This Perspective explores the contributions of the innate and adaptive immune systems to both viral control as well as toxicity during COVID-19 infections and offers suggestions to both understand and therapeutically modulate anti-COVID immunity.
Abstract
The novel 2019 strain of coronavirus is a source of profound morbidity and mortality worldwide. Compared with recent viral outbreaks, COVID-19 infection has a relatively high mortality rate, the reasons for which are not entirely clear. Furthermore, treatment options for COVID-19 infection are currently limited. In this Perspective, we explore the contributions of the innate and adaptive immune systems to both viral control as well as toxicity during COVID-19 infections and offer suggestions to both understand and therapeutically modulate anti-COVID immunity.
Introduction
Since its emergence, the 2019 strain of coronavirus (hereafter COVID-19) has been a rising international cause of morbidity and mortality. This global devastation is partly explained by the nature of viral transmission; the median incubation time from COVID-19 infection to the appearance of symptomatic dyspnea ranges from four to seven days, creating a large window of time for transmission during which patients have few symptoms (Guan et al., 2020; Huang et al., 2020). In addition, many infected patients remain completely asymptomatic and yet are fully capable of transmitting the virus (Bai et al., 2020; Rothe et al., 2020). Also contributing to the destructive power of this pandemic is the significantly higher rate of morbidity and mortality in patients who ultimately develop symptoms. The majority of patients with severe disease develop acute respiratory distress syndrome (ARDS), a clinical phenomenon marked by development of bilateral infiltrates and hypoxemia, defined as a decrease in the ratio of arterial PO2 to inhaled FiO2 (Thompson et al., 2017). Almost all COVID-19 patients who develop ARDS require mechanical ventilation; these patients tend to remain ventilator dependent for 10–14 d, and most ventilated patients ultimately succumb to the disease (Bhatraju et al., 2020; Wu et al., 2020).
Generally speaking, the most common therapeutic options for viral infections are directed at either blocking viral entry or replication or promoting durable cellular and humoral immunity for the uninfected population via vaccination. Unfortunately, there is no Food and Drug Administration–approved medication to block or limit COVID-19 entry or replication, and vaccine development remains in the early stages. Furthermore, we understand little regarding the factors that govern either development or remission of severe disease. To date, the most significant predictors of disease severity relate to either activation or suppression of the host immune response. In this Perspective, we will discuss the role of both innate and adaptive immune responses in contributing to the clinical course of COVID-19 infection and highlight potential strategies for therapeutic intervention.
COVID-19: The case for innate immune hyperactivation
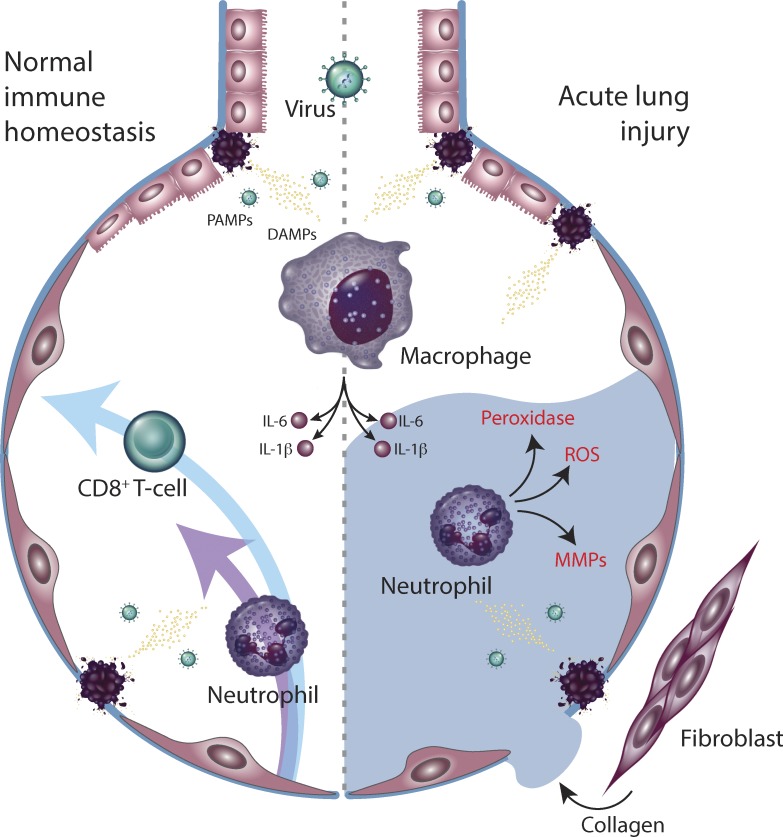
There is a compelling case for innate immune hyperactivity in driving the acute lung injury that defines severe COVID-19 infections. Tissue-resident macrophages have been implicated in the process of epithelial damage that initiates ARDS (Jacobs et al., 1989; Pison et al., 1988). Macrophages are activated by either damage-associated molecular patterns (DAMPs) such as intracellular contents released from dying cells and/or proteins released following tissue injury (such as heat-shock proteins, hyaluronan fragments, or heparin sulfate; Kuipers et al., 2011), or pathogen-associated molecular patterns (PAMPs) such as viral RNA or oxidized phospholipids (Diebold et al., 2004; Imai et al., 2008). Both DAMPs and PAMPs are likely generated during initial infection and lysis of pneumocytes by COVID-19. These molecules activate multiple innate immune pathways, through either TLRs (Medzhitov et al., 1997), NLRP3/inflammasome activation (Martinon et al., 2002), or triggering of cytoplasmic DNA sensors such as cGAS-STING and RIG-I-MAVS (Hornung et al., 2006; Pichlmair et al., 2006; Sun et al., 2013). The resultant signal transduction drives production of cytokines the exert both autocrine and paracrine effects, activating antiviral gene expression programs in neighboring cells as well as recruiting additional innate and adaptive immune cells with distinct roles in antiviral immunity and tissue homeostasis.
The inflammatory cascade initiated by macrophages contributes to both viral control and tissue damage. Production of type I and type III interferons promotes intracellular antiviral defenses in neighboring epithelial cells, which may limit viral dissemination, while release of IL-6 and IL-1β promotes recruitment of neutrophils and cytotoxic T cells (Fig. 1). Within the lung parenchyma, activated neutrophils release leukotrienes and reactive oxygen species that directly induce pneumocyte and endothelial injury, directly leading to acute lung injury. As local viral control is achieved, macrophage-derived IL-6 promotes T follicular helper differentiation as well as B cell germinal center formation and antibody production to confer long-term immunity (Harker et al., 2011). In severe or persistent viral infections, however, persistent neutrophil-mediated alveolar damage leads to interstitial flooding, ventilation/perfusion mismatching, and hypoxemic respiratory failure.
Figure 1.
Innate immune regulation of antiviral defense and tissue toxicity. Virally derived DAMPs and PAMPs activate tissue-resident macrophages. Downstream production of IL-6 and IL-1β recruit neutrophils and CD8+ T cells, which control viral growth (left) but also induce tissue damage, leading to alveolar flooding and fibrosis (right). MMP, matrix metalloproteases.
