Abstract
Antimicrobial peptides (AMPs) are a class of templates with application potential for drug development. Amphibians are important sources of AMPs. Duttaphrynus melanostictus is the main source of traditional Chinese medicine “Chansu”, which has anti-infection effect while without a clear mechanism. This study aimed to find the cathelicidin peptide in D. melanostictus and then investigate the activity in vivo and in vitro, and an AMP-encoding gene (cathelicidin-DM, GenBank: KJ820824.1) was obtained from the constructed cDNA library of D. melanostictus. The MIC test and SYTOX Green uptake were used for the evaluation of the bactericidal capacity and mechanisms. The serum stability tests were used for the evaluation of the application potential. The skin wound infection model and in vivo imaging were used for in vitro application of possibility evaluation. The results showed that cathelicidin-DM was a 37 amino acid AMP with good bactericidal ability, which was similar to melittin: both can kill bacteria within 15 min. Moreover, cathelicidin-DM exhibits good therapeutic potential in the mouse wound infection model, and it can be enriched to the site of infection for treatment. Thus, cathelicidin-DM could be a new template for antimicrobial drug development given its good antibacterial activity in vivo and in vitro.
Introduction
Antibiotics are among the most important discoveries in the history of chemotherapeutic drugs and have greatly reduced mortality rates from infectious diseases.1−3 However, the abuse of antibiotics has led to the outbreak of drug-resistant bacteria, especially multi-drug-resistant (MDR) and extensively drug-resistant (XDR) bacteria, which seriously threaten the life and health of human beings.4−7 Thus, novel antibacterial substances that mitigate severe antibiotic resistance are urgently needed. The naturally occurring host defense peptides (HDPs) may be the novel antibacterial substances. HDPs, sometimes called antimicrobial peptides (AMPs) because of their ability to kill microorganisms, are produced by almost all kinds of organisms as components of the innate immune system.8−10 Their key attributes are positive charge, hydrophobicity, and amphipathic structures linked to the antimicrobial potency of peptide candidates. Since the first cathelicidin AMP, Bac5, was discovered from bovine, a lot of cathelicidins AMPs have been identified from vertebrates, including fish, amphibians, birds, reptiles, and mammals.8,11−14 AMPs possess a broad spectrum of antibacterial activities and exert strong inhibitory effects on most Gram-positive bacteria, Gram-negative bacteria, and fungi (including MDR and XDR microorganisms).15−17 Other advantages of AMPs include anti-inflammatory and immunomodulatory activities, virulence factor neutralization, and slow resistance development. Given these characteristics, AMPs exhibit great potential in the development of anti-infection drugs.
Amphibians have naked, scaleless, and hairless skin and live in wet conditions. Wet conditions are very suitable for microbial reproduction.18,19 The anatomical characteristics and diverse living environments of amphibians render them capable of facing severe challenges from microorganisms.20 More than 1400 kinds of AMPs have been found in the skin secretions of amphibians, such as Paa yunnanensis, Limnonectes fragilis, Rana catesbeiana, and Tylototriton verrucosus, which cover more than 600 species of frogs.21−23 Bioactive substances protect amphibians from pathogenic microorganisms.24−26 However, only few cathelicidin AMPs have been identified in the amphibians. Cathelicidins play critical roles in the innate immune system of most vertebrates and can provide the first line of defense against various infectious factors.8 The precursors of cathelicidins comprise three parts, namely, N-terminal signal peptide (30 residues), highly conserved cathelin domain (99–114 residues), and C-terminal mature peptide (12–100 residues) with remarkable structural variety.27 In 2011, the first amphibian cathelicidin AMP (cathelicidin-AL) was identified from Amolops loloensis.28 Several amphibian cathelicidin AMPs were then identified from different amphibians, including BG-CATH37 from Bufo bufo gargarizans Cantor,16 cathelicidin-OA1 from Odorrana andersonii,29 cathelicidin-NV from plateau frog Nanorana ventripunctata,30 Lf-CATH1 from L. fragilis,21 and Temporin-1Ta from the European red frog Rana temporaria.31
In 2018, WHO included traditional Chinese medicine in the International Classification of Diseases; this event brought great opportunities for the development of traditional Chinese medicine materials.32 The traditional Chinese medicine material “Chansu” was a white serous substance secreted from the posterior ear and skin glands of Duttaphrynus melanostictus or Bufo bufo gargarizans Cantor; this substance has functions in detumescence and exerts analgesic, anti-inflammatory, and detoxification effects.33−35 Thus far, studies on the active components of “Chansu” have focused on small molecular compounds, but the research on protein/polypeptide bioactive substances is not enough.36,37 In 2010, researchers found that the skin secretions of D. melanostictus have strong bacteriostatic activity against Gram-negative and Gram-positive bacteria, although this activity may stop after trypsin treatment. Thus, the skin secretions of D. melanostictus may contain some peptides or proteins.38
In the present study, an AMP encoding gene (cathelicidin-DM) was isolated from the cDNA library of D. melanostictus dorsal skin. Then, the function of the synthesized cathelicidin-DM and the truncated peptides were determined.
Results
Molecular Cloning and Characterization of Cathelicidin-DM
Total RNA was successfully extracted from the dorsal skin of D. melanostictus and synthesized to cDNA library. A cathelicidin-encoded cDNA was identified and designated as cathelicidin-DM. The bioinformatics analysis of cathelicidin-DM is shown in Figure 1. The complete nucleotide sequence of cathelicidin-DM (GenBank accession number KJ820824) and the deduced amino acid sequence are shown in Figure 1A. The open reading frame was composed of 164 amino acid residues (aa), including a predicted signal peptide, conserved cathelin domain, and variable cationic C-terminus.27 The multisequence alignment of amphibian cathelicidins indicates that cathelicidin-DM is similar to other amphibian cathelicidins, including Xenopus tropicalis Cath-2, L. fragilis Cath-1 and Cath-2, Rana catesbeiana Cath-1 and Cath-2, and Nanorana yunnanensis Cath in the structure (Figure 1B). Elastase, which was generally considered to be responsible for the release of mature cathelicidins,43,44 was not observed in amphibian cathelicidins. The precursors of amphibian AMPs often possess a typical dibasic cleavage site for trypsin-like proteases.45 Moreover, a conserved dibasic cleavage site was found in all amphibian cathelicidins. The mature cathelicidin-DM was predicted as SSRRKPCKGWLCKLKLRGGYTLIGSATNLNRPTYVRA by Translate Tool, which contains 37 amino acids including nine basic amino acids (Arg + Lys) and has no acidic amino acid (Asp + Glu). The theoretical molecular weight is 4.1659 kDa, and the isoelectric point is 11.05. The secondary structure of cathelicidin-DM was predicted at Predicprotein (http://www.predicprotein.org) (Figure 2A), and the 3D structure was predicted at the Quark server (http://zhanglab.ccmb.med.umich.edu/QUARK/)12 (Figure 2B). The predicted structure of cathelicidin-DM contains three antiparallel β sheets combined with the random coil (Figure 2B) and is consistent with the result of the CD spectra (Figure S1). The physicochemical properties of the peptide showed that cathelicidin-DM is a stable polypeptide.
