672.2001.01470.x. [DOI] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. Wastewater-based epidemiology for early detection of viral outbreaks. In: O’Bannon D., editor. Women in Water Quality. Springer Nature Switzerland; 2020. pp. 75–97. [DOI] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. 2020. Inhibition of SARS-CoV-2 Infection (Previously 2019-nCoV) by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting its Spike Protein that Harbors a High Capacity to Mediate Membrane Fusion. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Tong J., Liu M., Shen Y., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020:1–6. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Li C., He J. 2020. Evidence for Gastrointestinal Infection of SARS-CoV-2. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020 doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science (80-. ) 2020;2762:1–10. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Bao L., Deng W., Xu L., Li F., Lv Q., Yu P., Chen T., Xu Y., Zhu H., Yuan J., Gu S., Wei Q., Chen H., Yuen K.Y., Qin C. An animal model of mers produced by infection of rhesus macaques with MERS coronavirus. J. Infect. Dis. 2014;209:236–242. doi: 10.1093/infdis/jit590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Kasuo S., Azegami Y., Uchiyama Y., Satsumabayashi K., Shiraishi T., Katayama K., Wakita T., Takeda N., Oka T. Characterization of sapoviruses detected in gastroenteritis outbreaks and identification of asymptomatic adults with high viral load. J. Clin. Virol. 2009;45:67–71. doi: 10.1016/j.jcv.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.C., Chan M., Vasoo S., Wang L.F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.S., Lye D.C. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. J. Am. Med. Assoc. 2020:1–7. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., Xu J., Xu W. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X. Lou, Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020 doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ni C., Gao R., Wang Y., Yang L., Wei J., Lv T. 2020. Recapitulation of Infection and Cholangiocyte Damage with Human Liver Organoids. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang W., Zhong Q., Hou W., Yang Z., Xiao S.Y., Zhu R., Tang Z., Wang Y., Xian Q., Tang H., Wen L. Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine. 2005;23:3202–3209. doi: 10.1016/j.vaccine.2004.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Zhao G., Chu H., Wang D., Yan H.H.N., Poon V.K.M., Wen L., Wong B.H.Y., Zhao X., Chiu M.C., Yang D., Wang Y., Au-Yeung R.K.H., Chan I.H.Y., Sun S., Chan J.F.W., To K.K.W., Memish Z.A., Corman V.M., Drosten C., Hung I.F.N., Zhou Y., Leung S.Y., Yuen K.Y. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3 doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G., Shi Z.-L. 2020. Discovery of a Novel Coronavirus Associated with the Recent Pneumonia Outbreak in Humans and its Potential Bat Origin. bioRxiv. [DOI] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2000231. [DOI] [PMC free article] [PubMed] [Google Scholar]