Significant evidence indicates that a dysregulated innate immune response contributes to the clinical presentation of patients with severe COVID-19 infections. COVID-19–infected patients harbor an expanded population of circulating monocytes that secrete both IL-6 and IL-1β (Wen et al., 2020 Preprint; Zhang et al., 2020 Preprint); as a result, patients with COVID-19 have elevated levels of serum IL-6, as well as lactate dehydrogenase levels, compared with healthy controls (Chen et al., 2020a). Circulating lactate dehydrogenase is a marker of pyroptosis—a form of nonprogrammed cell death driven primarily by inflammasome-mediated IL-1β production that results in release of cytoplasmic proteins and factors (Rayamajhi et al., 2013). In particular, the severity of IL-6 elevation correlates with the need for mechanical ventilation and ultimately with mortality, likely reflecting the distinct role that IL-6 plays in amplifying the innate immune response by recruiting additional immune mediators (Chen et al., 2020a). This is in contrast to IL-1β, which is generated as a precursor transcript and is cleaved in response to inflammasome activation, after which it acts locally to enhance neutrophil cytotoxicity (Martinon et al., 2002). These elevations in innate immune cytokines have led to the hypothesis that an innate immune-mediated “cytokine storm,” similar to the cytokine release syndrome (CRS) observed in patients receiving treatment with chimeric antigen receptor–transduced T cells (CAR-T), is primarily responsible for the toxicity and end-organ damage mediated by COVID-19 infections (Grupp et al., 2013; Mehta et al., 2020). A role for cytokine-driven neutrophil mobilization in COVID-associated lung toxicity may explain why neutrophilia, despite the absence of secondary bacterial infections, is associated with mortality (Lagunas-Rangel, 2020), and why administration of monoclonal antibodies targeting IL-6 has shown initial clinical promise (Gritti et al., 2020).
However, caution is warranted before invoking IL-6–mediated CRS as the sole pathological driver in severe COVID-19 infections. First, COVID-19 patients lacks most of the hallmarks of CRS, including hypotension, capillary leak syndrome, and neurotoxicity (Hay et al., 2017). Second, the clinical course of CRS is far more acute than that seen in COVID-19 infections, with fever occurring within 2 d and neurotoxicity within 5 d (Neelapu et al., 2018). Accordingly, serum IL-6 levels are far lower in COVID-19 infections than in CRS, with peak levels typically less than 100 pg/ml in COVID-19, compared with 1,000–10,000 pg/ml in CRS (Chen et al., 2020a; Maude et al., 2014). Third, deaths in COVID-19–infected patients appear to be due to primary respiratory failure, rather than from distributive shock or status epilepticus, as was predominantly seen in CRS-associated deaths (Lee et al., 2014).
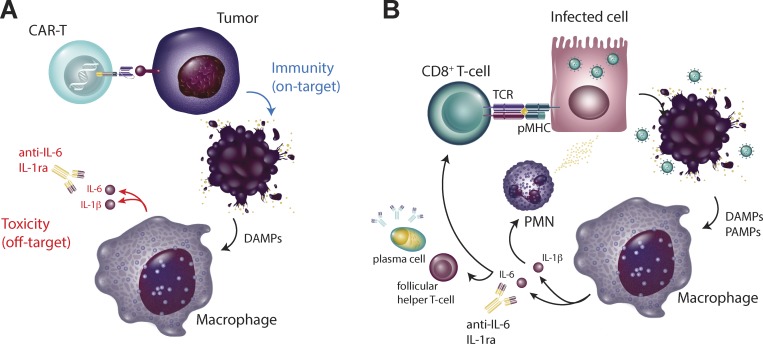
Perhaps most importantly, the clinical results of therapeutic blockade of circulating IL-6 with tocilizumab or siltuximab are thus far mixed; one clinical trial showed evidence of improvement in 33% of patients, at least suggesting that in many cases, a hyperinflammatory response is not primarily responsible for COVID-19–associated morbidity and mortality (Gritti et al., 2020). This may be in part due to ongoing IL-6 production that overcomes direct targeting and may explain the more favorable early data with IL-6 receptor blockade (Xu et al., 2020a Preprint). However, persistent symptoms in the face of IL-6 blockade also makes sense when one considers the underlying etiology of cytokine release in viral infections as compared with CAR-T cell therapy (Fig. 2). In CAR-T–mediated CRS, macrophage-dependent production of IL-1 and IL-6 occurs secondary to T cell–mediated killing of tumor cells (Giavridis et al., 2018; Norelli et al., 2018). Inhibiting this secondary innate response prevents immune-mediated toxicity while permitting ongoing CAR-T–dependent antitumor efficacy. In the case of viral infections such as COVID-19, however, macrophage activation is occurring as a primary response to viral infection (Tate et al., 2016). Dampening the innate immune response in this setting is likely to mitigate off-target toxicity but may be permissive for viral dissemination in the absence of an alternative source of viral control, either through pharmacologic therapy or alternative means of immune-mediated control. It is also worth noting that while many arms of the innate immune response are potently activated by COVID-19, the type I and type III interferon response appears to be muted in response to COVID-19 infection (Blanco-Melo et al., 2020 Preprint), suggesting that some aspects of the innate immune response to COVID-19 might actually benefit from careful amplification (Kotenko et al., 2003). Finally, IL-6 was initially described as a member of the type I interferon family (IFN-β2; Revel and Zilberstein, 1987) but was later found to have no intrinsic antiviral activity (Reis et al., 1988). This suggests that antiviral activity and host tissue toxicity might be at least partially uncoupled, and therefore inhibition of select arms of the innate immune response (such as IL-6–mediated neutrophil recruitment) could limit tissue toxicity while permitting ongoing antiviral T and B cell–mediated adaptive immunity and memory.
Figure 2.
Distinctions between CAR-T and virally mediated hypercytokinemia. (A and B) During CAR-T cell–driven CRS (A), blockade of macrophage-derived IL-1 and IL-6 limits tissue toxicity without interfering with antitumor immunity. However, during viral infections (B), blockade of macrophage function may impair both innate and adaptive viral control.
COVID-19: The case for adaptive immune dysregulation
Regardless of whether innate immune-mediated toxic inflammation contributes to COVID-19–related morbidity and mortality, it is clear that viral dissemination is a key driver of severe disease. The rare histopathologic specimens obtained either postmortem or via liver or kidney biopsy in COVID-19–infected patients have almost universally revealed the presence of inclusion bodies, consistent with viral persistence (Chen et al., 2020 Preprint; Diao et al., 2020 Preprint; Yao et al., 2020). Furthermore, detection of circulating viral RNA in the peripheral blood is strongly linked to disease severity (Chen et al., 2020 Preprint).