Figure 1.
Sequence analysis of cathelicidin-DM. (A) Nucleotide sequence encoding cathelicidin-DM and the deduced amino acid sequence. The putative TATA box, signal peptide, and polyadenylation signal are marked by italic, underline, and dashed underline, respectively. The mature peptide is boxed. The stop codon TAA is indicated by asterisks. (B) Alignment of cathelicidins identified in amphibians. The mature peptide of known amphibian cathelicidins is underlined. (*) Indicates positions which have a single, fully conserved residue. (:) Indicates conservation between groups of strongly similar properties. (.) Indicates conservation between groups of weakly similar properties.
Figure 2.
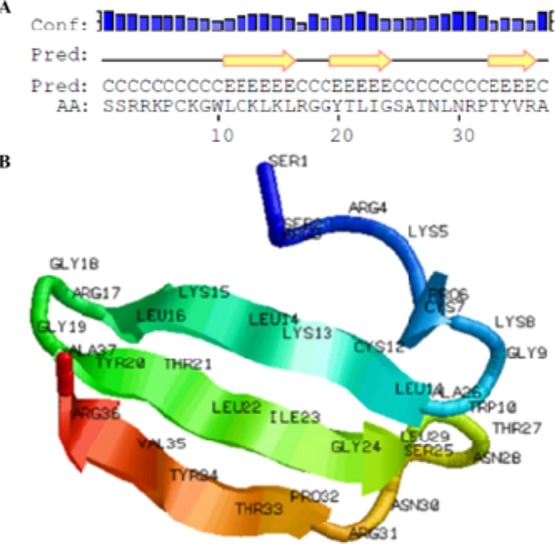
Predicted secondary structure and three-dimensional structure of cathelicidin-DM. (A) Predicted secondary structure of cathelicidin-DM and (B) de novo structure prediction of cathelicidin-DM. The model was produced by the QUARK server. Visualization of the structure was accomplished using RASMOL.
Expression Profile of Cathelicidin-DM
The expression profiles of cathelicidin-DM in healthy D. melanostictus tissues were evaluated by semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis (Figure 3). All of the tested tissues expressed cathelicidin-DM genes, including the small intestine, large intestine, liver, spleen, ovary, and skin. Cathelicidin-DM was highly expressed in the spleen and large intestine and had low expression in the small intestine, ovary, liver, and skin.
Figure 3.
Tissue expression profile of cathelicidin-DM. The sample from lane 1–6 was skin, small intestine, large intestine, liver, ovarium, and spleen, respectively. β-Actin as the internal reference.
Analysis of Antimicrobial Activity and SYTOX Green Uptake of Cathelicidin-DM
The antimicrobial activity of cathelicidin-DM and its derivatives (Cath-DM-CT1, Cath-DM-CT2, Cath-DM-CT3, Cath-DM-NT, Cath-DM-NCT1, and Cath-DM-NCT2; >95% purity; Figure 4) against the tested bacteria were determined as minimal inhibitory concentration (MIC). Cathelicidin-DM and its derivatives (except Cath-DM-NT and Cath-DM-NCT1) exhibited potent antimicrobial activity against common pathogens, including MDR and XDR clinical isolates (Table 1). The MIC of cathelicidin-DM against Staphylococcus haemolyticus (CI 1410970016), Enterococcus faecalis (MDR 14U0445), and Staphylococcus aureus ATCC25923 was 12 μg/mL. The MIC of cathelicidin-DM against Escherichia coli (MDR 13A10022), E. coli (MDR 13U1780), E. coli ATCC25922, and Citrobacter freundii (CI 1410UR0112) was 12 μg/mL, while that for Staphylococcus paratyphoid A (CI 1410UR0112), Klebsiella pneumoniae (MDR 13U1752), K. pneumoniae (MDR 13A11923), and K. pneumoniae (XDR 13A13361) was 6 μg/mL. All the derivatives except Cath-DM-NT and Cath-DM-NCT1 showed similar antimicrobial activity against partially tested strains, such as S. haemolyticus (CI 1410970016) and XDR K. pneumoniae but lost such activity against E. faecalis (MDR 14U0445) and two E. coli strains. The derivatives lost antimicrobial activity against the strains inhibited by cathelicidin-DM.
Figure 4.
Cathelicidin-DM and its truncated peptides.
Table 1. MICs of Cathelicidin-DM and Its Derivativesa.
| MICs (μg/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| bacterial strains | cathelicidin-DM | cath-DM-CT1 | cath-DM-CT2 | cath-DM-CT3 | cath-DM-NT | cath-DM-NCT1 | cath-DM-NCT2 | Amp | cathelicidin-DM after incubation in 50% serum |
| Gram-Positive Bacteria | |||||||||
| S. haemolyticus (CI 1410970016) | 12 | 6 | 3 | 3 | |||||
| E. faecalis (MDR 14U0445) | 12 | 2 | |||||||
| S. aureus ATCC25923 | 12 | 12 | 0.15 | ||||||
| Gram-Negative Bacteria | |||||||||
| E. coli ATCC25922 | 12 | 0.3 | |||||||
| E. coli (MDR 13U1780) | 12 | 12 | 12 | 32 | |||||
| E. coli (MDR 13A10022) | 12 | 32 | |||||||
| S. paratyphi A (CI 1410BL0513) | 6 | 6 | 12 | 6 | 32 | 6 | |||
| C. freundii (CI 1410UR0112) | 12 | 6 | 12 | 12 | |||||
| K. pneumoniae (MDR 13U1752) | 6 | 6 | 32 | 6 | |||||
| K. pneumoniae (MDR 13A11923) | 6 | 32 | 6 | ||||||
| K. pneumoniae (XDR 13A13361) | 6 | 6 | 3 | 6 | 32 | 6 | |||
Note: CI, clinically isolated strain; MDR, multi-drug-resistant clinical isolate; XDR, extensive drug-resistant clinical isolate; (-) no detectable activity at a dose of 48 μg/mL.
