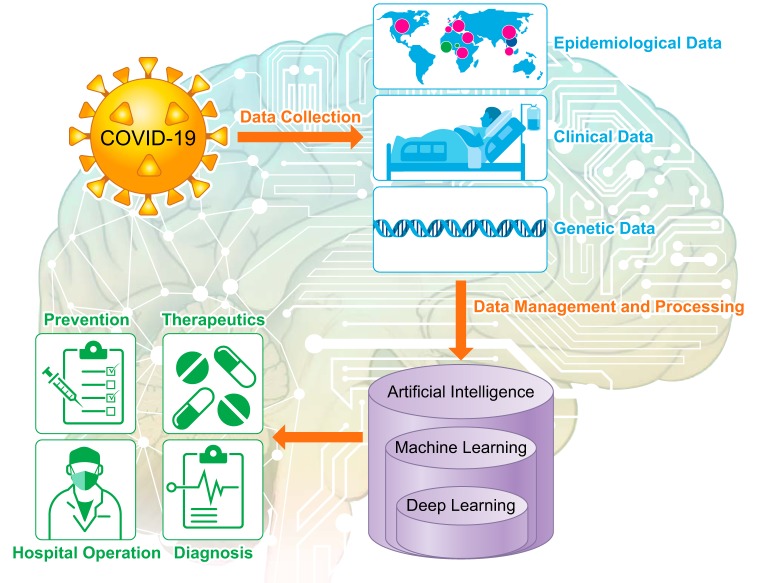
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (13), has become an unprecedented public health crisis. Coronavirus Resource Center at Johns Hopkins University of Medicine has reported a total of 23,638 deaths as worldwide COVID-19 infections surpass 500,000 (as of 5 PM EST on March 26, 2020). On March 16, 2020, the White House, collaborating with research institutes and tech companies, issued a call to action for global artificial intelligence (AI) researchers for developing novel text and data-mining techniques to assist COVID-19-related research. The Allen Institute for AI in partnership with leading research groups issued an open-source, weekly updated COVID-19 Open Research Dataset (2), which continuously documents COVID-19-related scholarly articles to accelerate novel research projects urgently requiring real-time data. The large-scale data of COVID-19 patients can be integrated and analyzed by advanced machine learning algorithms to better understand the pattern of viral spread, further improve diagnostic speed and accuracy, develop novel effective therapeutic approaches, and potentially identify the most susceptible people based on personalized genetic and physiological characteristics. Inspirationally, within a short period of time since COVID-19 outbreak, advanced machine learning techniques have been used in taxonomic classification of COVID-19 genomes (8), CRISPR-based COVID-19 detection assay (6), survival prediction of severe COVID-19 patients (11), and discovering potential drug candidates against COVID-19 (4).
Personalized protective strategies can greatly benefit from precise classifications of the population based on categorized COVID-19 susceptibility. The earlier observation that elderly people have a higher risk to COVID-19 is challenged by a recent finding that more and more young adults suffer from severe COVID-19 symptoms, indicating an urgent need of a comprehensive risk evaluation based on personalized genetic and physiological characteristics. Human angiotensin-converting enzyme 2 (ACE2), expressed in epithelial cells of lung, small intestines, heart, and kidneys, is an entry receptor for SARS-CoV-2 spike glycoprotein (3, 13). Fang et al. (3) hypothesized that increased expression of ACE2, from using ACE2-stimulating drugs to treat hypertension and diabetes, could actually worsen clinical outcomes of COVID-19 infection. Indeed, this hypothesis should be further tested with strict experimental designs and long-term clinical observations. Therefore, biochemistry (e.g., ACE2 expression level) and clinical data (e.g., age, respiratory pattern, viral load, and survival) of COVID-19 patients with underlying medical conditions can be analyzed by machine learning approaches to not only identify any reliable features (e.g., ACE2) for risk prediction, but also further perform risk classification and prediction for a balanced preparation of ongoing disease treatment and COVID-19 defense (Fig. 1). ACE2 genetic polymorphism, represented by diverse genetic variants in human genome, has been shown to affect virus-binding activity (1), suggesting a possible genetic predisposition to COVID-19 infection. Therefore, machine learning analysis of genetic variants from asymptomatic, mild or severe COVID-19 patients can be performed to classify and predict people based on their vulnerability or resistance to potential COVID-19 infection, by which the machine learning model can also return those prioritized genetic variants, such as ACE2 polymorphism, in their decision-making process as important features for functional and mechanistic studies (Fig. 1).
Fig. 1.
Application of artificial intelligence and machine learning in the fight against COVID-19.
Currently, ongoing efforts have been made to develop novel diagnostic approaches using machine learning algorithms. For example, machine learning-based screening of SARS-CoV-2 assay designs using a CRISPR-based virus detection system was demonstrated with high sensitivity and speed (6). Neural network classifiers were developed for large-scale screening of COVID-19 patients based on their distinct respiratory pattern (10). Similarly, a deep learning-based analysis system of thoracic CT images was constructed for automated detection and monitoring of COVID-19 patients over time (5). Rapid development of automated diagnostic systems based on artificial intelligence and machine learning not only can contribute to increased diagnostic accuracy and speed but will also protect healthcare workers by decreasing their contacts with COVID-19 patients (Fig. 1).