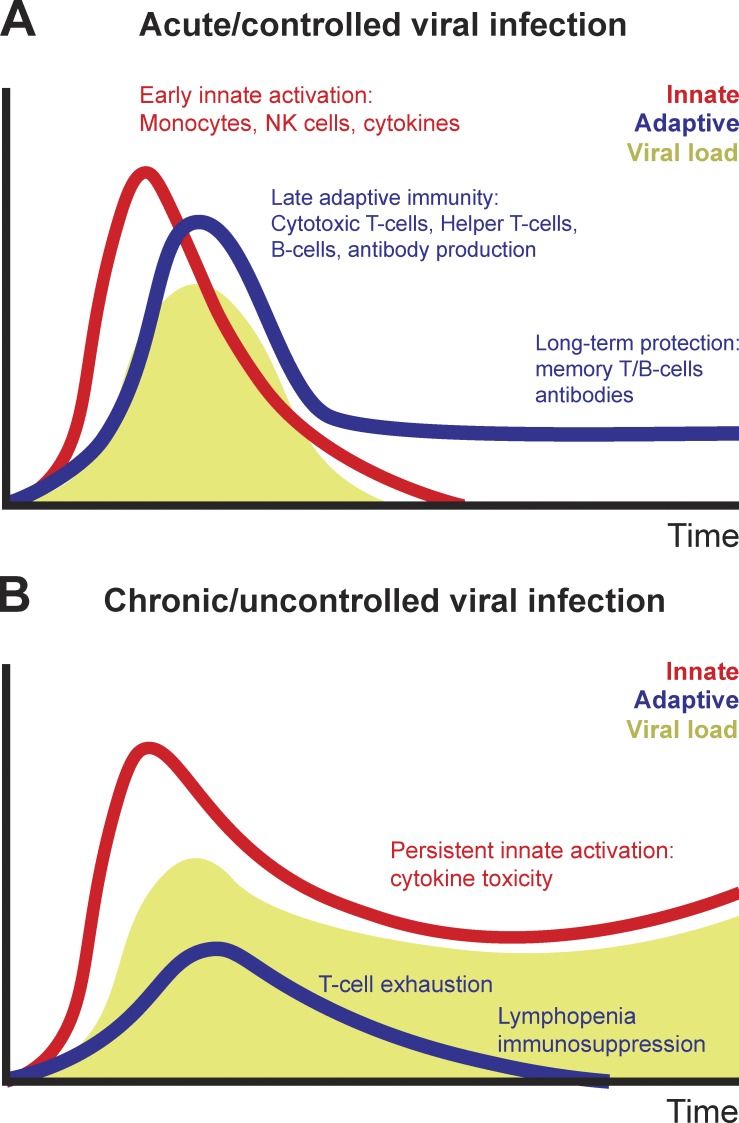
What is permitting viral dissemination in patients who succumb to COVID-19 infections? Insufficient activation of type I and type III interferons is undoubtedly a key contributor to innate immune failure to control viral persistence. In addition, decades of mechanistic work in immunology have demonstrated that an intact T cell–mediated adaptive immune response is essential for clearing and maintaining long-term suppression of viral infections; this is supported by the significantly increased risk of viral reactivation in patients whose adaptive immune system is suppressed (Broers et al., 2000; Shah et al., 1974). During acute viral infections, virally derived peptides activate both naive CD8+ and CD4+ T cell proliferation and differentiation. Effective viral clearance, which occurs within a week of initial infection, requires both CD8+ effector T cell–mediated killing of virally infected cells as well as CD4+ T cell–dependent enhancement of CD8+ and B cell responses (Ahmed et al., 1984). However, T cell–dependent cytokine release and direct cellular cytotoxicity can also contribute to tissue inflammation and toxicity and accelerate mortality particularly during severe viral infections. Indeed, the ability of T cell responses to cause tissue damage explains the importance of endogenous inhibitory immunoreceptors (otherwise known as immune “checkpoints”) in limiting effector T cell responses (Barber et al., 2006). Following viral clearance, the majority of virus-specific T cells undergo apoptosis; however, retention of a virus-specific memory T cell population is required for long-term antiviral immunity (Fig. 3 A). The importance of the adaptive immune response in viral clearance suggests that chronic viral infections must, by definition, either evade or suppress adaptive immunity. Over 80 yr ago, genetic variants of viral strains that persistently infected immunocompetent mice were identified (Traub, 1936). Rather than failing to activate T cell responses, these viral infections were characterized by persistent antigenic activation of T cells, ultimately driving a nonresponsive cell state known as T cell “exhaustion” (Shin and Wherry, 2007). This phenotype has been described in numerous chronic viral infections and is often accompanied by lymphopenia (Fig. 3 B; Moskophidis et al., 1993).
Figure 3.
Immune dysregulation during chronic viral infections. (A and B) During acute viral infections (A), early innate and adaptive immune function lead to viral suppression, followed by development of adaptive immunity. During chronic viral infections (B), persistent virus leads to T cell depletion and exhaustion while triggering ongoing innate immune inflammation. NK, natural killer.
Is the clinical toxicity observed during COVID-19 infection a product of adaptive immune hyperactivity or suppression? The clinical presentation of COVID-19 infection is more consistent with a subacute rather than an acute viral illness. Compared with H1N1 influenza infections, in which the median incubation time was 2 d and the majority of intensive care unit admissions occurred within 24–48 h of admission (Bautista et al., 2010), COVID-19–infected patients present to the hospital with a median incubation time of 5–7 d and are typically hospitalized for an additional 3–4 d before requiring intensive care unit admission and/or mechanical ventilation (Huang et al., 2020). This subacute pattern of progression raises the possibility that immunosuppression, due both to T cell depletion and exhaustion, contributes to COVID-19 viral persistence and mortality. Lymphopenia is the most consistent laboratory abnormality in COVID-19–infected patients. Notably, progressive lymphodepletion is observed in patients who clinically deteriorate during COVID-19 infection, whereas recovery of lymphocyte counts tends to directly precede clinical recovery (Chen et al., 2020 Preprint). Similar to T cells from both mice and humans with chronic viral infections (Day et al., 2006; Fisicaro et al., 2017; Wherry et al., 2007), both lung-resident and circulating T cells from COVID-19 patients potently up-regulate markers of T cell exhaustion, including PD-1 and Tim-3 (Diao et al., 2020 Preprint). Lung-infiltrating CD8+ T cells from severe COVID-19 patients exhibit transcriptional hallmarks of terminal T cell exhaustion including expression of CCL4 and GZMB; notably, the most expanded T cell population in the bronchoalveolar lavage of severe COVID-19 patients is marked by expression of MKI67 and TYMS, genes that are specifically up-regulated in terminally exhausted CD8+ T cells extracted from melanoma tumors (Sade-Feldman et al., 2018). Conversely, single-cell sequencing of peripheral blood mononuclear cells of patients recovering from COVID-19 infection shows signs of clonal expansion, T cell activation, and T cell memory formation, consistent with an effective adaptive immune response (Wen et al., 2020 Preprint). Notably, both lymphopenia and T cell exhaustion have been observed in recent viral pandemics (Box 1). This does not rule out a contribution of CD8+ T cells to inflammation and lung pathology; indeed, it is worth noting that many markers of T cell exhaustion are also up-regulated in effector T cells, and lung-infiltrating T cells from more mild COVID infections significantly up-regulated genes related to T cell activation. We suggest that CD8+ T cell inflammation–driven lung pathology occurs early in disease during the rapid expansion of short-lived effector CD8+ T cells after which persistent viral antigen may lead to T cell inactivation, exhaustion, and depletion. A role for the adaptive immune system in suppressing COVID-19 viral dissemination may explain the association of COVID-19 disease severity with age. Age-related waning of adaptive immune function, also known as “immunosenescence,” is characterized by a loss of T cell clonal diversity and a contraction of naive T cells with proliferative capacity (Youm et al., 2012). A smaller, more restricted T cell repertoire is likely more prone to antigen-mediated exhaustion during chronic viral infections; conversely, the relatively diverse and expanded pool of naive T cells may explain the relatively diminished severity of COVID-19 infections in children (Xu et al., 2020b).
Box 1. Are there immunological similarities between COVID-19 and recent pandemics?
In the 2003 SARS-CoV pandemic, single stranded RNA fragments from SARS-CoV viral particles were shown to markedly activate TLR-mediated innate immune responses (Li et al., 2013). Lymphopenia also occurred in the overwhelming majority of patients (Lee et al., 2003; Poutanen et al., 2003; Tsang et al., 2003) and was associated with severe disease (Peiris et al., 2003; Wong et al., 2003). SARS also significantly affected children, in whom lymphopenia was also a common feature (Hon et al., 2003). Conversely, development of virus-specific memory T cells was associated with resolution of disease and protection from subsequent infection (Channappanavar et al., 2014; Peng et al., 2006).