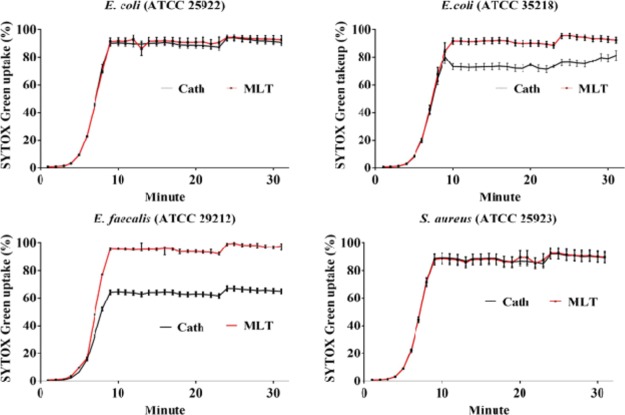
The action mode of cathelicidin-DM against the tested bacteria can be related to membrane permeabilization. This supposition was supported by the results of the SYTOX Green uptake experiment. SYTOX green is an excellent nuclear dye with green fluorescence, which can penetrate the fragile cell walls of dead cells. Thus, it is a useful indicator of dead cells. The bactericidal effect of cathelicidin-DM is consistent with that of melittin44 (Figure 5), an AMP that can lyse cell membranes. The bactericidal rate of cathelicidin-DM is also similar to that of melittin, killing more than 60% of E. faecalis (ATCC29212) and E. coli (ATCC35218) within 10 min and 80% of E. coli (ATCC25922) and S. aureus (ATCC25923) within 10 min. After mixing with a blood serum for 12 h, cathelicidin-DM lost its inhibitory activity against Gram-positive bacteria but retained its activity against Gram-negative bacteria (Table 1).
Figure 5.
SYTOX green uptake of cathelicidin-DM and melittin. Four tested bacteria including E. coli (ATCC 25922), E. coli (ATCC 35218), E. faecalis (ATCC 29212), and S. aureus (ATCC 25923) were diluted to 2 × 105 CFU/mL. After the addition of peptides to the final concentrations corresponding to their respective MIC, they were mixed with a final concentration of 50 mM SYTOX Green (Invitrogen) for 15 min in the dark and finally detected in wavelengths 485 and 523 nm filters for excitation and emission.
Therapeutic Effect of Cathelicidin-DM on Mouse Skin Infection Model
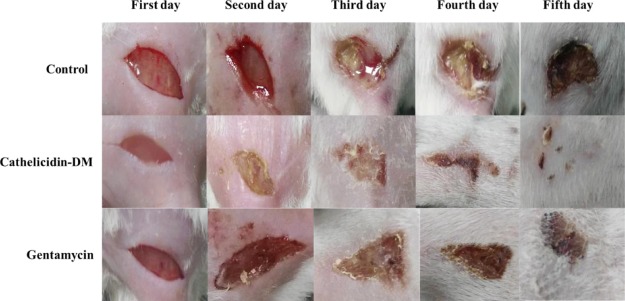
A mouse model of skin infection was constructed by daubing E. coli ATCC25922 (100 μL, 2 × 105 CFU/mL) in the 1 cm2 wound. One hour after model construction, the mice in the three groups were intravenously injected with cathelicidin-DM (10 mg/kg), gentamycin (5 mg/kg), or PBS (pH 7.4) as the control for 5 days. The mice were photographed every 24 h. The wound in the mouse that received cathelicidin-DM (Figure 6) healed slightly faster than the wounds in the control and gentamycin groups. Two days after the administration of cathelicidin-DM, the wound-healing effect was significantly different.
Figure 6.
Wound healing of mice after 5 days of continuous administration. All the three mice groups (three female mice/group) were administered with cathelicidin-DM (10 mg/kg), gentamycin (5 mg/kg), or PBS as control lasted for 5 days by tail vein injection. The mice were photographed every 24 h.
Detection of Cathelicidin-DM Distribution in the Target Site
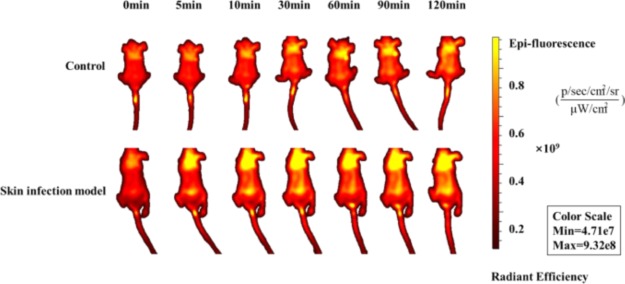
The hair was removed from two mice groups (skin infection and control groups, three female mice/group). After 1 h postinoculation of E. coli (ATCC25922), all the two groups received 10 mg/kg of FITC-labeled cathelicidin-DM by tail vein injection. Then, the mice were immediately anesthetized before they were transferred to the IVIS LT imaging system (IVIS spectrum) and measured at 0, 5, 10, 30, 60, 90, and 120 min. Significant fluorescence was observed in different regions, such as the chest, abdomen, and brain (Figure 7). FITC-cathelicidin-DM was abundant in infected areas. Hence, cathelicidin-DM was distributed to different tissues and exhibited enrichment effect on bacterial infection sites.
Figure 7.
In vivo imaging of FITC-labeled cathelicidin-DM. All the two mice groups (three female mice/group) were administered with cathelicidin-DM (10 mg/kg) and gentamycin (5 mg/kg) as control lasted for 5 days by tail vein injection. The mice were photographed every 24 h.