An effective therapeutic strategy is urgently needed to treat rapidly growing COVID-19 patients worldwide. As there is no effective drug proven to treat COVID-19 patients, it is critical to develop efficient approaches to repurpose clinically approved drugs or design new drugs against SARS-CoV-2. A machine learning-based repositioning and repurposing framework was developed to prioritize existing drug candidates against SARS-CoV-2 for clinical trials (4). Additionally, a deep learning-based drug discovery pipeline has been used to design and generate novel drug-like compounds against SARS-CoV-2 (12). AlphaFold (9), which is a deep learning system developed by Google DeepMind, has released predicted protein structures associated with COVID-19, which can take months by traditional experimental approaches, serving as valuable information for COVID-19 vaccine formulation. Moreover, COVID-19 vaccine candidates were proposed by a newly developed Vaxign reverse vaccinology tool integrated with machine learning (7). The tremendous amount of COVID-19 treatment data in worldwide hospitals also require advanced machine learning methods for analyzing personalized therapeutic effects for evaluating new patients, such as hospitalization prediction, which can not only provide better care for each patient but also contribute to local hospital arrangement and operation (Fig. 1).
As artificial intelligence and machine learning scientists have been eagerly searching and waiting for real-time data generated by this pandemic around the world, timely delivery of COVID-19 patient data, such as physiological characteristics and therapeutic outcome of COVID-19 patients, followed by subsequent data transformation for easy access, is extremely important, but challenging. Figure 1 is a schematic representation of the workflow, but there are several steps in the process that currently limit the application of machine learning and artificial intelligence to combat COVID-19. Availability of COVID-19-related clinical data, which can be managed and processed into easily accessible databases, is a key current barrier. Therefore, development of cyber-infrastructure to fuel world-wide collaborations is important. To this end, the US federal agencies are already promoting the formations of consortia and funding opportunities (https://www.nsf.gov/pubs/2020/nsf20055/nsf20055.jsp). In addition to these initiatives, integrating COVID-19-related clinical data with existing biobanks, such as the UK Biobank, with pre-existing data of those patients (if already in biobanks), such as their genotype and physiological characteristics, could maximize our efforts toward a faster and feasible approach for meaningful data-mining by bioinformaticians and computational scientists. A centralized collection of worldwide COVID-19 patient data will be beneficial for future artificial intelligence and machine learning research to develop predictive, diagnostic, and therapeutic strategies against COVID-19 and similar pandemics in future.
GRANTS
X. Cheng acknowledges funding support from the Dean’s Postdoctoral to Faculty Fellowship from the University of Toledo College of Medicine and Life Sciences and P30 Core Center Pilot Grant from NIDA Center of Excellence in Omics, Systems Genetics, and the Addictome. B. Joe acknowledges support from the National Heart Lung and Blood Institute Grant HL-143082.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.A., S.A., I.M., and X.C. prepared figure; A.A., S.A., I.M., and X.C. drafted manuscript; A.A., S.A., I.M., P.B.M., B.J., and X.C. edited and revised manuscript; A.A., S.A., I.M., P.B.M., B.J., and X.C. approved final version of manuscript.
REFERENCES
- 1.Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wen F, Huang X, Ning G, Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov 6: 11, 2020. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.COVID-19 Open Research Dataset (CORD-19). 2020, https://pages.semanticscholar.org/coronavirus-research.
- 3.Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med In press, 2020. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge Y, Tian T, Huang S, Wan F, Li J, Li S, Yang H, Hong L, Wu N, Yuan E, Cheng L, Lei Y, Shu H, Feng X, Jiang Z, Chi Y, Guo X, Cui L, Xiao L, Li Z, Yang C, Miao Z, Tang H, Chen L, Zeng H, Zhao D, Zhu F, Shen X, Zeng J. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv, 2020. doi: 10.1101/2020.03.11.986836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E. Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring using Deep Learning CT Image Analysis. arXiv2003.05037; 2020. [Google Scholar]
- 6.Metsky HC, Freije CA, Kosoko-Thoroddsen T-SF, Sabeti PC, Myhrvold C. CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach. bioRxiv, 2020. doi: 10.1101/2020.02.26.967026. [DOI] [Google Scholar]
- 7.Ong E, Wong MU, Huffman A, He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. bioRxiv; 2020. doi: 10.1101/2020.03.20.000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randhawa GS, Soltysiak MPM, El Roz H, de Souza CPE, Hill KA, Kari L. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID-19 case study. bioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K, Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature 577: 706–710, 2020. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Hu M, Li Q, Zhang X-P, Zhai G, Yao N. Abnormal respiratory patterns classifier may contribute to large-scale screening of people infected with COVID-19 in an accurate and unobtrusive manner. arXiv2002.05534; 2020. [Google Scholar]
- 11.Yan L, Zhang H-T, Xiao Y, Wang M, Sun C, Liang J, Li S, Zhang M, Guo Y, Xiao Y. Prediction of survival for severe Covid-19 patients with three clinical features: development of a machine learning-based prognostic model with clinical data in Wuhan. medRxiv; 2020. doi: 10.1101/2020.02.27.20028027. [DOI] [Google Scholar]
- 12.Zhavoronkov A, Aladinskiy V, Zhebrak A, Zagribelnyy B, Terentiev V, Bezrukov DS, Polykovskiy D, Shayakhmetov R, Filimonov A, Orekhov P. Potential COVID-2019 3C-like Protease Inhibitors Designed Using Generative Deep Learning Approaches. Insilico Med Hong Kong Ltd A 307: E1, 2020. [Google Scholar]
- 13.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273, 2020. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]