In the 2009 H1N1 influenza A pandemic, lymphopenia occurred in the majority of patients (Cao et al., 2009; Perez-Padilla et al., 2009). In stark contrast to COVID-19, H1N1 occurred frequently and caused significant morbidity in children; of note, H1N1 infection caused lymphopenia in the majority of pediatric patients (Rhim et al., 2011). Biologically, modeling of H1N1 infection in mice revealed high levels of oxidative stress within the lungs (Chandler et al., 2016); this was associated with marked elevation of systemic innate inflammatory factors including MCP-1 and IL-6 (Gao et al., 2013), and elevated IL-6 correlated with disease severity (Bradley-Stewart et al., 2013; Hagau et al., 2010). Notably, peripheral blood mononuclear cells from patients showed intact innate immune responses to heat-killed bacteria, but defective responses to T cell stimulation (Giamarellos-Bourboulis et al., 2009). This was associated with up-regulation of inhibitory immunoreceptors such as PD-1 and PD-L1 in the lungs and peripheral blood of patients (Erickson et al., 2012; Valero-Pacheco et al., 2013).
In the 2013 MERS-CoV pandemic, lymphopenia occurred somewhat less frequently (Assiri et al., 2013) but was also independently associated with disease severity, and recovery was associated with improved outcomes (Ko et al., 2016; Min et al., 2016). This defect was specific to MERS-CoV–specific CD8+ T cells, which inversely correlated with disease severity and duration of viral shedding (Zhao et al., 2017). Elevations in serum IL-6 levels were also observed in patients whose clinical course worsened (Kim et al., 2016).
Therapeutically, high-dose glucocorticoids are the only strategy used thus far to modulate the adaptive immune system of COVID-19 patients. While not studied in a controlled fashion, available data thus far suggest that high-dose systemic steroids are at best ineffective and possibly harmful. Clinical worsening following glucocorticoid administration is consistent with a contributing role for T cell suppression in viral persistence and is in line with evidence showing a similar lack of efficacy of glucocorticoids in other severe viral infections (Arabi et al., 2018; Lansbury et al., 2019). Conversely, treatments aimed at reducing T cell exhaustion or death have shown efficacy in promoting clearance of chronic viral infections. For example, both treatment with cytokines that enhance T cell self-renewal, such as IL-7, and blockade of inhibitory immunoreceptor-based interactions that suppress T cell proliferation, such as PD-1/PD-L1, have individually been shown to promote antiviral immunity (Barber et al., 2006; Pellegrini et al., 2011). One commonly used therapeutic strategy in severe viral infections, but not yet in COVID-19–infected patients, is the use of antioxidants such as N-acetylcysteine (N-Ac). N-Ac may be particularly beneficial in clinical syndromes driven by high redox stress, such as viral infections and ARDS (Lenz et al., 1999). Accordingly, N-Ac has shown modest benefit in ARDS (Suter et al., 1994; Zhang et al., 2017) as well in influenza (Lai et al., 2010), dengue (Abeysekera et al., 2012; Kumarasena et al., 2010; Senanayake et al., 2013), rotavirus (Guerrero et al., 2014), HIV (Akerlund et al., 1996; Eylar et al., 1993; Look et al., 1998), and viral hepatitis (Sotelo et al., 2009). While N-Ac is likely to have multiple benefits in patients with chronic viral infections, including protection of lung pneumocytes, hepatocytes, and colonic epithelial cells from virally driven apoptosis, it has the notable ability to reverse lymphopenia in this setting, suggesting that oxidative stress may contribute to lymphopenia during chronic viral infection.
An integrated immune-based approach
Is there a way to therapeutically balance immune toxicity and immunosuppression to improve outcomes in COVID-19–infected patients? To answer to this question, we require a deeper understanding of both innate and adaptive immune evolution over the course of COVID-19 illness. To this end, there is a need to obtain serial peripheral blood, and, when technically feasible and clinically appropriate, bronchoalveolar lavage samples over the course of COVID-19 infection and clearance. Longitudinal assessment of patients during the course of illness is essential, as it may help determine whether distinct clinical presentations are driven by differential immune responses. For example, it remains unclear whether the antiviral immune responses in patients who rapidly develop respiratory failure upon hospital admission are distinct from those in patients who develop respiratory failure 7–10 d after hospital admission. The answer to this question will have key implications for the appropriate design of clinical trials aimed as modulating the anti-COVID immune response. Longitudinal assessment of anti-COVID immune responses is particularly important in the setting of therapeutic interventions, both antiviral and immune-targeted. For example, patients who respond to IL-6 blockade have a parallel improvement in lymphocyte counts (Xu et al., 2020a Preprint), suggesting that in some cases, COVID-19–associated lymphopenia may be a product of bystander inflammation (Stelekati et al., 2014).
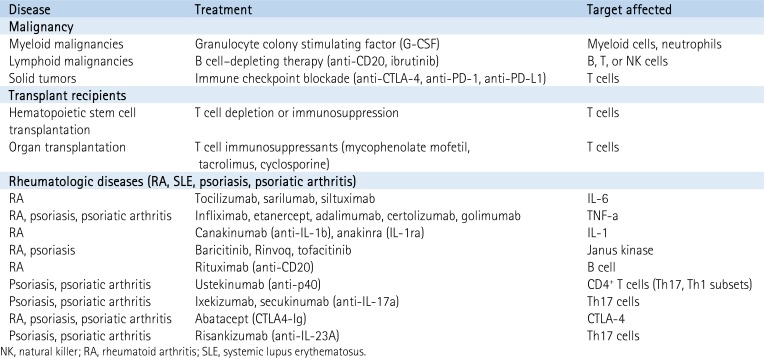
Longitudinal sampling of COVID-19–infected patients with either inherent or iatrogenic disruption of host immunity may help identify specific regulators of the anti-COVID immune response (Box 2). A few key patient cohorts are worth noting. First, patients with malignancies have in many cases discrete defects in B cell, T cell, and myeloid cell maturation and are often treated with agents that affect individual immune cell subpopulations. Second, patients with solid organ transplants require ongoing T cell immunosuppression, offering an opportunity to explore the contribution of the T cell–dependent immune response to COVID-19. Finally, patients with rheumatologic diseases often have dysregulation of innate immunity, due either to their disease itself or to the multiple distinct innate immune-targeted pharmacological agents that are Food and Drug Administration–approved for treatment for rheumatologic conditions.
Box 2. Monitoring the natural history of COVID-19 infection in immunologically distinct patient populations
An intriguing way to explore the contribution of individual components of the immune system to anti–COVID-19 immunity is to monitor outcomes and biomarkers in patients with either native defects or iatrogenic manipulation of their immune system.
In the absence of clear biological data, however, and given the urgent need for novel therapeutic options, we offer our perspective on the anti–COVID-19 immune response. While both innate and adaptive immune activation likely contribute to COVID-19–mediated toxicity, decades of mechanistic work support a requirement for a functional immune system to achieve durable antiviral control. Given the critical role of the immune response in this context, we would caution against immunosuppression without a mechanism of antiviral control, either pharmacologically or through activation of innate or adaptive immunity. We also suggest that adequate T cell homeostasis is not only predictive of but required for successful viral clearance and clinical improvement.
