Abstract
Using carbohydrate microarrays, we characterized the carbohydrate binding activity of SARS-CoV neutralizing antibodies elicited by an inactivated SARS-CoV vaccine. In these antibodies, we detected undesired autoantibody reactivity specific for the carbohydrate moieties of an abundant human serum glycoprotein asialo-orosomucoid (ASOR). This observation provides important clues for the selection of specific immunologic probes to examine whether SARS-CoV expresses antigenic structures that mimic the host glycan. We found that lectin PHA-L (Phaseolus vulgaris L.), which is specific for a defined complex carbohydrate of ASOR, stained the SARS-CoV-infected cells specifically and intensively. Taken together, we present immunologic evidence that a carbohydrate structure of SARS-CoV shares antigenic similarity with host glycan complex carbohydrates. The experimental approaches we applied in this study are likely applicable for the identification of immunologic targets of other viral pathogens.
Keywords: microarray, carbohydrate, autoantigen, antibody and lectin, asialo-orosomucoid
sars-cov is a newly identified human coronavirus that caused an outbreak of severe acute respiratory syndrome (SARS) (4, 8, 11). Although substantial efforts have been made to study the etiologic agent of the disease, the carbohydrate structures of SARS-CoV remain largely uncharacterized. In this study, we introduced a glycan array-based approach to probe the carbohydrate-based antigenic structures of SARS-CoV. More specifically, we constructed glycan arrays to display carbohydrate antigens of defined structures and then applied these tools to detect carbohydrate-specific antibody “fingerprints” that were elicited by a SARS vaccine. We reasoned that if SARS-CoV expressed antigenic carbohydrate structures, then immunizing animals using the whole virus-based vaccines would have the possibility to elicit antibodies specific for these structures. In addition, if SARS-CoV displayed a carbohydrate structure that mimics host cellular glycans, then vaccinated animals may develop antibodies with autoimmune reactivity to their corresponding cellular glycans.
EXPERIMENTS AND RESULTS
Our laboratory has established a practical bioarray platform for the construction of carbohydrate microarrays (14, 16). Using this technology, we constructed a glycan array (Fig. 1, A and B)1 to display a collection of 51 carbohydrate antigens, including both microbial polysaccharides and cellular glycan complex carbohydrates. Blood group substances A, B, O, Lewis, I, and i antigens, and their precursors and structural derivatives were specially included. These complex carbohydrates are covalently attached to cellular proteins by either O- or N-glycosylation linkages as protein posttranslational modifications or linked to a membrane-bound lipid molecule. Therefore, scanning the antibody fingerprints of immunized or infected subjects using this glycan array is a specific immunologic approach to exploring the evidence of viral expression of corresponding complex carbohydrates.
Fig. 1.
Glycan arrays to characterize antibody profiles of vaccinated animals (A and B) and to scan for asialo-orosomucoid (ASOR)-specific immunological probe (C and D). Glycan arrays I and II were constructed in our laboratory. As shown in the stained array images, each antigen preparation at a given concentration was spotted as triplet replicate spots in parallel. Antigen preparations spotted on each glycan array and their array location are summarized in Supplemental Tables S1 and S2 (available at the Physiological Genomics web site). Microarray images were captured using a ScanArray 5000 Standard BioChip Scanning System (GSI Lumonics, and Packard BioChip Technologies). Methods for carbohydrate microarray construction, array staining, image capturing, and data processing were described in our previous publications (14, 16). A and B: glycan array I contains 51 antigens, and each antigen was printed at 0.5 mg/ml (∼0.5 ng/microspot) without further dilution. This array was applied to scan horse anti-Pn18 antiserum (A), as well as anti-SARS neutralizing antibodies (B). One microliter of each