Discussion
The emergence of MDR and XDR pathogens is an urgent public problem to be solved all over the world, especially in intensive care units.46 Given that many patients have serious illnesses and lacked access to effective treatment conditions, mortality related to MDR and XDR pathogens is extremely high.47 Thus, novel antimicrobial agents are urgently needed. The skin of amphibians is exposed and moist, and the mucus it secretes contains many antibacterial substances with special molecular structures and complex functions.48 AMPs are defensive peptides produced by the immune defense system to resist the pathogenic effects of exogenous pathogens. AMPs contain 10–50 amino acid residues, most of which are rich in basic amino acids, such as lysine and arginine, and have positive charges as a whole. Many studies showed that the main mechanism of AMPs is to destroy membrane integrity. AMPs interfere with some important cellular processes and thus play a bactericidal role. Given that AMPs mainly target cell membrane structures, AMPs result in a generally low drug resistance and thus considered substitutes and synergists for antibiotics and used in addressing increasingly serious problem of bacterial drug resistance. AMPs affect the pathogens of some cash crops and are expected to become human or animal antimicrobial agents. D. melanostictus is an important traditional Chinese medicine in China, and its “Chansu” has important medicinal value.49 However, D. melanostictus, which is widely distributed in Yunnan, Guizhou, Sichuan province, has not yet attracted enough attention. Therefore, we attempted to screen cDNA-encoding AMPs from the cDNA library of the D. melanostictus skin. A cathelicidin AMP was identified and named cathelicidin-DM, which is the first identified cathelicidin AMP from D. melanostictus.50−52
According to the RT-PCR analysis results, the expression pattern of cathelicidin-DM is similar to the expression patterns of other amphibian cathelicidins.21−23,28 Notably, the expression of cathelicidin-DM in the skin is not high compared with that in other tissues. This finding suggests that cathelicidin-DM has other important functions in D. melanostictus in addition to its direct bactericidal effect. Especially, the high expression in the large intestine and kidney indicates that cathelicidin-DM may play an important role in some processes of digestion and processes in the urinary and endocrine systems.
Although the C-terminal mature peptide region of cathelicidin-DM significantly differed from those of other amphibian cathelicidins, it still possesses potent antibacterial activities. Six truncated peptides were designed from cathelicidin-DM according to its charge, hydrophilicity, and hydrophobicity for the evaluation of the potential of cathelicidin-DM as a template for the development of novel peptide antibiotics. Cathelicidin-DM has strong antibacterial activity against Gram-positive strains and Gram-negative strains (including clinically isolated MDR and XDR strains). After cathelicidin-DM mixed with serum for 12 h, it lost its antibacterial activity in some tested strains. The human serum is rich in a variety of proteases, such as thrombin and matrix metalloproteinases, which hydrolyze cathelicidin-DM. Whether or not affected by serum, cathelicidin-DM showed good activity against clinically isolated K. pneumoniae (MDR 13U1752), K. pneumoniae (MDR 13A11923), and K. pneumoniae (XDR 13A13361).
In the mouse models of skin infection, cathelicidin-DM showed stronger effect than gentamicin and significantly accelerated the healing of the wound. Using a small animal imaging system, we intuitively observed that cathelicidin-DM was distributed in many places in the body, especially in the brain and heart. In the case of trauma, enrichment was observed. This event promoted the wound-healing ability of cathelicidin-DM. Moreover, cathelicidin-DM can enter the blood–brain barrier into the brain, as shown in the in vivo imaging of FITC-labeled cathelicidin-DM. Hence, cathelicidin-DM is a potential drug carrier that targets the brain in the future.
Conclusions
Cathelicidin-DM was identified in the present work by molecular cloning from D. melanostictus and was used to treat skin wound infections. Cathelicidin-DM possesses potent antibacterial activities against many pathogens including MDR and XDR strains. It has a good therapeutic effect in the wound infection model. All the characteristics suggest that cathelicidin-DM from traditional Chinese medicine “Chansu” may be an ideal template peptide antibiotic in the development of novel antimicrobial agents.
Experimental Section
Ethical Approval
This study was carried out in accordance with the recommendations of the Animal Care and Utilization Committee of the Kunming University of Science and Technology, which also approved the study protocol.
cDNAs Synthesis and Cathelicidin-DM Cloning
Adult D. melanostictus (collected from Xishuangbanna, Yunnan Province, China) was housed (at 25 °C with a 12/12 h light/dark cycle and access to food and water ad libitum), maintained, and cared for in the Animal Experiment Center of Kunming University of Science and Technology. The cDNA library of the dorsal skin was constructed according to the instructions of SMARTer cDNA library construction kit (Clontech, USA), which was then used in the screening of cDNAs encoding cathelicidin-DM. The first strand was synthesized by CDS III/3′ primer and SMARTTM IV oligonucleotide. The CDS III/5′ primer provided by the kit was used in the synthesis of the second strand. Two oligonucleotide primers, S1 and S2, in the sense of direction were designed according to the conserved sequences of cathelicidins from other amphibians. Nested PCR was performed with CDS III/3′ primer in the antisense direction. The PCR conditions were as follows: 1 min at 95 °C, followed by 25 cycles of 30 s at 95 °C, 30 s at 58 °C, and 1 min at 72 °C, and final extension at 72 °C for 10 min. The PCR products of the second round were amplified by the primer S2 and the CDS III/3′ primer, inserted into the pMD 19-T simple vector, and transformed into DH5α competent cells. Sequencing primers M13F and M13R were used for positive clone selection. Another primer, R2, was designed according to the sequenced nucleotide sequence above and combined with the CDS III/5′ primer to amplify the 5′ ends of the cDNA. Sequencing was performed again as described above, and the cathelicidin-DM CDS sequence was finally obtained, which was used for the designing of specific primers (cath-F/R). All the primers used in this study are shown in Table 2.