Therefore, when exploring blockade of the innate immune response, we recommend approaching patients with early decompensation differently from patients who deteriorate later during their hospitalization, as these are likely to reflect immunologically distinct sets of patients. In patients with early, rapid deterioration and clinical and laboratory hallmarks of increased inflammation (fevers, shock, and elevations in IL-6 and C-reactive protein), attenuation of the peak immune response, either with corticosteroids or more specifically targeting of IL-6 or IL-1β, may limit bystander tissue toxicity during the early immune response. However, early immune hyperactivity may be a reflection of high viral burden, and blockade of the immune response may simply mask an appropriate response to a significant viral infection. We therefore advise that any immunosuppression be for a limited period of time, and that patients who receive early immunosuppression be monitored to ensure that they do not recrudesce with severe disease.
Furthermore, we recommend particular caution when considering early immunosuppression in patients with signs of underlying adaptive immune dysfunction, be it due to host risk factors or severe disease-related lymphopenia, as these patients may be at risk of viral dissemination. In patients who receive IL-6 blockade, careful follow-up and antibody serology should be considered to ensure development of anti-COVID IgG-mediated humoral immunity.
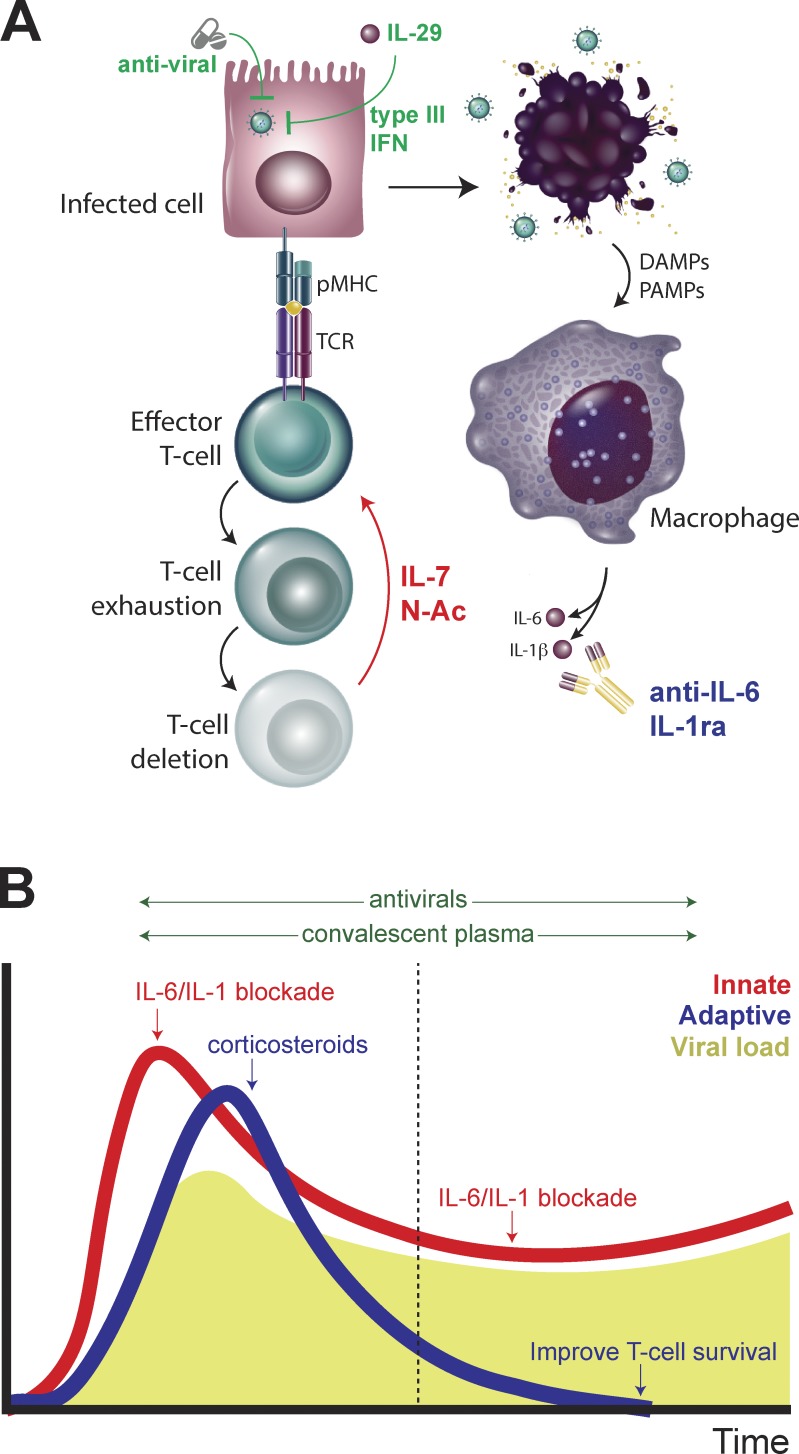
In patients who develop respiratory failure later in their hospital course, the negative consequences of ongoing innate immune activation may indeed support IL-6 blockade. However, the severe lymphopenia, T cell exhaustion, and consequent adaptive immunosuppression present in these patients may worsen the consequences of targeting innate immunity and, furthermore, may impede a much-needed protective antibody response. We urge caution when considering IL-6 blockade as monotherapy in this setting and suggest that it may best be paired with antiviral therapy. Furthermore, in these patients, as well as those with minimal or transient response to IL-6 blockade, we propose that treatments aimed at cautiously enhancing either innate immune-mediated antiviral immunity (Kotenko et al., 2003) or adaptive T cell–mediated immunity may be beneficial. These may include but are not limited to activation of type I or type III interferon responses, blocking reactive oxygen species–mediated T cell death, or promoting T cell homeostasis and/or proliferation. In either early or late disease, an effective humoral response is a key contributor to viral clearance; here, we draw particular attention to the promise of treating infected patients with serum from convalescent patients that is rich in immunoglobulins targeting COVID-19 (Duan et al., 2020). This may help overcome the impairment in humoral immunity imposed by IL-6 blockade. Development of monoclonal antibodies targeting the SPIKE glycoprotein may serve a similar purpose without subjecting patients to the risks of viral contamination from convalescent donors (Wrapp et al., 2020). These potential therapeutic approaches are described in Fig. 4.
Figure 4.
Enhancing innate and adaptive immunity to combat COVID-19 infections. (A) Enhancing antiviral sensing through activation of type I interferon responses or adaptive immunity by rescuing T cells from exhaustion-dependent cell death may improve anti–COVID-19 immunity, particularly in the context of blocking macrophage-dependent cytokine production. (B) Distinct strategies for modulating innate and adaptive immune responses during early and late COVID-19 infection may lead to more effective viral control.
Conclusions
We are in the very nascent stages of our understanding of how the interaction between innate and adaptive immunity mediates both viral control and host toxicity during severe COVID-19 and other pandemic viral infections. Despite limitations in our capacity to investigate the mechanistic underpinnings of COVID-19–driven pathology, we can draw upon knowledge gained from years of fundamental work in viral immunology to rapidly select and test treatment options that recruit both innate and adaptive immune mechanisms to prevent and treat this and future viral pandemics.
Acknowledgments
S.A. Vardhana is a Parker Fellow with the Parker Institute of Cancer Immunotherapy and is supported by a Burroughs Wellcome Fund Career Award for Medical Scientists. This work was additionally supported by the Memorial Sloan-Kettering Cancer Center support grant P30 CA008748.
Author contributions: S.A. Vardhana and J.D. Wolchok conceived and wrote the manuscript.