antiserum was applied to stain corresponding glycan array at 1:100 dilution in 1% BSA (wt/vol) phosphate-buffered saline (PBS), pH 7.4, containing 0.05% (wt/vol) Tween 20 and 0.025% (wt/vol) NaN3. The horse IgG antibodies captured by glycan arrays were visualized and quantified by using a rabbit anti-horse IgGFITC antibody. Locations of autoantigen ASOR were marked with an arrow key in each stained glycan array. The levels of IgG antibodies in the SARS-CoV vaccinated horse, as measured by the mean values of the fluorescence intensity minus background signal, are 9,043 and 1,494 for ASOR (B, coordinate D7) and agalacto-orosomucoid (AGOR) (B, coordinate D8), respectively. In the Streptococcus pneumoniae type 18 polysaccharide (Pn18) immunized horse, the values of anti-ASOR and anti-AGOR IgG activities are 448 (A, coordinate D7) and 501 (A, coordinate D8), respectively. This anti-ASOR-carbohydrate antibody reactivity was confirmed by multiple bioarray assays, as well as an antigen-specific ELISA assay (data not shown). C and D: glycan array II displays 24 antigens and was enriched with preparations of Gal-containing carbohydrate antigens. The initial antigen spotting concentration for glycan array II is 0.5 mg/ml (∼0.5 ng/microspot). These were further diluted at 1:3, 1:9, and 1:27. A panel of monoclonal antibodies and lectins with anti-carbohydrate activities were scanned using glycan array II. Two examples were shown in C and D. They were stained with corresponding biotinylated lectins at a concentration of 0.5 μg/ml. Streptavidin-Cy5 conjugate was applied to reveal specific staining signal. A: image of glycan array I stained by a horse antiserum elicited by Pn18 polysaccharide. This antibody preparation was adopted from the late Professor Elvin A. Kabat of Columbia University. B: image of glycan array I stained by a preparation of horse anti-SARS-CoV antisera. The antisera were prepared by J. Lu’s laboratory in the School of Public Health of Sun Yat-sen University, China. C: image of glycan array II stained by lectin PHA-L (Phaseolus vulgaris-L) staining. This lectin is specific for the tri- and tetra-antennary oligosaccharides containing Galβ1,4GlcNAc-linked units at nonreducing end (2). D: image of glycan array II stained by lectin GS-1-B4 (Griffonia simplicifolia I-B4 agglutinin). This lectin was considered to be highly specific for Galα1,3Gal, a xentogenic epitope of porcine tissues (17). Distance between adjacent microspots is 375 μm, center to center.
We applied this strategy to characterize horse antisera, including a preparation of anti-SARS-CoV neutralization antibodies with a viral neutralization titer of 1:10,240, a horse antiserum to Streptococcus pneumoniae type 18 (Pn18) polysaccharide, as well as preimmunized horse control serum specimens. As illustrated in Fig. 1A below, our bioarray scanning detected pathogen-specific antibody fingerprints in the Pn18 antiserum. A number of polysaccharide antigens with shared carbohydrate moieties were positively stained by this antiserum. As we expected, such anti-Pn polysaccharide reactivities were not seen in the horse neutralizing antibodies elicited by SARS-CoV. However, we detected significant levels of IgG antibodies to a human serum glycoprotein, asialo-orosomucoid (ASOR), in the anti-SARS-CoV antibodies (Fig. 1B).
This is an unexpected finding since ASOR is an abundant human serum glycoprotein. Previous investigations have identified a complex carbohydrate of ASOR, which is composed of the tri- and tetra-antennary oligosaccharides containing Galβ1,4GlcNAc-linked units at the nonreducing end (12, 13). The Galβ1,4GlcNAc sugar moiety is a core element of the blood group substance-related complex carbohydrates, being referred as type II sugar structures (17). There was no detectable antibody reactivity to agalacto-orosomucoid (AGOR), a degalactosyl derivative of ASOR, which differs from ASOR solely by the absence of the terminal galactosyl sugar residue. Therefore, the observed anti-ASOR antibody reactivity is likely specific for the sugar moieties of ASOR, and the terminal galactose (Gal) contributes significantly to the carbohydrate binding reactivity.