Table 2. Primers Used in This Study.
| primer | sequence (5′–3′) | size (bp) |
|---|---|---|
| CDS III/3′ primer | ATTCTAGAGGCCGAGGCGGCCGACATGd(T)30N-1N-3′(N = A, C, G or T; N-1 = A, G or C) | 27 |
| SMARTTM IV oligonucleotide | AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG | 39 |
| CDS III/5′-primer | AAGCAGTGGTATCAACGCAGAGT | 23 |
| S1 | CCYGCCACYSCAGARRTICA | 20 |
| S2 | CAAGTNGTBGCHGGRDIVA | 19 |
| M13 F | CGCCAGGGTTTTCCCAGTCACGAC | 24 |
| M13 R | GAGCGGATAACAATTTCACACAGG | 24 |
| R2 | GGACCTCCTTGTTCAGACTG | 20 |
| cath-F | GCCTGAGGTCCAAGATGGA | 19 |
| cath-R | GGCTTGAAATCACACTGGGTT | 21 |
| actin-F | AAATGGCTACAGCTGCTTCCT | 21 |
| actin-R | AAGGCTGGAAGAGGGCTTCT | 21 |
Sequence Analysis
The cathelicidin-DM nucleotide sequence was analyzed by BLAST and Translate Tool (http://web.expasy.org/translate/) for the deduction of amino acid sequences.39 Multiple sequence alignment was performed using Clustal Omega (https://predictprotein.org/) and then manually curated.40 Secondary structure was predicted at Predictprotein (http://www.predicprotein.org), and 3D structure was predicted at the Quark server (http://zhanglab.ccmb.med.umich.edu/QUARK/).41
Expression Profile Analysis by RT-PCR Assay
The RT-PCR assay was performed to analyze the gene expression of cathelicidin-DM in different tissues of D. melanostictus, including the skin, spleen, liver, large intestine, small intestine, and ovary. After the tissues were removed, they were immediately placed in liquid nitrogen, and the total RNA extraction of each tissue and organ was performed with Trizol Reagent (Invitrogen, USA) according to the manufacturer’s instructions. The first-strand cDNA was synthesized by M-MLV reverse transcriptase (Invitrogen, USA), which was used as the template for PCR amplification with specific primers cath-F/R of cathelicidin-DM. The total RNA was used as the template, and oligo dT(18) as a primer. The β-actin-specific primer actin-F/R was designed on the basis of the β-actin gene in the GenBank database, which was used as the RT-PCR internal reference (Table 2).
Peptide Synthesis
Cathelicidin-DM, FITC-labeled cathelicidin-DM, and six truncated cathelicidin-DMs (Cath-DM-CT1, Cath-DM-CT2, Cath-DM-CT3, Cath-DM-NT, Cath-DM-NCT1, and Cath-DM-NCT2) based on the deduced amino acid sequence are synthesized by the peptide synthesizer (APEX396, AAPPTec) in Phtd peptides Co., Ltd. (Zhengzhou, China) and used for bioassays. All the six peptides were purified with C18 RP-high-performance liquid chromatography (HPLC) column and 0.1% (v/v) trifluoroacetic acid/acetonitrile as an elution solution with a linear gradient from 30 to 100% acetonitrile in 20 min. The purity of the final product was higher than 95% [the HPLC and mass spectrometry (MS) report of the synthesized peptides were supplied in the Supporting Information]. The peptide was dissolved in sterilized ultrapure water and stored at −20 °C with a storage concentration of 2 mg/mL.
Antibacterial Assays
The synthetic peptides were tested for MICs against strains, including S. haemolyticus (CI 1410970016), E. coli (MDR 13A10022), E. coli (MDR 13U1780), E. coli ATCC25922, Salmonella Paratyphi A (CI 1410BL0513), E. faecalis (MDR 14U0445), C. freundii (CI 1410UR0112), K. pneumoniae (MDR 13U1752), K. pneumoniae (MDR 13A11923), K. pneumoniae (XDR 13A13361), and S. aureus (ATCC25923), as shown in Table 1 (all the tested bacteria were provided by the Clinical Laboratory of First People’s Hospital of Yunnan Province. The drug resistance information of the tested clinical strains is shown in Table S1). The strains were inoculated on a Luria–Bertani (LB) solid medium and cultured in a 37 °C incubator. The single colony was selected and cultured in the LB liquid medium in a 37 °C incubator to logarithmic growth phase. Then, the concentration of bacterial solution was detected with an ultraviolet spectrophotometer. According to 1 OD600 = 1 × 109 CFU/mL, the bacterial solution was diluted to 2 × 105 CFU/mL with an LB liquid medium. Then, 100 μL of the LB liquid medium was added to each well of aseptic 96-well plate, and then, 100 μL of the AMP solution (192 μg/mL) was added to the first well. After mixing, we transferred 100 μL of the solution from the first well to the second well to perform serial dilutions. The diluted bacterial solution (100 μL) was added to each well and mixed and cultured at 37 °C for 16 h. The absorbance at 600 nm was determined. Each peptide was repeated three times. The MIC was defined as the lowest peptide concentration that fully inhibited the bacterial growth.
SYTOX Green Uptake
Four tested bacterial strains (50 μL, 2 × 105 CFU/mL), namely, E. coli (ATCC25922), E. coli (ATCC35218), E. faecalis (ATCC29212), and S. aureus (ATCC25923), were cultured as previously described and then centrifuged. The supernatant was discarded, and the precipitate was washed with aseptic PBS three times and resuspended with 100 μL of aseptic PBS (the tested bacteria were provided by the Clinical Laboratory of First People’s Hospital of Yunnan Province). After the addition of the peptides to the final concentrations corresponding to their respective MIC, the suspension was mixed with the SYTOX Green (Invitrogen, USA) to a final concentration of 50 mM and static for 15 min in the dark. The fluorescence intensity was monitored continuously in the Multiskan Sky Microplate Spectrophotometer (ThermoFisher Scientific, USA) for 30 min at 37 °C with 485 and 523 nm wavelength filters for excitation and emission. Melittin was used as the positive control.42 The strongest fluorescence intensity was used as the denominator, and the others were used as numerators for the evaluation of bactericidal effect.
Serum Stability Test
The MIC method was used for serum stability test. Each blood serum (collected from the healthy volunteers and provided by the Clinical Laboratory of First People’s Hospital of Yunnan Province) was mixed with cathelicidin-DM to a final concentration of 2 × MIC. The mixture was cultured at 37 °C for 12 h. The tested strains included Gram-positive and Gram-negative bacteria, which were the same as those used in the MIC assay.