References
- Abeysekera R.A., Illangasekera U., Jayalath T., Sandeepana A.G., and Kularatne S.A.. 2012. Successful use of intravenous N-acetylcysteine in dengue haemorrhagic fever with acute liver failure. Ceylon Med. J. 57:166–167. 10.4038/cmj.v57i4.5085 [DOI] [PubMed] [Google Scholar]
- Ahmed R., Salmi A., Butler L.D., Chiller J.M., and Oldstone M.B.. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 160:521–540. 10.1084/jem.160.2.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerlund B., Jarstrand C., Lindeke B., Sönnerborg A., Akerblad A.C., and Rasool O.. 1996. Effect of N-acetylcysteine(NAC) treatment on HIV-1 infection: a double-blind placebo-controlled trial. Eur. J. Clin. Pharmacol. 50:457–461. 10.1007/s002280050140 [DOI] [PubMed] [Google Scholar]
- Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., Jose J., Pinto R., Al-Omari A., Kharaba A., et al. ; Saudi Critical Care Trial Group . 2018. Corticosteroid Therapy for Critically Ill Patients with Middle East Respiratory Syndrome. Am. J. Respir. Crit. Care Med. 197:757–767. 10.1164/rccm.201706-1172OC [DOI] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., et al. 2013. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 13:752–761. 10.1016/S1473-3099(13)70204-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., and Wang M.. 2020. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 323:1406 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., and Ahmed R.. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 439:682–687. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Bautista E., Chotpitayasunondh T., Gao Z., Harper S.A., Shaw M., Uyeki T.M., Zaki S.R., Hayden F.G., Hui D.S., Kettner J.D., et al. ; Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza . 2010. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 362:1708–1719. 10.1056/NEJMra1000449 [DOI] [PubMed] [Google Scholar]
- Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., Greninger A.L., Pipavath S., Wurfel M.M., Evans L., et al. 2020. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. N. Engl. J. Med. NEJMoa2004500 10.1056/NEJMoa2004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W., Moller R., Panis M., Sachs D., Albrecht R.A., and tenOever B.R.. 2020. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv https://www.biorxiv.org/content/10.1101/2020.03.24.004655v1 (Preprint posted March 24, 2020).
- Bradley-Stewart A., Jolly L., Adamson W., Gunson R., Frew-Gillespie C., Templeton K., Aitken C., Carman W., Cameron S., and McSharry C.. 2013. Cytokine responses in patients with mild or severe influenza A(H1N1)pdm09. J. Clin. Virol. 58:100–107. 10.1016/j.jcv.2013.05.011 [DOI] [PubMed] [Google Scholar]
- Broers A.E., van Der Holt R., van Esser J.W., Gratama J.W., Henzen-Logmans S., Kuenen-Boumeester V., Löwenberg B., and Cornelissen J.J.. 2000. Increased transplant-related morbidity and mortality in CMV-seropositive patients despite highly effective prevention of CMV disease after allogeneic T-cell-depleted stem cell transplantation. Blood. 95:2240–2245. 10.1182/blood.V95.7.2240 [DOI] [PubMed] [Google Scholar]
- Cao B., Li X.W., Mao Y., Wang J., Lu H.Z., Chen Y.S., Liang Z.A., Liang L., Zhang S.J., Zhang B., et al. ; National Influenza A Pandemic (H1N1) 2009 Clinical Investigation Group of China . 2009. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N. Engl. J. Med. 361:2507–2517. 10.1056/NEJMoa0906612 [DOI] [PubMed] [Google Scholar]
- Chandler J.D., Hu X., Ko E.J., Park S., Lee Y.T., Orr M., Fernandes J., Uppal K., Kang S.M., Jones D.P., et al. 2016. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 311:R906–R916. 10.1152/ajpregu.00298.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Zhao J., Meyerholz D.K., and Perlman S.. 2014. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 88:11034–11044. 10.1128/JVI.01505-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. 2020a Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest.:137244 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ling J., Mo P., Zhang Y., Jiang Q., Ma Z., Cao Q., Hu W., Zou S., Chen L., et al. 2020. b. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. medRxiv 10.1101/2020.03.03.20030437 (Preprint posted March 6, 2020). [DOI]
- Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. 2020. c. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely associated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. medRxiv 10.1101/2020.02.29.20029520 (Preprint posted March 3, 2020). [DOI] [PMC free article] [PubMed]
- Chen Y., Feng Z., Diao B., Wang R., Wang G., Wang C., Tan Y., Liu L., Wang C., Liu Y., et al. 2020. d. The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Directly Decimates Human Spleens and Lymph Nodes. medRxiv 10.1101/2020.03.27.20045427 (Preprint posted March 31, 2020). [DOI]
- Day C.L., Kaufmann D.E., Kiepiela P., Brown J.A., Moodley E.S., Reddy S., Mackey E.W., Miller J.D., Leslie A.J., DePierres C., et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 443:350–354. 10.1038/nature05115 [DOI] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., et al. 2020. a. Reduction and functional exhaustion of T-cells in patients with Coronavirus disease 2019 (COVID-19). medRxiv 10.1101/2020.02.18.20024364 (Preprint posted February 20, 2020). [DOI] [PMC free article] [PubMed]
- Diao B., Wang C., Wang R., Feng Z., Tan Y., Wang H., Wang C., Liu L., Liu Y., Liu Y., et al. 2020. b. Human Kidney is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. medRxiv 10.1101/2020.03.04.20031120 (Preprint posted April 10, 2020). [DOI]
- Diebold S.S., Kaisho T., Hemmi H., Akira S., and Reis e Sousa C.. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 303:1529–1531. 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., et al. 2020. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA. 202004168 10.1073/pnas.2004168117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.J., Gilchuk P., Hastings A.K., Tollefson S.J., Johnson M., Downing M.B., Boyd K.L., Johnson J.E., Kim A.S., Joyce S., et al. 2012. Viral acute lower respiratory infections impair CD8+ T cells through PD-1. J. Clin. Invest. 122:2967–2982. 10.1172/JCI62860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eylar E., Rivera-Quinones C., Molina C., Báez I., Molina F., and Mercado C.M.. 1993. N-acetylcysteine enhances T cell functions and T cell growth in culture. Int. Immunol. 5:97–101. 10.1093/intimm/5.1.97 [DOI] [PubMed] [Google Scholar]
- Fisicaro P., Barili V., Montanini B., Acerbi G., Ferracin M., Guerrieri F., Salerno D., Boni C., Massari M., Cavallo M.C., et al. 2017. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med. 23:327–336. 10.1038/nm.4275 [DOI] [PubMed] [Google Scholar]
- Gao R., Bhatnagar J., Blau D.M., Greer P., Rollin D.C., Denison A.M., Deleon-Carnes M., Shieh W.J., Sambhara S., Tumpey T.M., et al. 2013. Cytokine and chemokine profiles in lung tissues from fatal cases of 2009 pandemic influenza A (H1N1): role of the host immune response in pathogenesis. Am. J. Pathol. 183:1258–1268. 10.1016/j.ajpath.2013.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Raftogiannis M., Antonopoulou A., Baziaka F., Koutoukas P., Savva A., Kanni T., Georgitsi M., Pistiki A., Tsaganos T., et al. 2009. Effect of the novel influenza A (H1N1) virus in the human immune system. PLoS One. 4 e8393 10.1371/journal.pone.0008393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavridis T., van der Stegen S.J.C., Eyquem J., Hamieh M., Piersigilli A., and Sadelain M.. 2018. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 24:731–738. 10.1038/s41591-018-0041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritti G., Raimondi F., Ripamonti D., Riva I., Landi F., Alborghetti L., Frigeni M., Damiani M., Mico C., Fagiuoli S., et al. 2020. Use of siltuximab in patients with COVID-19 pneumonia requiring ventilatory support. medRxiv 10.1101/2020.04.01.20048561 (Preprint posted April 15, 2020). [DOI]