Expression of ASOR-like complex carbohydrates by coronaviruses is previous unrecognized. Like many mammalian viruses, SARS-CoV utilizes the host cellular machineries for protein glycosylation. Thus presence of viral carbohydrate moieties that share antigenic similarity with host glycans is theoretically possible. Examining whether SARS-CoV expresses such antigenic structures is important for us to understand the immune evasion mechanisms of SARS-CoV, as well as the carbohydrate-mediated viral-host interactions that may take place at various stages of the viral infection. Detection of anti-ASOR activity in SARS-CoV neutralizing antibodies provided us important clues for the selection of specific immunologic probes to monitor viral expression of such carbohydrate moiety.
Since terminal Gal residue is critical to the identified ASOR-carbohydrate epitope, we constructed the glycan array II (Fig. 1, C and D) to display a panel of Gal-related complex carbohydrates and applied this array to scan for the immunologic reagents that are highly specific for the Gal-related sugar chain cluster of ASOR. We discovered that lectin PHA-L (Phaseolus vulgaris L.; EY Laboratories, San Mateo, CA) is highly specific for ASOR (Fig. 1C, array location A6). In contrast, lectin GS-1 (Griffonia simplicifolia) showed no binding to ASOR, although this glycan array detected its selective binding of a number of Gal-containing complex carbohydrates (Fig. 1D).
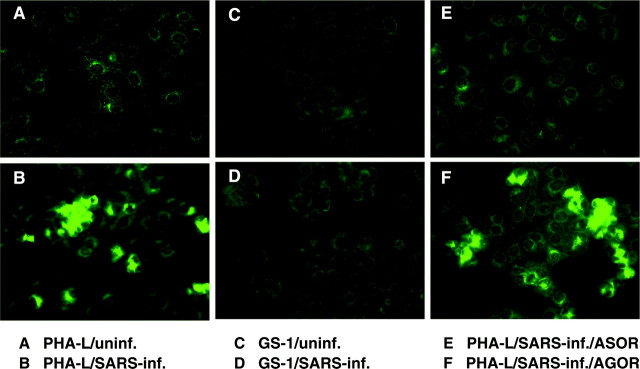
We further examined whether SARS-CoV expresses the sugar moieties that share antigenic similarity with the ASOR-associated complex carbohydrates, by using the above specific carbohydrate probes. A cell array was introduced to conduct this investigation (EUROIMMUN, Lübeck, Germany). This device was decorated with both SARS-CoV-infected and uninfected monkey Vero E6 cells at given array locations. Results illustrated in Fig. 2 showed that lectin PHA-L (Fig. 2B) but not GS-1 (Fig. 2D) positively stained the SARS-CoV-infected cells. Neither PHA-L (Fig. 2A) nor GS-1 (Fig. 2B) stained the uninfected cells under the same conditions. The staining specificity of PHA-L was further examined by a competition assay, whereby either ASOR (Fig. 2F) or AGOR (Fig. 2E) was applied as free inhibitor to compete with the binding. Only ASOR blocked completely the PHA-L staining of the SARS-CoV-infected cells. These results demonstrate that PHA-L recognition of SARS-CoV-infected cells is specific for the carbohydrate structures that share antigenic similarity with a complex carbohydrate of ASOR.
Fig. 2.
Lectin PHA-L specifically stains SARS-CoV-infected cells. Cell arrays from EUROIMMUN (Lübeck, Germany) were applied for this investigation. This array contains both SARS-CoV-infected Vero (Vero E6) cells and uninfected Vero cells and has been treated with a disinfecting fixing agent and was gamma-irradiated to inactivate infectious virus particles while preserving antigenic structures of SARS-CoV to ensure the test’s diagnostic sensitivity. Immunofluorescence staining was carried out as described (16). This experiment demonstrated that lectin PHA-L stained the SARS-CoV-infected Vero cells, whereas GS-1 did not. In addition, the PHA-L staining was completely blocked by ASOR. In contrast, AGOR did not inhibit the staining at the same concentration (50 μg/ml). A and C: noninfected Vero cells. B and D–F: SARS-CoV-infected Vero cells. A and B: PHA-LBI (0.5 μg/ml)/avidinFITC. C and D: GS-1BI (0.5 g/ml)/avidinFITC. E and F: PHA-LBI/avidinFITC in the presence of an inhibitor (50 μg/ml), either ASOR (E) or AGOR (F). BI, biotin; FITC, fluorescein.