Application of Cathelicidin-DM in a Mouse Skin Infection Model
Gentamicin is an aminoglycoside antibiotic, which is mainly used to treat bacterial infections, especially those caused by Gram-negative bacteria. Gentamicin can bind to the 30 s subunit of the bacterial ribosome and block the synthesis of bacterial proteins. Moreover, gentamicin is one of the few thermostable antibiotics, so it is widely used in the treatment. Thus, gentamicin was used in this study as the positive control. An overnight culture of E. coli ATCC25922 in LB liquid medium was subcultured in a fresh LB liquid medium to log phase, and then, the bacterial solution was centrifuged at 3000g for 10 min. The precipitate was washed three times with aseptic PBS buffer and then resuspended with aseptic PBS to a final concentration of 2 × 105 CFU/mL. Nine 6 week old female Kunming mice (weighing 27–30 g) were obtained from the Laboratory Animal Center of Kunming Medical University (Kunming, Yunnan) and housed (at 25 °C with a 12/12 h light/dark cycle and access to food and water ad libitum), maintained, and cared for in the Animal Experiment Center of Kunming University of Science and Technology. The mice were randomly divided into three groups (three mice/group). Hair at the back of each mouse was carefully shaved, and then, the back skin was scrubbed with warm water, dried, and wiped with alcohol. A 1 cm2 wound was generated with an incision using a surgical knife aseptically. Then, 100 μL 2 × 105 CFU/mL of E. coli (ATCC25922) liquid was daubed in the wound. All the mice were intravenously injected with cathelicidin-DM (10 mg/kg), gentamycin (5 mg/kg), or PBS as control 1 h postinoculation. The same dose was administered daily for 5 days. The wound-healing rate of the mice was measured every day.
In Vivo Imaging of FITC-Labeled Cathelicidin-DM
Six 6 week old female Kunming mice (weighing 27–30 g) were randomly divided into two groups (skin infection and control groups) for the evaluation of the tissue distribution of intravenously injected FITC-cathelicidin-DM. The hair of each mouse was removed, and the skin infection group was constructed as previously described. The control group had normal growth. One-hour postinoculation of E. coli, the two groups received 10 mg/kg of FITC-labeled cathelicidin-DM through tail vein injection. The mice were immediately anesthetized before transferring to the IVIS LT imaging system (IVIS spectrum) and then measured at 0, 5, 10, 30, 60, 90, and 120 min. The IVIS Lumina LT imaging system showed a region with strong yellow luminescence and low red luminescence. Increasing bioluminescence indicates elevated FITC-labeled cathelicidin-DM concentration. The images were an overlay of photographic images and bioluminescence generated by a computer-generated color scale.
Acknowledgments
The authors are very grateful to the First People’s Hospital of Yunnan Province for its great help on the strains.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00189.
Circular dichroism spectra and drug resistance information of the tested clinical strains, and HPLC and MS report of the synthesized peptides (PDF)
Author Present Address
∥ The College of Life Sience, Sichuan University, Chengdu, Sichuan, China.
Author Present Address
⊥ Dunwill Medical Technology Development Co., Ltd, Songjiang district, Shanghai, China.
Author Present Address
# Jiangsu Hansoh Pharmaceutical Group Co., Ltd, Lianyuangang, Jiangsu, China
Author Contributions
Y.Q.S. and C.L. are contributed equally to this work. Y.Q.S. and C.L. performed the experiments and data analysis. M.W., Z.J.C., and Y.L. assisted in collection and molecular cloning; and X.S.X., Y.Z.S., A.Z., and Y.S. conceived and designed the study. Y.Z.S., A.Z., and Y.S. conceived and supervised the project and prepared the manuscript. All authors read and approved the final version of the manuscript.
This research was funded by Yunnan Science and Technology Commission, grant number 2015BC001, and Yunnan Science and Technology Commission, grant number 2015DH010, and National Natural Science Foundation of China, grant number 31860607.
The authors declare no competing financial interest.
Supplementary Material
References
- Aminov R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 2017, 133, 4–19. 10.1016/j.bcp.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Moser C.; Lerche C. J.; Thomsen K.; Hartvig T.; Schierbeck J.; Jensen P. Ø.; Ciofu O.; Høiby N. Antibiotic therapy as personalized medicine - general considerations and complicating factors. APMIS 2019, 127, 361–371. 10.1111/apm.12951. [DOI] [PubMed] [Google Scholar]
- Tortorella E.; Tedesco P.; Palma Esposito F.; January G. G.; Fani R.; Jaspars M.; de Pascale D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 2018, 16, 355. 10.3390/md16100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan S.; Andrews I. W.; Collins J. J. Targeting Antibiotic Tolerance, Pathogen by Pathogen. Cell 2018, 172, 1228–1238. 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- Lim C.; Takahashi E.; Hongsuwan M.; Wuthiekanun V.; Thamlikitkul V.; Hinjoy S.; Day N. P.; Peacock S. J.; Limmathurotsakul D. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. Elife 2016, 5, e18082 10.7554/elife.18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan R.; Matsoso P.; Pant S.; Brower C.; Røttingen J.-A.; Klugman K.; Davies S. Access to effective antimicrobials: a worldwide challenge. Lancet 2016, 387, 168–175. 10.1016/s0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R.; Duse A.; Wattal C.; Zaidi A. K. M.; Wertheim H. F. L.; Sumpradit N.; Vlieghe E.; Hara G. L.; Gould I. M.; Goossens H.; Greko C.; So A. D.; Bigdeli M.; Tomson G.; Woodhouse W.; Ombaka E.; Peralta A. Q.; Qamar F. N.; Mir F.; Kariuki S.; Bhutta Z. A.; Coates A.; Bergstrom R.; Wright G. D.; Brown E. D.; Cars O. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. 10.1016/s1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Zanetti M. The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 2005, 7, 179–96. 10.21775/cimb.007.179. [DOI] [PubMed] [Google Scholar]