- Grupp S.A., Kalos M., Barrett D., Aplenc R., Porter D.L., Rheingold S.R., Teachey D.T., Chew A., Hauck B., Wright J.F., et al. 2013. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 368:1509–1518. 10.1056/NEJMoa1215134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. ; China Medical Treatment Expert Group for Covid-19 . 2020. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. NEJMoa2002032 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C.A., Torres D.P., García L.L., Guerrero R.A., and Acosta O.. 2014. N-Acetylcysteine treatment of rotavirus-associated diarrhea in children. Pharmacotherapy. 34:e333–e340. 10.1002/phar.1489 [DOI] [PubMed] [Google Scholar]
- Hagau N., Slavcovici A., Gonganau D.N., Oltean S., Dirzu D.S., Brezoszki E.S., Maxim M., Ciuce C., Mlesnite M., Gavrus R.L., et al. 2010. Clinical aspects and cytokine response in severe H1N1 influenza A virus infection. Crit. Care. 14:R203 10.1186/cc9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Lewis G.M., Mack L., and Zuniga E.I.. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 334:825–829. 10.1126/science.1208421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay K.A., Hanafi L.A., Li D., Gust J., Liles W.C., Wurfel M.M., López J.A., Chen J., Chung D., Harju-Baker S., et al. 2017. Kinetics and biomarkers of severe cytokine release syndrome after CD19 chimeric antigen receptor-modified T-cell therapy. Blood. 130:2295–2306. 10.1182/blood-2017-06-793141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon K.L., Leung C.W., Cheng W.T., Chan P.K., Chu W.C., Kwan Y.W., Li A.M., Fong N.C., Ng P.C., Chiu M.C., et al. 2003. Clinical presentations and outcome of severe acute respiratory syndrome in children. Lancet. 361:1701–1703. 10.1016/S0140-6736(03)13364-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H., Wang H., et al. 2008. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 133:235–249. 10.1016/j.cell.2008.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R.F., Tabor D.R., Burks A.W., and Campbell G.D.. 1989. Elevated interleukin-1 release by human alveolar macrophages during the adult respiratory distress syndrome. Am. Rev. Respir. Dis. 140:1686–1692. 10.1164/ajrccm/140.6.1686 [DOI] [PubMed] [Google Scholar]
- Kim E.S., Choe P.G., Park W.B., Oh H.S., Kim E.J., Nam E.Y., Na S.H., Kim M., Song K.H., Bang J.H., et al. 2016. Clinical Progression and Cytokine Profiles of Middle East Respiratory Syndrome Coronavirus Infection. J. Korean Med. Sci. 31:1717–1725. 10.3346/jkms.2016.31.11.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Park G.E., Lee J.Y., Lee J.Y., Cho S.Y., Ha Y.E., Kang C.I., Kang J.M., Kim Y.J., Huh H.J., et al. 2016. Predictive factors for pneumonia development and progression to respiratory failure in MERS-CoV infected patients. J. Infect. 73:468–475. 10.1016/j.jinf.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., and Donnelly R.P.. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69–77. 10.1038/ni875 [DOI] [PubMed] [Google Scholar]
- Kuipers M.T., van der Poll T., Schultz M.J., and Wieland C.W.. 2011. Bench-to-bedside review: Damage-associated molecular patterns in the onset of ventilator-induced lung injury. Crit. Care. 15:235 10.1186/cc10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasena R.S., Mananjala Senanayake S., Sivaraman K., de Silva A.P., Dassanayake A.S., Premaratna R., Wijesiriwardena B., and de Silva H.J.. 2010. Intravenous N-acetylcysteine in dengue-associated acute liver failure. Hepatol. Int. 4:533–534. 10.1007/s12072-010-9176-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunas-Rangel F.A. 2020. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. J. Med. Virol. 10.1002/jmv.25819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K.Y., Ng W.Y., Osburga Chan P.K., Wong K.F., and Cheng F.. 2010. High-dose N-acetylcysteine therapy for novel H1N1 influenza pneumonia. Ann. Intern. Med. 152:687–688. 10.7326/0003-4819-152-10-201005180-00017 [DOI] [PubMed] [Google Scholar]
- Lansbury L., Rodrigo C., Leonardi-Bee J., Nguyen-Van-Tam J., and Lim W.S.. 2019. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst. Rev. 2 CD010406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., et al. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986–1994. 10.1056/NEJMoa030685 [DOI] [PubMed] [Google Scholar]
- Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., Grupp S.A., and Mackall C.L.. 2014. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 124:188–195. 10.1182/blood-2014-05-552729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz A.G., Jorens P.G., Meyer B., De Backer W., Van Overveld F., Bossaert L., and Maier K.L.. 1999. Oxidatively modified proteins in bronchoalveolar lavage fluid of patients with ARDS and patients at-risk for ARDS. Eur. Respir. J. 13:169–174. 10.1034/j.1399-3003.1999.13a31.x [DOI] [PubMed] [Google Scholar]
- Li Y., Chen M., Cao H., Zhu Y., Zheng J., and Zhou H.. 2013. Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 15:88–95. 10.1016/j.micinf.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look M.P., Rockstroh J.K., Rao G.S., Barton S., Lemoch H., Kaiser R., Kupfer B., Sudhop T., Spengler U., and Sauerbruch T.. 1998. Sodium selenite and N-acetylcysteine in antiretroviral-naive HIV-1-infected patients: a randomized, controlled pilot study. Eur. J. Clin. Invest. 28:389–397. 10.1046/j.1365-2362.1998.00301.x [DOI] [PubMed] [Google Scholar]
- Martinon F., Burns K., and Tschopp J.. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F., et al. 2014. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371:1507–1517. 10.1056/NEJMoa1407222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., and Janeway C.A. Jr.. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 388:394–397. 10.1038/41131 [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., and Manson J.J.; HLH Across Speciality Collaboration, UK . 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 395:1033–1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min C.K., Cheon S., Ha N.Y., Sohn K.M., Kim Y., Aigerim A., Shin H.M., Choi J.Y., Inn K.S., Kim J.H., et al. 2016. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 6:25359 10.1038/srep25359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis D., Lechner F., Pircher H., and Zinkernagel R.M.. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 362:758–761. 10.1038/362758a0 [DOI] [PubMed] [Google Scholar]
- Neelapu S.S., Tummala S., Kebriaei P., Wierda W., Gutierrez C., Locke F.L., Komanduri K.V., Lin Y., Jain N., Daver N., et al. 2018. Chimeric antigen receptor T-cell therapy - assessment and management of toxicities. Nat. Rev. Clin. Oncol. 15:47–62. 10.1038/nrclinonc.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norelli M., Camisa B., Barbiera G., Falcone L., Purevdorj A., Genua M., Sanvito F., Ponzoni M., Doglioni C., Cristofori P., et al. 2018. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 24:739–748. 10.1038/s41591-018-0036-4 [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., et al. ; SARS study group . 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 361:1319–1325. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M., Calzascia T., Toe J.G., Preston S.P., Lin A.E., Elford A.R., Shahinian A., Lang P.A., Lang K.S., Morre M., et al. 2011. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell. 144:601–613. 10.1016/j.cell.2011.01.011 [DOI] [PubMed] [Google Scholar]