DISCUSSION
Taken together, we have demonstrated that 1) there is present in SARS-CoV neutralization antibodies an undesired autoantibody reactivity; 2) this autoimmune reactivity is directed at the complex carbohydrate of an abundant human serum glycoprotein ASOR; 3) a specific immunologic probe, lectin PHA-L, was identified to detect this complex carbohydrate; and 4) PHA-L stains the SARS-CoV-infected cells specifically and intensively. Therefore, we have obtained sufficient immunologic evidence that a viral-expressed carbohydrate structure is responsible for the induction of the anti-ASOR autoimmunity in vaccinated animals.
These observations raise concerns on human use of the whole virus-based SARS vaccine that is produced by the monkey Vero E6 cell. It is necessary to eliminate the undesired autoimmunogenic activity of this preparation of inactivated SARS-CoV. Given the complexity of carbohydrate synthesis and substantial sugar chain variation among species and cell types (7, 15), it is possible to identify an alternative cell line or to genetically modify the Vero E6 cell line by altering its glycosylation pathway and thereby producing vaccines with enhanced efficacy without autoimmunogenic activity (5, 6). Developing different types of viral-free vaccines (9) is an important complement to the whole viral-based vaccination strategy.
Our investigation also raises the question about the involvement of autoimmune responses in SARS pathogenesis. ASOR is an abundant human serum glycoprotein, and the ASOR-type complex carbohydrates are also expressed by other host glycoproteins (3, 10). Thus the human immune system is generally nonresponsive to these “self” carbohydrate structures. However, when similar sugar moieties were expressed by a viral glycoprotein, their cluster configuration could differ significantly from those displayed by a cellular glycan, thereby generating a novel “non-self” antigenic structure. A documented example of such antigenic structure is a broad-range HIV-1 neutralization epitope recognized by a monoclonal antibody 2G12. This antibody is specific for a unique cluster of sugar chains displayed by the gp120 glycoprotein of HIV-1 (1). It is, thus, important to examine whether naturally occurring SARS-CoV expresses the ASOR-type autoimmune reactive sugar moieties. During a SARS epidemic spread, the viruses replicate in human cells. Their sugar chain expression may differ from the monkey cell-produced viral particles. Scanning of the serum antibodies of SARS patients using glycan arrays or other specific immunologic tools may provide information to clarify this question.
Collaborative efforts must be made to elucidate the structure of sugar chains that are responsible for the observed autoimmunogenic activity of SARS-CoV, to determine the glycoproteins displaying such sugar moieties, and to identify cellular receptors that bind SARS-CoV via specific protein-carbohydrate interaction. Since a number of cellular receptors that bind the ASOR complex carbohydrate have been identified (10, 12), this study provided clues to explore the possible roles of carbohydrate-mediated receptor-ligand interactions in SARS-CoV infection, especially in determining host-range and tissue-tropic characteristics of the virus. The experimental approaches we present here are likely applicable for the immunologic characterization of other viral pathogens.
Acknowledgments
We are grateful to Drs. Wenhua Ling and Huanying Zheng for many helpful discussions and to Dr. Shaoyi Liu, Dr. Chao Deng, and Brian Trummer for excellent technical assistance to this work. We also acknowledge the late Professor Elvin A. Kabat and previous students, postdoctoral fellows, and collaborators for their contributions to the collection of carbohydrate antigens and antibodies that were applied in this study.
D. Wang and J. Lu contributed equally to this work.
Article published online before print. See web site for date of publication (http://physiolgenomics.physiology.org).