- Mygind P. H.; Fischer R. L.; Schnorr K. M.; Hansen M. T.; Sönksen C. P.; Ludvigsen S.; Raventós D.; Buskov S.; Christensen B.; De Maria L.; Taboureau O.; Yaver D.; Elvig-Jørgensen S. G.; Sørensen M. V.; Christensen B. E.; Kjærulff S.; Frimodt-Moller N.; Lehrer R. I.; Zasloff M.; Kristensen H.-H. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980. 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- Gomes B.; Augusto M. T.; Felício M. R.; Hollmann A.; Franco O. L.; Gonçalves S.; Santos N. C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. 10.1016/j.biotechadv.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Hong J.; Liu X.; Yang H.; Liu R.; Wu J.; Wang A.; Lin D.; Lai R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS One 2008, 3, e3217 10.1371/journal.pone.0003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson D. G. Pollution from drug manufacturing: review and perspectives. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130571. 10.1098/rstb.2013.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X.; Tang X.; Luo L.; Wang Y.; Lai R.; Lu Q. A novel ranacyclin-like peptide with anti-platelet activity identified from skin secretions of the frog Amolops loloensis. Gene 2016, 576, 171–175. 10.1016/j.gene.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Agerberth B.; Gunne H.; Odeberg J.; Kogner P.; Boman H. G.; Gudmundsson G. H. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 195–199. 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; Li Z.; Liu H.; He X.; Zhang Y.; Jin J.; Che J.; Li C.; Chen W.; Lai R.; Liu J. Novel analgesic peptides from the tree frog of Hyla japonica. Biochimie 2014, 99, 38–43. 10.1016/j.biochi.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Sun T.; Zhan B.; Gao Y. A novel cathelicidin from Bufo bufo gargarizans Cantor showed specific activity to its habitat bacteria. Gene 2015, 571, 172–177. 10.1016/j.gene.2015.06.034. [DOI] [PubMed] [Google Scholar]
- Parilti R.; Caprasse J.; Riva R.; Alexandre M.; Vandegaart H.; Bebrone C.; Dupont-Gillain C.; Howdle S. M.; Jérôme C. Antimicrobial peptide encapsulation and sustained release from polymer network particles prepared in supercritical carbon dioxide. J. Colloid Interface Sci. 2018, 532, 112–117. 10.1016/j.jcis.2018.07.125. [DOI] [PubMed] [Google Scholar]
- Bowdish D. M. E.; Davidson D. J.; Hancock R. E. W. Immunomodulatory properties of defensins and cathelicidins. Curr. Top. Microbiol. Immunol. 2006, 306, 27–66. 10.1007/3-540-29916-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bletz M. C.; Kelly M.; Sabino-Pinto J.; Bales E.; Van Praet S.; Bert W.; Boyen F.; Vences M.; Steinfartz S.; Pasmans F.; Martel A. Disruption of skin microbiota contributes to salamander disease. Proc. Biol. Sci. 2018, 285, 20180758. 10.1098/rspb.2018.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechas S. V.; Acosta-González A.; Escobar L. A.; Kueneman J. G.; Sánchez-Quitian Z. A.; Parra-Giraldo C. M.; Rollins-Smith L. A.; Reinert L. K.; Vredenburg V. T.; Amézquita A.; Woodhams D. C. Microbiota and skin defense peptides may facilitate coexistence of two sympatric Andean frog species with a lethal pathogen. ISME J. 2019, 13, 361–373. 10.1038/s41396-018-0284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Cai S.; Gao J.; Zhang S.; Lu Y.; Qiao X.; Yang H.; Wang Y. Identification and polymorphism discovery of the cathelicidins, Lf-CATHs in ranid amphibian (Limnonectes fragilis). FEBS J. 2013, 280, 6022–6032. 10.1111/febs.12521. [DOI] [PubMed] [Google Scholar]
- Wei L.; Yang J.; He X.; Mo G.; Hong J.; Yan X.; Lin D.; Lai R. Structure and function of a potent lipopolysaccharide-binding antimicrobial and anti-inflammatory peptide. J. Med. Chem. 2013, 56, 3546–3556. 10.1021/jm4004158. [DOI] [PubMed] [Google Scholar]
- Ling G.; Gao J.; Zhang S.; Xie Z.; Wei L.; Yu H.; Wang Y. Cathelicidins from the bullfrog Rana catesbeiana provides novel template for peptide antibiotic design. PLoS One 2014, 9, e93216 10.1371/journal.pone.0093216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H.; Maruoka T.; Aruga A.; Amano T.; Ohgo S.; Shiroishi T.; Tamura K. Prx-1 expression in Xenopus laevis scarless skin-wound healing and its resemblance to epimorphic regeneration. J. Invest. Dermatol. 2011, 131, 2477–2485. 10.1038/jid.2011.223. [DOI] [PubMed] [Google Scholar]
- Duda T. F. Jr.; Vanhoye D.; Nicolas P. Roles of diversifying selection and coordinated evolution in the evolution of amphibian antimicrobial peptides. Mol. Biol. Evol. 2002, 19, 858–864. 10.1093/oxfordjournals.molbev.a004143. [DOI] [PubMed] [Google Scholar]
- Campbell L. J.; Crews C. M. Wound epidermis formation and function in urodele amphibian limb regeneration. Cell. Mol. Life Sci. 2008, 65, 73–79. 10.1007/s00018-007-7433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M.; Gennaro R.; Scocchi M.; Skerlavaj B. Structure and biology of cathelicidins. Adv. Exp. Med. Biol. 2000, 479, 203–18. 10.1007/0-306-46831-X_17. [DOI] [PubMed] [Google Scholar]
- Hao X.; Yang H.; Wei L.; Yang S.; Zhu W.; Ma D.; Yu H.; Lai R. Amphibian cathelicidin fills the evolutionary gap of cathelicidin in vertebrate. Amino Acids 2012, 43, 677–685. 10.1007/s00726-011-1116-7. [DOI] [PubMed] [Google Scholar]