- Peng H., Yang L.T., Li J., Lu Z.Q., Wang L.Y., Koup R.A., Bailer R.T., and Wu C.Y.. 2006. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 8:2424–2431. 10.1016/j.micinf.2006.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Padilla R., de la Rosa-Zamboni D., Ponce de Leon S., Hernandez M., Quiñones-Falconi F., Bautista E., Ramirez-Venegas A., Rojas-Serrano J., Ormsby C.E., Corrales A., et al. ; INER Working Group on Influenza . 2009. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N. Engl. J. Med. 361:680–689. 10.1056/NEJMoa0904252 [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., and Reis e Sousa C.. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- Pison U., Brand M., Joka T., Obertacke U., and Bruch J.. 1988. Distribution and function of alveolar cells in multiply injured patients with trauma-induced ARDS. Intensive Care Med. 14:602–609. 10.1007/BF00256763 [DOI] [PubMed] [Google Scholar]
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., et al. ; Canadian Severe Acute Respiratory Syndrome Study Team . 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995–2005. 10.1056/NEJMoa030634 [DOI] [PubMed] [Google Scholar]
- Rayamajhi M., Zhang Y., and Miao E.A.. 2013. Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol. Biol. 1040:85–90. 10.1007/978-1-62703-523-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis L.F., Le J.M., Hirano T., Kishimoto T., and Vilcek J.. 1988. Antiviral action of tumor necrosis factor in human fibroblasts is not mediated by B cell stimulatory factor 2/IFN-beta 2, and is inhibited by specific antibodies to IFN-beta. J. Immunol. 140:1566–1570. [PubMed] [Google Scholar]
- Revel M., and Zilberstein A.. 1987. Interferon-beta 2 living up to its name. Nature. 325:581–582. 10.1038/325581b0 [DOI] [PubMed] [Google Scholar]
- Rhim J.W., Lee K.Y., Youn Y.S., Kang J.H., and Kim J.C.. 2011. Epidemiological and clinical characteristics of childhood pandemic 2009 H1N1 virus infection: an observational cohort study. BMC Infect. Dis. 11:225 10.1186/1471-2334-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., et al. 2020. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med. 382:970–971. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. 2018. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 175:998–1013.e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake M.P., Jayamanne M.D., and Kankananarachchi I.. 2013. N-acetylcysteine in children with acute liver failure complicating dengue viral infection. Ceylon Med. J. 58:80–82. 10.4038/cmj.v58i2.5684 [DOI] [PubMed] [Google Scholar]
- Shah K.V., Daniel R.W., Zeigel R.F., and Murphy G.P.. 1974. Search for BK and SV40 virus reactivation in renal transplant recipients. Transplantation. 17:131–134. 10.1097/00007890-197401000-00022 [DOI] [PubMed] [Google Scholar]
- Shin H., and Wherry E.J.. 2007. CD8 T cell dysfunction during chronic viral infection. Curr. Opin. Immunol. 19:408–415. 10.1016/j.coi.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Sotelo N., de los Angeles Durazo M., Gonzalez A., and Dhanakotti N.. 2009. Early treatment with N-acetylcysteine in children with acute liver failure secondary to hepatitis A. Ann. Hepatol. 8:353–358. 10.1016/S1665-2681(19)31749-1 [DOI] [PubMed] [Google Scholar]
- Stelekati E., Shin H., Doering T.A., Dolfi D.V., Ziegler C.G., Beiting D.P., Dawson L., Liboon J., Wolski D., Ali M.A., et al. 2014. Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity. 40:801–813. 10.1016/j.immuni.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter P.M., Domenighetti G., Schaller M.D., Laverrière M.C., Ritz R., and Perret C.. 1994. N-acetylcysteine enhances recovery from acute lung injury in man. A randomized, double-blind, placebo-controlled clinical study. Chest. 105:190–194. 10.1378/chest.105.1.190 [DOI] [PubMed] [Google Scholar]
- Tate M.D., Ong J.D.H., Dowling J.K., McAuley J.L., Robertson A.B., Latz E., Drummond G.R., Cooper M.A., Hertzog P.J., and Mansell A.. 2016. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci. Rep. 6:27912 10.1038/srep27912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.T., Chambers R.C., and Liu K.D.. 2017. Acute Respiratory Distress Syndrome. N. Engl. J. Med. 377:562–572. 10.1056/NEJMra1608077 [DOI] [PubMed] [Google Scholar]
- Traub E. 1936. Persistence of Lymphocytic Choriomeningitis Virus in Immune Animals and Its Relation to Immunity. J. Exp. Med. 63:847–861. 10.1084/jem.63.6.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., et al. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977–1985. 10.1056/NEJMoa030666 [DOI] [PubMed] [Google Scholar]
- Valero-Pacheco N., Arriaga-Pizano L., Ferat-Osorio E., Mora-Velandia L.M., Pastelin-Palacios R., Villasís-Keever M.A., Alpuche-Aranda C., Sánchez-Torres L.E., Isibasi A., Bonifaz L., et al. 2013. PD-L1 expression induced by the 2009 pandemic influenza A(H1N1) virus impairs the human T cell response. Clin. Dev. Immunol. 2013 989673 10.1155/2013/989673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. 2020. Immune Cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. medRxiv 10.1101/2020.03.23.20039362 (Preprint posted March 31, 2020). [DOI] [PMC free article] [PubMed]
- Wherry E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wong R.S., Wu A., To K.F., Lee N., Lam C.W., Wong C.K., Chan P.K., Ng M.H., Yu L.M., Hui D.S., et al. 2003. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 326:1358–1362. 10.1136/bmj.326.7403.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., and McLellan J.S.. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 367:1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., et al. 2020. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.H.M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., et al. 2020a Effective treatment of severe COVID-19 patients with tocilizumab. chinaxiv http://chinaxiv.org/abs/202003.00026 (Preprint posted March 5, 2020). [DOI] [PMC free article] [PubMed]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., et al. 2020b Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 26:502–505. 10.1038/s41591-020-0817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., Mou H.M., Wang L.H., Zhang H.R., Fu W.J., et al. 2020. [A pathological report of three COVID-19 cases by minimally invasive autopsies]. Zhonghua Bing Li Xue Za Zhi. 49 E009. [DOI] [PubMed] [Google Scholar]
- Youm Y.H., Kanneganti T.D., Vandanmagsar B., Zhu X., Ravussin A., Adijiang A., Owen J.S., Thomas M.J., Francis J., Parks J.S., et al. 2012. The Nlrp3 inflammasome promotes age-related thymic demise and immunosenescence. Cell Rep. 1:56–68. 10.1016/j.celrep.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ding S., Li C., Wang Y., Chen Z., and Wang Z.. 2017. Effects of N-acetylcysteine treatment in acute respiratory distress syndrome: A meta-analysis. Exp. Ther. Med. 14:2863–2868. 10.3892/etm.2017.4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Guo R., Lei L., Liu H., Wang Y., Wang Y., Dai T., Zhang T., Lai Y., Wang J., et al. 2020. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv 10.1101/2020.03.24.20042655 (Preprint posted March 26, 2020). [DOI]
- Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., Layqah L.A., Alghamdi M.G., Al Gethamy M.M., Dada A.M., et al. 2017. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2:2 10.1126/sciimmunol.aan5393 [DOI] [PMC free article] [PubMed] [Google Scholar]