Address for reprint requests and other correspondence: D. Wang, Dept. of Genetics, Stanford Univ. School of Medicine, Beckman Center B011, Stanford, CA 94305-5318 (E-mail: wangd@cgcmail.cpmc.columbia.edu or dwang1@stanford.edu).
10.1152/physiolgenomics.00102.2004
Footnotes
The Supplementary Material for this article (Supplemental Tables S1 and S2, see legend to Fig. 1) is available online at http://physiolgenomics.physiology.org/cgi/content/full/00102.2004/DC1.
REFERENCES
- 1.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, and Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science : 2065–2071, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Cummings RD and Kornfeld S. Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. J Biol Chem : 11230–11234, 1982. [PubMed] [Google Scholar]
- 3.Cummings RD and Kornfeld S. The distribution of repeating [Galβ1,4GlcNAcβ1,3] sequences in asparagine-linked oligosaccharides of the mouse lymphoma cell lines BW5147 and PHAR 2.1. J Biol Chem : 6253–6260, 1984. [PubMed] [Google Scholar]
- 4.Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, and Osterhaus AD. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature : 240, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galili U, Chen ZC, Manches O, Plumas J, and Preisler H. Preparation of autologous leukemia and lymphoma vaccines expressing alpha-gal epitopes. J Hematother Stem Cell Res : 501–511, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Galili U, Repik PM, Anaraki F, Mozdzanowska K, Washko G, and Gerhard W. Enhancement of antigen presentation of influenza virus hemagglutinin by the natural human anti-Gal antibody. Vaccine : 321–328, 1996. [DOI] [PubMed] [Google Scholar]
- 7.Galili U, Shohet SB, Kobrin E, Stults CL, and Macher BA. Man, apes, and Old World monkeys differ from other mammals in the expression of alpha-galactosyl epitopes on nucleated cells. J Biol Chem : 17755–17762, 1988. [PubMed] [Google Scholar]
- 8.Ksiazek TG et al. (SARS Working Group). A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med : 1953–1966, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Marshall E and Enserink M. Medicine. Caution urged on SARS vaccines. Science : 944–946, 2004. [DOI] [PubMed] [Google Scholar]
- 10.Pacifico F, Montuori N, Mellone S, Liguoro D, Ulianich L, Caleo A, Troncone G, Kohn LD, Di Jeso B, and Consiglio E. The RHL-1 subunit of the asialoglycoprotein receptor of thyroid cells: cellular localization and its role in thyroglobulin endocytosis. Mol Cell Endocrinol : 51–59, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, and Bellini WJ. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science : 1394–1399, 2003. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz AL, Fridovich SE, Knowles BB, and Lodish HF. Characterization of the asialoglycoprotein receptor in a continuous hepatoma line. J Biol Chem : 8878–8881, 1981. [PubMed] [Google Scholar]
- 13.Wall DA, Wilson G, and Hubbard AL. The galactose-specific recognition system of mammalian liver: the route of ligand internalization in rat hepatocytes. Cell : 79–93, 1980. [DOI] [PubMed] [Google Scholar]
- 14.Wang D. Carbohydrate microarrays. Proteomics : 2167–2175, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Wang D. Carbohydrate antigens. In: Encyclopedia of Molecular Cell Biology and Molecular Medicine, edited by Meyers RA. Wiley-VCH, 2004, vol. II, chapt. 11, p. 277–301.
- 16.Wang D, Liu S, Trummer BJ, Deng C, and Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol : 275–281, 2002. [DOI] [PubMed] [Google Scholar]
- 17.Wu AM, Song S, Tsai M, and Herp A. A guide to the carbohydrate specificities of applied lectin-2 (updated in 2000). In: Molecular Immunology of Complex Carbohydrates 2. Advances in Experimental Medicine and Biology (2nd ed.), edited by Wu AM. New York: Kluwer Academic/Plenum, 2001, vol. 491, p. 551–585. [DOI] [PubMed]