- Cao X.; Wang Y.; Wu C.; Li X.; Fu Z.; Yang M.; Bian W.; Wang S.; Song Y.; Tang J.; Yang X. Cathelicidin-OA1, a novel antioxidant peptide identified from an amphibian, accelerates skin wound healing. Sci. Rep. 2018, 8, 943. 10.1038/s41598-018-33558-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes N. J.; Cruce C. E.; O’Donnell J. N.; Wunderink R. G.; Hauser A. R. Resistance Trends and Treatment Options in Gram-Negative Ventilator-Associated Pneumonia. Curr. Infect. Dis. Rep. 2018, 20, 3. 10.1007/s11908-018-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieco P.; Luca V.; Auriemma L.; Carotenuto A.; Saviello M. R.; Campiglia P.; Barra D.; Novellino E.; Mangoni M. L. Alanine scanning analysis and structure-function relationships of the frog-skin antimicrobial peptide temporin-1Ta. J. Pept. Sci. 2011, 17, 358–365. 10.1002/psc.1350. [DOI] [PubMed] [Google Scholar]
- Cyranoski D. Why Chinese medicine is heading for clinics around the world. Nature 2018, 561, 448–450. 10.1038/d41586-018-06782-7. [DOI] [PubMed] [Google Scholar]
- Shimada K.; Fujii Y.; Yamashita E.; Niizaki Y.; Sato Y. Studies on cardiotonic steroids from the skin of Japanese toad. Chem. Pharm. Bull. 1977, 25, 714–730. 10.1248/cpb.25.714. [DOI] [PubMed] [Google Scholar]
- Ma H.; Zhou J.; Guo H.; Shang E.; Zhu Z.; Kuzmanov U.; Lv X.; Di L.; Yu B.; Wu Q.; Duan J. A strategy for the metabolomics-based screening of active constituents and quality consistency control for natural medicinal substance toad venom. Anal. Chim. Acta 2018, 1031, 108–118. 10.1016/j.aca.2018.05.054. [DOI] [PubMed] [Google Scholar]
- Ma H.; Niu H.; Cao Q.; Zhou J.; Gong Y.; Zhu Z.; Lv X.; Di L.; Qian D.; Wu Q.; Duan J. a. Metabolomics method based on ultra high performance liquid chromatography with time-of-flight mass spectrometry to analyze toxins in fresh and dried toad venom. J. Sep. Sci. 2016, 39, 4681–4687. 10.1002/jssc.201600827. [DOI] [PubMed] [Google Scholar]
- Wu J.; Yang J.; Wang X.; Wei L.; Mi K.; Shen Y.; Liu T.; Yang H.; Mu L. A frog cathelicidin peptide effectively promotes cutaneous wound healing in mice. Biochem. J. 2018, 475, 2785–2799. 10.1042/bcj20180286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L.; Zhou L.; Yang J.; Zhuang L.; Tang J.; Liu T.; Wu J.; Yang H. The first identified cathelicidin from tree frogs possesses anti-inflammatory and partial LPS neutralization activities. Amino Acids 2017, 49, 1571–1585. 10.1007/s00726-017-2449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen-Ting H. U.; Zhang Y. X.; Ri-Ga M.; Zhang K.; Huang X.; Chen M. J. Preparation and properties of antimicrobial peptides from skin secretion of Bufo melanostictus. Chin. J. Bioprocess Eng. 2010, 8, 21–24. [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D.; Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 2012, 80, 1715–1735. 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-L.; Su P.-Y.; Chang Y.-S.; Wu S.-Y.; Liao Y.-D.; Yu H.-M.; Lauderdale T.-L.; Chang K.; Shih C. Identification of a novel antimicrobial peptide from human hepatitis B virus core protein arginine-rich domain (ARD). PLoS Pathog. 2013, 9, e1003425 10.1371/journal.ppat.1003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scocchi M.; Skerlavaj B.; Romeo D.; Gennaro R. Proteolytic cleavage by neutrophil elastase converts inactive storage proforms to antibacterial bactenecins. Eur. J. Biochem. 1992, 209, 589–595. 10.1111/j.1432-1033.1992.tb17324.x. [DOI] [PubMed] [Google Scholar]
- Panyutich A.; Shi J.; Boutz P. L.; Zhao C.; Ganz T. Porcine polymorphonuclear leukocytes generate extracellular microbicidal activity by elastase-mediated activation of secreted proprotegrins. Infect. Immun. 1997, 65, 978–985. 10.1128/iai.65.3.978-985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.; Liu H.; Yang X.; Che Q.; Liu R.; Yang H.; Liu X.; You D.; Wang A.; Li J.; Lai R. Bi-functional peptides with both trypsin-inhibitory and antimicrobial activities are frequent defensive molecules in Ranidae amphibian skins. Amino Acids 2012, 43, 309–316. 10.1007/s00726-011-1079-8. [DOI] [PubMed] [Google Scholar]
- Chmielarczyk A.; Pobiega M.; Ziółkowski G.; Pomorska-Wesołowska M.; Romaniszyn D.; Krawczyk L.; Wójkowska-Mach J. Severe infections caused by multidrug-resistant non-fermentative bacilli in southern Poland. Adv. Clin. Exp. Med. 2018, 27, 401–407. 10.17219/acem/68545. [DOI] [PubMed] [Google Scholar]
- Koutserimpas C.; Samonis G.; Plataki M. N.; Bikis C.; Kontakis G.; Kofteridis D. P. Multidrug-resistant Gram-negative osteomyelitis: a 10-year study. G Chir 2018, 34, 284–290. [PubMed] [Google Scholar]
- Liu H.-h.; Sun Q.; Jiang Y.-t.; Fan M.-h.; Wang J.-x.; Liao Z. In-depth proteomic analysis of Boleophthalmus pectinirostris skin mucus. J Proteomics 2019, 200, 74–89. 10.1016/j.jprot.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Li Z. Y.; Qu T.; Wang P. F.; Chen L. M.; Wang Z. M.; Gao H. M. [Advance on quality control of toad venom and its key influence factors]. Zhongguo Zhong Yao Za Zhi 2017, 42, 863–869. 10.19540/j.cnki.cjcmm.20170121.006. [DOI] [PubMed] [Google Scholar]
- Cho J. H.; Sung B. H.; Kim S. C. Buforins: histone H2A-derived antimicrobial peptides from toad stomach. Biochim. Biophys. Acta 2009, 1788, 1564–1569. 10.1016/j.bbamem.2008.10.025. [DOI] [PubMed] [Google Scholar]
- Park C. B.; Kim M. S.; Kim S. C. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. Commun. 1996, 218, 408–413. 10.1006/bbrc.1996.0071. [DOI] [PubMed] [Google Scholar]
- Kim H. S.; Park C. B.; Kim M. S.; Kim S. C. cDNA cloning and characterization of buforin I, an antimicrobial peptide: a cleavage product of histone H2A. Biochem. Biophys. Res. Commun. 1996, 229, 381–387. 10.1006/bbrc.1996.1814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.